Abstract
Aberrant gene expression plays critical roles in the development of colorectal cancer (CRC). Here we show that POTEE, which was identified as a member E of POTE ankyrin domain family, was significantly upregulated in colorectal tumors and predicted poor overall survival of CRC patients. In CRC cells, POTEE could act as an oncogene and could promote cell growth, cell-cycle progression, inhibit apoptosis, and elevates xenograft tumor growth. Mechanically, we used microarray analysis and identified a POTEE/SPHK1/p65 signaling axis, which affected the biological functions of CRC cells. Further evaluation showed that overexpression of POTEE could increase the protein expression of SPHK1, followed by promoting the phosphorylation and activation of p65 protein. Altogether, our findings suggested a POTEE/SPHK1/p65 signaling axis could promote colorectal tumorigenesis and POTEE might potentially serve as a novel biomarker for the diagnosis and an intervention of colorectal cancer.
Subject terms: Colorectal cancer, Predictive markers
Introduction
Colorectal cancer is the third most commonly diagnosed cancer (10.2% of the total cases) and the second leading cause of cancer related deaths (9.2% of the total cancer deaths) in 2018 globally1. Both incidence and death rates of colorectal cancer are increasing rapidly and maintain an upward trend in Asian countries2. The global burden of colorectal cancer (CRC) is expected to increase by 60% to more than 2.2 million new cases and 1.1 million deaths by 20302–4. Exploring related genes in the development of CRC and finding important links that affect the biological characteristics of CRC are critical ways to understand the malignancy of tumors and to improve the survival and prognosis of CRC patients5.
POTE (Prostate, Ovary, Testes, and Embryo) is a newly detected gene family that contains ankyrin and spectrin domains and express in a variety of human cancers6–8. This family has 11 exons and 10 introns and spans 32 kb of chromosome 21q11.2 region, which consists of at least 10 highly homologous genes located on chromosomes 2, 8, 13, 14, 15, 18, 21, and 227. POTEE is a paralogs located at chromosome 2 that contains three distinct regions: N-terminal cysteine-rich domains followed by seven ankyrin repeats and C-terminal spectrin-like helices9,10. Previous studies have shown that POTEE was only weakly expressed in normal tissues of prostate and breast, but its expression was significantly elevated in their tumor counterparts8,11. It was also reported that serum POTEE level in non-small cell lung cancer (NSCLC) patients was associated with advanced TNM stage and might serve as a potential prognostic indicator of NSCLC patients12. In addition, upregulation of POTEE also indicated poorer prognosis of ovarian cancer patients13. Recently, a study showed that overexpression of POTEE in macrophages and its subtype could provide a platform for mTOR and Rictor binding thereby resulting in activation of mTORC210. Although above-mentioned studies suggested a potential oncogenic role of POTEE in various cancer types, its biological functions and tumorigenesis mechanisms remains largely unknown. In colorectal cancer, the dysregulation of POTEE are still undefined to our knowledge.
Here, we conducted researches on detecting the expression status and clinical characteristics of POTEE in colorectal cancer tumor samples and cells, with the aim to elucidate the oncogenic roles and potential mechanisms of POTEE both in vitro and in vivo. Our study provides new mechanistic insights into the roles of POTEE in promoting SPHK1/p65 signaling, which might server as a potential biomarker and a novel intervention target for colorectal neoplasia.
Results
POTEE is upregulated and predicts poor clinical outcome in CRC patients
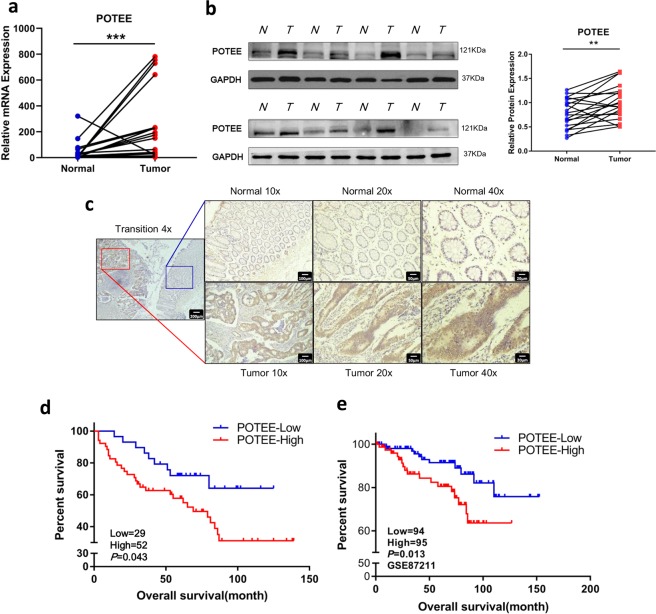
To explore the expression of POTEE in CRC, we firstly carried out quantitative real-time polymerase chain reaction (qRT-PCR) to analyze the messenger RNA (mRNA) expression of POTEE in 20 pairs of CRC samples and their normal counterparts. The results showed that POTEE was significantly upregulated in tumors (19/20, 95%) in comparison with their paired normal mucosa (Fig. 1a). Consistent with mRNA level, the protein expression analyzed by western blot and immunohistochemistry (IHC) also verified the elevated expression of POTEE in colorectal tumor tissues (Fig. 1b, c). What’s more, ours results further revealed the intense nuclear and weak cytoplasmic staining of POTEE in the epithelial component of carcinomas (Fig. 1c; Supplementary Fig. 1a. b).
Fig. 1. POTEE is upregulated and predicts poor clinical outcome in CRC patients.
a The relative mRNA levels of POTEE in 20 paired CRC and adjacent normal tissues. Results are shown as mean ± SD. *P < .05, **P < .01, ***P < .001, based on Paired t-test. b Western blots of POTEE protein in 20 pairs of CRC tissues, GAPDH was loaded as a control. Relative protein expression were measured by Image J and shown as mean ± SD. *P < .05, **P < .01, ***P < .001, based on Paired t-test. c Representative staining of POTEE with immunohistochemistry (IHC) analysis. d Overall survival rates of the CRC patients (n = 81) grouped by POTEE protein expression levels. e Overall survival of patients with CRC stratified by POTEE mRNA expression from GSE87211 dataset
Next, we went on investigating the clinical relevance of POTEE expression with CRC patients and finally 81 samples with visible POTEE staining by IHC and complete follow-up data from our center were included for our following analysis. We detected strong POTEE expression in almost 65% of colorectal tumor samples examined, and most of adjacent normal mucosa showed weak or absent POTEE signal. Kaplan–Meier survival analysis showed high POTEE expression predicted poorer overall survival (OS) than those with weak POTEE expression (Fig. 1d). Clinicopathological characteristics of 81 CRC patients showed that high POTEE expression was obviously associated with more lymph nodal metastasis, higher clinical stage, and larger tumor size (Table 1). External dataset also validated that high POTEE mRNA level could also predicted significantly poorer overall survival in GSE87211 with 189 rectal cancer patients involved (Fig. 1e). Together, these data indicated that POTEE is upregulated in human CRC tissues and overexpression of POTEE correlates with malignant phenotypes as well as unfavorable prognosis of colorectal cancer.
Table 1.
