Abstract
The high rate of intensive care unit false arrhythmia alarms can lead to disruption of care and slow response time due to desensitization of clinical staff. We study the use of machine learning models to detect false ventricular tachycardia (v-tach) alarms using ECG waveform recordings. We propose using a Supervised Denoising Autoencoder (SDAE) to detect false alarms using a low-dimensional representation of ECG dynamics learned by minimizing a combined reconstruction and classification loss. We evaluate our algorithms on the PhysioNet Challenge 2015 dataset, containing over 500 records (over 300 training and 200 testing) with v-tach alarms. Our results indicate that using the SDAE on Fast Fourier Transformed (FFT) ECG at a beat-by-beat level outperforms several competitive baselines on the task of v-tach false alarm classification. We show that it is important to exploit the underlying known physiological structure using beat-by-beat frequency distribution from multiple cardiac cycles of the ECG waveforms to obtain competitive results and improve over previous entries from the 2015 PhysioNet Challenge.
1. Introduction
Intensive care units (ICU) false arrhythmia alarm rates have been reported to be as high as 88.8%, and can lead to disruption of care and slow response time (Drew et al., 2014). Detecting and suppressing false arrhythmia alarms could potentially have high impact on the quality of patient care, reducing the chance of missing a life-threatening true alarm due to staff desensitization. We investigate representation learning approaches to finding discriminative features of ECG dynamics for false alarm reduction. We focus on the problem of detecting false alarms for one of the life-threatening arrhythmias, ventricular tachycardia (v-tach), defined as five or more ventricular beats with a heart rate higher than 100 bpm (Clifford et al., 2015). Of all the life-threatening arrhythmia alarm types, false v-tach alarms have proven to be the hardest to detect (Clifford et al., 2015), and remain an open challenge.
This work investigates the utility of both linear and non-linear embeddings of ECG for detecting v-tach false alarms. We present a supervised generative model, Supervised De-noising Autoencoders (SDAE), to classify ventricular tachycardia alarms using non-linear embeddings of ECG dynamics. The model, a variant of a Denoising Autoencoder (Vincent et al., 2008), is learned using a combination of discriminative and generative losses.
Furthermore, we explore feature transformations that utilize known physiological structure within ECG signals to enable learning under the constraints of limited labeled data. To this end, we propose a multi-stage approach that utilizes the FFT-transform of consecutive (heart) beats. We compare the SDAE against several baseline approaches that use a wide range of time and frequency domain ECG features.
Technical Significance
2The application of neural networks based representation learning techniques in arrhythmia analysis and false alarm reduction in critical care has had limited success, partly due to sparse availability of labeled data (Clifford et al., 2017; Schwab et al., 2018). The best performing approaches for false v-tach alarm detection in the 2015 PhysioNet Challenge rely on a combination of expert-defined rule-based logic analysis and simple machine-learning models (Clifford et al., 2016; Kalidas and Tamil, 2016; Plesinger et al., 2016). To make representation learning practical, in the low labeled data setting considered here, we leverage FFTs applied to the ECG signal spanned between consecutive heart beats. We then leverage the SDAE to learn nonlinear embeddings which are used for the task of false v-tach alarm detection. Tests on a real-world ICU dataset from 2015 PhysioNet Challenge containing over 500 records indicate that the proposed approach leads to improved performance over several baselines, including previous entries from the 2015 PhysioNet Challenge. The use of beat level information enables scalable learning of models that performs well even when labels are scarce.
Clinical Relevance
In modern ICUs, where critically-ill patients are closely monitored, as many as 187 audible alarms have been reported per ICU bed per day (Drew et al., 2014). Arrhythmia alarms contribute to approximately 45% of the overall ICU alarms, and have a high false alarm rate of 88.8%, which can lead to lower patient care quality (Drew et al., 2014; Clifford et al., 2015). Detecting and minimizing false alarms could potentially reduce the chance of missing a life-threatening true alarm due to staff desensitization.
2. Overview
2.1. Background and Motivation
Erroneous triggers of v-tach alarms typically occur due to noise and ECG-related artifacts (such as electrode movements) (Clifford, 2006). Ventricular arrhythmias are characterized by distortion of beat morphology with a broader QRS complex1. Our approach leverages the fact that the broader QRS complexes during v-tach often cause a shift in the spectral peak of the ECG to a lower frequency. However, a challenge to be overcome is that noise and artifacts can exhibit similar morphologies as abnormal QRS complexes, almost indistinguishable from the periodic anomalies from a true v-tach episode (Clifford, 2006). To tackle this, our approach leverages data from multiple heartbeats. The hypothesis we test is whether the changes in the frequency spectrum across multiple beats provide a sufficiently discriminative signal. To take into account that the discriminative function may potentially be non-linear, we leverage a neural network based representation learning approach to learn a lower-dimensional embedding of the ECG’s spectral dynamics which we use to distinguish between true and false alarms.
