SUMMARY
Most human cancers originate from high-turnover tissues, while low-proliferating tissues, like skeletal muscle, exhibit a lower incidence of tumor development. In Duchenne muscular dystrophy (DMD), which induces increased skeletal muscle regeneration, tumor incidence is increased. Rhabdomyosarcomas (RMSs), a rare and aggressive type of soft tissue sarcoma, can develop in this context, but the impact of DMD severity on RMS development and its cell of origin are poorly understood. Here, we show that RMS latency is affected by DMD severity and that muscle stem cells (MuSCs) can give rise to RMS in dystrophic mice. We report that even before tumor formation, MuSCs exhibit increased self-renewal and an expression signature associated with RMSs. These cells can form tumorspheres in vitro and give rise to RMSs in vivo. Finally, we show that the inflammatory genes Ccl11 and Rgs5 are involved in RMS growth. Together, our results show that DMD severity drives MuSC-mediated RMS development.
In Brief
Boscolo Sesillo et al. describe how Duchenne muscular dystrophy severity increases rhabdomyosarcoma (RMS) formation and reduces latency. This study shows that muscle stem cells give rise to tumorspheres in vitro and RMS tumors in vivo and identify genes involved in RMS growth.
Graphical Abstract

INTRODUCTION
Skeletal muscle is a low-turnover tissue maintained by muscle stem cells (MuSCs), which are required for tissue growth and repair (Brack and Rando, 2012; Tierney and Sacco, 2016). In adult tissues, MuSCs exist in a quiescent state and become activated in response to injury to generate progenitors that differentiate to repair the damaged myofibers (Almada and Wagers, 2016; Yin et al., 2013). The low turnover of skeletal muscle is associated with a low frequency of tumor formation and growth compared to highly proliferative tissues such as epithelia (Bar-Yehuda et al., 2001; Parlakian et al., 2010; Tomasetti and Vogelstein, 2015). Rhabdomyosarcoma (RMS) is a rare soft tissue sarcoma that can develop throughout life but has a higher incidence in young children (Sultan et al., 2009; https://nccd.cdc.gov/uscs/childhoodcancerdetailedbyICCC.aspx). RMS is characterized by the expression of the myogenic genes DESMIN, MYOD1, and MYOGENIN (Morotti et al., 2006), and this has led to the notion that RMS can originate from myogenic progenitors. Indeed, mouse models with genetic modifications associated with human RMS (Chen et al., 2013; Shern et al., 2014), which include PAX3:FOXO1 translocation and loss of function of p53 or Ink4a/ARF locus, as well as alterations of Hippo, Sonic hedgehog, and Kras pathways, in myogenic progenitors manifest a high frequency of RMS development (Blum et al., 2013; Keller et al., 2004; Rubin et al., 2011; Tremblay et al., 2014). However, it was recently shown that RMS can also originate from endothelial progenitors that transdifferentiate into myogenic cells in response to hyperactivation of the Sonic hedgehog pathway during development (Drummond et al., 2018; Hatley et al., 2012), suggesting that transformation of non-muscle cells coincides with acquisition of myogenic features and RMS formation. These findings indicate that the cell of origin of RMS is poorly understood and may vary depending on the context.
The mdx mouse, a model for Duchenne muscular dystrophy (DMD), a progressive muscle degenerative disease that places a strong regenerative pressure on the MuSC compartment (Duchenne, 1867; Hoffman et al., 1987), shows an increased incidence of spontaneous RMS development (Chamberlain et al., 2007; Fernandez et al., 2010; Hosur et al., 2012). Increased susceptibility to RMS has been attributed to multiple features of the dystrophic disease, including accumulation of DNA damage, inflammation, and reactive oxygen species (Fanzani et al., 2013; Hanahan and Weinberg, 2000; Latella et al., 2017; Schmidt et al., 2011; Wang et al., 2014). This suggests that a highly regenerative pathological muscle microenvironment is more permissive to tumor development. In addition, recent studies reported that genetic deletion of p53 in chronically regenerating mdx mice led to RMS development (Camboni et al., 2012; Fernandez et al., 2010). Consistent with these mouse studies, DMD has been associated with the development of RMS in four separate clinical case reports (Bűget et al., 2014; Jakab et al., 2002; Rossbach et al., 1999; Saldanha et al., 2005), further suggesting a correlation between RMS and DMD. However, RMS cell of origin in the DMD context and how the severity of the dystrophic disease influences tumor development are still poorly understood.
Here, we crossed the mdx/mTR mouse, a model that mimics severe human dystrophic disease progression through progressive telomere shortening in MuSCs, with p53Het mice (Jacks et al., 1994; Sacco et al., 2010). We show that DMD disease severity reduces mouse lifespan through early tumor formation. We further show that in this mouse model, MuSCs have a gene expression signature associated with RMS even before tumor development, and they exhibit increased self-renewal, impaired differentiation, and accumulation of DNA damage, indicating increased tissue turnover and genomic instability. We provide evidence that MuSCs from this mouse model have tumorigenic potential and are capable of forming tumorspheres in vitro and solid tumors in vivo when transplanted into immunodeficient mice, indicating that RMS in dystrophic contexts can originate from MuSCs. Finally, we show that Ccl11 and Rgs5, two genes involved in the inflammatory response and stem cell fate decisions during wound healing (Bahrami et al., 2014; Kindstedt et al., 2017; Rees et al., 2015), are downregulated in MuSCs from mdx/mTR mice lacking p53, and their overexpression leads to decreased tumorsphere size. Together, these findings demonstrate that MuSCs can give rise to RMS in high-turnover conditions, highlighting the importance of assessing MuSC behavior after treatments aimed at increasing their self-renewal ability for treating DMD.
RESULTS
Severity of Muscular Dystrophy Decreases the Latency of RMS Formation and Mouse Lifespan
In order to assess whether RMS formation in a dystrophic context is dependent on increased tissue turnover, we employed a severe DMD mouse model (mdx/mTR) and compared these mice to mdx mice (Camboni et al., 2012; Sacco et al., 2010). p53Het mice were crossed with mdx/mTR (STAR Methods) to enhance tumor development, consistent with a previous report (Camboni et al., 2012). Tumor frequency was significantly increased in mdx/mTR mice that partially or completely lack p53 when compared to non-dystrophic p53 mice and mdx/mTR mice with intact p53 (Figure 1A; Table S1). This observation indicates that the absence of p53 and the presence of a highly regenerative environment synergistically contribute to increased tumor frequency. To determine the specific contribution of the disease in tumor formation, we focused on healthy and dystrophic mice within discrete p53 status (p53Het VS p53Het mdx/mTR and p53KO VS p53KO mdx/mTR) and observed both a significant increase in tumor frequency and a reduction in tumor latency in dystrophic mice, indicating that DMD plays a major role in tumor formation (Figure 1B; Table S1). Our data show that p53WT mdx/mTR mice have a tumor frequency comparable to what has previously been reported for mdx mice (Camboni et al., 2012; Fernandez et al., 2010); however, the age of onset is dramatically accelerated (9 months) compared to the less severe DMD mouse model (18 months), suggesting that increased disease severity mediates early tumor development (Figures 1A, 1B, S1A, and S1B). Consistent with this finding, we observed that in both p53Het mdx/mTR and p53KO mdx/mTR mice, the latency of tumor formation decreases along with increased disease severity, starting as early as 2 months of age in mdx/mTRG2 mice (Figures 1B, 1C, and S1C–S1F). Moreover, the average latency of tumor formation in p53KO mdx/mTR mice is significantly reduced when compared to p53Het mdx/mTR (Figures 1B, 1C, S1E, and S1F). In addition, in p53Het mdx/mTR mice, the progression of tumor formation is faster when compared to p53Het mdx mice; indeed, by 10 months, 85% of the mice develop tumors. Together, these results show that the synergistic effect of DMD and p53 impacts tumor frequency and that disease severity plays a major role in the age of tumor onset.
Figure 1. p53Het/KO mdx/mTR Mice Have a Higher Incidence and Earlier Onset of RMS Tumor Development than p53KO or Dystrophic Mice.
(A) Bar graph representing frequency of tumor formation for each genotype (n = 10−57).
(B) Graphical representation of age of tumor onset for p53 and p53 mdx/mTR mice. Dots represent individual mice that developed solid tumors (n = 1–36 animals). **p < 0.01; ***p < 0.001 (unpaired t test).
(C) Kaplan-Meier survival curve for p53WT/Het/KO mdx/mTR mice (n = 39−133 animals). ****p < 0.0001 (Mantel-Cox test).
(D) Pie chart representing anatomical location of tumors in p53 mdx/mTR mice (n = 159 tumors). Percentage for each location is indicated.
(E) Representative H&E staining of tumor tissues. Top left: from p53Het mdx/mTRG1 mouse shoulder; top right: from p53Het mdx/mTRG1 mouse leg; bottom left: from p53Het mdx/mTRHet mouse leg; bottom right: from p53KO mdx/mTRG2 mouse leg. White arrows indicate myofibers (left images), spindle-like cells (top right), and an anaplastic cell (bottom right). Scale bar, 200 μm.
(F) Representative images (top) and zoomed area (bottom) from a p53KO mdx/mTRG2 mouse tumor (desmin in green; DAPI in blue). Scale bars represent 200 μm (top) and 50 μm (bottom).
(G) Representative images (top) and zoomed area (bottom) from a p53Het mdx/mTRG1 mouse tumor (myogenin in green; DAPI in blue). Scale bars represent 100 μm (top) and 25 μm (bottom).
(H) Quantification of tumors expressing desmin and myogenin.
