Abstract
Carbapenem resistance in gram-negative bacteria has caused a global epidemic that continues to grow. Although carbapenemase-producing Enterobacteriaceae have received the most attention because resistance was first reported in these pathogens in the early 1990s, there is increased awareness of the impact of carbapenem-resistant nonfermenting gram-negative bacteria, such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia. Moreover, evaluating the problem of carbapenem resistance requires the consideration of both carbapenemase-producing bacteria as well as bacteria with other carbapenem resistance mechanisms. Advances in rapid diagnostic tests to improve the detection of carbapenem resistance and the use of large, population-based datasets to capture a greater proportion of carbapenem-resistant organisms can help us gain a better understanding of this urgent threat and enable physicians to select the most appropriate antibiotics.
Keywords: Acinetobacter baumannii, carbapenemases, carbapenem-resistant Enterobacteriaceae, gram-negative bacteria, Pseudomonas aeruginosa
Carbapenem resistance in gram-negative bacteria has become a worldwide problem. The 2017 World Health Organization (WHO) global priority list of pathogens ranks carbapenem-resistant Enterobacteriaceae (CRE), carbapenem-resistant Pseudomonas aeruginosa, and carbapenem-resistant Acinetobacter baumannii in the highest priority category (ie, critical) [1]. To address this global epidemic, identification and ongoing surveillance of carbapenem-resistant gram-negative bacteria are needed.
Evidence suggests that patients who are infected by carbapenem-resistant pathogens have an increased likelihood of morbidity and mortality compared with those infected by susceptible pathogens [2–4], which is likely due to administration of antibiotics with suboptimal or no activity against these organisms [5]. Thus, recognizing the risk of carbapenem resistance [6], particularly in the most vulnerable patient populations [5, 7–9], and/or early detection of specific carbapenem resistance mechanisms [10] are critical to reduce the risk of mortality, length of hospitalization, and associated costs [2]. The alarming level of carbapenem resistance has presented particular challenges for the management of a variety of infections caused by nonfermenters because of the low permeability of the outer bacterial membrane to several antibiotics, including, but not limited to, the carbapenems [11, 12].
The concerns surrounding CRE-related infections [2, 13] have recently been mitigated to some degree by the approval of new β-lactam–β-lactamase inhibitor combination therapies, which demonstrate activity against strains with specific underlying resistance mechanisms [14]; however, on-therapy resistance has already been reported [15]. The use of older agents, such as tigecycline or colistin, is frequently associated with unclear efficacy and/or toxicity issues [11]. It is clear that understanding specific mechanisms underlying carbapenem resistance and monitoring local epidemiology would lead to more effective treatment of infections caused by carbapenem-resistant gram-negative bacteria.
MECHANISMS OF CARBAPENEM RESISTANCE
Enzymatic Hydrolysis
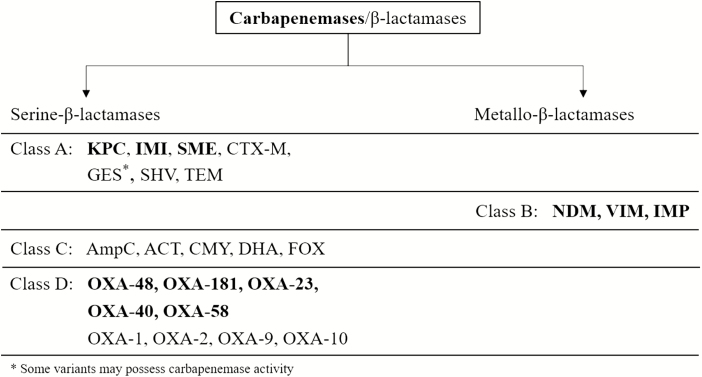
One key mechanism of carbapenem resistance is hydrolysis of carbapenems by carbapenemase enzymes, which are encoded mainly on plasmids and are highly transmissible [16]. The Ambler classification system categorizes β-lactamase enzymes into 4 groups (ie, A, B, C, D) based on their central catalytic domain and substrate preference (Figure 1) [17]. Of these, classes A, B, and D include carbapenemases, whereas class C enzymes hydrolyze primarily cephalosporins [18]. Enzymes in classes A, C, and D have serine in the active catalytic site, whereas class B enzymes are metallo-β-lactamases (MBLs) with zinc in the active site [18]. Among the newer agents, avibactam inhibits class A (eg, Klebsiella pneumoniae carbapenemase [KPC]), class C (eg, ampicillin chromosomal cephalosporinase [AmpC]), and only some class D (eg, oxacillin carbapenemase/oxacillinase [OXA]–48) serine-β-lactamases, but does not significantly inhibit the activity of class B MBLs (eg, imipenemase metallo-β-lactamase [IMP], Verona integron-encoded metallo-β-lactamase [VIM], New Delhi metallo-β-lactamase [NDM]) [19]. Similarly, vaborbactam inhibits class A and C enzymes but not those belonging to class B and D [20].
Figure 1.
