Abstract
Lung cancer is the leading cause of cancer-related death worldwide yet in vitro disease models have been limited to traditional 2D culture utilizing cancer cell lines. In contrast, recently developed 3D models (organoids) have been adopted by researchers to improve the physiological relevance of laboratory study. We have hypothesized that 3D hydrogel-based models will allow for improved disease replication and characterization over standard 2D culture using cells taken directly from patients. Here, we have leveraged the use of 3D hydrogel-based models to create lung cancer organoids using a unique cell source, pleural effusion aspirate, from multiple lung cancer patients. With these 3D models, we have characterized the cell populations comprising the pleural effusion aspirate and have tracked phenotypic changes that develop during short-term in vitro culture. We found that isolated, patient cells placed directly into organoids created anatomically relevant structures and exhibited lung cancer specific behaviors. On the other hand, cells first grown in plastic dishes and then cultured in 3D did not create similar structures. Further, we have been able to compare chemotherapeutic response of patient cells between 2D and 3D cell culture systems. Our results show that cells in 2D culture were more sensitive to treatment when compared with 3D organoids. Collectively, we have been able to utilize tumor cells from pleural effusion fluid of lung cancer patients to create organoids that display in vivo like anatomy and drug response and thus could serve as more accurate disease models for study of tumor progression and drug development.
Keywords: pleural effusion, 3D culture, disease modeling, chemotherapy
Graphical Abstract
Introduction
Lung cancer remains the leading cause of cancer-related death worldwide1-2. Specifically, adenocarcinoma of the lung is the most common histological type and is directly related to smoking and other environmental exposures. Advances in early diagnosis and treatment of patients is of primary interest since the 5 year survival rate of patients with lung cancer is only 15%1. One of the symptoms of lung adenocarcinoma is the development of malignant pleural effusion (PE) which is a build-up of aspirate between the parietal and visceral pleural layers3. Because advanced-stage lung cancer causes fluid imbalances which force fluid into the pleural space, PE has been related to inflammation and infection, which increase capillary permeability and can lead to subsequent metastasis 4. Lung cancer patients frequently have PE drained to relieve multiple outward symptoms such as cough, shortness of breath, or discomfort in the chest through two primary methods: thoracentesis or thoracoscopy 5.
One major challenge in studying lung adenocarcinoma is the low quantity of cells that can be isolated from lung tissue biopsies. PE can act as a liquid biopsy offering information about lung adenocarcinoma prognosis. PE samples contain a multitude of cell types which are representative of the heterogeneity present in lung adenocarcinoma. After being utilized for diagnostic purposes, excess PE is often discarded, unlike solid tumor biopsies which are commonly stored as a paraffin-embedded block for future analysis 6. In this study we isolated and identified viable cells that originated from lung adenocarcinoma in PE aspirates. PE samples are a readily-available, cost-effective source of tumor cells for monitoring of disease progression and further studying the effects of treatments, as well as for creating better lung cancer models to support precision medicine applications in the future.
Current models for lung adenocarcinoma include both two-dimensional (2D) and three-dimensional (3D) cell culture systems, as well as patient-derived mouse xenografts (PDX) 7. Patient derived cells can be isolated from tumor biopsies and used in in vitro tumor models to track disease progression and screen chemotherapeutics, which represents an ideal platform for precision medicine applications. However, isolating pure populations of cancer cells eliminates many of the non-tumor cell components, such as stromal cells or extracellular matrix components (ECM), that maintain or support tumor progression. PDX models aim to alleviate these concerns by creating microenvironments in which cells may behave more similarly to physiologic conditions8-9 however they lack both human and cancer-specific components, which may alter the human cancer cell behavior10. Considering these limitations, ex-vivo 3D cell culture models have aimed to create microenvironments that incorporate specific ECM and stromal components to best replicate disease behavior and progression. Most current 3D models are formed either by creating cell aggregates (spheroids) or by encapsulating cells in hydrogels, surrounding them in ECM components similar to their native microenvironment11-12. Hydrogels can be chemically modified to vary internal porosity, stiffness, and component availability for tissue-specific customization. Lung-specific 3D tissue models have thus far included decellularized lung extracellular matrices and scaffolds, naturally based hydrogels such as collagen and alginate, and polymers13-16. Previous studies have shown clear changes in cell phenotype and genotype over time between 2D and 3D methods17.
In this study, we have isolated tumor and stromal cells from PE aspirate biopsies, cultured those cells in 3D model systems (organoids) and in standard 2D tissue culture plastic dishes (Table S1). Our goal for the current study was to determine maintenance of PE-derived cells in vitro and to further characterize interactions between tumor and stromal cells, ultimately to develop a more accurate model for lung adenocarcinoma (Figure 1). We were able to document morphological changes in cells over time and apply chemotherapeutic treatments in 3D and 2D culture systems. The results show differential responses between each of the systems, which provides insight into the importance of 3D over 2D cultures. Collectively, our data suggest that PE liquid biopsies are suitable for studying lung adenocarcinoma and the importance of using 3D over 2D culture for disease modeling and drug efficacy screening studies.
