Abstract
Background
The latent human immunodeficiency virus type 1 (HIV-1) reservoir represents a major barrier to a cure. Based on the levels of HIV-1 DNA in naive (TN) vs resting memory CD4+ T cells, it is widely hypothesized that this reservoir resides primarily within memory cells. Here, we compared virus production from TN and central memory (TCM) CD4+ T cells isolated from HIV-1–infected individuals on suppressive therapy.
Methods
CD4+ TN and TCM cells were purified from the blood of 7 HIV-1–infected individuals. We quantified total HIV-1 DNA in the CD4+ TN and TCM cells. Extracellular virion-associated HIV-1 RNA or viral outgrowth assays were used to assess latency reversal following treatment with anti-CD3/CD28 monoclonal antibodies (mAbs), phytohaemagglutinin/interleukin-2, phorbol 12-myristate 13-acetate/ionomycin, prostratin, panobinostat, or romidepsin.
Results
HIV-1 DNA was significantly higher in TCM compared to TN cells (2179 vs 684 copies/106 cells, respectively). Following exposure to anti-CD3/CD28 mAbs, virion-associated HIV-1 RNA levels were similar between TCM and TN cells (15 135 vs 18 290 copies/mL, respectively). In 4/7 donors, virus production was higher for TN cells independent of the latency reversing agent used. Replication-competent virus was recovered from both TN and TCM cells.
Conclusions
Although the frequency of HIV-1 infection is lower in TN compared to TCM cells, as much virus is produced from the TN population after latency reversal. This finding suggests that quantifying HIV-1 DNA alone may not predict the size of the inducible latent reservoir and that TN cells may be an important reservoir of latent HIV-1.
Keywords: HIV-1, latent, reservoir, naive, memory
In this study, we show that although the frequency of human immunodeficiency virus type 1 infection is lower in naïve compared to central memory cells CD4+ T cells, as much virus is produced from the naive population following latency reversal.
The reservoir of latent human immunodeficiency virus type 1 (HIV-1) that resides in resting CD4+ T cells constitutes a major barrier to a cure [1–5]. This reservoir is unaffected by the immune system or by antiretroviral therapy (ART) but has the potential to produce infectious virus, which may contribute to persistent plasma viremia during ART or viral rebound if ART is interrupted. The resting CD4+ T-cell population, however, is heterogeneous, and HIV-1 DNA has been detected in a number of different T-cell subsets, including naive (TN), stem cell-like memory (TSCM), central memory (TCM), transitional memory (TTM), effector memory (TEM), and terminally differentiated (TTD) cells [6–13]. Several studies have tried to define the relative contributions of each of these subsets to the total pool of latent HIV-1 infection by comparing total viral DNA in the sorted CD4+ T-cell subsets in peripheral blood [6–8, 12]. In general, these studies found that the TCM, TTM, and TEM cell populations consistently contained higher levels of HIV-1 DNA than TN cells. Consequently, reservoir studies have primarily focused on the memory CD4+ T-cell compartment, and the TN cell population has been largely overlooked.
In our laboratory, we recently used a primary cell model of HIV-1 latency to address differences in the establishment and reversal of viral latency in highly purified TN and TCM cells [14]. Consistent with previously published studies [15–18], we found that HIV-1 infected TN cells less efficiently than TCM cells. However, when the infected TN cells were treated with latency reversing agents (LRAs), including anti-CD3/CD28 monoclonal antibodies (mAbs), phorbol-myristate-acetate (PMA)/phytohaemagglutinin (PHA), and prostratin, as much, if not more, extracellular virion-associated HIV-1 RNA was produced per infected TN cell compared to infected TCM cell. There were no major differences in the genomic distribution of HIV-1 integration sites between TN and TCM cells that accounted for these observed differences [14]. These findings suggested that TN cells may be an important reservoir of latent HIV-1 infection and should not be overlooked simply because the frequency of infection of these cells is lower than in the memory T-cell subsets. To validate these in vitro findings, in this study, we quantified the inducible latent HIV-1 reservoirs in CD4+ TN and TCM cells from infected individuals on suppressive ART.