2.2. Approach Overview
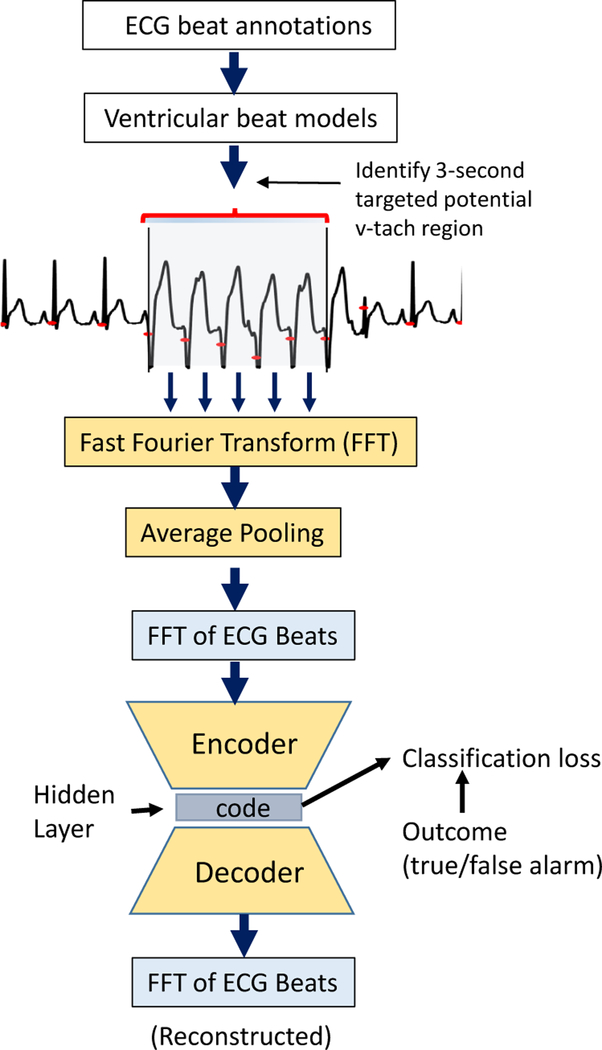
We take a multi-stage approach to false alarm detection from ECG segments. Figure 1 illustrates our approach from the ECG data processing pipeline to deriving FFT-transformed ECG features on a beat-by-beat basis as input to the model for false alarm detection.
Figure 1: Approach Overview:
ECG data processing pipeline and the SDAE model which uses FFT-transformed ECG beats as input for v-tach false alarm classification.
We use the MIT-BIH database, with annotated beat labels, to build a ventricular beat (v-beat) classifier that classifies whether the FFT transform of a beat is a ventricular or non-ventricular beat.
We apply a peak-detection algorithm to multi-channel ECG signals in the PhysioNet 2015 Challenge dataset to identify heartbeats. For each ECG record, in the last 25 seconds before alarm onset, we apply the v-beat classifier to estimate the probability of a ventricular beat. The goal is to obtain a small region in the ECG recordings to focus on when predicting alarm outcomes. This is done by identifying a 3-second target interval with the highest v-beat probability (averaged over consecutive beats) among beats where the heart rate exceeds 100bpm.
From the targeted region, we extract the following features: (i) FFT of beats: obtained by performing an FFT transform on each ECG beat from all ECG beats in the three-second target interval prior to the alarm onset. Each beat is represented as a 41-dimensional FFT-transformed feature vector, and (ii) FFT transform on the entire 3-second targeted ECG segment. Figure 1 illustrates the data processing and FFT-transformation pipeline.
We use the data from the previous step to classify (with different models) the probability of a true or false alarm. In Figure 1, we show how the best performing approach, the SDAE, is used to predict false alarms using the FFT-transformed features extracted from the target intervals of the training record.
3. Datasets
Our approach (with ECG data transformation pipeline outlined in Figure 1) requires the use of two datasets which we describe in this section.
3.1. MIT-BIH Arrhythmia Dataset
We use the MIT-BIH Arrhythmia Database (Moody and Mark, 2001; Goldberger et al., 2000) to train a ventricular beat identification model. The MIT-BIH dataset contains 48 half-hour excerpts of two-channel ambulatory ECG recordings (360 Hz), obtained from 47 subjects studied by the BIH Arrhythmia Laboratory between 1975 and 1979. All data are re-sampled to 250 Hz to match the sampling frequency of the PhysioNet Challenge data.
3.2. PhysioNet Challenge 2015 Dataset
The 2015 PhysioNet Challenge event (Clifford et al., 2015, 2016) focused on five types of life-threatening arrhythmias, including ventricular tachycardia (v-tach), asystole, extreme bradycardia, extreme tachycardia, and ventricular fibrillation/flutter. The goal of the challenge was to reduce the number of false alarms, while avoiding suppression of true alarms.
