Abstract
Antimicrobial resistance is an increasing problem worldwide, and Salmonella spp. resistance to quinolone was classified by WHO in the high priority list. Recent studies in Europe and in the US reported the presence of small plasmids carrying quinolone resistance in Enterobacteriaceae isolated from poultry and poultry products. The aims of this study were to identify and characterize plasmid-mediated quinolone resistance in Salmonella spp. and to investigate transduction as a possible mechanism associated to its dissemination. First, we assessed resistance to nalidixic acid and/or ciprofloxacin in 64 Salmonella spp. and detected resistance in eight of them. Genomic analyses determined that six isolates of different serotypes and sources carried an identical 2.7-kb plasmid containing the gene qnrB19 which confers quinolone resistance. The plasmid detected also has high identity with plasmids reported in the US, Europe, and South America. The presence of similar plasmids was later surveyed by PCR in a local Salmonella collection (n = 113) obtained from diverse sources: food (eggs), wild and domestic animals (pigs, horse, chicken), and human clinical cases. qnrB19-carrying plasmids were found in 8/113 Salmonella tested strains. A bioinformatics analysis including Chilean and previously described plasmids revealed over 95.0% of nucleotide identity among all the sequences obtained in this study. Furthermore, we found that a qnrB19-carrying plasmid can be transferred between Salmonella of different serotypes through a P22-mediated transduction. Altogether our results demonstrate that plasmid-mediated quinolone resistance (PMQR) is widespread in Salmonella enterica of different serotypes isolated from human clinical samples, wild and domestic animals, and food in Chile and suggest that transduction could be a plausible mechanism for its dissemination. The occurrence of these antimicrobial resistance elements in Salmonella in a widespread area is of public health and food safety concern, and it indicates the need for increased surveillance for the presence of these plasmids in Salmonella strains and to assess their actual impact in the rise and spread of quinolone resistance.
Keywords: antimicrobial resistance, foodborne diseases, plasmid, quinolones, qnrB19, Salmonella spp., Chile, plasmid-mediated quinolone resistance
Introduction
Antimicrobial resistance is an increasing worldwide problem and a global concern that involves significant health and economic burden (Naylor et al., 2016). Latin America is not an exception: the Pan American Health Organization (PAHO) declared that the spread of pathogens carrying antimicrobial traits challenges both, disease control and disease treatment, and it significantly impacts public health in the region [Pan American Health Organization (PAHO), 2014].
The emergence of antimicrobial resistance in microorganisms occurs naturally; however, the increasing use of antimicrobials promotes the natural selection of resistant bacteria (Holmes et al., 2016). Quinolones are one of the groups of antimicrobials used in humans for the treatment of bacterial infections, and they are also widely used in animal production (Millanao et al., 2011; Singer et al., 2016). Quinolone resistance in enteric pathogens has been described in Latin America [González and Araque, 2013; Pan American Health Organization (PAHO), 2014], and an increase in Salmonella spp. resistance to quinolones has been recently reported in Chile [Instituto de Salud Pública de Chile (ISP), 2015] and in the United States (Medalla et al., 2013; Karp et al., 2018).
Antimicrobial resistance to quinolones can be the result of target mutations reducing the drug’s binding to the enzymes gyrase or topoisomerase IV (Hooper and Jacoby, 2016). Additionally, genes harbored in plasmids—such as qnr genes—codify for proteins that protect the target enzymes from quinolone action in the phenomena known as plasmid-mediated quinolone resistance (PMQR) (Hooper and Jacoby, 2016). The presence of antimicrobial resistance genes in plasmids is of great concern from a public health perspective because they can easily spread from one bacterium to another through horizontal gene transfer (Rozwandowicz et al., 2018). Three small plasmids carrying the gene qnrB have been described since 2010 in South America (Pallecchi et al., 2009; Tran et al., 2012; Cordeiro et al., 2016). The plasmids were obtained from bacteria isolated in Colombia, Peru, and Argentina, and their sizes ranged from 2,699 to 2,750 bp (Karczmarczyk et al., 2010; Pallecchi et al., 2010; Tran et al., 2012). Moreover, some of them can be transferred by conjugation (Andres et al., 2013). Recently, similar plasmids have also been reported in Europe and North America in Salmonella isolated from poultry (Fiegen et al., 2017; Tyson et al., 2017).
The aims of this study were to investigate the presence and characteristics of plasmid-mediated quinolone resistance in Salmonella spp. of different serotypes and sources, and to investigate whether transduction could be a potential mechanism associated to its dissemination.
Materials and Methods
Strains
Salmonella enterica (S. enterica; n = 64) from different serotypes were isolated from wild birds (n = 28), human clinical cases (n = 23), eggs (n = 9), and sea lions (n = 4). Isolates were collected between 2009 and 2012 from different locations along Chile. Isolate’s information, including genomic sequences, was reported in previous publications (Toro et al., 2015, 2016) (Supplementary Table S1). Additionally, 113 S. enterica strains from our historic collection were recovered to survey for the presence of qnrB19-carrying plasmids through PCR as described below. These S. enterica belong to different serotypes and were originally isolated from pigs (n = 9), horses and their environment (n = 25), cattle (n = 3), poultry and poultry farm environments (n = 29), wild birds (n = 18), wild reptiles (n = 17), and Chilean mouse opossum (n = 12) (Supplementary Table S2). Strains were recovered in trypticase soy agar (TSA) agar (BD, Franklin Lakes, NJ) and after an overnight incubation, DNA was purified with the Qiagen DNeasy Blood and Tissue kit (Qiagen, Valencia, CA).
