Abstract
Potato (Solanum tuberosum) is one of the most important crops in the world. However, it is generally difficult to breed a new variety of potato crops because they are highly heterozygous tetraploid. Steroidal glycoalkaloids (SGAs) such as α-solanine and α-chaconine found in potato are antinutritional specialized metabolites. Because of their toxicity following intake, controlling the SGA levels in potato varieties is critical in breeding programs. Recently, genome-editing technologies using artificial site-specific nucleases such as TALEN and CRISPR-Cas9 have been developed and used in plant sciences. In the present study, we developed a highly active Platinum TALEN expression vector construction system, and applied to reduce the SGA contents in potato. Using Agrobacterium-mediated transformation, we obtained three independent transgenic potatoes harboring the TALEN expression cassette targeting SSR2 gene, which encodes a key enzyme for SGA biosynthesis. Sequencing analysis of the target sequence indicated that all the transformants could be SSR2-knockout mutants. Reduced SGA phenotype in the mutants was confirmed by metabolic analysis using LC-MS. In vitro grown SSR2-knockout mutants exhibited no differences in morphological phenotype or yields when compared with control plants, indicating that the genome editing of SGA biosynthetic genes such as SSR2 could be a suitable strategy for controlling the levels of toxic metabolites in potato. Our simple and powerful plant genome-editing system, developed in the present study, provides an important step for future study in plant science.
Keywords: genome editing, potato, steroidal glycoalkaloids, TALEN
Introduction
Potato (Solanum tuberosum) is the fourth most important crop in the world. Breeding of potato is challenging due to its polyploidy and heterogeneity. Generally, it takes more than 10 years to realize a new cultivar during potato breeding (Asano and Tamiya 2016; Tiwari et al. 2013). Steroidal glycoalkaloids (SGAs) are specialized metabolites that are majorly found in solanaceous plants including potato. Since the major SGAs in potato, α-solanine and α-chaconine, are toxic and bitter, potato breeders have been attempting to generate SGA-free potato cultivars for a long time. However, such efforts have not been a complete success. Recently, we identified sterol side chain reductase 2 (SSR2) encoding a key enzyme for SGA biosynthesis in solanaceous plants (Sawai et al. 2014). SSR2 catalyzes the conversion of cycloartenol and desmosterol to cycloartanol and cholesterol, respectively. Since cycloartanol and cholesterol are the common precursors of SGAs in potato, we attempted to disrupt the SSR2 gene to obtain toxic SGA-free potato plants using an artificial nuclease, transcription activator-like effector (TALE) nuclease (TALEN) with constant TALE repeats. Among twenty-nine transgenic potato plants, two transgenic lines exhibit targeted mutagenesis in SSR2 and one seems to be SSR2-disrupted since no intact SSR2 sequence has been detected in the plant. As expected, SSR2-disrupted potato plants show an SGA-reduced phenotype. However, the efficiency of obtaining mutants in transgenic potatoes remains still low. In addition, based on sequencing analysis, the knockout mutant seems to be a genetic chimera/mosaic. Highly active TALENs with variable TALE repeats harboring non-repeat-variable di-residue (non-RVD) variations called Platinum TALENs have been developed (Sakuma et al. 2013a). Platinum TALENs have been applied to introduce targeted mutagenesis in cultured human cell lines (Sakuma et al. 2013a), frogs (Sakane et al. 2014; Sakuma et al. 2013a), rats (Sakuma et al. 2013a), fungi (Mizutani et al. 2016), and plant tissues (Kusano et al. 2016). However, there have been no reports regarding the application of Platinum TALEN for genome editing in whole regenerated plants.
Generally, the artificial site-specific nuclease expression cassettes (transgenes) in the genome-edited plants are removed by crossing to generate the null-segregants. Since multiple transgene insertions in parent lines reduce obtaining null-segregants in the progenies, it is important to select plants carrying a single copy of the transgenes. The Southern blotting analysis is a conventional method for estimating copy numbers of transgene in transgenic plants. In the present study, we developed a Platinum TALEN expression vector construction system, generated transgenic potatoes with the vector, and determined copy numbers of transgene in them by a simple quantitative real-time PCR based method instead of the Southern blotting analysis. Our study will provide new options for effective procedures of plant genome editing.