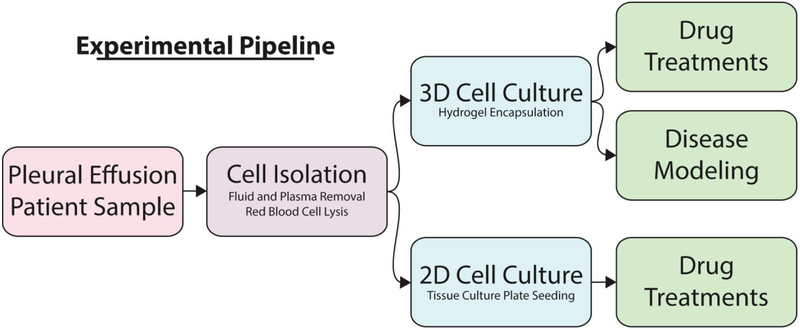
Figure 1. Experimental Pipeline for Pleural Effusion.
Pleural effusion cells are separated from the PE liquid through centrifugation and cells are plated in tissue culture dishes (2D) or encapsulated in hydrogels (3D). Both 2D and 3D cultures allow for comparative drug treatment in order to determine differences in predicted drug response. The 3D culture method further allows for disease modeling over a longer period of time.
Materials and Methods
Cell Isolation
PE samples were obtained from Wake Forest Baptist Hospital under IRB#BG04-104 with consent from patients previously diagnosed with lung adenocarcinoma and contained between 500 mL and 1 L of fluid each yielding between 4 and 7.5 million cells for use in culture. Samples were transferred from the primary container in which they were delivered into 50 mL centrifuge tubes and centrifuged to separate cells from fluid and plasma. Once cells were pelleted, BD PharmLyse was used following manufacturer’s instructions (1:10, lysing buffer:deionized water) to lyse and subsequently remove red blood cells (BD Biosciences, San Jose, CA). Cells were centrifuged, cell lysate was removed, and the cells were resuspended in RPMI 1640 medium with 5% fetal bovine serum and 200 U/mL penicillin and streptomycin (RPMI-5) (ThermoFisher Scientific, Waltham, MA). Cells were counted and seeded into 3D hydrogel cultures (106 cells/mL), also referred to as organoids, or onto tissue culture plastic. From samples, approximately fifty organoids, one 6-well tissue culture plastic plate, and fifty 96-well tissue culture plastic (TCP) plates were made at P0.
Cell Culture
Cells were isolated from two patient samples (Sample 1 – isolated on 1-4-2017; Sample 2 – isolated on 7-23-2017) and placed into 3D and 2D culture. Hydrogels were prepared using three parts methacrylated type I collagen (6 mg/mL) to one part thiolated hyaluronan (1 mg/mL) (Advanced Biomatrix, Carlsbad, CA; ESI Bio, Almeda, CA). Cells were resuspended in hydrogel at 106 cells/mL. In suspension, organoids were made by depositing 10 μL of gel into each well of 48-well TCP plates that were previously coated with a thin layer of polydimethylsiloxane (PDMS), to reduce cell adherence and outgrowth. Once plated, hydrogels were crosslinked with UV light (880 mW/cm2) for 1 second. Media was added to the wells and organoids were allowed to grow for 3 weeks for 1-4-17 samples and 3-6 weeks for 7-23-17 samples prior to fixation (referred to as P0 or passage 0, 3 weeks and P0 or passage 0, 6 weeks) (Figure 2). Cells placed into 2D culture were either plated in 6-well or 96-well TCP plates. Those in 6-well plates were grown for 2 weeks and then passaged into organoids (referred to as P1 or passage 1 organoids). Those in 96-well plates were initially seeded at 1,000 cells per well and grown for 3 weeks before drug treatment. RPMI-5 was used for all cultures and media was changed ever 3-4 days. PE lung adenocarcinoma cell line H460 (pleural effusion, lung adenocarcinoma) was also utilized for experimentation and placed in 2D culture using 96-well TCP seeded at 1,000 cells per well. Cells were grown for 7 days and then drug treatment was administered as described below. H460s were also placed into hydrogels using the methods described at a density of 7 × 106 cells/mL. A mixed 3D culture was also created using a combination of H460 and normal human lung fibroblasts (NHLF) to determine if there were any differences in drug response (3.5 × 106 cells/mL) of each cell type.
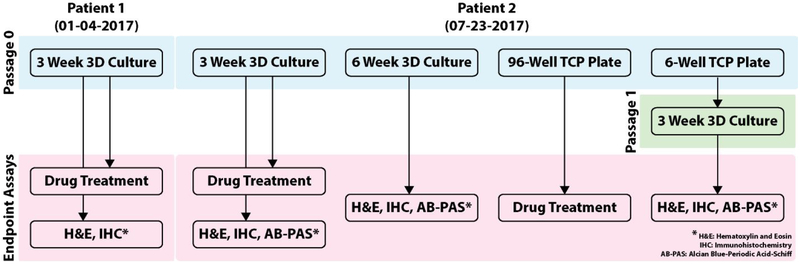
Figure 2. Cell Usage Schematic.
Of the patient samples collected, two (referred to as samples 1-4-17 and 7-23-17) were cultured and subsequently characterized or treated with chemotherapy. Isolated cells from Sample 1-4-17 were placed directly into organoids and grown for 3 weeks at the end of which they were characterized using IHC and H&E. Sample 7-23-17 cells were isolated and placed directly into organoids grown for either 3 or 6 weeks, or grown on TCP. Organoids grown for 3 weeks were either characterized using IHC and H&E or used for drug treatment. Organoids grown for 6 weeks were used for characterization. Sample 7-23-17 cells grown on TCP were initially placed in either 96-well or 6-well TCP plates. Those in the 96-well plate were used for drug treatment. Those in the 6-well plate were passaged at approximately 3 weeks and those cells were made into organoids. The organoids from passaged TCP wells were grown for 3 weeks and characterized.