In vitro Determination of Quinolone Resistance
Salmonella isolates (n = 64) were tested for antimicrobial susceptibility against the quinolones nalidixic acid (NAL) and ciprofloxacin (CIP), and to other 12 drugs included in the NARMS program at the US FDA Center for Veterinary Medicine (CVM) following their standard protocols (Karp et al., 2017). In brief, minimum inhibitory concentrations (MICs) were determined by the microdilution method through the Sensititre automated microbial susceptibility system (Thermo Fisher Scientific, Waltham, MA) at the Laboratory of the Center for Veterinary Medicine, U.S. Food and Drug Administration. Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and Pseudomonas aeruginosa ATCC 27853 were used as controls for antimicrobial MIC determinations. The results were interpreted according to the Clinical and Laboratory Standard Institute MIC standards [Clinical Laboratory Standards Institute (CLSI), 2015].
In silico Determination of Antimicrobial Resistance
Genomic data for Salmonella (n = 64) isolates was previously reported (Toro et al., 2015, 2016). In this study, we investigated the presence of acquired antimicrobial resistance genes and known chromosomal point mutations linked to antimicrobial resistance in quinolone-resistant strains using the ResFinder 3.0 server [Center for Genomic Epidemiology (CGE), Technical University of Denmark, Lyngby, Denmark, https://cge.cbs.dtu.dk/services/ResFinder/] (Zankari et al., 2012). Gene identification threshold was set at 60% identity and 60% minimum length.
Plasmid Sequence Study
Nucleotide sequences of contigs containing quinolone antimicrobial resistance genes—as identified with ResFinder (Supplementary Table S5)—were used to search for identities at the NCBI database with the BLAST algorithm using the highly similar sequences setting (megablast). Previously reported plasmids with high identity values (n = 6) were found and used for genetic comparison (Supplementary Table S3). The plasmid set was compared by aligning their nucleotide sequences in Geneious Prime 2019.1.1 (Biomatters, New Zealand) with the ClustalW (Larkin et al., 2007) plug-in with default parameters. A phylogenetic reconstruction was crafted from the alignment with the RAxML plug-in using the “Rapid bootstrapping and search for the best scoring Maximum-likelihood tree” option with 1,000 replicates (Stamatakis et al., 2008), and a consensus tree was obtained.
Screening Study for Small Plasmids Carrying the qnrB19 Gene
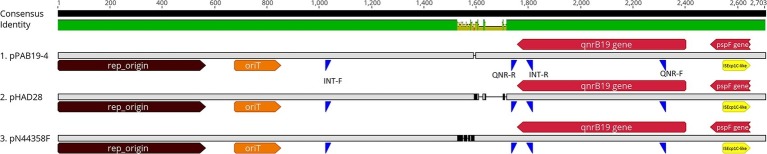
We determined the presence of plasmids similar to pPAB19-4 in 113 different Salmonella isolates from our historic collection through PCR. For primer design, three similar but distinct plasmids were aligned (i.e., pPAB19-4, pHAD28, and pN44358F; Supplementary Figure S1, Supplementary Table S3) with ClustalW in Geneious using default parameters, and potential target regions were identified (Figure 1). Finally, two sets of primers were designed: the first set of primers amplified a 589-bp region from the 5′ end of the qnrB19 gene to the immediate intergenic region (QNR-F: 5′ ACTGCGATTTTTCAGGTGCC 3′; QNR-R: 5’CATCTCCCGGTGTAAACGCT 3′), and the second set of primers amplified a variable-size plasmid backbone region (704–790 bp) from the distant intergenic region to the 3’end of the qnrB19 gene (Int-F 5′ CTGACAAACTTGACGCCTGC 3′; Int-R5′ GACAGCTACCAGGCATCGTT 3′) (Figure 1). GoTaq DNA Polymerase (Promega, San Luis Obispo, CA) was used for each PCR, and the following conditions were used: initial denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, extension of 72°C for 1 min, and a final extension of 72°C for 7 min. PCRs were amplified using an Axygen MaxyGene II Thermal Cycler (Corning, Corning, NY).
Figure 1.
Linear representation of plasmids carrying the qnrB19 gene. Plasmids pPAB19-4, pHAD28, and pN44358F were selected to represent different clusters resulting from the phylogenetic analysis (Supplementary Figure S1). Alignment was crafted with the ClustalW plug-in in Geneious Prime 2019 using default parameters. Consensus identity bar: green represents 100% identity among all sequences, green-brown indicates 30 to <100% identity, and red shows <30% identity. Genomic features are represented by different colors; brown: plasmid origin of replication (rep_origin; nt 1-564); orange: Origen of transfer (oriT; nt 677-851); red: genes qnrB-19 (nt 2,402-1,758) and gene pspF (nt2,647-2,496); yellow: ISEcp1C-like insertion sequence (nt 2,512-2,409). Primer alignment sites are represented in blue: INT-F and INT-R represent primers for the intergenic region. QNR-F and QNR-R represent primers for the qnrB-19 gene region.