METHODS
Isolation of CD4+ TN and TCM Cells From HIV-1–infected Individuals on ART
Leukapheresis was performed on 7 HIV-1–infected donors who were on suppressive ART (<20 copies of HIV-1 RNA/mL plasma) for ≥5 years (Table 1). We obtained approximately 2×109 peripheral blood mononuclear cells (PBMCs) from each leukapheresis to complete the studies described. All donors provided written informed consent, and the University of Pittsburgh Institutional Review Board approved the blood donation protocol. CD4+ TN cells were isolated as previously described [14]. In 3 donors, the TSCM cells were removed from the TN population by depletion of CD95+ cells. TCM cells were isolated as previously described [14], with the addition of positive selection for CD62L+ cells after isolating the CD45RA- cells. The CD62L- fraction was also saved as the TTM+TEM cell population. In Supplementary Figure 1, we provide a detailed schematic that outlines our purification strategy. All magnetic bead purification kits and antibodies were from Miltenyi Biotec (Auburn, CA). The purity of the TN and TCM cells was assessed by flow cytometry (LSR II, BD Biosciences) using the following antibodies: CD3-V450, CD4-PerCP-Cy5.5, CD45RA-FITC, CCR7-PE, CD27-APC-H7, and CD62L-APC (BD Biosciences). Data were analyzed using FlowJo vX.0.7. The TN and TCM cell surface phenotypes were as follows: TN cells (CD45RA+, CCR7+, CD27+, CD62L+, [CD95-]) and TCM cells (CD45RA-, CCR7+, CD27+, CD62L+). TN and TCM cell purity was routinely found to be ≥98 % and ≥96 %, respectively (Supplementary Figure 2).
Table 1.
Characteristics of Study Participants
| Donor No. | Sex | Age (y) | Race | Current CD4 Count, cells/mm3 | Pre-ART Viral Load, copies of HIV-1 RNA/mL | Years of Suppression | Current ART Regimen |
|---|---|---|---|---|---|---|---|
| 1 | F | 36 | Black | 809 | 802 000 | 10.2 | TDF/FTC/RPV |
| 2 | F | 57 | Black | 380 | 143 000 | 5.2 | TDF/FTC/ATZ/r |
| 3 | M | 57 | White | 656 | 520 000 | 12 | TDF/FTC/EFV |
| 4 | M | 51 | White | 603 | Unknown | 7.3 | TAF/FTC/EVG/c |
| 5 | M | 46 | Black | 714 | 41 952 | 6.0 | ABC/3TC/EFV |
| 6 | F | 58 | Black | 1426 | 366 200 | 15 | TDF/FTC/RPV |
| 7 | F | 62 | Black | 1033 | Unknown | 10.7 | TDF/FTC/DTG |
| Mean | 52 | 803 | 9.5 |
Abbreviations: 3TC, lamivudine; ABC, abacavir; ATZ/r, atazanavir/ritonavir; DTG, dolutegravir; EFV, efavirenz; EVG/c, elvitegravir/cobicistat; FTC, emtricitabine; RPV, riplivirine; TAF, tenofovir alafenamide; TDF, tenofovir disproxil fumarate.
Ex vivo Reactivation of Latent HIV-1 From CD4+ TN and TCM Cells
Purified TN and TCM cells were plated in 24-well plates at a density of 106 cells/well and cultured in the presence of 300 nM efavirenz and 300 nM raltegravir to prevent viral spread. Cells were activated for 7 days in duplicate wells with anti-CD3/CD28 mAbs (3 beads per cell; Life Technologies), 10 µg/mL PHA (Remel) + 100 U/mL interleukin 2 (IL-2; Roche), 5 nM PMA + 500 μg/mL ionomycin (Sigma), 5 µM prostratin, 17 nM panobinostat (Selleckchem; pulsed for 30 minutes), or 50 nM romidepsin (Selleckchem; pulsed for 4 hours). Unstimulated cells were used as a negative control. Two and 4 days post-activation, IL-2 (10U/mL), efavirenz (300nM), and raltegravir (300nM) were added to each well.
Quantification of Total HIV-1 DNA
Total HIV-1 DNA was quantified in freshly isolated T-cell subsets as defined in Supplementary Figure 3, as well as in pooled duplicate culture wells. Total cellular DNA was extracted and was assayed for total HIV-1 DNA by quantitative polymerase chain reaction assay (PCR), as described previously [19, 20]. Each sample was run in triplicate using the LightCycler 480 System (Roche). DNA standards were included, as described previously [21]. HIV-1 DNA was normalized to the total number of cells assayed by quantitative PCR amplification of the CCR5 gene [22].
Quantification of Extracellular Virion-associated HIV-1 RNA
Extracellular virion-associated HIV-1 RNA was extracted and quantified as previously described [14].
Quantitative Viral Outgrowth Assay
The quantitative viral outgrowth assay was carried out as previously described [23]. Infectious units per million cells (IUPM) were calculated as described previously [23, 24]. Outgrowth positive wells were determined using a p24 enzyme-linked immunosorbent assay (ZeptoMetrix Corporation).
Flow Cytometry
T-cell activation was assessed by flow cytometry using the following antibodies (from BD Biosciences): CD3-V450, CD4-PerCP-Cy5.5, CD25-PE-Cy7, CD69-PE, and HLA-DR-FITC. Cell viability was determined using a LIVE/DEAD fixable cell viability dye for flow cytometry (Invitrogen). All samples were run on an LSRII, and the data were analyzed using FlowJo vX.0.7.