Drug Treatment
Drug treatments were administered to cells isolated from the second patient in both 3D and 2D. At 3 weeks, P0 2D cultures were treated with three drugs, each at three concentrations as follows: cisplatin and pemetrexed (0.2 μM, 0.2 μM; 2 μM, 2 μM; 20 μM, 20 μM), carboplatin and pemetrexed (0.2 μM, 0.2 μM; 2 μM, 2 μM; 20 μM, 20 μM), and crizotinib (1 μM; 10 μM; 100 μM) in RPMI-5. After 3 days of treatment, an MTS assay was performed (n=3) following manufacturer’s instructions with incubation duration of 30 minutes. Two additional wells were fixed with 4% paraformaldehyde (PFA). This drug treatment was replicated in 2D using H460 cells. At 5 weeks 4 days, P0 6-week organoids were treated with cisplatin and pemetrexed (2 μM, 2 μM), carboplatin and pemetrexed (2 μM, 2 μM), and crizotinib (10 μM) in RPMI-5. After 3 days, an MTS assay was performed (n=3) following manufacturer’s instructions with incubation duration of 2 hours. Additional organoids (n=2) were fixed for histological processing with 4% PFA. H460 cells in 2D culture alone, H460 in 3D organoids alone, and H460 with NHLFs in 3D organoids were all treated as previously described 2D and 3D studies for patient samples.
Immunohistochemistry
Cultures in 3D were fixed with 4% PFA for 30 minutes and then paraffin processed as appropriate for histology. Paraffin sections were cut at 5 μm thick sections for staining. Hematoxylin and eosin (H&E) staining was done via autostainer for each of the samples shown. Alcian blue – periodic acid-Schiff stain (AB-PAS) was carried out per manufacturer’s instructions for P0 3-week, P0 6-week, and P1 3-week organoids. For H&E stained P0 3-week, P0 6-week, and P1 3-week structures within organoids were quantified using ImageJ. Representative images from three separate organoids for each condition were quantified using the drawing tool and collecting the area from each structure. Number of structures for each condition ranged from 65 to 110. Immunohistochemistry was performed to further characterize samples 1-4-17 and 7-23-17 organoids. Sample 1-4-17 organoids were stained using primary antibodies α-smooth muscle actin (αSMA; 1:200, ab7817, Abcam, Cambridge, MA), CD45 (1:200, ab10558, Abcam), pan-cytokeratin (CK, 1:200, ab215838, Abcam), P63 (1:200, CM163, Biocare Medical, Pacheco, CA), and DAPI (1:300, ab228549, Abcam). For sample 7-23-17, primary antibodies Mucin5AC (1:200, ab3649), CK7(1:200, ab9021), and CK20 (1:200, ab76126) stains were utilized for characterization and were incubated on sections overnight at 4°C (Abcam). CK7 and CK20 were carried out as co-stains and DAPI was used amongst all stains. Secondary antibodies utilized for sample 7-23-17 were administered to slides and incubated for 1 hour at room temperature as follows: Texas Red, goat anti-rabbit (1:200, TI-1000) and FITC horse anti-mouse (1:200, FI-2000) (Vector Labs, Burlingame, CA). Each of the samples was finally stained with DAPI (1:300, ab228549, Abcam) for 15 minutes at room temperature. Sample 7-23-17 slides were imaged using an Olympus BX63 motorized upright microscope for both brightfield and fluorescence microscopy (Olympus, Shinjuku, Japan).
Results
Cells from the drained PE of lung adenocarcinoma patients (~1 L) were used to produce lung adenocarcinoma 2D and 3D cultures. Multiple PE samples were obtained, resulting in viable cell cultures on TCP and in 3D organoids (Table S1). Cells on TCP were successfully sub-cultured (passaged) up to 4 times with decreasing cellular proliferation with increasing passage. Preliminary immunohistochemistry of 3D organoids revealed the presence of both tumor and non-tumor cells. Of the different samples, we chose two (1-4-17 and 7-23-17) for thorough histological and immunohistochemical characterizations and compared the 2D and 3D cultures (Fig 2).
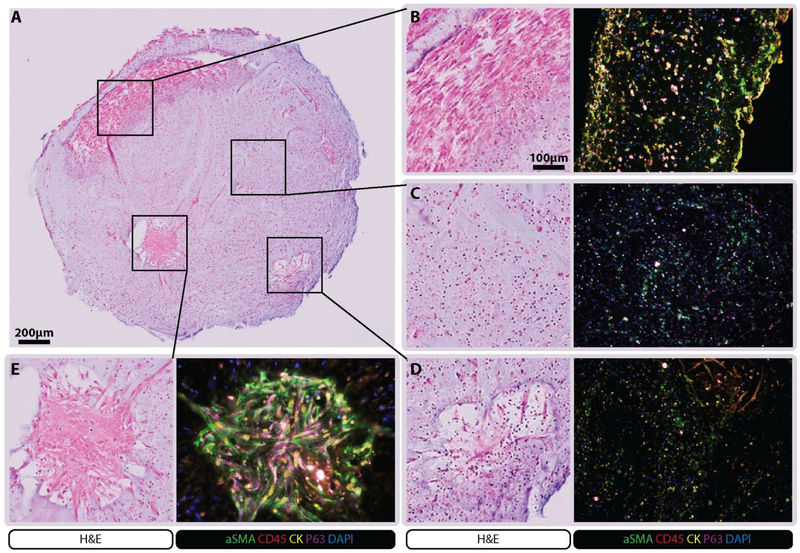
The 1-4-17 sample yielded enough material for initial characterization of the PE samples and to optimize the encapsulation procedure using hydrogels. Organoids containing cells from the 1-4-17 sample were cultured for 3 weeks then analyzed. H&E staining revealed various structures within the organoids including aligned stromal cells and clusters of cancer cells with both immune and stromal components (Figure 3). Immunostaining showed the presence of cells expressing αSMA (fibroblasts), CD45 (hematopoietic cells), cytokeratin (tumor cells), and P63 (potentially cancer stem cells) in the different structural regions of the organoids (Figure 3). These cells were maintained within the organoids and may have co-localized during the 3-week culture period. For example, two regions within a single organoid were shown to have cellular structures that stained with tumor cell markers (CK+, P63+) and fibroblast like cells (αSMA+) extending from their periphery, indicating outgrowth and interaction with the matrix (Figure 3D-E). Hematopoietic cells (CD45+) were observed primarily within the core of these structures and also dispersed within other regions. A large region near the edge of the organoid showed fibroblast-like cells that stained consistently for both CK and αSMA, indicating co-localization of fibroblasts and tumor cells (Figure 3B). In regions without apparent structures, staining indicated sporadic presence of CK+, P63+, and αSMA+ cell populations although elongation, co-localization, and interaction with matrix were not apparent (Figure 3C).
