Abstract
BACKGROUND
Fetal akinesia deformation sequence (FADS) is a broad spectrum disorder with absent fetal movements as the unifying feature. The etiology of FADS is heterogeneous and mostly still unknown. A prenatal diagnosis of FADS relies on clinical features obtained by ultrasound and fetal muscle pathology. However, the recent advances of next-generation sequencing (NGS) can effectively provide a definitive molecular diagnosis.
CASE SUMMARY
A fetus presented after 24 wk and 6 d of gestation with absent fetal movements and multiple abnormal ultrasonographic signs. The mother had had a previous abortion due to a similarly affected fetus a year before. A clinical diagnosis of FADS was made. The parents refused cord blood examination and chose abortion. A molecular diagnosis of fetal muscle using NGS of genes found a compound heterozygous mutation in the MUSK gene: c.220C > T (chr9: 113449410 p.R74W) and c.421delC (chr9: 113457745 p.P141fs).
CONCLUSION
To our knowledge, this is the first report in China showing that a mutation in MUSK is associated with FADS. This supports previous finding that a lethal mutation of MUSK will cause FADS. A precise molecular diagnosis for genetic counseling and options for a prenatal diagnosis of FADS are very important, especially for recurrent FADS; this may also provide evidence for both prenatal and preimplantation genetic diagnoses.
Keywords: MUSK gene, Fetal akinesia deformation sequence, Joint contractures, Case report
Core tip: Fetal akinesia deformation sequence (FADS) is a broad spectrum disorder with absent fetal movements, and its etiology is heterogeneous. Mutations in genes expressed at the neuromuscular junction (NMJ) are increasingly recognized as important causes of FADS. MUSK is required for the formation and maintenance of the NMJ. Here we describe a compound heterozygous mutation of the MUSK gene that caused FADS in a Chinese fetus.
INTRODUCTION
Moessinger proposed that decreased or absent fetal movements, independent of the cause, can lead to a predictable series of secondary anomalies[1]. Clinical symptoms of fetal akinesia deformation sequence (FADS) include joint contractures, subcutaneous edema, fetal hydrops, polyhydramnios, pulmonary hypoplasia, intrauterine growth restriction, micrognathia, cleft palate, hypoplasia of the limb muscles, short umbilical cord, decreased intestinal motility, and shortened bowel, with a phenotype that may be complicated by brain anomalies or restrictive dermopathy. The etiology of FADS is heterogeneous: Both genetic and environmental factors may affect normal develop-mental processes in the fetus and lead to FADS[2]. Mutations in genes expressed at the neuromuscular junction (NMJ) are increasingly recognized as important causes of FADS[3]. MUSK is required for the formation and maintenance of the NMJ. To date, two homozygous mutations of MUSK have been reported to cause FASD: a c.40dupA mutation[4] and a missense variant [c.1724T4C; p. (Ile575Thr)][5]. Here we describe a compound heterozygous mutation of the MUSK gene that caused FADS in a fetus in China and possibly in her sibling.
CASE PRESENTATION
Chief complaints
Menopause for 6 mo and fetal abnormality for 13 d.
History of present illness
A 34-year-old woman, gravida 2, para 0, abortus 1, was referred to our department because of fetal abnormality. She had regular menstrual cycle before pregnancy, the last menstrual period was February 27, 2017, and the expected date of childbirth was December 3, 2017. She went to prenatal examination regularly, the noninvasive prenatal test result was low risk, the prenatal ultrasound examination showed abnormal ultrasonographic signs and lack of fetal movement. The mother had not felt any fetal movement during pregnancy. The parents decided to terminate the second pregnancy at the gestational age of 24 wk and 3 d of gestation.
History of past illness
A previous pregnancy showed a similarly affected fetus electively aborted at 25 wk of gestation. However, only ultrasound information about the fetus was available with no postnatal findings (case 1 in Table 1).
Table 1.
