Abstract
Hybrid Oncidium orchids, such as Oncidium Gower Ramsey and Oncidium “Honey Angel,” are popular cut flowers in Japan and Taiwan. Due to pollen sterility, no new varieties have been created by conventional breeding methods. Recently, we employed RNA interference (RNAi) technology to suppress phytoene synthase and successfully modified floret hue from yellow to white (Liu et al. 2019). Transgenic white Oncidium orchids, Honey Snow MF-1, have been grown to test their genetic stability, and their environmental biosafety was assessed for approximately one year under government regulatory instructions from the Council of Agriculture, Taiwan. In the present study, pollen sterility was demonstrated by cytological observation of the microsporogenesis step, pollen morphology abortion, and failure of pollen germination. Assays on allelopathic effect on the other plants and the soil rhizospheric microbial flora-revealed that transgenic Oncidium orchids are potentially safe with regard to environmental biodiversity. Therefore, the general release permissions have been granted and an application for licensing for commercial production is under way.
Keywords: allelopathy, environmental biosafety assessment, Oncidium orchid, RNA interference (RNAi) technology
Introduction
Oncidium is an orchid genus of more than 330 species in the subtribe Oncidiinae of the Orchidaceae family. The plants are native to Central and South America. Breeding efforts in the past have produced many commercial cultivars, such as Oncidium Sharry Baby, Oncidium Sweet Sugar, and Oncidium Gower Ramsey, which are widespread in the world market. Among them, Oncidium Gower Ramsey has been the most favorite cut flower in Asia. Thus far, these commercialized hybrids of Oncidium orchids have been the second-favorite orchids commercially, next to Phalaenopsis spp. Oncidium Gower Ramsey was introduced as a crop in Taiwan in 1986. It is well accepted in the cut flower markets of Taiwan and Japan, with its brilliant yellow florets and pretty appearance, and has been Taiwan’s most exported cut flower. Until 2006, a new somaclonal variety, Oncidium “Honey Angel,” was introduced to Taiwan and soon replaced Oncidium Gower Ramsey. Flower customers prefer Oncidium “Honey Angel” over Oncidium Gower Ramsey because of the former’s shinier yellow pigment. The disappearance of brownish spots in its petal tissue owes to the gene silencing of chalcone synthase by promoter methylation (Liu et al. 2010). Currently, Oncidium “Honey Angel” is the main Oncidium orchid cultivar cultivated in Taiwan and exported to Japan; in fact, it accounts for about 90% of the annual yield.
In spite of its popular acceptance, commercial Oncidium orchid cut flowers have been only yellow hues thus far. Such a lack of variety causes market growth hysteresis. Although many breeders and researchers have been devoted to creating new varieties, progress has not materialized. This is due to the reproduction sterility of Oncidium Gower Ramsey, Oncidium “Honey Angel” and related cultivars (Supplementary Table S1). To fulfill market anticipation, we attempted to generate new Oncidium varieties with white florets. In the past, we employed an RNA-mediated gene-silencing method accompanied by a petal-specific expressed promoter, the promoter of the chromoplast-specific carotenoid-associated gene (Pchrc) derived from Oncidium Gower Ramsey (Chiou et al. 2008), to engineer Oncidium “Honey Angel” by specifically blocking phytoene synthase transcripts in petal tissues (Liu et al. 2019). The transgenic lines, with white florets, were selected for cultivation testing. Molecular analysis has shown the downregulation of phytoene synthase transcripts in petal tissues, and biochemical analysis has revealed that levels of xanthophyll compounds were only 5–10% of those of the control plant, Oncidium “Honey Angel” (Chiou et al. 2010; Liu et al. 2019). Those results indicated that RNAi-based genetic modification is effective for creating the expected variety featuring white flowers for the Oncidium orchid industry.
In order to obtain environmental biosafety approval from a government, the possible impacts of transgenics varieties on the native ecosystem must be clarified. This is a general rule for every country (Watanabe et al. 2005). Therefore, we assessed the environmental biosafety risk of transgenic Oncidium orchids under the regulatory instructions of the Taiwanese government. This assessment comprises the reproduction of pollen viability; intra-species, inter-species, and inter-genus pollen hybridization; an allelopathic test; assessment of the risk to the rhizospheric soil microbe community, etc. In this study, we evaluated and compared the pollen viability and the potential impacts on other plants and rhizospheric soil microbes between transgenic and non-transgenic Oncidium orchids and tried to assess the potential risks of transgenic Oncidium orchids on the biodiversity.