Correlation between expression of POTEE and clinical characteristics of CRC patients
| Characteristics | Cases | POTEE Immunostaining | χ2 value | P | |
|---|---|---|---|---|---|
| Decreased expression | High expression | ||||
| Gender | 0.200 | 0.655 | |||
| Male | 42 | 16 (38.1%) | 26 (61.9%) | ||
| Female | 39 | 13 (33.3%) | 26 (66.7%) | ||
| Age (years) | 1.454 | 0.228 | |||
| <60 | 32 | 14 (43.8%) | 18 (56.3%) | ||
| ≥60 | 49 | 15 (30.6%) | 34 (59.4%) | ||
| Lymph nodal metastasis | 4.464 | 0.029 | |||
| N0 | 49 | 22 (44.8%) | 27 (45.2%) | ||
| N1/N2 | 32 | 7 (21.8%) | 25 (78.1%) | ||
| TNM | 5.901 | 0.013 | |||
| I/II | 47 | 22 (46.8%) | 25 (53.2%) | ||
| III/IV | 34 | 7 (20.6%) | 27 (79.4%) | ||
| Tumor sizea | 4.500 | 0.042 | |||
| <5 cm | 46 | 21 (45.7%) | 25 (54.3%) | ||
| ≥5 cm | 34 | 8 (23.5%) | 26 (76.5%) | ||
a1cases missing Tumor size data
POTEE plays oncogenic roles in CRC cells
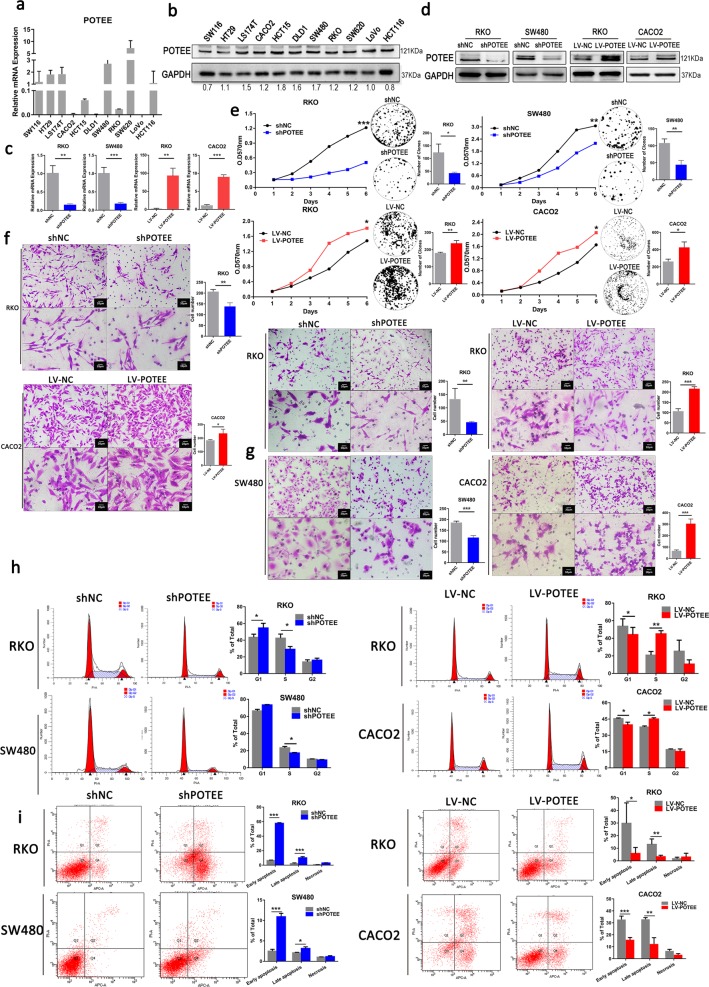
Given the significant expression difference and clinical relevance of POTEE, we further evaluated the functional roles of POTEE in CRC cells. We firstly examined endogenous expression of POTEE in 11 colorectal cancer cell lines by qRT-PCR and western blot analysis (Fig. 2a, b). Then, POTEE-targeting shRNA (shPOTEE) or corresponding controls (shNC) were used to establish stable POTEE-knockdown cell lines in SW480 and RKO cells with relatively high and moderate endogenous POTEE expression, respectively. While GFP-tagged POTEE-expressing recombinant lentivirus (LV-POTEE) or control vectors (LV-NC) were introduced into RKO and CACO2 cells. The transfection efficiency was verified by qRT-PCR and western blot analysis (Fig. 2c, d). Results showed that POTEE knockdown in RKO and SW480 cells significantly inhibited cell proliferation and colony formation, while ectopic expression of POTEE in RKO and CACO2 cells significantly promoted cell growth (Fig. 2e). In addition, wound-healing assays and transwell migratory and invasive assays revealed that upregulated POTEE could remarkably enhanced the migratory and invasive ability of RKO and CACO2 cell lines, while knockdown POTEE in RKO and SW480 exerted opposite effects and weaken the metastasis of cells. (Fig. 2f, g; Supplementary Fig. 2a, b). As cell proliferation, cell-cycle regulation and apoptosis are tightly bound14,15, so we next evaluated the effect of POTEE on cell-cycle regulation in cells with different POTEE status. Results showed POTEE overexpression resulted in decreased cell proportions in the G1 phase while significantly elevated cell fractions in the S phase in RKO and CACO2 cells, but knockdown of POTEE reversed cell-cycle progression in SW480 and RKO cells (Fig. 2h). Moreover, to further identify whether POTEE could play a role in apoptosis control, we treated cells with 5-Fluorouracil (5-Fu) to induce cell apoptosis, results revealed that knockdown of POTEE led to a sharp increase of apoptosis as compared with their negative controls. However, apoptosis induced by 5-Fu treatment was significantly rescued after POTEE overexpression (Fig. 2i). Collectively, these data supported the oncogenic role of POTEE in inducing proliferation, migration and invasion, inhibiting apoptosis and promoting cell-cycle progression in CRC cells in vitro.
Fig. 2. POTEE enhances tumorigenesis and inhibits apoptosis of CRC cells.