Materials and methods
TALEN expression vector construction
MultiSite Gateway system adaptive capture vectors for Platinum Gate TALEN system were constructed as follows. The region containing 6-module Golden Gate TALEN scaffold (NC-type which have N- and C-terminal truncated TALEN scaffold, Sakuma et al. 2013b) was amplified from pGW-TAL-NC (Zeo) (Sawai et al. 2014) using primer sets 5′-AAA GCA GGC TTA ACT AGT AAA AAT GGC TTC CTC CCC-3′/5′-AAA AGT TGG GTG GGC GCG CCC ACC CTT T-3′ and 5′-AAT AAA GTT GTA ACT AGT AAA AAT GGC TTC CTC C-3′/5′-GAA AGC TGG GTT GGC GCG CCC ACC CTT T-3′, and attB adapter primers 5′-GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT A-3′/5′-GGG GAC AAC TTT GTA TAG AAA AGT TGG GTG-3′ and 5′-GGG GAC AAC TTT GTA TAA TAA AGT TGT A-3′/5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTT-3′. The attB surrounding PCR products (attB1-attB4 and attB3-attB2) were cloned into pDONR221-P1P4 and pDONR221-P3-P2 by Gateway BP reaction to create pENTR-attL1-TAL-NC-attL4 and pENTR-attL3-TAL-NC-attL2, respectively. To alter the bacterial selection marker from kanamycin resistance gene to zeocin resistance gene, pENTR-attL1-TAL-NC-attL4 and pENTR-attL3-TAL-NC-attL2 were digested using AflII and EcoRV and ligated with pDONR/Zeo vector treated with the same restriction enzymes. The resulting plasmid vectors were named pYS_016 (pENTR-attL1-TAL-NC-attL4 [Zeo]) and pYS_017 (pENTR-attL3-TAL-NC-attL2 [Zeo]).
The region containing Platinum Gate TALEN scaffold was amplified from ptCMV-153/47VR-XX (XX=HD, NG, NI or NN) (Sakuma et al. 2013a) using primer set 5′-CAC CAC TAG TAA AAA TGG CTT CCT CCC CTC CAA AGA AAA-3′/5′-TTA AAA GTT TAT CTC ACC GTT ATT A-3′. pGW-TAL-NC (Zeo), pYS_016, pYS_017, and the PCR amplicons were digested using SpeI and MfeI and ligated to create pYS_021-XX (attL1-attL2), pYS_022-XX (attL1-attL4) and pYS_023-XX (attL3-attL2).
To construct a pRI 101-AN-GW vector, Gateway reading flame cassette was inserted into the SmaI site of pRI 101 AN (Takara Bio, Japan). The region containing NOS promoter-Hpt-Rbcs terminator in pH35GC (Inplanta Inovations Inc., Japan) was amplified using 5′-ATT CCC GGG TTT CTG GAG TTT AAT GAG CTA AGC-3′/5′-GCG ATC GCG GGG ACC CAT CGA TGC ATC GAT-3′. The plasmid backbone of pRI 101-AN-GW was amplified using 5′-GGC CGG CCA TGT TAA TTA AGG GGA CCT GCA GGC ATG CAA-3′/5′-CAG AAA CCC GGG AAT TTG GGC CAT CGC CC-3′, CaMV35SP-AtADH 5′-UTR in pRI 101-AN-GW was amplified using 5′-GGT CCC CGC GAT CGC TGC CTG CAG GTC CCC AGA TTA-3′/5′-GGC GCG CCG CGG CCG CAC TAG TCT CGA TAT CAA CAG TGA AGA ACT TGC TTT T-3′, while HSP terminator in pESTRA (Sawai et al. 2014) was amplified using 5′-CGG CCG CGG CGC GCC TCG CGA GCT CTA TGA AGA TGA AGA TGA A-3′/5′-TTA ACA TGG CCG GCC GAA TTC GCC CTT GGG TTT AAA CCC-3′. The four PCR amplicons were fused using an In-Fusion HD Cloning Kit (Takara Bio) to generate a pYS_001 vector. The region containing RbcsT-35SP-5′UTR in pYS_001 was amplified using 5′-TAC AAA GTT GTA AGT AGA TGC CGA CCG GG-3′/5′-ATA CAA AGT TGT TAT CAA CAG TGA AGA ACT TGC TTT-3′ and further amplified using 5′-GGG GAC AAC TTT TCT ATA CAA AGT TGT A-3′/5′-GGG GAC AAC TTT ATT ATA CAA AGT TGT-3′. It was subsequently transferred to pDONR 221 P4r-P3r using a BP reaction to construct pYS_018 (attR4-RbcsT-35SP-5′UTR-attR3).