Materials and methods
Plant material and cultivation
The plants used in this study are Oncidium “Honey Angel” and Oncidium “Honey Snow” MF-1. The former are recipient plants with yellow florets used as controls, and the latter are RNAi genetically modified orchids with white florets. Both orchid plants, 50 pot seedlings each, were selected for testing at the bolting stage and were grown in a confined greenhouse at the Biotechnology Center in the Southern Taiwan, Academia Sinica, Tainan, Taiwan. Also, plant materials were assayed for parts of experiments performed in University of Tsukuba. The growth conditions mimicked natural conditions, 25–30°C/day, 20–25°C/night; light intensity 250–350 µm/m2 s/14 h. Growth characteristics and flowering parameters were recorded. The risk assessment was performed following the protocol described by Yu et al. (2013a, b).
Cytological study of microsporogenesis in pollen formation
The anther treatment and microscopic technique were performed as described by Chung (2015). In brief, anthers were removed from young flower buds and placed on glass slides, a drop of solution containing 1% carmine and 45% acetic acid was dropped onto each slide, the anthers were crushed completely with forceps and covered with a cover slide, and then the slides were gently heated on an alcohol lamp. The meiotic phase of microspore formation was observed by light microscope (Olympus BH-2, Japan).
Allelopathic tests
To assess the difference in the effects on surrounding plants between transgenic and non-transgenic Oncidium orchids, their allelopathic activities were assayed by two methods: the sandwich method and the soil-mixed method were employed to assess the allelopathic activities of leaves and exudates from roots on peripheral plants, respectively (Tran et al. 2018; Yu et al. 2013a; Yu et al. 2013b). In the sandwich method, leaf samples were taken and dried at 60°C for 24 h, 5 ml of low-melting-point agar (LMP; 0.5% w/v) was put in each well of a 6-well multi-dish plate (34 mm in diameter), and a specific quantity of about 0.1 g of dry leaf was put on the surface of the agar layer. After solidification, another 5 ml of LMP agar was poured on top of the first layer. Five sterilized lettuce seeds (Lactuca sativa L. var. capitate) were spread on the solid agar surface in each well. Seedlings were grown at 25°C for 3 days in the dark. The germination rate and the radicle and hypocotyl lengths were measured. The longest and shortest seedlings in each well were excluded. Nine wells were prepared for each evaluation.
In the soil-mix method to assay the allelopathic impact of the leachates released from the transgenic Oncidium orchid, about 1 g of dried leaf was mixed with 100 g of sterilized soil (Kureha mix soil) and equally distributed into 6 plastic cells (3.5 cm high×2.5 cm bottom×3.5 cm top). Five lettuce seeds were placed on the surface of the leaf–soil mixture in each cell. After incubation for 5 days, 25°C in the dark, the germination rate and the radicle and hypocotyl lengths were measured.
Assaying rhizosphere microbe community
Pot substrates cultivated with Oncidium orchids were sampled. Of this, about 30 g was dried at 80°C for 1 day as a dry weight sample, and the other 30 g soil sample was immersed in 270 ml of 15 mM phosphate buffer (pH 7.0). The diluted soil solution (100 ml) was spread onto each of 2 kinds of agar media: OGYE for fungal incubation and PTYG for actinomycetes and bacterial cultivation. OGYE medium comprises (per liter) yeast extract 5 g, glucose 20 g, biotin 0.1 mg, tetracycline 50 mg. PTYG medium per liter comprises peptone 0.25 g, tryptone 0.25 g, yeast extract 0.5 g, glucose 0.5 g, MgSO4·7H2O 30 mg, and CaCl2·2H2O 3.5 mg. Three replicates of each medium were tested. Cultures were incubated in the dark at room temperature. After incubation for 3 or 7 days in the dark at 25°C for OGYE or PTYG, the formatted colonies were counted. The number of colonies on PTYG medium after wiping the surface with 70% ethanol was taken as the number of actinomycetes.
Statistical analysis
Data shown are the means with standard errors of at least 5 independent biological samples. Two-way analysis of variance (ANOVA) was used to evaluate differences among the samples.