a, b qRT-PCR (a) and western blot (b) were performed to examine POTEE endogenous expression in 11 colon cancer cell lines. c, d Transfection efficiency was detected by qRT-PCR (c) and western blot (d). Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test. e MTT (left) and colony formation assays (right) in indicated cell lines with different POTEE expression. Statistics were measured by 2-way ANOVA or Student t, *P < .05, **P < .01, ***P < .001. f, g Migration (f) and invasion (g) assay in indicated cell lines with different POTEE expression. Statistics were measured by Student t, *P < .05, **P < .01, ***P < .001. h Cell-cycle distribution analysis. Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test. i Apoptosis analysis with annexin V-APC/PI staining. Cells were exposed to 50 µg/ml 5-Fluorouracil for 36–48 h. Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test
RNA-Seq reveals POTEE-regulated signaling pathways in CRC cells
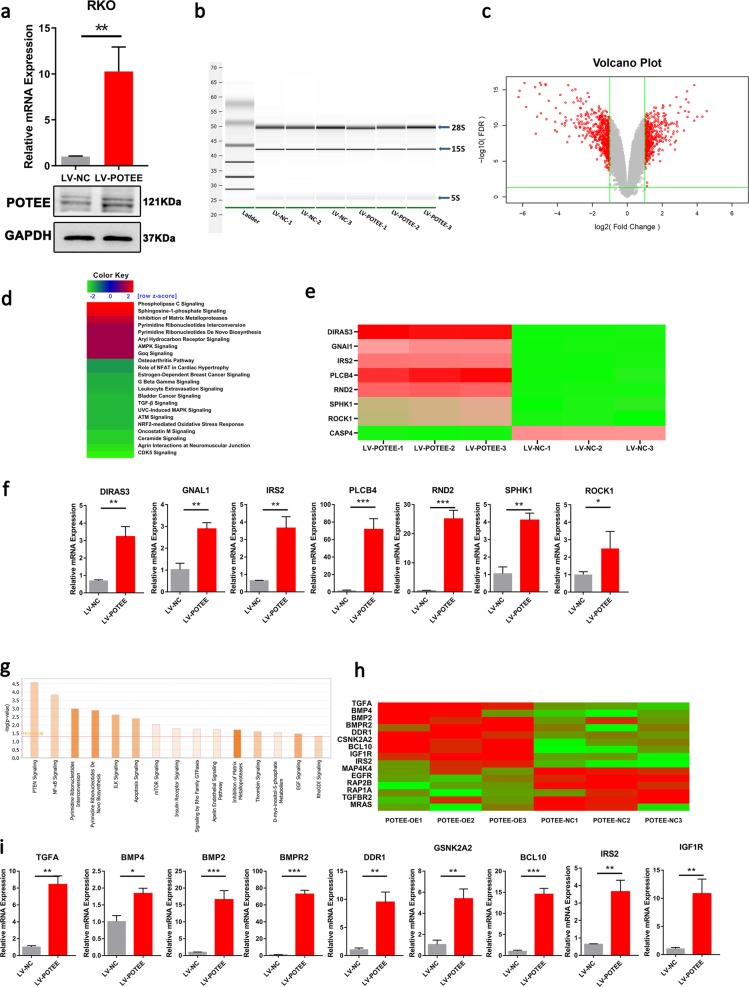
To better detect POTEE-mediated molecular pathways in CRC cells, we conducted a genome-wide analysis to globally characterize POTEE-regulated transcriptome changes. Total RNAs from RKO cells with LV-NC or LV-POTEE were subjected to transcriptomic sequencing (RNA-Seq). The expressions of POTEE mRNA and protein were validated prior to RNA-Seq (Fig. 3a). After assessing the intactness of the samples and verifying rare degraded mRNA (Fig. 3b), microarray analysis was performed and a total of 638 genes were identified to be significantly regulated in POTEE-overexpressed cells with respect to control cells (Fig. 3c; Supplementary Fig. 3). Subsequent Ingenuity Pathway Analysis (IPA) of the significantly regulated genes revealed that the top-three ranked molecular pathways based on IPA z-score included “Phospholipase C”, “Sphingosine-1-phosphate (S1P)”, and “Inhibition of Matrix Metalloproteases”. In the meantime, when analyzed according to different P-value, the most enriched pathways were “PTEN Signaling”, “NF-κB Signaling”, and “Pyrimidine Ribonucleotides Interconversion” in sequence (Fig. 3d, g).
Fig. 3. RNA-seq reveals POTEE-regulated signaling pathways in CRC.
a qRT-PCR (left) and western blot (right) showing POTEE expression in RKO cells with POTEE overexpression or mock cells. Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test. b The total mRNA integrity was tested on the Agilent 2100 bioanalyzer used the Eukaryote Total RNA Nano assay. c Differentially expressed genes (2-fold) between mock and LV-POTEE RKO cells were determined by RNA-Seq and shown by volcano plot. d IPA z-score analysis. IPA z-score > 2 or < -2 was considered to be significant activation or inhibition. Analysis result showed that the IPA z-score of S1P signaling was 2.449. e Heat map showing the microarray result for the differentially expression genes involved in S1P pathway. Scale bar: red (upregulated), green (downregulated). f qRT-PCR was performed to examine seven upregulated genes involved in S1P molecules network in RKO cells with POTEE overexpression or mock cells. Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test. g Histogram sort based on P-value, and the horizontal line represented actively classic pathway. h Heat map showing the microarray result for the differentially expression genes involved in NF-κB pathway. Scale bar: red (upregulated), green (downregulated). i Indicated upregulated 9 genes involved in NF-κB pathway measured by qRT-PCR. Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test
According to the results of microarray analysis, genes involved in S1P molecules network was most significantly enriched and we identified seven genes (DIRAS3, GNAL1, IRS2, PLBC4, RND2, SPHK1, ROCK1, fold change > 1.5) were sharply upregulated and one gene (CASP4, fold change > 1.5) was shown to be downregulated (Fig. 3e) in S1P signaling and.the seven upregulated genes were chosen for further validation with qRT-PCR (Fig. 3f). As was reported, intracellular activation of S1P signaling was catalyzed by the key and rate-limiting sphingosine kinase isoform, SPHK116, which was also found to be significantly increased in POTEE-overexpressed RKO cells and was focused in our following detection.
As NF-κB pathway was also the top activated POTEE-mediated carcinogenic pathways ranked by P-value (Fig. 3g), and it was reported that NF-κB pathway was a main downstream of S1P signaling17–19, so we further did heat map according to the nine genes significantly upregulated and five downregulated genes involved in NF-κB pathway, and qRT-PCR analysis was also undertook to identify the accuracy of the microarray sequence (Fig. 3h, i).
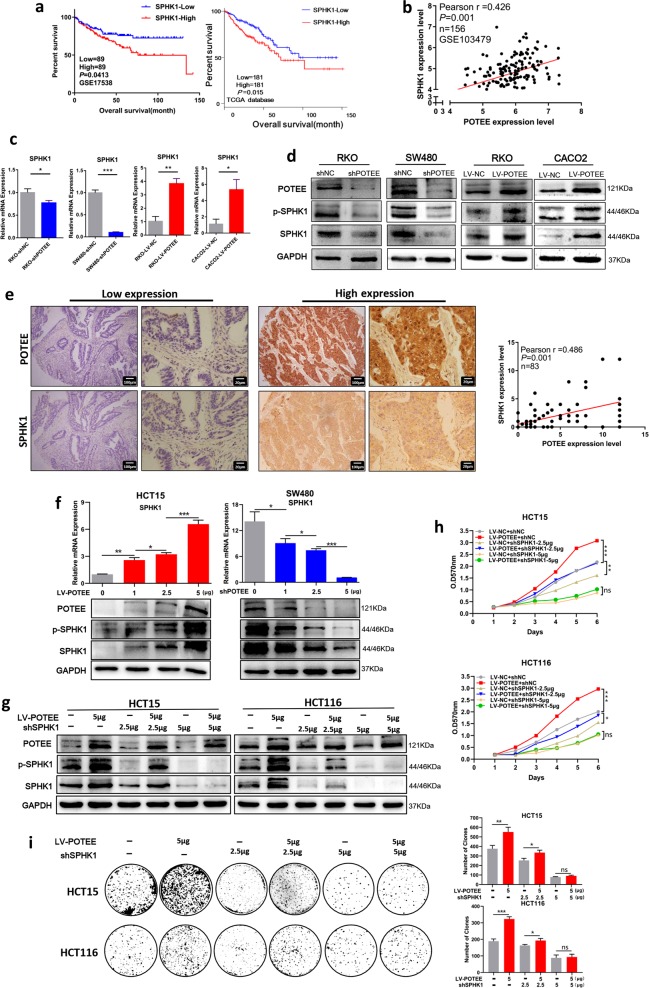
POTEE facilitates proliferation through upregulating SPHK1
As SPHK1 is the key sphingosine kinase that drives S1P signaling20,21, we then went on detecting the possible involvement of SPHK1 in POTEE-mediated molecular downstream. It’s reported that high expression level of SPHK1 promoted the development of tumor malignancy and significantly correlated with occurrence and poor prognosis of cancer patients16,20, so we firstly analyzed the correlation between SPHK1 expression and the survival of CRC patients. Results showed that increased SPHK1 mRNA level could predict poorer overall survival than those patients with relatively low SPHK1 expression in GSE17538 dataset with 178 CRC samples and in TCGA database with 362 colorectal cancer patients involved (Fig. 4a). We then analyzed the correlation of POTEE and SPHK1 in GSE103479 dataset, and results revealed that the mRNA expression of POTEE and SPHK1 were positively correlated (r = 0.426, P = 0.001) (Fig. 4b). To further verify SPHK1 expression under different POTEE status, we performed qRT-PCR and western blot analysis and found that both the mRNA and protein level of SPHK1 were significantly upregulated in CACO2 and RKO cells with POTEE-overexpression, while knockdown of POTEE repressed SPHK1 expression (Fig. 4c, d), and the protein level of phosphorylated SPHK1 showed the same change tendency according to different POTEE expression (Fig. 4d). In addition, we performed IHC staining with both anti-POTEE and anti-SPHK1 antibodies on our tissue microarrays with 83 CRC patients involved to measure the correlation between them, and results also revealed a significantly positive correlation between POTEE and SPHK1 on protein level (Fig. 4e).