To construct destination vector pYS_015, pRI 101-AN-GW was digested with NdeI and SacI and a Gateway cassette was inserted in the NdeI and SacI sites of pRI 201-AN (Takara Bio).
To construct SSR2_C targeting Platinum TALEN entry vectors (Sawai et al. 2014), we followed Platinum Gate TALEN construction system (Sakuma et al. 2013a). During the second assembly, pYS_022-NI and pYS_023-NG vectors were used as capture vectors for the left and right TALEN monomers, respectively. pYS_022-SSR2_C_Left, pYS_023-SSR2_C_Right, pYS_018 digested by NruI, and pYS_015 were mixed with LR Clonase II Plus enzyme mix (Invitrogen) to obtain a plant expression vector, pYS_026-SSR2_C (Supplementary Figure S1).
Plant materials and bacterial strain
A potato plant cultivar, Sassy, was cultured in vitro on Murashige and Skoog (1962) medium, without phytohormones (pH 5.7) or grown in soil, with a photoperiod of 16 h at 23°C. For genetic transformation, Agrobacterium tumefaciens GV3101 with pMP90 was used.
Transformation and plant regeneration
Internodal explants (4–6 mm) in vitro plants were used for the transformation. The agrobacteria with pYS_026-SSR2_C were incubated for 15 h at 28°C in YEB medium (1 g/l yeast extract, 5 g/l beef extract, 5 g/l peptone, 5g/l sucrose, 0.5 g/l MgSO4 7H2O, pH 7.0) with 50 mg/l kanamycin, prior to co-cultivation with the explants. The explants were incubated on Growth Medium (MS medium with salts and vitamins, 3% sucrose, pH 5.7) containing 1 : 10 vol. of bacterial suspension for 30 s, blotted dry on a filter paper, and cultured on Co-cultivation Medium (Growth medium containing 2 mg/l trans-zeatin (Tokyo Kasei), 0.05 mg/l indole-3-acetic acid (IAA, Wako) and 100 µM acetosyringone (Sigma)). After 3 days of co-cultivation, the explants were placed on Selection medium (Growth Medium containing trans-zeatin 2 mg/l, 0.05 mg/l IAA, 25 mg/l kanamycin, and 250 mg/l carbenicillin). When they were well developed, shoots were sub cultured on MS medium containing 100 mg/l kanamycin and 250 mg/l carbenicillin.
Confirmation of targeted mutagenesis in the SSR2 gene and off-target activity against the SSR1 gene
Genomic regions surrounding the SSR2 target sites were amplified by TaKaRa Ex Taq (Takara Bio) with the primer set 5′-GCT ATT CCG TGG TCT CAA GG-3′/5′-TGG ACC ATA AAT CAT GCC TTC-3′ or 5′-ACC CTA GGA GGA AGA TCC AG-3′/5′-TGG ACC ATA AAT CAT GCC TTC-3′. PCR amplicons were cloned using TOPO TA Cloning Kit (Invitrogen). Randomly selected clones were selected for the Sanger sequencing analysis. The potential off-target region of the SSR1 gene was also amplified with the primer set 5′-CAC CAT GAC AGA TGT TCA GGC TCC-3′/5′-TCA ATC TTC AGG CTC ATC AAC T- 3′ and the amplicons were used for template DNA in direct sequencing analysis using primer 5′-GAT GAT CTT ACT GTT GGT GG-3′.
