Abstract
Mitochondrial Fe-S cluster biosynthesis is accomplished within yeast utilizing the biophysical attributes of the “Isu1” scaffold assembly protein. As a member of a highly homologous protein family, Isu1 has sequence conservation between orthologs and a conserved ability to assemble [2Fe-2S] clusters. Regardless of species, scaffold orthologs have been shown to exist in both “disordered” and “structured” conformations, a structural architecture is directly related to conformations utilized during Fe-S cluster assembly. During assembly, the scaffold helps direct the delivery and utilization of Fe(II) and persulfide substrates to produce [2Fe-2S] clusters, however Zn(II) binding alters the activity of the scaffold while at the same time stabilizing the protein in its structured state. Additional studies confirm Zn binds to the scaffold’s Cys rich active site, and has an impact on the protein’s ability to make Fe-S clusters. Understanding the interplay between Fe(II) and Zn(II) binding in vitro may help clarify metal loading events that occur during Fe-S cluster assembly in vivo.
Here we determine the metal:protein stoichiometry for Isu1 Zn and Fe binding to be 1:1 and 2:1, respectively. As expected, while Zn binding shifts the Isu1 to its structured state, however folding is not influenced by Fe(II) binding. X-ray absorption spectroscopy (XAS) confirms Zn(II) binds to the scaffold’s cysteine rich active site but Fe(II) binds at a location distinct from the active site. XAS results show Isu1 binding initially of either Fe(II) or Zn(II) does not perturb the metal site structure of alternate metal. XAS confirmed that four scaffold orthologs bind iron as high-spin Fe(II) at a site composed of ca. 6 oxygen and nitrogen nearest neighbor ligands. Finally, in our report Zn binding dramatically reduces the Fe-S cluster assembly activity of Isu1 even in the presence of frataxin. Given the Fe-binding activity we report for Isu1 and its orthologs here, a possible mechanism involving Fe(II) transport to the scaffold’s active site during cluster assembly have been considered.
Model for Zn association to the active site of yeast Fe-S cluster assembly scaffold protein Isu1
INTRODUCTION
Metal homeostasis is central for the viability of nearly every cell.1 Iron homeostasis in particular is critical in biology; iron’s varied redox capabilities and high prevalence within nature make it often the metal of choice to assist metalloproteins in performing complex chemistry that cells cannot perform using only an organic toolkit.2, 3 Iron, often incorporated into Fe-S clusters, plays a pivotal role in several essential biochemical pathways, including: sensing oxidative stress in the cytosol, directing DNA repair within the nucleus and driving cellular respiration inside the mitochondria.4–6 Fe-S cluster biogenesis in eukaryotes occurs predominately within the mitochondria.7, 8 In yeast, Fe-S cluster assembly is accomplished directly on the scaffold protein (Isu1), using sulfur in the form of a persulfide provided by the cysteine desulfurase (Nfs1) enzyme.9, 10 Nfs1 works in complex with the yeast accessory protein (Isd11) and the acetyl carrier protein (Acp1) to accomplish its enzymatic activity.11–14 Electrons, essential for forming and stabilizing Fe-S clusters produced on the Isu1 template, are provided by the yeast ferredoxin (Fdx1).15 Yeast frataxin (Yfh1), an iron binding protein that regulates cysteine desulfurase activity, also plays a pivotal in regulating Nfs1 activity and an unconfirmed role in Fe delivery to Isu1.16–25 During Fe-S cluster assembly, several scaffold orthologs have been shown to interconvert between disordered and structured states, indicating a dynamic interplay within the protein itself and/or between scaffold and protein partners is may occur during assembly; this dynamic mobility on Isu1 may help drive substrate delivery to and [2Fe-2S] assembly by the scaffold.18, 26–28
Several studies have probed the biophysical aspects of iron binding to different scaffold orthologs.27, 29–31 The yeast and fly scaffolds (Isu1 and fIscU, respectively), which have high sequence homology, have been shown to bind two Fe(II) atoms with affinities in the μM to sub-μM range;30 similar Fe(II) binding affinities have been reported for the human and bacterial scaffold orthologs.32 In the yeast and fly orthologs, Fe(II) has been shown to coordinate to the scaffold in an environment dominated by oxygen and nitrogen based ligands, suggesting ferrous ions are coordinating at a site on the protein distinct from its Cys rich active site.30, 31 While the protein partner(s) used to accomplish Fe(II) delivery to the scaffold during in vivo Fe-S cluster assembly remains elusive, a recent report indicates that human frataxin promotes delivery of Fe(II) to the human scaffold (ISCU) to activate Fe-S cluster assembly.33 In this system, frataxin-mediated Fe delivery occurred only when both L-cysteine and reductant (human ferredoxin “FDX2” or DTT) were provided, and these data would be consistent with Fe(II)-loaded frataxin being essential for iron delivery to the scaffold during mitochondrial Fe-S cluster assembly.33 Possible candidates that deliver Fe(II) to the scaffold remain unclear, however a low-molecular mass iron complex found within mitochondria could be a source for the iron utilized during Fe-S assembly.34, 35 Given the ca. 10 μM affinity of Fe(II) to Yfh1 and an estimated concentration of xsxsxsxsxsxs~ 150 μM for the nonheme high-spin Fe(II) pool in mitochondria,36 it seems reasonable that frataxin would be predominately iron loaded when within the organelle and therefore positioned to help direct the transfer of Fe(II) to the scaffold when part of a complex with the scaffold.16 Finally, in yeast, Yfh1 binds directly to Isu1 in an iron dependent manner at a 1:1 stoichiometry at micromolar binding affinity, however human frataxin has been reported to only bind to the human scaffold (ISCU) when associated as part of a multi-protein assembly complex, so additional clarification related to scaffold metal loading and protein complexation formation events within the Fe-S cluster assembly pathway need to be explored.18, 33
Zinc stabilizes the scaffold in its structured state and also inhibits scaffold activity during Fe-S cluster assembly. Structural data from bacterial and human orthologs confirm zinc coordinates directly to cysteine residues at the scaffold’s active site.29, 37–39 Furthermore, zinc has been used to stabilize human ISCU’s structure when the scaffold was part of the human Fe-S cluster assembly complex, which included the bacterial acetyl carrier protein within the assembly [NFS1]2[ISD11]2[Acp]2[ISCU]2 protein complex stoichiometry (referred from here on as “NIAU”).29 Biochemically, zinc coordination was suggested to be an artifact of bacterial IscU overexpression, and the extent of zinc loading was shown to be variable in relation to changes in the cell growth conditions.40 Zinc coordination to ISCU negatively inhibits the cysteine desulfurase activity within the human NIAU complex, as a result of zinc ligating to ISCU active site residues (Asp 71, Cys95 and His137) and the NFS1 active site persulfide forming/transfer residue (Cys381); Zn coordination was suggested to likely inhibit persulfide production by reducing the availability of essential active-site residues from both proteins.40 Interestingly, in this study it was also shown that human frataxin restores cysteine desulfurase activity by the NIAU complex once frataxin was coordinated by upon binding, liberating the NFS1 Cys sulfur from Zn coordination.41
Here we perform a biophysical characterization of both Fe(II) and Zn(II) binding to the yeast scaffold. Iron and zinc binding affinities to Isu1 were determined using an in vitro metal binding competition assay developed to measure Fe(II) binding for proteins under conditions that closer resemble in vivo competitive binding environments experienced by apo-proteins within cells.31 The impact of Fe and/or Zn binding relative to coordination by the alternate metal, the influence of metal binding on the scaffold’s structure, and the impact of both metals individually and in combination on the scaffold’s cluster assembly activity abilities have been characterized in this report. Furthermore, we performed a structural characterization comparison of Fe(II) bound to the yeast, fly, human and bacterial scaffold orthologs. Our overall objective was to provide some of the molecular details needed to better understand scaffold iron-loading events prior to and during Fe-S cluster assembly. We also wanted to confirm zinc’s ability to inhibit Fe-S cluster assembly in yeast and to investigate if zinc can serve as a tool to help us evaluate not just the structure but the functionality of the scaffold orthologs.
METHODS
Sc Isu1 Expression and Purification
The wild type Isu1 (S. cerevisiae) gene was transformed into C41(DE3) cells and incorporated into a pET21b-mIsu1 plasmid for protein expression (Lucigen), as outlined previously.31 Cells were grown with Ampicillin (0.1μg/μL) in Luria-Bertani broth at 37°C and shaken at 250 rpm. Cells were induced with a final concentration of 0.8 mM IPTG at an OD600 of 0.6. Following induction, cells were grown for an additional 3 hr and harvested by centrifugation at 8000 rpm for 20 min at 4˚C. Cells were stored as a pellet at −80˚C until lysing. All isolation steps were performed at 4˚C following our published protocol outlined below.30, 31 Frozen cells were resuspended in 50 mM NaPO4, 300 mM NaCl, 20 mM Imidazole and 5 mM 2-Mercaptoethanol (β-Me) pH 7.5 as a binding buffer, in the presence of Complete EDTA free Protease inhibitor cocktail tablets (Roche). Cells were lysed by three passes through an Emulsaflex (ATA Scientific), followed by centrifugation at 21,000 rpm for 60 min.30 Crude fractions were filtered first through a 0.4 μm then 0.2 μm sterile filter. Soluble fractions were loaded onto a 5 mL His-Trap purification column (GE Healthcare) pre-equilibrated with binding buffer outlined above. Protein was eluted using an imidazole gradient between 20 – 500 mM imidazole concentrations. Isu1 fractions were pooled, concentrated and dialyzed (2 buffer exchanges for 3 hr each in 2L volume) against 20 mM HEPES, 300 mM NaCl, 10 mM EDTA and 5 mM β-Me (pH 7.5) buffer. Dialyzed samples were passed through a Sephadex 75 (GE Healthcare) size exclusion column equilibrated with 20 mM HEPES, 300 mM NaCl, 10 mM EDTA and 5 mM β-Me (pH 7.5) buffer. Isu1 eluted from the column as a single species at an estimated 95% purity, as judged by the presence of only a single A280 peak in our purification column chromatograph, and by SDS-page gel analysis from a 4–20% polyacrylamide gel.18 Protein samples were dialyzed into storage buffer (20 mM HEPES, 300 mM NaCl and 5 mM TCEP (pH 7.5)), concentrated to 600 μM and stored at 4°C within an anaerobic chamber. Isolation protocols for the fly, bacterial and human scaffold orthologs have been published previously.28, 30, 42, 43
To verify the metal loaded content for as isolated Isu1, we performed ICP-MS analysis of possible bound metal concentrations on our apo-Isu1 samples prior to use for cluster assembly activity measurements and on samples before and following metal loading for the XAS analysis. Analysis was performed at the University of Utah Health Sciences Protein Core Facility (http://cores.utah.edu/mass-spectrometry-proteomics/) to determine the average metal contaminations (Fe or Zn) for Isu1 following our isolation protocol. Average metal concentrations for what we call as isolated “apo”-Isu1 at 346.5 ± 163.5 μM protein concentrations (using 4 samples from 2 independent preparations) were 2.0 ± 1.1 μM Fe and 2.2 ± 1.2 μM Zn, representing an average of ca. 0.6% metal contaminant for both Fe and Zn bound to our apo-Isu1 samples (Supplemental Table 1) despite extensive dialysis against EDTA during our isolation protocol. Iron and zinc concentrations were also measured following metal loading and spin concentrating for al XAS sample outlined below.