Figure 3. 1-4-17 Sample Characterization.
Organoids made from cells from of the 1-4-17 sample were cultured for 3 weeks and then processed for histology and immunohistochemistry. H&E staining revealed many unique structures characterized using αSMA, CD45, pan cytokeratin, P63 and DAPI. It was revealed that the structures retained a mixed cell population.
This patient was a 66-year-old female at time of sample collection with pathology indicating primary lung adenocarcinoma. Genetic testing revealed PIK3CA E545K and p53 V272L mutations. The patient was initially treated with carboplatin and pemetrexed two years previous to sample collection, and received nivolumab throughout the year previous to sample collection. The patient was ultimately unresponsive to these treatments.
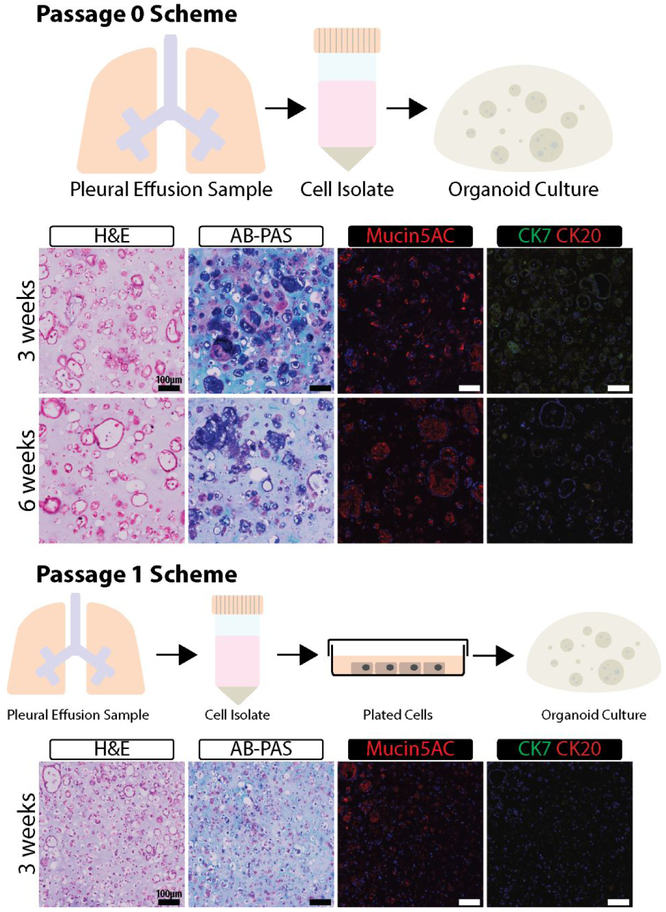
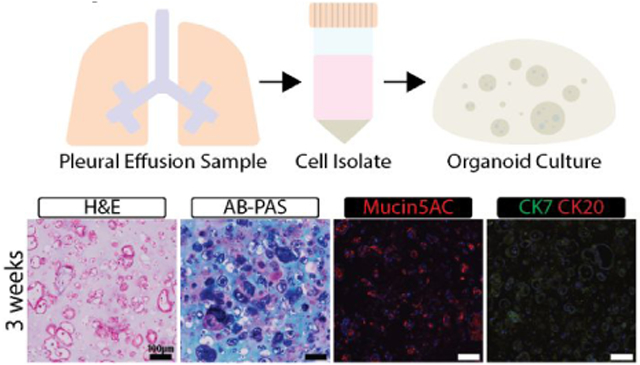
The 7-23-17 sample yielded an increased number of cells, allowing for histological. analysis of organoids and for 2D versus 3D drug testing. Organoids made from freshly isolated PE-derived cells (Passage 0 or P0) were initially stained with H&E and showed large acini structures with empty lumen after 3- and 6- weeks incubation (Figure 4). Organoid hydrogels remained the size they were created at (~10 μL) for the entire 6-weeks they were in culture, with cellular reorganization changes observed within the organoids, and no signs of necrosis were noted. Cells that were sub-cultured once in TCP and then encapsulated into organoids (P1 organoids) formed smaller acini structures after 3-weeks in culture (Figure S1). The smallest structures were formed within the center of the organoids and the largest were observed only near the edges of each of the organoids with size being approximately the same as those from P0. AB-PAS staining of mucins showed the presence of acidic mucins within the large structures. The magenta stain also within or directly next to the acini structures indicated the presence of neutral mucins18. The staining appeared to decrease significantly in the P1 organoids. To determine which specific mucin type is present, we stained the organoids with anti-mucin 5AC antibodies. This mucin, not produced by cancer cells but related to disease outcome, was found to be correlated to both primary and metastatic lung cancer in previous studies 19. The immunostaining results indicated that mucin 5AC is present in organoids made with P0 cells and its signal is substantially reduced in P1 organoids. The staining is most prominent along the edges of the organoid where more considerably sized structures are present (Figure 4). These results are consistent with AB-PAS staining. With the presence of disease state-related mucin presenting cells, the identification of cancer within the cultures was necessary to guarantee the model was representative of lung adenocarcinoma. For each of the organoids, expression of CK7 and CK20 was evaluated. Previous studies have demonstrated that lung adenocarcinoma is CK7+/CK20− 20. The PE cell organoids were positive for CK7 and negative for CK20, similar to these studies positive stains were most prominent within the P0 organoids and less so in the P1 organoids and appeared to be integrated into the acini structures, suggesting colocalization and interaction with stromal cells to support cancer growth (Figure 4).