Task:
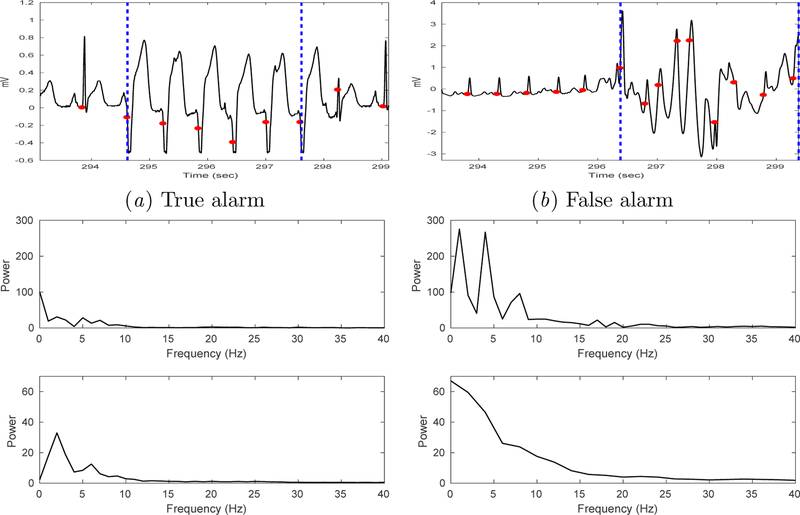
The PhysioNet 2015 Challenge (Clifford et al., 2015; Goldberger et al., 2000) consists of two events: (1) real-time classification using only data up to the alarm onset; (2) retrospective analysis in which the contestants are allowed to use the 10-second data after the alarm onset for classification. Here, we focus strictly on the real-time setting where only data prior to the alarm onset time is used. Figure 2 shows an example of true and false v-tach alarm each from the PhysioNet Challenge 2015 training dataset.
Figure 2:
Example true vs. false v-tach alarms (from PhysioNet Challenge 2015). Each plot shows data in the 25-second interval immediately prior to the v-tach alarm onset (at the end of the interval).
Statistics:
The dataset comprises 1250 records (750 train, 500 hidden test), each containing one of the five life-threatening arrhythmia alarms. Data is recorded on 3 to 4 channels, including two channels of ECG and one or two of the following channels: arterial blood pressure (ABP), respiration, or photoplethysmography (PPG). Each record contains 5-minute recordings of multi-channel physiological waveforms (250 Hz) immediately prior to the alarm onset. A subset of the records also contain 10-second recordings of the waveforms after the alarm onset. Among the 1250 records, 562 records contain v-tach alarms, and overall more than 75% of these v-tach alarms are false. The training and test sets of our study are extracted from the 562 records (train N = 341, test N = 221) corresponding to the v-tach alarms from the PhysioNet Challenge 2015 dataset. Among the 341 v-tach alarms in the training records, 73.3% are false (91 true alarms, and 250 false alarms). Among the 221 v-tach alarms in the hidden test set 79.6% are false (45 true alarm, 176 false alarms). We used 337 records from the training set containing either lead II (N = 331) or V (N = 312) ECGs. The hidden test set contains all 221 records with v-tach alarms. We extract the last 25-seconds ECG segments from each record prior to the alarm onset for analysis.
Visualization:
Figure 3 shows a plot of ECG waveform segments, their respective FFT from 3-sec targeted interval (top panel Figure 3c and d), and beat-level FFT-transformed features averaged over 3-seconds (bottom panel Figure 3c and d) from a true and false v-tach alarm records from the training set. Figure 4 compares the spectral content of ECG beats extracted from the targeted region of the true versus false alarm records from the dataset (lead II) at the population-level. Population median in each frequency bin is plotted as a solid line, with the interquartile range (IQR) as dashed lines. Note that the power spectral distribution of the true alarms peaks at the 4 Hz location. It is known that peaks at 1, 4, 7 and 10 Hz, in a normal ECG (with QRS width 80–100 msec), correspond approximately to the heart rate (at 60 bpm), T wave, P wave, and the QRS complex respectively (Clifford, 2006).
Figure 3:
Example ECG segments which triggered true (a) and false (b) v-tach alarms. Two vertical blue dashed lines demarcate the targeted 3-second segment identified by our algorithm. (c) and (d) show FFT from an entire 3-sec targeted segment (top) vs. average FFT from individual beats in the 3-second targeted interval (bottom).
Figure 4:
FFT of beats (lead II) - population median and IQR.
3.3. Targeting ECG Segments with the Ventricular Beat Identification Model
We used the peak detection algorithm developed by Johnson et al. (2015) and the Pan Tompkins algorithm (Pan and Tompkin, 1985) to identify each ECG beat for the MIT-BIH Arrhythmia database and the PhysioNet Challenge 2015 dataset. A model for ventricular beat identification was trained using the MIT-BIH database. We applied the v-beat classifier learned from the MIT-BIH data to identify a potential v-tach episode as a 3-second ECG segment from each record in the 2015 PhysioNet Challenge. We detail the algorithm used to identify the 3-second targeted interval in Appendix A.
3.4. Transforming from the Time-Domain to the Frequency-Domain
Due to heart rate variability, durations of RR-intervals (i.e. intervals between R-peaks) are different. It is a common approach to nondimensionalize time so that all the beats are aligned. Here, we adopt a different approach using frequency distribution of the ECG beat to avoid rescaling the time axis. FFT is applied to every beat within the selected 3-second range in order to obtain the frequency spectrum between 0.1Hz and 40Hz. The beat-level FFT of all beats from a 3-second targeted window of each record are then averaged.
4. Predictive Models for False Alarm Detection
4.1. Baselines
Across different inputs, for prediction, we study the use of: i) Logistic Regression (LR), ii) feed-forward neural network in the original space (MLP), iii) a feedforward neural network applied to the projection of data onto the principal components (PCA/MLP), and iv) Supervised Denoising Autoencoder (SDAE). For the baselines, a three-layer MLP with Rectified Linear Units (ReLU) as non-linearities between intermediate layers is used. The MLPs parameters were selected using a grid search performed over the layer size (16, 32, 64, 128, 256), and dropout (0.2, 0.4, 0.5). The dimensionality of the PCA had a significant impact on performance; it was selected using cross validation with a grid search over dimensions (5, 10, 15, 25).