Fig. 4. POTEE facilitates proliferation through up-regulating SPHK1.
a Kaplan–Meier survival curves depicting overall survival of patients with CRC grouped by SPHK1 mRNA expression levels. Data were deposited to GSE17538 dataset (left) and TCGA database (right). b Correlation between mRNA expression of POTEE and SPHK1 based on Pearson χ2 test, data was deposited to GSE103479 dataset. c qRT-PCR was performed to detect mRNA level of SPHK1 in indicated cell lines. Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test. d Total and phosphorylated levels of SPHK1 (p-SPHK1) were analyzed by western blotting in the cell lines with different POTEE expression. e. Identification of the correlation between protein levels of POTEE and SPHK1 in CRC tissues with IHC analysis. Statistics were measured by Pearson χ2 test. f qRT-PCR (up) and western blot (down) showing POTEE and SPHK1 expression in HCT15 and SW480 with different doses of overexpression and knockdown in POTEE. Results are shown as mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001 based on Student t-test. g Western blot detected indicated proteins with different doses of knockdown in POTEE. h, i MTT (h) and colony formation assays (i) in cells with different POTEE and SPHK1 expression. Statistics were measured by 2-way ANOVA or Student t, *P < .05, **P < .01, ***P < .001
In order to further determine the role of SPHK1 in the regulation of POTEE-mediated oncogenesis, we firstly detected endogenous expression of SPHK1 in 11 colorectal cancer cell lines by qRT-PCR and western blot analysis to analyze the endogenous consistency between POTEE and SPHK1, and results showed that the endogenous expression level of SPHK1 was finely accordant with POTEE in quite a part of these cell lines, and we additionally chose HCT15 and HCT116 cells in our following experiments for their moderate expression of both POTEE and SPHK1 (Supplementary Fig. 4a, b). To explore the role of SPHK1 in POTEE-mediated tumor promoting functions, we firstly transfected gradient increased doses of POTEE-expressing recombinant lentivirus or POTEE-targeting shRNA into HCT15 and SW480 cells, respectively. Results showed that gradient overexpression or knockdown of POTEE led to a progressive upregulation or loss of downstream SPHK1 expression, which firmly indicated the key role of POTEE in regulating SPHK1 in CRC cells (Fig. 4f). Then, we used POTEE-overexpressed cells to generate cells with different levels of SPHK1 knockdown, and results revealed that different doses of knockdown in SPHK1 could progressively block POTEE-mediated SPHK1 elevation (Fig. 4g; Supplementary Fig. 4c). To better elucidate the functional changes, we conducted MTT tests and colony formation assays to determine whether suppressed SPHK1 could reverse POTEE-mediated cell proliferation. Results revealed that low and moderate inhibition of SPHK1 was capable enough to partly rescue the proliferative ability of indicated cell lines, and CRC cells with POTEE overexpression was more sensitive to SPHK1 knockdown for significantly severer inhibition of cell viability than their negative controls, and the cell growth was extremely suppressed after extensive SPHK1 knockdown (Fig. 4h, i; Supplementary Fig. 4d, e). Hence, our results suggested SPHK1 was an oncogene and was indispensable for POTEE-mediated oncogenic functions in CRC cells.
p65 plays as a functional downstream molecular of POTEE/SPHK1 axis
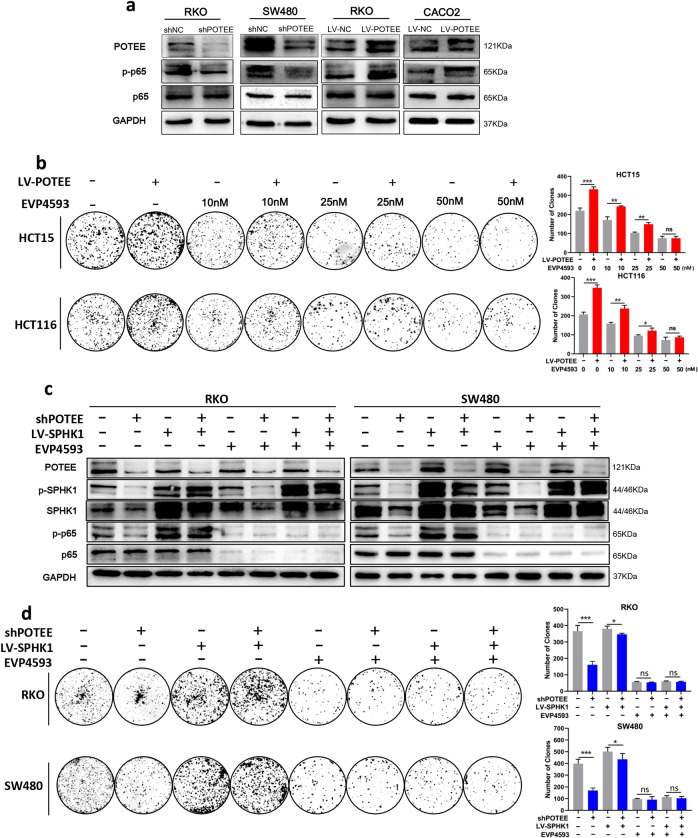
As previous studies have verified that SPHK1-mediated carcinogenesis via the activation of NF-κB pathway, and in light of our microarray results earlier (Fig. 3b), we went on detecting the activation of NF-κB signaling in CRC cells with different POTEE overexpression. Due to the expression of p65 and phosphoinositide-p65 (p-p65) are key mediators of NF-κB activation22,23, we firstly analyzed their expressions and found that although there showed no significant difference in p65 protein expression, the phosphorylated levels of p65 (p-p65) decreased in POTEE-knockdown cells and increased in POTEE-overexpressed cells (Fig. 5a).