Classification of carbapenemases/β-lactamases depending on their central catalytic domain. Adapted from [17]. Abbreviations: ACT, AmpC type β-lactamase; AmpC, ampicillin chromosomal cephalosporinase; CMY, cephamycin-hydrolyzing β-lactamase; CTX-M, cefotaxime-hydrolyzing β-lactamase–Munich; FOX, plasmid-mediated class C β-lactamase; GES, Guiana extended-spectrum β-lactamase; IMI, imipenem-hydrolyzing β-lactamase; IMP, imipenemase metallo-β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillin carbapenemase/oxacillinase; SHV, sulfhydryl variant of the TEM enzyme; SME, Serratia marcescens enzyme; TEM, Temoneira class A extended-spectrum β-lactamase; VIM, Verona integron-encoded metallo-β-lactamase.
Although most class A enzymes do not exhibit intrinsic carbapenemase activity, this group of enzymes includes the prevalent KPC [18]. All (class B) MBLs possess carbapenemase activity, and this group includes the acquired VIM, IMP, and NDM enzymes that may be found in many gram-negative species [18]. Class C includes AmpC β-lactamase enzymes that are not carbapenemases per se, as their hydrolytic activity against carbapenems is very weak or nonexistent, but that can play a role in resistance to carbapenems in the context of permeability defects [21]. This is true, in particular, for many enterobacterial species that naturally produce a class C cephalosporinase (such as Enterobacter species, Serratia marcescens, Proteus species, Providencia species, Morganella morganii, and Hafnia alvei) and P. aeruginosa [22, 23]. Class D (also termed oxacillin carbapenemase [OXA enzymes]) enzymes constitute a heterogeneous group of β-lactamases with significant carbapenemase activity, especially OXA-48–type enzymes in Enterobacteriaceae and OXA-23 [24, 25], frequently found in A. baumannii [26, 27]. Stenotrophomonas maltophilia has intrinsic carbapenem resistance due to the presence of a chromosomally encoded MBL, namely L1 [28].
Other Carbapenem Resistance Mechanisms
Nonenzymatic carbapenem resistance mechanisms include loss of expression of porin-encoding genes, mutations in chromosomally encoded porin genes (such as OprD), and overexpression of genes encoding efflux pumps (such as MexAB-OprM, MexXY-OprM, or MexCD-OprJ), particularly in P. aeruginosa [25, 29, 30]. Porins are nonspecific channels in the outer membrane of gram-negative bacteria that permit the passive transport of hydrophilic small molecules and nutrients (and also some antibiotics) across the otherwise impermeable membrane [30]. Porin loss and efflux pump overexpression associated with carbapenem resistance may also contribute to cross-resistance to other β-lactams and other antibiotic classes [31]. This is commonly observed in association with carbapenemase production in A. baumannii. Some Enterobacteriaceae, such as Proteus species, Providencia species, and M. morganii, have intrinsic resistance to imipenem and require resistance to other carbapenems to be classified as CRE [23]. Carbapenem resistance can also be attributed to mutations or other modifications that alter the production level or the binding affinity of penicillin-binding proteins, mechanisms that have been observed rarely in Escherichia coli [32], P. aeruginosa [31], and A. baumannii [33].
DIAGNOSTICS
Both the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) annually define the susceptibility breakpoints to commercially available carbapenems, including doripenem, ertapenem, imipenem, and meropenem for gram-negative species, although EUCAST no longer provides doripenem breakpoints [34, 35]. When a strain is found to be nonsusceptible to carbapenems (ie, intermediate or resistant), the mechanism of resistance is still unknown [13, 36, 37]. Thus, to confirm the production of carbapenemases and/or presence of other mechanisms, further biochemical assays and/or gene-based tests must be performed [13, 16, 22, 23]. Determining the mechanism of carbapenem resistance can help in the selection of the most appropriate antibiotic therapy early in the treatment of gram-negative infections. For therapeutic decision making, the rapid turnaround time (defined as 1 day or as short as <2 hours) would be particularly beneficial in reducing length of hospitalization and/or time spent in the intensive care unit [13, 37, 38]. Both biochemical and molecular technologies are widely available, with endorsement from CLSI, EUCAST, and/or the US Food and Drug Administration.
The biochemical assays include the Carba NP [37, 39], its derivative Blue Carba [40], and β Carba [22] tests, which are inexpensive and confirm phenotypically carbapenemase-producing organisms (but not other resistance mechanisms). These methods are based on the expression of any carbapenemase enzyme during bacterial growth in culture (ie, up to 24–48 hours), and use imipenem or meropenem as a substrate, which is then hydrolyzed by the carbapenemase. The colorimetric positive signal may be obtained in <1 hour (eg, Carba NP) and can be used directly from clinical samples (blood cultures, infected urine). Furthermore, specific inhibitors of carbapenemase activity can be included, such as avibactam, vaborbactam, or ethylenediaminetetraacetic acid [22, 38, 41]. Further biochemical assays include the carbapenemase inactivation method, which is also inexpensive, and the matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF) technology, which may be cost-effective in large centers and hospitals. However, all of these methods described above, besides having some specificity or sensitivity issues, are also unable to identify the exact carbapenemase enzyme and require growth of bacteria [13, 38, 42].