Statistical Analyses
Statistical comparison between paired samples was performed using a Wilcoxon matched-pairs signed rank test. For all unpaired samples, statistics were determined using a Mann-Whitney test. For statistical comparisons between unpaired samples where N <6, statistics were determined using an unpaired t test. For all statistical analyses, P < .05 was considered significant. All statistics were calculated in GraphPad Prism v6.0.
RESULTS
Donor Characteristics
Experiments were performed using PBMC obtained from 7 (4 females, 3 males) chronically HIV-1–infected donors on suppressive ART who met the eligibility criterion of having plasma HIV-1 RNA ≤20 copies/mL for ≥5 years, with a median of 9.5 years (Table 1). The median age was 52 years. Five of the donors were black and 2 were white. The median CD4+ T-cell count at the time of leukapheresis was 803 cells/mm3.
CD4+ TN Cells Harbor Less Total HIV-1 DNA Than TCM Cells
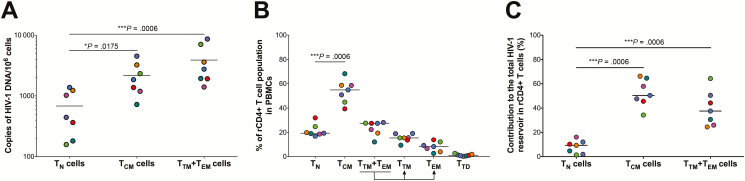
HIV-1 DNA was detectable in both the TN and TCM subsets in all 7 donors (Figure 1A, Supplementary Table 1). However, consistent with prior studies [6–8, 25], the levels of total HIV-1 DNA were significantly higher (median fold change, 5.4; range, 1.2–14.7; P = .0175) in the TCM cells (mean, 2179 copies/106 cells; range, 723–4533) compared to TN cells (mean, 684 copies/106 cells; range, 158–1380). We also quantified total HIV-1 DNA in the combined CD4+ TTM/TEM cell population (Figure 1A). These cells harbored slightly higher levels of HIV-1 DNA compared to the TCM cells (mean fold change, 1.8; range, 1.1–3.0); however, this increase was not statistically significant. The TTM/TEM cell population, however, harbored significantly higher levels of total HIV-1 DNA vs the TN cells (P = .0006). Next, we determined the contribution of each T-cell subset to the total HIV-1 reservoir in resting CD4+ T cells as previously described [12]. First, we estimated the frequency of each T-cell subset in the resting CD4+ T cells from each donor (Figure 1B, Supplementary Figure 3). We then calculated the contribution of each CD4+ T-cell subset to the overall reservoir of HIV-1 DNA in the resting CD4+ T-cell population by taking into consideration both the frequency of each T-cell subset in the peripheral blood, as well as the frequency of the total HIV-1 DNA in that subset. We found that the CD4+ TCM population harbored the highest levels of total HIV-1 DNA (Figure 1C), consistent with previously published studies [12].
Figure 1.
Quantification of the total human immunodeficiency virus type 1 (HIV-1) DNA reservoir in CD4+ T naive (TN), T central memory (TCM), transitional memory (TTM)+effector memory (TEM) cells purified from HIV-infected individuals on antiretroviral therapy. A, Quantification of total HIV-1 DNA in CD4+ TN, TCM, and TTM+TEM cells. Total HIV-1 DNA was normalized to the cell number (determined by quantification of the CCR5 gene). B, The frequency of different resting CD4 T-cell subsets in the peripheral blood of HIV-1–infected individuals. C, Contribution (%) of the CD4+ TN, TCM, and TTM+TEM cells to the total HIV-1 DNA reservoir, taking into account both the frequency of the CD4+ T-cell subset and the frequency of HIV-1 DNA in that subset. In the plots, each colored dot represents a unique donor. All P values were determined using a Mann–Whitney test. Abbreviation: PBMC, peripheral blood mononuclear cell.