Clinical characteristics of the two study fetuses and fetuses with fetal akinesia deformation sequence syndrome in the literature
|
Article |
1 |
2 |
3 |
|||||||||||||||
|
Fetus number |
2 |
5 |
11 |
|||||||||||||||
|
Nationality |
China |
Sweden |
The Netherlands |
|||||||||||||||
|
Molecular analysis |
c.220C >T
c.421delC |
c.40dupA, g.113 431 224 dupA |
c.1724T > C p.Ile575Thr |
|||||||||||||||
| Number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
| GW at birth | 25 | 24 + 6 | 27 | 30 | 18 | 18 | 17 | 41 | 33 + 4 | 38 + 1 | 38 + 4 | 33 + 1 | 35 | 23 | 32 + 2 | 22 | 31 | 23 |
| Pregnancy outcome | TOP | TOP | Died | IUFD | TOP | TOP | TOP | Died <2 h | Died <1 h | Died 5 d | Died <1 h | IUFD | Died <1 h | TOP | Died <1 h | TOP | Died <2 h | TOP |
| Prenatal findings | ||||||||||||||||||
| Reduced/absent fetal movement | + | + | + | + | + | + | + | NA | NA | NA | - | + | NA | + | + | + | NA | + |
| Joint contractures | + | + | + | + | + | + | + | NA | NA | NA | NA | + | NA | + | + | + | NA | + |
| Polyhydramnios | + | + | + | + | - | - | - | NA | + | + | + | + | + | - | + | - | + | + |
| FGR | + | + | - | - | - | - | - | NA | + | - | - | - | - | - | + | - | - | - |
| Subcutaneous edema | + | + | + | + | - | - | - | NA | NA | NA | NA | - | NA | - | - | - | NA | - |
| Fetal stomach not visualized | + | + | + | + | + | + | + | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Hydrothorax | - | - | + | + | - | - | - | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Micrognathia | + | + | - | - | - | - | - | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Postnatal findings | ||||||||||||||||||
| Birth weight | NA | 890 | NA | NA | NA | NA | NA | 2700 | 1870 | 3470 | 3250 | 1363 | 1910 | 551 | 1610 | 408 | 1250 | 490 |
| Sex of fetus | F | F | F | F | M | M | F | F | F | M | M | F | M | M | M | F | F | M |
| Flex hips | NA | + | + | + | + | + | - | + | + | - | + | + | - | + | + | + | NA | + |
| Hyperextended knees | NA | + | + | + | + | + | - | NA | + | - | + | + | - | + | + | - | NA | - |
| Flexed elbows | NA | + | + | + | + | + | + | NA | + | - | - | + | - | + | + | - | NA | - |
| Clenched fingers | NA | + | + | + | + | + | + | NA | + | + | + | + | + | + | + | + | NA | + |
| Club/rocker feet | NA | + | + | + | + | +1 | + | + | + | - | + | - | + | + | + | + | - | + |
| Low set ear | NA | + | + | + | - | - | + | NA | + | - | - | + | NA | + | + | + | + | - |
| Hypertelorism | NA | - | NA | NA | NA | NA | NA | NA | + | + | - | + | NA | + | + | + | - | + |
| Micrognathia | NA | + | + | - | + | + | + | + | - | + | - | + | - | + | + | + | - | + |
| Cleft palate | NA | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Edema | NA | + | + | - | - | - | + | NA | - | - | + | - | - | + | + | - | - | + |
| Pulmonary hypoplasia | NA | + | + | + | - | - | + | + | + | + | + | + | + | + | + | + | NA | + |
The fetus had feet deformity. +: Present; -: Absent. Pedigree 1: The two fetuses in our article. Pedigree 2: The five fetuses reported by Wilbe et al[4]. Pedigree 3: The 11 fetuses diagnosed with a c.1724T > C mutation reported by Tan-Sindhunata et al[5]. F: Female; M: Male; GW: Gestational week; NA: Not assessed; FGR: Fetal growth restriction; TOP: Termination of pregnancy; IUFD: Intrauterine fetal death.
Personal and family history
The parents were non-consanguineous Chinese, and there was no family history of the parents.
Physical examination upon admission
General physical examination was normal. Fatal heart rate was 146 bpm, the height of the uterine fundus was 24 cm, and there was fetal head presentation.
Laboratory examinations
The results of routine blood test were: WBC 1.1 × 1010/L, NE 71.0%, RBC 3.56 × 1012/L, HGB 115 g/L, and PLT 2.79 × 1011/L. K+ was 3.99 mmol/L, Na+ was 136.3 mmol/L, and Ca2+ was 2.32 mmol/L. Routine urine test was normal.