Harvest and SGA measurements of the tubers of SSR2-edited potato plants
In vitro SSR2-edited potato plants and non-transgenic Sassy were planted in soil. After about five months of cultivation in a green house, tubers were harvested and the weights were measured. Extraction and quantification of SGAs contained in the SSR2-edited transgenic plants were performed as previously described (Nakayasu et al. 2017).
Estimation of transgene copy number in genome-edited potato plants
Genomic DNA was extracted and purified from each edited genome and non-transgenic potato plants using NucleoSpin Plant II (Takara Bio) according to the manufacturer’s protocol. Quantitative real time PCR (qPCR) analysis was performed using FastStart Essential Master Mix (Roche) reagent and LightCycler Nano system (Roche). To construct the standard plasmid for relative quantification, a partial sequence of potato adenine phosphoribosyl transferase (APRT: accession number CK270447) was amplified from Sassy genomic DNA using 5′-GAA CCG GAG CAG GTG AAG AA-3′/5′-GAA GCA ATC CCA GCG ATA CG-3′ (Nicot et al. 2005) and cloned into pCR4 Blunt TOPO vector (Invitrogen). The resulting vector was digested with EcoRI and the fragment containing a partial APRT was inserted into the EcoRI site of pRI 201-AN to generate the standard plasmid, pRI 201-APRT. The plasmid DNA contained one copy of partial APRT and the NPTII gene (Supplementary Figure S2A). Approximately 50 ng genomic DNA from potatoes or 0.25–50 pg pRI 201-APRT were used as template DNA for the qPCR. The primer sets 5′-TCT CCT GTC ATC TCA CCT TGC-3′/5′-CTT CCA TCC GAG TAC GTG CT-3′ and 5′-GAA CCG GAG CAG GTG AAG AA-3′/5′-GAA GCA ATC CCA GCG ATA CG-3′ were used for amplification of NPTII and APRT, respectively, in the qPCR. The amounts of NPTII and APRT were calculated using pRI 201-APRT as standard DNA. Copy numbers of NPTII in each transgenic potato were calculated as 4× relative NPTII / relative APRT (Supplementary Figure S2B).
Results
Platinum TALEN induced targeted mutagenesis
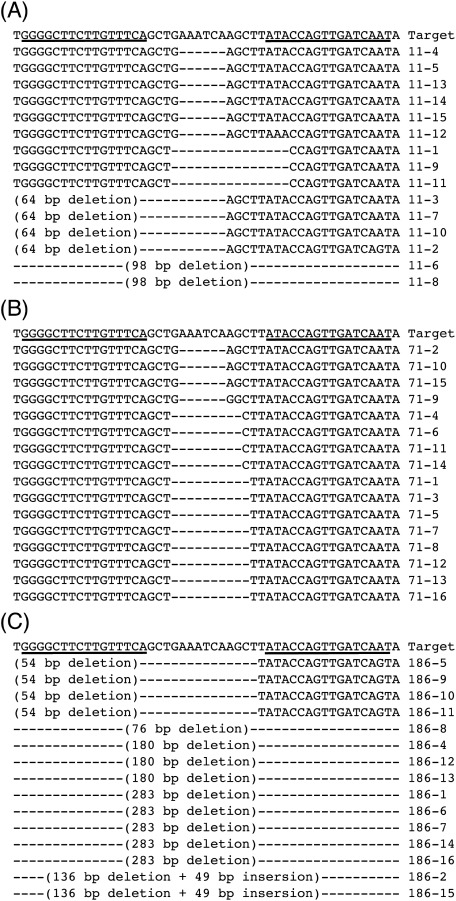
We developed the Platinum TALEN expression vector construction system to facilitate plant science research. In the system, a TALEN expression vector was easily constructed using Golden Gate and MultiSite Gateway® LR reaction (Supplementary Figure S1). The constructed Platinum TALEN expression vector targeting previously reported regions of SSR2 (Sawai et al. 2014) was used for Agrobacterium-mediated potato transformation, and three independent transgenic lines were generated. The target sequences in each transgenic line were confirmed by Sanger sequence after cloning into a TOPO vector. For the three transgenic lines, at least 15 randomly selected clones are shown in Figure 1. We could not detect their intact SSR2 sequences. Almost all the detected mutations of the clones were simple deletions when compared with the sequence of the SSR2 gene. The potential off-target sequences in SSR1, paralog of SSR2, had a 7-bp difference from the 32-bp target site of the SSR2 (Supplementary Figure S3A). Based on direct sequencing analysis, we observed no mutations in SSR1 in all the three transgenic lines (Supplementary Figure S4).
