Abstract
γ-aminobutyric acid receptors (GABAARs) are vital for controlling excitability in the brain. This is emphasized by the numerous neuropsychiatric disorders that result following receptor dysfunction. A critical component of most native GABAARs is the α subunit. Its transmembrane domain is the target for many modulators, including endogenous brain neurosteroids that impact on anxiety, stress and depression, and for therapeutic drugs such as general anaesthetics. To understand the basis for modulating GABAAR function, high-resolution structures are required. Here we present the first atomic structures of a GABAAR chimera at 2.8Å resolution, including those bound with potentiating and inhibitory neurosteroids. These define new allosteric binding sites for these modulators that are associated with the α-subunit transmembrane domain. Our findings will enable neurosteroids to be exploited for therapeutic drug design to regulate GABAARs in neurological disorders.
The GABAAR is a pre-eminent member of the pentameric ligand-gated ion channel (pLGIC) superfamily comprising amongst others, nicotinic acetylcholine (nAChR), glycine (GlyR) and serotonin type-3 (5-HT3R) receptors1, 2. GABAARs possess an anion-selective ion channel that, following agonist activation, enables Cl- flux to shunt and often hyperpolarise the membrane. This reveals their primary task in the brain, which is to inhibit neuronal excitation3 and it is widely acknowledged that dysfunctional GABA signalling results in neurological disorders4, 5.
GABA signalling via type A receptors occurs by a combination of rapid phasic and persistent tonic inhibition. The former requires the activation of synaptic GABAA receptors composed of αβγ subunits that are clustered at inhibitory synapses. The latter involves diffusely-located extrasynaptic αβγ and αβδ subunit-containing GABAARs3, 6. Whilst varying the receptor subunit composition confers distinctive physiological and pharmacological properties7, all GABAARs share core fundamental properties. Both synaptic and extrasynaptic receptor classes desensitize8–10 upon prolonged agonist exposure, and both are modulated by naturally-occurring brain neurosteroids11 that ‘fine-tune’ the time-course and extent of postsynaptic inhibition.
An important and common structural denominator for most GABAARs is the α-subunit. There are six isoforms (α1-6) and of these α1 is the most widely expressed in the brain12. In combination with β2/3 and γ2 subunits this forms the prototypic synaptic GABAAR. The extracellular domain (ECD) of the α-subunit is vital in forming part of the interfacial GABA binding sites between β-α subunits. Notably, structural elements within the α subunit transmembrane domain (TMD) shape GABA channel architecture and also strongly influence the biophysical and pharmacological properties of individual GABAAR subtypes7, 13, including their modulation by neurosteroids14 and general anaesthetics15. Naturally occurring neurosteroids are synthesised in the brain from cholesterol and represent a potent endogenous modulator of GABAergic transmission. These compounds can modulate both synaptic and extrasynaptic receptors11 and are classified on their ability to potentiate or inhibit GABAAR activity. Significantly the GABAAR α-subunit TMD confers sensitivity to neurosteroids such as allopregnanolone and its derivatives14.
To further our understanding of GABAAR function, and how this depends on modulation by allosteric ligands, requires the generation of new structural information. This will also provide insight into the disruptive effects of receptor mutations that are associated with neuropsychiatric diseases16. To date, structural studies have been limited mainly to a reliance on homology modelling and structural comparisons between homologous pLGIC members for which atomic level resolution is available2.
Here, we describe structural details of the GABAAR at atomic resolution based on using a new chimera-based modular construct. We present crystal structures of the GABAAR α1-subunit transmembrane domain alone, and in complex with the two classes of brain neurosteroids: the potentiating stress hormone-derived tetrahydro-deoxycorticosterone (THDOC), and the inhibitory neurosteroid, pregnenolone sulfate (PS). These new receptor structures also allow clear observation of the GABA ion channel for which we can resolve the recently defined desensitization gate that lies deep within the pore10.
Results
Designing a functional GABAA receptor chimera
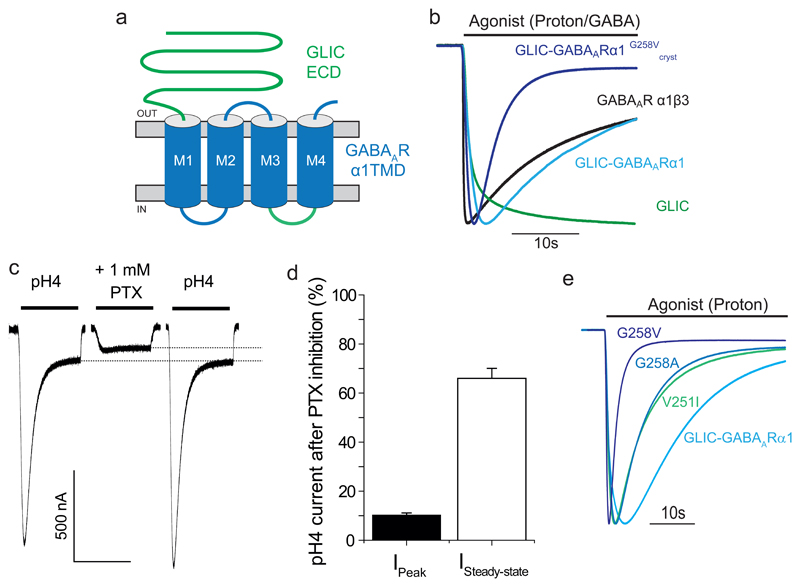
To provide high-resolution X-ray crystallographic structural information for the GABAAR TMD, we developed a new ‘prokaryotic-eukaryotic’ chimera. This involved fusing the ECD of the prokaryotic homolog GLIC from Gloeobacter violaceus17, 18, with the TMD (comprising the α-helices M1-M4 and associated linkers) from the GABAAR α1-subunit (Fig 1a, Supplementary Fig. 1a). GLIC was selected for constructing the chimera since it readily crystallises as a homomer17, 18, with principal crystal contacts contributed by the ECD. Moreover, it forms a functional chimera when fused to the TMD of a similar pLGIC, the GlyR19. Finally, the kinetic profiles of proton-activated currents for wild-type (WT) GLIC are slow and distinct from that for most GABAARs (Fig. 1b). This enables the functional and pharmacological characteristics of the chimeric GABAAR α1-subunit TMD to be readily identified.
Figure 1. Structure and function of the GLIC-GABAARα1 chimera.
(a) Schematic representation of the GLIC-GABAARα1 chimera. The ECD is contributed by the GLIC subunit (green, residues 1-194) and the TMD is from the GABAAR α1 subunit (blue, residues 223-428, excluding the intracellular domain between M3 and M4, which derives from GLIC, green loop). (b) Peak-scaled currents induced by applied (bar) orthosteric agonists for: α1β3 GABAAR (10 mM GABA); wild-type (WT) GLIC (protons - pH 4); and chimera constructs with and without the G258V mutation (proton – pH 4). GLIC-GABAARα1G258Vcryst was used for crystallization experiments. (c) The GABAA channel blocker picrotoxin (PTX; 1 mM) inhibits proton-activated currents (pH 4) in the chimera voltage-clamped at -60 mV. Dotted lines show the extent of steady-state current inhibition. (d) Bar-graph showing current remaining after PTX inhibition of peak and steady-state pH4 currents. Values are means ± sem (n = 4 for both, independent experiments). Note the peak currents are more profoundly inhibited by PTX compared to steady-state currents. (e) Peak-scaled proton-activated (pH 4 – 4.5) currents (VH = -60 mV) for chimeras with gain-of-desensitization mutations in the α1-subunit TMD 10. Increased residue side-chain volume at the -3’ Val and 2’ Gly in M2 increases the rate of desensitization.