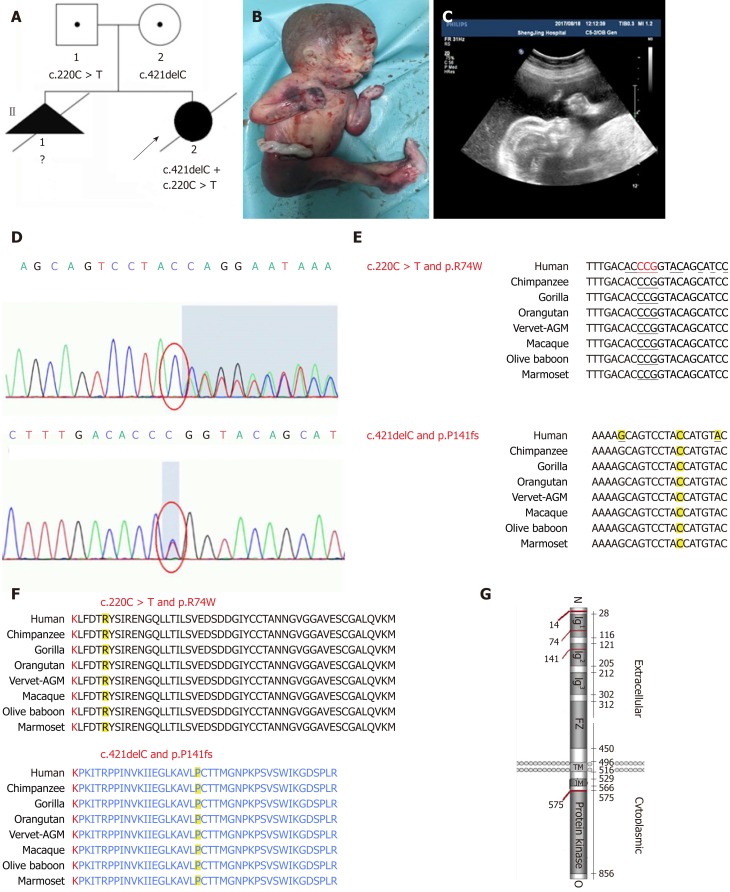
Total genomic DNA was extracted from the aborted fetus’ muscular tissue and peripheral blood leukocytes were isolated from the parents according to standard protocols. Targeted enrichment of whole exome DNA was performed using a Nextera Rapid Capture Exome kit (Illumina, San Diego, CA, United States) according to manufacturers’ protocols. Whole exome libraries were then sequenced on an Illumina HiSeq platform using a 2 × 100 bp sequencing protocol. Potentially homozygous, compound heterozygous, and de novo variants were filtered out. An identified compound heterozygous mutation in the MUSK gene was confirmed using standard Sanger sequencing. For amplification of the genomic region that includes the mutations identified in this study, the following two primer pairs were used: (1) GTGGTCGGGATTGACAGCA (forward) and CACAGCTGAAGACCCTGGG (reverse); and (2) CCCAGGGTCTTCAGCTGTG (forward) and CCTCTGTCATGCTGCCCAA (reverse). We found that the fetus carried both a frameshift mutation, c.421delC (p. Pro141Hisfs*15), and a missense mutation, c.220C > T (p. R74W) in the MUSK gene, involving the same transcript (ENST00000374448.8; Figure 1D). After examining the mutation sites of family members, it was determined that fetal mutations were inherited from both parents. The fetus’ mother carried the c.220C > T mutation and the c.421delC mutation was detected in the fetus’ father; thus the fetus received two different MUSK mutations, one from each parent, resulting in a compound heterozygous mutation.
Figure 1.
An affected family history pedigree. Ultrasound and genetic data revealed a frameshift mutation in muscle, skeletal receptor tyrosine kinase. A: Pedigree of affected family highlights two affected fetuses; B: Photograph of a fetus with fetal akinesia deformation sequence (FADS) from the second pregnancy after an abortion at 25 gestational weeks; C: The second pregnancy displayed polyhydramnios fetal hydrops and micrognathia; D: Sanger sequencing revealed that the mother and father had different heterozygous mutations, and the fetus had a compound heterozygous mutation; E: Mutation sites of c.421delC and c.220C > T in each species and conserved region; F: Mutation sites of Pro141Hisfs*15 and p. R74W in each species and conserved region; G: Domain structure of muscle, skeletal receptor tyrosine kinase (MUSK) protein and other mutations in MUSK previously reported to cause FADS.
Hematoxylin and eosin staining and immunohistochemical staining were performed on muscle biopsies from the affected fetus, and a dead fetus after spontaneous abortion at the same gestational weeks. Immunohistochemical analysis was performed using mouse monoclonal antibodies against slow myosin heavy chain (NCL MHCs; 1:500 dilution), and fast myosin heavy chain (MHCf; 1:500 dilution), both of them were purchased from Abcam (Cambridge, United Kingdom). Significant atrophy was demonstrated in all muscle biopsies. Only a few scattered fibers that were analysed expressed slow myosin. The vast majority of fibers expressed fast myosin. Increased amounts of loose connective tissue were found around and within muscle fascicles. A comparison of MHCf in samples from normal and affected fetuses showed a statistically significant difference between the two, with the affected fetus showing more MHCf (Figure 2).