Similar Total Virus Recovery From CD4+ TN and TCM Cells Following Exposure to LRAs
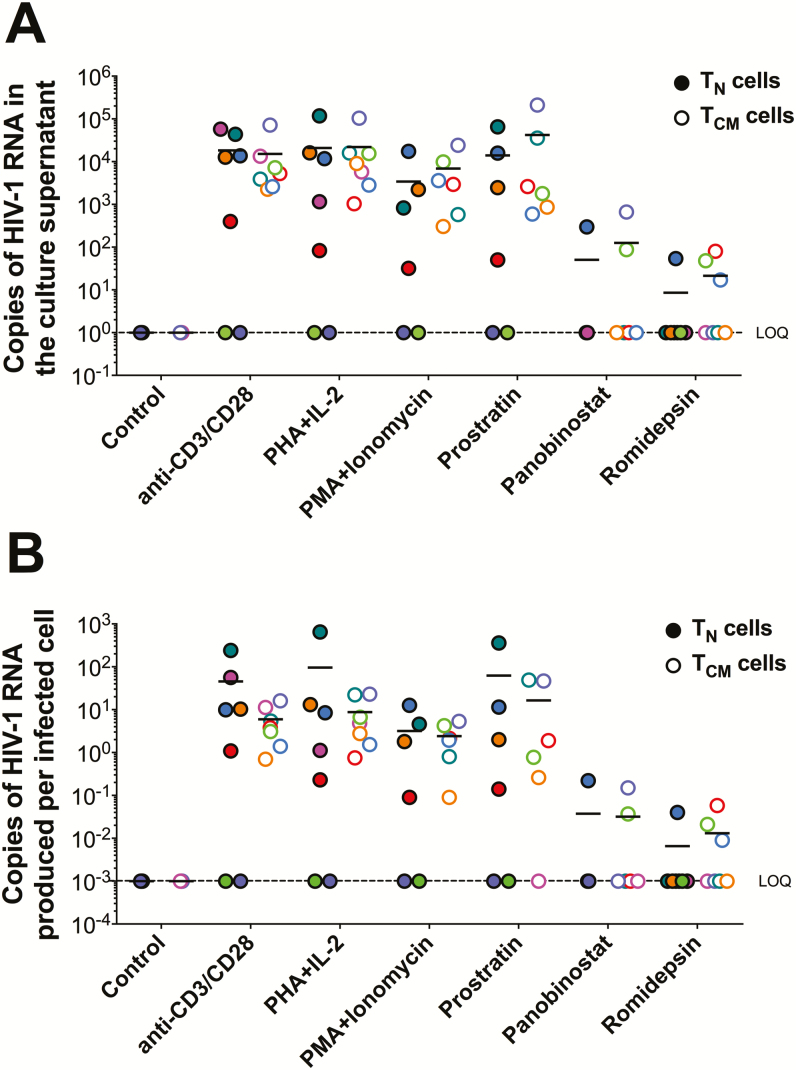
We quantified total virus recovery (ie, extracellular virion-associated HIV-1 RNA in the culture supernatant) from the donor-derived TN and TCM cells after exposure to 6 different LRAs using an ultrasensitive assay capable of single-copy detection of HIV-1 RNA (Figure 2A, Supplementary Table 1). The mean number of copies of HIV-1 RNA produced per 106 cells was not significantly different between the TN and TCM subsets following treatment for any of the LRAs tested. Interestingly, in cells from 4 of the 7 donors (donors 1, 3, 5, and 7), the virion-associated HIV-1 RNA levels produced were higher for TN cells compared to TCM cells, independent of the LRA used (Supplementary Table 1). When the extracellular virion-associated RNA copies were normalized to the number of infected cells (Figure 2B), we found that more HIV-1 RNA was produced per infected TN cell than TCM cell when stimulated with anti-CD3/CD28 mAb, PHA+IL-2, PMA/PHA, or prostratin, although this did not reach statistical significance. In contrast, both romidepsin and panobinostat failed to increase virus production in either cell type in the majority of donors. This finding is consistent with other studies that demonstrated that histone deacetylase inhibitors do not consistently increase virus production ex vivo in resting CD4+ T cells from HIV-1–infected individuals on suppressive ART [26–31].
Figure 2.
Total virus recovery from CD4+ T naive (TN) and T central memory (TCM) cells from human immunodeficiency virus type 1 (HIV-1)–infected individuals on antiretroviral therapy following latency reversal. A, Total copies of extracellular virion-associated HIV-1 RNA produced from TN or TCM cells after exposure to the latency reversing agents indicated. B, Copies of HIV-1 RNA produced per infected CD4+ TN or TCM cell. Each colored dot represents a unique donor. Solid dots represent data from CD4+ TN cells, while open dots represent data from CD4+ TCM cells. N = 7; except for phorbol-myristate-acetate +Iono, prostratin, and panobinostat, where N = 6. Abbreviations: HIV-1, human immunodeficiency virus type 1; lono, ionomycin; LOQ, limit of quantification; PHA, phytohaemagglutinin; PMA, phorbol-myristate-acetate; TEM, T effector memory cell.