Previous studies of pLGICs have revealed that the interfacial loops at the base of the ECD, and the extracellular segments of the TMD, are important for transmitting the process of agonist-binding to ion channel opening1. In the chimera, loops 2, 7 and 9 at the base of the ECD β-sheets, which are important for transmitting the agonist binding signal1, are contributed by GLIC, while the M2-M3 linker of the TMD derives from the GABAAR α1-subunit (Supplementary Fig. 1a). In optimising the chimera for crystallization, the large intracellular M3-M4 domain was replaced with the shorter linker from GLIC, -SQPARAA- (Fig. 1a, Supplementary Fig. 1a).
Functional properties of the GABAA receptor chimera
To ensure that the GLIC-GABAARα1 chimera was functional, receptors were expressed in Xenopus oocytes and assessed using two-electrode voltage clamp (TEVC) electrophysiology. As predicted for a chimera incorporating the ECD from GLIC and the TMD from the GABAAR α1-subunit, it was activated by protons in a concentration-dependent manner, yet formed a Cl- selective ion channel characteristic of GABAA receptors (Supplementary Fig. 1b-d). Accordingly, proton-activated currents were inhibited by the GABA channel blocker picrotoxin (Fig. 1c,d). Furthermore, prolonged proton applications evoked desensitizing Cl- currents, similar to response profiles for α1β3 GABAARs activated by high GABA concentrations, and therefore distinct from the slower response of WT GLIC channels (Fig. 1b). Protons can modulate native GABAARs, but this is critically dependent on a histidine residue in the TMD of the β-subunit 20. This is absent in our chimera and therefore suggests that residues in the ECD determine the sensitivity to protons.
To further validate the chimera as a new model for α1 subunit-containing GABAARs we mutated key residues within the TMD that underpin gating transitions that affect receptor desensitization (Supplementary Fig. 1e). Consistent with results from α1β2 GABAARs10, mutating valine 251 at -3’ in M2 (numbered from a conserved arginine at the base of M2 defined as position 0’) to isoleucine (V251I), or by mutating glycine 258 (4’) to valine or alanine (G258V, A), yielded chimeras exhibiting profound desensitization following proton activation (Fig. 1e). Thus, the characteristics of the chimera TMD are in accord with those expected of the GABA α1-subunit TMD.
Crystal structure of the GABAA receptor chimera
After screening for the expression and purification of various receptor chimeras in Sf9 insect cells, we noted that purification of GLIC-GABAARα1 was markedly improved by including the desensitizing G258V mutation (Supplementary Fig. 2a,b). Significantly, this receptor (GLIC-GABAARα1G258Vcryst) retained its functionality (Fig. 1b) and was thermostable as a pentamer in detergent micelles (Supplementary Fig. 2c,d).
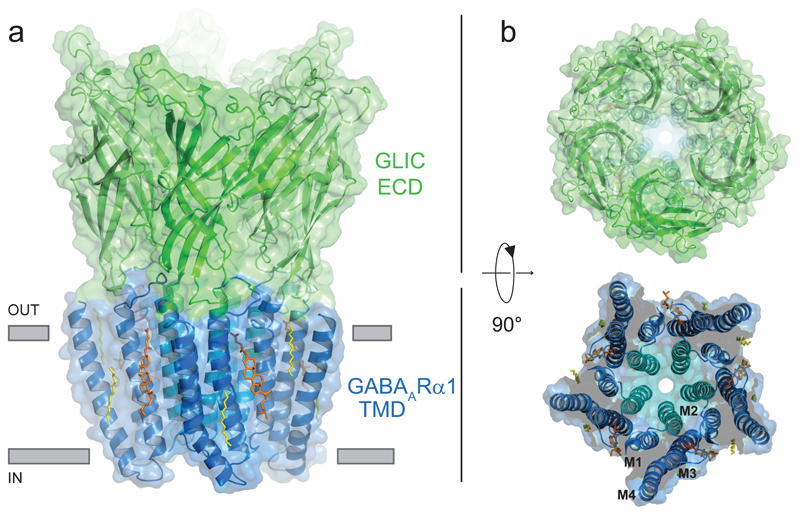
We determined the crystal structure of GLIC-GABAARα1G258Vcryst at 2.8Å resolution under low pH, to capture the receptor in an agonist-bound, desensitized conformation (Supplementary Fig. 3, Table 1). The chimera forms a homo-pentamer with each subunit composed of an ECD, comprising inner and outer layers of β-sheets from GLIC, coupled to four α-helices in the TMD with an integral pore from the α1-subunit (Fig. 2). A continuous solvent accessible pathway (characteristic of all pLGICs) follows a 5-fold symmetry axis through the centre of the ECD and TMD. A positive electrostatic surface potential extending the length of the TMD is conducive to Cl- permeation (Supplementary Fig. 4). The ECD structure is compact21, 22, with bound acetate ions contributed by the crystallisation solution, consistent with the agonist-bound state for WT GLIC23, 24.
Table 1. Data collection and refinement statistics (molecular replacement).
| GLIC-GABAAR alpha1 at pH4.6 (PDB 5OSA) |
GLIC-GABAAR alpha1 in complex with THDOC at pH4.5 (PDB 5OSB) |
GLIC-GABAAR alpha1 in complex with PS at pH4.5 (PDB 5OSC) |
|
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 185.0/ 133.9/ 162.7 | 185.2/133.5/162.3 | 183.3/133.2/162.0 |
| α, β, γ (º) | 90/ 103.5/ 90 | 90/ 103.4/ 90 | 90/ 102.7/ 90 |
| Resolution (Å) | 95.28-2.75 (2.8-2.75)a | 95.03-3.8 (3.97-3.80) | 83.52-2.97 (3.05-2.97) |
| Rmerge | 0.068 (0.772) | 0.184 (1.184) | 0.098 (1.367) |
| Rpim | 0.066 (0.73) | 0.112 (0.698) | 0.093 (1.279) |
| I/σ(I) | 10.3 (1.2) | 4.6 (1.5) | 7.9 (1.0) |
| CC1/2 | 0.946 (0.868) | 0.996 (0.982) | 0.997 (0.546) |
| Completeness (%) | 100 (99.9) | 99.9 (99.8) | 99.4 (99.4) |
| Redundancy | 3.5 (3.5) | 7(7.2) | 3.4 (3.5) |
| Refinement | |||
| Resolution (Å) | 29.98 - 2.75 | 29.93 – 3.8 | 29.92 – 3.1 |
| No. reflections | 99761 | 36915 | 68399 |
| Rwork / Rfree | 20.2/23.1 | 24.6/29.2 | 21.2/24.4 |
| No. atoms | 12802 | 12561 | 1278 |
| Protein | 12457 | 12419 | 12436 |
| Ligand/ion | 310 | 142 | 347 |
| Water | 35 | ||
| B factors | |||
| Protein | 87.02 | 189.95 | 108.11 |
| Ligand/ion | 122.76 | 190.94 | 181.28 |
| Water | 71.18 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.001 | 0.003 |
| Bond angles (º) | 0.94 | 0.44 | 0.51 |
Data for crystal 2 were merged from 2 data sets collected from the same crystal
Values in parentheses are for the highest-resolution shell.