We next assessed the anatomical distribution of the tumors developed in this mouse model. The majority of tumors (~80%) were localized in the limbs and abdomen (Figures 1D and S1G), consistent with what has been previously reported in both RMS mouse models and human patients (Bűget et al., 2014; Camboni et al., 2012; Chamberlain et al., 2007; Fernandez et al., 2010; Jakab et al., 2002; Rossbach et al., 1999; Saldanha et al., 2005), and were tightly associated with the musculature (Figures 1D and S1G). Moreover, ~30% of the p53Het/KO mdx/mTR mice developed more than one tumor, consistent with what has previously been reported in other dystrophic mouse models (Figure S1H) (Fernandez et al., 2010). Phenotypically, the isolated tumors appeared composed by solid and highly vascularized tissue with tan- and white-colored areas. Histological analysis showed muscle myofibers embedded in highly undifferentiated areas of the tissue, spindle-like cells, and anaplastic cells (Figure 1E). Furthermore, the majority of the analyzed tumors expressed desmin (96%) and myogenin (77%), the myogenic markers used for RMS diagnosis (Figures 1F–1H; Table S2). Together, these results show that mdx/mTR mice that do not express p53 develop RMS earlier in life and at a higher incidence. The age of tumor onset decreases with DMD disease severity, indicating that a more degenerative environment promotes the development of RMS.
Expansion of MuSCs and Accumulation of DNA Damage in the p53KO mdx/mTR RMS Mouse Model
As RMS in p53Het/KO mdx/mTR mice mainly developed in close proximity to muscles and increased tissue turnover decreases the age of onset of tumor formation, we next asked whether alterations of the mature muscle and/or the MuSC compartment, the main player in muscle regeneration, contribute to tumor development. To this aim, we isolated gastrocnemius muscles from 2-month-old p53KO mdx/mTRG2 mice and p53WT mdx/mTRG2 controls before tumor development, and we observed a significant increase in the number of Pax7+ MuSC in p53KO mdx/mTRG2 mice versus controls (Figures 2A and 2B). While we did not observe significant differences in the number of MyoD+ cells (marker of myogenic commitment) between p53WT mdx/mTRG2 and p53KO mdx/mTRG2 mice (Figures S2A and S2B), we observed an increase in the percentage of areas with newly regenerated myofibers by embryonic myosin heavy chain (eMyHC) expression (Figures 2C and 2D), suggesting increased muscle turnover. Supporting this finding, we also observed a significant increase in the number of smaller myofibers in muscles of p53KO mdx/mTRG2 mice, another hallmark associated with muscle regeneration (Figures S2C and S2D).
Figure 2. Deletion of p53 in mdx/mTRG2 Mice Induces an Increase in the MuSCs Pool and Tissue Turnover.
(A) Representative images from p53WT (top) and p53KO (bottom) mdx/mTRG2 mouse tissues (Pax7 in green; laminin in white; DAPI in blue). Scale bar, 50 μm.
(B) Quantification of Pax7+ cells (n = 5–8 animals). ***p < 0.001 (unpaired t test).
(C) Representative images from p53WT and p53KO mdx/mTRG2 mouse tissues (eMyHC in green; laminin in red; DAPI in blue). Scale bar, 200 μm.
(D) Quantification of eMyHC+ areas in p53WT and p53KO mdx/mTRG2 mouse tissues (n = 3−5 animals). *p < 0.05 (unpaired t test).
(E) Representative images from p53WT and p53KO mdx/mTRG2 mouse tissues (Pax7 in green; pH2AX in red; laminin in white; DAPI in blue). White arrows indicate MuSCs positive for the DNA damage marker. Scale bar, 100 μm.
(F) Quantification of Pax7+/pH2AX+ cells over total Pax7+ cells (n = 3 animals). *p < 0.05 (unpaired t test).
(G) Quantification of pH2AX+ centrally nucleated myofibers over total centrally nucleated myofibers (n = 3 animals). *p < 0.05 (unpaired t test).
It has previously been reported that muscular dystrophy activates DNA damage response pathways (Schmidt et al., 2011; Tichy et al., 2017), and we recently showed that MuSCs from mdx/mTRG2 mice accumulate DNA damage when compared to mdx/mTRHet/G1 mice (Latella et al., 2017). Thus, we assessed whether knockout of p53 in mdx/mTRG2 mice could accumulate further DNA damage and whether this could lead to tissue instability, favoring RMS development. Co-immunostaining of the DNA damage markers gH2AX or 53BP1 and Pax7 in muscle tissues showed a significant increase in the number of MuSCs positive for DNA damage (Figures 2E, 2F, S2E, and S2F). Interestingly, a significant increase in DNA damage was also detected in myonuclei of centrally nucleated myofibers, suggesting that DNA damage accumulates not only in MuSCs but also in differentiated muscle tissue (Figure 2G and S2G).
Together, our results show that muscles isolated from p53KOmdx/mTRG2 mice before tumor formation exhibit a significant increase in MuSC number that is associated with an increase in regenerating areas, suggesting accelerated muscle turnover. In addition, both mature muscle and MuSCs from p53KO mdx/mTRG2 mice accumulate DNA damage, indicating that the tissue is more genetically unstable and thus more susceptible to transformation.
MuSCs from p53KOmdx/mTRG2 Mice Exhibit Increased Self-Renewal and Impaired Differentiation Ability
The mouse model utilized for this study is a germline knockout for p53, dystrophin, and the telomerase RNA component (mterc); thus, although MuSCs from p53KO mdx/mTRG2 mice show a significant expansion in vivo, we cannot rule out the possibility that other muscle resident cells involved in the process of regeneration, such as fibroadipogenic progenitor cells (FAPs) (Joe et al., 2010; Uezumi et al., 2010), exhibit similar behavior. To this aim, we isolated MuSCs and FAPs through flow cytometry, using as positive markers CD34 and α7-integrin and CD34 and Sca1, respectively, as previously described (Joe et al., 2010; Sacco et al., 2008), and assessed them through clonal analysis (Figure 3A). We first observed that clonogenicity is significantly increased in MuSCs from p53KO mdx/mTRG2 mice compared to their wild-type (WT) counterparts, whereas there is no difference when p53WT mdx/mTRG2 and p53KO mdx/mTRG2 FAPs were compared (Figures 3B and 3C). Moreover, p53KO mdx/mTRG2 MuSCs developed significantly larger clones compared to all other cellular populations under analysis, consistent with what was observed in vivo, while p53KO mdx/mTRG2 FAPs did not exhibit any difference in clone size compared to their control counterparts (Figures 3D and 3E). These results suggest that MuSCs from p53KO mdx/mTRG2 mice have an increased ability to develop clones and can also preferentially expand in culture when compared to FAPs. We next asked whether a difference in cell fate dynamics could determine the observed phenotype in MuSCs. Within p53KO mdx/mTRG2 clones, we observed a significant increase in the percentage of Pax7+/MyoD− cells and a decrease of Pax7−/MyoD+ cells, which suggests that p53KO mdx/mTRG2 MuSCs have an increased ability to expand in an undifferentiated manner and lower myogenic commitment when compared to their wild-type counterparts (Figures 3F and 3G). Thus, we asked whether this behavior would also be validated at the population level through assessment of myogenic differentiation (Figure 3H). In this context, we observed a significant decrease in the differentiation index in p53KO mdx/mTRG2 MuSCs when compared to their wild-type counterparts (Figures 3I and 3J), as shown by the lower percentage of p53KO mdx/mTRG2 nuclei within myosin heavy chain (MyHC)-positive cells (a marker associated with muscle cell terminal differentiation). Despite this significant difference in differentiation ability, p53KO mdx/mTRG2 MuSCs did not show any defects in cell fusion, suggesting that those cells that are capable of differentiation can efficiently form myotubes (Figure S3F). The decrease in the differentiation index was associated with a significant expansion of Pax7+ nuclei in p53KO mdx/mTRG2 differentiated cultures (Figures 3K, 3L, and S3B), suggesting that p53KOmdx/mTRG2 MuSCs have increased self-renewal capacity, consistent with what we observed through clonal analysis.
Figure 3. MuSCs from p53KO mdx/mTRG2 Mice Display Increased Self-Renewal and Impaired Differentiation Ability.
(A) Experimental design for (B)–(G).
(B) Bar graph representing percentage of clones developed from p53WT and p53KO mdx/mTRG2 MuSCs. *p < 0.05 (unpaired t test). n = 2 technical replicates for 3 animals.
(C) Bar graph representing the percentage of clones developed from p53WT and p53KO mdx/mTRG2 FAPs. n = 3−6 technical replicates for two animals.
(D) Representative images for clones from MuSCs and FAPs. Left: p53WT mdx/mTR samples. Right: p53KO mdx/mTR samples. DAPI in blue. Scale bar, 100 μm.
(E) Graphical representation of clone size from p53WT mdx/mTR and p53KO mdx/mTR MuSCs and FAPs. ****p < 0.0001 (one-way ANOVA). n = 13−87 clones for two animals per genotype.
(F) Representative images from p53WT and p53KO mdx/mTRG2 MuSCs clones (Pax7 in green; MyoD in red; DAPI in blue). Scale bar, 100 μm.
(G) Quantification of Pax7+ and MyoD+ cells in clones from p53WT and p53KO mdx/mTR MuSCs. *p value < 0.05; ****p < 0.0001 (two-way ANOVA). n = 13−26 clones from two animals per genotype).
(H) Experimental design for (I)–(L).
(I) Representative images from p53WT and p53KO mdx/mTRG2 MuSCs 3 days after differentiation (myosin heavy chain in green; DAPI in blue). Scale bar, 100 μm.
(J) Quantification of MuSCs differentiation index (number of nuclei within Mf20+ cells over total number of nuclei). **p < 0.01 (unpaired t test). n = 3−4 animals.
