Abstract
Heparin-induced thrombocytopenia (HIT) is a life-threatening, immune-mediated adverse reaction to heparin anticoagulants. The inability to predict HIT represents a considerable liability associated with heparin administration. Genetic studies of HIT are challenging due to the scarcity of true HIT cases, potential for misclassification, and many environmental risk factors. Genetic studies have not consistently identified risk alleles for HIT, the production of platelet factor 4/heparin antibodies or the thromboembolic complications of HIT. Genes implicated in HIT and platelet factor 4/heparin antibody levels include FCGR2A, TDAG8, HLA-DR and others. Compelling evidence also suggests that the FCGR2A H131R polymorphism is associated with HIT-related thrombosis. There is a need for well-powered, multiethnic studies with laboratory confirmation of HIT, detailed patient- and drug-specific data, and inclusion of both serologic and thromboembolic outcomes. Genomic biomarkers identified from such studies offer the possibility of shifting current clinical practice paradigms from early detection and treatment to prevention.
Keywords: : anticoagulant, biomarker, genetics, genome-wide association study, heparin, heparin-induced thrombocytopenia, low molecular weight heparin, pharmacogenomics
Heparin-induced thrombocytopenia (HIT) is an antibody-mediated platelet activation condition in patients receiving unfractionated heparin (UFH) or low molecular weight heparin [1]. HIT develops in up to 2.4% of patients treated with heparin anticoagulants, has a >30% mortality rate and results in catastrophic thromboembolic complications, including life- and limb-threatening thrombosis [2–5]. Approximately 12 million individuals, or a third of all hospitalized patients, are exposed to heparins each year [6]. Although newer anticoagulants are available that carry little or no HIT risk, UFH is likely to remain a mainstay of prevention and treatment of thrombosis due to its immediate onset of action, short half-life, low cost, lack of renal adjustments, ease and inexpensiveness of reversal and laboratory monitoring, and a wealth of evidence supporting its use [7].
Despite the widespread use of heparin anticoagulants and high morbidity of HIT, clinical tools are not available to evaluate HIT risk prior to heparin administration. Several risk factors for HIT have been identified, including increased patient age, dose and duration of heparin, severity of trauma, intravenous versus subcutaneous administration, and UFH versus low molecular weight heparin formulations [8,9]. However, prevention of HIT-related thrombosis is only possible after manifestations of HIT are evident [6,10]. The inability to predict HIT represents a liability associated with heparin administration. Genomic biomarkers offer the potential to distinguish patients who are predisposed to HIT and could shift current clinical practice paradigms from early detection and treatment to prevention. Such biomarkers would enable clinicians to avoid heparin in at-risk patients, strategically monitor patients at high risk for HIT and prevent potentially catastrophic HIT-related thromboses. This special report will review the available evidence for genomic influences on HIT, evaluate the mechanistic insights that this evidence provides and provide a perspective on the potential for prevention of HIT using genomic biomarkers.
Unmet clinical needs for heparin-induced thrombocytopenia
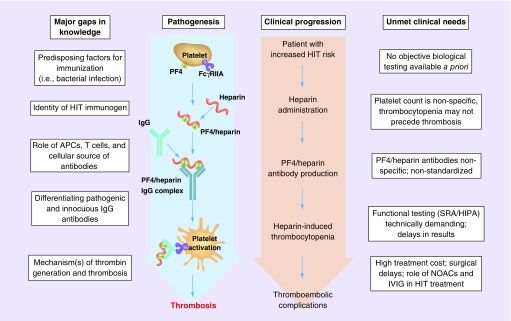
Two major types of HIT exist. Type I HIT is a transient, non-immune complication of heparin treatment with few clinical consequences that is caused by agglutination effects of heparin on platelets [11]. In contrast to type I HIT, type II HIT is an antibody-mediated platelet activation condition associated with life- and limb-threatening thrombosis [2–5]. Classic type II HIT manifests as an approximate 50% fall in platelet count within 5–10 days of heparin administration. The timing and magnitude of platelet count decrease are central to the estimation of HIT likelihood using the 4Ts score [8,12]. This score also includes the presence of thrombosis and other potential causes of thrombocytopenia, and is used to estimate the likelihood of HIT when clinically suspected. Platelet count monitoring is the primary mode of detecting HIT, but can result in a number of false-positive diagnoses (Figure 1). Decreasing platelet counts can be due to a number of causes, especially for the post-surgical and intensive care populations in which HIT is common. In a substantial number of HIT patients, thrombocytopenia may not occur at all, or thrombotic events may occur before thrombocytopenia is evident [3].