The specific assays used to detect the presence of known carbapenemase genes located on plasmids, or porin channel or efflux pump mutations, are normally gene based and amplify the potential genes present by the use of oligomer primers and probes [13, 16, 38, 43]. Commercially available polymerase chain reaction (PCR) tests include Check-Direct carbapenemase-producing Enterobacteriaceae (CPE) assays (Check-Points, Wageningen, the Netherlands), Xpert Carba-R (Cepheid, Sunnyvale, California), EazyPlex SuperBug ID complete A/B (Amplex, Giessen, Germany), and the very recent point-of-care GenePOC technology (GenePOC, Quebec City, Canada). All 4 methods can detect KPC, NDM, and VIM encoding genes with 100% sensitivity, and OXA-48–type carbapenemases (including OXA-181) with 83%–100% sensitivity; however, only Xpert Carba-R detects IMP-1 [44]. Turnaround time is usually the same day [13, 38]. The commercial microarrays allow for the detection of a much higher number of target genes than PCR with 100% sensitivity and typically include bacterial identification targets as well as resistance markers (eg, KPC, NDM, OXA, VIM, IMP, Guiana extended-spectrum β-lactamase [GES], German imipenemase [GIM], and São Paulo metallo-β-lactamase [SPM] carbapenemases). Currently available systems include Verigene (Luminex, Austin, Texas) [45], BioFire FilmArray (Salt Lake City, Utah) [46], and the Check-Points systems [47]. Whole genome sequencing allows detection of either carbapenemase genes or other resistance-associated mutations and may also play a role as the technology becomes less expensive and more widespread [38]. However, such an approach requires a significant expertise and adequate equipment, which is not systematically available, and a precise knowledge of combined resistance mechanisms (eg, mutations, level of expression).
Some of these rapid gene-based assays, such as the Xpert Carba-R platform or BioFire FilmArray, have the potential for direct specimen sampling (eg, nasal swab, rectal swab, sputum, wound specimen, blood, urine) without the need for culturing, allowing appropriate treatment to be initiated as soon as the carbapenemase resistance mechanism has been identified and minimizing the risk of treatment failure associated with empiric antimicrobial therapy [10, 44].
Despite the technological advances in molecular and biochemical rapid diagnostics, there are 2 fundamental considerations: (1) a negative test does not imply that the organism is carbapenem susceptible, as it may still be resistant due to nonenzymatic mechanisms; (2) conversely, the presence of a gene does not systematically imply the organism is carbapenem resistant, owing to the level of expression of the resistance gene; and (3) a positive biochemical test will not identify the specific carbapenemase enzyme. Consequently, only phenotypic tests relying on actual growth inhibition provide a full susceptibility picture.
GLOBAL EPIDEMIOLOGY OF CARBAPENEM-RESISTANT PATHOGENS
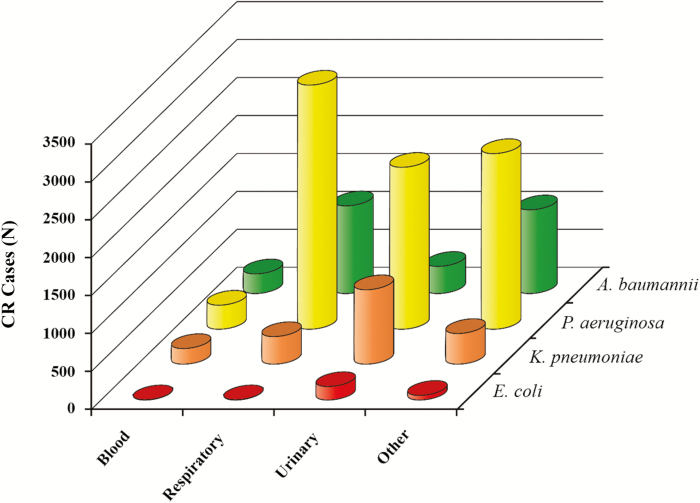
Although data are limited for some regions, the overall burden of disease caused by carbapenem-resistant pathogens is similar in most regions (ie, Asia-Pacific, the Indian continent, Europe, North America, and Latin America), with nonfermenters being the most problematic pathogens followed by a relatively lower proportion of CREs (Table 1) [3, 5, 21, 26, 41, 48–54]. Data of both large surveillance studies and smaller hospital investigations demonstrate similarity in carbapenem resistance rates irrespective of the methodology used to detect the mechanism of resistance or the antibiotic used. The reported rates of carbapenem resistance seem to be considerably higher for nonfermenters (frequently >60%) than for fermenters (frequently <10%) across regions [3, 21, 26, 41, 48–57]. Specifically, in the US based study from the Premier Healthcare Database, which collects data on both hospital- and community-acquired infections, 44.8% of A. baumannii and 14.2% of P. aeruginosa isolates were carbapenem resistant, compared with only 1% of Enterobacteriaceae [3, 58]. Of note, this was applicable in all infection types investigated (ie, bloodstream, respiratory, urinary, and other) (Figure 2) [3, 58]. Importantly, 82.3% of all carbapenem-resistant infections were caused by A. baumannii or P. aeruginosa, whereas only 17.7% were caused by K. pneumoniae or E. coli [3, 58]. Carbapenem resistance rates by pathogen differ depending on the site of infection [3]. For example, rates for both P. aeruginosa and A. baumannii are much lower in bloodstream infections (BSIs) than respiratory infections [3]. The implication of this finding is that epidemiological studies that track only BSI isolates probably underreport carbapenem resistance rates. Additionally, S. maltophilia, which is intrinsically carbapenem resistant, was isolated at the highest rate from hospitalized patients with nosocomial pneumonia in Asia-Pacific, Europe, and North America (range, 51.7%–62.6%) or BSIs in Latin America (56.8%) in the SENTRY surveillance program between 1997 and 2016 [53]. The Japan Nosocomial Infections Surveillance (JANIS) 2016 report, which included data from 1653 facilities, found that the rates of imipenem and meropenem nonsusceptibility according to CLSI 2012 breakpoints were 0.1% and 0.2% for E. coli, 0.2% and 0.5% for K. pneumoniae, 17.9% and 12.3% for P. aeruginosa, and 3.1% and 1.9% for Acinetobacter species, respectively [59].