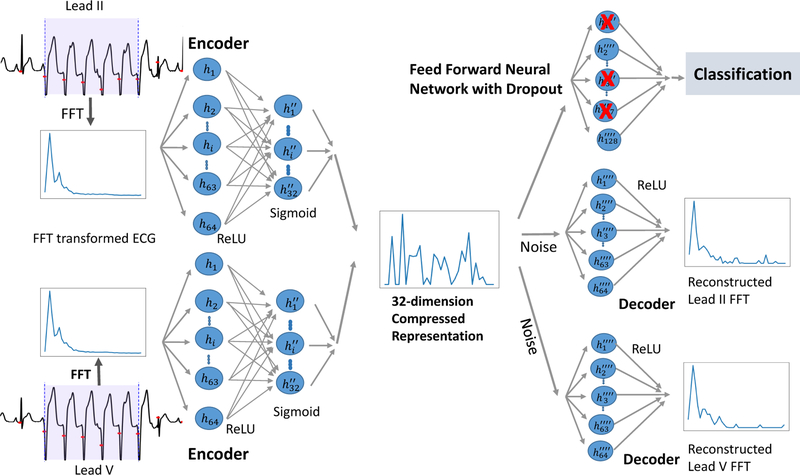
4.2. Supervised Denoising Autoencoder (SDAE)
To study how generative approaches affect this domain, we use a supervised denoising autoencoder on FFT transformed ECG waveforms. Denoising autoencoders (Vincent et al., 2010, 2008; Lovedeep and Gondara, 2016; Bengio et al., 2013) are a class of autoencoders where the goal of the model is to reconstruct the input given a noisy version of it. The model is trained to perform the reconstruction using an intermediate low-dimensional representation. Here, we use this representation to predict the label corresponding to whether or not the beats represented in the input correspond to a true alarm. Figure 5 shows the SDAE architecture when the model is applied to multi-channel ECG data. The SDAE may be viewed as learning a nonlinear projection akin to the linear one learned by PCA. The SDAE is trained using the ADAM optimizer (Kingma and Ba, 2014) with two loss functions: (1) a reconstruction loss (in our case the mean-squared error) that encourages the model to learn a representation capable of reconstructing the input and (2) a prediction loss that encourages the low-dimensional representation to be well suited to predicting the likelihood of a true alarm. The model parameters were selected using a grid search performed over the layer size (16, 32, 64, 128, 256), dropout (0.2, 0.4, 0.5), and variance of the noise added to the hidden state (0, 0.001, 0.01). A ten-fold-cross-validation was used to compute the validation accuracy, which in turn was used to determine the best model parameters. The parameters and architecture of the model are discussed in Appendix C.
Figure 5:
SDAE model diagram for v-tach false alarm detection using two leads of ECG. Each of these input ECG channels is run through two perception layers and then combined to form a 32-dimension compressed hidden-layer representation. The data from individual leads are then reconstructed through separate decoding layers, while an outcome and classification loss are gathered using the combined layer.
4.3. Evaluation
Learning from ECG waveforms:
We study predicting false alarms from the following types of ECG features i) default 10-second raw ECG waveform intervals extracted from 10-seconds prior to the alarm onset (denoted waveform) ii) targeted 3-second raw ECG waveform segments identified using the MIT-BIH ventricular beat model (denoted 3-sec interval) and iii) beat-level spectral representation from the targeted region (denoted beats).
Experimental Procedure:
The training data was split 10 different times, each resulting in 70% used for training and 30% used for validation. We report the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) and Area under the Curve (AUC) from the receiver operating characteristic (ROC) curve. Confidence intervals (CI) for area under the receiver operating curves (AUCs) were based on the method described in DeLong et al. (1988). For our final model, our primary performance metric when comparing with previous approaches is the PhysioNet Challenge 2015 scores, defined as (TP + TN)/(TP + TN + FP + 5 * FN), a function of the following variables: true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN).
5. Results
5.1. Classification Using Single-Lead ECG Waveforms
Table 1 compares the performance of various representation learning techniques and classifiers when using different feature representations of single-lead ECG. In particular, our goal is to characterize and compare the performance of classifiers built on linear (PCA) vs. non-linear (autoencoder-based) representations of various time and frequency domain features of ECG signals. We experiment with (i) the raw ECG waveform from targeted 3-second segments, (ii) frequency content from targeted 3-second ECG segments, and (iii) averaged beat-level frequency content from targeted ECG segments. Additionally, we also evaluated the performance of classifiers that predict false alarms using the 10-second raw waveform immediately prior to the alarm onset; the performance (not shown in Table 1) of all classifiers investigated in this study was significantly worse (AUCs mostly in the 0.60’s range or lower) than using the 3-second targeted waveform.