Protein Metal Binding Competition Assays
Individual Fe(II) or Zn(II) metal binding affinities to Isu1 were determined according to our recently reported competition assay, a procedure that uses mag-fura-2 (Molecular Probes) as a metal chelator which competes with the protein during affinity characterization but provides a chromophore for competition metal binding.30, 31 As previously outlined, mag-fura-2 forms a 1:1 complex with divalent cations (Fe and Zn) displaying a maximum absorbance at 325 nm when metal is bound or a feature at 366 nm when in its apo state.31 The progressive reduction of absorbance at 366 nm, indicative of metal loading, was measured using a Shimadzu UV-1800 spectrophotometer housed within a wet anaerobic chamber (Coy Lab Products). Titration data, obtained by adding divalent metal ions in sub-stoichiometric equivalents into an Isu1/mag-fura-2 mixture, were collected anaerobically at room temperature using a 1 cm quartz cuvette. Due to overlap between the Fe- and Zn-loaded mag-fura-2 signals, we could not distinguish specific changes in metal binding activity when both metals were added, so we were limited to measuring metal binding affinities only for individual metals interacting with Isu1. All samples were prepared anaerobically in 20 mM HEPES and 300 mM NaCl (pH 7.5) running buffer for use during binding analysis.
Specific details for the experiments and protocols for anaerobic preparation of the samples are outlined below. All septa capped reagents/buffers were bubbled with hydrated Ar(g) while stirring for 30 min, and these were then stored capped within the Coy anaerobic wet chamber overnight prior to experimentation. Several independent apo-protein samples, incubated with 5 mM TCEP in running buffer prior to titrations, were dialyzed (centrifugation, 6 buffer exchanges) within the anaerobic chamber along with running buffer to remove TCEP prior to metal loading. Metal and chelator samples were prepared and data collected using only anaerobic running buffer within the oxygen free environment. During the titration, mag-fura-2 concentrations were varied between 0 and 8 μM, while the protein concentration was held constant at 4 μM, and multiple independent data sets on independent samples were collected. An anaerobic solution of 2.0 mM ammonium ferrous sulfate hexahydrate (Sigma), prepared in running buffer was added in progressive sub-stoichiometric increments to the protein/mag-fura-2 mixture until signal saturation was reached. After each addition, an absorption spectrum was collected between the wavelength range of 200 – 800 nm. Zn titrations were performed in the identical manner but using a 2.0 mM zinc sulfate (Sigma) solution to substitute for iron. Initial apo mag-fura-2 concentrations were determined using the molar absorptivity (ε) value of 29,900 M−1cm−1 for the compound, and values were measured at the wavelength of 366 nm.31 The absorbance at 366 nm, corrected for dilution, was then used to calculate binding parameters for Isu1 relative to the amount of metal added. Binding data were simulated with the program DYNAFIT44 using a non-linear least squares analysis script to identify the binding capacity and metal stoichiometry for the protein distinct from mag-fura-2, in a manner previously outlined.30, 31 Each titration experiment was simulated using both one and two-site metal binding models for comparison.
Circular Dichroism of Fe or Zn Bound Isu1
The impact of metal binding on Isu1’s secondary structure was measured in the presence of bound Fe or Zn by circular dichroism (CD) spectroscopy. Samples of apo- and holo-Isu1 were prepared within a Coy wet anaerobic chamber from the identical anaerobic apo-protein stock solution. CD spectra of apo- and metal loaded Isu1 samples were collected using a 0.1 cm anaerobic quartz cuvette on a benchtop Jasco 1500 spectrophotometer, equipped with a Peltier VT unit set at 27 °C. Apo- and Fe-loaded Isu1 samples were prepared in 1 mM NaPO4 buffer (pH 7.5), whereas Zn-loaded Iscu1 samples were prepared in 5mM NaPO4 buffer (pH 7.5); the buffer change was to improve the stability of Isu1 since lower salt caused the protein to ppt when loading Zn. For holo-Isu1 samples, 1 equivalent of Zn(II)(aq) or 2 equivalents of Fe(II)(aq) were added to the anaerobic protein solution. Each sample had a final protein concentration of 10 μM; this protein concentration was chosen, given the Kd’s measured for metal binding listed above, to ensure that the majority of the protein in our CD samples were metal loaded. Twenty scans total were collected and averaged for each final spectrum, and spectra were collected on independent duplicate samples to ensure data reproducibility. Before protein spectral collection, a baseline was collected at all wavelengths and used for subtraction for protein data to eliminate buffer signals. Spectra were analyzed using the Jasco CD Pro analysis software and simulated using the CONTIN method using the SP29, SP37, SP43, SMP50 and SMP56 spectral reference sets.45 Values obtained from simulations from each database were averaged to obtain final CD simulation analysis parameters.
X-ray Absorption Spectroscopy
X-ray absorption spectroscopy (XAS) was used to characterize the structure of metal bound to the scaffold in solution. Metal loaded protein XAS samples were prepared anaerobically within a Coy chamber using solution samples dialyzed in buffer prepared with hydrated Ar(g) obtained by bubbling gas through a hydrator. Yeast Isu1 XAS samples were prepared at a 1.0 mM final protein concentration in 20 mM HEPES (pH 7.0), 300 mM NaCl and 5 mM β-Me buffer. Isu1 XAS spectra were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) on beamline 9–3. Duplicate independent samples were loaded with 1.9 equivalents of Fe(II) per 1 equivalent of protein using an ammonium ferrous sulfate hexahydrate solution and/or with 0.9 equivalents of Zn per 1 equivalent of protein using a zinc sulfate solution; protein was in slight excess and samples were prepared at concentrations well above the metal binding affinity to ensure all metal was bound to protein. XAS data for scaffold orthologs from H. sapien, D. melanogaster, and T. maritima, as well as an additional verification of the S. cerevisiae Isu1 control sample, were collected on independent duplicate samples at SSRL on beamline 7–3 and at the National Synchrotron Radiation Laboratory on beamline X3b. Ortholog comparison samples were loaded with only 0.95 equivalents of ferrous salt per 1 equivalent protein for consistency. All XAS samples were diluted to 0.7 mM final protein concentrations by addition 30% glycerol as a glassing agent. Samples were then loaded into XAS Lucite sample cells wrapped with Kapton tape, flash frozen in liquid nitrogen, removed from the glove box and finally stored in liquid nitrogen until data collection. SSRL beamlines 7–3 and 9–3 were equipped with a Si[220] double-crystal monochromator, while NSLS beamline X3b was equipped with a Si[111] single-crystal monochromator. All beamlines are equipped with focusing mirrors for harmonic rejection. Fluorescence spectra were collected using a Canberra 100-element on beamline 9–3 or 30-element Ge solid state detectors at beamlines 7–3 and X3b. During data collection, samples were maintained at 10 K using an Oxford Instruments continuous flow liquid helium cryostat at both SSRL beamlines or at 24 K using a helium Displex Cryostat at NSLS. Beamline 9–3 XAS data were collected with a 6 μm Mn or a 6 μm Cu filter placed between the cryostat and the detector to reduce low energy scattering in the Fe and the Zn XAS, respectively. During data collection, a Fe or Zn foil spectrum were collected simultaneously with the protein data for use in spectral energy calibration. XAS spectra were recorded using 5 eV steps within the pre-edge region, 0.25 eV steps within the edge region and 0.05 Å−1 increments within the extended X-ray absorption fine structure (EXAFS) region. Data were collected to k = 13 Å−1 for both elements, integrating from 1 to 25 s in a k3-weighted manner. An average of 6 (beamline 9–3) to 10 (beamlines 7–3 and X3b) individual scans were collected for each sample and these were averaged for analysis. Each scan lasted approximately 40 minutes. Each spectrum was closely monitored for X-ray induced radiation damage to the sample.