Table 1.
Reported Carbapenem Nonsusceptibility or Resistance Rates by Region
| Nonsusceptibility or Resistance Rate | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | Asia-Pacific | Ref. | India, Nepal, Pakistan, Vietnam | Ref. | Europe | Ref. | North America | Ref. | Latin America | Ref. |
| Acinetobacter baumannii | 55.7%–56.2% | [53]a | 0%–100% | [26]d | 58.1%–60.5% | [53]a | 32.0%–36.5% | [53]a | 53.1%–54.6% | [53]a |
| 78.4%–79.00% | [53]a | … | … | 76.3%–77.8% | [53]a | 42.3%–45.1% | [53]a | 85.6%–86.3% | [53]a | |
| 71.4%–71.9% | [49]c | … | … | 2.5%–81.5% | [41]b | 40.1%–50.4% | [3]c | 57.5% | [51]c | |
| 25.0%–90.5% | [48]b | … | … | 65.8%–84.6% | [21]c | 11.4% | [55]c | 21%–90% | [52]d | |
| … | … | … | … | 90.7%–100% | [54]c | … | … | 79.3%–89.2% | [56]a | |
| Pseudomonas aeruginosa | 17%–50% | [50]b | … | … | 0%–35.6% | [41]b | 10.3%–19.4% | [3]c | 64.6% | [51]c |
| 25.4%–34.6% | [49]c | … | … | 24.2%–56.4% | [21]c | 58.5% | [55]c | 14%–57% | [52]d | |
| 10.3%–46.7% | [48]b | … | … | 14.4% | [57]a | 26.1% | [57]a | 38.1%–45.8% | [56]a | |
| 16.8% | [57]a | … | … | … | … | … | … | 24.8% | [57]a | |
| Klebsiella pneumoniae | 5%–25% | [50]b | 0%–52% | [26]d | 0.2%–33.4% | [41]b | 3.1%–4.9% | [3]c | 1.3%–28.6% | [56]a |
| 1.6%–19.1% | [49]c | … | … | 31.7%–58.6% | [21]c | 12.9% | [55]c | 16.0% | [57]a | |
| 3.8% | [57]a | … | … | 15.7% | [57]a | 6.0% | [57]a | … | … | |
| Escherichia coli | 0%–3% | [50]b | 0%–34% | [26]d | 0%–7.0% | [41]b | 0.2%–0.4% | [3]c | 0.4%–9.0% | [56]a |
| 1.6%–7.1% | [49]c | … | … | … | … | 4.3% | [55]c | … | … | |
| Enterobacteriaceae (other) | 0% | [50]b | 2.7%–21.3% | [26]d | … | … | 2.10% | [55]c | … | … |
| 2.4%–32.1% | [49]c | … | … | … | … | … | … | … | … | |
| 0.4%–12.5% | [48]b | … | … | … | … | … | … | … | … | |
Figure 2.
Number of infections caused by carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli based on the Premier Healthcare Database. Adapted from [3]. Abbreviation: CR, carbapenem resistant.
Information provided by such epidemiological databases must be viewed with caution because the sites and the sample collection methodology may vary [41]; in addition, resistance or nonsusceptibility rates may depend on the antibiotic tested [49]. The ongoing European multicenter COMBACTE surveillance program is a comprehensive program that collects information on the methodology used to detect carbapenem resistance mechanisms in multiple gram-negative pathogens as well as resistance rates, which are determined by both CLSI and EUCAST breakpoints [41]. Results of the COMBACTE study may clarify the most optimal methodology for detection of carbapenem resistance and the timings for interventions, both of which will aid physicians in the management of resistant infections [41].
GEOGRAPHIC DISTRIBUTION OF CARBAPENEM RESISTANCE MECHANISMS
Asia-Pacific
In contrast to North America and Europe, NDM and other MBLs (eg, IMP, VIM), and OXA-48–type, rather than KPC, were the predominant carbapenemases in CRE in Southeast Asia [60]. A 2013–2016 study of 130 carbapenem-resistant isolates in the Philippines identified 45 (35%) carbapenemase-producing bacterial isolates with 43 (33%) testing positive for NDM and 2 (1.5%) for VIM, both of which were P. aeruginosa [61]. Surveillance reports in India and surrounding countries reveal that the most frequent carbapenemase enzymes remain NDM in Enterobacteriaceae and OXA-23 in A. baumannii [26].