Figure 1. . The pathogenesis of heparin-induced thrombocytopenia is characterized by gaps in our knowledge.
Genomic biomarkers have the potential to answer critical questions at every stage of HIT pathogenesis, including the predisposing immunogen, the cellular source of antibodies, the identification of pathogenic IgG antibodies and the mechanisms of thrombosis. Similarly, the clinical progression of patient who will eventually develop HIT is characterized by various unmet clinical needs. Genomic biomarkers have the potential to meet many of the unmet clinical needs for HIT, including limitations in the clinical utility of platelet count monitoring, PF4/heparin antibody testing, functional assay testing and limited treatment options.
APC: Antigen-presenting cell; FcγRIIA: Platelet FcγRIIa receptor; HIPA: Heparin-induced platelet aggregation; HIT: Heparin-induced thrombocytopenia; IVIG: Intravenous immunoglobulin; NOAC: New oral anticoagulant; PF4: Platelet factor 4; SRA: Serotonin release assay.
After HIT is suspected based on decreasing platelet counts, clinical guidelines recommend that heparin be discontinued and an alternative anticoagulant started in patients with 4Ts scores greater than or equal to four [6]. Alternative anticoagulants options for HIT in the USA are few, including argatroban, bivalirudin and fondaparinux, and often carry significant additional drug and monitoring costs [13]. Argatroban is the only available US FDA-approved agent for prevention of thrombosis related to HIT. Danaparoid is a unique agent that suppresses platelet activation by replacing PF4/heparin antibody complexes on the platelet surface [14]. However, danaparoid is no longer available in the USA and manufacturing shortages are common with this agent [15]. Direct oral anticoagulants (DOACs) such as rivaroxaban have an emerging role in the treatment of HIT and are an attractive option due to their oral administration and the limited treatment options for HIT. However, the use of DOACs to treat HIT is primarily backed by case studies and little prospective evidence is available to support DOAC use in HIT [16,17].
In suspected HIT patients, clinical guidelines recommend acquiring a platelet factor 4 (PF4)/heparin IgG immunoassay. Up to 50% of heparin-treated patients will develop PF4/heparin antibodies, but only a fraction of those patients will develop full-blown HIT and its thromboembolic complications [1,18,19]. Although PF4/heparin antibody tests have a near 100% negative predictive value, they are not able to differentiate pathogenic IgG antibodies from those without clinical consequences. PF4/heparin antibody tests are also not standardized across laboratories [20]. Finally, functional assay confirmation of HIT in patients with a positive HIT antibody test is needed to conclusively diagnose HIT. These tests are technically demanding, usually restricted to specialized laboratories, and have the potential to result in significant delays in diagnosis.
Genomic biomarkers have the potential to be used prior to heparin administration as a screening tool to reduce heparin administration to patients with a high HIT risk. Such biomarkers might also be helpful as a risk stratification tool, identifying patients who would benefit from additional monitoring and early treatment to combat thromboembolic complications. Such approaches would reduce over diagnosis and over treatment of HIT, and combat the need for high cost alternative anticoagulants while waiting for laboratory test results. Genomic studies may identify new drug targets, leading to an increase in the clinician's armamentarium for HIT treatment. RNA signatures such as proliferating T-cell receptor clonotypes might also address the critical unmet clinical need for high accuracy diagnostic tools that can confirm HIT early in the disease process.