Figure 1. Target SSR2 sequence in transgenic potato plants. Multiple alignment of SSR2 target sequence and PCR products amplified from transgenic potato lines #11 (A), #71 (B), and #186 (C). TALEN-recognized sites were underlined in the target sequences. Dashes indicate deletions.
Tuber yield and SGA contents of SSR2 knockout potato generated by Platinum TALEN
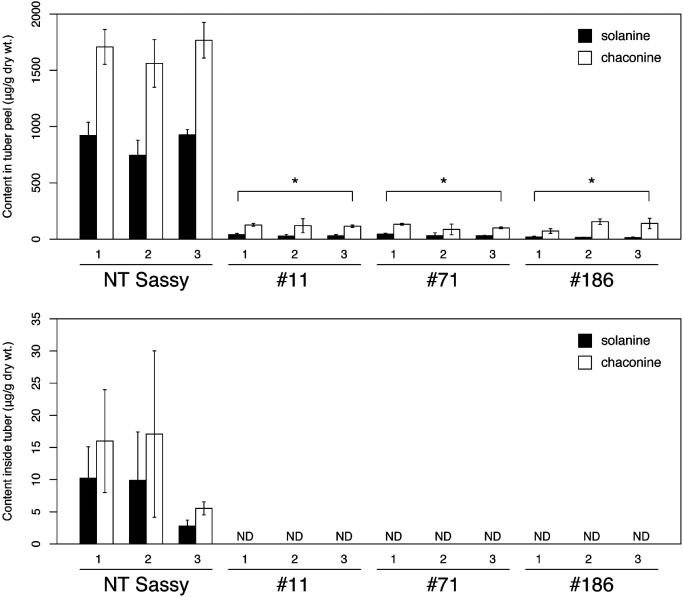
We did not observe any difference in growth among the plants of the three SSR2-edited lines and the control plants. The three lines had similar tuber yields to the control in the green house (Figure 2). The levels of SGA, α-solanine, and α-chaconine in the SSR2-edited potato tuber peels and the inner tissues of the tuber were quantified using High Performance Liquid Chromatography-Mass Spectrometry (Figure 3). In the tuber peels, only minimal amounts of SGA were detected in the SSR2-edited potato compared with the amounts detected in the non-transgenic samples. In the non-transgenic samples, lower amounts of SGA existed in the inner tissues of the tuber when compared to the amounts in the tuber peel. The levels of SGA in the SSR2-edited in the tissues of the potatoes were below detectable levels.
Figure 2. Phenotypes of SSR2-edited transgenic potatoes. (A) The yield of the potato tubers from control (non-transformed Sassy, NT Sassy) and SSR2-edited lines (#11, #71, #186) shows the mean from three plants with error bars indicating SD. (B) Tubers collected from one plant.