Figure 2.
Histological, immunohistochemical, and Western blot findings. A: Tissues from the affected fetus; A i: Hematoxylin and eosin staining showed a large variation in muscle fiber size, with many atrophic fibers and increased amounts of loose connective tissue; A ii: Immunohistochemistry with antibody against slow myosin demonstrated only a few scattered type 1 fibers; A iii: Immunohistochemistry with antibody against fast myosin demonstrated that the majority were muscle fibers; B: The results for a control fetus (dead fetus after a spontaneous abortion after the same number of gestational weeks); C: Western blot indicating that the amount of fast myosin heavy chain was significantly higher in the muscles of the affected fetus compared to the control fetus. The loading control is GAPDH. P < 0.05.
Imaging examinations
Prenatal ultrasound signs for the second female fetus included pronated club hands (Figure 1B and C) and rocker-bottom feet. In addition, a micromandible, increased nuchal skin thickness (1.05 cm), and fetal hydrops were also noted. Fetal growth was not appropriate for the gestational age and fetal growth restriction was diagnosed. A clinical diagnosis of FADS was made (case 2 in Table 1).
FINAL DIAGNOSIS
FADS syndrome; G1 P0 G24 wk, delivery of a dead fetus.
TREATMENT
We inducted the labor after routine examinations and consultation of relevant experts.
OUTCOME AND FOLLOW-UP
The patient was discharged on the third day after abortion and was followed for preimplantation genetic diagnosis (PGD).
DISCUSSION
The unifying feature of FADS is a reduction or lack of fetal movement. The clinical symptoms of the two affected fetuses above and two reviews of several affected fetuses described in the medical literature, including prenatal and postnatal findings, are summarized in Table 1. Nearly all cases with an affected fetus chose to terminate the pregnancy in the face of reduced/absent fetal movements and joint contractures. Eight neonates died within a few hours or days after birth and none survived during the neonatal period. Six fetuses did not undergo a prenatal assessment and died within a few hours after birth. In this regard, a prenatal examination is always important if FADS is suspected.
Although etiologies are heterogeneous, the prevailing view is that more than half of all FADS cases are of neuromuscular origin. Several causative gene mutations have been identified because of the development of NGS technologies. Gene mutations of proteins at the NMJ are presently recognized as an important cause of FADS, including CHRNA1 (OMIM 100690)[6], CHRND (OMIM 100720)[6], CHRNG (OMIM 100730)[7], DOK7 (OMIM 610285)[8], RAPSN (OMIM 601592)[9], and MUSK (OMIM601296)[4]. Up to December 31, 2017, MUSK mutations have been identified in three families with FADS involving 18 patients, including the present case (Table 1). The four mutations in the three pedigrees are located in exons (Figure 1G). The MUSK protein is highly conserved among different species (Figure 1F), and hence the full-length protein is important for its function. In this case, the c.421delC frameshift mutation led to the premature termination of MUSK protein translation and the generation of a truncated, 154-amino acid protein, which resulted in MUSK loss of function. A c.220C > T missense mutation caused an amino acid substitution at residue 74 from arginine to tryptophan. This site is also highly conserved among different species (Figure 1E).
MUSK is required for the formation and maintenance of the NMJ. The MUSK protein contains three Ig-like domains: A frizzled-like cysteine-rich domain, a transmembrane helix, and a cytoplasmic tyrosine kinase domain[10] (Figure 1G). In this case, c.421delC and c.220C > T mutations are located in Ig-like 1/2 domains, which are important for the agrin activation of MUSK. Histological and immuno-histochemical findings revealed fiber atrophy and a predominance of type II fibers. We proposed that the compound heterozygous mutation caused a loss of the normal function of MUSK.
CONCLUSION
In conclusion, we herein have reported the first Chinese case of a compound heterozygous mutation of MUSK that caused FADS as an important addition to the MUSK gene mutation database. Further investigation of the relationship between the phenotype and genotype of the compound heterozygous mutation will enable a better understanding of this rare disease.
ACKNOWLEDGEMENTS
We thank the family who participated in this study and who supplied the samples and medical histories.
Footnotes
Informed consent statement: Written informed consent was obtained from the patient for publication of this case report and accompanying images.
Conflict-of-interest statement: The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data sharing statement: No additional data are available.
CARE Checklist (2016) statement: The authors have read the CARE Checklist statement, and the manuscript was prepared and revised according to the CARE Checklist statement.