Figure 2. Structure of the GABAA receptor chimera.
Crystal structure of the chimera showing side (a) and plan (b) views with the ECD from GLIC (green) and the TMD of GABAARα1 (blue). M2 helices (cyan) line the ion channel. Cholesteryl hemisuccinate molecules (orange) and the detergent acyl chains (yellow) are bound at the periphery of the TMD and shown in stick form.
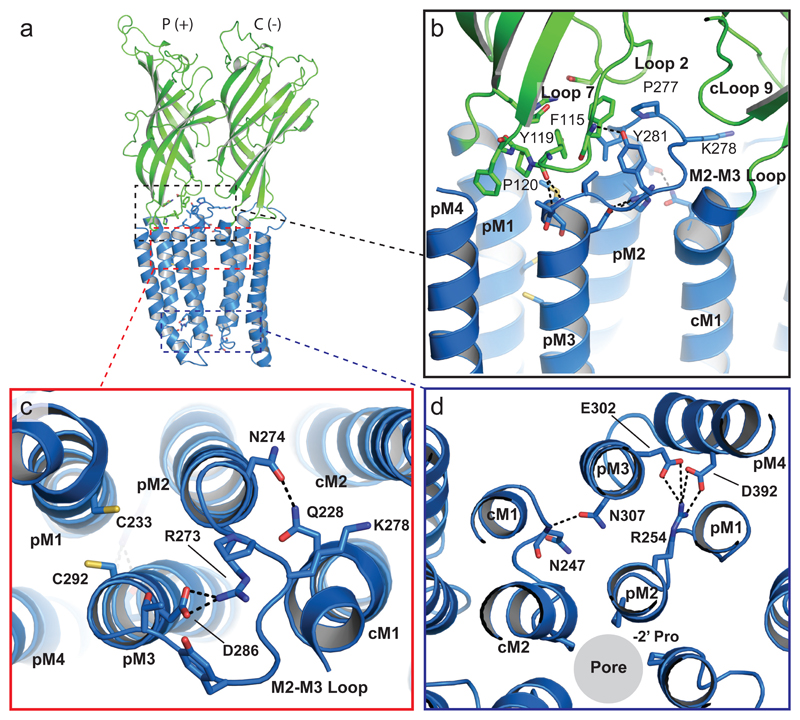
The ECD (Supplementary note 1) is connected to the GABA α1-subunit TMD through extensive interactions between ECD loops 2, 7 and 9 and the M2-M3 linker of the TMD (Fig. 3). Crucially, by adopting a cis conformation, the conserved proline (P120) at the tip of loop 723 enables hydrogen (H)-bonding between the backbone-carbonyl of Y119 and residues at the tip of M3. The hydroxyl group of Y281 (equivalent to Y251 in GLIC) in the M2-M3 linker also forms an H-bond with the backbone amino-group of F115 on loop 7. This interaction further stabilizes domain coupling (Fig 3b). The conserved nature of these ECD-TMD interactions ensures ion channel gating follows agonist binding. It also highlights the critical role of the highly conserved K278 in the M2-M3 linker25 (Fig 3b,c), which is also conserved in GLIC (K248). Previous studies of α1β2γ2 GABAARs reveal a role for this residue in the initiation of channel gating26, where it likely stabilises an open state of the GABAAR27. Furthermore, disrupting these interactions by mutating in α1-subunits, causes familial epilepsies28.
Figure 3. Coupling at the receptor chimera ECD-TMD interface.
(a) Side-view of the receptor showing two subunits forming the principal (p, +) and complementary (c, -) inter-subunit interfaces. The dashed boxes are magnified in panels b-d. (b) Residues that interact at the coupling interface between the ECD and TMD are shown. Identified residues (in stick form) are broadly conserved across GLIC and GABAA receptor subunits, and putative H-bonding is shown by black dashed lines. (c) Residues involved in putative inter-subunit H-bonding and intra-subunit salt-bridge interactions in the upper half of the TMD are shown. The Cys residues (yellow) in M1 and M3 do not form a disulfide bridge. (d) Residues involved in putative inter-subunit H-bonding and intra-subunit salt-bridge interactions in the lower half of the TMD are shown.
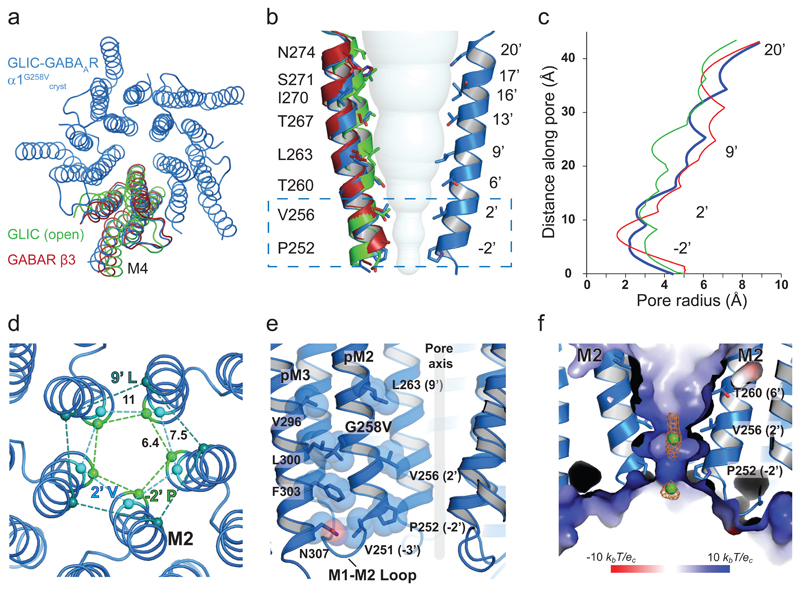
The geometry of the GABAAR α1-subunit pore (Fig. 4) exhibits structural similarities with other pLGICs under non-resting conditions. These include: GLIC at acidic pH in an ‘agonist-bound’ state17, 18; glutamate-activated Cl- channels (GluClcryst) bound to ivermectin29; GABA β3 homomers bound with benzamidine30; and ligand-bound GlyRα131 and GlyRα332. The channel lining M2 α-helices reveal an expanded pore at the extracellular portal (at 20’; Fig. 4a,b) that gradually tapers towards the intracellular exit at -2’, which is characteristic of a desensitized conformation (Fig. 4b,c). This structure is distinct from that for GLIC-GlyRα1, which adopts a locally-closed conformation when crystallized at low pH19 (Supplementary Fig. 5). In addition to M2, both the M1 and M3 α-helices of the GLIC-GABAARα1 chimera also superimposed onto related pLGIC TMD structures. However, the extracellular end of M4 was rotated around a highly conserved proline (P400), compared to M4 in GABA β3 homomers (Fig 4a).
Figure 4. Structure of the GABAAR chimera channel in a desensitized state.