XAS spectra were processed and analyzed using the EXAFSPAK program suite written for Macintosh OS X.46 Fluorescence scans corresponding to each channel were examined for anomalies. A Gaussian function was fit to the pre-edge region and a cubic spline was fit to the EXAFS region of the data for baseline subtraction. Spectra were simulated using single and multiple scattering amplitude and phase functions generated by the Feff v7.2 (7–3 and X3b data) and v8.0 (for 9–3 data) software packages; Feff was integrated directly within the EXAFSPAK software.47 Calibrations of the theoretical scale factors (Sc) and energy shift (E0) values were obtained by fitting crystallographically characterized compounds to provide the Sc and E0 values utilized during data simulations.48 Calibrated Sc and E0 values were held static during empirical data simulations, using the dimensionless Sc value of 0.95 for both Zn and Fe samples. E0 values for protein Fe-O/N/C simulations were set at −11.5 eV.48 E0 values used to fit protein Zn-O/N/C/S interactions were set at −15.25 eV.49 The final Fe and Zn EXAFS data were fit over a k range of 1 to 13.0 and to 1 to 12.5 Å−1, respectively. EXAFS spectra were simulated using both filtered and unfiltered data, however only simulations of unfiltered data are presented in this report. Simulation protocols and criteria for determining the best fit to the empirical data have been described previously.48
Fe-S Cluster Formation Assay
Fe-S cluster assembly was measured for the yeast NIaUF protein complex (all yeast proteins except bacterial Acp) following our previously published protocol.30 All solutions and reagents were made anaerobic by bubbling with hydrated Ar(g), while protein was prepared anaerobic by extensive dialysis within hydrated Ar(g) bubbled buffer. Protein was dialyzed and then stored on ice within our Coy anaerobic chamber until used in the assembly assay. The reaction buffer used during cluster assembly contained 20 mM HEPES and 300 mM NaCl (pH 7.5). Reagents and apo-Isu1 samples were mixed within the Coy chamber in the following amounts: 10 μM Nfs1-Isd11 complex, 50 μM Isu1, 10 μM Yfh1, 500 μM L-Cys, 5 mM DTT and 75 μM Fe(II)(aq). Fe-S cluster assembly was measured in the presence and absence of 1 equivalent of buffered Zn(II)(aq); Zn was preloaded onto Isu1 before combining the reaction mixture. Following addition of Fe(II), samples were loaded into a 1 cm pathlength anaerobic CD cuvette while the samples were in the glove box, capped and then quickly transferred to a benchtop Jasco-1500 Spectrophotometer for data collection (with an average of a 30 sec dead-time during transfer). CD spectra were measured at 430 nm for 60 minutes to monitor cluster formation activity, as previously reported.30 Data were signal averaged from 3 independent measurements and the experiment was performed in duplicate on proteins isolated from independent preparations.
Sequence Alignment of Scaffold Orthologs and the Modeled Structure of Zn-Isu1
Amino acid sequences of the scaffold orthologs were obtained from the GeneBank depository (yeast #KZV07380.1, Fly #NP_649840, Human #ACA52543 and Thermatoga #Q9X192, respectively) and all sequences were aligned using the Clustal Omega software package.50 Since there is no structure of Isu1, a modeled Zn-Isu1 structure, constructed using the program I-Tasser, was based on the human ISCU crystal structure (PDB #5WLW) as the reference template for the initial molecular architecture.29, 51, 52 The NCBI-yeast Isu1 sequence was used as the query protein. Zinc placement within the structure was modelled using the Zn XAS metrical parameters obtained from Zn bond lengths and coordination characterization information outlined by XAS within this report. The modeled structure was colored using PyMOL software.53
RESULTS & CONCLUSIONS
Iron and Zinc-Binding Characteristics of Isu1
To better understand Isu1’s iron and zinc binding characteristics, we determined metal binding affinities and the metal:protein stoichiometries for the yeast scaffold using our competition metal binding assay. We believe this competition assay better reflects the heterogeneous binding environment experienced by proteins in vivo.54 Mag-fura-2 is a useful chelator for measuring binding events under competition conditions for both Fe(II) and Zn(II) metals since it’s affinity is tuned closely to that seen for the scaffold protein family members.31, 55 Apo-mag-fura-2 has a well-established spectroscopic signal at 366 nm (peak maximum) that shifts to 325 nm upon Fe(II) or Zn(II) binding (Figure 1A & B, respectively). Progressive iron or zinc binding causes a decay in the 366 nm absorbance maximum (Figure 1C & D, respectively); this decay profile can be simulated to obtain protein metal-binding characteristics (Table 1).31 Simulations of our Fe(II) titrations into the mag-fura-2/Isu1 competitive mixture indicated an optimal binding stoichiometry of ca. 2 iron ions to 1 Isu1, with relative dissociation constants of Kd1 = 3.5 ± 1.8 and Kd2 = 10 ± 8.6 μM. Simulations of our Zn titrations into the mag-fura-2/Isu1 mixture indicated an optimal binding stoichiometry of 1 zinc ion to 1 Isu1 molecule, with a single dissociation constant of Kd = 0.44 ± 0.19 μM. The Zn binding value for Isu1 is tighter than that reported for bacterial IscU (5.8 μM) but slightly weaker than that measured for the human ortholog (0.16 μM).40, 56 We were unable to measure specific metal binding in the presence of both Fe and Zn since spectroscopic signals from both Fe and Zn-loaded mag-fura-2 significantly overlap, hence preventing deconvolution of each binding interaction within a mixed metal environment. All binding data were collected under anaerobic conditions to avoid protein oxidation and to stabilize the redox state of aqueous Fe(II).
Figure 1. Fe and Zn Binding Affinity to Isu1 Measured using Mag-Fura-2 within a Competition Based Assay.
(A) Representative titration spectra of iron into Isu1 and (B) zinc into Isu1. Initial scan for metal titrations is shown in red with subsequent titrations in black. Mag-fura-2 to protein ratio was varied from 1:2 (red), 1:1 (black), 2:1 (blue) for Fe (C) and Zn titrations (D), respectively. Spectra were collected in duplicate using independent samples to ensure spectral reproducibility.
Table 1. Summary of Binding Affinity Measured in Mag-Fura-2 Competition Assay.
Assay was performed at 25.0 °C under anaerobic conditions, with binding parameters determined by simulating the signal values at 366 nm using DynaFit.43 Metal to protein stoichiometry was best fit using a 2:1 binding model for Fe-Isu1 and a 1:1 binding model for Zn-Isu1.
| Sample | Kd1 | Kd2 |
|---|---|---|
| Fe:Mag-Fura-2 | 3.5 μM ± 1.8 μM | 10 μM ± 8.6 μM |
| Zn: Mag-Fura-2 | 0.44 μM ± 0.19 μM | ---- |
In a previous report, we characterized the iron binding affinity for D37A- and wt-Isu1 using Isothermal Titration Calorimetry (ITC).18, 31 The D37A-Isu1 mutation was shown in orthologs to stabilize bound Fe-S clusters against release from the scaffold.57 These metal binding affinities, obtained by ITC, are compared to values obtained from our competition assay for wt-Isu1 from this report. All measurements were performed at the same pH and buffering capacity, however the salt concentration in the previous reports were with lower salt (150 mM NaCl). The higher salt concentration in this report was used to better stabilize both the Fe and Zn-loaded protein. Using ITC, we determined D37A-Isu1 binds 2 Fe(II) atoms with Kd values of 2.0 and 0.006 μM, while for wt-Isu1 measurements, affinities were 1.0 and 0.02 μM; the 2 Fe binding affinities in this report using our competition assay were 3.5 and 10 μM. The primary binding affinity in all measurements were similar (ranging between 3.5 and 1.0) and these values are close to Kd’s measured for the human and bacterial orthologs at 2.0 ± 0.2 μM and 2.7 μM, respectively.42 The second affinity measured by ITC was consistently much lower than the second value measured using the competition assay within this report, possibly indicating real differences in the second metal binding under homogeneous vs. heterogeneous binding environments. Alternatively, the second affinity measured by ITC may represent a thermodynamic event unrelated to metal binding.
Structural Changes on Isu1 Concurrent with Fe and Zn Binding
Circular dichroism (CD) spectroscopy was used to determine if the secondary structure of Isu1 is altered during iron or zinc binding. Published Fe(II) and Zn(II) titrations into bacterial IscU showed that while Fe(II) binding caused only a minimal impact on the IscU fold, Zn(II) binding dramatically increased the helical fold of the metal-loaded protein.56 In our study, all CD spectra were collected under anaerobic conditions to avoid metal/protein oxidation. Averaged CD spectra comparing apo-Isu1 with Isu1 loaded with either Fe or Zn are shown in Figure 2A and B, respectively. Average secondary structural fold parameters obtained by simulating these data are listed in Table 2. Comparison of spectra for apo- vs. Fe(II)-loaded Isu1 show a high similarity in overall spectral signature. In contrast, addition of a single equivalent of Zn(II) into Isu1 induces a noticeable change in the overall spectrum, with a relative increase in the 223 nm signal and a shift in the apo-protein’s 203 nm signal towards 208 nm, consistent with an increase in α-helical content in the Zn loaded sample. Simulation parameters for the apo- and Fe(II)-loaded sample are nearly identical, however parameters from the Zn-Isu1 CD data confirm an overall enhancement (ca. 12% in overall fold) in the helical content relative to the apo- and Fe(II)-Isu1 samples, and a relative decrease (ca. 11% in overall fold) in β-sheet content for the Zn sample. These structural parameters are consistent with measured values obtained from Zn(II) bound human ISCU.40 A high helical content in Zn-Isu1 is also consistent with the high helicity observed in the human Zn-ISCU crystal structure.29 Overall, these data confirm Zn drives Isu1 into an enhanced structured state while Fe alone does not.
Figure 2. Circular Dichroism of Apo, Fe- and Zn-Loaded Isu1.
Representative spectra of apo- (solid line) and 2Fe-loaded (dashed line) Isu1 in panel (A). Representative spectra of apo- (solid) and Zn-loaded (dashed) Isu1 in panel (B). An average of 30 scans were collected for each representative spectrum displayed; trials were performed in duplicate with independent samples to ensure spectral reproducibility.
Table 2. Circular Dichroism Fitting Percent Parameters of Apo, Fe and Zn-Loaded Isu1.
Secondary structural abbreviations include α (alpha-helix), β (beta-sheet), O (structured other) and D (disordered).
| Sample | α | β | O | D | Fit a |
|---|---|---|---|---|---|
| apo-Isu1 | 16.5 ± 8.4 | 26.7 ± 10.5 | 23.8 ± 2.3 | 33.0 ± 0.7 | 0.04 |
| 2Fe-Isu1 | 16.0 ± 3.1 | 26.7 ± 3.3 | 23.5 ± 0.9 | 34.0 ± 2.8 | 0.05 |
| 1Zn-Isu1 | 27.9 ± 0.5 | 15.0 ± 0.7 | 24.6 ± 0.1 | 32.5 ± 0.1 | 0.05 |
| 1Zn-ISCU b | 44 | 20 | 35 | 21 | NA |
Fit: goodness of fit parameter expressed as normalized spectral fit standard deviation (nm).