Genomic Comparison of Small Plasmids Carrying the qnrB19 Gene
Newly identified qnrB-19-carrying isolates (n = 4/113) were submitted for whole genome sequencing (WGS). WGS of two of these isolates (DR-021 with number CFSAN035154 and DR-022 with number CFSAN035155) was described in a previous publication (Toro et al., 2018). The other two isolates (i.e., DR-039 and DR-040; acc. Numbers: SAMN11569606 and SAMN11569607, respectively) were sequenced at MicrobesNG at the School of Biosciences, University of Birmingham (Birmingham, United Kingdom) (Supplementary Table S2). Libraries were created with the nextera-XT library prep kit, and sequencing was performed in an Illumina HiSeq plaform using 250-bp paired end protocol to a minimum coverage of 30x. Genomes were de novo assembled from raw reads with SPAdes version 3.1.0 (Bankevich et al., 2012), and annotation was done at the RAST annotation server (Aziz et al., 2008). Plasmid sequences were obtained from assembled genomes using the “map to reference” utility in Geneious Prime, and pPAB19-4 sequence (acc. Nbr: JN995611.1) was selected as a reference. Later, a comprehensive comparative analysis including all qnrB19-carrying plasmids was run in Geneious Prime 2019.1.1 (Biomatters, New Zealand) with the ClustalW (Larkin et al., 2007) and with MAUVE (Darling et al., 2004) plug-ins using default parameters. Additionally, plasmid sequences were screened for phage DNA using PHASTER 2.7 (Arndt et al., 2016), and we also searched for type IV secretion system (T4SS) elements for all 12 genomes containing pPAB-19-like Chilean plasmids using ConjScan in the Pasteur Galaxy platform v1.0.2 (Abby et al., 2016).
Plasmid Transduction Assays
Transduction experiments were performed according to the protocol previously described by Maloy et al. (1996). Salmonella Heidelberg SAL4674 (CFSAN024772)—a qnrB19-plasmid carrier strain sequenced in this study—was used as donor, and phage P22HT (high transduction frequency) was used as transducing phage. Briefly, the donor was grown overnight in Luria Bertani broth (LB) broth (Difco; Detroit, MI) supplemented with 15 μg/ml of NAL (Sigma Chemical Co, Saint Louis, MO). Then, 200 μl of the Salmonella culture was mixed with 1 ml of P22 broth (LB broth containing 0.2 mg/ml MgSO4 × 7H2O, 2 mg/ml citric acid, 13.1 mg/ml K2HPO4 × 3H2O, 3.5 mg/ml NaNH4HPO4 × 4H2O, 2 mg/ml glucose, and 100 μl of P22 phage lysate), and the mixture was incubated overnight at 37°C with agitation. To obtain the phage lysate, the culture was centrifuged at 13,000 rpm for 2 min, and 100 μl of chloroform was added to the supernatant. Phage lysates were kept at 4°C until its use. For transduction experiments, Salmonella Typhimurium 14,028 s marked with a chloramphenicol resistance cassette in the phoN gene (recipient strain) was grown overnight at 37°C with agitation, and 200 μl of this culture was mixed with 20 μl of the P22 phage lysate obtained from the donor strain. Then, the mixture was incubated at room temperature for 15 min to allow phage adsorption, and 1 ml of LB broth was added. The mixture was incubated at 37°C for 60 min to allow phenotypic expression of the antibiotic marker. Later, the mixture was centrifuged at 13,000 rpm for 2 min, and the supernatant was discarded. The cell pellet was suspended in 1 ml of LB broth, and serial 10-fold dilutions were spread on LB agar plates containing NAL (15 μg/ml) and chloramphenicol (20 μg/ml) for determination of transductants. Negative controls, i.e., experiments run without the phage or bacteria, were included as described by Maloy et al. (1996). The transduction frequency was calculated as the ratio of the number of transductants (CFU) obtained in the receptor strain to the number of plaque-forming units (PFUs) in the transduction mixture of three independent experiments. The plasmid transduction to the recipient strain was verified by PCR amplification with the same two sets of primers mentioned in the previous section.
Results
In vitro Determination of Antimicrobial Resistance
The antimicrobial susceptibility was evaluated for 64 Salmonella strains. In vitro tests showed that eight Salmonella spp. had reduced susceptibility to quinolones: of those, two strains (SAL4629 and SAL4630) were resistant to CIP (MIC ≥ 1.0 μg/ml), and five (SAL 4629, SAL4630, SAL4675, SAL4676, and SAL4679) were resistant to NAL (MIC ≥32 μg/ml) (Table 1). In addition, strain SAL4630, isolated from eggs, was also resistant to tetracycline, trimethoprim-sulfamethoxazole and sulfidoxazole. Antimicrobial susceptibility results for strains SAL4629 and SAL4630 were reported in Toro et al., 2016.
Table 1.
Minimum inhibitory concentration (MIC) to quinolones of Salmonella strains isolated in Chile.
| Strain | Acc. number | Serotype | Sequence type | Source | MIC CIP (μg/ml) | MIC NAL (μg/ml) | Antimicrobial resistance genes |
|---|---|---|---|---|---|---|---|
| SAL4629¥ | LILV00000000 | Enteritidis | 11 | Poultry | 1.0 | 32 | qnrB19* |
| SAL4630¥ | LILU00000000 | Enteritidis | 11 | Poultry | 1.0 | 32 | qnrB19* |
| SAL4674 | JWQH00000000 | Heidelberg | 15 | Kelp Gull | 0.5 | 16 | qnrB19* |
| SAL4675 | JWQG00000000 | Heidelberg | 15 | Kelp Gull | 0.25 | > 32 | gyrA aa 83 F/S** |
| SAL4676 | JWQF00000000 | Heidelberg | 15 | Kelp Gull | 0.25 | > 32 | gyrA aa 83 F/S** |
| SAL4678 | JWQE00000000 | Heidelberg | 15 | Human | 0.5 | 16 | qnrB19* |
| SAL4679 | JWQD00000000 | Heidelberg | 15 | Human | 0.5 | 32 | qnrB19* |
| SAL4688 | JWRD00000000 | Senftenberg | 14 | Kelp Gull | 0.5 | 16 | qnrB19* |
CIP, ciprofloxacin; NAL, nalidixic acid.