(a) Plan view superimposing WT GLIC and GABA β3 subunit TMDs on GLIC-GABAARα1G258Vcryst revealing conformational changes to the TMD principally by tilting of M2 and rotation of M4. (b) Two M2 α-helices of the GLIC-GABAARα1G258Vcryst (blue) are shown with equivalent M2 helices from WT GLIC (green) and GABA β3 (red) subunits. Note the tilting of the helices to form a constriction in the lower part of the pore (box). The solvent accessible volume of the channel is represented by spheres. (c) Pore radius profiles through the channel. The ordinate directly relates to (b) for GLIC-GABAARα1G258Vcryst, GABA β3 and WT GLIC open state channels.(d) Pore constrictions formed by M2 lining residues at the level of -2’ Pro, 2’ Val (desensitization gate) and 9’ Leu (activation gate; all shown as Cα-spheres with distances in angstroms (Ǻ)). (e) Residues lining the M2-M3 interface and M1-M2 linker form the components of a desensitization gate. (f) Positive electrostatic surface potential of the chimera at the cytoplasmic portal of the ion channel. Cl- ions are represented as green spheres and omit style map is calculated when ions were excluded from the refinement (contoured at 2σ, orange).
At the extracellular end, the expanded pore is stabilized by an intra-subunit salt bridge between R273 (19’ in M2) and D286 (in M3), and by inter-subunit H-bonding between N274 (20’ in M2) and Q228 (in M1, which is also highly conserved across most GABAAR subunits; Fig 3c). Notably, although two cysteines on M1 (C233) and M3 (C292) are in close proximity, they do not form a disulfide bridge (as proposed from homology modelling studies33). Looking deeper into the pore, the leucine ring at 9’ associated with the activation gate34 is open (pore radius ~5Å) with side-chains rotated out towards M2 of the adjacent subunit. This is also observed for structures of the GABA receptor β3 homomer, GlyRα1, and GlyRα3 bound to ivermectin (Fig 4b, Supplementary Fig. 5)30, 31, 35.
Descending further into the pore, two constrictions are evident formed by rings of residues at 2’ (Val) and -2’ (Pro; Fig. 4d,e, Supplementary Fig. 5). This region forms the ‘desensitization gate’ recently described for GABAARs and GlyRs10, and the ion selectivity filter23. The pore is narrowest at the -2’ proline (2.1 Å radius) allowing the passage of Cl- lacking a hydration shell (1.8 Å radius), whilst precluding hydrated Cl- (~3.3 Å radius; Fig. 4b-d, Supplementary Fig. 5). Analysing the surface potential electrostatics of the channel reveals an electropositive region at the intracellular end (Fig. 4f, Supplementary Fig. 4), which would facilitate anion-selective permeation. This potential likely arises from side-chain dipoles in M2 α-helices29. Given that the chimera channel is anion-selective, peaks in electron density maps in the pore can be tentatively assigned to Cl- ions (~6σ in Fo-Fc maps). This location is in spatial proximity to the anion-binding sites proposed for anion-selective GluCl29 and GlyRα3 channels36. Moreover, a self-stabilized arrangement of water molecules, similar to those in GLIC23, is apparent at the level of 6’ Thr (Supplementary Fig. 4). Interestingly, at the cytosolic face, the chimera TMD exhibits ‘pockets’ of positive electrostatic potential, reminiscent of those in anion-selective GluCl channels 29 (Supplementary Fig. 4).
Previous patch-clamp studies of GLIC at pH < 4.5, revealed that entry into a desensitized state occurs within ~1.5 - 10 s37, 38. Here, introducing G258V near to the physically-constricted desensitization gate between 2’ and -2’ of GLIC-GABAARα1 resulted in currents that rapidly declined (Fig. 1b,e), suggesting that GLIC-GABAARα1G258Vcryst adopts a desensitized conformation at pH 4.5. Indeed, residues known to affect GABAAR desensitization10 similarly affected proton-activated responses for the chimera (Fig. 1e), and formed steric interactions at the interface between M2 and M3, near the base of the TMD (Fig. 4e). Our structure reveals that this putative desensitized state is stabilized by intra-subunit salt bridges between R254 (0’ in M2) and E302 (M3) and D392 (M4) at the base of the helical bundle, and by inter-subunit H-bonding between N307 (M3) and the backbone amino group of N247 (in the M1-M2 loop; Fig. 3d). This provides structural evidence for a pLGIC in a desensitized state.
Cholesterol binding to the GABAA receptor chimera
A common structural feature of the proton-bound chimera was electron density (~5σ in Fo-Fc maps) at a cavity between M3 in the principal (p, +) subunit and M1 from the complementary (c, -) subunit (Supplementary Fig. 6a,b). This was assigned to cholesteryl hemisuccinate (CHS) used during purification. The site partially overlaps with sites for lipids and ivermectin in GluCl29, 39 and for ivermectin in GlyRs31, 35 (Supplementary Fig. 6). Modelling and refinement of CHS indicated that its orientation best fit the electron density when cholesterol was tilted away from the receptor with the hemisuccinate moiety protruding between M3 and M1. This group could H-bond with S269 (15’ in M2), which is a key determinant for allosteric modulation of GABAARs by volatile anaesthetics, and of GLIC by ethanol22, 40, 41(Supplementary Fig. 6). This location for CHS is analogous to that proposed for cholesterol at GABAARs from homology modelling and molecular dynamic (MD) simulations33. This suggests that without the bulky hemisuccinate group, cholesterol could penetrate deeper into this interface, forming an H-bond between the hydroxyl group of the cyclohexanol ring and 15’ serine33.
Location of the potentiating neurosteroid binding site
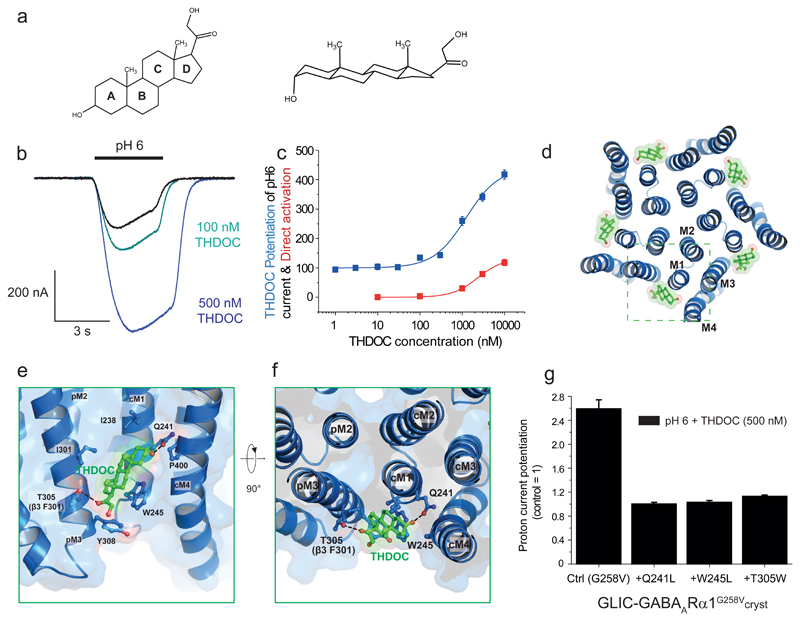
GABAARs are major targets in the brain for naturally-occurring neurosteroids derived from stress (e.g., tetrahydro-deoxycorticosterone (THDOC: Fig. 5a)) or sex (e.g., allopregnanolone) hormones11. These molecules potentiate GABAAR function at physiologically-relevant nanomolar concentrations, while at higher (micromolar) concentrations they cause direct receptor activation. Given their high potency at GABAARs, the potential for therapeutic application of neurosteroids is particularly appealing42. A highly conserved binding site underlying the neurosteroid potentiating action is considered, from homology modelling and mutagenesis, to be located within the α-helices of GABAAR α subunits14.