Data obtained from the crystal structure of Zn loaded ISCU (PDB 5WLW).29
XAS Analysis for Fe and Zn Sites Coordinated to Isu1
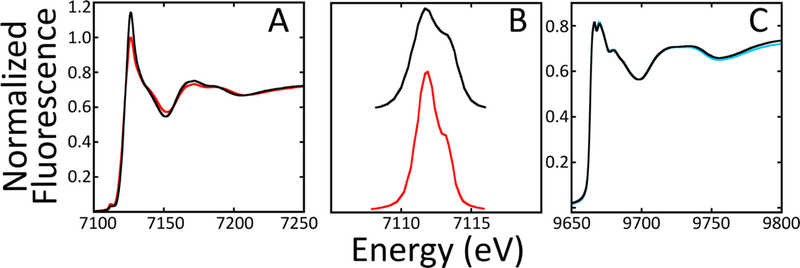
X-ray absorption spectroscopy (XAS) was used to characterize metal electronic and structural parameters for iron and zinc loaded individually and in combination onto Isu1. X-ray absorption near edge spectroscopy (XANES) provides insight into the metal oxidation state, iron spin state and metal-ligand structural symmetry. Iron XANES for Fe(II)-Isu1 are consistent with high-spin ferrous metal bound to protein in a pseudosymmetric 6-coordinatel ligand environment constructed only by oxygen and nitrogen ligands (Figure 3A).21, 56, 57 Iron XANES for both the Fe- and Zn/Fe- loaded Isu1 samples are similar in overall spectral nature, suggesting the Fe structural environment does not change significantly when zinc is also added to the protein. First inflection edge energies in the Fe XANES, which is a measurement of the average iron oxidation state, for the 2Fe-Isu1 and 1Zn/2Fe-Isu1 samples were measured to occur at 7121.4 ± 0.1 and 7122.7 ± 1.3 eV (Table 3), respectively, consistent with iron existing as Fe(II) within both samples; these values are similar to those previously reported for Fe(II) bound to D37A, wt-Isu1 and fly IscU.18, 30, 31 Analysis of the pre-edge 1s→3d electronic transitions in the Fe XANES, a feature that can be used to confirm metal-ligand symmetry and also metal oxidation state, for the Fe- and Zn/Fe-Isu1 samples appear at similar maximum energies, and peak areas are consistent with 6 coordinate octahedral high-spin Fe(II) model compounds in both samples (Figure 3B).58 However, addition of Zn to the Fe-Isu1 sample caused the already low 1s→3d transition area to decrease by ca. 50%, consistent with a increase in Fe-ligand coordination symmetry when zinc is added to the sample (Table 3). Analysis of the Zn XANES for the zinc-only loaded (1Zn-Isu1) and the 1Zn/2Fe-Isu1 samples are identical, indicating the Zn(II) coordination environment on Isu1 is unperturbed by Fe(II) binding. The Zn XANES edge max signal shows a duel nature suggesting metal is bound in a mixed Zn-S and Zn-O/N ligand environment.59, 60 Finally, Fe XANES spectra were used to test for differences in the Fe and Zn loading sequence when forming 1Zn/2Fe-Isu1 complex. These data, provided in Supplemental Figure 1 with edge fitting analysis provided in Supplemental Table 2, show there is no significant structural difference from the perspective of the iron when Zn(II) is loaded prior to or following Fe(II) loading onto Isu1.
Figure 3. Normalized XANES for Fe- and Zn-Loaded Isu1.
(A) Representative spectra for Fe XANES for 2Fe-Isu1 (red) and 1Zn/2Fe-Isu1 (black). (B) Expended display of the Fe-XANES pre-edge features for 2Fe-Isu1 (red) and 1Zn/2Fe-Isu1 (black). (C) Representative Zn XANES for 1Zn-Isu1 (blue) and 1Zn/2Fe-Isu1 (black).
Table 3. Analysis of Pre-edge and Edge First-Inflection Point Features from Fe-XANES for 2Fe-Isu1 and 1Zn/2Fe-Isu1.
Pre-edge analysis of the 1s→3d transition in Fe(II) Isu1 samples. Peak inflection values were calculated by taking the first derivate of the edge feature for each.
| Pre-Edge Area a | Peak-First Inflection Energy (eV) |
|
|---|---|---|
| 2Fe- Isu1 | 8.46 ± 2.94 | 7121.4 ± 0.1 |
| 1Zn/2Fe-Isu1 | 4.04 ± 1.34 | 7122.7 ± 1.3 |
Dimensionless area values.
Simulations of the X-ray absorption fine structure (EXAFS) region of the XAS spectrum provides direct metrical characteristics for ligands coordinated to the metal bound to a metalloprotein.61 Fourier transform (FT) of EXAFS provides a visualization of the pseudo radial distribution for metal-ligand coordination environments in the sample. Fe and Zn EXAFS analysis were used to characterize the metal site structure for Isu1 when metals are coordinated separately and in combination. The Fe-EXAFS spectra for 2Fe-Isu1 and 1Zn/2Fe-Isu1, and their subsequent FT spectra, are presented in Figure 4 (A, B, C & D). The single large feature in each FT suggests a highly symmetric metal-ligand coordination environment for Fe(II) in both samples (Figure 4B and D). Simulations of the Fe-EXAFS for 2Fe-Isu1 suggests iron is coordinated completely by oxygen and nitrogen-based ligands, with ca. 5 ligands centered at an average Fe-O/N bond length of 2.19 Å and an additional nearest-neighbor Fe-O/N ligand centered at 2.04 Å; long-range Fe•••C scattering is also observed at distances of 3.16 Å and 4.17 Å (Table 4). Simulations of the Fe-EXAFS for 1Zn/2Fe-Isu1 are consistent with only a single resolvable ca. 5 ± 1 coordinate Fe-O/N ligand environment centered at an average bond-length of 2.13 Å; long-range Fe•••C scattering is observed in this sample at 3.15 Å and 4.18 Å. The high Debye-Waller factor, a measure of the metal-ligand bond disorder, seen in the fit for the Fe-O/N environment in the 1Zn/2Fe-Isu1 sample suggests a change in the symmetry of the Fe-nearest neighbor ligands as compared to the 2Fe-Isu1 sample, however attempts to deconvolute two possible distinct nearest neighbor environments for the 1Zn/2Fe-Isu1 sample failed, subjective to the spectral resolution of 0.13 Å for the data, likely indicating a condensation of the two ligand environments in the Fe only sample as it moved to a more organized Fe-ligand environment when Zn is added.61 Bond lengths from the iron nearest neighbor ligand environments in both samples are consistent with distances reported for six coordinate Fe(II)-O/N models found within the Cambridge Structural Database.62
Figure 4: Raw EXAFS, Fourier Transforms of EXAFS and Spectral Simulations for Fe- and Zn-Isu1.
Full Fe EXAFS for 2Fe-Isu1 and 1Zn/2Fe-Isu1 are shown in (A) and (C), respectively. Full Zn EXAFS for 1Zn-Isu1 and 1Zn/2Fe-Isu1 are shown in (E) and (G), respectively. Fourier transforms for the Fe EXAFS 2Fe-Isu1 and 1Zn/2Fe-Isu1 are shown in (B) and (D), respectively. Fourier transforms for the Zn EXAFS for 1Zn-Isu1 and 1Zn/2Fe-Isu1 are shown in (F) and (H), respectively. Raw data is shown in black while the best-fit simulations for each data set are shown in green.
Table 4: Summary of Fe and Zn EXAFS Best Fit Simulation Parameters for Metal Loaded Isu1.
EXAFS data were simulated over the k-range of 1.0 to 13.0 k−1.
| Nearest-Neighbor Ligand Environment a |
Long-Range Ligand Environment a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Atom b | R(Å) c | C.N. d | σ2 e | Atom b | R(Å) c | C.N. d | σ2 e | F’ f |
| Fe EXAFS | |||||||||
| 2Fe-Isu1 | O/N | 2.04 | 1.0 | 1.07 | C | 3.16 | 2.0 | 2.81 | 0.15 |
| O/N | 2.19 | 5.0 | 3.42 | C | 4.17 | 4.0 | 5.22 | ||
| 1Zn/2Fe-Isu1 | O/N | 2.13 | 5.0 | 5.18 | C | 3.15 | 2.0 | 4.78 | 0.31 |
| C | 4.18 | 4.0 | 2.60 | ||||||
| Zn EXAFS | |||||||||
| 1Zn-Isu1 | O/N | 2.00 | 2.0 | 5.20 | C | 3.12 | 1.0 | 1.78 | 0.29 |
| S | 2.30 | 2.0 | 3.41 | C | 4.06 | 2.0 | 5.82 | ||
| 1Zn/2Fe-Isu1 | O/N | 2.00 | 2.0 | 4.66 | C | 3.13 | 2.0 | 5.56 | 0.47 |
| S | 2.30 | 2.0 | 3.40 | C | 4.07 | 2.0 | 3.02 | ||
Independent metal-ligand scattering environment.
Scattering atoms: N (nitrogen), O (oxygen), C (carbon), S (sulfur).
Average metal-ligand bond length (+/−) 0.12 Å−1.
Average metal-ligand coordination number (+/−) 1.0.
Average Debye-Waller factor (Å × 103).
Number of degrees of freedom weighted mean square deviation between data and fit.