Europe
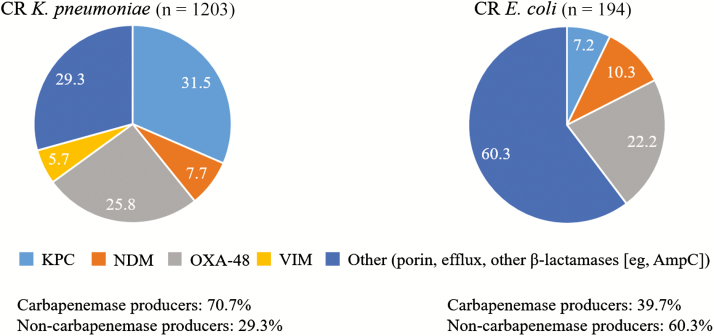
In the prospective, multinational European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) study, 37% of carbapenem-nonsusceptible K. pneumoniae and 19% of carbapenem-nonsusceptible E. coli were confirmed to possess a carbapenemase gene, with those encoding KPC (42%) and OXA-48 (38%) carbapenemases being found most frequently [62]. However, 29.3% (353/1203) of K. pneumoniae and 60.3% (117/194) of E. coli isolates were confirmed to also have other resistance mechanisms (Figure 3), suggesting that a large proportion of CRE infections currently lack effective and relatively safe antibiotic treatment options [62]. Although the EuSCAPE study focused on CRE infections and did not collect information on nonfermenters, carbapenem-resistant nonfermenters have been reported in some countries (eg, Germany: outbreak by GIM-1 MBL-producing P. aeruginosa; Greece: emergence of P. aeruginosa concurrently producing VIM and KPC) [63, 64].
Figure 3.
Distribution of carbapenem resistance mechanisms in Enterobacteriaceae species in the European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) study. Adapted from [62]. Abbreviations: AmpC, ampicillin chromosomal cephalosporinase β-lactamase; CR, carbapenem resistant; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillin carbapenemase/oxacillinase; VIM, Verona integron-encoded metallo-β-lactamase.
North America
Based on North American datasets, approximately 50% of all CRE isolates tested appear to be CPE. In the US Centers for Disease Control and Prevention’s Multisite Gram-Negative Surveillance Initiative, a population-based surveillance system from 7 US communities, 47.9% (range, 15.4%–76.5%) of CRE isolates were confirmed as CPE (all KPC) by PCR [65]. Other carbapenemases (NDM, VIM, and OXA-48) are also being detected in the United States [23]. Among CRE strains in the Canadian Nosocomial Infection Surveillance Program, the most common carbapenemases were KPC-type (66.9% per year) and NDM-1 (17.3% per year), with a significant increase in S. marcescens enzyme family carbapenemase and OXA-48 over the 5-year period [66]. In Acinetobacter species, the most prevalent mechanism of resistance to carbapenems is associated with specific carbapenemases such as OXA-23 and other OXA-type carbapenemases (eg, OXA-40, OXA-58) [67].
Latin America
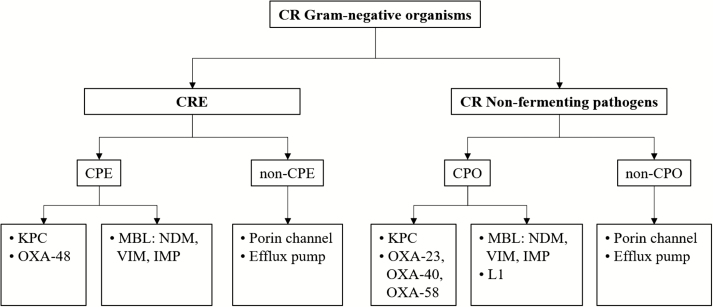
Investigations into specific mechanisms have revealed the spread of virtually all resistance mechanisms across the region. The first case of IMP-1–expressing K. pneumoniae was reported in Brazil in 2005 [68]. In a Mexican hospital, a significant increase in carbapenem resistance was found between 2011 and 2015, and 96% of carbapenem-resistant K. pneumoniae expressed KPC [69]. In a study by López-García, detection of IMP and GES enzymes was reported in carbapenem-resistant P. aeruginosa; however, some of the strains had multiple mechanisms present simultaneously, such as MBLs and loss of porin expression, resulting in extremely high meropenem minimum inhibitory concentrations [70]. Among A. baumannii strains, the most common carbapenemase enzymes corresponded to OXA enzymes in this region [27], including OXA-23, OXA-58, OXA-72, OXA-143, and OXA-253; however, NDM-1, VIM-1, IMP-1, and IMP-10 have also been detected. These epidemiological studies are crucial in understanding the evolution and spread of these strains, and also highlight that molecular characterization of the clones spreading across hospitals may support infection control as well as physicians’ decisions with regard to selection of the best available antibiotic therapy. The broad range of mechanisms (Figure 4) in both Enterobacteriaceae and nonfermenters has an impact on the selection of the most appropriate antibiotic for carbapenem-resistant infections because their spectrum of activity greatly depends on the presence of these resistance mechanisms. For example, some of the new β-lactam–β-lactamase inhibitor combination antibiotics have limited activity against MBLs and nonfermenting pathogens, as described in the manuscript by Lee & Doi [71].
Figure 4.