Antimicrobial susceptibility ranges. CIP: susceptible (≤ 0.06); non-susceptible (0.06–1); resistant (≥ 1).
NAL: susceptible (≤ 16); resistant (≥ 32).
Acquired resistance genes searched using ResFinder tool, DTU. qnrB19: acquired antimicrobial resistance gene, plasmid borne.
Genomic gyrA gene: mutation in aminoacid 83.
Data published in Toro et al., 2016.
In silico Determination of Quinolone Resistance
We identified genes or known gene mutations for antimicrobial resistance in all eight strains displaying quinolone reduced susceptibility or resistance (Table 1). The mutation gyrA S83F, previously linked to quinolone resistance (Piddock et al., 1998; Reche et al., 2002; Tyson et al., 2015), was found in two of the eight later isolates (Table 1). Interestingly, all six strains that lacked the gyrA mutation but were resistant or had reduced susceptibility to quinolones carried the gene qnrB19, which has been linked to reduced susceptibility to quinolones in Gram negative bacteria (Tran et al., 2012). In silico sequence typing and serotyping analysis showed that these six strains belonged to different sequence type (ST) and to different serotypes (Table 1).
Plasmid Sequence Study
Our analysis revealed that the qnrB19 gene was harbored in a 2,702-bp plasmid 100% identical among all six sequenced strains (Table 1; Supplementary Figure S1). A search within the NCBI database also identified that the sequence was 100% identical to plasmid pPAB19-4 (accession number: JN995611), which was first described by Tran et al. (2012) in a clinical Salmonella sp. isolate obtained in Argentina. Several other plasmids from the database showed high identity values, and six were selected for phylogenetic analysis (Supplementary Figures S1, S2, Supplementary Table S3). The analysis revealed that 12 plasmid sequences (six from our study and six from the NCBI database) grouped in three clusters: one cluster included plasmid pPAB19-4, all six Chilean plasmids, and plasmid pMK101 reported in Colombia (Karczmarczyk et al., 2010). Plasmids pECY6-7 (Peru), pSGI5 (The Netherlands), and pN44358F (US) clustered together (Hammerl et al., 2010; Pallecchi et al., 2010; Tyson et al., 2017). Finally, plasmid pHAD28 reported recently in Germany did not cluster with other plasmids (Fiegen et al., 2017) (Supplementary Figure S1). Differences among these three clusters were located in an intergenic region upstream of the qnrB gene (Supplementary Figures S1, S2). Finally, plasmids pPAB19-4, pN44358F, and pHAD28 were selected as representative of each cluster and used for primer design (Figure 1).
Plasmid Screening by PCR in a Historic Salmonella Collection
PCR amplification with the two sets of primers (Figure 1) identified the presence of the qnrB19-carrying plasmids in 8/113 Salmonella strains isolated from chicken (1/29), pigs (3/9), horse (1/25), wild birds (2/18), and one from a chicken farm environment (1/1). qnrB-19-carrying plasmids were detected in eight Salmonella isolates of serotypes Hadar, Typhimurium, and serogroup B Salmonella, and were obtained in Chile from 2012 to 2017 (Supplementary Table S2).
Genomic Comparison of Small Plasmids Carrying the qnrB19 Gene
We sequenced four of the isolates containing qnrB-19 plasmids from our historic collection and added their plasmid sequences (pDR-021, pDR-022, pDR-039, and pDR-040) to the general analysis. The complete set of 16 plasmid sequences (six from the NCBI database, six from Salmonella genomes previously reported in Toro et al., 2015, and Toro et al., 2016, and four plasmid sequences from our historic collection) was compared showing that nucleotide differences were of a few nucleotides and hosted in the same intergenic region (Figures 2, 3; Supplementary Table S4). All Chilean plasmids clustered together with plasmid pPAB-19. Newly identified plasmid sequences had over 95% identity to pPAB19-4: plasmids pDR-039 and pDR-040 had 95.5% identity to pPAB19-4, and plasmids pDR-021 and pDR-022 had 97.2% identity (Figure 2; Supplementary Table S4). Differences detected among plasmid sequences were 50-bp or 127-bp insertions in the intergenic region starting at nucleotide 1,486 of pPAB19-4 (Figure 3; Supplementary Figure S3). Additional characterization showed that none of plasmids was identified as similar to phage or prophage sequences at BLAST, and the PHASTER screening showed absence of phage DNA in these sequences. No T4SS elements were found either in the Salmonella genomes carrying pPAB19-4-like plasmids. Presence of other plasmid replicons varied (Supplementary Table S4).
Figure 2.