Analysis of the Zn-Isu1 EXAFS in the absence (1Zn-Isu1) and presence of iron (1Zn/2Fe-Isu1) was used to characterize ligand metrical parameters for zinc bound to the protein, as well as to determine if the zinc structure is perturbed upon addition of iron. The Zn EXAFS for Zn-Isu1 and 1Zn/2Fe-Isu1 are shown in Figure 4 (panels E and G, respectively). The Fourier transforms of the Zn EXAFS in both samples show a nearest neighbor ligand pattern split into two independent environments (Figure 4F and H, respectively). Simulations of the Zn EXAFS in both samples are most consistent with a 4 coordinate Zn system with ca. 2 oxygen/nitrogen ligands centered at 2.00 Å and ca. 2 sulfur ligands centered at 2.30 Å. Debye-Waller (DW) factor values from simulations to the Zn-O/N and Zn-S nearest neighbor environments in both samples are very similar, indicating only minimal changes in the Zn-ligand bond disorder when Fe(II) is either present or absent. Bond lengths from Zn-O/N and Zn-S ligand average values in both samples are similar to those reported for 4 coordinate Zn-N2S2 models compounds (2.07 Å and 2.26 Å, respectively) published within the Cambridge Structural Database.62 Finally, in both samples, long-range Zn•••C scattering is observed at ca. 3.12 and 4.06 Å.
To compare iron coordination environments across orthologs, iron loaded Hs, Dm, Sc and Tm scaffold samples were prepared and characterized by Fe XAS along under identical conditions, loading only a single Fe(II) atom onto each protein. Full Fe EXAFS and Fourier transforms for the iron loaded orthologs are presented in Figure 5. Similarities in the Fourier transforms of the Fe-EXAFS between all samples suggest a conserved Fe-nearest neighbor ligand environment constructed by a ligand environment centered at a bond length of ca. 2.1 Å (displayed R + Δ is at 1.7Å), as shown in Figure 5B, D, F & H. Simulations of the Fe-EXAFS for the iron atom coordinated to yeast, human and fly scaffold orthologs suggest an overall 6 coordinate Fe-nearest-neighbor ligand environment constructed by two resolvable Fe-O/N ligand sets centered at an average bond length between 1.99 and 2.01 Å and the second between 2.14 and 2.15 Å (Table 5); the ligand set composition between these three samples is very similar. The Fe-nearest neighbor coordination environment for the bacterial scaffold ortholog is more symmetric, based on the fitting analysis, with only a single six coordinate Fe-nearest neighbor ligand environment constructed only by oxygen and nitrogen ligands centered at 2.13 Å. Long-range Fe•••C scattering is observed in all 4 samples, suggesting a complex backscattering pattern due to the carbon atoms attached to coordinating ligand atoms that exist as part of the amino acid side chains providing the direct metal ligand; the presence of these features confirm metal is bound to the protein and not simply existing as adventitious metal. Sulfur scattering was not observed for iron in any of the scaffold samples, consistent with our previous reports.18, 31 Finally, there is a subtle shift in the Fe-O/N bond lengths reported for 1Fe-Isu1 (Table 5) vs. 2Fe-Isu1 (Table 4), possibly suggesting the first iron atom that coordinates to Isu1 binds in a slightly different orientation. This would be consistent with the two binding affinities measured for Isu1 Fe binding we observed during our competition assay. However, differences between the one or two Fe(II) loaded Isu1 samples are at the limits of the uncertainty for the XAS technique, so these may be an artifact of data collection.
Figure 5. EXAFS, Fourier Transforms of EXAFS and Spectral Simulations for Fe-Loaded Isu1 orthologs.
Fe-EXAFS spectra and Fourier transforms of the Fe-EXAFS are shown respectively for a single iron atom bound to the following Isu1 orthologs: H. sapiens (panels A and B), D. melanogaster (panels C and D), S. cerevisiae (panels E and F) and T. maritima (panels G and H). Raw data is shown in black while simulated data is shown in green.
Table 5: Summary of Isu1 Ortholog Fe-EXAFS Simulation Best Fit Parameters.
Spectra were fit over a k-range of 1.0 to 13.0 k−1.
| Nearest-Neighbor Ligand Environment a |
Long-Range Ligand Environment a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Atom b | R(Å) c | C.N. d | σ2 e | Atom b | R(Å) c | C.N. d | σ2 e | F’ f |
| S. cerevisiae | O/N | 2.01 | 1.0 | 3.78 | C | 3.01 | 1.5 | 1.14 | 0.99 |
| O/N | 2.14 | 4.5 | 2.94 | C | 4.13 | 3.0 | 4.77 | ||
| H. sapiens | O/N | 2.01 | 1.5 | 2.02 | C | 3.13 | 3.0 | 5.83 | 0.45 |
| O/N | 2.14 | 4.0 | 1.28 | C | 3.53 | 1.0 | 1.00 | ||
| C | 4.13 | 6.5 | 5.95 | ||||||
| D. melanogaster | O/N | 1.99 | 3.0 | 3.78 | C | 3.13 | 1.5 | 1.14 | 0.99 |
| O/N | 2.15 | 2.0 | 2.94 | ||||||
| T. maritima | O/N | 2.13 | 6.0 | 4.30 | C | 3.10 | 3.0 | 1.56 | 0.99 |
| C | 3.27 | 4.5 | 1.20 | ||||||
| C | 3.49 | 4.5 | 1.66 | ||||||
| C | 3.71 | 3.5 | 3.90 | ||||||
Independent metal-ligand scattering environment.
Scattering atoms: N (nitrogen), O (oxygen), C (carbon), S (sulfur).
Average metal-ligand bond length (+/−) 0.12 Å−1.
Average metal-ligand coordination number (+/−) 1.0.
Average Debye-Waller factor (Å × 103).
Number of degrees of freedom weighted mean square deviation between data and fit.
Cluster Assembly Assay with Zn-Loaded Isu1
To clarify the impact zinc binding has on the ability of Isu1 to perform Fe-S cluster assembly, we screened the effect Zn has on the yeast NIaUF (complex with yeast Nfs1, Isu1, Isd11, Yfh1 and bacterial Acp) activity using our cluster assembly protocol.30 Fe-S cluster assembly activity by NIaUF was measured in the presence and absence of Zn. In the absence of zinc, the yeast NIaUF complex rapidly made Fe-S clusters when provided with both aqueous L-Cys and Fe(II) as substrates (Supplemental Figure 2); cluster assembly was measured by an increase in intensity of the 2Fe-2S cluster specific 430 nm CD spectral signal.30, 63 Following addition of a stoichiometric amount of Zn loaded onto Isu1 prior to NIaUF complex assembly, Fe-S cluster assembly activity was dramatically reduced (to ca. 10% of uninhibited as seen in the absence of Zn), indicating zinc significantly inhibits cluster assembly by the yeast NIaUF complex. Interestingly, a previous report using the human NIaU complex indicated complete inhibition of cysteine desulfurase activity by Zn, which could be reversed upon addition of frataxin.40 Here we measure only Fe-S cluster assembly, and our data shows the impact of Zn on inhibiting the entire pathway is dramatic even in the presence of yeast frataxin.
Sequence Alignment of Isu1 Orthologs
The amino acid sequence alignment for the yeast, fly, human and bacterial scaffolds are given in Figure 6 for use in comparison of conserved residues the might help illuminate which regions of the protein could be used for iron binding at a potential site distinct from the Cys rich protein active site. There is a high overall sequence homology between orthologs (ca. 94%, 99% and 90% between Sc and the Hs, Dm and Tm orthologs, respectively) and also a high sequence identity (67% and 61% between Sc and the Hs or the Dm orthologs, respectively) for the eukaryotic proteins. The sequence identity between the yeast and the prokaryotic samples are lower (23% between Sc and Tm). The three conserved active site cysteine residues (Isu1 residues Cys69, Cys96, Cys139) are highlighted in pink, the conserved active site aspartic acid (Isu1 residue Asp71) is highlighted in yellow, and the histidine residue conserved in eukaryotes also at the active site (Isu1 residue His138) is highlighted in orange within Figure 6. All these identified residues play key roles in scaffold Fe-S cluster assembly and constitute residues at the Zn binding sites in the human through bacterial scaffolds.29, 37–39 Met141, shown in red as the Isu1 suppressor mutant identified by the Dancis Lab (U Penn) that confers cluster assembly activity in yeast under frataxin depleted conditions,64 is in close proximity to the Isu1 residue Cys138. Folding in the C-terminal helical region where this methionine is located is variable within different scaffold structures depending on Zn or Fe-S cluster loading at the protein’s active site,65 suggesting dynamic transitions of structure in this region may play a role in metal delivery and/or Fe-S cluster assembly and release. Conserved residues observed in the primary sequence of ortholog family members that may contribute to an initial iron binding site unique can be identified through conservation of the acidic residues and also conserved histidines. Specific residues that have high sequence identity based using the Isu1 numbering scheme include: Glu42, Asp53, Asp81, Asp89, Glu108, Asp116/Asp117, and Glu129; each residue is conserved across both Prokaryotes and Eukaryotes.
Figure 6. Sequence alignment of Isu1 orthologs.
Mature protein sequences from H. sapiens, D. melanogaster, S. cerevisiae and T. maritima. Identical residues are shown in blue and conserved/semi conserved residues are shown in green. Conserved 3-Cys active site consists of C69, C96, and C139 (magenta) and D71 (yellow) or H138 (orange) according to the consensus ruler. The methionine conserved only in Eukaryotes (M141) is highlighted in red.
Modeling of Zn-Isu1 Metal Site Structure
Since the Isu1 structure has not been solved, we modeled the Zn-Isu1 structure and compared it to previously published Zn-bound ortholog structures.29, 39 The NCBI sequence for Isu1 was used to predict the protein’s structure.52, 56 The Isu1 Zn-site structure was energy minimized and the overall modelled structure was compared to known PDB crystallographic structures.56 Based on the modeled structure, shown in Figure 7, the single Zn atom binds to the Isu1 Fe-S cluster assembly active site using a subgroup of the conserved cysteine residues possibly including Cys69, Cys96 and Cys139. In this calculated structure, acidic residue Asp71 is positioned to provide additional O/N ligands to the bound metal, while a slight unfolding of the C-terminal helix would be required for His138 to participate as a ligand to coordinate Zn.
Figure 7. Sequence Alignment of Isu1 Orthologs.