Algorithm to assess potential carbapenem resistance mechanisms in Enterobacteriaceae and nonfermenter species. Abbreviations: CPE, carbapenemase-producing Enterobacteriaceae; CPO, carbapenemase-producing organism; CR, carbapenem resistant; CRE, carbapenem-resistant Enterobacteriaceae; IMP, imipenemase metallo-β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; L1, a class B metallo-β-lactamase; MBL, metallo-β-lactamase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillin carbapenemase/oxacillinase; VIM, Verona integron-encoded metallo-β-lactamase.
CONCLUSIONS
Carbapenem resistance affects both nonfermenters and fermenters in all regions, and mechanisms appear to vary geographically. However, the rates of carbapenem resistance were consistently higher in nonfermenters than fermenters. The complexity of the overall problem is reflected by the use of different carbapenems in hospitals, differences in susceptibility breakpoints, inadequate level of infection control, and low availability of rapid diagnostic methods to facilitate early appropriate interventions in patients who are either colonized or infected by carbapenem-resistant pathogens. Overall, we observe a growing spread of carbapenemase producers (OXA-23) in A. baumannii, mostly in patients hospitalized in the intensive care unit. Carbapenemase types in Enterobacteriaceae are more variable, with a trend toward dissemination of KPC producers in hospital-acquired pathogens (mostly K. pneumoniae) and dissemination of OXA-48 and NDM producers in community-acquired Enterobacteriaceae (mostly E. coli), particularly in Europe. The mechanism of resistance varies according to geographic location, and this should guide the choice of carbapenem resistance testing method. As some β-lactamase inhibitors have weak or no inhibitory activity against carbapenemases, such as MBLs or OXA, identifying the mechanism of resistance at the genomic level and the susceptibility of the pathogen are equally important when choosing the appropriate antibiotic. Rapid diagnostic tests that provide results which can be properly interpreted for the detection of carbapenem resistance can facilitate both therapeutic decision making and infection control measures. Until hospitals have better rapid diagnostic methods, clinicians should make use of national surveillance studies, regional databases, and local hospital- and ward-level susceptibility data to help guide their antibiotic treatment decisions when their patients are at risk of being infected by a carbapenem-resistant pathogen.
Notes
Acknowledgments. Editorial support was provided by Highfield (Oxford, United Kingdom), sponsored by Shionogi Inc (Florham Park, New Jersey).
Financial support. This review article was sponsored by Shionogi & Co, Ltd (Osaka, Japan), but the authors did not receive any fee for their authorship.
Supplement sponsorship. This supplement is sponsored by Shionogi & Co., Ltd.
Potential conflicts of interest. P. N. and L. P. have codeveloped the Rapid Carba NP test marketed by bioMérieux Ltd (Marcy-l’Etoile, France), under the trade name Rapidec Carba NP. P. N. has received speaker’s fees from Shionogi. Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics.2017. Available at: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. Accessed 1 February 2019.
- 2. van Duin D, Kaye KS, Neuner EA, Bonomo RA. Carbapenem-resistant Enterobacteriaceae: a review of treatment and outcomes. Diagn Microbiol Infect Dis 2013; 75:115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai B, Echols R, Magee G, et al. Prevalence of carbapenem-resistant gram-negative infections in the United States predominated by Acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infect Dis 2017; 4:ofx176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cai B, Echols R, Morgan G, et al. Carbapenem-resistant gram-negative pathogens among hospitalized patients in the United States between 2010 and 2015. Poster presented at: IDWeek 2016, New Orleans, LA,25–30 October 2016; Poster 362. [Google Scholar]
- 5. Kim T, Lee EJ, Park SY, et al. Natural prognosis of carbapenem-resistant Acinetobacter baumannii bacteremia in patients who did not receive appropriate antibiotic treatment: a retrospective multicenter study in Korea. Medicine (Baltimore) 2018; 97:e12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. World Health Organization. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities.2017. Available at: https://www.who.int/infection-prevention/publications/guidelines-cre/en/. Accessed 1 February 2019.
- 7. Andria N, Henig O, Kotler O, et al. Mortality burden related to infection with carbapenem-resistant gram-negative bacteria among haematological cancer patients: a retrospective cohort study. J Antimicrob Chemother 2015; 70:3146–53. [DOI] [PubMed] [Google Scholar]
- 8. Pouch SM, Satlin MJ. Carbapenem-resistant Enterobacteriaceae in special populations: solid organ transplant recipients, stem cell transplant recipients, and patients with hematologic malignancies. Virulence 2017; 8:391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018; 7:212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banerjee R, Humphries R. Clinical and laboratory considerations for the rapid detection of carbapenem-resistant Enterobacteriaceae. Virulence 2017; 8:427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. El Chakhtoura NG, Saade E, Iovleva A, et al. Therapies for multidrug resistant and extensively drug-resistant non-fermenting gram-negative bacteria causing nosocomial infections: a perilous journey toward ‘molecularly targeted’ therapy. Expert Rev Anti Infect Ther 2018; 16:89–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen S, Hackel M, Hayes J, et al. In vitro antibacterial activity of cefiderocol against an international collection of carbapenem-non-susceptible gram-negative bacteria isolated from respiratory, blood, skin/soft tissue and urinary sources of infection: SIDERO-WT-2014–2016. Poster presented at: 29th European Congress of Clinical Microbiology and Infectious Diseases, Amsterdam, Netherlands,13–16 April 2019; Poster 1855. [Google Scholar]
- 13. Bonomo RA, Burd EM, Conly J, et al. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 2018; 66:1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Duin D, Bonomo RA. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis 2016; 63:234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shields RK, Chen L, Cheng S, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne bla(KPC-3) mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 2017; 61 pii:e02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nordmann P, Poirel L. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 2013; 68:487–9. [DOI] [PubMed] [Google Scholar]
- 17. Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 1980; 289:321–31. [DOI] [PubMed] [Google Scholar]
- 18. Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 2010; 54:969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abboud MI, Damblon C, Brem J, et al. Interaction of avibactam with class B metallo-β-lactamases. Antimicrob Agents Chemother 2016; 60:5655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lomovskaya O, Sun D, Rubio-Aparicio D, et al. Vaborbactam: spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob Agents Chemother 2017; 61:e01443–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis 2016; 3:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. European Committee on Antimicrobial Susceptibility Testing. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 2.0.2017. Available at: http://www.eucast.org/resistance_mechanisms/. Accessed 1 February 2019.