Manuscript source: Unsolicited manuscript
Peer-review started: December 27, 2018
First decision: March 10, 2019
Article in press: September 9, 2019
Specialty type: Medicine, research and experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Karatza AA S-Editor: Tang JZ L-Editor: Wang TQ E-Editor: Xing YX
Contributor Information
Na Li, Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning Province, China; Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang 110004, Liaoning Province, China.
Chong Qiao, Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning Province, China; Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang 110004, Liaoning Province, China.
Yuan Lv, Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning Province, China; Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang 110004, Liaoning Province, China.
Tian Yang, Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning Province, China; Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang 110004, Liaoning Province, China.
Hao Liu, Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning Province, China; Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang 110004, Liaoning Province, China.
Wen-Qian Yu, Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning Province, China; Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang 110004, Liaoning Province, China.
Cai-Xia Liu, Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning Province, China; Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang 110004, Liaoning Province, China. liucx@sj-hospital.org.
References
- 1.Moessinger AC. Fetal akinesia deformation sequence: an animal model. Pediatrics. 1983;72:857–863. [PubMed] [Google Scholar]
- 2.Hall JG. Pena-Shokeir phenotype (fetal akinesia deformation sequence) revisited. Birth Defects Res A Clin Mol Teratol. 2009;85:677–694. doi: 10.1002/bdra.20611. [DOI] [PubMed] [Google Scholar]
- 3.Ravenscroft G, Sollis E, Charles AK, North KN, Baynam G, Laing NG. Fetal akinesia: review of the genetics of the neuromuscular causes. J Med Genet. 2011;48:793–801. doi: 10.1136/jmedgenet-2011-100211. [DOI] [PubMed] [Google Scholar]
- 4.Wilbe M, Ekvall S, Eurenius K, Ericson K, Casar-Borota O, Klar J, Dahl N, Ameur A, Annerén G, Bondeson ML. MuSK: a new target for lethal fetal akinesia deformation sequence (FADS) J Med Genet. 2015;52:195–202. doi: 10.1136/jmedgenet-2014-102730. [DOI] [PubMed] [Google Scholar]
- 5.Tan-Sindhunata MB, Mathijssen IB, Smit M, Baas F, de Vries JI, van der Voorn JP, Kluijt I, Hagen MA, Blom EW, Sistermans E, Meijers-Heijboer H, Waisfisz Q, Weiss MM, Groffen AJ. Identification of a Dutch founder mutation in MUSK causing fetal akinesia deformation sequence. Eur J Hum Genet. 2015;23:1151–1157. doi: 10.1038/ejhg.2014.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michalk A, Stricker S, Becker J, Rupps R, Pantzar T, Miertus J, Botta G, Naretto VG, Janetzki C, Yaqoob N, Ott CE, Seelow D, Wieczorek D, Fiebig B, Wirth B, Hoopmann M, Walther M, Körber F, Blankenburg M, Mundlos S, Heller R, Hoffmann K. Acetylcholine receptor pathway mutations explain various fetal akinesia deformation sequence disorders. Am J Hum Genet. 2008;82:464–476. doi: 10.1016/j.ajhg.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan NV, Brueton LA, Cox P, Greally MT, Tolmie J, Pasha S, Aligianis IA, van Bokhoven H, Marton T, Al-Gazali L, Morton JE, Oley C, Johnson CA, Trembath RC, Brunner HG, Maher ER. Mutations in the embryonal subunit of the acetylcholine receptor (CHRNG) cause lethal and Escobar variants of multiple pterygium syndrome. Am J Hum Genet. 2006;79:390–395. doi: 10.1086/506256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt J, Morgan NV, Marton T, Maxwell S, Harrison BJ, Beeson D, Maher ER. Germline mutation in DOK7 associated with fetal akinesia deformation sequence. J Med Genet. 2009;46:338–340. doi: 10.1136/jmg.2008.065425. [DOI] [PubMed] [Google Scholar]
- 9.Vogt J, Harrison BJ, Spearman H, Cossins J, Vermeer S, ten Cate LN, Morgan NV, Beeson D, Maher ER. Mutation analysis of CHRNA1, CHRNB1, CHRND, and RAPSN genes in multiple pterygium syndrome/fetal akinesia patients. Am J Hum Genet. 2008;82:222–227. doi: 10.1016/j.ajhg.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stiegler AL, Burden SJ, Hubbard SR. Crystal structure of the frizzled-like cysteine-rich domain of the receptor tyrosine kinase MuSK. J Mol Biol. 2009;393:1–9. doi: 10.1016/j.jmb.2009.07.091. [DOI] [PMC free article] [PubMed] [Google Scholar]