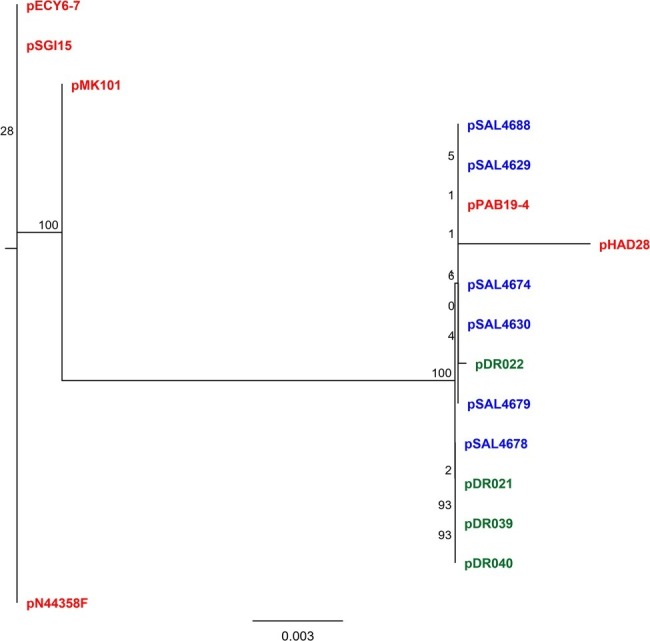
Phylogenetic tree of 16 pPAB-19-like plasmids inferred by maximum-likelihood analysis The analysis included 16 plasmids including six previously described plasmids and 10 plasmids described in the present study. Phylogeny was inferred with RAxML in Geneious Prime (Biomatters, New Zealand) with 1,000 replicates. Colored names represent plasmid sequence source. Red: data from NCBI; blue: genomes reported in Toro et al. (2015) and Toro et al. (2016); green: this study.
Figure 3.

Whole plasmid sequence comparison of 16 qnrB-19-like plasmids. The analysis included 16 plasmids including six previously described plasmids and 10 plasmids described in the present study. Whole plasmid sequence comparison was performed with the Mauve plug-in in Geneious Prime with default settings. White box represents location of the qnrB-19 gene in each plasmid sequence. Red filled bars represent identity among tested sequences.
Phage-Mediated Dissemination of the pPAB19-4 Plasmid
It has been reported that P22- and P22-like bacteriophages are capable of efficiently transferring both chromosomal fragments and low-copy number plasmids in Salmonella (Mann and Slauch, 1997; Schmieger and Schicklmaier, 1999). Therefore, to investigate a potential mechanism by which pPAB19-4 disseminates through different Salmonella serotypes, P22 transduction assays were performed. The transduction frequency was determined as 1.63 × 10−7 cfu/pfu (SD ± 6.5 × 10−6) and none of the negative controls showed transductants. These results showed that P22 bacteriophage efficiently transfers the plasmid from S. Heidelberg donor strain to the recipient S. Typhimurium strain.
Discussion
Antimicrobial resistance is one of the main concerns in public health. WHO defined this problem as high priority and promotes improving research and surveillance as measures to attack the problem (Tacconelli et al., 2017). Quinolones are a group of antimicrobials that are used to treat human and animal bacterial infections; therefore, resistance to these drugs is of great concern (Tacconelli et al., 2017). In Chile, recent studies have determined that quinolone resistance is on the rise [Instituto de Salud Pública de Chile (ISP), 2015], and a recent study reported that Salmonella susceptibility to quinolones is decreasing in the US (Karp et al., 2018). In the present study, we found that a small plasmid harboring the gene qnrB19 was the main responsible for quinolone antimicrobial resistance in six Salmonella spp. isolates. Strains carrying the plasmid showed reduced susceptibility or resistance to CIP, and three of them were also resistant to NAL (Table 1). Traditionally, plasmid-mediated quinolone resistance (PMQR) has been linked to low-level quinolone resistance, promoting the selection of quinolone-resistant strains (Rodríguez-Martínez et al., 2016). Also, recent studies have described the presence of plasmids carrying the qnrB19 gene as responsible for quinolone resistance in Salmonella spp. isolated in Europe and the US from poultry and swine sources (Fiegen et al., 2017; Tyson et al., 2017). Similar plasmids have been found in E. coli from human and food animals from different geographic regions as well (Hammerl et al., 2010; Jones-Dias et al., 2013; Cummings et al., 2017).
Genes of the qnr family can be hosted in small and large plasmids (Hammerl et al., 2010; Tran et al., 2012). The gene qnrB19 was first described in a E. coli isolated from pigs in Guandong, China (Yue et al., 2008), and different plasmids carrying the same gene were reported in Latin-American countries such as Peru, Bolivia, and Colombia (Pallecchi et al., 2010; Tran et al., 2012). Furthermore, an identical plasmid (pPAB19-4) to the one we found in this work was first isolated in Argentina (Tran et al., 2012) from a Salmonella sp. clinical isolate, and a very similar plasmid (pSGI15; FN428572) was also found in Salmonella Typhimurium from a human clinical case in the Netherlands (Hammerl et al., 2010). Moreover, another identical plasmid was recently isolated from pork products and swine in the US (Tyson et al., 2017). We found also two other highly similar qnrB-19 gene-carrying plasmids; identity to pPAB19-4 was tested through different software, and we detected that when differences existed, they were concentrated in the intergenic region and were indels of 77–137 bp, which were not coding regions, and all plasmid features were the same with pPAB19-4 (Supplementary Figure S3).