Figure 4. 7-23-17 Sample Characterization.
Organoids made from cells from of the 7-23-17 sample were cultured for the indicated times and then processed for histology and immunohistochemistry. H&E, AB-PAS, Mucin5A, CK7, and CK20 were used to characterize the organoids.
This patient was a 41-year-old male at time of sample collection and pathology indicated adenocarcinoma which was possibly metastasis from primary pancreatic cancer. Genetic testing revealed TERT L38L mutation. The patient initially received 5-fluoraurcil, irinotecan and oxaliplatin treatment. Disease progressed and the patient was subsequently treated with gemcitabine and paclitaxel. All treatments were given prior to sample collection. The patient was ultimately unresponsive to these treatments.
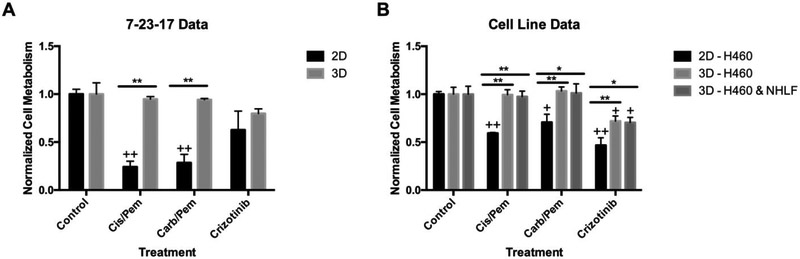
With data showing cancer cells were present within the organoids and expressed markers of lung adenocarcinoma, we proceeded to evaluate the response of the cultures to chemotherapeutic drugs. Response to chemotherapeutics used for treating lung adenocarcinoma was assessed using freshly isolated PE cells in both organoids and on TCP (Figure 5 and Figure S2)21. We based drug concentrations on previously cited values and on our preliminary testing of patient and H460 cells cultured on TCP (Figure S2)22-23. Our results showed statistically significant differences between the response of tumor cells on TCP and in the organoids; specifically, the cells on TCP showed statistically greater responses to cisplatin/pemetrexed and carboplatin/pemetrexed (cis/pem and carb/pem, respectively) combinations than the organoids (Figure 5A and Figure S2). Tumor cell viability in the organoids (as measured by mitochondrial metabolism by MTS) was not significantly different than in untreated organoids. On the other hand, crizotinib, an ALK (anaplastic lymphoma kinase) and ROS1 (c-ros oncogene 1) inhibitor, used in lung adenocarcinoma patients, had a moderate effect on tumor cell viability both in TCP cultures and organoids. To validate these results we created tumor organoids with H460, a cell line derived from PE of patient with lung adenocarcinoma. The results with H460 cells were similar to those of cells from the 7-23-17 sample with low sensitivity to chemotherapy treatment (Figure 5B and Figure S2). We also tested if the addition of normal human lung fibroblasts (NHLF) to the H460 cells in the organoids will change their response to the drug treatments (Figure 5B and Figure S2). We found that the addition of fibroblasts did not significantly change chemotherapeutic response. H&E staining further confirmed these results (Figure S3). The addition of fibroblasts did not have an effect on organoids’ structure or drug response. Collectively, the differences indicate that 1) tumor cells in 2D and 3D differ in their response the chemotherapies, 2) these differences can vary based on the particular drug and 3) the presence of non-cancer cells such as fibroblasts had no significant effect on cancer cell drug response in the organoids.
Figure 5. Organoid and TCP Chemotherapy Screening.
Cell from the 7-23-17 sample (A) and H460 PE-derived lung adenocarcinoma cells (B) were cultured in organoids for 5 weeks and in TCP and then treated with the indicated chemotherapy drugs for 3 days. MTS assay was carried out to determine cell viability and results were normalized to untreated cells (Control). Some of the organoids made with H460 cells contained in addition normal human lung fibroblasts (NHLF) (* p<0.05, ** p<0.005) (+ p<0.05, ++ p<0.005 with control).