Table 1:
Single lead ECG. Lead II (N=214), V (N=203). AUC (95% CI) shown.
| Data | LR | MLP | PCA+MLP | SDAE | |||
|---|---|---|---|---|---|---|---|
| 1 | 3-sec waveform | II | 0.54 (0.44, 0.64) | 0.77 (0.67, 0.86) | 0.83 (0.75, 0.92) | 0.77 (0.68, 0.86) | |
| 2 | 3-sec waveform | V | 0.57 (0.47, 0.67) | 0.74 (0.65, 0.84) | 0.78 (0.69, 0.87) | 0.76 (0.67, 0.85) | |
| 3 | FFT 3-sec interval | II | 0.70 (0.60, 0.80) | 0.84 (0.76, 0.92) | 0.85 (0.77, 0.93) | 0.87 (0.80, 0.94) | |
| 4 | FFT 3-sec interval | V | 0.73 (0.63, 0.82) | 0.81 (0.73, 0.90) | 0.85 (0.77, 0.93) | 0.86 (0.77, 0.93) | |
| 5 | FFT of beats | II | 0.76 (0.67, 0.86) | 0.89 (0.86, 0.98) | 0.88 (0.84, 0.97) | 0.87 (0.80, 0.94) | |
| 6 | FFT of beats | V | 0.69 (0.60, 0.79) | 0.87 (0.79, 0.94) | 0.87 (0.79, 0.94) | 0.88 (0.80, 0.94) | |
We find that FFT-transforming the ECG signal prior to predicting false alarms performs significantly better than using the raw ECG waveforms directly. Furthermore, we find that learning a non-linear representation of the ECG frequency content using SDAE achieves even better performance, with an AUC of 0.87 (0.80, 0.94) and 0.86 (0.77, 0.93) using lead II and V ECG channels respectively, than other baselines that include LR, MLP and PCA/MLP. Using FFT-transformed beat-level representations from single lead ECG, we note that MLP and PCA-MLP performed better than when FFT features from entire waveform segments were used (Table 1). Table 1 demonstrates that exploiting beat-level information in ECG dynamics generally leads to a boost in performance results.
5.2. Classification with Two-Channels of ECG Waveforms
Table 2 compares the performance of different classifiers using two-lead ECGs. We list the best known results from the 2015 PhysioNet Challenge as a comparison. The first-place entry by Kalidas and Tamil (2016) achieved a Challenge score of 75.10 in the real-time event and 76.70 when considering both real-time and retrospective events. The second-place entry in v-tach alarm by Plesinger et al. (2016) used ECG, ABP, and PPG waveforms, and achieved a Challenge score of 72.73 in real-time event, and 75.07 when considering both real-time and retrospective events. The performance is generally better in retrospective events, since data post alarm onset can be used for classification.
Table 2:
Performance Using Two-Channel ECG (N=221). RT (realtime), Retro (retrospective).
| Event | Features | Sens. | Spec. | Prec. | F1 | AUC | Score | ||
|---|---|---|---|---|---|---|---|---|---|
| 1 | MLP | RT | FFT 3-sec interval | 0.89 | 0.73 | 0.45 | 0.60 | 0.87 (0.80, 0.94) | 67.98 |
| 2 | PCA-MLP | RT | FFT 3-sec interval | 0.87 | 0.78 | 0.48 | 0.62 | 0.89 (0.83, 0.96) | 69.29 |
| 3 | SDAE | RT | FFT 3-sec interval | 0.84 | 0.79 | 0.51 | 0.63 | 0.89 (0.82, 0.95) | 68.49 |
| 4 | MLP | RT | FFT of beats | 0.89 | 0.67 | 0.52 | 0.64 | 0.89 (0.83, 0.96) | 71.65 |
| 5 | PCA-MLP | RT | FFT of beats | 0.89 | 0.80 | 0.50 | 0.65 | 0.88 (0.82, 0.95) | 73.82 |
| 6 | SDAE | RT | FFT of beats | 0.89 | 0.86 | 0.62 | 0.73 | 0.91 (0.85, 0.97) | 77.59 |
| 7 | Challenge,1st | RT | ECG | 0.89 | 0.80 | - | - | - | 75.10 |
| 8 | Challenge, 2nd | RT | ECG/ABP | 0.82 | 0.84 | - | - | - | 72.73 |
| 9 | Challenge, 1st | RT/Retro | ECG | 0.90 | 0.82 | - | - | - | 76.75 |
| 10 | Challenge, 2nd | RT/Retro | ECG/ABP | 0.85 | 0.84 | - | - | - | 75.07 |
We find that using SDAE to learn non-linear embeddings of beat-by-beat FFT-transformed ECG achieved the best performance in comparison to other baselines. The SDAE achieved an AUC of 0.91 (0.85, 0.97) and a higher F1 score (0.73) and Challenge score (77.59) than other baselines, including MLP and PCA/MLP. We also note that SDAE achieved a higher Challenge score (77.59) than the winning Challenge 2015 entry (score 75.10) by Kalidas and Tamil in the real-time event (using data only from prior to the alarm onset). SDAE reduced the v-tach false alarm rate of the test set from 79.64% to 11.3% (suppressed 151 false v-tach alarms), at the cost of missing 11.1% (5 out of 45) true alarms. We report the specificity of SDAE when sensitivity is set at 0.89, and note that SDAE has a higher specificity (0.86) than the other baselines for the real-time event.