T-Cell Activation and Total Virus Recovery in CD4+ TN and TCM Cells
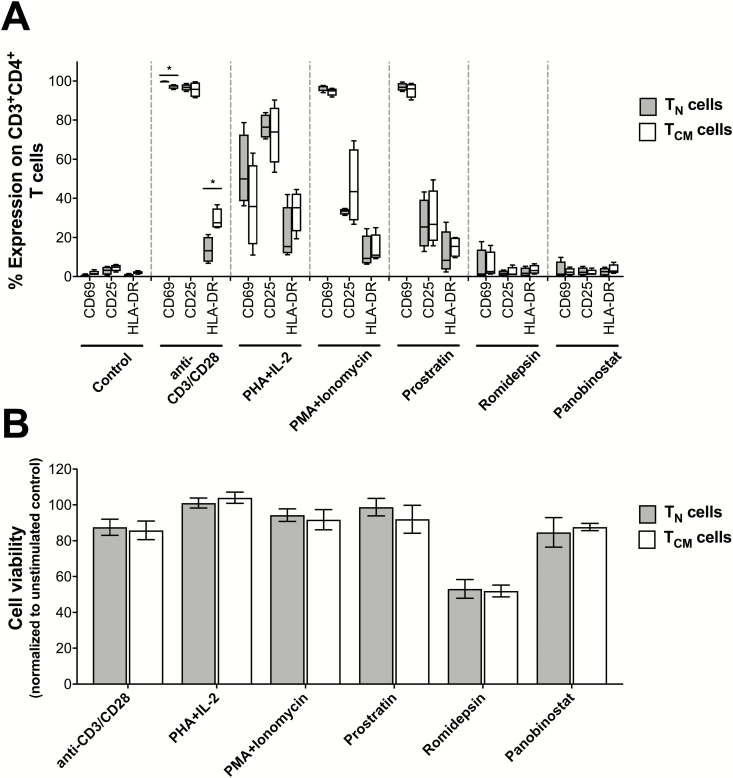
As described above, more HIV-1 RNA was produced per infected TN cell compared to TCM cell after exposure to anti-CD3/CD28 mAb, PHA+IL-2, PMA/PHA, or prostratin. Each of these LRAs induces T-cell activation. Therefore, we next assessed whether differences in T-cell activation between the TN and TCM cells, at the time of harvest, could potentially account for differences in total virus recovery. T-cell activation was assessed by measuring surface expression of CD25, CD69, and HLA-DR by flow cytometry (Figure 3A). We also evaluated cell viability by LIVE/DEAD staining (Figure 3B). As expected, anti-CD3/CD28 mAbs, PHA+IL-2, PMA+ionomycin, and prostratin induced T-cell activation in both TN and TCM cells. However, there were no significant differences between TN and TCM cells that could adequately account for the observed differences in total virus recovery (Figure 2A). Similarly, there were no significant differences in cell viability between TN and TCM cells following LRA stimulation (Figure 3B) or in controls (90.3% vs 89.5%, respectively).
Figure 3.
Impact of the latency reversing agent (LRA) on global T-cell activation and cell viability on CD4+ T naive (TN) and T central memory (TCM) cells purified from human immunodeficiency virus type 1 (HIV-1)–infected individuals on antiretroviral therapy. A, Expression of the T-cell activation markers CD25, CD69, and HLA-DR on CD4+ TN and TCM cells 7 days post-LRA exposure. All data are shown as the mean ± standard error of the mean (SEM; N = 7). B, CD4+ TN or TCM cell viability assessed 7 days post-LRA exposure. All data are shown as the mean ± SEM (N = 4). No significant differences were noted between the 2 cell types, as measured by a Mann–Whitney test. Abbreviations: lono, ionomycin; PHA, phytohaemagglutinin; PMA, phorbol-myristate-acetate; TEM, T effector memory cell.
Frequency of Replication-competent HIV-1 Is Similar in CD4+ TN and TCM Cells
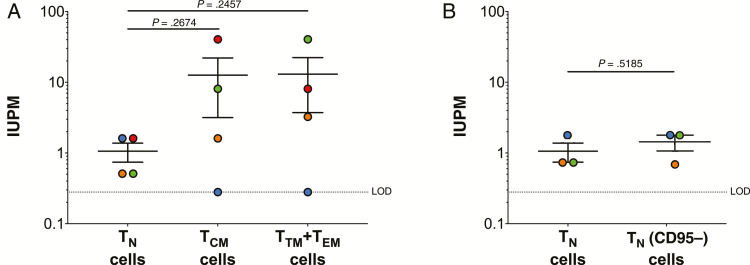
The total virus recovery assay does not inform as to whether the HIV-1 is replication competent. Therefore, we also performed a quantitative viral outgrowth assay on a subset of the donors (donors 4–7) as previously described [23]. In these experiments, we included the TTM+TEM cell subset in addition to the TN and TCM cells. We detected replication-competent HIV-1 in the TN cells from all 4 donors, but only 3 of the 4 donors in TCM and TTM+TEM cells (Figure 4A). While on average higher IUPM values were determined in the TCM and TTM+TEM cells compared to the TN cells (Figure 4A), if we normalized these values to the number of infected cells, we found minimal differences in IUPM between the different subsets (Table 2).