Pathophysiology of heparin-induced thrombocytopenia
The pathogenesis of HIT begins when heparin, a linear polyanion, binds to the positively charged PF4, an endogenous chemokine stored in α granules of platelets. Neoepitopes on PF4/heparin complexes are recognized by IgG antibodies, forming PF4/heparin–IgG complexes [21]. These PF4/heparin antibodies activate platelets and monocytes via the platelet Fcγ-receptor (FcγRIIa), causing thrombin generation and HIT-associated thrombotic complications [22–24]. Interactions between PF4 and heparin are dependent on charge and stoichiometric ratios, such that an excess of either PF4 or heparin will prevent assembly of PF4/heparin antibody complexes [25,26]. This property of PF4/heparin antibody complexes is exploited in laboratory assays such as the serotonin release assay [27].
HIT has characteristics of both innate and adaptive immunity. IgG antibodies are typically produced within 5–10 days, suggesting a secondary immune response, and are required to elicit an immune response [28,29]. However, these antibodies are typically transient, disappearing after approximately 90 days [30]. Recently, a direct role of PF4/heparin complexes in innate immunity has been described [31]. Heterologous immunity models have also been described, in which immune cells are primed by a bacterial antigen to stimulate an immune response upon subsequent heparin exposure [24,32]. Interestingly, PF4 has a role in innate host defense through binding to negatively charged lipopolysaccharides on bacterial cell walls, a defense mechanism which may be misdirected at linear polyanions such as heparin [33,34]. A severe form of HIT, autoimmune HIT, occurs without prior heparin administration and commonly develops after major surgery or infection [35]. Autoimmune HIT may be the result of these misdirected bacterial defense mechanisms against endogenous linear polyanions such as DNA, RNA, polyphosphates or bacterial wall components [36,37].
Despite decades of research into the immunopathology of HIT, the HIT immunogen, the roles of antigen-presenting cells and T cells, and the B-cell subtypes that produce PF4/heparin antibodies are unknown [7,38]. The clinical significance of nonpathogenic PF4/heparin antibodies and the molecular basis that distinguishes them from pathogenic PF4/heparin antibodies remain unclear. Although informative, prior targeted molecular approaches have not fully identified the mechanisms of HIT, likely due to the complicated and unusual nature of the HIT immune response as well as the relatively narrow focus of these targeted approaches. Genetic studies such as genome-wide association studies (GWAS) have the potential to answer many of these questions and further inform the pathophysiology of this adverse drug reaction.
Genetic studies of HIT
Genetic studies of HIT are challenging due to the scarcity of true HIT cases, the potential for misclassification and the many known patient- and drug-specific environmental factors that affect HIT risk. As with other uncommon pharmacogenomic phenotypes, studies aimed at heritability or those require samples before and after drug exposure are burdensome. This difficulty in sample collection and accurate phenotyping is reflected in the disparate associations and lack of replication observed in genetic studies of HIT. This work is largely limited by a small number of HIT cases and often by a lack of key considerations, such as confirmation of HIT cases by functional assay, rigorously defined thromboembolic outcomes, and a PF4/heparin antibody-positive group to differentiate associations with HIT from associations with seroconversion and antibody production. Furthermore, the evidence is almost entirely generated in individuals of European ancestry, inviting the potential for racial and ethnic disparities if translational tools are developed. There is an ongoing need for well-powered, multiethnic studies with laboratory confirmation of HIT, detailed patient- and drug-specific data, and inclusion of both PF4/heparin antibody test results and thromboembolic outcomes to investigate the genetic basis of HIT. Such studies have the potential to identify clinically implementable biomarkers for HIT and generate key mechanistic insights into HIT pathophysiology.