Discussion
The goal for the current study was to use a unique cell source, PE aspirate, to biofabricate a 3D lung adenocarcinoma model for disease modeling and chemotherapy screening. By encapsulating isolated, primary cells into specialized hydrogels, we have produced lung cancer organoids that preserve cancer cell characteristics and tumor heterogeneity. We have observed the presence of adenocarcinoma, stromal, and immune cells that drive cellular reorganization and co-localization that cannot be recapitulated in simple 2D culture.
In sample 1-4-17, we identified large branching structures containing CK+ tumor cells and αSMA+ fibroblasts (Figure 3) suggesting that tumor cells within the organoid are supported by the stromal fraction. Cancer-associated fibroblasts (CAFs) have previously been linked to poor prognosis24. CAFs have been previously shown to support the cancer microenvironment which can induce drug resistance25. However, for the current study CAFs were not added since the samples already contained stromal fibroblasts. However, αSMA+ fibroblasts were present at the periphery of the organoids, suggesting that they may be active and acting as CAFs. They could also form a fibrotic capsule around the tumor organoid, similar to many solid tumors. The CK+ cells were identified with a broad range antibody that detects CK 4- 10, 13, 14, 18, and 19, indicating that cancer cells remained within the patient derived tumor organoids. Finally, the presence of CD45+ cells indicates that hematopoietic cells such as leukocytes can be preserved within the hydrogel-based 3D tumor organoids. In sample 7-23-17, we observed mucin-containing acini structures. This finding shows the tumor organoids provide a supporting microenvironment that facilitates formation of bronchiole-like structures that produce mucin, similar to lung adenocarcinoma in vivo, and these acinar structures were preserved over the course of 6 weeks with minimal changes. On the other hand, if the tumor cells were first grown on tissue culture plastic dishes and then encapsulated in hydrogel to form 3D organoids, these structures were significantly smaller and produced less mucin. This indicates when tumor cells are removed from their natural environment and cultured on non-physiological substrates (plastic), they lose their ability of tissue-like re-organization. In contrast, the 3D tumor organoids provide a proper physical-chemical environment that serves as a physiologically relevant model of in vivo tumor tissue which maintains phenotypic and re-organization features. Overall, this study indicates that samples from PE aspirate yield adequate cell numbers and diversity to create tumor organoids that better represent lung carcinomas compared with standard cell culture models. Within the scope of our study, we have not been able to control cell fate in 2D to yield the phenotypes seen in 3D over time indicating that 3D conditions are more appropriate for preserving cell phenotypes that allow for acini formation and cellular colocalization. The organoid system demonstrated capability of preserving structures and heterogeneity for up to 6 weeks and suggest that it can model the interactions between different cell types within the tumor. Current lung adenocarcinoma disease models are commonly 2D cells cultures or involve in vivo xenografts whereas, models that have integrated the use of 3D cultures primarily use cell lines 26-27. The tumor organoid technology described here uniquely combines flexible and easily assayed in vitro culture with an in vivo-like environment that can be used to study disease progression and assess treatments.
An essential function of tumor modeling is testing the response of the cancer cells to treatments such as chemotherapies. A comparison in the response to compounds commonly used to treat lung adenocarcinoma28-30 revealed a significant difference between PE-derived cells in 2D and 3D cultures. The cells in TCP were significantly more sensitive to cis/pem and carb/pem combinations compared with the cancer cells in the organoids. On the other hand, the response to crizotinib, an ALK (anaplastic lymphoma kinase) and ROS1 (c-ros oncogene 1) inhibitor, was more similar (yet with a significant difference) between PE-derived cancer cells in 2D and 3D cultures. These results could probably not be explained by diffusion since the organoids were only 10μl in size. On the other hand, it may be due to loss of cancer cell sensitivity to a specific drug after in vitro culture expansion. It has been reported that cancer cells cultured in plastic dishes for extended periods of time significantly change their phenotype and response to drugs17. Furthermore, our results have shown a phenotypic change of the cancer cells, in forming smaller acinar structures and lower production of mucin, even after one passage in plastic culture dishes. Together, these results suggest that cancer cells in our organoids have a more physiologic response to chemotherapeutic drugs however, the differences between the two culture systems is dependent on the specific target of the drug. As such, it is important to document the drug sensitivity of cancer cells in the tumor organoids and ensure the expression levels of targeted proteins and pathways are sufficient to predict drug response in vivo31-32. Accurate expression of oncogenic pathways in organoid models is essential for precision medicine in order to prescribe an optimal treatment to each patient based on the genetic profiles of their tumor cells. Our model has been able to use PE aspirate removed by a routine, required, and relatively non-invasive procedure for the creation of 3D organoid models that better represent lung tumor microenvironment. The use of regularly removed PE fluid provide a unique opportunity to researchers and clinicians to monitor disease progression and response to treatment in vitro. Our data shows that there is a substantial difference between 2D cultures and 3D organoids in phenotype and drug response indicating that these models yield different results and researchers should look for the best representation of the disease state of interest.
Collectively, this study highlights a unique lung cancer model system, using PE aspirate for lung cancer cell isolation and the creation of 3D organoids that preserve the phenotype of the cancer cells as well as maintain cancer and stromal cell interaction for an extended period in vitro. Our data supports the use of 3D over 2D culture models for lung adenocarcinoma drug testing. Although we were limited by the sample size of patients, our data supports the necessity to continue to pursue the use of PE-derived 3D tumor organoid cultures in replacement of 2D cultures to best model lung cancer disease progression and treatments.