Interestingly, strains isolated in this study and carrying pPAB19-4-like plasmids were isolated from Salmonella of diverse sources and different serotypes (Table 1; Supplementary Tables S1, S2): S. Enteritidis strains were isolated from eggs (chicken), S. Heidelberg and S. Senftenberg strains were isolated from Kelp gull, and S. Heidelberg strain was isolated from human clinical cases. S. Typhimurium from pigs and S. Hadar from chicken also contained similar plasmids. This suggests that this plasmid is probably being transmitted among diverse Salmonella lineages by HGT. Supporting this hypothesis, isolates of Salmonella Heidelberg and Senftenberg carrying this plasmid have been isolated from swine and pork products in the United States (Tyson et al., 2017).
The widespread presence of pPAB19-4-like plasmids among diverse Salmonella serotypes, hosts, years, and geographic locations poses a risk for global human and animal populations. A better understanding of the mechanism involved in the spread of these plasmids could be used to understand their dissemination in the environment. Since unrelated Salmonella serotypes and E. coli have carried identical plasmids, it was plausible to think that horizontal gene transfer mechanisms were involved on their dissemination. The pPAB19-4 plasmid is small (2.7 kb) and lacks mob and tra genes, therefore, self-conjugation is not possible (Tran et al., 2012); for this reason, we did not include DNAse treatment in our experiments. A similar plasmid (pPAB19-2) was transferred by conjugation (Andres et al., 2013), suggesting that more than one mechanism of horizontal gene transfer is possible in these types of plasmids. Our results demonstrated that pPAB19-4 plasmids can be transferred from S. Heidelberg to S. Typhimurium by transduction assisted by a P22 bacteriophage. Transduction frequency reported in the current study (1 transducent in 106 phage) is similar to that reported in previous studies (Mašlanová et al., 2016; Varga et al., 2016). Importantly, our study shows transduction in experimental conditions, indicating that transduction is another plausible mechanism for pPAB19-4-like plasmids spread in the environment.
Our data demonstrates that the pPAB19-4 plasmid confers antimicrobial resistance to Salmonella able to cause human disease (Table 1); therefore, surveillance of the genetic trait in different hosts and environments—including wild and domestic animals, foods and the environment—will be fundamental to understand its relevance in the spread of quinolone resistance. This will be of special importance in regions of the world where the plasmid has not been described yet. Additionally, we believe that it will be important to assess the role of migratory birds and wild fauna in spreading these elements and the associated antimicrobial resistance phenotypes since we found the plasmid in Salmonella isolated from gulls. Likewise, we detected the plasmid in Salmonella isolated from 2009 to 2015 in Chile, and it was also identified in the US in Salmonella isolated in 2013 and 2014 (Tyson et al., 2017). Taken together, evidence suggests that this plasmid has been circulating for at least 8 years in several countries, and that it is spreading to other parts of the world.
In conclusion, the pPAB19-4-like plasmids could play an important role in antimicrobial resistance worldwide. Since the plasmid can be transferred between different salmonellae circulating in the food chain, wild animals, and human clinical cases, it is important to start its active surveillance and to explore the presence of diverse mobile genetic elements carrying this and other resistance genes impacting public health.
Data Availability Statement
The datasets generated for this study can be found in the NCBI accession numbers: SAMN11569606, SAMN11569607.
Bioethics Statements
Salmonella enterica (S. enterica; n = 64) isolates from wild birds (n = 28), human clinical cases (n = 23), eggs (n = 9), and sea lions (n = 4) were collected between 2009 and 2012 from different locations along Chile. Isolate’s information, including genomic sequences, was reported in previous publications (Toro et al., 2015, 2016) (Supplementary Table S1). The historic strain collection used in this study was obtained in a previous study by AM-S, approved by the University Andrés Bello Bioethics Committee, Santiago, Chile.
Author Contributions
AM-S wrote the manuscript, analyzed data, and designed the study. DP, VS, and IG performed plasmid transduction assays. DR performed laboratory experiments. PR provided material and wrote the manuscript. PN and AR-J critically reviewed the manuscript. MT wrote the manuscript, performed data analysis, and conceived the study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the kind support of Dr. Narjol Gonzalez-Escalona (CFSAN/FDA) for sequencing Salmonella strains and of Ms. Sherry Ayers (CVM/FDA) for performing the antimicrobial resistance test.
Footnotes
Funding. Genome sequencing of two isolates was provided by MicrobesNG (http://www.microbesng.uk), which is supported by the BBSRC (grant number BB/L024209/1). MT thanks CONICYT/FONDECYT (grant N°11150491) for financial support, and AM-S thanks CONICYT/FONDECYT (grants N°11140108 and N°1181167) and Millennium Science Initiative of the Ministry of Economy, Development and Tourism, Government of Chile for the grant “Millennium Initiative for Collaborative Research on Bacterial Resistance (MICROB-R).” PN thanks Millennium Science Initiative, Ministry of Economy, Development and Tourism of Chile for the grant “Millennium Nucleus in the Biology of the Intestinal Microbiota.”