Figure 5. Interfacial subunit binding site for the neurosteroid THDOC.
(a) Chemical structure of THDOC in 2D and 3D. (b) Membrane currents for GLIC-GABAARα1G258Vcryst (expressed in Xenopus oocytes) activated by protons (EC10-15) in the absence and presence of THDOC revealing profound potentiation. (c) Proton and THDOC concentration-response curves for the chimera. Normalized plots represent fits to mean ± sem data points with the Hill equation for potentiation (blue) by THDOC of the pH6 (EC10) current (= 100 %), or direct activation (red) of the chimera by THDOC. EC50 value for potentiation is 1.23 ± 0.09 µM (n = 4), and for direct activation, 2.30 ± 0.09 µM (n = 3). (d) For GLIC-GABAARα1G258Vcryst, THDOC (green sticks) binds across each subunit-subunit interface (box) in the pentamer. (e,f) THDOC binding at interfaces between principal (p) and complementary (c) subunits. Side-views (in the membrane, e) and plan views (extracellular, f) are shown. Dashed lines indicate H-bonding (distances; Q241-steroid, 2.4 - 2.9 Å and T305-steroid, 3.1 - 3.4 Å). Putative hydrophobic interactions (<4 Å) are formed between W245 and rings C and D of THDOC. Labelled residues contribute directly to neurosteroid binding or line the binding pocket. (g) Relative effects of Q241L, W245L and T305W mutations on THDOC (500 nM) potentiation of GLIC-GABAARα1G258Vcryst proton-activated responses. Data shown are means ± sem of biological replicates (Ctrl (G258V) n = 4 oocytes; +Q241L n = 3; +W245L n = 4; +T305W n = 6). Results are representative of injections into oocytes taken from 3 Xenopus laevis performed over 5 separate days.
To identify the neurosteroid binding site, we used the GLIC-GABAARα1G258Vcryst chimera, which is sensitive to THDOC, evident from the markedly potentiated proton-activated currents using TEVC and increased thermal stabilisation of the detergent-solubilized receptor (Fig. 5b,c, Supplementary Fig. 2e). Moreover, as for native GABAARs, THDOC could also directly activate the chimera in the absence of protons. These effects of THDOC are comparable to those observed with native GABAARs (α1β3 EC50s for potentiation and direct activation: 0.57 ± 0.1 μM; 2.46 ± 0.02 μM (n = 4 and 3 independent experiments) respectively; Fig. 5c). We observed no sensitivity to 1 – 3 μM THDOC for GABA β3 homomers (EC10 pentobarbitone-gated current 99.3 ± 2.0 % of control, n = 3) or WT GLIC receptors (EC20 proton-activated current 93.9 ± 1.7 %, n = 7).
To explore the basis for neurosteroid binding to GABAARs, we determined the structure of GLIC-GABAARα1G258Vcryst bound to THDOC at 3.8 Å following co-crystallization. Neurosteroid molecules were bound to each subunit TMD. These were not located within the α1-subunit α-helices, as previously proposed14, but unambiguously across the subunit-subunit interface (Fig. 5d, Supplementary Fig. 7a,b). The electron density maps allowed confident positioning of the β-face of the neurosteroid backbone and orientation of the critical A-ring hydroxyl group at position C3.
At this new site, the ring core of THDOC binds in a ‘hydrophobic groove’ that runs between juxtaposed subunits, anchored by H-bonding at each end of the molecule. This conforms to a canonical steroid binding site (Fig. 5e,f, Supplementary Fig. 7). The architecture of this site and the strong electron density for bound THDOC confirmed the pivotal binding role previously assigned to Q241 in M1 of the α1 subunit14. A single H-bond is formed between Q241 in the complementary (c, -) subunit and the C3α hydroxyl in ring A of THDOC (Fig. 5e,f). The importance of Q241 was demonstrated by its mutation (Q241L), which ablated neurosteroid sensitivity (Fig. 5g) without affecting the proton sensitivity of the chimera (Supplementary Fig. 1d). The complementary subunit also contributes a tryptophan (W245) to the neurosteroid binding site, which is critical for neurosteroid potentiation at recombinant GABAARs43. The indole side-chain of W245 is orientated parallel to THDOC rings C and D, presumably interacting via hydrophobic stacking (Fig. 5e,f).
As predicted, its mutation (W245L) ablated neurosteroid potentiation (Fig. 5g). The orientation of the neurosteroid is supported by serial mutations of Q241 revealing that potentiation is maintained if the substituent is an H-bond acceptor (e.g., Q, N and H) but not if it only engages as an H-bond donor (e.g., W, R and K)14. Given that the C20 ketone of THDOC can only act as an H-bond acceptor, this argues for a molecular orientation in the binding site whereby the C3α hydroxyl of THDOC H-bonds to Q241.
Across the subunit interface, THDOC’s ring D ketone forms another H-bond with T305 in M3 of the principal (p, +) subunit (Fig. 5e,f), and, consistent with a binding role for this residue, its substitution by tryptophan (T305W) ablated THDOC potentiation (Fig. 5g). The corresponding residue in the β-α interface of native β3-containing GABAARs is F301. Interestingly, this is photolabelled by a neurosteroid-analogue in GABA β3 homomers, in accord with a binding role44. The base of the binding pocket in the principal subunit is formed by the aromatic ring of Y308, a residue highly conserved across GABAARs subunits. In the physiological context of binding at the β-α interface of GABAARs, it is apparent that the steroid molecule is also physically supported by aromatic residues contributed by both subunits. Whilst there is strong conservation of these aromatic residues across inhibitory pLGICs (Supplementary Fig. 1e & 7), modulation by THDOC is primarily dependent upon H-bonding at the complimentary face to Q241 in M1. This is only provided by the α-subunit containing receptors and not present in β subunits (Supplementary Fig. 7e).
We performed MD simulations to corroborate our crystallographic interpretation for the orientation of THDOC in the binding site. Of many potential docking poses, only one consistently mapped onto the crystal structure. In this position Q241 and W245 would coordinate with the A-ring of THDOC, while T305 coordinates the D-ring (Supplementary Fig. 8a,b & Supplementary Movie 1). The THDOC position remained stable during MD simulation. Notably, the C3α hydroxyl group donated an H-bond to Q241, and the C21 hydroxyl of THDOC was orientated towards the membrane where it could donate an H-bond to the lipid head groups.
Interestingly, the effects of THDOC on receptor structure are subtle. Although such small movements are insufficient to provide extensive insight, it is apparent that the overall structural geometry conforms to that observed for the proton-bound chimera structure in a desensitized state. The THDOC-bound structure thus resembles that of GlyRα1 bound to ivermectin31 and GlyRα3 bound to the analgesic, AM-360736, which both potentiate the agonist response. Considering the physical contours and binding interactions of the neurosteroid binding site described here, and from previous electrophysiological studies14, we would expect this site to accommodate potentiating neurosteroids of distinct stereochemistry exhibiting high efficacy modulation of GABAARs45, 46.