Results and discussion
Growth characters and flower morphology
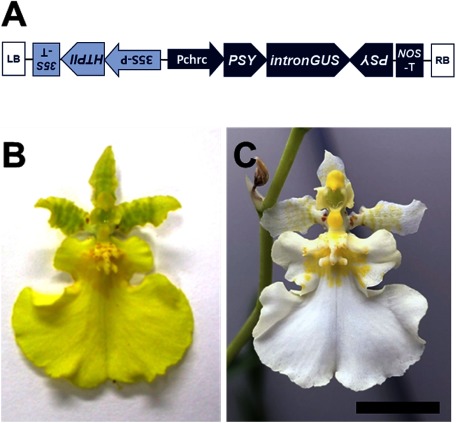
Growth rate, flowering time, and flower morphology of Oncidium “Honey Angel” (control) and Oncidium “Honey Snow” MF-1 (transgenic) in the confined greenhouse were observed from April to December 2014. Therefore, a similar cultivation technology was used for both plants, and no differences were found between the varieties, except that the pigment was yellow in Oncidium “Honey Angel” and white in Oncidium “Honey Snow” MF-1 (Figure 1). In the T-DNA in the transgenic Oncidium “Honey Snow”, the PSY siRNA construct was aligned in the region downstream of the petal-specific promoter, Pchrc, and we have reported that the PSY siRNAs were expressed in the floral organs, but few were expressed in leaves (Liu et al. 2019). This indicated that the genetically modified point is restricted to petal tissues and that no negative collateral effects occur in other tissues. It also suggests that the activity of the Pchrc promoter is precisely functioning in petal tissues.
Figure 1. T-DNA transformed in Oncidum “Honey Snow” MF-1 and floret appearance. (A) Schematic diagram of T-DNA transformed in transgenic Oncidium “Honey Snow” MF-1.PSY, a 150-bp fragment of the phytoene synthase gene, intronGUS, a β-glucuronidase gene containing an intron of the castor bean catalase gene; HPTII, hygromycin phosphotransferase II derived from E. coli; Pchrc, promoter of the OgCHRC gene; 35S-P, CaMV35S promoter; NOS-T, terminator of the nopalin synthase gene derived from Rhizobium radiobacter; RB and LB, right and left border. (B) Typical floret of non-transgenic Oncidium “Honey Angel”. (C) Typical floret of Oncidium “Honey Snow” MF-1.
Pollen formation in microsporogenesis
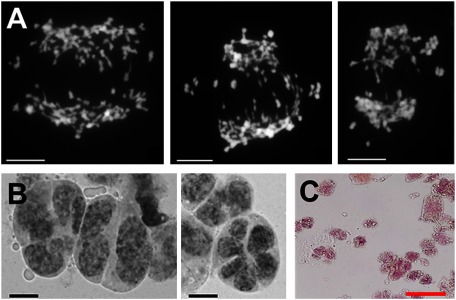
To investigate the cytological meiosis of microsporogenesis (the process of pollen formation), we microscopically observed the meiotic phase of anther. We found that the chromosome complements at the anaphase and telophase showed laggard and bridged chromosomes during segregation. Uneven segregation was observed (Figure 2A). This resulted from the abnormal pairing between chromosomes. It may owe to the hetero hexaploid eukaryotes of Oncidium Gower Ramsey that were generated through a multistep breeding process. The multistep breeding process and multiancestor have been considered the reasons that Oncidium Gower Ramsey and Honey Angel lost pollen fertility and pollination. The transgenic Oncidium Honey Snow MF-1 was generated by RNAi-engineering on Oncidium Honey Angel, which is a somaclonal variant of Oncidium Gower Ramsey (Liu et al. 2012). Theoretically, therefore, Honey Snow and Honey Angel inherits the same genetic system. Furthermore, we observed the end of the microsporogenesis stage. Normally, a tetrad formation of 4 microspores is the typical structure in the completeness of microsporogenesis. However, abnormal end products were generated (Figure 2B). Only abnormal types of diad, triad, hexad, etc., were formed (Figure 2B). This suggests that multiple ancestors of chromosome origin caused chaotic pairing and unequal segregation. Consequently, microspores were not generated normally. Furthermore, the microscopic observations also showed that the pollen grains irregularly shaped and of varying size, unlike the uniform appearance of pollen grains in wild orchids of Oncidium sphacelatum (Figure 2C).
Figure 2. Microsporogenesis and pollen grain of Oncidium “Honey Snow” MF-1. (A) Chromosomal laggard at anaphase I stage during microsporogenesis. The laggard caused some chromatids to separate unequally into daughter cells. (B) Microspore formation showing abnormal types of diads and hexads (right panel), as well as triads and tetrads of various sizes (left panel). Unequal chromatid separation caused the abnormal microspore formation. (C) Pollen grains showing irregular shape, abnormal development of cell shape, and cell content. Acetocarmine staining was not homogenous in pollen cells. Scale indicates 50 µm.