Figure 3. SGA contents of tubers from SSR2-edited transgenic potatoes. The predominant SGA levels in the tuber peels and in the tissues of the non-transgenic (NT) Sassy and SSR2-edited transgenic lines (#11, #71, #186). Three potato tubers were analyzed for each line. Error bars indicate the SD of three technical replicates. Asterisks indicate values significantly different from the NT (Dunnett’s test, p<0.001). ND, Not detected.
Estimation of copy numbers of NPTII in the transgenic potato genome
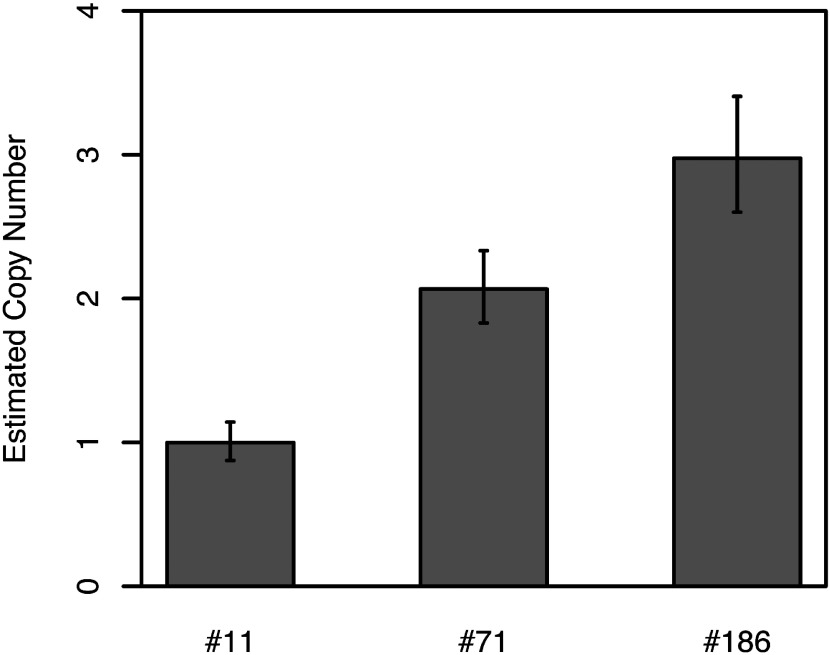
The copy numbers of the NPTII gene in transgenic potato genome were estimated using qPCR technology (Supplementary Figure S2). The relative amounts of exogenous NPTII were divided by the relative amounts of endogenous APRT. Subsequently, the copy numbers of NPTII in the genome were calculated by quadrupling the value since the potatoes used in the present study were tetraploid. Based on the appropriate calculations, copy numbers of NPTII in SSR2-knockout lines #11, #71, and #186 were estimated to be 1, 2, and 3, respectively (Figure 4).
Figure 4. Estimated copy number of the NPTII gene in each transgenic potato. Each transgenic potato genomic DNA sample was tested with triple reactions. The copy numbers of NPTII were estimated as the 4× ratio between the relative amounts of NPTII versus those of endogenous APRT genes (Supplementary Figure S4). Error bars indicate SD (n=3).