(K) Representative immunofluorescence images from p53WT and p53KO mdx/mTRG2 differentiated MuSCs (Pax7 in green; pH2AX in red; DAPI in blue). Scale bar, 200 μm.
(L) Quantification of percentage of Pax7+ cells in p53WT and p53KO mdx/mTRG2 differentiating cultures. *p < 0.05 (unpaired t test). n = 4−5 animals.
Together, these results show that p53KO mdx/mTRG2 MuSCs expand both in vivo and in vitro due to increased self-renewal ability and impaired differentiation capacity. In line with our findings, it has been previously shown that defective progression through the myogenic lineage is a feature of RMS, suggesting a possible involvement of MuSCs in RMS development in a dystrophic context (Puri et al., 2000; Tapscott et al., 1993; Yang et al., 2009).
Freshly Isolated MuSCs from p53 mdx/mTR Mice Form Tumorspheres In vitro and Give Rise to Tumors In vivo
To test which muscle-resident cell type has tumorigenic potential, we isolated MuSCs, FAPs, and lineage+ (Lin+) (CD45+CD31+CD11b+) endothelial cells and macrophages from p53WT and p53KO mdx/mTRG2 2-month-old mice via flow cytometry (Figure S4A), and we performed a tumorsphere assay in vitro, where cells are grown in anchorage-independent manner to enrich for tumor-propagating cells (TPCs) (Johnson et al., 2013; Shin et al., 1975; Walter et al., 2011) (Figure 4A). As a positive control for the assay, we employed cells isolated from tumors developed in the mdx/mTR mouse model (RMS cells). Cells were plated (at 100 cells per well) in serum-free media, supplemented only with growth factors (fibroblast growth factor [FGF] and epidermal growth factor [EGF]) on low-attachment plates and grown in suspension culture. After 30 days, Lin+ cells were no longer detectable in the culture dishes, suggesting they could not survive in these growing conditions. Both MuSCs and FAPs survived through formation of cellular aggregates that favor cell survival in suspension conditions (Figure S4B) (Johnson et al., 2013; Weiswald et al., 2015). Strikingly, MuSCs, but not FAPs, isolated from p53KO mdx/mTRG2 mice formed tumorspheres with a morphology and size similar to the spheroid structures formed by RMS cells (Figures 4B and 4C). The frequency of tumorsphere development was lower in p53KO mdx/mTRG2 MuSCs than in RMS cells (0.3% versus 9.3%, respectively). Out of the six p53KO mdx/mTRG2 mice tested for this assay, three (50%) were able to form tumorspheres, consistent with the tumor development frequency of p53KO mdx/mTRG2 mice (see Figure 1A). These results indicate that MuSCs from these mice have tumorigenic potential. To demonstrate the ability of MuSC-derived tumorspheres to develop RMS in vivo, we performed allografts through subcutaneous injection (20,000 cells) into immunodeficient mice (Figure 4A). As a positive control, we transplanted, in parallel, tumorspheres developed from RMS cells (Figure S4C). In RMS cell allografts, all the transplanted mice presented with visible tumors within 6 weeks after the original injection. Analysis of these tumors revealed the presence of desmin-positive cells and high levels of gH2AX staining, confirming the development of malignant RMS (Figure S4D). MuSC-derived tumorsphere allografts developed into solid tumors 4 months after cell injection (Figure 4D). In isolated tumors, we detected rhabdomyoblasts and spindle-like cells, desmin expression, and DNA damage, confirming that masses generated from MuSC-derived tumorspheres are indeed RMS (Figures 4E and 4F).
Figure 4. MuSCs Isolated from p53KO mdx/mTRG2 Mice Can Form Tumorspheres In vitro and RMS after Transplantation In vivo.
(A) Experimental design for (B)–(F).
(B) Representative images of tumorspheres from p53KO mdx/mTRG2 MuSCs and RMS cells. Scale bar, 500 μm.
(C) Quantification of tumorspheres size. ***p < 0.001 (one-way ANOVA). n = 0−87 tumorspheres from one to five animals.
(D) Image of a tumor that developed in an immunodeficient mouse injected with 20,000 tumorsphere-derived cells before (top) and after (bottom) explant.
(E) Representative H&E staining of tumor tissue. Top: undifferentiated areas of the tumor; bottom: spindle-like cell structure. Scale bar, 100 μm.
(F) Representative images from tumor tissue isolated from an immunodeficient mouse injected with 20,000 MuSC-derived tumorsphere cells (desmin in green; γH2AX in red; DAPI in blue). Scale bars represent 100 μm (top) and 50 μm (bottom).
(G) Experimental design for (H)–(J).
(H) Image of a tumor that developed after 200,000 freshly isolated MuSCs were allografted into an immunodeficient mouse before (top) and after (bottom) explant.
(I) Representative H&E staining of tumor histology. Top: myofibers embedded in undifferentiated areas; bottom: multinucleated cells. Scale bar, 100 μm.
(J) Representative images of tumor tissue isolated from immunodeficient mice injected with a 200,000 MuSC solution (desmin in green; γH2AX in red; DAPI in blue). Scale bar, 100 μm.
To further demonstrate the tumorigenic potential of MuSCs and rule out the possibility that FAPs can also generate RMS in vivo, we conducted allograft experiments employing both freshly isolated MuSCs and FAPs (200,000 cells) (Figure 4G). None of the immunodeficient mice injected with FAPs developed any visible or palpable mass. Conversely, 50% of the mice injected with MuSCs presented with tumors within 22 weeks (Figure 4H). Histological analysis of the tumors highlighted the present of muscle fibers in close contact with undifferentiated areas and of anaplastic cells, hallmarks of RMS (Figure 4I). Tumor identity was further confirmed through immunofluorescent staining for desmin and gH2AX (Figure 4J). As a positive control, we transplanted RMS cells (50,000 and 5,000,000 cells) (Figure S4E). RMS cells developed tumors within 21 and 40 days, respectively, from the injection and presented both morphological and histological features associated with RMS (Figures S4F and S4G). Tumors derived from RMS cells were highly positive for the DNA damage marker γH2AX, indicating tissue instability (Figure S4G). These results indicate that MuSCs have tumorigenic potential both in vitro and in vivo and confirmed that MuSCs, but not FAPs, are capable of generating RMS in dystrophic conditions.
RNA Sequencing of MuSCs from the mdx/mTR Mouse Model Identifies Genes Involved in Cellular Transformation and RMS Development
Our findings showed that p53KO mdx/mTRG2 muscles exhibited an increase in MuSC number characterized by increased self-renewal, impaired differentiation, and accumulation of DNA damage and that these cells have tumorigenic capacity both in vitro and in vivo. In order to identify candidate genes involved in MuSC-mediated RMS formation, we first sought to determine whether pre-tumorigenic p53KO mdx/mTRG2 MuSCs already possess a gene signature associated with mouse RMS. To this aim, we performed RNA-sequencing analysis of freshly isolated cells derived from 2-month-old tumor-free adult p53WT mdx/mTRG2 and p53KO mdx/mTRG2 male mice. Differentially expressed genes were identified by cufflink analysis and subjected to meta-analysis through Correlation Engine (data were analyzed against disease atlas, filtered into the cancer subset, and for studies in mouse models) (Figure S5). Our data correlated with two previously published studies that examined RMS allografts derived from MuSCs (Table S3) (Blum et al., 2013; Nishijo et al., 2009). These identified correlations confirmed that p53KO mdx/mTRG2 MuSCs isolated from mice before tumor formation already carry a genetic signature associated with RMS, making these cells optimal candidates for the identification of genes that could be involved in the process of cellular transformation and tumor development. To enhance the chance of identifying relevant genes correlated with RMS development in a dystrophic context, we compared RNA-sequencing data of freshly isolated cells derived from p53WT mdx/mTRG2 and p53KO mdx/mTRG2 mice with those of MuSCs from p53WT and p53KO (wild-type for dystrophin and mTerc) from both uninjured mice and mice injured 3 days before (Figure 5A). Differential expression analysis was performed and allowed the identification of 145 genes uniquely associated with p53KO mdx/mTRG2 MuSCs (Figure 5A; Table S4). We utilized two separate approaches to identify candidate genes. One approach aimed at examining the top differentially expressed genes, while the second utilized Correlation Engine to visualize functional relationships among genes (through “Pathway viewer”) (Figures 5B–5D). The obtained lists were then filtered based on their expression in human RMS or human cancers (Hayes et al., 2018; Shern et al., 2014). This resulted in the identification of two (Rgs5 and Lars2) and four candidate genes (SerpinE2, Ccl11, Flt3g, and Edntb), respectively, that were further validated through qPCR analysis in both MuSCs and FAPs (Figures 5B, 5E, and 5F). None of the tested genes where differentially expressed between p53WTmdx/mTRG2 and p53KO mdx/mTRG2 FAPs, whereas four of them were significantly downregulated in p53KO mdx/mTRG2 MuSCs (Rgs5, SerpinE2, Ccl11, and Flt3g) (Figures 5E and 5F). These results show that MuSCs isolated before tumor development already exhibit a tumor-associated gene signature, making them a useful tool for the identification of genes involved in cellular transformation and RMS development.
Figure 5. Identification of Candidate Genes Involved in Tumor Development through RNA-Sequencing Analysis of p53KO mdx/mTRG2 and Non-dystrophic p53KO MuSCs.
(A) Venn diagram representing overlap of differential expression analysis performed on three sets of samples: p53WT and p53KO MuSCs from healthy mice, p53WT and p53KO MuSCs from mice injured 3 days before isolation, and p53WT mdx/mTRG2 and p53KO mdx/mTRG2 MuSCs.