Mature protein sequences from H. sapiens, D. melanogaster, S. cerevisiae and T. maritima. Identical residues are shown in blue and conserved/semi conserved residues are shown in green. Conserved 3-Cys active site consists of C69, C96, and C139 (magenta) and D71 (yellow) or H138 (orange) according to the consensus ruler. The methionine conserved only in Eukaryotes (M141) is highlighted in red.
DISCUSSION
Iron binding by the scaffold protein is an initial event that must occur during mitochondrial Fe-S cluster biosynthesis.37,40 In this report, we show Isu1 binds up to two iron atoms required for Fe-S cluster assembly at similar but distinct μM Fe(II) binding affinities. Here we utilized our competition Fe(II) binding assay to measure Isu1’s Fe(II) affinity in a manner that closer represents the solution environment experienced by the protein in vivo (i.e., in the presence of many Fe(II) binding molecules).31 As shown previously, iron binds to Isu1 as a stable high-spin symmetric 6 coordinate Fe(II) complex using only oxygen and nitrogen based ligands. The implication of having an Fe-binding ligand architecture constructed exclusively by oxygen and nitrogen ligands, in a manner shown here to be conserved across both prokaryotes and eukaryotes, is that the scaffold likely initially must bind Fe(II) at a site distinct from the protein’s sulfur rich active site before Fe-S cluster assembly can occur. Here we used CD spectroscopy to confirm that Isu1 binds Fe(II) while the protein exists in its disordered structural form, and our data indicate iron does not drive the protein into a structured state. While scaffold binding to the cysteine desulfurase forms a stable protein complex in the absence of iron,41, 66, 67 yeast pulldown analysis indicated Yfh1 was also a primary binding partner to Isu117, 68 and we have shown Yfh1 binds to Isu1 in an iron dependent manner.18 The dynamic structural characteristics seen for scaffold orthologs, either with regard to their intramolecular26, 69 or intermolecular protein flexibility,30 has been shown to be a factor for the scaffold when performing Fe-S cluster biosynthesis. These data therefore suggest that while the recent structural report showing atomic details of how human ISCU interacts with NIAUF, providing significant details regarding the multiprotein complex orientation,27, 29, 37, 69, 70 these structures do provide only a single snapshot of the cluster assembly apparatus at one state in time. Details suggesting an unstructured Fe(II)-loaded Isu1 homodimer or a Fe(II)-loaded Isu1/protein heterodimer therefore may exist prior to Nfs1 coordination at some during in vivo Fe-S cluster biosynthesis, and the dynamic aspects of the proteins should also be considered when discussing a cluster assembly mechanism.
In this report we reconfirm our previous result showing Fe(II) bound to Isu1 can be used to directly produce Fe-S clusters when Fe-Isu1 is part of the yeast NIaUF protein complex, assuming reductant and L-Cys are also provided.18, 30 This was previously confirmed using XAS, showing the Fe(II) bound to Isu1 in the O/N based ligand site, is redirected to a low-spin ca. 3 sulfur / 1 O/N nearest neighbor ligand site with a pronounced iron•••iron vector at 2.7Å ; these structural characteristics along with a CD signature consistent with a 2Fe-2S cluster indicates the bound iron is converted into a cluster.18, 31 To accomplish cluster assembly, the iron would therefore need to be redirected from the initial loading to Isu1 active site, or possibly the initial site consists of a subset of O and N based ligands at the protein’s active site. Mutagenesis studies identifying the initial Fe-binding site residues will be imperative for understanding this process. A comparison of amino acid sequences between orthologs suggests several potential Fe-binding residues on Isu1 could participate at this initial Fe binding site (i.e., Isu1 residues Glu42, Asp53, Asp81, Asp89, Glu108, Asp116/Asp117, and Glu129); mutagenesis experiments on Isu1 are underway to test each residue’s significance to Fe binding. We speculate that the scaffolds having an initial Fe(II) binding site separate from their cluster assembly site may help orchestrate the coordinated delivery of both substrates simultaneously or in series in order to drive cluster assembly. Furthermore, a scaffold structural transition that drives Fe delivery to the active site for use during Fe-S cluster assembly could be triggered when the scaffold associates with protein partners (i.e., Nfs1, Yah1 and/or Yfh1) post or concurrent with sulfur delivery, as recently suggested for the human system.41 In this way, scaffold dynamics contribute to substrate delivery and scaffold folding may provide the energy needed to promote the Fe(II) intramolecular translocation event that drives Fe(II) from initial to active site. And given that we see Fe(II) binding to similar sites in the 4 orthologs, metal transfer between sites it likely mechanistically important across family members.
Zinc binding has shown to stabilize the fold of several scaffold orthologs, allowing for their macromolecular structural determination.27, 29, 37, 69, 70 In this report, we show a single Zn ion binds directly to the Isu1 active site at sub μM binding affinity (Kd = 0.44 μM), a value an order of magnitude tighter than Isu1’s Fe binding affinity. These affinity differences between Fe(II) and Zn(II) are not surprising since zinc is predicted to bind more tightly in accordance to its position in the Irving-Williams series.71 Following our standard purification protocol, we show by ICP-MS that as-isolated “apo”-Isu1 is consistently partially loaded with only very low levels of contaminant iron and zinc (ca. 0.6% metal to protein stoichiometry), even after continuous exposure to EDTA during isolation. Our CD data confirms stoichiometric Zn drives Isu1 into its structured conformation, a transition observed for several scaffold orthologs.27,29,37,68,69 Our structural analysis also confirms Zn ions bind to Isu1 in the manner similar to that reported for Zn-loaded orthologs, i.e. through direct coordination in part to active site Cys residues.40 Regarding Fe-S cluster assembly activity, Zn binding to Isu1 when part of the yeast NIaUF complex dramatically inhibits cluster biosynthesis even when Zn-Isu1 is in the presence of frataxin and iron. Interestingly, it was recently shown that zinc binding to human ISCU inhibits cysteine desulfurase activity by the human NIaU complex, however this inhibition was reduced when frataxin was added.40 We believe both results may however be consistent. In the human report, while frataxin binding to the Zn-NIaU complex at the Zn-ISCU/NFS1 interface liberates the NFS1 active site Cys for participation in Zn coordination, hence allowing for cysteine desulfurase enzymatic activity to occur, the Zn in this structure remains stably bound to the ISCU active site.41 The zinc bound at the ISCU active site will certainly be expected to inhibit Fe-S assembly in a manner identical to what we see using the yeast proteins within this study. However, there could also differences that exist between the activities of the ortholog proteins, so verification of Fe-S assembly by human Zn-NIaU complex needs to be tested.
In this report, we also explore the impact on Isu1 when it binds both Zn and Fe ions. Our goal was to further explore the physiological impact of Zn binding to the scaffold and see if these data could supply additional mechanistic details related to iron delivery to the scaffold active site. Since Zn-Isu1 binding drives the Fe-loaded scaffold into its structured conformation, and Zn binds at the Isu1 active site in a manner similar to that observed when the scaffold has a Fe-S cluster loaded, these data support a molecular folding transition that is likely required that drives or is concurrent to cluster assembly. Analysis of the Fe XANES 1s→3d pre-edge feature for Fe-Isu1 indicates that binding zinc after the Fe(II) slightly alters the iron-ligand coordination symmetry but does not change the ligands to the metal or the metal spin state, and it indicates there is flexibility in the protein structure to allow for the subtle change in iron ligand environment after the additional metal binds. In contrast, iron binding to the Zn-loaded Isu1 where zinc is coordinated to the protein’s active site cysteine residues does not alter the protein’s overall fold or perturb the Zn structure, indicating Zn loaded protein is in a stable conformation. Our XAS results suggest the binding sequence of either Zn or Fe first does not preclude the final metal site structures. Combined, these data suggest a model where Fe and Zn binding occurs at independent sites on Isu1, and that while Zn binding favors/influences the structured state of Isu1, it does so without preventing Fe binding (Figure 8). However, only Zn free Isu1, when part of the NIaUF complex, is active for Fe-S cluster assembly, confirming Zn binding prevents Fe-S cluster assembly activity on Isu1.
Figure 8. Modeled Zinc Binding Site on Zn-Isu1.