Supplementary Material
Acknowledgements
The authors acknowledge funding through the NIH NCI grant 5R33CA202822. The authors wish to acknowledge the support of the Wake Forest Baptist Comprehensive Cancer Center Tumor Tissue and Pathology Shared Resource supported by the National Cancer Institute’s Cancer Center Support Grant award number P30CA012197. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.”
Footnotes
Supporting Information
Additional information describing other samples collected and processed utilizing pleural effusion aspirate as well as characterization of 2D cell culture drug treatment and 3D cell culture drug treatments are included.
References
- 1.Siegel RL; Miller KD; Jemal A, Cancer statistics, 2018. CA Cancer J Clin 2018, 68 (1), 7–30. DOI: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research, N., Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511 (7511), 543–50. DOI: 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Held-Warmkessel J; Schiech L, Caring for a patient with malignant pleural effusion. Nursing 2008, 38 (11), 43–7; quiz 48. DOI: 10.1097/01.NURSE.0000341079.53082.b1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch T; Kalish L; Mentzer S; Decamp M; Strauss G; Sugarbaker D, Optimal therapy of malignant pleural effusions. Int J Oncol 1996, 8 (1), 183–90. DOI: 10.3892/ijo.8.1.183 [DOI] [PubMed] [Google Scholar]
- 5.Shafiq M; Frick KD; Lee H; Yarmus L; Feller-Kopman DJ, Management of Malignant Pleural Effusion: A Cost-Utility Analysis. J Bronchology Interv Pulmonol 2015, 22 (3), 215–25. DOI: 10.1097/LBR.0000000000000192 [DOI] [PubMed] [Google Scholar]
- 6.Wahidi MM; Reddy C; Yarmus L; Feller-Kopman D; Musani A; Shepherd RW; Lee H; Bechara R; Lamb C; Shofer S; Mahmood K; Michaud G; Puchalski J; Rafeq S; Cattaneo SM; Mullon J; Leh S; Mayse M; Thomas SM; Peterson B; Light RW, Randomized Trial of Pleural Fluid Drainage Frequency in Patients with Malignant Pleural Effusions. The ASAP Trial. Am J Respir Crit Care Med 2017, 195 (8), 1050–1057. DOI: 10.1164/rccm.201607-1404OC [DOI] [PubMed] [Google Scholar]
- 7.Stock K; Estrada MF; Vidic S; Gjerde K; Rudisch A; Santo VE; Barbier M; Blom S; Arundkar SC; Selvam I; Osswald A; Stein Y; Gruenewald S; Brito C; van Weerden W; Rotter V; Boghaert E; Oren M; Sommergruber W; Chong Y; de Hoogt R; Graeser R, Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci Rep 2016, 6, 28951 DOI: 10.1038/srep28951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight E; Przyborski S, Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J Anat 2015, 227 (6), 746–56. DOI: 10.1111/joa.12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siolas D; Hannon GJ, Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res 2013, 73 (17), 5315–9. DOI: 10.1158/0008-5472.CAN-13-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassidy JW; Caldas C; Bruna A, Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res 2015, 75 (15), 2963–8. DOI: 10.1158/0008-5472.CAN-15-0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skardal A; Devarasetty M; Kang HW; Mead I; Bishop C; Shupe T; Lee SJ; Jackson J; Yoo J; Soker S; Atala A, A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomater 2015, 25, 24–34. DOI: 10.1016/j.actbio.2015.07.030 [DOI] [PubMed] [Google Scholar]
- 12.Edmondson R; Broglie JJ; Adcock AF; Yang L, Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 2014, 12 (4), 207–18. DOI: 10.1089/adt.2014.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stratmann AT; Fecher D; Wangorsch G; Gottlich C; Walles T; Walles H; Dandekar T; Dandekar G; Nietzer SL, Establishment of a human 3D lung cancer model based on a biological tissue matrix combined with a Boolean in silico model. Mol Oncol 2014, 8 (2), 351–65. DOI: 10.1016/j.molonc.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godugu C; Patel AR; Desai U; Andey T; Sams A; Singh M, AlgiMatrix based 3D cell culture system as an in-vitro tumor model for anticancer studies. PloS one 2013, 8 (1), e53708 DOI: 10.1371/journal.pone.0053708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunphy SE; Bratt JA; Akram KM; Forsyth NR; El Haj AJ, Hydrogels for lung tissue engineering: Biomechanical properties of thin collagen-elastin constructs. J Mech Behav Biomed Mater 2014, 38, 251–9. DOI: 10.1016/j.jmbbm.2014.04.005 [DOI] [PubMed] [Google Scholar]
- 16.Roudsari LC; Jeffs SE; Witt AS; Gill BJ; West JL, A 3D Poly(ethylene glycol)-based Tumor Angiogenesis Model to Study the Influence of Vascular Cells on Lung Tumor Cell Behavior. Sci Rep 2016, 6, 32726 DOI: 10.1038/srep32726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skardal A; Devarasetty M; Rodman C; Atala A; Soker S, Liver-Tumor Hybrid Organoids for Modeling Tumor Growth and Drug Response In Vitro. Ann Biomed Eng 2015, 43 (10), 2361–73. DOI: 10.1007/s10439-015-1298-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mall M; Grubb BR; Harkema JR; O’Neal WK; Boucher RC, Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice. Nat Med 2004, 10 (5), 487–93. 10.1038/nm1028 [DOI] [PubMed] [Google Scholar]