Fig. 5. p65 plays as a functional downstream molecular of POTEE/SPHK1 axis.
a Total and phosphorylated levels of p65 (p-p65) were analyzed by western blotting in the cell lines with POTEE knockdown or POTEE overexpression. b Colony formation assays with indicated concentration of EVP4593 or DMSO. Statistics were measured by Student t, *P < .05, **P < .01, ***P < .001. c Western blot analysis in indicated cell lines with EVP4593 (0 nM, 10 nM, 25 nM, 50 nM, 48 h) or DMSO treatment. d Colony formation assays with indicated treatment. Statistics were measured by Student t, *P < .05, **P < .01, ***P < .001
We then tested whether NF-κB activation was required for POTEE-mediated proliferation of CRC cells. We used a pharmacologic approach to inhibit the activation of NF-κB with EVP4593 (Selleck Chemicals, America), which targeted p6524,25 at different concentration. Results showed EVP4593 could suppress POTEE-mediated cell proliferation in a dose-dependent manner, and although EVP4593 could inhibit the cells growth of both control and POTEE-overexpressed cells, stronger repression was seen in cells with POTEE-overexpression, similarly, high concentration of EVP4593 could completely blocked cell growth induced by POTEE overexpression (Fig. 5b; Supplementary Fig. 5b, c, d). Additionally, we used RKO and SW480 with POTEE knockdown to overexpress SPHK1 and found that SPHK1 overexpression could rescue the protein level of p-p65 in cell lines with POTEE knockdown, but high concentration of EVP4593 treatment could inhibited this upregulation. Likewise, knockdown SPHK1 was also capable of reducing p-p65 levels in POTEE-overexpressed cell lines (Fig. 5c; Supplementary Fig. 5e). What’s more, the results of colony formation assays suggested that gradient inhibition of p-p65 could progressively suppress cell proliferation promoted by POTEE/SPHK1 axis, and excessive inhibition eliminated the difference of cell number affected either by POTEE or SPHK1 (Fig. 5d). Collectively, these results implied that POTEE contributed to the proliferation of CRC cells through the SPHK1-NF-κB signaling axis.
Knockdown POTEE could decrease tumor growth in vivo
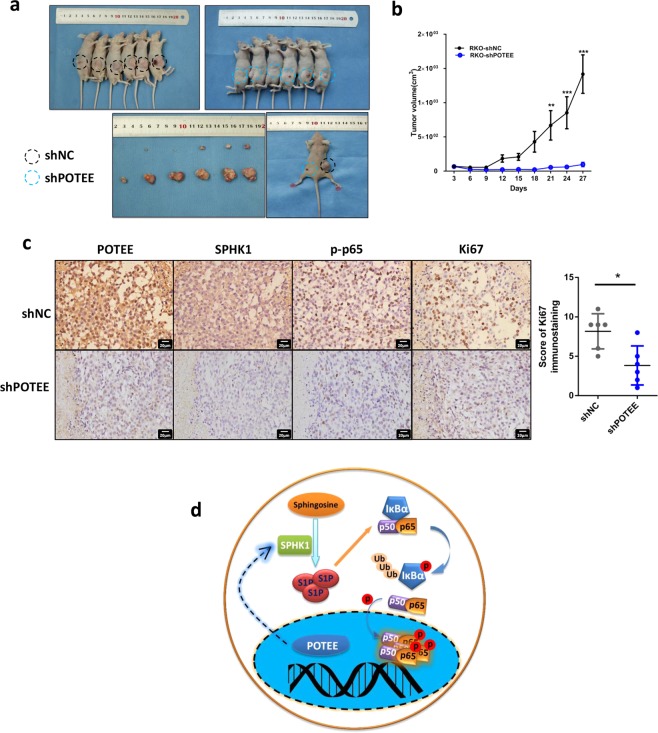
Considering our in vitro findings, we further tested the functional roles of POTEE in vivo with subcutaneous xenograft models. Results revealed that RKO cells with POTEE-knockdown suppressed tumor growth by 93.2% when compared to their negative control cells (Fig. 6a, b). POTEE silence significantly inhibited cell proliferation in the tumor sections as determined by Ki67 staining (Fig. 6c). Furthermore, knockdown of POTEE also led to decreased expression of SPHK1 and p-p65 in the tumors (Fig. 6c). These findings highlighted an essential role of POTEE/SPHK1/p65 signaling in promoting colorectal tumor growth (Fig. 6d).
Fig. 6. Knockdown POTEE could decrease tumor growth in vivo.
a Representative photographs showing the tumors in mice after the subcutaneous transplantation of RKO cells with POTEE knockdown or control cells. b Tumor growth curves measured at the indicated time points between POTEE knockdown and control groups. Statistics were measured by 2-way ANOVA, *P < .05, **P < .01, ***P < .001. c Representative IHC photographs showed positive relationship between POTEE, SPHK1, p-p65, Ki67 expression in tumor tissues (left). Results of Ki67 immunohistochemistry staining were evaluated by the staining scores (right). Results are shown as mean ± SD (n = 6). *P < .05 based on Paired t-test. d. Schematic depiction of the mechanisms underlying CRC proliferation via POTEE/SPHK1/p65 signaling
Discussion
POTE is a gene family and several studies have demonstrated that POTE genes were associated with a variety of human cancers8. POTEE was firstly detected in prostate cancer and its aberrant expressions was then identified in other cancer types6,7. Based on omics-based whole-genome transcriptome and whole-proteomic profiling, researchers observed that POTEE was closely related to breast cancer and might be involved in disease progression11. In Tripathi’s research, POTEE could interact with mTOR and Rictor (Rapamycin-insensitive companion of mTOR) proteins in tumor associated macrophages of prostate cancer, and then enhanced cell growth and proliferation through immune modulation10. Furthermore, recent findings indicated the roles of POTEF, one of homologous protein of POTEE, in the regulation of apoptosis in prostate cancer cells26. Despite the possible oncogenic roles of POTEE in different cancer types, its expression pattern, functions, mechanism largely remain obscure and need further study.
In this work, we conducted a more detailed investigation of the expression of POTEE and the relationships among POTEE expression and the prognosis and clinicopathological parameters of CRC patients. Specifically, the mRNA and protein expression of POTEE were significantly upregulated in human CRC tumor samples and cell lines. High POTEE level was positively associated with poor outcomes and malignant phenotypes of CRC patients. Our gain-of-function and loss-of-function experiments in vitro and in vivo clearly suggested a oncogenic role of POTEE in regulation of cell proliferation, migration and invasion, cell-cycle progression and apoptosis repression from both in vitro and in vivo evidence.
Since the molecular pathways associated with POTEE still remained obscure, we performed an mRNA microarray between POTEE-overexpressed RKO cells and control cells, and Sphingosine-1-phosphate (S1P) signaling was one of the most enriched pathways. S1P signaling is widely involved in many diseases, like inflammatory diseases and cancer progression, for its important and fundamental regulations of cell growth, apoptosis, and cell migration21,27,28. S1P is a bioactive type of sphingolipid, which is converted from ceramide and intracellular S1P was catalyzed mainly by the key sphingosine kinase isoforms, SPHK129. Previous studies have demonstrated that high expression of SPHK1 in tumors was associated with poor prognosis of pulmonary, gastric, pancreatic and colonic cancer patients30–33 and high expression of phosphorylated SPHK1 could promote cell growth and metastasis34. In our work, we consistent expression of SPHK1 with POTEE from both endogenous and exogenous evidence, and they are also positively correlated in the tumor samples of CRC patients. On functional rescue experiments, we further identified SPHK1 was an indispensable downstream of POTEE-mediated cell growth. However, the mechanism for transcription induction between POTEE and SPHK1 remain unknown. As some research suggested that loss of SPHK1 was critical to p53-dependent tumor suppression mechanism, and SPHK1 inhibition combined with p53 overexpression played an important role in inducing cell death35,36; however, in our work, the expression level of p53 seemed to be little affected by different POTEE expression (Supplementary Fig. 5a), which indicated POTEE-mediated SPHK1 changes was independent of p53 status.