(B) Schematic representation of the approach utilized for target gene and pathway identification.
(C) Table for the top 10 differentially expressed genes unique to dystrophic MuSCs.
(D) Table for the canonical pathways relevant for the 145 genes unique to dystrophic MuSCs.
(E and F) qRT-PCR validation in MuSCs (E) and FAPs (F) of the genes differentially expressed uniquely in dystrophic mice.
****p < 0.0001; **p < 0.01; *p < 0.05 unpaired t test). n = 3−7 animals.
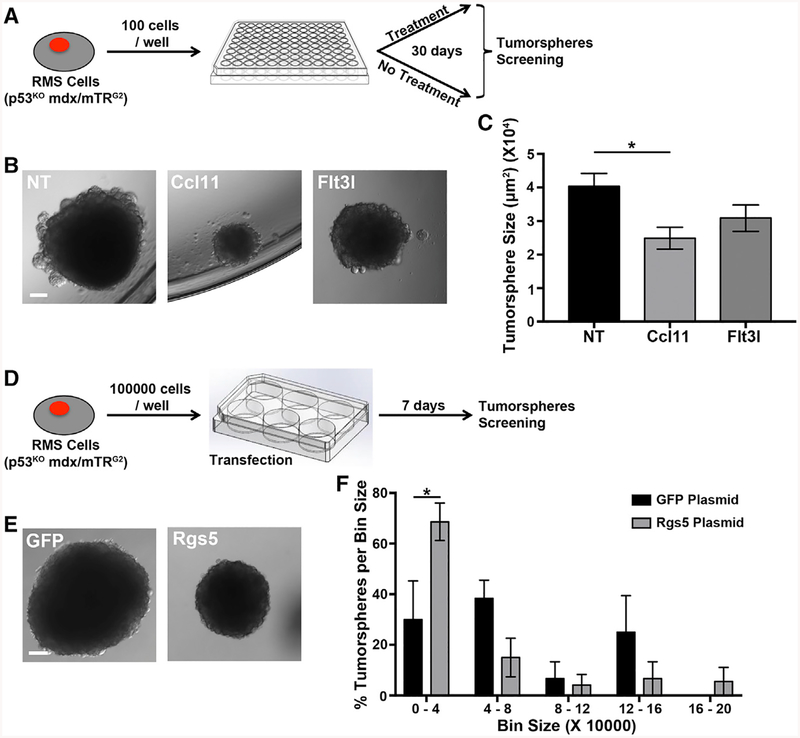
RMS Cells Treated with Ccl11 and Rgs5 Develop Smaller Tumorspheres than Controls
To determine if any of the identified genes could be involved in RMS development, we employed RMS cells from p53KO mdx/mTRG2 mouse tumors and either treated them with Ccl11 or Flt3l recombinant proteins or transfected them with Rgs5 plasmid (Figures 6A and 6D). After treating the cells once per week for 30 days with either recombinant protein, we observed a significant reduction in tumorsphere size in the samples exposed to Ccl11 (Figures 6B and 6C). Moreover, we observed a trend in reduction of tumorspheres frequency (from 1% in non-treated cells to 0.8% in Ccl11-treated cells). No difference was observed in tumorsphere size or number when cells were treated with Flt3l, even though tested downstream targets (Cdkn1a and Soc3) were upregulated in response to treatment (Figures 6B, 6C, and S6A). In parallel, RMS cells were transfected with Rgs5 plasmid and then placed in suspension culture for 7 days in order to allow for tumorsphere formation (Figure 6D). All of the tumorspheres that formed in the control plasmid showed GFP expression (Figure S6B). Tumorspheres that developed in cells transfected with the Rgs5 plasmid were visibly reduced in size, indeed showing a significant increase in the number of smaller-sized spheres when compared to controls (Figures 6A, 6F, and S6C).
Figure 6. Tumor Cells Treated with Ccl11 Recombinant Protein or Transfected with Rgs5 Plasmid Produce Smaller Tumorspheres.
(A) Experimental design for (B) and (C).
(B) Images of tumorspheres that originated from RMS cells (p53KO mdx/mTRG2) in control conditions (NT) and in response to treatment with Ccl11 or Flt3l recombinant proteins. Scale bar, 500 μm.
(C) Quantification of tumorspheres size. *p < 0.05 (one-way ANOVA). n = 41−50 tumorspheres from three biological replicates.
(D) Experimental design for (E) and (F).
(E) Images of tumorspheres from RMS cells (p53KOmdx/mTRG2) transfected with either a GFP plasmid (left) or an Rgs5 plasmid (right). Scale bar, 500 μm.
(F) Distribution of tumorsphere size for RMS cells (p53KO mdx/mTRG2).
* p < 0.05 (one-way ANOVA). n = 3 biological replicates for the experiment.
Taken together, these results show Ccl11 and Rgs5 downregulation observed in p53KO mdx/mTRG2 MuSCs contributes to the process of tumor growth.
DISCUSSION
Our findings demonstrate that DMD disease severity reduces RMS latency, indicating that the increased degenerative environment promotes the development of RMS. Moreover, the high degenerative environment increases tissue turnover and places a selective pressure on the MuSC compartment, leading to cellular transformation. Indeed, p53KO mdx/mTRG2 MuSCs isolated before tumor development already present both phenotypic and molecular features of RMS cells, and they are capable of forming tumorspheres in vitro and giving rise to RMS tumors in vivo when injected into immunodeficient mice.
The mouse model employed in this study is a triple germline knockout (KO); thus, the phenotype we describe is caused by the synergistic effect of three genetic modifications. p53, dystrophin, and telomerase have all been implicated in cancer development (Blasco et al., 1997; Donehower et al., 1992; Jacks et al., 1994; Wang et al., 2014). p53 is a well-known tumor suppressor gene that is activated in response to stress signals, such as DNA damage, and starts a specific transcriptional program to maintain genomic stability (Charni et al., 2017). Dystrophin has been recently identified as a tumor suppressor protein in cancers of mesenchymal origin and has a role in modulating tumor invasiveness (Wang et al., 2014). The prolonged absence of telomerase expression causes increased tumor susceptibility due to DNA damage accumulation (Blasco et al., 1997; Rudolph et al., 1999).
The type of tumors that developed from mice carrying single genetic deletions of these three genes is heterogeneous, as p53KO mice mainly develop lymphomas and sarcomas, mTRKO mice develop carcinomas and lymphomas, and dystrophic mice exhibit RMSs formation (Donehower et al., 1992; Fernandez et al., 2010; Jacks et al., 1994; Rudolph et al., 1999; Xiong et al., 2007). p53KO mice crossed to mTRKO mice mainly develop lymphomas and carcinomas, while p53KO mdx mice only develop RMSs (Artandi et al., 2000; Camboni et al., 2012). Lack of expression of dystrophin causes the development of DMD and thus continuous activation of the muscle regenerative program. In our study, spontaneous tumor development occurred in 10% mdx/mTR mice at ~9 months of age (Het to G2), consistent with the low incidence of tumor formation in mdx mice, but with an earlier age of onset, suggesting involvement of disease severity and increased tissue turnover in tumor formation (Chamberlain et al., 2007; Chang et al., 2016; Fernandez et al., 2010; Latella et al., 2017; Sacco et al., 2010). In our study, we employed mice in a C57BL/6 background and compared their tumor phenotype to both C57BL/10 and C57BL/6 × C57BL/10 mixed-background mice utilized in previous studies (Camboni et al., 2012; Fernandez et al., 2010). While we cannot rule out the possibility that genetic background impacts RMS formation, our results show that an increase in DMD severity both increases RMS formation and reduces latency.
DMD has been associated with increased genetic instability in whole muscle and with accumulation of DNA damage in MuSCs, leading to the hypothesis that these changes could have a significant impact on RMS development in DMD mice (Latella et al., 2017; Schmidt et al., 2011; Tichy et al., 2017). Compared to other dystrophic mouse models, p53KO mdx/mTRG2 mice show a higher accumulation of DNA damage in vivo, downregulation of DNA repair pathways in MuSCs, and a concomitant increase in regeneration, suggesting a potential effect of these changes on the more rapid and aggressive RMS development in these mice. Indeed, most common cancers occur in high-turnover tissues, such as intestine, skin, and prostate (https://www.cancer.gov/about-cancer/understanding/statistics).
Tumor formation and growth are mediated by a population of cancer stem cell-like cells; thus, their characterization and identification are of fundamental importance in the design of targeted therapies (Tysnes and Bjerkvig, 2007). It has been shown that for multiple cancer types, increased self-renewal within the cancer-initiating population drives tumor development (Pardal et al., 2003). Multiple studies in the zebrafish RMS model and RMS cell lines identified, also in this context, self-renewal and impairment in differentiation as the cellular causes leading to expansion of TPCs and tumor growth. Sonic hedgehog (SHH), Notch, and both canonical and non-canonical Wnt signaling have been implicated in these cellular mechanisms (Chen et al., 2014; Hayes et al., 2018; Ignatius et al., 2017; Morotti et al., 2006; Satheesha et al., 2016). Consistent with what has been previously reported, here we show that MuSCs in dystrophic mice have increased self-renewal capacity and decreased differentiation ability, suggesting that these cellular mechanisms could be involved in transformation and RMS development.