Genome-wide association studies
A recent GWAS identified a SNP in chromosome 5 in AC106799.2 as a risk allele for HIT (Table 1) [39]. This SNP was associated with HIT in both discovery and replication cohorts with a combined odds ratio (OR) of 2.77 (combined p = 2.7 × 10-8). However, there were only 96 suspected HIT cases, the authors used a liberal threshold for significance (α = 1 × 10-4), and this SNP was not the most significantly associated SNP in the discovery cohort. This variant is located in the lincRNA AC106799.2, which does not have functional annotation, and is situated in an intergenic region without protein coding genes, making speculation on the pathophysiologic implications of this association difficult.
Table 1. . Published literature identifying significant genetic associations with heparin-induced thrombocytopenia.
| Study | Locus | Phenotype(s) | Variant(s) | Effect size(s) | Reported p-value(s) | Study description and limitations | Ref. |
|---|---|---|---|---|---|---|---|
| Witten et al. | Chromosome 5 near AC106799.2 | HIT | rs1433265 | 2.79 (1.69–4.65; discovery) 2.77 (1.64–4.68) (replication) |
p = 6.47 × 10-5 (discovery) p = 1.5 × 10-4 (replication) |
GWAS; 96 HIT (discovery) and 86 HIT (replication) and equal number heparin-treated controls; subset of cases HIPA confirmed; liberal discovery α = 1 × 10-4 | [39] |
| Karnes et al. | TDAG8 (GPR65) | HIT, Ab production | rs1887289 | 16.83 (5.90–48; discovery) | p = 1.01 × 10-8 | GWAS; 67 HIT cases and 884 matched, heparin-treated controls (discovery); no functional assay; SNP replicated for Ab production but not HIT | [40] |
| rs3742704 | 33.48 (8.01–140; discovery) | p = 2.04 × 10-7 | |||||
| Rollin et al. | PTPRJ (CD148) | HIT, platelet activation | rs1503185 | 0.37 (0.20–0.68) | p = 0.002 | 179 Abneg, 160 Abpos and 97 HIT patients; SRA confirmation; no replication; functional data support effect of SNPs on platelet activation | [41] |
| rs1566734 | 0.36 (0.20–0.67) | p = 0.001 | |||||
| Gruel et al. | FCGR3A (CD16A) | HIT | rs396991 | 21.5% (HIT) vs 9.5% (Abpos) | p = 0.02 | 86 Abneg, 84 Abpos and 102 HIT patients; SRA confirmation; no replication | [42] |
| Burgess et al. | FCGR2A (CD32A) | HIT | H131R (rs1801274) | 0/19 RR (HIT) 7/22 RR (healthy) | p < 0.01 | 20 HIT and 24 healthy control patients; SRA confirmation; opposite direction of previous association; no replication | [43] |
| Carlsson et al. | FCGR2A | HIT | H131R (rs1801274) | HIT vs Abneg (χ2) | p < 0.001 | 389 HIT and 351 Abneg patients; HIPA confirmation; no replication | [44] |
| Karnes et al. | HLA-DR | HIT | DRB3*01:01 | 2.81 (1.57–5.02) | p = 2.1 × 10-4 (q = 0.02) |
65 HIT and 350 heparin-treated, matched controls; no functional assay; no replication | [45] |
| Paparella et al. | HLA-DR | Ab production | HLA-DR3 serotype | Relative risk 5.3 for Abpos vs Abneg | p = 0.01 | 10 Abpos and 59 Abneg consecutive patients undergoing CPB; no replication | [46] |
| Pouplard et al. | IL10 | Ab production | IL10G G20 microsatellite | 0.29 (0.12–0.70) | p = 0.006 | 85 Abneg, 84 Abpos and 82 HIT patients; SRA confirmation; no replication; no multiple comparisons adjustment | [47] |
| Rollin et al. | ACP1 | Ab production | Haplotype: rs11553742/rs11553746 | 1.8 (1.2–2.6) | p = 0.005 | 179 Abneg, 160 Abpos and 89 HIT patients; SRA confirmation; no replication; no multiple comparisons adjustment | [48] |