NF-κB signaling pathway contains five structurally related proteins, which form homodimers and heterodimers to upregulate or suppress genes expression by binding to κB elements (target DNA sequences)37. The release of p65/p50 complex from the inhibitor complex requires the activation of classical NF-κB pathway and physiologically linked to the regulation of cellular growth and survival38–40. The regulation of cell growth depends on the phosphorylation status of the p65 NF-κB subunit22. It had been pointed out that p65 could regulate cell-cycle progression, cell apoptosis and cell growth in various cancer types41,42. It is reported that TNF receptor-associated factor 2 (TRAF2) binding to SPHK1 is required for TNF-α-induced nuclear translocation of p6543. And SPHK1 is also been identified as a target of miR-506, and knockdown miR-506 could resulted in upregulation of SPHK1 and followed by activation of NF-κB pathway in pancreatic cancer44. Combine with our microarray result, we wondered whether NF-κB signaling could act as a downstream effector of POTEE/SPHK1 axis. With this hypothesis, we used p65 inhibitor to verified that gradually suppressed p-p65 could progressively abrogated the effects of cell growth induced by POTEE overexpression and knockdown SPHK1 could also rescue the increase of p-p65 mediated by POTEE overexpression. Collectively, we speculated that POTEE could affect p65 activation by regulating SPHK1 expression.
Nevertheless, since researches on POTEE are extremely limited and the application of POTEE antibody could only be used for western blot and immunohistochemistry, so the precise interaction between POTEE and SPHK1 is still unknown. From our current data, we could only figure out that large amount of POTEE locate in cell nucleus and a small part of it lie in the cytoplasm, and POTEE could activate the mRNA and protein expression of SPHK1. In our hypothesis, POTEE may act as a transcription factor, or directly interact with the upstream signal molecule of SPHK1, or directly interact with SPHK1 for induction of phosphorylation, which might be more complicated and needs further investigation.
In conclusion, our present study sheds light on the oncogenic functions of POTEE in CRC progression. Mechanistically, our results also showed that POTEE might play a critical role in regulating the expression of SPHK1 and followed activation of NF-κB singling, which might act as a novel biomarker and a potential intervention of CRC patients.
Materials and methods
Patients and samples
The Institute Research Medical Ethics Committee of Nanfang Hospital (Guangzhou, China) granted approval for this study. Fresh and formalin-fixed tissue samples from patients with CRC were collected from the Department of General Surgery, Nanfang Hospital, affiliated to Southern Medical University. 20 pairs of CRC and non-tumor-adjacent tissue samples from the patients who received curative surgery in the Nanfang Hospital were used for quantitative real-time polymerase chain reaction (qRT-PCR) or western blot analysis and 81 CRC surgical specimens were used for immunohistochemistry (IHC) and prognosis analysis. In addition, a tissue microarray (TMA) contained tumor resections from 83 CRC patients were used to analyze the correlation of protein expression between POTEE and SPHK1.
Western blot analysis
Cells or tissues were lysed with RIPA buffer (Amresco, America) and total proteins were separated by 10% SDS-PAGE (Amresco, America) and transferred to PVDF membranes. Membranes were incubated with respective primary antibodies overnight at 4 °C and washed three times. Then, these blots were incubated with HRP-conjugated secondary antibody (Cell Signaling Technology #7074, America) for 1 h at room temperature. After washed three times the blots were visualized by enhanced chemiluminescence (FDbio-pico ECL, China). Primary antibodies used for western blot were listed as follows: POTEE (Biorbyt #orb312653, Britain), SPHK1 (Proteintech #10670-1-AP, America), p-SPHK1 (Proteintech #19561-1-AP, America), p65 (Cell Signaling Technology #8402, America), p-p65 (Cell Signaling Technology #3033, America), GAPDH (Cell Signaling Technology #5170, America).
Quantitative real-time PCR (qRT-PCR)
Total RNAs from cells or tissues were extracted using TRIzol reagent (TaKaRa, Japan) and reverse transcribed into cDNA using a PrimeScript RT-PCR Kit (TaKaRa, Japan) according to the manufacturer’s instructions. Briefly, we analyzed the expressions of mRNA using SYBR™ Premix Ex Taq™ (TaKaRa, Japan) under the LightCycler 96 Detection System (Roche) and GAPDH was used for normalization. Primer sequences for quantitative real-time PCR were listed in Table 2.
Table 2.
Primer sequences for qRT-PCR in this study
| GAPDH-FP | 3′-TGACTTCAACAGCGACACCCA-5′ | GAPDH-RP | 3′-CACCCTGTTGCTGTAGCCAAA-5′ |
| POTEE-FP | 3′-CTGCACTACGCTATCTATAA-5′ | POTEE-RP | 3′-TCAAGTAGAAGGCTGACTAT-5′ |
| TGFA-FP | 3′-AGGTCCGAAAACACTGTGAGT-5′ | TGFA-RP | 3′-AGCAAGCGGTTCTTCCCTTC-5′ |
| DDR1-FP | 3′-CCGACTGGTTCGCTTCTACC-5′ | DDR1-RP | 3′-CGGTGTAAGACAGGAGTCCATC-5′ |
| GNAI1-FP | 3′-CTGCGCTGGATGCTTGATTTT-5′ | GNAI1-RP | 3′-GATTCACCAGCACCTGGCAA-5′ |
| SPHK1-FP | 3′-GCTGCGAAGTTGAGCGAAAA-5′ | SPHK1-RP | 3′-GGCTGGACCCAGTCGGCG-5′ |
| BMP2-FP | 3′-ACTACCAGAAACGAGTGGGAA-5′ | BMP2-RP | 3′-GCATCTGTTCTCGGAAAACCT-5′ |
| BMPR2-FP | 3′-GACAGGAGACCGTAAACAAGG-5′ | BMPR2-RP | 3′-CCATATCGACCTCGGCCAATC-5′ |
| CSNK2A2-FP | 3′-AAAAGCTGCGACTGATAGATTGG-5′ | CSNK2A2 -RP | 3′-GAGGCTACACGAACATTGTACTC-5′ |
| RND2-FP | 3′-AAGATCGTGGTGGTGGGAG-5′ | RND2-RP | 3′-CATAACTCCCGGGATAGGCG-5′ |
| IRS2-FP | 3′-GGCCACCATCGTGAAAGAGTG-5′ | IRS2-RP | 3′-GTGACCTTGCCTTGTTGGTG-5′ |
| DIRAS3-FP | 3′-AGCTTATTCCAACAGATGCCA-5′ | DIRAS3-RP | 3′-AACCAACTGGACACCCCAAG-5′ |
| PLCB4-FP | 3′-ACATGCCTCAAGAAACACTGGA-5′ | PLCB4-RP | 3′-GCTGTGGGCTCAATTTCATCA-5′ |
| BMP4-FP | 3′-ATGATTCCTGGTAACCGAATGC-5′ | BMP4-RP | 3′-CCCCGTCTCAGGTATCAAACT-5′ |
| BCL10-FP | 3′-GCAGTTGTGAACCTTTTCCAGA-5′ | BCL10 -RP | 3′-TGGATGCCCTCAGTTTTTCAG-5′ |
| ROCK1-FP | 3′-GGTCTAAATAGGTCGTCGTACAA-5′ | ROCK1-RP | 3′-TCCGTACCATGCTACCATATGT-5′ |
Immunohistochemistry (IHC)
Slides were stained using the MaxVision TM2 Kit (MXB® Biotechnologies, China) according to the manufacturer’s instructions. At least three individual fields (20×) were chosen to estimate the score of each slide. Immunostaining intensity was divided into 4 grades: 0, negative; 1, weak; 2, moderate; 3, strong. As well as the proportion of staining-positive cells: 0, < 5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; 4, >75%. Each slide was scored by the cross product of the value of proportion of staining-positive cells and the value of immunostaining intensity. Two independent and experienced pathologists were invited to assess and confirm the results. Primary antibodies were POTEE (Biorbyt #orb312653, Britain), SPHK1 (Proteintech #10670-1-AP, America), p-p65 (Cell Signaling Technology #3033, America), Ki67 (Cell Signaling Technology #9449, America).