Analysis of transcriptional profiling of p53KO mdx/mTRG2 MuSCs and further in vitro validation allowed us to identify two genes associated with RMS growth. Ccl11 (C-C motif chemokine ligand 11) is a chemokine expressed in multiple cell types, and it is required for recruitment of immune cells in response to inflammatory signals (Kindstedt et al., 2017). Rgs5 (regulator of G protein signaling 5) is expressed in multiple organs, including skeletal muscle, and its overexpression has been involved the inhibition of the Sonic hedgehog signaling pathway in multiple cellular compartments (Liu et al., 2017; Mahoney et al., 2013). Moreover, as previously discussed, inflammation, changes in immune response, and overexpression of Sonic hedgehog are causative factors for cancer development (Coussens and Werb, 2002; Grivennikov et al., 2010; Hanna and Shevde, 2016). Inthe context of DMD, decreased expression of Ccl11could impair the recruitment of immune cells in the muscle and deregulate the temporal coordination of the inflammatory response and tissue repair that contributes to DNA damage accumulation within the MuSCs compartment. Concurrent changes in Sonic hedgehog regulatory genes, such as Rgs5, could further contribute to cellular transformation by boosting MuSCs self-renewal capacity and decreasing differentiation ability, DNA damage, and consequent cellular transformation. Here, we show that rescued expression of these genes in RMS cells induces a reduction in tumorsphere size, indicating their involvement in RMS development.
In conclusion, we demonstrate that in DMD, RMS can originate from MuSCs and that accelerated tumor onset is dependent on dystrophic disease severity. Moreover, we identified two genes, Ccl11 and Rgs5, that, when downregulated, favor the process of RMS growth. Interestingly, in DMD, MuSCs progressively lose their ability to repair degenerating muscle, and thus, multiple studies have focused on finding ways to extend the self-renewal capacity of this cell population (Dumont and Rudnicki, 2016; Sacco et al., 2010). In order to ensure effective and long-lasting development of novel treatments targeting MuSCs in DMD, our findings highlight the need to assess the health-span and genetic stability of this cell population.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alessandra Sacco, Ph.D. (asacco@sbpdiscovery.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All protocols were approved by the Sanford Burnham Prebys Medical Discovery Institute Animal Care and Use Committee. Mice were housed according to institutional guidelines, in a controlled environment at a temperature of 22°C ± 1°C, under a 12-h dark-light period and provided with standard chow diet and water ad libitum. Male and female p53WT mdx/mTRHet/G1/G2, p53Het mdx/mTRHet/G1/G2, p53KO mdx/mTR Het/G1/G2, p53WT, p53Het, and p53KO (between 1 and 25-month-old) were used. Specifically, male and female mice were used for evaluation of tumor frequency, age of tumor occurrence and generation of survival curve. Analysis of muscle tissues and MuSCs and FAPs isolation were performed on 2 to 3 months old male mice. All mice were maintained in C57BL/6J background. NOD/SCID mice used were all males between 2 and 3 months of age at the beginning of the experiment.
Cultured cells
All cells were cultured in incubators at 37°C and 5% CO2. Freshly isolated MuSCs, derived from 2 to 3 months old male mice, were plated on tissue culture plates coated with laminin and maintained in growth media (45% DMEM, 40% F10, 15% FBS, and 1:10 dilution of 25 mg/mL β -FGF). Terminal myogenic differentiation was induced with DMEM and 2% horse serum for three days. Clonal analysis was performed on 50% growing media and 50% conditioned media. Tumorsphere assay was performed in tumorsphere media (DMEM/F12, 1% N2, 10 ng/mL EGF, and 10 ng/mL β-FGF). RMS cells were maintained in growth media (DMEM High Glucose and 10% FBS). Primary tumor cells were isolated from 2 mice: a 3.9 months old p53KO mdx/mTRHet and a 3.3 months old male p53KO mdx/mTRG2 mouse (the first developed a single tumor in the forelimb, while the second 2 tumors, on the hind limb and on the forelimb). Cells were plated either in adhesion (DMEM High Glucose and 20% FBS) or in suspension culture (employing tumorspheres media) depending on the performed assay.
METHOD DETAILS
Animals
All protocols were approved by Sanford Burnham Prebys Medical Discovery Institute (SBPMDI) Animal Care and Use Committee. Heterozygous B6.129S2-Trp53tm1Tyj/J were purchased from Jackson Laboratories and used to generate the p53 colony. The mdx/mTR line was kindly donated by H. M. Blau (Sacco et al., 2010), and crossed with p53 colony, generating the p53 mdx/mTR colony. Maintenance of the p53 colony was done using heterozygous breeders. p53 mdx/mTR colony was originated by crossing a p53 heterozygous (p53Het) male mouse to an mdx/mTR heterozygous (mdx/mTRHet) female. The resulting p53Het/mdx/mTRHet male progeny was backcrossed the mdx/mTRHet female. p53Het/mdx/mTRHet progeny was then used to generate 1st breeding generation (identified as G1) and p53Het/mdx/mTRG1 progeny originated the 2nd generation breeding (identified as G1). The line was maintained with three concurrently existing generations of breeding. The generated colonies were maintained at the SBPMDI Animal Facility. The mice used for the present study range from 1 to 26 months of age. Nod.scid mice were purchase from the Sanford Burnham Prebys Medical Discovery Institute Animal Facility.
Skeletal muscle injury
Muscle injury was performed in tibialis anterior and extensor digitorium longus. The protocol employed has been previously described in Shea et al., 2010 (Shea et al., 2010), with minor revisions. Mice were anesthetized with isofluorane and injected in 10 sites with 50 mL of barium chloride (catalog number: 202738, Sigma) resuspended in PBS (1.2% w/v). MuSCs were isolated 3 days after performing the injury.
Cells Isolation
Muscle stem cells (MuSCs) were isolated as describes in Gromova et al. (2015) with minor revisions (Gromova et al., 2015; Tierney and Sacco, 2017). For dystrophic and uninjured muscles we started the isolation process from tibialis anterior, gastrocnemius and quadriceps, whereas for injured muscle we employed only injured tibialis anterior. Muscles were minced and sequentially incubated for 1 hour and 30 minutes in 700 units/ml collagenase type II solution (catalog number: 17101–015, Life technologies, GIBCO®) and collagenase and dispase II (catalog number: 04942078001, Roche) solution (100 units/mL and 2 units/mL respectively) for 30 more minutes. Muscle tissues were then passed through a 10 mL syringe with 20 G needle for 10 times and further filtered with a 70 μm nylon filter. Antibodies incubation was performed in a 1 mL volume. First incubation was performed using biotin labeled cells (CD45+, CD11b+, CD31+, Sca1+ cells) to identify non-myogenic cells that were then depleted using streptavidin beads (catalog number: 130-048-101, Miltenyi Biotec) through magnetic field. The second incubation was performed using CD34+ and integrin α−7 antibody to label the population of interest. MuSCs were isolated with BD Biosciences FACSAria II cell sorter as CD45−, CD11b−, CD31−, Sca1−, CD34+ and integrin α −7+ population.
Fibro/adipogenic progenitors (FAPs) and Lineage positive cells (Lin+) were isolated using minor modifications of the previous protocol. No depletion for biotinilated cells was performed and FAPs were identified as Sca1+, CD34+ cells, while Lin+ cells express CD45+, CD11b+, CD31+.
Flow cytometry data were analyzed using FlowJo
RMS cells were isolated from tumors developed in p53KO mdx/mTRG2 mice using the same digestion protocol as MuSCs. After digestion RMS cells were filtered on 70 μm nylon filters, and plated. No markers were used for identification of RMS cells population.
MuSCs Culture and Differentiation
After isolation MuSCs were plated on Laminin (catalog number: 11243217001, Roche, 1:4 dilution in PBS) in growing media (45% DMEM (catalog number: 11885–092, Invitrogen), 40% Ham’s F-10 (catalog number: 11550–043, Life technologies, GIBCO®), 15% FBS (catalog number: FB-11, Omega Scientific), 1% Pen/Strep (catalog number: 15140163, Life technologies, GIBCO®), 1:10 dilution of 25 μg/mL β-FGF (catalog number: 100–18B, Peprotech)). After 6 hours MuSCs were either fixed, or induced to differentiate in serum free media (DMEM, 2% Hourse serum (catalog number: 16050114, Life technologies, GIBCO®),1% Pen/Strep) for 3 days. In each condition cells were further processes for immunostaining analysis.
RMS Cells Treatment
RMS cells (p53KO mdx/mTRG2) were grown in DMEM high glucose (catalog number: 11965092, Life technologies, GIBCO®), 20% FBS, and 1% Pen/Strep. Before treatment the cells were plated, 100 cells per well, in 96 wells plates and treated either with 100 ng/mL of Ccl11 recombinant protein (catalog number: 420-ME-020, R&D Systems) or with 50 ng/mL of Flt3l recombinant protein (catalog number: 427-FL-005, R&D Systems). Treatement with recombinant proteins was repeated once a week until the 30 day end point of the experiment. RMS cells (p53WT mdx/mTRG2 and p53KO mdx/mTRG2) were plated, 200000 cells per well, in 6 wells plates and transfected using Lipofectamine 3000 with either GFP plasmid or Rgs5 plasmid (catalog number: MR201631, Origene) (Cormack et al., 1996). Transfection was performed following manufacturer instructions on adherent cells. Two days after transfection, the cells were enzymatically detached from the plate and placed in suspension conditions for 7 days (100,000 cells per well).
Clonal Analysis
MuSCs were plated in laminin coated 96-well plates (1 cell per well) through BD Biosciences FACSAria II cell sorter. The assay was performed in 50% growing media and 50% conditioned media (growing media collected and filtered from plates cultured with proliferating myoblasts for 24 hours). Cells were grown for 10 days. A clone is considered more than 1 cell per well. Assessment of clonogenicity was performed as percentage of wells containing clones over the total number of wells.