Discussion
The gene encoding the cholesterol biosynthetic enzyme, SSR2, has been previously targeted using TALEN technology (Sawai et al. 2014). In a previous study, 29 individual transgenic potato plants harboring the TALEN expression vector were generated, and only two plants contained the desired mutagenesis in SSR2 following treatment with estradiol. In the present study, we used highly active TALENs with variable TALE repeats harboring non-RVD variations called Platinum TALENs (Sakuma et al. 2013a) to target the same regions of the SSR2 gene targeted in the previous report and constitutive cauliflower mosaic virus 35S promoter with an Arabidopsis ADH 5′ non-coding sequence (Sugio et al. 2008) instead of estradiol inducible promoter. As a result, all the three transgenic potato lines had the targeted mutations in SSR2 and no intact SSR2 sequence was detected following a sequence analysis (Figure 1). We designed that the TALEN target site in SSR2 gene was in conserved domain of FAD/FMN-containing dehydroganase in the enzyme (Supplementary Figure S3B). Some mutated SSR2 alleles were 3n base deletions (6-bp, 9-bp, and 12-bp), not frameshift mutation. But the deletions of indispensable amino acids would cause the disruption of enzymatic activities and all mutated SSR2 alleles could indicate non-functional mutations. Since less than four types of mutated SSR2 sequences were detected in each transgenic tetraploid potato except single nucleotide polymorphisms potentially generated during PCR reactions, we assumed that all the transformants were SSR2-knockout plants and not genetically mosaic. Generally, a sequence recognized by TALEN is long enough to genome edit with accuracy. We confirmed that no detectable mutations in potential off-target sites in SSR1 (7-bp difference of 32-bp recognized sequence) were found in a direct sequencing analysis (Supplementary Figure S4).
Recently, genome-editing technology using site-specific nucleases such as TALEN and CRISPR/Cas9 have been applied to potatoes to improve their agricultural characteristics (Andersson et al. 2017; Butler et al. 2015; Clasen et al. 2016; Kusano et al. 2016; Nicolia et al. 2015; Sawai et al. 2014). It is difficult to compare the genome editing efficiency between different studies, since varying target sequences, artificial nucleases, and transformation/transfection methods are employed in different studies. The Platinum TALEN system developed in the present study could be one of the highest efficiency genome-editing systems for introducing targeted mutations into potato genome with accuracy because all potatoes transformed with our binary vector seemed to have mutations in all four SSR2 alleles of a tetraploid and had no off-target mutations in the paralogous gene, SSR1 (Figure 1, Supplementary Figure S4). Since our vector construction system relied on Gateway Cloning technology, any expedient destination vectors which harbor other selection markers, promoters, and terminators for genes of interest, could be used for Platinum TALEN expression in any plant species.
Since SSR2 encodes a key enzyme for SGA biosynthesis in potato (Sawai et al. 2014), SSR2-knockout plants generated in the present study exhibited the expected SGA-reduced phenotype with a tuber yield similar to that of the non-edited plants under the controlled environments (Figures 2, 3). The low amounts of SGAs in SSR2-knockout potato tuber peels could be explained based on the previously reported in vitro enzymatic activities of SSR1 and SSR2. SSR1 mainly catalyzes the reduction of Δ24(28)-sterol. However, it also exhibited weak Δ24(25) reduction activity in, the major enzymatic activity of SSR2, and could contribute to the biosynthesis of common SGA precursors, cycloartanol and cholesterol, in SSR2-knockout potatoes (Sawai et al. 2014). Recently, many genes responsible for SGA biosynthesis have been reported in potato and tomato (Itkin et al. 2013; Nakayasu et al. 2017; Umemoto et al. 2016). The recently identified genes could be suitable targets for the generation of SGA-free potatoes using genome-editing technology.
Plants that have targeted mutation without transgenes could be treated as non-transgenic plants. To obtain such null-segregants through genetic segregation of transgenes and targeted mutations following the introduction of mutations in target loci by transformed site directed nucleases, it is necessary that copy numbers of transgenes in transgenic genome-edited plants are as low as possible to increase the rate of null-segregants following the crossing. The copy numbers in each transgenic plant is generally analyzed by the Southern blotting analysis, a method that requires relatively large amounts of purified genomic DNA and is time consuming and labor intensive. In the present study, we used real time qPCR techniques to easily evaluate the copy numbers of NPTII. Using a plasmid DNA, which harbors an NPTII (transgene) and a partial APRT (endogenous single-copy gene in potato) as standard, relative amounts (numbers) of NPTII in potato genome were evaluated (Figure 4, Supplementary Figure S2). This simple procedure could help the selection of plants carrying a single copy of the transgenes for parents of null-segregants in the future.
