Fig. 2.
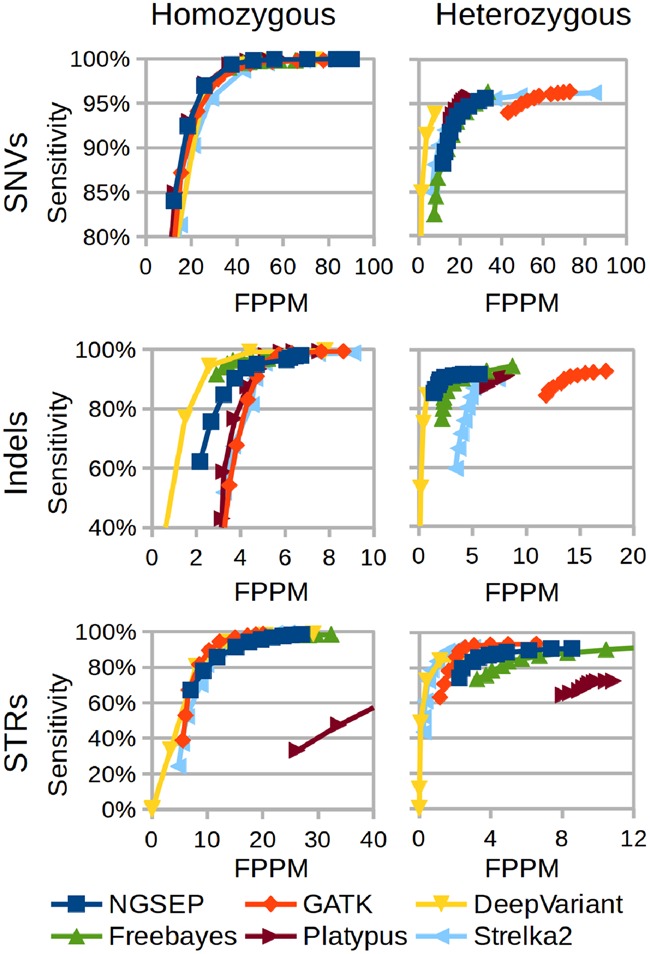
Comparison of different tools for variants discovery from reads taken from an F1 pool of segregants derived from two yeast haploid strains. Results are discriminated by variant type (SNVs, Indels and STRs) and gold standard genotype (homozygous variant or heterozygous). The proportion of FPPM is used as a measure of specificity. Curves are obtained varying the filter of minimum genotype quality (GQ field in the VCF file) from 0 to 90