| Rollin et al. | FCGR2A | HITT, IgG binding | H131R (rs1801274) | 5.9 (1.7–20) | p = 0.008 | 35 HITT, 54 HIT, 160 Abpos and 174 Abneg patients; SRA confirmation; no replication | [49] |
| Carlsson et al. | FCGR2A | HITT | H131R (rs1801274) | 37% (HITT) vs 17% (HIT) | p = 0.036 | 68 HITT and 54 HIT patients, HIPA confirmation; no replication | [44] |
| Harris et al. | ITGB3 (GPIIIa) | HITT | PIA2 | 4.68 (1.39–15.72) | p = 0.009 | 39 HIT and 27 HITT patients; no replication; no Abneg or Abpos groups; no functional assay | [50] |
| Pamela et al. | FCGR2A, ITGB3 (GPIIIa), PECAM1 | HITT | 3 SNP risk score (H131R, PIA2, L125V) | 8.00 (4.59–13.93) | p = 0.012 | 50 HIT, 53 HITT and 51 Abpos patients; HIPA confirmation; no single SNP association; no replication | [51] |
Ab: PF4/heparin antibody; Abneg: Patient with negative PF4/antibody test result; Abpos: Patient with positive PF4/antibody test result but negative functional assay result; CPB: Cardiopulmonary bypass; GWAS: Genome-wide association study; HIPA: Heparin-induced platelet aggregation assay; HIT: Heparin-induced thrombocytopenia; HITT: Heparin-induced thrombocytopenia with thrombosis; RR: Recessive genotype for H131R R allele; SRA: Serotonin release assay.
Another GWAS included 67 suspected HIT cases and observed significant SNP associations with HIT near the TDAG8 (or GPR65) gene [40]. These associations included an intronic SNP rs1887289 (OR: 16.83 [5.90–47.97]; p = 1.01 × 10-8) in moderate linkage disequilibrium (LD) with a missense SNP rs3742704 (OR: 33.48 [8.01–140]; p = 2.04 × 10-7). Two replication cohorts were used, including a German cohort of PF4/heparin antibody-tested patients and a French cohort of patients with three groups: patients with HIT, patients testing positive for PF4/heparin antibodies but negative on functional assay (Abpos patients) and patients testing negative for PF4 antibodies (Abneg patients). Whereas rs3742704 showed a significant association with PF4/heparin IgG titer levels in the German cohort (β = 0.07 [standard error: 0.03]; p = 0.03), this SNP did not show association with HIT in the French cohort (OR: 1.90 [0.16–22.76]; p = 0.61). This association of TDAG8 SNPs implies an important mechanistic role of T cells in HIT. TDAG8 SNPs might decrease TDAG8 function or abundance, leading to overactive or unregulated T cells in HIT or PF4/heparin antibody production.
In both GWAS, a low number of HIT cases were available for genome-wide analysis. In addition, many of the HIT cases in both studies were determined through antibody testing and 4Ts scores, and were not functional assay confirmed. This approach invites the potential for both misclassification and false-positive associations. Furthermore, discovery cohorts in both studies did not include both HIT cases and Abpos patients. Inclusion of an antibody-positive group is important to distinguish genetic influences on HIT development versus PF4/heparin antibody production. The mechanistic insight of these studies is also rather unclear. Whereas the association of TDAG8 SNPs suggests T-cell help as part of pathogenesis, the chromosome 5 association has an unclear biological role in HIT.