5.3. Varying the Number of Training Examples
We investigate how the SDAE behaves under different feature representations (beat-by-beat vs. FFT-transformed 3-second waveform segments) as a function of the labeled data size. Table 3 shows the classification performance of SDAE as the training data size varies from 25 to over 300 using FFT transformation of 3-second segment versus beat-by-beat data respectively. Using FFT-transformed beat-by-beat data, SDAE scales gracefully as the training sample size is reduced to 25.
Table 3:
SDAE performance as a function of training size using two ECG channels. (Test set N=221)
| Training Size | 25 | 50 | 150 | 250 | 337 | |
|---|---|---|---|---|---|---|
| 1 | FFT waveform | 0.63 (0.53, 0.72) | 0.63 (0.53, 0.72) | 0.87 (0.80, 0.94) | 0.88 (0.81, 0.95) | 0.89 (0.82, 0.95) |
| 2 | FFT B2B | 0.86 (0.79, 0.93) | 0.87 (0.80, 0.94) | 0.91 (0.85, 0.97) | 0.91 (0.85, 0.97) | 0.91 (0.85, 0.97) |
Using 25 training samples, SDAE with beat-by-beat data achieved an AUC of 0.86 (0.79, 0.93) which is only slightly worse than when using the full training data size (N=337) (p = 0.035). Using 50 or more training samples, SDAE with beat-by-beat data achieved similar performance as using the full data set (AUC of 0.87 [0.80, 0.94] trained with 50 samples vs.0.91 [0.85, 0.97] with full dataset, p-val = 0.058). This showcases that leveraging beat level information is crucial for ensuring good predictive performance when labels are scarce.
6. Discussion and Related Work
Human heart beats generate a wide-range of complex ECG dynamics, which have been studied extensively under both healthy and pathological conditions. Classical ECG analyses are based on hand-crafted features obtained from temporal and/or frequency analyses, which are then used as inputs in a machine learning classifier. Recent advances on deep learning inspire new models where features are learned from segments of ECG signals. While deep learning has made significant advances in the domains of image and voice analysis, the application of deep learning in physiological waveform analysis has had limited success, partly due to limited availability of labeled data. Expert-defined rule-based approaches or simple machine learning models (such as gradient boosting, or random forest) combined with hand-crafted features often outperform more complex models, such as deep neural networks (Clifford et al., 2017).
Here, our results indicate that direct application of several machine learning techniques on raw waveforms performed poorly (with the current training sample size). We show that learning representations of the FFT-transformed ECG waveforms results in significantly better performance than using raw waveforms. We study both linear and non-linear embeddings of ECG for the purpose of detecting false v-tach alarms. Tests on PhysioNet Challenge 2015 dataset indicate that, for both linear and non-linear embeddings, representation learning approaches that exploit the underlying known physiological structure, specifically, using FFT-transformed ECG beats, averaged over multiple cardiac cycles, lead to higher Challenge scores compared to models that use the entire ECG waveform segments. In the case of SDAE, this procedure is key to out-performing previous known best results from PhysioNet 2015 Challenge. We conjecture that this averaged beat-by-beat Fourier feature representation significantly simplifies the learning task (relative to models that use the entire ECG waveform segments) under the constraint of limited labeled data.
When comparing the performance of linear vs. non-linear embeddings using various ECG feature transformation, we observe that non-linear embeddings from SDAE achieved slightly better performance than PCA/MLP, when beat-level frequency features from two-channels of ECG were used. Further investigation is required to characterize and improve the performance of these approaches as the sample size increases and to avoid suppressing any true positive alarms. An avenue of future work is to leverage unlabeled data to improve the quality of the nonlinear embeddings in the SDAE generative framework. We leave taking advantage of this to future work.
The PhysioNet Challenge 2015 event focused on five types of life-threatening arrhythmias, including asystole, extreme bradycardia, extreme tachycardia, ventricular tachycardia, and ventricular fibrillation/flutter. More than 200 entries were submitted for this event (see the collection of articles in the special issue of Physiological Measurement (Clifford et al., 2015)). Among all the arrhythmia alarm types, v-tach has proven to be the hardest to classify, and received the lowest prediction accuracy (Clifford et al., 2015). While machine learning approaches have been proposed (Eerikinen et al., 2016; Ansari et al., 2016), the winning entry in v-tach alarm classification by Kalidas and Tamil (2016) used logical analysis to improve classification results from SVM classification, and Plesinger et al. (2016) used signal processing and rule-based reasoning to achieve the second best performance in v-tach alarm classification in the 2015 PhysioNet Challenge.
Early work on v-tach false alarm reduction by Aboukhalil et al. (2008) used a data fusion approach with rule-based logic to reduce 5 types of false arrhythmia alarms from 42.7% to 17.2% (on average) when simultaneous ECG and arterial blood pressure waveform were available. They reported false v-tach alarm suppression as the most challenging task (with the lowest suppression rate among all alarm categories tested), with a reduced false v-tach alarm rate from 46.6% to 30.8%, at the cost of suppressing 14.5% and 4% of the true alarms in the train and test set respectively. In (Schwab et al., 2018), a supervised multitask learning approach was proposed to reduce the number of training labels required to suppress general ICU false alarms. Our work, in contrast, focuses on v-tach false alarms, and uses FFT-transformation of individual ECG beats for scalable learning. Rajpurkar et al. (2016) used a deep convolutional neural network for arrhythmia detection, and obtained good performance with a significantly larger dataset.