Zn ion (green) coordinated by C69, C96, C139 (magenta) and H138 (orange) or D71 (yellow). Modeled using I-Tasser server.51,52
There are still several questions that remain regarding the physiological relevance or significance of Zn binding to scaffold orthologs. The biophysical impact of Zn loading onto Isu1’s structure is clear; Zn stabilizes the structured conformation of the protein, driving equilibrium from the disordered (D) to structured (S) protein conformation.26, 27, 37, 69 Zinc binding is obviously beneficial if one wants to study the structured state of the scaffold. Assuming the D→S structured transition relates directly to protein conformational changes seen during Fe-S cluster assembly, Zn loading also helps stabilize the homogeneous structured form of the scaffold, allowing for biochemical analysis of ortholog in this state. In our in vitro studies, where we strip zinc from our samples by chelation with EDTA, any unintended Zn binding to Isu1 and the subsequent inhibition of the NIaUF complex is certainly an artifact of handling conditions. Since zinc binds tighter than iron to Isu1, and it inhibits NIaUF activity, a normal in vivo activity for the metal during Fe-S cluster assembly seems unlikely. However, it is possible that a role for Zn in regulating ISC assembly under perturbed conditions in vivo may be of interest. In E. coli, under conditions of zinc toxicity, excess Zn ions disrupt Fe-S cluster biosynthesis on the scaffold by binding irreversibly to IscU, indicating a direct consequence for Zn binding and inhibition of the scaffold under Zn toxicity conditions.72 Furthermore, a close correlation between Zn and Fe-S clusters binding at similar sites on zinc-finger proteins suggest the selective placement of the correct metal (species) must be tightly regulated to ensure normal cell function, so these mechanisms need to be further investigated.73
Following the initial review of our manuscript, a detailed and highly informative article was published evaluating Zn binding to human ISCU; this rich paper nicely discussed the impact of Zn-loaded ISCU on cluster assembly in relation to direct Fe exchange at the scaffold’s active site, protein partner activation related to Fe-S assembly and it provided several details related to the cluster assembly mechanism that is performed by ISCU and its protein partners.74 There were several similarities between results outlined in this publication and ours, as well as several differences. Similarities include: their result indicate Zn binding precludes Fe loading at the ISCU active site; frataxin alone does not promote exchange of Fe for Zn bound to the ISCU active site; their appearance that a Fe-ISCU species with only N/O ligands (albeit at only 15% of their total sample) exists following Fe loading, and their result showing Zn blocks Fe-S cluster assembly. Key differences include: they show iron binding drives ISCU into the structured conformation (our CD analysis suggests this is not the case for Isu1); they show Fe loaded ISCU, in the presence of frataxin but the absence of ferredoxin, does not make clusters, our cluster activity assay by CD in combination with our XAS analysis (appearance of a ca. 2.7Å Fe•••Fe vector and Fe-S nearest neighbor ligands) shows that a 2Fe-2S cluster are formed on Isu1; they show Zn-loaded ISCU in the absence of ferredoxin but with frataxin will produce clusters only at elevated (7X) stoichiometric concentrations of L-Cys, our Zn-loaded sample does not, even at 50X relative L-Cys concentrations. Their CD, Mossbauer and MS analysis clearly show in their samples that bulk Fe(II) will bind and replace Zn(II) at the ISCU active site.74 In our hands, we have consistently shown in the yeast and fly orthologs18, 30, 31 that Fe(II) binds completely at a ligand site consistent of only O/N based ligands, and we have further demonstrated in this report that binding is conserved in the human and bacterial systems. As a point of clarification, they acknowledge our Fe-O/N exclusive ligand binding articles but indicate we have not shown this Fe(II) is directed to form Fe-S clusters. We have, in fact, illustrated this using yeast Isu1 early on, showing by XAS the formation of Fe-S and 2.7Å Fe•••Fe structured iron species following the chemical addition of activated sulfur,18 and again through enzymatic addition of sulfur by the cysteine desulfurase that drives the Fe in the O/N ligand site to make Fe-S clusters on the fly Isu1 ortholog.30 A limitation in others and our previous work, accurately pointed out in the Gervason paper, is that we used DTT instead of the ferredoxin to provide reducing equivalents and we do this again in this report; however our results confirm, at the DTT concentrations utilized in our activity assay, we are still making 2Fe-2S clusters as our major species and not producing the Fe-S aggregates cautiously reported to be produced with DTT by the Barondeau lab.63 A limitation in the XAS technique is that while the method is highly accurate for measuring metal-ligand bond lengths (accuracy of ± 0.02Å), it is in fact an averaging technique; so while XANES analysis provides direct insight into the oxidation and spin state of bound iron, and EXAFS analysis provides insight into the iron-ligand bond lengths, coordination number and ligand identity, these analysis are for the averaged species found for the protein in solution. We concede the Fe-O/N species we observe in all orthologs within this report are therefore only the predominate species, and we cannot exclude the possibility that a small portion of Fe that exists in our samples is not ligated to sulfur atoms. However, we believe the appearance of a Fe-O/N only ISCU species in the Gervason report (albeit at only 15%) and the fact that we observe this species as the predominate form in our report (likely at above 90% of our species) may suggest a shift in equilibrium between a Fe-O/N and a Fe-S dominated species occurs depending on the handling and environmental conditions. Without identification of residues that provide the O/N ligands by our lab or others, it is also equally impossible for us to conform if the O/N based ligands we observe bound to Fe(II) in Isu1 its orthologs do not originate from the scaffold active site His and Asp residues. Therefore, we believe further exploration directed at understanding the differences between the Gervason report and ours/others will help provide additional insight related to the Fe binding and transfer processes in relation to Fe-S cluster assembly, so this will be a directive for our future research.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by funds from the National Institutes of Health for B.L. and T.L.S. (R01 DK068139) and J.A.C. (R01 AI072443). B.L. was also supported by a NIH-Detroit Cardiovascular Training Grant to Wayne State University (T32HL12082205). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL) on beamlines 7-3 and 9-3, and at the National Synchrotron Light Source (NSLS) on beamline X3b. SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program.
ABBREVIATIONS
- XAS
X-ray absorption spectroscopy
- XANES
X-ray absorption near edge spectroscopy
- EXAFS
extended X-ray absorption fine structure
- ITC
isothermal titration calorimetry
- CD
circular dichroism
- NFU
normalized fluorescence units
- NIaUF
yeast protein complex including Nfs1 (N), Isd11 (I), bacterial Acp1 (a), Isu1 (U) and Yfh1 (F)
BIBLIOGRAPHIC REFERENCES
- 1.Banci L and Bertini I, in Metallomics and the Cell. Metal Ions in Life Sciences, ed. Banci L, Springer, Dordrecht, 2013, vol. 12, pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 2.Ward RJ and Crichton RR, Ironing out the Brain, Met Ions Life Sci, 2019, 19. [DOI] [PubMed] [Google Scholar]
- 3.Sherman HG, Jovanovic C, Stolnik S, Baronian K, Downard AJ and Rawson FJ, New Perspectives on Iron Uptake in Eukaryotes, Front Mol Biosci, 2018, 5, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lill R, Broderick JB and Dean DR, Special issue on iron-sulfur proteins: Structure, function, biogenesis and diseases, Biochim Biophys Acta, 2015, 1853, 1251–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wachnowsky C, Fidai I and Cowan JA, Iron-sulfur cluster biosynthesis and trafficking - impact on human disease conditions, Metallomics : integrated biometal science, 2018, 10, 9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barupala DP, Dzul SP, Riggs-Gelasco PJ and Stemmler TL, Synthesis, delivery and regulation of eukaryotic heme and Fe-S cluster cofactors, Arch Biochem Biophys, 2016, 592, 60–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melber A and Winge DR, Steps Toward Understanding Mitochondrial Fe/S Cluster Biogenesis, Methods Enzymol, 2018, 599, 265–292. [DOI] [PubMed] [Google Scholar]
- 8.Braymer JJ and Lill R, Iron-sulfur cluster biogenesis and trafficking in mitochondria, J Biol Chem, 2017, 292, 12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber J, Neumann K, Prohl C, Muhlenhoff U and Lill R, The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins, Mol Cell Biol, 2004, 24, 4848–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerber J, Muhlenhoff U and Lill R, An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1, EMBO Rep, 2003, 4, 906–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Vranken JG, Jeong MY, Wei P, Chen YC, Gygi SP, Winge DR and Rutter J, The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis, Elife, 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiedemann N, Urzica E, Guiard B, Muller H, Lohaus C, Meyer HE, Ryan MT, Meisinger C, Muhlenhoff U, Lill R and Pfanner N, Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins, Embo J, 2006, 25, 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adam AC, Bornhovd C, Prokisch H, Neupert W and Hell K, The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria, Embo J, 2006, 25, 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cory SA, Van Vranken JG, Brignole EJ, Patra S, Winge DR, Drennan CL, Rutter J and Barondeau DP, Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions, Proc Natl Acad Sci U S A, 2017, 114, E5325–E5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R and Lill R, Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis, Proc Natl Acad Sci U S A, 2010, 107, 11775–11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M and Kaplan J, Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin, Science, 1997, 276, 1709–1712. [DOI] [PubMed] [Google Scholar]
- 17.Ramazzotti A, Vanmansart V and Foury F, Mitochondrial functional interactions between frataxin and Isu1p, the iron-sulfur cluster scaffold protein, in Saccharomyces cerevisiae, FEBS Lett, 2004, 557, 215–220. [DOI] [PubMed] [Google Scholar]
- 18.Cook JD, Kondapalli KC, Rawat S, Childs WC, Murugesan Y, Dancis A and Stemmler TL, Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe-S cluster assembly, Biochemistry, 2010, 49, 8756–8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foury F, Pastore A and Trincal M, Acidic residues of yeast frataxin have an essential role in Fe-S cluster assembly, EMBO Rep, 2007, 8, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bou-Abdallah F, Adinolfi S, Pastore A, Laue TM and Chasteen N. Dennis, Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli, J Mol Biol, 2004, 341, 605–615. [DOI] [PubMed] [Google Scholar]
- 21.He Y, Alam SL, Proteasa SV, Zhang Y, Lesuisse E, Dancis A and Stemmler TL, Yeast frataxin solution structure, iron binding, and ferrochelatase interaction, Biochemistry, 2004, 43, 16254–16262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park S, Gakh O, O’Neill HA, Mangravita A, Nichol H, Ferreira GC and Isaya G, Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation., J. Biol. Chem, 2003, 278, 31340–31351. [DOI] [PubMed] [Google Scholar]