Cell culture and stable cell lines construction
The CRC cell lines RKO, SW480, and CACO2 were purchased from the American Type Culture Collection and maintained at 37°C in an atmosphere containing 5% CO2. The cells were cultured in 1640 medium supplemented with 10% fetal bovine serum (Gbico, America). Lentiviral vectors were designed and constructed by GENECHEM Biotech at Shanghai, China (http://genechem.bioon.com.cn/). Flag-tagged POTEE-overexpressed vectors (LV-POTEE) and control vectors (LV-NC) were transfected into RKO and CACO2 cells, while POTEE shRNA (shPOTEE) and control short hairpin RNA (shNC) were transfected into RKO and SW480 cells to generate POTEE-knockdown cells. Moreover, SPHK1 shRNA (shSPHK1) and control short hairpin RNA (shRNA) were transfected into LV-NC or LV-POTEE cells above mentioned. Transfection procedures were performed according to the manufacturer’s instruction.
MTT and colony formation assay
For MTT assays, stably transfected cells (1000 cells per well) were cultivated on 96-well plates and cell proliferation were detected for 6 days with MTT (Beyotime, China) at 570 nm. For the colony formation assays, cells (500 cells per well) were cultivated in six-well plates and maintained with 1640 medium containing 10% FCS at 5% CO2, 37 °C for 8–12 days. At the end of experiments, colonies formed were washed with phosphate buffer (PBS), fixed in methanol and stained with 0.1% crystal violet. Colonies containing more than 50 cells for each well were counted (in triplicate).
Cell migration and invasion assay
For scratch test, cells was cultural about 70–80% confluence in six-well plate. Scratches were made by 20 µl pipette tip. Then the cells were cultural under serum-starved condition. Images were taken at 0 h, 24 h, and 48 h to evaluate the wound-healing. Eight micrometer pore insets transwell chambers were used for migration and invasion assay. For migration assay, 1 × 105 cells were plated on uncoated insets with serum-starved condition. For invasion assay, uncoated insets were filled with 100 µl 1:20 diluted matrigel and 1 × 106 cells were plated on the above insets with serum-starved condition. The lower chamber contained 600 μl culture medium containing 20% FBS. After 36–48 h incubation, invaded cells were fixed with methanol and stained with 5% crystal violet.
In vivo subcutaneous xenograft models
All nude mice were purchased from Laboratory Animal Center of Southern Medical University and were humanly treated under the guidelines of the Treatment Committee of Southern Medical University. Stable transfection cells with POTEE knockdown and control cells (5 × 107) were transplanted subcutaneously into the bilateral flanks of 3–4 weeks old nude mice. Tumor diameters were measured every 3 days, and the formula Tumor Volume (mm3) = (length × width2)/2 was used to evaluated the tumor volumes. Animals were sacrificed 27 days after injection and the tumor tissues were fixed in formalin for IHC analysis.
Flow cytometry
For cell-cycle analysis, cells under detection were collected and then fixed in 70% ethanol overnight. Next day, the cells were washed with PBS and incubated with RNase A solution and PI staining. The detailed procedures were performed according to the manufacturer’s instructions (KeyGEN BioTECH, China). For apoptosis assay, stable transfected cells under detection were collected after 5-Fu treatment for 36–48 h. Cells were then collected and apoptosis was then detected with Annexin V-FITC/PI cell apoptosis detection kit (KeyGEN BioTECH, China).
Microarray analysis
POTEE-overexpressed RKO cells and negative control cells were used for gene expression profiling. Intactness of the samples was assessed by the Agilent 2100 bioanalyzer (Agilent, America). Identification of differentially expressed genes was performed by Shanghai Genechem Co., LTD (Shanghai, China). Microarray analysis of mRNA profiles using Gene Matrix platforms (http://gcloud.taogene.com) based on IPA analysis.
Statistical analysis
The results were shown as the mean ± SD and P < 0.05 was considered to be statistically significant. The differences in survival rates were assessed by Kaplan–Meier analysis. Pearson χ2 test was used to analyze the correlation between the expression of POTEE and SPHK1. All of the statistical analyses were carried out with SPSS software (version 16.0) or GraphPad Prism (version 6.0).
Supplementary information
Acknowledgements
This work was supported by the Natural Science Foundation of Guangdong Province, China (2017A030313588, 2018A030313547, 2016A030310387, 2017A030313154); Science and Technology Planning Project of Guangdong Province (2014A020212629); Foundation of President of The Nanfang Hospital (No. 2015C020); Medical Scientific Research Foundation of Guangdong Province, China (A2016237); Science and Technology Planning Project of Guangzhou, China (No. 201707010214); and National Natural Science Foundation of China (No. 31601023).
Author contributions
Study concept and design: Z.S., X.F., Y.F., Y.D., H.D. Development of methodology: Z.S., X.F., Y.F. Draft of the manuscript and data acquisition: Z.S., X.F., Y.F., Y.L., Z.Z., Z.L., M.L. Analysis and interpretation of data: Z.S., X.F., Y.F., Y.D., H.D. Administrative, technical, or material support: Z.S., G.L., Y.D., H.D. Study funding and supervision: Z.S., Y.D., H.D.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by N. Barlev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhiyong Shen, Xiaochuang Feng, Yuan Fang.
Contributor Information
Yi Ding, Phone: +86-20-61642136, Email: dingyi197980@126.com.
Haijun Deng, Phone: +86-20-61642136, Email: navyd999@163.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-2046-7).