Figure 4.
Quantification of replication-competent human immunodeficiency virus type 1 (HIV-1) in CD4+ T naive (TN), T central memory (TCM), transitional memory (TTM)+effector memory (TEM) cells from HIV-1–infected donors on antiretroviral therapy. A, Infectious units per million cells (IUPM) values determined from TN, TCM, and TTM+TEM cells. B, IUPM values determined for TN cells with (TN cells) and without (TNCD95-) T stem cell-like memory cells. Each colored dot represents a unique donor. Data are shown as the mean ± standard error of the mean. P values were determined using an unpaired t test.
Table 2.
Differences in Infectious Units per Million Cells and Total Human Immunodeficiency Virus Type 1 (HIV-1) DNA Measured in CD4+ TN, TCM, and TTM+TEM Cells Isolated from HIV-1–infected Individuals on Antiretroviral Therapy
| Virological parameter | TCM vs TN | TTM+TEM vs TN | TCM vs TTM+TEM |
|---|---|---|---|
| Fold change in IUPM | 11.90 | 12.29 | 1.03 |
| Fold change in HIV-1 DNA | 5.60 | 13.70 | 1.75 |
| Difference in IUPM when corrected for difference in HIV-1 DNA | 2.13 | 0.90 | 0.59 |
Abbreviations: IUPM, infectious units per million cells; TCM, T central memory cell; TEM, T effector memory cell; TN, T naive cell; TTM, T transitional memory cell.
Collectively, these data highlight that TN cells are not only able to produce virus following latency reversal, but a portion of the virus produced from these cells is replication competent.
Depletion of TSCM Cells From TN Population Has No Impact on the HIV-1 DNA or Virus Recovery
CD4+ TSCM are thought to be a reservoir of latent HIV-1 in vivo [8]. TSCM and TN cells express many of the same characteristic surface markers, including CD45RA, CCR7, CD62L, CD28, CD27, IL-7Rα (CD127), and CD95. Therefore, based on our purification protocol, our TN population would include TSCM cells. We therefore removed TSCM cells from the TN population by depletion of CD95, which is expressed on TSCM but not TN cells, in 3 donors (Table 3). Consistent with previous reports, we found that the TSCM population accounted for roughly 5% of the TN cell population (range, 3.26%–6.28%) [8, 10, 32–35]. We observed no significant differences in total HIV-1 DNA (Table 3), total virus recovery (Table 3), or replication-competent HIV-1 (Figure 4B) in the total TN population vs the TN population that was lacking the TSCM cells (TN [CD95-]).
Table 3.
Quantification of Human Immunodeficiency Virus Type 1 (HIV-1) DNA and Inducible Extracellular HIV-1 RNA in TN and TN(CD95-) Cells Purified From HIV-1–Infected Individuals on Antiretroviral Therapy
| Donor No. | HIV-1 DNA (copies/106 cells) | HIV-1 RNA in the Supernatant (copies/mL) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TN | TN CD95- | Anti-CD3/CD28 | Phytohaemagglutinin + Interleukin 2 | Phorbol-myristate- acetate + ionomycin | Prostratin | Panobinostat | Romidepsin | |||||||
| TN | TN CD95- | TN | TN CD95- | TN | TN CD95- | TN | TN CD95- | TN | TN CD95- | TN | TN CD95- | |||
| 5 | 1232 | 1249 | 12 670 | 5460 | 16 100 | 5930 | 2220 | 1042 | 2465 | 1615 | 0 | 0 | 0 | 0 |
| 6 | 158 | 173 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 1380 | 1112 | 13 700 | 12 100 | 11 700 | 9050 | 17 400 | 15 250 | 15 860 | 12 280 | 298 | 61 | 54 | 144 |
| Mean | 923 | 845 | 8790 | 5853 | 9267 | 4993 | 6540 | 5431 | 6108 | 4632 | 99 | 20 | 18 | 48 |
Abbreviations: TN, T naive cell.