Candidate gene or candidate SNP studies
The majority of studies on the genetic determinants of HIT have investigated specific genes or SNPs with putative roles in HIT immunology. A number of early studies investigated the influence of genomic variation in FCGR2A. FCGR2A (also known as CD32) carries a functionally relevant H131R polymorphism (rs1801274) in the IgG binding region and this H131R polymorphism has been inconsistently associated with HIT in several populations of primarily European ancestry [43,44,52–55]. The inconsistent association results are summarized succinctly in a meta-analysis, which found no consistent effect of the SNP on HIT using a random effects model (OR: 1.11 [0.56–2.19]; p = 0.77) [56,57]. Several subsequent studies have found no association of FCGR2A H131R with HIT [42,58]. In one candidate SNP study, the influence of the FCGR2A H131R and the FCGR3A (also known as CD16) 158V/F (rs396991) polymorphisms was investigated. Although no differences in genotype frequency were found between HIT and Abneg patients, rs396991 VV homozygotes were significantly higher in HIT patients versus Abpos patients (21.5 vs 9.5%, p = 0.02), a difference that was more pronounced among patients with high PF4/heparin antibody titers (optical density >1.5) [42]. The association of FCGR2A and FCGR3A polymorphisms with HIT reinforces the role of platelet receptors in HIT pathogenesis. SNPs at critical residues in these receptors might affect binding or activation by PF4/heparin antibody complexes.
In the same cohort, three missense polymorphisms in PTPRJ (also known as CD148) were tested for association with HIT [41]. Two missense SNPs in strong LD were associated with HIT compared with both Abpos and Abneg patients (ORs for rs1566734 were 0.47 [0.25–0.89], p = 0.03, and 0.36 [0.20–0.67], p = 0.001, respectively). Although this study did not include a replication cohort, the authors performed several functional studies and observed associations between these SNPs and multiple indices of platelet activation. Yet another study in the same cohort found that an IL10 promoter microsatellite polymorphism IL10G G20 was protective for production of PF4/heparin antibodies (OR: 0.29 [0.12–0.70]; p = 0.006) [47]. However, the authors did not adjust their significance level for multiple testing and, like many previous studies of HIT, the results have not been replicated in an independent cohort. Similarly, a haplotype in ACP1 was found to associate with PF4/heparin antibody production (OR: 1.8 [1.2–2.6]; p = 0.005) in this cohort, which had been increased to 179 Abneg, 160 Abpos and 89 HIT patients [48]. This study also observed no association between the PTPN22 SNP rs2476601 and HIT or PF4/heparin antibody production. PTPRJ, PTPN22 and ACP1 code various protein tyrosine phosphatases, which are critical for immune response and platelet activation signaling pathways. Association of polymorphisms in protein tyrosine phosphatase genes as well as IL10, a key regulatory cytokine in antibody-mediated immunity, implicates variation in these genes in immune regulation of PF4/heparin antibody production. Variation in the human leukocyte antigen (HLA) region of the genome has a central role in T-cell-mediated secondary immune responses and has been strongly associated with immune-mediated disease and adverse drugs reactions [59]. The HLA-DR3 antigen was first associated with the presence of PF4/heparin antibodies in a smaller study with 10 Abpos and 59 Abneg patients (relative risk = 5.3, p = 0.01) [46]. The GWAS studies which identified a TDAG8 SNP as a potential HIT susceptibility allele also interrogated the HLA region using a candidate gene approach [40]. The analysis identified rs4348358 upstream of HLA-DRA as the strongest association for the candidate gene study (OR: 0.25 [CI 0.15–0.44]; p = 2.06 × 10-6). In a subsequent study of the same cohort, sequenced HLA alleles and killer cell immunoglobulin-like receptors (KIR) types imputed from GWAS array data were tested for association with HIT [45]. The authors found that the HLA-DRB3*01:01 allele was significantly associated with HIT (OR: 2.81 [1.57–5.02]; p = 2.1 × 10-4, q = 0.02). Although no KIR types were associated with HIT, a significant interaction was observed between KIR2DS5 and the HLA-C1 KIR binding group (p = 0.03). Together, these studies suggest a role of the HLA region in the pathogenesis of HIT, specifically an influence of class II HLA variation and CD4+ T cells in HIT pathogenesis. Because these cohorts did not include functional assay confirmation of HIT cases, it is difficult to discern whether variation in HLA-DR is associated with HIT or with the production of PF4/heparin antibodies.