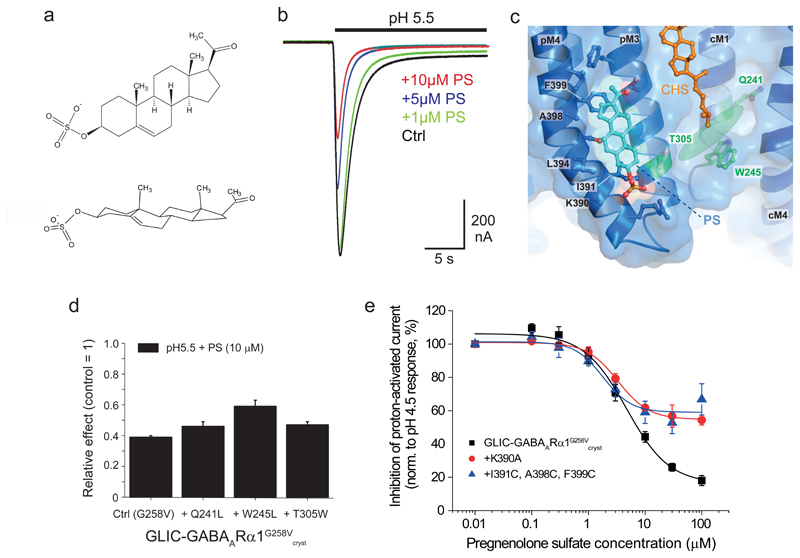
Inhibitory neurosteroid binding site involves M3 and M4
GABAARs are also modulated by naturally-occurring inhibitory neurosteroids in the brain, exemplified by pregnenolone sulfate (PS; Fig. 6a). Their binding site on GABAARs has remained elusive although the consensus view is that PS binds at a discrete site from that for potentiating neurosteroids47. Expressing GLIC-GABAARα1G258Vcryst in Xenopus oocytes and using TEVC indicated a PS binding site was present with an apparent affinity comparable to that for native GABAARs (Fig 6b). To establish its location, we co-crystallized the chimera with PS (Supplementary note 2). Electron density maps for PS-bound crystals (at 3.0 Å) revealed distinctive peaks of density along the bilayer exposed face of M3 and M4 (Fig. 6c, Supplementary Fig. 8c). Modelling of PS suggests it aligns perpendicularly to the membrane at this intra-subunit site possibly engaging in van der Waals interactions principally along the outer face of M4. Although we cannot precisely orientate PS at this resolution, MD simulations indicated that the ring A sulfate group points towards the base of the TMD, potentially interacting with K390 (side-chain density for which was missing in diffraction data; Fig. 6c, Supplementary Fig. 8c-e & Supplementary Movie 2). The PS ring structure most likely interacts with several residues including I391, A398 and F399 in pM4.
Figure 6. Inhibitory neurosteroid pregnenolone sulfate binds at a distinct site.
(a) Chemical structure of pregnenolone sulfate (PS) in 2D and 3D. (b) Inhibition of submaximal (pH 5.5) membrane currents for GLIC-GABAARα1G258Vcryst by PS. (c) Intra-subunit binding site for PS (cyan sticks/spheres) located at the lipid face of M3 and M4 α-helices, viewed from the plane of the membrane. Hydrophobic and aromatic residues that line the bilayer-exposed face of M3 and M4 are labelled. These form a smooth groove at the protein surface, with PS bound alongside the α-helices. Residues that bind THDOC (forming the potentiating neurosteroid binding site) are labeled in green, and THDOC orientation is shown by the transparent green oval shape. Cholesteryl hemisuccinate binding is also indicated (orange sticks). (d) Relative effects of Q241L, W245L, and T305W mutations on PS (10 μM) inhibition of GLIC-GABAARα1G258Vcryst proton–activated responses. Values are means ± sem (n = 4, 4, 6 and 3, respectively from independent experiments). (e) Proton-activated (pH4.5) steady-state current inhibition by PS for GLIC-GABAARα1G258Vcryst, and for the chimeras containing either K390A or I391C, A398C, F399C mutations. The curve fits were generated by the inhibition model equation. Note the reduced inhibition for the mutant receptors. Data shown are means of biological replicates (Ctrl (G258V) n = 4 oocytes; +Q241L n = 4; +W245L n = 4; +T305W n = 6; +K390A n = 4; + I391C, A398C, F399C n = 4). Results are representative of injections into oocytes taken from 8 Xenopus laevis performed over 18 separate days.
PS poses from MD simulations were less stable compared to those for THDOC, possibly reflecting the labile nature of this site. From different starting orientations, PS had a tendency to transition rapidly into an alternative pose in its proposed binding site. In this position, the β-surface-protruding methyl groups are oriented towards the hydrophobic environment of the membrane, the sulfate group forms a salt bridge with K390, and the PS ring core interacts with subsequent turns of the M4 α-helix up to L402.
This discrete site and the varied nature of the residues involved, may explain the enigmatic nature of PS modulation, and account for the difficulties in identifying its binding site47. In accord with a discrete site, PS inhibition was unaffected by mutating Q241, W245 or T305, which ablated THDOC potentiation (Fig. 6d, Supplementary Fig. 8f)48. However, mutating charged (K390A) or hydrophobic (I391C, A398C and F399C) pM4 residues in the chimera significantly reduced the inhibition of steady-state currents by PS (Fig. 6e). To investigate further, we developed a kinetic model (Supplementary Methods) to simulate the effects of PS and the M4 mutations. By assuming PS binds preferentially to the activated/desensitized states of GLIC-GABAARα147, the model suggested that the mutations caused small changes (3-4-fold) promoting receptor entry into a desensitized state and increasing entry into agonist bound-blocked states. There was minimal effect on the PS dissociation constant and overall, the mutated residues had more profound effects on PS efficacy than potency, which may partly explain its mechanism of inhibition (Supplementary Fig. 8g).
Discussion
Here we present a new GABAAR chimera that allows the structural and functional examination of the α1-subunit transmembrane domain for one of the brain’s most prevalent inhibitory neurotransmitter receptors.
Our description of a novel crystal structure for a functional GLIC-GABAARα1 chimera has enabled the structural interrogation of the GABAAR α subunit TMD. This domain contains numerous modulator binding sites, including the GABA ion channel. We reveal several important facets. Firstly, a precise structural location for the binding site for the potentiating neurosteroids. The GABAAR subunit interface is evidently critical for modulation by potentiating neurosteroids. Previously, we had proposed that two sites were necessary to explain the direct activation of the receptor by neurosteroids and potentiation of receptor function. However, with the new structure presented here both activation and potentiation apparently proceed from a single interfacial binding site, a feature that is explicable by using a Monod-Wyman-Changeux model of receptor operation49. Secondly, whereas potentiating neurosteroids discretely bind between subunits near the lipid interface, inhibitory neurosteroids seemingly bind to a discrete intra-subunit TMD site.