Evaluation of potential allelopathic impact on peripheral plants
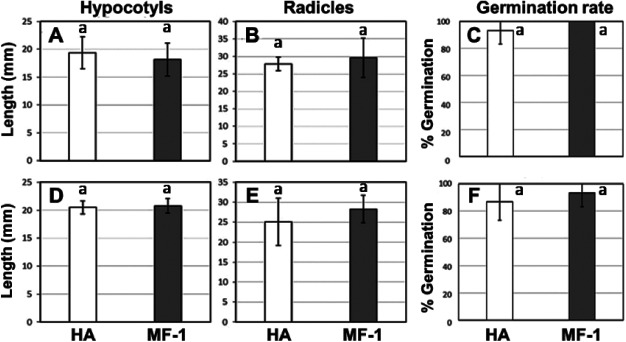
Allelopathic impact assays are used in biosafety assessments to assay the allelopathic impacts of the leachates released from dried leaves (Fujii et al. 2003, 2004). Two methods were employed to test whether transgenic Oncidium orchids released any metabolites to damage surrounding vegetation. By monitoring the growth and germination rates of lettuce seeds, we found that data from both the soil-mixed method and the sandwich method revealed no significant difference between the non-transgenic Oncidium “Honey Angel” and the transgenic Oncidium “Honey Snow” MF-1 in seedling growth and germination rates (α=0.05, Figure 3). The results suggested that there was no significant difference in the potential impact on the growth of peripheral plants between the transgenic Oncidium “Honey Snow” MF-1 and non-transgenic Oncidium “Honey Angle”.
Figure 3. Allelopathic evaluation using the sandwich method and soil-mixed method for transgenic Oncidium “Honey Snow” MF-1. (A–C) Results of sandwich method. The hypocotyl and radicle growth rates and the germination rate of lettuce seedling grown in agar sandwiching 100 mg of dried leave were shown in A, B, and C, respectively. (D–E) Results of soil-mixed method. The hypocotyl and radicle growth rates and the germination rate of lettuce seedling grown in 100 g of soil mixing containing 1 g of dried leave were shown in D, E, and F, respectively. Dark grey bar represents transgenic Oncidium “Honey Snow” MF-1, while light bars represent non-transgenic Oncidium “Honey Angel.” Each graph shows the mean and S.E. of the measurements obtained from 10 lettuce seedlings. ANOVA did not show any significant difference between non-transformants and transformants.
Evaluation of potential impact on microbial populations
To evaluate the potential impact on microorganisms adjacent to the root of transgenic Oncidium “Honey Snow” MF-1, we checked number of culturable microorganisms in sphagnum after cultivation of the Oncidium orchids. The investigation of culturable microorganisms was performed by the plate culture method widely used for the environmental risk assessment of transgenic plants in Japan (Shiomi et al. 1992). The results revealed no significant differences in the colony formation units (CFUs) of filamentous fungi, actinomycetes, and bacteria, except for a difference in actinomycetes CFUs between the transgenic Oncidium “Honey Snow” MF-1 and the non-transgenic orchid (α=0.05, Figure 4). These findings suggested that there was no significant difference in potential impact on the microorganism population between the transgenic orchid, Oncidium “Honey Snow” MF-1 and non-transgenic Oncidium “Honey Angel.”
Figure 4. Survey of rhizosphere microbial population. (A–D) Rhizosphere microbes of transgenic and non-transgenic Oncidium orchids were cultivated by OGYE plate cultivation for fungi (A and B) and PTYG plate cultivation for bacteria including actinomycetes (C and D). (E–G) Calculated colony-forming units of fungi, actinomycetes, and bacteria were shown in E, F and G, respectively. The error bars indicate standard error. Five individuals of each orchid were used repeatedly. ANOVA did not show any significant difference between the transgenic and non-transgenic orchids (E–G). HA, Oncidium “Honey Angel”; MF-1, Oncidium “Honey Snow” MF-1. There were no significant differences between the transgenic and non-transgenic Oncidium orchids by ANOVA (p>0.05).
Conclusion
Currently, Oncidium Gower Ramsey and Oncidium “Honey Angel” are two important ornamental Orchid crops for cut flower exportation from Taiwan. Their floral characteristics have been popularly accepted in the flower market, but customers have longed for an Oncidium orchid with a different petal color. The present transgenic Oncidium “Honey Snow” MF-1 with white hue florets could be a satisfactory variety to meet customers’ desire.