DISCUSSION
The latent HIV-1 reservoir is frequently described as residing within resting memory CD4+ T cells. This is largely due to the consistent finding that memory CD4+ T cells harbor the highest levels of HIV-1 DNA in individuals on ART [10–13]. This has yielded little research into the contribution of CD4+ TN cells to the latent reservoir. In 2013 Sáez-Cirión et al reported that in some infected individuals who received ART within 10 weeks of primary infection in the French Virological and Immunological Studies in Controllers After Treatment Interruption cohort, viremia could be controlled for at least 24 months post-treatment interruption [6]. In this patient population, HIV-1 DNA was only detected in CD4+ TN cells in 2 of 11 individuals, whereas the resting memory CD4+ T-cell subsets (TCM, TTM, and TEM) harbored comparable levels of HIV-1 DNA [6]. This finding suggested to us that the latent HIV-1 reservoir in CD4+ TN cells may be more important than previously considered and could be a factor that contributes to viral control in these individuals.
Using a primary cell model of HIV-1 latency [14], we previously reported that although TN cells harbored significantly lower levels of HIV-1 DNA, they produced as many virions as did the TCM cells following latency reversal. Consistent with this in vitro data [14], in this study we show that although the frequency of HIV-1 DNA is lower in TN compared to TCM cells purified from HIV-1–infected individuals on ART, as much, if not more, virus is produced from the TN cells following exposure to LRAs (Figure 2). Importantly, we recovered replication-competent HIV-1 from the CD4+ TN, TCM, and TTM+TEM subsets. While on average higher IUPM values were determined in the TCM and TTM+TEM cells compared to the TN cells (Figure 4A), once we normalized these values to the number of infected cells, we found minimal differences in IUPM between the different subsets (Table 2). This finding suggests that a higher proportion of the HIV-1 DNA in the TN cells, compared to the TCM and TTM+TEM cells, may be intact and replication competent. Recently, Hiener et al quantified the frequency of intact proviruses in different T-cell subsets from HIV-1–infected individuals on ART [36] and found a higher proportion of intact proviruses in TN cells compared to TCM cells. Interestingly, in that study, they found that the highest frequency of intact provirus was in the TEM compartment and that no intact provirus could be recovered from the TCM compartment. However, others have found varying levels of replication-competent virus across CD4+ T-cell subsets, generally finding that TCM cells contain the highest IUPM on a population level; however, this is not consistent across all donors [12, 37]. These differences could be reflective of differences in time of infection prior to ART initiation, time on ART, or other immunological disparities. Further investigation is warranted to better understand differences in T-cell reservoirs between individuals.
In conclusion, our data highlight that quantifying HIV-1 DNA alone may not be predictive of the size of the inducible latent reservoir and further reinforce that CD4+ TN cells, which are abundant and have exceptionally long half-lives (1–8 years) [38, 39], are an important cellular reservoir of latent HIV-1 infection. There are, however, limitations to our work. First, our sample size was relatively small (n = 7). Second, our work only provided insight into the latent viral reservoir in peripheral blood and not tissue. In regard to the latter, Mavigner et al [40] recently reported that CD4+ TN cells substantially contributed (approximately 70%–80%) to the total CD4+ reservoir of Simian immunodeficiency virus (SIV) infection in both blood and lymph nodes of ART-suppressed rhesus macaques infants. In contrast, the SIV reservoir in CD4+ TN cells in the peripheral blood of ART-suppressed adult rhesus macaques was much lower (14%) but still contributed significantly to the reservoir in lymph nodes (approximately 40%–60%).
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. J. M. Z., J. W. M., and N. S. C designed the study. J. M. Z., D. M., and M. S collected the data. J. M. Z. Manuscript did the statistical analysis. J. M. Z. and N. S. C. wrote the manuscript. All authors reviewed and approved the final manuscript.
Financial support. This work was supported by the National Institutes of Health (National Institute of Allergy and Infectious Diseases; grants R56AI139010 to N. S. C, T32AI065380 to J. M. Z., and funds from the National Cancer Institute under contract HHSN261200800001E to J.W.M) and the Bill & Melinda Gates Foundation (grant OPP1115715 to J.W.M.).