Thirdly, we definitively describe the desensitization gate for GABAARs, caused by constriction at the base of the ion channel. This structural feature is conserved across anion-permeable pLGICs captured in desensitized-like states (e.g. GABA receptor β3 and GlyRα1 bound to ivermectin). It is also notable in the recent crystal structure of a heteromeric α4β2 nAChR50 that a similar constriction of the ion channel is evident at the cytosolic portal, consistent with a non-conductive, desensitized state. This, together with the structure presented here, suggests common structural features underpin desensitization in both anion- and cation-permeable pLGICs.
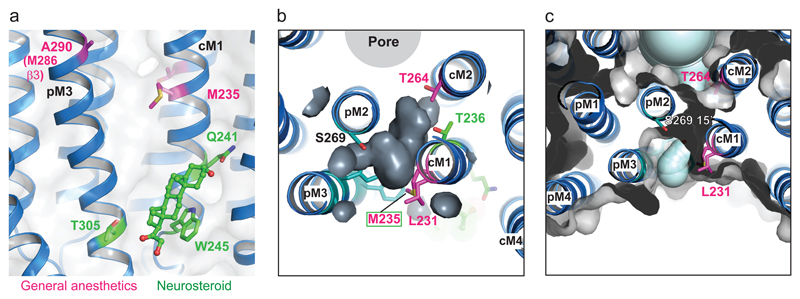
Finally, it is clear that the α-subunit interface in GABAARs also forms a binding site for another important class of modulators, the intravenous general anaesthetics (e.g., etomidate), which can photolabel the β+-α- interface at a methionine (M235) in the α1-subunit51 (Fig. 7a). Our crystal structures reveal that the etomidate and neurosteroid binding sites are non-overlapping, such that the same interface can accommodate both molecule types binding simultaneously (Fig. 7a). This likely explains why neurosteroids will enhance potentiation by etomidate52.
Figure 7. Inter-subunit anaesthetic binding cavity and aqueous tunnel.
(a) Structure of the GLIC-GABAARα1G258Vcryst chimera showing the location of residues involved in anaesthetic binding (magenta, M235 & A290 (equivalent to M286 in β3) and the binding site for potentiating neurosteroids (shown in stick representation, green). Note their accommodation at the same subunit interface. (b) Transverse-view of an aqueous tunnel reveals that it runs close to residues implicated in both anaesthetic (magenta), CHS (teal) and neurosteroid (green) binding sites and would be accessible from the channel pore or the membrane-exposed face of the TMD. (c) Another transverse view of the aqueous tunnel showing the proximity of the CHS binding site (teal) and key residues involved in anesthetic binding (magenta). The tunnel runs from the lipid interface at L231 in M1, through to the back of the ion channel at T264 (10’) in M2, near the 9’ activation gate, and exits into the pore at S269 (15’).
Further interrogation of the chimera structure at the subunit-subunit interface also revealed a lateral aqueous tunnel. This originates from the lipid bilayer and opens into the ion channel at 15’ level. This tunnel is part of a larger inter-subunit cavity lined by residues implicated in anaesthetic binding; it is also partly-occupied by CHS (Fig. 7b,c, Supplementary Fig. 6d).
Given the significant role neurosteroids play in anxiety, stress and other neurological disorders11, these modular structures provide a new template for exploring how allosteric sites can be used by drugs to modulate GABAARs - an area of significant physiological, pathological and future therapeutic relevance for the brain.
Methods
All details regarding the methods used can be found in the online version of the paper.
Supplementary Material
Acknowledgements
This work was supported by the Medical Research Council (MR/K005537/1). DL was in receipt of a MRC PhD studentship. OJA was supported by the Carlsberg Foundation. MGG was funded by the Wellcome Trust and Royal Society (104194/Z/14/Z). We thank the beamline staff at Diamond Light Source and ESRF for assistance and advice; Ambrose Cole for technical support (crystallization, data collection and processing); Pierre-Jean Corringer for technical advice; Charlotte Jones for additional mutagenesis and functional electrophysiology. We also thank the Advanced Research Computing (ARC) facility, the EPSRC UK National Service for Computational Chemistry Software (NSCCS) at Imperial College London (grant no. EP/J003921/1) and the ARCHER UK National Supercomputing Services for computer time granted via the UK High-End Computing Consortium for Biomolecular Simulation, HECBioSim (http://www.hecbiosim.ac.uk), supported by EPSRC (grant no. EP/L000253/1).
Footnotes
Author Contributions
DL and TGS jointly planned the project, performed data analyses and wrote the manuscript. DL generated the receptor constructs, optimized receptor expression, purification and crystallization, carried out structure solution and model building (assisted by MGG with structural analysis). DL and PT performed electrophysiology experiments. MF carried out site-directed mutagenesis. TGS performed the kinetic modelling. OA and PB performed the molecular dynamics simulations and docking. MG helped plan and design desensitization experiments. All authors contributed towards editing the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Reference List
- 1.Miller PS, Smart TG. Binding, activation and modulation of Cys-loop receptors. Trends in Pharmacological Sciences. 2010;31:161–174. doi: 10.1016/j.tips.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Corringer PJ, et al. Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure. 2012;20:941–956. doi: 10.1016/j.str.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Farrant M, Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABAA receptors. Nat Rev Neurosci. 2005;6:215–229. doi: 10.1038/nrn1625. [DOI] [PubMed] [Google Scholar]
- 4.Braat S, Kooy R. The GABAA Receptor as a Therapeutic Target for Neurodevelopmental Disorders. Neuron. 2015;86:1119–1130. doi: 10.1016/j.neuron.2015.03.042. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph U, Mohler H. GABAA Receptor Subtypes: Therapeutic Potential in Down Syndrome, Affective Disorders, Schizophrenia, and Autism. Annual Review of Pharmacology and Toxicology. 2014;54:483–507. doi: 10.1146/annurev-pharmtox-011613-135947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mody I. Distinguishing between GABAA receptors responsible for tonic and phasic conductances. Neurochem Res. 2001;26:907–913. doi: 10.1023/a:1012376215967. [DOI] [PubMed] [Google Scholar]
- 7.Smart TG, Paoletti P. Synaptic Neurotransmitter-Gated Receptors. In: Sheng M, Sabatini BL, Sudhof TC, editors. The Synapse. Cold Spring Harbor Laboratory Press; New York: 2012. pp. 191–216. [Google Scholar]
- 8.Jones MV, Westbrook GL. Desensitized states prolong GABAA channel responses to brief agonist pulses. Neuron. 1995;15:181–191. doi: 10.1016/0896-6273(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 9.Bright DP, et al. Profound Desensitization by Ambient GABA Limits Activation of δ-Containing GABAA Receptors during Spillover. Journal of Neuroscience. 2011;31:753–763. doi: 10.1523/JNEUROSCI.2996-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gielen M, Thomas P, Smart TG. The desensitization gate of inhibitory Cys-loop receptors. Nat Commun. 2015;6 doi: 10.1038/ncomms7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belelli D, Lambert JJ. Neurosteroids: endogenous regulators of the GABAA receptor. Nat Rev Neurosci. 2005;6:565–575. doi: 10.1038/nrn1703. [DOI] [PubMed] [Google Scholar]
- 12.Fritschy JM, Brunig I. Formation and plasticity of GABAergic synapses: physiological mechanisms and pathophysiological implications. Pharmacol Ther. 2003;98:299–323. doi: 10.1016/s0163-7258(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 13.Picton AJ, Fisher JL. Effect of the α subunit subtype on the macroscopic kinetic properties of recombinant GABAA receptors. Brain Research. 2007;1165:40–49. doi: 10.1016/j.brainres.2007.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosie AM, Wilkins ME, da Silva HMA, Smart TG. Endogenous neurosteroids regulate GABAA receptors through two discrete transmembrane sites. Nature. 2006;444:486–489. doi: 10.1038/nature05324. [DOI] [PubMed] [Google Scholar]