The present transgenic Oncidium orchid is the first ornamental crop produced in Taiwan. As we know, it is also the first transgenic Oncidium orchid in the world. Its potential value in the future floral market is much anticipated. However, it is essential to assess its environmental biosafety before further field cultivation can begin. The present biosafety assays systematically following the GM regulations of the Taiwan government demonstrated that transgenic Oncidium “Honey Snow” MF-1 will not impact the natural environment or pose risks to it, including the risks of gene flow and contamination to affect biodiversity. Basically, solid evidence of pollen sterility strongly confirms its biosafety (Figure 2 and Supplementary Table S1). Therefore, the introduction of this plant should be socially acceptable when it is released for field cultivation and commercialization in the near future.
Acknowledgments
The work of assessing environmental biosafety was financially supported by the Council of Agriculture (COA), Taiwan, under a project led by Dr. Kai-Wun Yeh, and by the Taiwan–Japan bilateral project supported by COA, Taiwan to Dr. Kai-Wun Yeh and Dr. Kazuo N. Watanabe working at Tsukuba University, Japan. We also thank the technical support of Academia Sinica-Biotechnology Center in Southern Taiwan greenhouse core facility for technical assistance. The risk assessment was carried out following the GM regulations from COA, Taiwan. This Evironmental Risk Assessment certificate of Taiwan was approved by ERA, COA Taiwan on March 2018.
Abbreviations
- RNAi
RNA interference
- ANOVA
analysis of variance
Supplementary Data
References
- Chiou CY, Pan HA, Chuang YN, Yeh KW (2010) Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 232: 937–948 [DOI] [PubMed] [Google Scholar]
- Chiou CY, Wu K, Yeh KW (2008) Characterization and promoter activity of chromoplast specific carotenoid associated gene (CHRC) from Oncidium Gower Ramsey. Biotechnol Lett 30: 1861–1866 [DOI] [PubMed] [Google Scholar]
- Chung MC (2015) Chromosome techniques and FISH. In: ECT Yeung et al. (eds) Plant Microtechniques and Protocols. Springer International Publishing, Switzerland, pp 287–309
- Fujii Y, Parvez MM, Ohmae Y, Iida O (2003) Screening of 239 medicinal plant species for allelopathic activity using the sandwich method. Weed Biol Manage 3: 233–241 [Google Scholar]
- Fujii Y, Shibuya T, Nakatani K, Itani T, Hiradate S, Parvez MM (2004) Assessment method for allelopathic effect from leaf litter leachates. Weed Biol Manage 4: 19–23 [Google Scholar]
- Liu XJ, Chuang YN, Chiou CY, Chin DC, Shen FQ, Yeh KW (2012) Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in flora tissues of two Oncidium orchid cultivars. Planta 236: 401–409 [DOI] [PubMed] [Google Scholar]
- Liu YC, Yeh CW, Chung JD, Tsai CY, Chiou CY, Yeh KW (2019) Petal-specific RNAi-mediated silencing of the phytoene synthase gene reduces xanthophyll levels to generate new Oncidium orchid varieties with white-colour blooms. Plant Biotechnol J pbi.13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi M, Asakawa Y, Fukumoto F, Hamaya E, Hasebe A, Ichikawa H, Matsuda I, Muramatsu T, Okada M, Sato M, et al. (1992) Evaluation of the impact of the release of transgenic tomato plants with TMV resistance on the environment. Bull Natl Inst Agro-Environ Sci Jpn 8: 1–51 [Google Scholar]
- Tran NHT, Oguchi T, Matsunaga E, Kawaoka A, Watanabe KN, Kikuchi A (2018) Environmental risk assessment of impacts of transgenic Eucalyptus camaldulensis events highly expressing bacterial Choline Oxidase A gene. Plant Biotechnol 35: 393–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe KN, Sassa Y, Suda E, Chen CH, Inaba M, Kikuchi A (2005) Global political, economic, social and technological issues on transgenic crops. Plant Biotechnol 22: 515–522 [Google Scholar]
- Yu X, Kikuchi A, Matsunaga E, Shimada T, Watanabe KN (2013a) Environmental biosafety assessment on transgenic Eucalyptus globulus harboring the choline oxidase (codA) gene in semi-confined condition. Plant Biotechnol 30: 73–76 [DOI] [PubMed] [Google Scholar]
- Yu X, Kikuchi A, Shimazaki T, Yamada A, Ozeki Y, Matsunaga E, Ebinuma H, Watanabe KN (2013b) Assessment of the salt tolerance and environmental biosafety of Eucalyptus camaldulensis harboring a mangrin transgene. J Plant Res 126: 141–150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.