7. Conclusion
We developed a supervised representation learning approach to detect false v-tach alarms from two leads of ECG waveforms, and obtained improved performance over several base-lines, including previous results in the same task using the PhysioNet 2015 Challenge dataset. Our final best-performing model used a supervised denoising autoencoder, SDAE, to learn non-linear embeddings of spectral dynamics, averaged over multiple cardiac cycles. These results suggest that generative modeling may play a role in tackling the problem of v-tach false alarm detection. Future work will extend the current approach to combine information from multi-channel physiological waveforms (PPG, ABP) for false arrhythmia alarm reduction. We expect the full potential of such representational learning methods will lead to more significant results when the sample sizes are greatly increased. We also expect that representation learning will play an important role in analyzing other biomedical waveforms, such as time series of blood pressure, respiration, electroencephalogram (EEG), and photoplethysmogram (PPG).
ACKNOWLEDGMENTS
The authors thank Dr. Alistair Johnson for his assistance and generous contribution of his ECG beat annotation software. We thank Dr. Gari Clifford and the 2015 PhysioNet/CinC Challenge organizing team, especially Mr. Benjamin Moody for his help in providing the realtime Challenge scores for Challenge entries. We thank Dr. Wei Zong for valuable discussion. This work was in part supported by the National Institutes of Health (NIH) grant R01GM104987 and the National Science Foundation (NSF) grant CMMI-1661615. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, or the NSF.
Appendix A: Identify Potential V-Tach Episodes
A model for ventricular beats identification was trained using a total of 107,129 annotated beats from lead II of the MIT-BIH database (7,104 ventricular beats). A model for lead was also trained using 96,189 annotated beats from lead V in MIT-BIH database (7,010 ventricular beats). We extracted the FFT features from all beats, and trained a ventricular-beat logistic classifiers to classify a beat as either a ventricular or non-ventricular beat based on the FFT transformed features. The 10-fold cross validated median training AUC (and inter-quartile range) of this ventricular beat classifier is 0.94 (0.93, 0.94) using the MIT-BIH arrhythmia dataset.
For each beat (in the last 25-seconds of each record) in the PhysioNet Challenge dataset, we used the MIT-BIH ventricular beat model to estimate the probability of each beat being a ventricular beat based on the FFT of individual beats. At each beat j, with onset time tj, we estimated the probability of five consecutive beats starting at time tj being ventricular beats, by averaging v-beat probabilities over five consecutive beats. The onset time Ti of the potential v-tach region of each record i is identified as the onset time of the beat with the highest running 5-beat ventricular-beat probability (from among the beats with RR-intervals shorter than 600 ms). For each record i, we extracted its FFT features by averaging FFT of all individual beats in a K-second window starting at time Ti. We chose K to be 3 (seconds), since the v-tach is defined as 5 consecutive ventricular beats with heart rate of over 100 bpm, and therefore a 3-second of ECG interval should be sufficient in length to identify potential v-tach episodes (5 beats at less than 600 ms RR-interval per beat).
Appendix B: MLP Parameter Settings
The first layer of both input channels is a 64 neuron layer, with ‘ReLU’-activation, followed by a dropout of 0.5. The channels are then added together and given to 128-neuron ‘ReLU’ layer. Finally, a final sigmoid layer is applied for prediction.
Appendix C: SDAE Parameter Settings
The SDAE uses the ‘ADAM’ optimizer in attempt to minimize ‘mean-squared-error’ and ‘binary cross-entropy’ loss. Each input channel sequentially leads into a layer of size 64, each using a ‘ReLU’ activation function. Afterwards, the output of the size 64 hidden layer is given to a layer size of 32, which uses a ‘sigmoid’ activation function. The output of both of the 32 unit hidden layers are added together in order to find an underlying representation of the combined data. Depending on the results of the grid search, Gaussian noise is applied to the summed layer. The leads are once again separated and sent into a decoding 64 unit ‘ReLU’ layer, and a 41 unit ‘ReLU’ layer, which attempts to reconstruct the original signal from its respective lead. The output of summed layer is fed into a simple feed forward neural network. This simple feed forward neural network consisting of only 1 hidden layer with a hidden unit size of 128 and a ‘ReLU’ activation function; the output is then fed into a sigmoid layer to obtain a prediction. The model utilizes the ADAM optimizer, and a batch size of 64.
Footnotes
Research performed while at MIT.
Research performed while at MIT.
The width of a QRS complex corresponds to the time for the ventricles to depolarize.
Contributor Information
Eric P. Lehman, College of Computer and Information Science, Northeastern University, Boston, MA.
Rahul G. Krishnan, CSAIL & Institute for Medical Engineering & Science, Massachusetts Institute of Technology, Cambridge, MA
Xiaopeng Zhao, Department of Mechanical, Aerospace, and Biomedical Engineering, University of Tennessee, Knoxville, TN.