- 15.Franks NP. General anaesthesia: from molecular targets to neuronal pathways of sleep and arousal. Nat Rev Neurosci. 2008;9:370–386. doi: 10.1038/nrn2372. [DOI] [PubMed] [Google Scholar]
- 16.Kang JQ, Macdonald RL. Making sense of nonsense GABAA receptor mutations associated with genetic epilepsies. Trends in Molecular Medicine. 2009;15:430–438. doi: 10.1016/j.molmed.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilf RJC, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 18.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 19.Moraga-Cid G, et al. Allosteric and hyperekplexic mutant phenotypes investigated on an α1 glycine receptor transmembrane structure. PNAS. 2015;112:2865–2870. doi: 10.1073/pnas.1417864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkins ME, Hosie AM, Smart TG. Identification of a β subunit TM2 residue mediating proton modulation of GABA type A receptors. J Neurosci. 2002;22:5328–5333. doi: 10.1523/JNEUROSCI.22-13-05328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nury H, et al. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 22.Sauguet L, et al. Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat Commun. 2013;4:1697–1707. doi: 10.1038/ncomms2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauguet L, et al. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 2013;32:728–741. doi: 10.1038/emboj.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fourati Z, Sauguet L, Delarue M. Genuine open form of the pentameric ligand-gated ion channel GLIC. Acta Crystallogr D Biol Crystallogr. 2015;71:454–460. doi: 10.1107/S1399004714026698. [DOI] [PubMed] [Google Scholar]
- 25.Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. Coupling of agonist binding to channel gating in the GABAA receptor. Nature. 2003;421:272–275. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- 26.Sigel E, Buhr A, Baur R. Role of the conserved lysine residue in the middle of the predicted extracellular loop between M2 and M3 in the GABAA receptor. J Neurochem. 1999;73:1758–1764. doi: 10.1046/j.1471-4159.1999.731758.x. [DOI] [PubMed] [Google Scholar]
- 27.Hales TG, et al. An asymmetric contribution to gamma-aminobutyric type A receptor function of a conserved lysine within TM2-3 of alpha1, beta2, and gamma2 subunits. J Biol Chem. 2006;281:17034–17043. doi: 10.1074/jbc.M603599200. [DOI] [PubMed] [Google Scholar]
- 28.Kodera H, et al. De novo GABRA1 mutations in Ohtahara and West syndromes. Epilepsia. 2016;57:566–573. doi: 10.1111/epi.13344. [DOI] [PubMed] [Google Scholar]
- 29.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller PS, Aricescu AR. Crystal structure of a human GABA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du J, Lu W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, Chen H, Michelsen K, Schneider S, Shaffer PL. Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature. 2015;526:277–280. doi: 10.1038/nature14972. [DOI] [PubMed] [Google Scholar]
- 33.Henin J, Salari R, Murlidaran S, Brannigan G. A Predicted Binding Site for Cholesterol on the GABAA Receptor. Biophys J. 2014;106:1938–1949. doi: 10.1016/j.bpj.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unwin N. Neurotransmitter action: opening of ligand-gated ion channels. Cell. 1993;72(Suppl):31–41. doi: 10.1016/s0092-8674(05)80026-1. [DOI] [PubMed] [Google Scholar]
- 35.Huang X, Chen H, Shaffer PL. Crystal Structures of Human GlyRα3 Bound to Ivermectin. Structure. 2017;25:945–950. doi: 10.1016/j.str.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, et al. Crystal structures of human glycine receptor α3 bound to a novel class of analgesic potentiators. Nat Struct Mol Biol. 2017;24:108–113. doi: 10.1038/nsmb.3329. [DOI] [PubMed] [Google Scholar]
- 37.Velisetty P, Chakrapani S. Desensitization Mechanism in Prokaryotic Ligand-gated Ion Channel. Journal of Biological Chemistry. 2012;287:18467–18477. doi: 10.1074/jbc.M112.348045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Gutierrez G, Grosman C. Bridging the Gap between Structural Models of Nicotinic Receptor Superfamily Ion Channels and Their Corresponding Functional States. Journal of Molecular Biology. 2010;403:693–705. doi: 10.1016/j.jmb.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512:333–337. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mihic SJ, et al. Sites of alcohol and volatile anaesthetic action on GABAA and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 41.Howard RJ, et al. Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. PNAS. 2011;108:12149–12154. doi: 10.1073/pnas.1104480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy DS, Rogawski MA. Neurosteroid replacement therapy for catamenial epilepsy. Neurotherapeutics. 2009;6:392–401. doi: 10.1016/j.nurt.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bracamontes JR, Li P, Akk G, Steinbach JH. A neurosteroid potentiation site can be moved among GABAA receptor subunits. The Journal of Physiology. 2012;590:5739–5747. doi: 10.1113/jphysiol.2012.237255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen ZW, et al. Neurosteroid Analog Photolabeling of a Site in the Third Transmembrane Domain of the β3 Subunit of the GABAA Receptor. Mol Pharmacol. 2012;82:408–419. doi: 10.1124/mol.112.078410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosie AM, Wilkins ME, Smart TG. Neurosteroid binding sites on GABAA receptors. Pharmacology & Therapeutics. 2007;116:7–19. doi: 10.1016/j.pharmthera.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Covey DF, et al. Enantioselectivity of pregnanolone-induced gamma-aminobutyric acid(A) receptor modulation and anesthesia. J Pharmacol Exp Ther. 2000;293:1009–1016. [PubMed] [Google Scholar]
- 47.Seljeset S, Laverty D, Smart TG. Inhibitory neurosteroids and the GABAA receptor. Adv Pharmacol. 2015;72:165–187. doi: 10.1016/bs.apha.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Akk G, et al. Mutations of the GABA-A Receptor α1 Subunit M1 Domain Reveal Unexpected Complexity for Modulation by Neuroactive Steroids. Mol Pharmacol. 2008;74:614–627. doi: 10.1124/mol.108.048520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rusch D, Zhong H, Forman SA. Gating allosterism at a single class of etomidate sites on α1β2γ2L GABAA receptors accounts for both direct activation and agonist modulation. J Biol Chem. 2004;279:20982–20992. doi: 10.1074/jbc.M400472200. [DOI] [PubMed] [Google Scholar]
- 50.Morales-Perez CL, Noviello CM, Hibbs RE. X-ray structure of the human α4β2 nicotinic receptor. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li GD, et al. Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J Neurosci. 2006;26:11599–11605. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li GD, Chiara DC, Cohen JB, Olsen RW. Neurosteroids Allosterically Modulate Binding of the Anesthetic Etomidate to γ-Aminobutyric Acid Type A Receptors. Journal of Biological Chemistry. 2009;284:11771–11775. doi: 10.1074/jbc.C900016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.