- 23. Centers for Disease Control and Prevention. Facility guidance for control of carbapenem-resistant Enterobacteriaceae—November 2015 update. Available at: https://www.cdc.gov/hai/pdfs/cre/CRE-guidance-508.pdf. Accessed 1 February 2019.
- 24. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011; 17:1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quale J, Bratu S, Gupta J, Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 2006; 50:1633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA. Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev 2017; 30:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodríguez CH, Nastro M, Famiglietti A. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Rev Argent Microbiol 2018; 50:327–33. [DOI] [PubMed] [Google Scholar]
- 28. Chang YT, Lin CY, Chen YH, Hsueh PR. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front Microbiol 2015; 6:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53:4783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 2003; 67:593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moyá B, Beceiro A, Cabot G, et al. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 2012; 56:4771–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamachika S, Sugihara C, Kamai Y, Yamashita M. Correlation between penicillin-binding protein 2 mutations and carbapenem resistance in Escherichia coli. J Med Microbiol 2013; 62(Pt 3):429–36. [DOI] [PubMed] [Google Scholar]
- 33. Cayô R, Rodríguez MC, Espinal P, et al. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 2011; 55:5907–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 29th informational supplement. M100-S29. Wayne, PA: CLSI, 2019. [Google Scholar]
- 35. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. Available at: http://www.eucast.org. Accessed 1 February 2019.
- 36. Gniadek TJ, Carroll KC, Simner PJ. Carbapenem-resistant non-glucose-fermenting gram-negative bacilli: the missing piece to the puzzle. J Clin Microbiol 2016; 54:1700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2012; 18:1503–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lutgring JD, Limbago BM. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol 2016; 54:529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poirel L, Nordmann P. Rapidec Carba NP test for rapid detection of carbapenemase producers. J Clin Microbiol 2015; 53:3003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pires J, Novais A, Peixe L. Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 2013; 51:4281–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kostyanev T, Vilken T, Lammens C, Timbermont L, Van’t Veen A, Goossens H. Detection and prevalence of carbapenem-resistant gram-negative bacteria among European laboratories in the COMBACTE network: a COMBACTE LAB-Net survey. Int J Antimicrob Agents 2019; 53:268–74. [DOI] [PubMed] [Google Scholar]
- 42. van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 2015; 10:e0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chea N, Bulens SN, Kongphet-Tran T, et al. Improved phenotype-based definition for identifying carbapenemase producers among carbapenem-resistant Enterobacteriaceae. Emerg Infect Dis 2015; 21:1611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Findlay J, Hopkins KL, Meunier D, Woodford N. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J Antimicrob Chemother 2015; 70:1338–42. [DOI] [PubMed] [Google Scholar]
- 45. Ledeboer NA, Lopansri BK, Dhiman N, et al. Identification of gram-negative bacteria and genetic resistance determinants from positive blood culture broths by use of the verigene gram-negative blood culture multiplex microarray-based molecular assay. J Clin Microbiol 2015; 53:2460–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salimnia H, Fairfax MR, Lephart PR, et al. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 2016; 54:687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cunningham SA, Vasoo S, Patel R. Evaluation of the check-points check MDR CT103 and CT103 XL microarray kits by use of preparatory rapid cell lysis. J Clin Microbiol 2016; 54:1368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kiratisin P, Chongthaleong A, Tan TY, et al. Comparative in vitro activity of carbapenems against major gram-negative pathogens: results of Asia-Pacific surveillance from the COMPACT II study. Int J Antimicrob Agents 2012; 39:311–6. [DOI] [PubMed] [Google Scholar]
- 49. Jean SS, Hsueh PR, Lee WS, et al. Carbapenem susceptibilities and non-susceptibility concordance to different carbapenems amongst clinically important gram-negative bacteria isolated from intensive care units in Taiwan: results from the Surveillance of Multicentre Antimicrobial Resistance in Taiwan (SMART) in 2009. Int J Antimicrob Agents 2013; 41:457–62. [DOI] [PubMed] [Google Scholar]