Tumorsphere Assay
MuSCs, FAPS and Lin+ primary cells were plated in low attachment 96 well plates (100 cells per well) through BD Biosciences FACSAria II cell sorter. The sphere-forming assay was performed in DMEM/F12 (catalog number: 11320033, GIBCO) supplemented with 1% N2 supplement (catalog number: 17502048, GIBCO), 10 ng/mL EGF (catalog number: PMG8041, GIBCO), 10 ng/mL β-FGF (Shaheen et al., 2016). Cells were grown for 30 days and the media was supplemented once per week. Discrimination of tumorspheres and cellular aggregates was performed utilizing morphological parameters: tumorspheres were identified as spherical solid formation where individual cells are not distinguishable; cellular aggregates contained easily distinguishable cells.
Allografts
Tumorspheres, MuSCs and control RMS cells were both injected subcutaneously on the side of 2 months old nod.scid mice. Allorgrafts were performed diluting the selected number of cells first in 50 μL of PBS and then in 50 μL of Matrigel (catalog number: CB40234, Corning), and injecting a total of 100 μL of cells containing solution. Tumorshperes obtained by each mouse where pooled together, digested in Accutase (catalog number: A1110501, GIBCO) until a single cells solution was obtained and then counted before injection. Allografts were performed with 20,000 cells from tumorspheres and with either 50,000 or 5,000,000 RMS cells.
Immunofluorescence
Muscle tissues were isolated from healthy and injured mice at different time points. Tissues were embedded in OCT (catalog number: 4583, VWR) and frozen in 2-methyl butane. Tissues were sectioned in 10 μm thick slices and further processed through immunostaining. Fixation was performed either with 4% PFA (catalog number: 43368, Alfa Aesar) or with 50% Acetone/50% Methanol according to the primary antibody. Tissue sections were washed in PBS and then blocked and permeabilized in blocking buffer (0.3% Triton 100-X (catalog number: H5142, Promega) and 20% Goat Serum (catalog number: 16210–072, Life technologies, GIBCO®) in PBS). For Pax7, myogenin, and Ki67 staining antigen retrieval was performed. Incubation with the primary antibodies was conducted at room temperature overnight in blocking buffer.
Isolated cells were fixed in 4% PFA, washed in PBS and incubated 1 hour in blocking buffer. Primary antibody incubation was performed at room temperature overnight in 4% BSA (catalog number: SH30574.02, HyClone).
The antibodies used are the following: mous anti-Pax7 (catalog number: Pax7-c, Developmental Studies Hybridoma Bank (DSHB), 1:100 dilution), rabbit anti-laminin (catalog number: L9393, Sigma, 1:100 dilution), rat anti-laminin (catalog number: 05–206, Millipore, 1:100 dilution) mouse anti-myosin (embryonic) (catalog number: F1.652, DSHB, 1:100 dilution), rabbit anti-MyoD (catalog number: sc-760, Santa Cruz, 1:100 dilution), mouse anti-myogenin (catalog number: 556358, BD Biosciences; 1:100 dilution), mouse anti-desmin (D33) (catalog number: MS376S1, Thermo Scientific, 1:100 dilution) mouse anti-MHC (catalog number: Mf20-c, DSHB,1:50 dilution), rabbit anti-p-histone H2A.X (S139) (30E3) (catalog number: 9718S, Cell Signaling, 1:100 dilution), rabbit anti-53BP1 (catalog number: NB100–305, Novus Biologicals, 1:100 dilution), Alexa-conjugated secondary antibodies (Invitrogen, 1:250 dilution). Nuclear DNA was stained with Hoechst 33342 (Catalog number: H3570, Invitrogen).
Images were acquired with Inverted IX81 Olympus Compound Fluorescence Microscope, XYZ Automated stage - ASI 2000 (Applied Scientific Instrumentation Inc.), with Color/monochrome cooled CCD camera - Spot RT3 and MetaMorph 7.11 Software (UIC, Molecular Devices) at 10× or 20× magnification or using confocal scanning through Leica TCS SP8 and LAS × software at 20× magnification.
All images were edited and modified through Photoshop CS4 (Adobe).
RNA Isolation and Quantitative PCR
Total RNA was isolated with RNeasy Micro Kit (catalog number: 74004, QIAGEN) following the manufacturer instruction. RNA quantification was performed with Qubit RNA HS Assay Kit (catalog number: Q32852, Invitrogen). The RNA samples for RNA Sequencing analysis were aliquoted and further processed by SBPMDI Genomic Facility. The samples for qPCR analysis were further converted into cDNA with SuperScript® VILO cDNA Synthesis Kit and Master Mix (catalog number: 11754050, Invitrogen) following manufacturer instructions. Real time PCR was performed on LightCycler® 96 System (Roche) with Power SYBR® Green PCR Master Mix (catalog number: 4367659, Applied Biosystems), primers concentration and 0.5 ng of cDNA. Relative gene expression was calculated dividing the Ct value of each gene by the Ct value of the control (Large Ribosomal Protein, Rplpo). The used primers are listed in Table S5.
RNA Sequencing
PolyA RNA was isolated using the NEBNext® Poly(A) mRNA Magnetic Isolation Module and barcoded libraries were made using the NEBNext® Ultra II Directional RNA Library Prep Kit for Illuminaâ (NEB, Ipswich MA). Libraries were pooled and single end sequenced (1×75) on the Illumina NextSeq 500 using the High output V2 kit (Illumina Inc., San Diego CA).
Read data was processed in BaseSpace (https://basespace.illumina.com). Reads were aligned to to Mus musculus genome (mm10) using STAR aligner (https://code.google.com/p/rna-star/) with default settings. Differential transcript expression was determined using the Cufflinks Cuffdiff package (Trapnell et al., 2010).
Differentially expressed genes were further analyzed using three different approaches: Ingenuity Pathway Analysis (IPA), Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005), and Correlation Engine. Data were imported and analyzed in IPA using these parameter cut-offs: FPKM > 1; LogFoldChange > 2; p value < 0.01.
Quantification of Muscle Tissues
Cell numbers in vitro and in vivo were obtained through manual quantification utilizing Adobe Photoshop CS4 as a tool. Images were not modified before count was performed. eMyHC areas were manually quantified employing the measuring tool available in ImageJ64 and normalized to the total muscle area (Schneider et al., 2012). Cross-sectional area quantification was performed in automated manner using and internally developed Macro through ImageJ64.
STATISTICAL ANALYSIS
Error bars in the figures represent standard error of the mean and number of experiments is indicated by n in figure legends. n indicates animals employed for the experiment, clones quantified, tumorspheres, or times an experiment was performed. Specifics are indicated in the figure legends. Statistical significance (two-tails) was tested with Student’s T-Test for two groups comparison (qRT-PCR), and by either Fisher exact test for multiple groups with different size or one-way ANOVA for multiple groups with similar size. Tukey’s multiple comparison test was employed for post hoc analysis (mice life-span, myofibers CSA), Mantel-Cox test for survival curves.
All the statistical analysis was performed with Prism 7 program (GraphPad).
DATA AND SOFTWARE AVAILABILITY
Data were deposited on GEO (accession number: GSE123423).