Genetic studies of thrombosis in heparin-induced thrombocytopenia
Since thrombosis is the most clinically significant outcome related to HIT, there is much interest in identifying genomic predictors of HIT complicated by thrombosis (HITT). A candidate SNP study investigated the influence of selected SNPs in genes related to platelet glycoprotein function and clotting risk, finding no significant associations in a cohort of 63 patients with isolated HIT and 79 patients with HIT-related thromboembolic complications [60]. Similarly, factor V Leiden was not associated with thrombotic events in patients with HIT [61]. Gene polymorphisms that are well characterized long-term thrombosis risk factors may not be thrombosis risk factors in HIT and other relatively short-term clinical situations such as post-surgical thrombosis, cancer-associated hypercoagulability and HIT [60,62–64].
The FCGR2A H131R polymorphism has the most robust evidence for association with HITT. In a study of 35 HITT, 54 HIT, 160 Abpos and 174 Abneg patients, 131RR homozygous patients were at increased risk for thrombosis relative to HIT patients without thrombosis (OR: 5.9 [1.7–20]; p = 0.008) [49,65]. The authors provide an explanation for their findings with studies suggesting that plasma IgG plays a key role in modulating thrombosis in HIT. In another study of 68 HITT and 54 HIT patients, the RR genotype groups was more common in HITT patients versus HIT patients (37 vs 17%, p = 0.036) [44]. A review by Rollin et al. combined the evidence for the FCGR2A H131R RR genotype group and HITT based on five studies [44,49,51–53]. They observed an overall association between thrombosis and the RR genotype group among HIT patients (OR: 2.86 [1.7–4.8]; p < 0.0001) [66]. While there is moderate evidence for this association, more rigorous methods of combining these studies through meta-analysis is needed.
Other studies on the genetic determinants for HITT include investigations of the PIA2 polymorphism (also known as HPA-1) in ITGB3 (also known as GPIIIa), which was found to be associated with HITT in a small cohort of HIT patients (OR: 4.68 [1.39–15.72]; p = 0.009) [50]. In addition, a polygenic risk score incorporating three SNPs, including FCGR2A H131R, ITGB3 HPA-1 (PIA2) and the PECAM1 SNP L125V, was modestly associated with HITT (OR: 8.00 [4.59–13.93]; p = 0.012), despite no single SNP associations with HITT [51]. Repeated association of the FCGR2A H131R polymorphism with HITT suggests that platelets with FCGR2A carrying 131R are more intensely activated by PF4/heparin antibody complexes, causing an increase in thrombotic risk. The associations of PECAM1 and ITGB3 SNPs underscore the importance of platelet activation in HIT and related thrombosis.
Conclusion
While variation in multiple genes has been implicated in the pathogenesis of HIT, genetic studies have not consistently identified risk alleles for HIT, the production of PF4/heparin antibodies, or the thromboembolic complications of HIT. The association of the FCGR2A H131R polymorphism with HITT is a possible exception. The disparate associations from this body of literature are reflective of the difficulty in clinically diagnosing HIT and the inconsistency of study approaches. There is an ongoing need for well-powered studies that overcome the various limitations of previous literature. Such studies will be critical to assess whether genetic influences on HIT and HIT-related outcomes exist and whether variants can be identified of sufficient effect size to be clinically implementable biomarkers. Considering the widespread use of heparin and the high rate of morbidity and mortality, such biomarkers would be extremely valuable and could shift current clinical practice paradigms in HIT from early detection and treatment to prevention.