Potential conflicts of interest. J. W. M. is a consultant to Gilead Sciences and a shareholder of Co-Crystal, Inc. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. [DOI] [PubMed] [Google Scholar]
- 2. Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94:13193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wong JK, Hezareh M, Günthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–5. [DOI] [PubMed] [Google Scholar]
- 4. Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999; 5:512–7. [DOI] [PubMed] [Google Scholar]
- 5. Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–8. [DOI] [PubMed] [Google Scholar]
- 6. Sáez-Cirión A, Bacchus C, Hocqueloux L, et al. ; ANRS VISCONTI Study Group Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 2013; 9:e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bacchus C, Cheret A, Avettand-Fenoël V, et al. ; OPTIPRIM ANRS 147 Study Group A single HIV-1 cluster and a skewed immune homeostasis drive the early spread of HIV among resting CD4+ cell subsets within one month post-infection. PLoS One 2013; 8:e64219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buzon MJ, Sun H, Li C, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 2014; 20:139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ganesan A, Chattopadhyay PK, Brodie TM, et al. ; Infectious Disease Clinical Research Program HIV Working Group Immunologic and virologic events in early HIV infection predict subsequent rate of progression. J Infect Dis 2010; 201:272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jaafoura S, de Goër de Herve MG, Hernandez-Vargas EA, et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4⁺ memory T cells. Nat Commun 2014; 5:5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gibellini L, Pecorini S, De Biasi S, et al. HIV-DNA content in different CD4+ T-cell subsets correlates with CD4+ cell : CD8+ cell ratio or length of efficient treatment. AIDS 2017; 31:1387–92. [DOI] [PubMed] [Google Scholar]
- 12. Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. von Stockenstrom S, Odevall L, Lee E, et al. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis 2015; 212:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zerbato JM, Serrao E, Lenzi G, et al. Establishment and reversal of HIV-1 latency in naive and central memory CD4+ T cells in vitro. J Virol 2016; 90:8059–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai J, Agosto LM, Baytop C, et al. Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. J Virol 2009; 83:4528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eckstein DA, Penn ML, Korin YD, et al. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity 2001; 15:671–82. [DOI] [PubMed] [Google Scholar]
- 17. Helbert MR, Walter J, L’Age J, Beverley PC. HIV infection of CD45RA+ and CD45RO+ CD4+ T cells. Clin Exp Immunol 1997; 107:300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schnittman SM, Lane HC, Greenhouse J, Justement JS, Baseler M, Fauci AS. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type 1: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proc Natl Acad Sci U S A 1990; 87:6058–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hong F, Aga E, Cillo AR, et al. Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol 2016; 54:902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cillo AR, Vagratian D, Bedison MA, et al. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol 2014; 52:3944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cillo AR, Krishnan A, Mitsuyasu RT, et al. Plasma viremia and cellular HIV-1 DNA persist despite autologous hematopoietic stem cell transplantation for HIV-related lymphoma. J Acquir Immune Defic Syndr 2013; 63:438–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malnati MS, Scarlatti G, Gatto F, et al. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc 2008; 3:1240–8. [DOI] [PubMed] [Google Scholar]
- 23. Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 2005; 304:3–15. [DOI] [PubMed] [Google Scholar]
- 24. Rosenbloom DI, Elliott O, Hill AL, Henrich TJ, Siliciano JM, Siliciano RF. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis 2015; 2:ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brenchley JM, Hill BJ, Ambrozak DR, et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 2004; 78:1160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cillo AR, Sobolewski MD, Bosch RJ, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 2014; 111:7078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blazkova J, Chun TW, Belay BW, et al. Effect of histone deacetylase inhibitors on HIV production in latently infected, resting CD4(+) T cells from infected individuals receiving effective antiretroviral therapy. J Infect Dis 2012; 206:765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 2014; 20:425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laird GM, Rosenbloom DI, Lai J, Siliciano RF, Siliciano JD. Measuring the frequency of latent HIV-1 in resting CD4⁺ T cells using a limiting dilution coculture assay. Methods Mol Biol 2016; 1354:239–53. [DOI] [PubMed] [Google Scholar]
- 30. Spivak AM, Bosque A, Balch AH, Smyth D, Martins L, Planelles V. Ex vivo bioactivity and HIV-1 latency reversal by ingenol dibenzoate and panobinostat in resting CD4(+) T cells from aviremic patients. Antimicrob Agents Chemother 2015; 59:5984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Archin NM, Kirchherr JL, Sung JA, et al. Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J Clin Invest 2017; 127:3126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lugli E, Gattinoni L, Roberto A, et al. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat Protoc 2013; 8:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flynn JK, Paukovics G, Cashin K, et al. Quantifying susceptibility of CD4+ stem memory T-cells to infection by laboratory adapted and clinical HIV-1 strains. Viruses 2014; 6:709–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tabler CO, Lucera MB, Haqqani AA, et al. CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J Virol 2014; 88:4976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klatt NR, Bosinger SE, Peck M, et al. Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals. PLoS Pathog 2014; 10:e1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hiener B, Horsburgh BA, Eden JS, et al. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep 2017; 21:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soriano-Sarabia N, Bateson RE, Dahl NP, et al. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 2014; 88:14070–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Boer RJ, Perelson AS. Quantifying T lymphocyte turnover. J Theor Biol 2013; 327:45–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Macallan DC, Wallace D, Zhang Y, et al. Rapid turnover of effector-memory CD4(+) T cells in healthy humans. J Exp Med 2004; 200:255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mavigner M, Habib J, Deleage C, et al. Simian immunodeficiency virus persistence in cellular and anatomic reservoirs in antiretroviral therapy-suppressed infant rhesus macaques. J Virol 2018; 92:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.