Previously published datasets utilized in this manuscript are available in GEO (accession number: GSE108022).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-CD45 | BD Biosciences | Cat# 553078;RRID:AB_394608 |
| Rat monoclonal anti-CD11b | BC Biosciences | Cat# 553309;RRID:AB_394773 |
| Rat monoclonal anti-CD31 | eBioscience | Cat# 5011513 |
| Rat monoclonal anti-Sca1 | BD Biosciences | Cat# 553334;RRID:AB_394790 |
| Rat monoclonal anti-α7-integrin, phycoerythrin conjugated | Ablab | Cat# 67-0010-05 |
| Rat monoclonal anti-CD34, Alexa 647 conjugated | BD Biosciences | Cat# 560230;RRID:AB_1645200 |
| Streptavidin APC-Cy7 | BD Biosciences | Cat# 554063;RRID:AB_10054651 |
| Rabbit polyclonal anti-Laminin A1 | Sigma-Aldrich | Cat# L9393;RRID:AB_477163 |
| Rat monoclonal anti-Laminin B2 | Millipore | Cat# 05–206;RRID:AB_309655 |
| Mouse monoclonal anti-Myogenin | BD Biosciences | Cat# 556358;RRID:AB_396383 |
| Mouse monoclonal anti-Pax7 | Developmental Studies Hybridoma Bank | Cat# Pax7-c;RRID:AB_528428 |
| Mouse monoclonal anti-embryonic myosin heavy chain | Developmental Studies Hybridoma Bank | Cat# F1.652;RRID:AB_528358 |
| Rabbit polyclonal anti-Myod1 | Santa Cruz | Cat# sc-760;RRID:AB_2148870 |
| Mouse monoclonal anti-myosin heavy chain | Developmental Studies Hybridoma Bank | Cat# Mf20-c;RRID:AB_2147781 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat# ab15580;RRID:AB_443209 |
| Rabbit monoclonal anti-phospho-histone H2A.X | Cell Signaling | Cat# 9718S;RRID:AB_2118009 |
| Rabbit polyclonal anti-53BP1 | Novus Biologicals | Cat# NB100–305;RRID: AB_10001695 |
| Mouse monoclonal anti-desmin | Thermo Fisher Scientific | Cat# MS376S1;RRID:AB_61166 |
| Alexa fluor 488 goat anti-mouse IgG | Thermo Fisher Scientific | Cat# A11001;RRID:AB_2534069 |
| Alexa fluor 546 goat anti-rabbit IgG | Molecular Probes | Cat# A11035;RRID:AB_143051 |
| Alexa fluor 647 goat anti-rat IgG | Molecular Probes | Cat# A21247;RRID:AB_141778 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM Media | GIBCO | Cat# 11885092 |
| DMEM/F12 Media | GIBCO | Cat# 11320033 |
| DMEM High Glucose Media | GIBCO | Cat# 11965092 |
| Ham’s F10 Media | Life Technologies | Cat# 11550043 |
| Penicillin - Streptomycin | Life Technologies | Cat# 15140163 |
| Fetal Bovine Serum | Omega Scientific | Cat# FB-11 |
| Goat Serum | Life Technologies | Cat# 16210072 |
| Horse Serum | Life Technologies | Cat# 16050114 |
| Recombinant human βFGF-basic | Peprotech | Cat# 10018B |
| EGF recombinant mouse protein | GIBCO | Cat# PMG8041 |
| N-2 Supplements | GIBCO | Cat# 17502048 |
| Recombinant mouse Flt-3 Ligand Protein | R&D Systems | Cat# 427-FL-005 |
| Recombinant mouse CCL11/Eotaxin Protein | R&D Systems | Cat# 410-ME-020 |
| Accutase cell dissociation reagent | GIBCO | Cat# A1110501 |
| Matrigel membrane matrix | Corning | Cat# CB40234 |
| Collagenase, Type II | Life Technologies | Cat# 17101015 |
| Dispase II, protease | Life Technologies | Cat# 17105041 |
| Streptavidin microbeads | Miltenyi | Cat# 130-048-101 |
| Laminin from mouse EHS sarcoma | Roche | Cat# 11243217001 |
| Triton 100-X | Promega | Cat# H5142 |
| Hoechst 33342 | Invitrogen | Cat# H3570 |
| SYBR Green PCR Master Mix | Life Technologies | Cat# 4367659 |
| Bovine Serum Albumin | GE Healthcare Life Sciences | Cat# SH3057402 |
| OCT compound | VWR | Cat# 4583 |
| Paraformaldehyde 16% w/v aqueous solution | Alfa Aesar | Cat# 43368 |
| Barium chloride | Sigma-Aldrich | Cat# 202738 |
| PBS | GIBCO | Cat# 10010023 |
| Fluriso (Isofluorane) anesthetic agent | MWI Vet Supply | Cat# 502017 |
| Critical Commercial Assays | ||
| Lipofectamine 3000 transfection reagent | Thermo Fisher | Cat# L3000015 |
| miRNeasy Micro Kit | QIAGEN | Cat# 217084 |
| Qubit RNA HS Assay Kit | Invitrogen | Cat# Q32852 |
| SuperScript VILO cDNA Synthesis Kit | Invitrogen | Cat# 11754050 |
| NEBNext® Poly(A) mRNA Magnetic Isolation Module | New England Biolabs | Cat# E7490 |
| NEBNext® Ultra II Directional RNA Library Prep Kit for Illumina® | New England Biolabs | Cat# E7760 |
| Deposited Data | ||
| RNaseq data (raw and processed) | This paper | GEO: GSE123423 |
| Experimental Models: Organisms/Strains | ||
| Mouse: p53 mdx/mTR | This paper | N/A |
| Mouse: B6.129S2-Trp53tm1Tyj/J | The Jackson Laboratory | RRID:IMSR_JAX:002101 |
| Mouse: mdx/mTR | Sacco et al., 2010 | NA |
| Mouse: NOD.CD17-Prkdc < scid >/J | The Jackson Laboratory | RRID:IMSR_JAX:001303 |
| Oligonucleotides | ||
| pEGFP-C1 | Cormack et al., 1996 | Addgene 6084–1 |
| Rgs5 Mouse Tagged ORF Clone | Origene | Cat# NM_009063 |
| Primers for qRT-PCR, see Table S5 | This paper | N/A |
| Software and Algorithms | ||
| BeadSpace | Illumina | https://login.illumina.com/platform-services-manager/ |
| STAR aligner | https://code.google.com/p/rna-star | |
| Cufflinks Cuffdiff package | Trapnell et al., 2012 | https://github.com/cole-trapnell-lab/cufflinks |
| Ingenuity Pathway Analysis (IPA) | QIAGEN | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| Correlation Engine | Illumina | https://www.illumina.com/products/by-type/informatics-products/basespace-correlation-engine.html |
| Gene Set Enrichment Analysis (GSEA) | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| Adobe Photoshop CS4 | Adobe | https://www.adobe.com/es/products/photoshop.html |
| MetaMorph 7.11 Software | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| ImageJ | NIH | https://imagej.nih.gov/ij/download.html |
| FlowJo | FlowJo | https://www.flowjo.com |
| Graphpad - Prism 7 | GraphPad | https://www.graphpad.com |
| Others | ||
| 20G needles | BD | Cat# 305175 |
| 70 um nylon filter | Fisher Scientific | Cat# 22363548 |
| Gene expression in human rhabdomyosarcoma | Shern et al., 2014; Hayes etal., 2018 | GEO: GSE108022 |
Highlights.
Dystrophic disease progression accelerates rhabdomyosarcoma (RMS) formation
MuSCs carry a gene signature characteristic of RMSs before tumor formation
Increased self-renewal contributes to dystrophic MuSCs transformation
Dystrophic MuSCs induce the development of RMS in vivo
ACKNOWLEDGMENTS
We thank the following people at the SBP Core Facilities for technical support: B. Charbono, T. Omel, D. Sandoval, and A. Vasquez from the Animal Facility; A. Cortez and Y. Altman from the Flow Cytometry Core Facility; L. Boyd from the Cell Imaging Facility; and B. James from the Genomics Core Facility. This work was supported by the Ellison Medical Foundation (grant AG-NS-0843–11), the Glenn Foundation for Medical Research, the NIH (grant P30 AR061303), the NCI (Pilot Grant within the NCI Cancer Center Support Grant P30CA030199), and SBPMDI start-up funds to A.S.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and five tables can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.089.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Almada AE, and Wagers AJ (2016). Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat. Rev. Mol. Cell Biol 17, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, and DePinho RA (2000). Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406, 641–645. [DOI] [PubMed] [Google Scholar]
- Bahrami AJ, Gunaje JJ, Hayes BJ, Riehle KJ, Kenerson HL, Yeung RS, Stempien-Otero AS, Campbell JS, and Mahoney WM Jr. (2014). Regulator of G-protein signaling-5 is a marker of hepatic stellate cells and expression mediates response to liver injury. PLoS ONE 9, e108505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yehuda S, Barer F, Volfsson L, and Fishman P (2001). Resistance of muscle to tumor metastases: a role for a3 adenosine receptor agonists. Neoplasia 3, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, and Greider CW (1997). Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34. [DOI] [PubMed] [Google Scholar]
- Blum JM, Añó L, Li Z, Van Mater D, Bennett BD, Sachdeva M, Lagutina I, Zhang M, Mito JK, Dodd LG, et al. (2013). Distinct and overlapping sarcoma subtypes initiated from muscle stem and progenitor cells. Cell Rep. 5, 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, and Rando TA (2012). Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell 10, 504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bűget MI, Eren i. ,and Kűçűkay S (2014). Regional anaesthesia in a Duchenne muscular dystrophy patient for upper extremity amputation. Agri 26, 191–195. [DOI] [PubMed] [Google Scholar]
- Camboni M, Hammond S, Martin LT, and Martin PT (2012). Induction of a regenerative microenvironment in skeletal muscle is sufficient to induce embryonal rhabdomyosarcoma in p53-deficient mice. J. Pathol 226, 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JS, Metzger J, Reyes M, Townsend D, and Faulkner JA (2007). Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 21, 2195–2204. [DOI] [PubMed] [Google Scholar]
- Chang AC, Ong SG, LaGory EL, Kraft PE, Giaccia AJ, Wu JC, and Blau HM (2016). Telomere shortening and metabolic compromise underlie dystrophic cardiomyopathy. Proc. Natl. Acad. Sci. USA 113, 13120–13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charni M, Aloni-Grinstein R, Molchadsky A, and Rotter V (2017). p53 on the crossroad between regeneration and cancer. Cell Death Differ. 24, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project (2013). Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 24, 710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, DeRan MT, Ignatius MS, Grandinetti KB, Clagg R, McCarthy KM, Lobbardi RM, Brockmann J, Keller C, Wu X, and Langenau DM (2014). Glycogen synthase kinase 3 inhibitors induce the canonical WNT/b-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 111, 5349–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, and Falkow S (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Coussens LM, and Werb Z (2002). Inflammation and cancer. Nature 420, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr., Butel JS, and Bradley A (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221. [DOI] [PubMed] [Google Scholar]
- Drummond CJ, Hanna JA, Garcia MR, Devine DJ, Heyrana AJ, Finkelstein D, Rehg JE, and Hatley ME (2018). Hedgehog pathway drives fusion-negative rhabdomyosarcoma initiated from non-myogenic endothelial progenitors. Cancer Cell 33, 108–124.e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchenne. (1867). The pathology of paralysis with muscular degeneration (Paralysie Myosclerotique), or paralysis with apparent hypertrophy. Br. Med. J 2, 541–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont NA, and Rudnicki MA (2016). Targeting muscle stem cell intrinsic defects to treat Duchenne muscular dystrophy. NPJ Regen. Med 1, 16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanzani A, Monti E, Donato R, and Sorci G (2013). Muscular dystrophies share pathogenetic mechanisms with muscle sarcomas. Trends Mol. Med 19, 546–554. [DOI] [PubMed] [Google Scholar]