The development of reliable genome-editing systems is critical for plant genome modifications in basic and applied plant research. Currently, numerous CRISPR-Cas9 based genome-editing systems for plants are available. However, they require the protospacer adjacent motif sequence near their target sites and use shorter recognized sequences than the TALEN system. Our Platinum TALEN based system, developed in the present study, provides an additional choice for high efficiency plant genome editing.
Acknowledgments
This work was supported by the Cross-ministeral Strategic Innovation Promotion Program (SIP), Japan. We thank Chihoko Oishi, Keiko Miyamae and Chika Shimazu for technical assistance. We thank Kenji Asano for useful suggestions for the estimation of copy number of NPTII in transgenic potatoes by real time PCR.
Supplementary Data
References
- Andersson M, Turesson H, Nicolia A, Fält AS, Samuelsson M, Hofvander P (2017) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep 36: 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Tamiya S (2016) Breeding of pest and disease resistant potato cultivars in Japan by using classical and molecular approaches. Jpn Agric Res Q 50: 1–6 [Google Scholar]
- Butler NM, Atkins PA, Voytas DF, Douches DS (2015) Generation and inheritance of targeted nutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 10: e0144591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen BM, Stoddard TJ, Luo S, Demorest ZL, Li J, Cedrone F, Tibebu R, Davison S, Ray EE, Daulhac A, et al. (2016) Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 14: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179 [DOI] [PubMed] [Google Scholar]
- Kusano H, Onodera H, Kihira M, Aoki H, Matsuzaki H, Shimada H (2016) A simple Gateway-assisted construction system of TALEN genes for plant genome editing. Sci Rep 6: 30234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani O, Arazoe T, Toshida K, Hayashi R, Ohsato S, Sakuma T, Yamamoto T, Kuwata S, Yamada O (2016) Detailed analysis of targeted gene mutations caused by the Platinum-Fungal TALENs in Aspergillus oryzae RIB40 strain and a ligD disruptant. J Biosci Bioeng 123: 287–293 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assay with tobacco tissue cultures. Physiol Plantarum 15: 473–497 [Google Scholar]
- Nakayasu M, Umemoto N, Ohyama K, Fujimoto Y, Lee HJ, Watanabe B, Muranaka T, Saito K, Sugimoto Y, Mizutani M (2017) A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol 175: 120–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolia A, Proux-Wéra E, Åhman I, Onkokesung N, Andersson M, Andreasson E, Zhu LH (2015) Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J Biotechnol 204: 17–24 [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914 [DOI] [PubMed] [Google Scholar]
- Sakane Y, Sakuma T, Kashiwagi K, Kashiwagi A, Yamamoto T, Suzuki K (2014) Targeted mutagenesis of multiple and paralogous genes in Xenopus laevis using two pairs of transcription activator-like effector nucleases. Dev Growth Differ 56: 108–114 [DOI] [PubMed] [Google Scholar]
- Sakuma T, Ochiai H, Kaneko T, Mashimo T, Tokumasu D, Sakane Y, Suzuki K, Miyamoto T, Sakamoto N, Matsuura S, et al. (2013a) Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep 3: 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Hosoi S, Woltjen K, Suzuki K, Kashiwagi K, Wada H, Ochiai H, Miyamoto T, Kawai N, Sasakura Y, et al. (2013b) Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18: 315–326 [DOI] [PubMed] [Google Scholar]
- Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, et al. (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26: 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio T, Satoh J, Matsuura H, Shinmyo A, Kato K (2008) The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J Biosci Bioeng 105: 300–302 [DOI] [PubMed] [Google Scholar]
- Tiwari JK, Siddappa S, Singh BP, Kaushik SK, Chakrabarti SK, Bhardwaj V, Chandel P (2013) Molecular markers for late blight resistance breeding of potato: An update. Plant Breed 132: 237–245 [Google Scholar]
- Umemoto N, Nakayasu M, Ohyama K, Yotsu-yamashita M, Mizutani M, Seki H, Saito K, Muranaka T (2016) Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol 171: 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.