Roger G. Mark, Institute for Medical Engineering & Science, Massachusetts Institute of Technology, Cambridge, MA
Li-wei H. Lehman, Institute for Medical Engineering & Science, Massachusetts Institute of Technology, Cambridge, MA
References
- Aboukhalil A, Nielsen L, Saeed M, Mark RG, and Clifford GD. Reducing false alarm rates for critical arrhythmias using the arterial blood pressure waveform. J Biomed Inform, 41 (3):442–51, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari Sardar, Belle Ashwin, Ghanbari Hamid, Salamango Mark, and Najarian Kayvan. Suppression of false arrhythmia alarms in the ICU: a machine learning approach. Physiological Measurements, 37(8):1186–203, 2016. [DOI] [PubMed] [Google Scholar]
- Bengio Yoshua, Yao Li, Alain Guillaume, and Vincent Pascal. Generalized denoising autoencoders as generative models In Burges CJC, Bottou L, Welling M, Ghahramani Z, and Weinberger KQ, editors, Advances in Neural Information Processing Systems 26, pages 899–907. Curran Associates, Inc., 2013. [Google Scholar]
- Clifford Gari. Advanced Methods and Tools for ECG Data Analysis. Artech House, 2006. [Google Scholar]
- Clifford Gari, Silva Ikaro, Moody Benjamin, Li Qiao, Kella Danesh, Shahin Abdullah, Kooistra Tristan, Perry Diane, and Mark Roger. The PhysioNet/Computing in Cardiology PhysioNet challenge 2015: Reducing false arrhythmia alarms in the ICU. In Proceedings of Computing in Cardiology, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford Gari, Liu Chengyu, Moody Benjamin, Lehman Li-wei, Silva Ikaro, Li Qiao, Johnson Alistair, and Mark Roger. AF classification from a short single lead ECG recording: the PhysioNet Computing in Cardiology Challenge 2017. In Proceedings of Computing in Cardiology, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford GD, Silva I, Moody B, Li Q, Kella D, Chahin A, T Kooistra, Perry D, and Mark RG. False alarm reduction in critical care. Physiological Measurements, 37(8):E5–E23, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong Elizabeth R, DeLong David M, and Clarke-Pearson Daniel. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics, pages 837–845, 1988. [PubMed] [Google Scholar]
- Drew Barbara J, Harris Patricia, Zgre-Hemsey Jessica K., Mammone Tina, Schindler Daniel, Salas-Boni Rebeca, Bai Yong, Tinoco Adelita, Ding Quan, and Hu Xiao. Insights into the problem of alarm fatigue with physiologic monitor devices: A comprehensive observational study of consecutive intensive care unit patients. PLOSONE, 9(10), 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eerikinen Linda, Vanschoren Joaquin, Rooijakkers Michael J., Vullings Rik, and Aarts Ronald M. Reduction of false arrhythmia alarms using signal selection and machine learning. Physiological Measurements, 2016. [DOI] [PubMed] [Google Scholar]
- Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng C-K, and Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation, 101 (23):e215–e220, June 2000. [DOI] [PubMed] [Google Scholar]
- Johnson AEW, Behar J, Andreotti F, Clifford GD, and Oster J. Multimodal heart beat detection using signal quality indices. Physiological Measurements, 36:1665–1677, 2015. Code available at https://github.com/alistairewj/peak-detector/. [DOI] [PubMed] [Google Scholar]
- Kalidas V and Tamil LS. Cardiac arrhythmia classification using multi-modal signal analysis. Physiological Measurements, 37(8):1253–72, 2016. [DOI] [PubMed] [Google Scholar]
- Kingma Diederik P and Ba Jimmy. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014. [Google Scholar]
- Lovedeep and Gondara. Medical image denoising using convolutional denoising autoencoders. IEEE 16th International Conference on Data Mining Workshops (ICDMW), pages 241–246, 12 2016. [Google Scholar]
- Moody GB and Mark RG. The impact of the MIT-BIH arrhythmia database. IEEE Eng in Med and Biol, 20(3):45–50, 2001. [DOI] [PubMed] [Google Scholar]
- Pan Jiapu and Tompkin Willis. A real-time QRS detection algorithm. IEEE transactions on biomedical engineering, 3(4–5):230–236, 1985. [DOI] [PubMed] [Google Scholar]
- Plesinger F, Klimes P, Halamek J, and Jurak P. Taming of the monitors: reducing false alarms in intensive care units. Physiological Measurements, 37:1313–1325, 2016. [DOI] [PubMed] [Google Scholar]
- Rajpurkar Pranav, Hannun Awni, Haghpanahi Masoumeh, Bourn Codie, and Ng Andrew. Cardiologist-level arrhythmia detection with convolutional neural networks. In https://arxiv.org/pdf/1707.01836.pdf, 2016.
- Schwab Patrick, Keller Emanuela, Muroi Carl, Mack David J., Christian Strässle, and Walter Karlen. Not to cry wolf: Distantly supervised multitask learning in critical care. In Proceedings of the 35th International Conference on Machine Learning, 2018. [Google Scholar]
- Vincent Pascal, Larochelle Hugo, Bengio Y, and Manzagol Pierre-Antoine. Extracting and composing robust features with denoising autoencoders. In Proceedings of the 25th International Conference on Machine Learning, pages 1096–1103, 01 2008. [Google Scholar]
- Vincent Pascal, Larochelle Hugo, Lajoie Isabelle, Bengio Yoshua, and Manzagol Pierre-Antoine. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. Journal of Machine Learning Research, 2010. [Google Scholar]