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sung JJY, et al. An updated Asia Pacific Consensus Recommendations on colorectal cancer screening. Gut. 2014;64:121–132. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 4.Rebecca L. Siegel M, et al. Colorectal Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang K, et al. CDK5 functions as a tumor promoter in human colorectal cancer via modulating the ERK5–AP-1 axis. Cell Death Dis. 2016;7:e2415. doi: 10.1038/cddis.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bera TK, et al. Five POTE paralogs and their splice variants are expressed in human prostate and encode proteins of different lengths. Gene. 2004;337:45–53. doi: 10.1016/j.gene.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Tapan K. Bera DBZN, Byungkook Lee AIP. POTE, a highly homologous gene family located on numerous chromosomes and expressed in prostate, ovary, testis, placenta, and prostate cancer. Med. Sci. 2002;26:16975–16980. doi: 10.1073/pnas.262655399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bera TK, et al. POTE paralogs are induced and differentially expressed in many cancers. Cancer Res. 2006;66:52–56. doi: 10.1158/0008-5472.CAN-05-3014. [DOI] [PubMed] [Google Scholar]
- 9.Tapan K. Bera XLMY, Yoonsoo Hahn BLAI. A model for obesity and gigantism due to disruption of the Ankrd26gene. PNAS. 2007;1:270–275. doi: 10.1073/pnas.0710978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vekariya, U., Rawat, K., Saxena, R. & Tripathi, R. K. Identification of MΦ specific POTEE expression: Its role in mTORC2 activation via protein-protein interaction in TAMs. Cell Immunol.335, 30–40 (2018). [DOI] [PubMed]
- 11.CINE N, et al. Identification of ApoA1, HPX and POTEE genes by omic analysis in breast cancer. Oncol. Rep. 2014;32:1078–1086. doi: 10.3892/or.2014.3277. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, et al. Serum levels of the cancer-testis antigen POTEE and its clinical significance in non-small-cell lung cancer. PLoS One. 2015;10:e122792. doi: 10.1371/journal.pone.0122792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barger, C. J. et al. Expression of the POTE gene family in human ovarian cancer. Sci. Rep.-UK8, 17136 (2018). [DOI] [PMC free article] [PubMed]
- 14.Tang, W. et al. The p300/YY1/miR-500a-5p/HDAC2 signalling axis regulates cell proliferation in human colorectal cancer. Nat, Commun.10, 663 (2019). [DOI] [PMC free article] [PubMed]
- 15.Fang, Y. et al. CD36 inhibits β-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4 to repress colorectal tumorigenesis. Nat. Commun.10, 3981 (2019). [DOI] [PMC free article] [PubMed]
- 16.Zheng X, et al. The sphingosine kinase-1/sphingosine-1-phosphate axis in cancer: potential target for anticancer therapy. Pharmacol. Therapeut. 2019;195:85–99. doi: 10.1016/j.pharmthera.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Elbakyan A. Driver of NF κ B and STAT3 persistent activation in chronic intestinal inflammation and colitis-associated cancer. Arianne L Theiss. 2013;2:e24150. doi: 10.4161/jkst.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 2017;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang J, et al. Sphingosine-1-phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis-associated cancer. Cancer Cell. 2013;23:107–120. doi: 10.1016/j.ccr.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- 21.Kihara A, Mitsutake S, Mizutani Y, Igarashi Y. Metabolism and biological functions of two phosphorylated sphingolipids, sphingosine 1-phosphate and ceramide 1-phosphate. Prog. LIPID Res. 2007;46:126–144. doi: 10.1016/j.plipres.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez H, Davies AM. Regulation of neural process growth, elaboration and structural plasticity by NF-κB. Trends Neurosci. 2011;34:316–325. doi: 10.1016/j.tins.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YXGKF. Citrobacter rodentium-induced NF-kappaB activation in hyperproliferating colonic epithelia: role ofp65 (Ser536 phosphorylation. Br. J. Pharmacol. 2006;148:814–824. doi: 10.1038/sj.bjp.0706784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, et al. GSDMB promotes non-canonical pyroptosis by enhancing caspase-4 activity. J. Mol. Cell Biol. 2019;11:496–508. doi: 10.1093/jmcb/mjy056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X, et al. The outer membrane protein Tp92 ofTreponema pallidum induces human mononuclear cell death and IL-8 secretion. J. Cell Mol. Med. 2018;22:6039–6054. doi: 10.1111/jcmm.13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misawa A, et al. Androgen-induced lncRNA POTEF-AS1 regulates apoptosis-related pathway to facilitate cell survival in prostate cancer cells. Cancer Sci. 2017;108:373–379. doi: 10.1111/cas.13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hisano, Y. & Hla, T. Bioactive lysolipids in cancer and angiogenesis. Pharmacol. Therapeut.193, 91–98 (2018). [DOI] [PMC free article] [PubMed]
- 28.Brinkmann V. Sphingosine 1-phosphate receptors in health and disease: mechanistic insights from gene deletion studies and reverse pharmacology. Pharmacol. Therapeut. 2007;11.5:84–105. doi: 10.1016/j.pharmthera.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Xiong Y, Yang P, Proia RL, Hla T. Erythrocyte-derived sphingosine 1-phosphate is essential for vascular development. J. Clin. Invest. 2014;124:4823–4828. doi: 10.1172/JCI77685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pyne S, Adams DR, Pyne NJ. Sphingosine 1-phosphate and sphingosine kinases in health and disease: Recent advances. Prog. Lipid Res. 2016;62:93–106. doi: 10.1016/j.plipres.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Li WYCXJ. Progression and poor survival of patients sphingosine kinase 1 is associated with gastric cancer. Clin. Cancer Res. 2009;4:1393–1399. doi: 10.1158/1078-0432.CCR-08-1158. [DOI] [PubMed] [Google Scholar]
- 32.Johnson KR, et al. Immunohistochemical distribution of sphingosine kinase 1 in normal and tumor lung tissue. J. Histochem. Cytochem. 2005;53:1159–1166. doi: 10.1369/jhc.4A6606.2005. [DOI] [PubMed] [Google Scholar]
- 33.Shi W, et al. Resveratrol inhibits monocrotaline-induced pulmonary arterial remodeling by suppression of SphK1-mediated NF-κB activation. Life Sci. 2018;210:140–149. doi: 10.1016/j.lfs.2018.08.071. [DOI] [PubMed] [Google Scholar]
- 34.Speirs MMP, et al. Imbalanced sphingolipid signaling is maintained as a core proponent of a cancerous phenotype in spite of metabolic pressure and epigenetic drift. Oncotarget. 2019;10:449–479. doi: 10.18632/oncotarget.26533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heffernan-Stroud LA, et al. Defining a role for sphingosine kinase 1 in p53-dependent tumors. Oncogene. 2012;31:1166–1175. doi: 10.1038/onc.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lima S, et al. TP53 is required for BECN1- and ATG5-dependent cell death induced by sphingosine kinase 1 inhibition. Autophagy. 2018;14:942–957. doi: 10.1080/15548627.2018.1429875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 38.Chen, T. et al. PKCε phosphorylates MIIP and promotes colorectal cancer metastasis through inhibition of RelA deacetylation. Nat. Commun.8, 939 (2017). [DOI] [PMC free article] [PubMed]
- 39.Karin, M. How NF-kB is activated: the role of the IkB kinase (IKK) complex. Oncogene18, 6867–6874 (1999). [DOI] [PubMed]
- 40.Wang, Y. et al. STX2 promotes colorectal cancer metastasis through a positive feedback loop that activates the NF-κB pathway. Cell Death Dis.9, 664 (2018). [DOI] [PMC free article] [PubMed]
- 41.Handschick K, et al. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-κB-dependent gene expression. Mol. Cell. 2014;53:193–208. doi: 10.1016/j.molcel.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz ML, Kracht M. Cyclin-dependent kinases as coregulators of inflammatory gene expression. Trends Pharmacol. Sci. 2016;37:101–113. doi: 10.1016/j.tips.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez SE, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, et al. Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene. 2016;35:5501–5514. doi: 10.1038/onc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.