- 23.Pandey A, Gordon DM, Pain J, Stemmler TL, Dancis A and Pain D, Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly, J Biol Chem, 2013, 288, 36773–36786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bridwell-Rabb J, Fox NG, Tsai CL, Winn AM and Barondeau DP, Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry, Biochemistry, 2014, 53, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adinolfi S, Iannuzzi C, Prischi F, Pastore C, Iametti S, Martin SR, Bonomi F and Pastore A, Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS, Nat Struct Mol Biol, 2009, 16, 390–396. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Fuzery AK, Tonelli M, Ta DT, Westler WM, Vickery LE and Markley JL, Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB, Biochemistry, 2009, 48, 6062–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markley JL, Kim JH, Dai Z, Bothe JR, Cai K, Frederick RO and Tonelli M, Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery, FEBS Lett, 2013, 587, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansy SS and Cowan JA, Iron-sulfur cluster biosynthesis: toward an understanding of cellular machinery and molecular mechanism, Acc Chem Res, 2004, 37, 719–725. [DOI] [PubMed] [Google Scholar]
- 29.Boniecki MT, Freibert SA, Muhlenhoff U, Lill R and Cygler M, Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex, Nat Commun, 2017, 8, 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dzul SP, Rocha AG, Rawat S, Kandegedara A, Kusowski A, Pain J, Murari A, Pain D, Dancis A and Stemmler TL, In vitro characterization of a novel Isu homologue from Drosophila melanogaster for de novo FeS-cluster formation, Metallomics : integrated biometal science, 2017, 9, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues AV, Kandegedara A, Rotondo JA, Dancis A and Stemmler TL, Iron loading site on the Fe-S cluster assembly scaffold protein is distinct from the active site, Biometals, 2015, 28, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nuth M, Yoon T and Cowan JA, Iron-sulfur cluster biosynthesis: characterization of iron nucleation sites for assembly of the [2Fe-2S]2+ cluster core in IscU proteins, Journal of the American Chemical Society, 2002, 124, 8774–8775. [DOI] [PubMed] [Google Scholar]
- 33.Cai K, Frederick RO, Tonelli M and Markley JL, ISCU(M108I) and ISCU(D39V) Differ from Wild-Type ISCU in Their Failure To Form Cysteine Desulfurase Complexes Containing Both Frataxin and Ferredoxin, Biochemistry, 2018, 57, 1491–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick SP, Moore MJ and Lindahl PA, Detection of Labile Low-Molecular-Mass Transition Metal Complexes in Mitochondria, Biochemistry, 2015, 54, 3442–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore MJ, Wofford JD, Dancis A and Lindahl PA, Recovery of mrs3Deltamrs4Delta Saccharomyces cerevisiae Cells under Iron-Sufficient Conditions and the Role of Fe580, Biochemistry, 2018, 57, 672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holmes-Hampton GP, Miao R, Morales J. Garber, Guo Y, Munck E and Lindahl PA, A nonheme high-spin ferrous pool in mitochondria isolated from fermenting Saccharomyces cerevisiae, Biochemistry, 2010, 49, 4227–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iannuzzi C, Adrover M, Puglisi R, Yan R, Temussi PA and Pastore A, The role of zinc in the stability of the marginally stable IscU scaffold protein, Protein Science : A Publication of the Protein Society, 2014, 23, 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Oganesyan N, Shin DH, Jancarik J, Yokota H, Kim R and Kim SH, Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form, Proteins, 2005, 59, 875–881. [DOI] [PubMed] [Google Scholar]
- 39.Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT and Kennedy MA, Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site, J Mol Biol, 2004, 344, 567–583. [DOI] [PubMed] [Google Scholar]
- 40.Fox NG, Martelli A, Nabhan JF, Janz J, Borkowska O, Bulawa C and Yue WW, Zinc(II) binding on human wild-type ISCU and Met140 variants modulates NFS1 desulfurase activity, Biochimie, 2018, 152, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox NG, Yu X, Feng X, Bailey HJ, Martelli A, Nabhan JF, Strain-Damerell C, Bulawa C, Yue WW and Han S, Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism, Nat Commun, 2019, 10, 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foster MW, Mansy SS, Hwang J, Penner-Hahn JE, Surerus KK and Cowan JA, A Mutant Human IscU Protein Contains a Stable [2Fe-2S]2+ Center of Possible Functional Significance, J. Am. Chem. Soc, 2000, 122, 6805–6806. [Google Scholar]
- 43.Huang J, Dizin E and Cowan JA, Mapping iron binding sites on human frataxin: implications for cluster assembly on the ISU Fe-S cluster scaffold protein, J Biol Inorg Chem, 2008, 13, 825–836. [DOI] [PubMed] [Google Scholar]
- 44.Kuzmic P, Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase, Anal. Biochem, 1996, 237, 260–273. [DOI] [PubMed] [Google Scholar]
- 45.Sreerama N and Woody RW, Computation and analysis of protein circular dichroism spectra, Methods Enzymol, 2004, 383, 318–351. [DOI] [PubMed] [Google Scholar]
- 46.Rehr JJ and Ankudinov AL, Progress and challenges in the theory and interpretation of X-ray spectra, J Synchrotron Radiat, 2001, 8, 61–65. [DOI] [PubMed] [Google Scholar]
- 47.Pickering IJ, George GN, Yu EY, Brune DC, Tuschak C, Overmann J, Beatty JT and Prince RC, Analysis of sulfur biochemistry of sulfur bacteria using X-ray absorption spectroscopy, Biochemistry, 2001, 40, 8138–8145. [DOI] [PubMed] [Google Scholar]
- 48.Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR and Stemmler TL, Monomeric Yeast Frataxin Is an Iron-Binding Protein, Biochemistry, 2006, 45, 7767–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith AT, Barupala D, Stemmler TL and Rosenzweig AC, A new metal binding domain involved in cadmium, cobalt and zinc transport, Nature chemical biology, 2015, 11, 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sievers F and Higgins DG, Clustal Omega for making accurate alignments of many protein sequences, Protein Sci, 2018, 27, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy A, Kucukural A and Zhang Y, I-TASSER: a unified platform for automated protein structure and function prediction, Nat Protoc, 2010, 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J and Zhang Y, Protein Structure and Function Prediction Using I-TASSER, Curr Protoc Bioinformatics, 2015, 52, 5 8 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLano WL, PyMOL.Journal, 2002. [Google Scholar]
- 54.Koebke K, Batelu S, Koandegedara A, Smith SM and Stemmler TL, Refinement of Protein Fe(II) Binding Characteristics Utilizing a Competition Assay Exploiting Small Molecule Ferrous Chelators, Journal of Inorganic Biochemistry, 2019, Submitted. [DOI] [PubMed] [Google Scholar]
- 55.Shen H, Wang F, Zhang YX, Xu JJ, Long JG, Qin HG, Liu F and Guo JS, Zinc distribution and expression pattern of ZnT3 in mouse brain, Biological Trace Element Research, 2007, 119, 166–174. [DOI] [PubMed] [Google Scholar]
- 56.Roy P, Bauman MA, Almutairi HH, Jayawardhana WG, Johnson NM and Torelli AT, Comparison of the Response of Bacterial IscU and SufU to Zn(2+) and Select Transition-Metal Ions, ACS Chem Biol, 2018, 13, 591–599. [DOI] [PubMed] [Google Scholar]
- 57.Bandyopadhyay S, Chandramouli K and Johnson MK, Iron-sulfur cluster biosynthesis, Biochem Soc Trans, 2008, 36, 1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Randall CR, Shu L, Chiou Y-M, Hagen KS, Ito M, Kitajima N, Lachicotte RJ, Zang Y and Que L Jr., X-ray absorption pre-edge studies of high-spin iron(II) compounds, Inorg. Chem, 1995, 34, 1036–1039. [Google Scholar]
- 59.Wang B, Alam SL, Meyer HH, Payne M, Stemmler TL, Davis DR and Sundquist WI, Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4, J Biol Chem, 2003, 278, 20225–20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sazinsky MH, LeMoine B, Orofino M, Davydov R, Bencze KZ, Stemmler TL, Hoffman BM, Arguello JM and Rosenzweig AC, Characterization and structure of a Zn2+ and [2Fe-2S]-containing copper chaperone from Archaeoglobus fulgidus, J Biol Chem, 2007, 282, 25950–25959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bencze KZ, Kondapalli KC and Stemmler TL, in Applications of Physical Methods in Inorganic and Bioinorganic Chemistry: Handbook, Encyclopedia of Inorganic Chemistry, 2nd Edition, eds. Scott RA and Lukehart CM, John Wiley & Sons,LTD, Chichester, UK, 2007, pp. 513–528. [Google Scholar]
- 62.Groom CR, Bruno IJ, Lightfoot MP and Ward SC, The Cambridge Structural Database, Acta Crystallogr B Struct Sci Cryst Eng Mater, 2016, 72, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox NG, Chakrabarti M, McCormick SP, Lindahl PA and Barondeau DP, The Human Iron-Sulfur Assembly Complex Catalyzes the Synthesis of [2Fe-2S] Clusters on ISCU2 That Can Be Transferred to Acceptor Molecules, Biochemistry, 2015, 54, 3871–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon H, Knight SA, Pandey A, Pain J, Turkarslan S, Pain D and Dancis A, Turning Saccharomyces cerevisiae into a Frataxin-Independent Organism, PLoS Genet, 2015, 11, e1005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimomura Y, Wada K, Fukuyama K and Takahashi Y, The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis, J Mol Biol, 2008, 383, 133–143. [DOI] [PubMed] [Google Scholar]
- 66.Schmucker S, Martelli A, Colin F, Page A, Wattenhofer-Donze M, Reutenauer L and Puccio H, Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex, PLoS One, 2011, 6, e16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai CL and Barondeau DP, Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex, Biochemistry, 2010, 49, 9132–9139. [DOI] [PubMed] [Google Scholar]
- 68.Wang T and Craig EA, Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu, J Biol Chem, 2008, 283, 12674–12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim JH, Tonelli M, Kim T and Markley JL, Three-dimensional structure and determinants of stability of the iron-sulfur cluster scaffold protein IscU from Escherichia coli, Biochemistry, 2012, 51, 5557–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan R, Kelly G and Pastore A, The scaffold protein IscU retains a structured conformation in the Fe-S cluster assembly complex, Chembiochem, 2014, 15, 1682–1686. [DOI] [PubMed] [Google Scholar]
- 71.Irving H and Williams RJP, Order of Stability of Metal Complexes, Nature, 1948, 162, 746–747. [Google Scholar]
- 72.Li J, Ren X, Fan B, Huang Z, Wang W, Zhou H, Lou Z, Ding H, Lyu J and Tan G, Zinc Toxicity and Iron-Sulfur Cluster Biogenesis in Escherichia coli, Appl Environ Microbiol, 2019, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shimberg GD, Michalek JL, Oluyadi AA, Rodrigues AV, Zucconi BE, Neu HM, Ghosh S, Sureschandra K, Wilson GM, Stemmler TL and Michel SL, Cleavage and polyadenylation specificity factor 30: An RNA-binding zinc-finger protein with an unexpected 2Fe-2S cluster, Proc Natl Acad Sci U S A, 2016, 113, 4700–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gervason S, Larkem D, Mansour AB, Botzanowski T, Muller CS, Pecqueur L, Le Pavec G, Delaunay-Moisan A, Brun O, Agramunt J, Grandas A, Fontecave M, Schunemann V, Cianferani S, Sizun C, Toledano MB and D’Autreaux B, Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin, Nat Commun, 2019, 10, 3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.