- 19.Lakshmanan I; Rachagani S; Hauke R; Krishn SR; Paknikar S; Seshacharyulu P; Karmakar S; Nimmakayala RK; Kaushik G; Johansson SL; Carey GB; Ponnusamy MP; Kaur S; Batra SK; Ganti AK, MUC5AC interactions with integrin beta4 enhances the migration of lung cancer cells through FAK signaling. Oncogene 2016, 35 (31), 4112–21. DOI: 10.1038/onc.2015.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su YC; Hsu YC; Chai CY, Role of TTF-1, CK20, and CK7 immunohistochemistry for diagnosis of primary and secondary lung adenocarcinoma. Kaohsiung J Med Sci 2006, 22 (1), 14–9. DOI: 10.1016/S1607-551X(09)70214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travis WD; Brambilla E; Noguchi M; Nicholson AG; Geisinger K; Yatabe Y; Powell CA; Beer D; Riely G; Garg K; Austin JH; Rusch VW; Hirsch FR; Jett J; Yang PC; Gould M; American Thoracic, S., International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc 2011, 8 (5), 381–5. DOI: 10.1513/pats.201107-042ST [DOI] [PubMed] [Google Scholar]
- 22.Wang S; Zhang H; Scharadin TM; Zimmermann M; Hu B; Pan AW; Vinall R; Lin TY; Cimino G; Chain P; Vuyisich M; Gleasner C; McMurry K; Malfatti M; Turteltaub K; de Vere White R; Pan CX; Henderson PT, Molecular Dissection of Induced Platinum Resistance through Functional and Gene Expression Analysis in a Cell Culture Model of Bladder Cancer. PloS one 2016, 11 (1), e0146256 DOI: 10.1371/journal.pone.0146256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friboulet L; Li N; Katayama R; Lee CC; Gainor JF; Crystal AS; Michellys PY; Awad MM; Yanagitani N; Kim S; Pferdekamper AC; Li J; Kasibhatla S; Sun F; Sun X; Hua S; McNamara P; Mahmood S; Lockerman EL; Fujita N; Nishio M; Harris JL; Shaw AT; Engelman JA, The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014, 4 (6), 662–673. DOI: 10.1158/2159-8290.CD-13-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y; Zou L; Zhang Y; Chen Y; Xing P; Yang W; Li F; Ji X; Liu F; Lu X, Transforming growth factor-beta1 and alpha-smooth muscle actin in stromal fibroblasts are associated with a poor prognosis in patients with clinical stage I-IIIA nonsmall cell lung cancer after curative resection. Tumour Biol 2014, 35 (7), 6707–13. DOI: 10.1007/s13277-014-1908-y [DOI] [PubMed] [Google Scholar]
- 25.Shiga K; Hara M; Nagasaki T; Sato T; Takahashi H; Takeyama H, Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers (Basel) 2015, 7 (4), 2443–58. DOI: 10.3390/cancers7040902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meylan E; Dooley AL; Feldser DM; Shen L; Turk E; Ouyang C; Jacks T, Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature 2009, 462 (7269), 104–7. DOI: 10.1038/nature08462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tammela T; Sanchez-Rivera FJ; Cetinbas NM; Wu K; Joshi NS; Helenius K; Park Y; Azimi R; Kerper NR; Wesselhoeft RA; Gu X; Schmidt L; Cornwall-Brady M; Yilmaz OH; Xue W; Katajisto P; Bhutkar A; Jacks T, A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 2017, 545 (7654), 355–359. DOI: 10.1038/nature22334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw AT; Kim DW; Nakagawa K; Seto T; Crino L; Ahn MJ; De Pas T; Besse B; Solomon BJ; Blackhall F; Wu YL; Thomas M; O'Byrne KJ; Moro-Sibilot D; Camidge DR; Mok T; Hirsh V; Riely GJ; Iyer S; Tassell V; Polli A; Wilner KD; Janne PA, Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013, 368 (25), 2385–94. DOI: 10.1056/NEJMoa1214886 [DOI] [PubMed] [Google Scholar]
- 29.Sequist LV; Yang JC; Yamamoto N; O'Byrne K; Hirsh V; Mok T; Geater SL; Orlov S; Tsai CM; Boyer M; Su WC; Bennouna J; Kato T; Gorbunova V; Lee KH; Shah R; Massey D; Zazulina V; Shahidi M; Schuler M, Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013, 31 (27), 3327–34. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 30.Solomon BJ; Mok T; Kim DW; Wu YL; Nakagawa K; Mekhail T; Felip E; Cappuzzo F; Paolini J; Usari T; Iyer S; Reisman A; Wilner KD; Tursi J; Blackhall F; Investigators, P., First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014, 371 (23), 2167–77. DOI: 10.1200/JCO.2017.77.4794 [DOI] [PubMed] [Google Scholar]
- 31.Pauli C; Hopkins BD; Prandi D; Shaw R; Fedrizzi T; Sboner A; Sailer V; Augello M; Puca L; Rosati R; McNary TJ; Churakova Y; Cheung C; Triscott J; Pisapia D; Rao R; Mosquera JM; Robinson B; Faltas BM; Emerling BE; Gadi VK; Bernard B; Elemento O; Beltran H; Demichelis F; Kemp CJ; Grandori C; Cantley LC; Rubin MA, Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov 2017, 7 (5), 462–477. DOI: 10.1158/2159-8290.CD-16-1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weeber F; van de Wetering M; Hoogstraat M; Dijkstra KK; Krijgsman O; Kuilman T; Gadellaa-van Hooijdonk CG; van der Velden DL; Peeper DS; Cuppen EP; Vries RG; Clevers H; Voest EE, Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A 2015, 112 (43), 13308–11. DOI: 10.1073/pnas.1516689112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.