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02503/full#supplementary-material
References
- Andres P., Lucero C., Soler-Bistué A., Guerriero L., Albornoz E., Tran T., et al. (2013). Differential distribution of plasmid-mediated quinolone resistance genes in clinical enterobacteria with unusual phenotypes of quinolone susceptibility from Argentina. Antimicrob. Agents Chemother. 57, 2467–2475. 10.1128/AAC.01615-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. 10.1093/nar/gkw387, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abby S. S., Cury J., Guglielmini J., Néron B., Touchon M., Rocha E. P. C. (2016). Identification of protein secretion systems in bacterial genomes. Sci. Rep. 6:23080. 10.1038/srep23080, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI) (ed.) (2015). Performance standards for antimicrobial susceptibility testing, 25th Edn Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cordeiro N. F., Nabón A., García-Fulgueiras V., Álvez M., Sirok A., Camou T., et al. (2016). Analysis of plasmid-mediated quinolone and oxyimino-cephalosporin resistance mechanisms in Uruguayan Salmonella enterica isolates from 2011-2013. J. Glob. Antimicrob. Resist. 6, 165–171. 10.1016/j.jgar.2016.06.002, PMID: [DOI] [PubMed] [Google Scholar]
- Cummings K. J., Rodriguez-Rivera L. D., Norman K. N., Ohta N., Scott H. M. (2017). Identification of a plasmid-mediated quinolone resistance gene in Salmonella isolates from Texas dairy farm environmental samples. Zoonoses Public Health 64, 305–307. 10.1111/zph.12318, PMID: [DOI] [PubMed] [Google Scholar]
- Darling A. C. E., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. 10.1101/gr.2289704, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegen U., Klein G., de Jong A., Kehrenberg C. (2017). Detection of a novel qnrB19-carrying plasmid variant mediating decreased Fluoroquinolone susceptibility in Salmonella enterica Serovar Hadar. Microb. Drug Resist. 23, 280–284. 10.1089/mdr.2016.0067, PMID: [DOI] [PubMed] [Google Scholar]
- González F., Araque M. (2013). Association of transferable quinolone resistance determinant qnrB19 with extended-spectrum β -lactamases in Salmonella give and Salmonella Heidelberg in Venezuela. Int. J. Microbiol. 2013:628185. 10.1155/2013/628185, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerl J. A., Beutlich J., Hertwig S., Mevius D., Threlfall E. J., Helmuth R., et al. (2010). pSGI15, a small ColE-like qnrB19 plasmid of a Salmonella enterica serovar Typhimurium strain carrying Salmonella genomic island 1 (SGI1). J. Antimicrob. Chemother. 65, 173–175. 10.1093/jac/dkp383 [DOI] [PubMed] [Google Scholar]
- Holmes A. H., Moore L. S., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. 10.1016/S0140-6736(15)00473-0, PMID: [DOI] [PubMed] [Google Scholar]
- Hooper D. C., Jacoby G. A. (2016). Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 6, 1–21. 10.1101/cshperspect.a025320, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto de Salud Pública de Chile (ISP) (2015). Boletin de Resistencia Antimicrobiana. [Spanish]. Available at: http://www.ispch.cl/sites/default/files/BoletinRam-30112015A_0.pdf
- Jones-Dias D., Manageiro V., Francisco A. P., Martins A. P., Domingues G., Louro D., et al. (2013). Assessing the molecular basis of transferable quinolone resistance in Escherichia coli and Salmonella spp. from food-producing animals and food products. Vet. Microbiol. 167, 523–531. 10.1016/j.vetmic.2013.08.010, PMID: [DOI] [PubMed] [Google Scholar]
- Karczmarczyk M., Martins M., McCusker M., Mattar S., Amaral L., Leonard N., et al. (2010). Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol. Lett. 313, 10–19. 10.1111/j.1574-6968.2010.02119.x, PMID: [DOI] [PubMed] [Google Scholar]
- Karp B. E., Campbell D., Chen J. C., Folster J. P., Friedman C. R. (2018). Plasmid-mediated quinolone resistance in human non-typhoidal Salmonella infections: an emerging public health problem in the United States. Zoonoses Public Health 65, 838–849. 10.1111/zph.12507, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp B. E., Tate H., Plumblee J. R., Dessai U., Whichard J. M., Thacker E. L., et al. (2017). National antimicrobial resistance monitoring system: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog. Dis. 14, 545–557. 10.1089/fpd.2017.2283, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404, PMID: [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Stewart V. J., Taylor R. K., Miller S. I. (1996). Genetic analysis of pathogenic bacteria: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 603. [Google Scholar]
- Mann B. A., Slauch J. M. (1997). Transduction of low-copy number plasmids by bacteriophage P22. Genetics 146, 447–456. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mašlanová I., Stríbná S., Doškar J., Pantuček R. (2016). Efficient plasmid transduction to Staphylococcus aureus strains insensitive to the lytic action of transducing phage. FEMS Microbiol. Lett. 363, 1–7. 10.1093/femsle/fnw211, PMID: [DOI] [PubMed] [Google Scholar]
- Medalla F., Hoekstra R. M., Whichard J. M., Barzilay E. J., Chiller T. M., Joyce K., et al. (2013). Increase in resistance to ceftriaxone and nonsusceptibility to ciprofloxacin and decrease in multidrug resistance among Salmonella strains, United States, 1996-2009. Foodborne Pathog. Dis. 10, 302–309. 10.1089/fpd.2012.1336, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millanao B. A., Barrientos H. M., Gómez C. C., Tomova A., Buschmann A., Dölz H., et al. (2011). Injudicious and excessive use of antibiotics: public health and salmon aquaculture in Chile. Rev. Med. Chil. 139, 107–118. [PubMed] [Google Scholar]
- Naylor N. R., Silva S., Kulasabanathan K., Atun R., Zhu N., Knight G. M., et al. (2016). Methods for estimating the burden of antimicrobial resistance: a systematic literature review protocol. Syst. Rev. 5:187. 10.1186/s13643-016-0364-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallecchi L., Riccobono E., Mantella A., Bartalesi F., Sennati S., Gamboa H., et al. (2009). High prevalence of qnr genes in commensal enterobacteria from healthy children in Peru and Bolivia. Antimicrob. Agents Chemother. 53, 2632–2635. 10.1128/AAC.01722-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallecchi L., Riccobono E., Sennati S., Mantella A., Bartalesi F., Trigoso C., et al. (2010). Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob. Agents Chemother. 54, 678–682. 10.1128/AAC.01160-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan American Health Organization (PAHO) (2014). Technical advisory group on antimicrobial resistance and infection prevention and control. Final Report. 2014 ed. Washington DC. Available at: https://www.paho.org/hq/dmdocuments/2014/2014-cha-tag-antimicrobial-resistance-ipc.pdf (Accessed October 21, 2019).