- 50. Mendes RE, Mendoza M, Banga Singh KK, et al. Regional resistance surveillance program results for 12 Asia-Pacific nations (2011). Antimicrob Agents Chemother 2013; 57:5721–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oliveira VD, Rubio FG, Almeida MT, Nogueira MC, Pignatari AC. Trends of 9,416 multidrug-resistant gram-negative bacteria. Rev Assoc Med Bras (1992) 2015; 61:244–9. [DOI] [PubMed] [Google Scholar]
- 52. Labarca JA, Salles MJ, Seas C, Guzmán-Blanco M. Carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter baumannii in the nosocomial setting in Latin America. Crit Rev Microbiol 2016; 42:276–92. [DOI] [PubMed] [Google Scholar]
- 53. Gales AC, Seifert H, Gur D, Castanheira M, Jones RN, Sader HS. Antimicrobial susceptibility of Acinetobacter calcoaceticus–Acinetobacter baumannii complex and Stenotrophomonas maltophilia clinical isolates: results from the SENTRY Antimicrobial Surveillance Program (1997–2016). Open Forum Infect Dis 2019; 6(Suppl 1):S34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strateva T, Sirakov I, Stoeva T, et al. Carbapenem-resistant Acinetobacter baumannii: current status of the problem in four Bulgarian university hospitals (2014–2016). J Glob Antimicrob Resist 2019; 16:266–73. [DOI] [PubMed] [Google Scholar]
- 55. McCann E, Srinivasan A, DeRyke CA, et al. Carbapenem-nonsusceptible gram-negative pathogens in ICU and non-ICU settings in US hospitals in 2017: a multicenter study. Open Forum Infect Dis 2018; 5:ofy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sader HS, Castanheira M, Farrell DJ, Flamm RK, Mendes RE, Jones RN. Tigecycline antimicrobial activity tested against clinical bacteria from Latin American medical centres: results from SENTRY Antimicrobial Surveillance Program (2011–2014). Int J Antimicrob Agents 2016; 48:144–50. [DOI] [PubMed] [Google Scholar]
- 57. Sader HS, Castanheira M, Mendes RE, Flamm RK, Jones RN. Antimicrobial activity of high-proportion cefepime-tazobactam (WCK 4282) against a large number of gram-negative isolates collected worldwide in 2014. Antimicrob Agents Chemother 2017; 61 pii:e02409–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cai B, Echols R, Magee G, et al. Geographic distribution of carbapenem-resistant gram-negative infections in adult patients in US hospitals. Poster presented at: American Society for Microbiology [Microbe 2016], Boston, MA, 16–20 June2016; Poster 268. [Google Scholar]
- 59. Japan Nosocomial Infections Surveillance. Annual open report 2016. Available at: https://janis.mhlw.go.jp/english/report/open_report/2016/3/1/ken_Open_Report_Eng_201600_clsi2012.pdf. Accessed 1 February 2019.
- 60. Suwantarat N, Carroll KC. Epidemiology and molecular characterization of multidrug-resistant gram-negative bacteria in Southeast Asia. Antimicrob Resist Infect Control 2016; 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Velasco J, Valderama M, Peacock T, et al. Carbapenemase-producing Enterobacteriaceae and nonfermentative bacteria, the Philippines, 2013–2016. Emerg Infect Dis 2017; 23:1597–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grundmann H, Glasner C, Albiger B, et al. European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 2017; 17:153–63. [DOI] [PubMed] [Google Scholar]
- 63. Wendel AF, Kolbe-Busch S, Ressina S, et al. Detection and termination of an extended low-frequency hospital outbreak of GIM-1-producing Pseudomonas aeruginosa ST111 in Germany. Am J Infect Control 2015; 43:635–9. [DOI] [PubMed] [Google Scholar]
- 64. Karampatakis T, Tsergouli K, Politi L, et al. Molecular epidemiology of endemic carbapenem-resistant gram-negative bacteria in an intensive care unit. Microb Drug Resist 2019; 25:712–6. [DOI] [PubMed] [Google Scholar]
- 65. Guh AY, Bulens SN, Mu Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 2015; 314:1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mataseje LF, Abdesselam K, Vachon J, et al. Results from the Canadian Nosocomial Infection Surveillance Program on carbapenemase-producing Enterobacteriaceae, 2010 to 2014. Antimicrob Agents Chemother 2016; 60:6787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 2015: 45:568–85. [DOI] [PubMed] [Google Scholar]
- 68. Lincopan N, McCulloch JA, Reinert C, Cassettari VC, Gales AC, Mamizuka EM. First isolation of metallo-beta-lactamase-producing multiresistant Klebsiella pneumoniae from a patient in Brazil. J Clin Microbiol 2005; 43:516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bartolleti F, Seco BM, Capuzzo Dos Santos C, et al. Polymyxin B resistance in carbapenem-resistant Klebsiella pneumoniae, São Paulo, Brazil. Emerg Infect Dis 2016; 22:1849–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. López-García A, Rocha-Gracia RDC, Bello-López E, et al. Characterization of antimicrobial resistance mechanisms in carbapenem-resistant Pseudomonas aeruginosa carrying IMP variants recovered from a Mexican hospital. Infect Drug Resist 2018; 11:1523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee CS, Doi Y. Therapy of infections due to carbapenem-resistant gram-negative pathogens. Infect Chemother 2014; 46:149–64. [DOI] [PMC free article] [PubMed] [Google Scholar]