- Fernandez K, Serinagaoglu Y, Hammond S, Martin LT, and Martin PT (2010). Mice lacking dystrophin or alpha sarcoglycan spontaneously develop embryonal rhabdomyosarcoma with cancer-associated p53 mutations and alternatively spliced or mutant Mdm2 transcripts. Am. J. Pathol 176, 416–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, and Karin M (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova A, Tierney MT, and Sacco A (2015). FACS-based satellite cell isolation from mouse hind limb muscles. Bio. Protoc 5, e1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2000). The hallmarks of cancer. Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Hanna A, and Shevde LA (2016). Hedgehog signaling: modulation of cancer properies and tumor mircroenvironment. Mol. Cancer 15, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, Graff J, Galindo RL, and Olson EN (2012). A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell 22, 536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MN, McCarthy K, Jin A, Oliveira ML, Iyer S, Garcia SP, Sindiri S, Gryder B, Motala Z, Nielsen GP, et al. (2018). Vangl2/RhoA signaling pathway regulates stem cell self-renewal programs and growth in rhabdomyosarcoma. Cell Stem Cell 22, 414–427.e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH Jr., and Kunkel LM (1987). Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928. [DOI] [PubMed] [Google Scholar]
- Hosur V, Kavirayani A, Riefler J, Carney LM, Lyons B, Gott B, Cox GA, and Shultz LD (2012). Dystrophin and dysferlin double mutant mice: a novel model for rhabdomyosarcoma. Cancer Genet. 205, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Hayes MN, Lobbardi R, Chen EY, McCarthy KM, Sreenivas P, Motala Z, Durbin AD, Molodtsov A, Reeder S, et al. (2017). The NOTCH1/SNAIL1/MEF2C Pathway Regulates Growth and Self-Renewal in Embryonal Rhabdomyosarcoma. Cell Rep. 19, 2304–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, and Weinberg RA (1994). Tumor spectrum analysis in p53-mutant mice. Curr. Biol 4, 1–7. [DOI] [PubMed] [Google Scholar]
- Jakab Z, Szegedi I, Balogh E, Kiss C, and Oláh E (2002). Duchenne muscular dystrophy-rhabdomyosarcoma, ichthyosis vulgaris/acute monoblastic leukemia: association of rare genetic disorders and childhood malignant diseases. Med. Pediatr. Oncol 39, 66–68. [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, and Rossi FM (2010). Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol 12, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Chen H, and Lo PK (2013). In vitro tumorsphere formation assays. Bio. Protoc 3, e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, and Capecchi MR (2004). Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 18, 2614–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindstedt E, Holm CK, Sulniute R, Martinez-Carrasco I, Lundmark R, and Lundberg P (2017). CCL11, a novel mediator of inflammatory bone resorption. Sci. Rep 7, 5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella L, Dall’Agnese A, Boscolo FS, Nardoni C, Cosentino M, Lahm A, Sacco A, and Puri PL (2017). DNA damage signaling mediates the functional antagonism between replicative senescence and terminal muscle differentiation. Genes Dev. 31, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Hu Q, Jing J, Zhang Y, Jin J, Zhang L, Mu L, Liu Y, Sun B, Zhang T, et al. (2017). Regulator of G protein signaling 5 (RGS5) inhibits sonic hedgehog function in mouse cortical neurons. Mol. Cell. Neurosci 83, 65–73. [DOI] [PubMed] [Google Scholar]
- Mahoney WM Jr., Gunaje J, Daum G, Dong XR, and Majesky MW (2013). Regulator of G-protein signaling - 5 (RGS5) is a novel repressor of hedgehog signaling. PLoS ONE 8, e61421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotti RA, Nicol KK, Parham DM, Teot LA, Moore J, Hayes J, Meyer W, and Qualman SJ; Children’s Oncology Group (2006). An immuno-histochemical algorithm to facilitate diagnosis and subtyping of rhabdomyosarcoma: the Children’s Oncology Group experience. Am. J. Surg. Pathol 30, 962–968. [DOI] [PubMed] [Google Scholar]
- Nishijo K, Chen QR, Zhang L, McCleish AT, Rodriguez A, Cho MJ, Prajapati SI, Gelfond JA, Chisholm GB, Michalek JE, et al. (2009). Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer Res. 69, 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, and Morrison SJ (2003). Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer 3, 895–902. [DOI] [PubMed] [Google Scholar]
- Parlakian A, Gomaa I, Solly S, Arandel L, Mahale A, Born G, Marazzi G, and Sassoon D (2010). Skeletal muscle phenotypically converts and selectively inhibits metastatic cells in mice. PLoS ONE 5, e9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Wu Z, Zhang P, Wood LD, Bhakta KS, Han J, Feramisco JR, Karin M, and Wang JY (2000). Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev. 14, 574–584. [PMC free article] [PubMed] [Google Scholar]
- Rees PA, Greaves NS, Baguneid M, and Bayat A (2015). Chemokines in wound healing and as potential therapeutic targets for reducing cutaneous scarring. Adv. Wound Care (New Rochelle) 4, 687–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach HC, Lacson A, Grana NH, and Barbosa JL (1999). Duchenne muscular dystrophy and concomitant metastatic alveolar rhabdomyosarcoma. J. Pediatr. Hematol. Oncol 21, 528–530. [PubMed] [Google Scholar]
- Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, Prajapati SI, Abraham J, Arenkiel BR, Chen QR, et al. (2011). Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell 19, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, and DePinho RA (1999). Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, and Blau HM (2008). Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, and Blau HM (2010). Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha RM, Gasparini JR, Silva LS, de Carli RR, de Castilhos VU, das Neves MM, Araújo FP, Sales PC, and das Neves JF (2005). Anesthesia for Duchenne muscular dystrophy patients: case reports. Rev. Bras. Anestesiol 55, 445–449. [DOI] [PubMed] [Google Scholar]
- Satheesha S, Manzella G, Bovay A, Casanova EA, Bode PK, Belle R, Feuchtgruber S, Jaaks P, Dogan N, Koscielniak E, and Schäfer BW (2016). Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma. Oncogene 35, 2020–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WM, Uddin MH, Dysek S, Moser-Thier K, Pirker C, Höger H, Ambros IM, Ambros PF, Berger W, and Bittner RE (2011). DNA damage, somatic aneuploidy, and malignant sarcoma susceptibility in muscular dystrophies. PLoS Genet. 7, e1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen S, Ahmed M, Lorenzi F, and Nateri AS (2016). Spheroid-formation (colonosphere) assay for in vitro assessment and expansion of stem cells in colon cancer. Stem Cell Rev. 12, 492–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, and Brack AS (2010). Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell 6, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. (2014). Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 4, 216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SI, Freedman VH, Risser R, and Pollack R (1975). Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl. Acad. Sci. USA 72, 4435–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan I, Qaddoumi I, Yaser S, Rodriguez-Galindo C, and Ferrari A (2009). Comparing adult and pediatric rhabdomyosarcoma in the surveillance, epidemiology and end results program, 1973 to 2005: an analysis of 2,600 patients. J. Clin. Oncol 27, 3391–3397. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Thayer MJ, and Weintraub H (1993). Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science 259, 1450–1453. [DOI] [PubMed] [Google Scholar]
- Tichy ED, Sidibe DK, Tierney MT, Stec MJ, Sharifi-Sanjani M, Hosalkar H, Mubarak S, Johnson FB, Sacco A, and Mourkioti F (2017). Single stem cell imaging and analysis reveals telomere length differences in diseased human and mouse skeletal muscles. Stem Cell Reports 9, 1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney MT, and Sacco A (2016). Satellite cell heterogeneity in skeletal muscle homeostasis. Trends Cell Biol. 26, 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney M, and Sacco A (2017). Engraftment of FACS isolated muscle stem cells into injured skeletal muscle. Methods Mol. Biol 1556, 223–236. [DOI] [PubMed] [Google Scholar]
- Tomasetti C, and Vogelstein B (2015). Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay AM, Missiaglia E, Galli GG, Hettmer S, Urcia R, Carrara M, Judson RN, Thway K, Nadal G, Selfe JL, et al. (2014). The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell 26, 273–287. [DOI] [PubMed] [Google Scholar]
- Tysnes BB, and Bjerkvig R (2007). Cancer initiation and progression: involvement of stem cells and the microenvironment. Biochim. Biophys. Acta 1775, 283–297. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, and Tsuchida K (2010). Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol 12, 143–152. [DOI] [PubMed] [Google Scholar]
- Walter D, Satheesha S, Albrecht P, Bornhauser BC, D’Alessandro V, Oesch SM, Rehrauer H, Leuschner I, Koscielniak E, Gengler C, et al. ; CWS Study Group (2011). CD133 positive embryonal rhabdomyosarcoma stem-like cell population is enriched in rhabdospheres. PLoS ONE 6, e19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Marino-Enriquez A, Bennett RR, Zhu M, Shen Y, Eilers G, Lee JC, Henze J, Fletcher BS, Gu Z, et al. (2014). Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat. Genet 46, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiswald LB, Bellet D, and Dangles-Marie V (2015). Spherical cancer models in tumor biology. Neoplasia 17, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Xiao S, Peng F, Zheng H, Yu M, Ruan Y, Li W, Shang Y, Zhao C, Zhou W, et al. (2007). Herpes simplex virus VP22 enhances adenovirus-mediated microdystrophin gene transfer to skeletal muscles in dystrophin-deficient (mdx) mice. Hum. Gene Ther 18, 490–501. [DOI] [PubMed] [Google Scholar]
- Yang Z, MacQuarrie KL, Analau E, Tyler AE, Dilworth FJ, Cao Y, Diede SJ, and Tapscott SJ (2009). MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev. 23, 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, and Rudnicki MA (2013). Satellite cells and the muscle stem cell niche. Physiol. Rev 93, 23–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were deposited on GEO (accession number: GSE123423).
Previously published datasets utilized in this manuscript are available in GEO (accession number: GSE108022).