- Piddock L. J., Ricci V., McLaren I., Griggs D. J. (1998). Role of mutation in the gyrA and parC genes of nalidixic-acid-resistant Salmonella serotypes isolated from animals in the United Kingdom. J. Antimicrob. Chemother. 41, 635–641. 10.1093/jac/41.6.635, PMID: [DOI] [PubMed] [Google Scholar]
- Reche M. P., García de los Ríos J. E., Jiménez P. A., Rojas A. M., Rotger R. (2002). gyrA mutations associated with nalidixic acid-resistant salmonellae from wild birds. Antimicrob. Agents Chemother. 46, 3108–3109. 10.1128/AAC.46.9.3108-3109.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Martínez J. M., Machuca J., Cano M. E., Calvo J., Martínez-Martínez L., Pascual A. (2016). Plasmid-mediated quinolone resistance: two decades on. Drug Resist. Updat. 29, 13–29. 10.1016/j.drup.2016.09.001, PMID: [DOI] [PubMed] [Google Scholar]
- Rozwandowicz M., Brouwer M. S. M., Fischer J., Wagenaar J. A., Gonzalez-Zorn B., Guerra B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. 10.1093/jac/dkx488, PMID: [DOI] [PubMed] [Google Scholar]
- Schmieger H., Schicklmaier P. (1999). Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol. Lett. 170, 251–256. 10.1111/j.1574-6968.1999.tb13381.x, PMID: [DOI] [PubMed] [Google Scholar]
- Singer A. C., Shaw H., Rhodes V., Hart A. (2016). Review of antimicrobial resistance in the environment and tts relevance to environmental regulators. Front. Microbiol. 7:1728. 10.3389/fmicb.2016.01728, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. 10.1080/10635150802429642, PMID: [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2017). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Toro M., Retamal P., Allard M., Brown E. W., Evans P., Gonzalez-Escalona N. (2015). Draft genome sequences of 33 Salmonella enterica clinical and wildlife isolates from Chile. Genome Announc. 3, pii: e00054-15. 10.1128/genomeA.00054-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro M., Retamal P., Ayers S., Barreto M., Allard M., Brown E. W., et al. (2016). Whole genome sequencing analysis of Salmonella enteritidis isolated in Chile provides insights about possible transmission between gulls, poultry and humans. Appl. Environ. Microbiol. 82, 6223–6232. 10.1128/AEM.01760-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro M., Rivera D., Toledo V., Campos-Vargas R., Allard M. W., Hamilton-West C., et al. (2018). Genomics of Salmonella contaminating backyard production systems reveals persistence and transmission of genetically related salmonella on a farm basis. Zoonoses Public Health 65, 1008–1014. 10.1111/zph.12526, PMID: [DOI] [PubMed] [Google Scholar]
- Tran T., Andres P., Petroni A., Soler-Bistué A., Albornoz E., Zorreguieta A., et al. (2012). Small plasmids harboring qnrB19: a model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob. Agents Chemother. 56, 1821–1827. 10.1128/AAC.06036-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G. H., McDermott P. F., Li C., Chen Y., Tadesse D. A., Mukherjee S., et al. (2015). WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 70, 2763–2769. 10.1093/jac/dkv186, PMID: [DOI] [PubMed] [Google Scholar]
- Tyson G. H., Tate H. P., Zhao S., Li C., Dessai U., Simmons M., et al. (2017). Identification of plasmid-mediated quinolone resistance in Salmonella isolated from swine ceca and retail pork chops in the United States. Antimicrob. Agents Chemother. 61, pii: e01318-17. 10.1128/AAC.01318-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga M., Pantuček R., Ružičková V., Doškar J. (2016). Molecular characterization of a new efficiently transducing bacteriophage identified in meticillin-resistant Staphylococcus aureus. J. Gen. Virol. 97, 258–268. 10.1099/jgv.0.000329, PMID: [DOI] [PubMed] [Google Scholar]
- Yue L., Jiang H. X., Liao X. P., Liu J. H., Li S. J., Chen X. Y., et al. (2008). Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet. Microbiol. 132, 414–420. 10.1016/j.vetmic.2008.05.009, PMID: [DOI] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. 10.1093/jac/dks261, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI accession numbers: SAMN11569606, SAMN11569607.