Future perspective
Sufficiently powered GWAS with functional assay confirmation of HIT cases are currently underway, which will likely identify moderate to high effect size variants associated with HIT. Such data will aid in developing pathogenic models of HIT and will likely indicate whether clinically implementable biomarkers for HIT and HIT-related outcomes exist. Considering the lack of identification of such biomarkers from the previous literature, it is doubtful that risk alleles with high enough sensitivity and specificity to warrant a priori screening will be present as with HLA screening for other immune-mediated adverse drug reactions [67–69]. HIT is a complex condition related to aberrant immunologic and coagulation pathways, and many genetic associations with small to moderate effects sizes are likely to cumulatively influence HIT risk.
Genetic risk alleles are likely to be identified through studies of gene by environment interactions. Such studies will introduce additional challenges with regard to capturing key variables but would likely identify predisposing factors for HIT related to the secondary immune response, such as bacterial infection or colonization. Genetic predisposition to HIT and PF4/heparin antibody production is likely dependent on environmental factors rather than an independent genetic effect. Other omic approaches such as single immune cell sequencing are also likely to generate valuable insights into the disease and better define HIT endophenotypes such as autoimmune HIT. Such discoveries would have widespread clinical implications and could shift clinical practice paradigms, enabling clinicians to avoid heparin in at-risk patients, strategically monitor patients at high risk for HIT and/or prevent potentially catastrophic thromboembolic complications.
Executive summary.
Significance of heparin-induced thrombocytopenia
Heparin-induced thrombocytopenia (HIT) is a potentially catastrophic immune-mediated adverse drug reaction that represents a clinical liability for treatment with heparin anticoagulants.
Unfractionated heparin is likely to remain a mainstay of prevention and treatment of thrombosis due to its pharmacologic and cost benefits.
Genomic biomarkers for HIT have the potential to prevent HIT through a priori genotyping and to generate mechanistic insights into the biological basis of HIT.
Unmet clinical needs for heparin-induced thrombocytopenia
The inability to predict HIT represents a considerable liability associated with heparin administration.
Genomic biomarkers have the potential to meet many of the unmet clinical needs for HIT, including limitations in the clinical utility of platelet count monitoring, platelet factor 4 (PF4)/heparin antibody testing, functional assay testing and limited treatment options.
Pathophysiology of heparin-induced thrombocytopenia
Despite decades of research into the immunopathology of HIT, the exact cellular and molecular mechanisms underlying HIT have yet to be identified.
HIT has characteristics of both innate and adaptive immunity, and may be a secondary immune response following exposure to a yet unknown antigen.
Genetic studies of heparin-induced thrombocytopenia
Genetic studies of HIT are challenging due to the scarcity of true HIT cases, the potential for misclassification, and the many known environmental factors that affect HIT risk.
Genetic studies have not consistently identified risk alleles for HIT, the production of PF4/heparin antibodies or the thromboembolic complications of HIT.
The strongest evidence from this body of literature supports an association between the FCGR2A H131R polymorphism and thromboembolic complications of HIT.
These studies have key limitations, including small numbers of HIT cases, a lack of functional assay confirmation of HIT and an absence of rigorously defined thromboembolic and serological outcomes.
Conclusion
There is an ongoing need for well-powered, multiethnic studies with laboratory confirmation of HIT, detailed patient- and drug-specific data, and inclusion of serological and thromboembolic outcomes to investigate the genetic basis of HIT.
Genomic biomarkers identified from such studies offer the possibility of shifting current clinical practice paradigms, enabling clinicians to avoid heparin in at-risk patients, strategically monitoring patients at high risk for HIT, and/or prevent potentially catastrophic HIT-related thromboses.
Footnotes
Financial & competing interests disclosure
JH Karnes receives support from the American Heart Association (16SDG29090005) and the American College of Clinical Pharmacy Research Institute (Futures Grant). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Warkentin TE, Levine MN, Hirsh J, et al. Heparin-induced thrombocytopenia in patients treated with low-molecular-weight heparin or unfractionated heparin. N. Engl. J. Med. 1995;332(20):1330–1335. doi: 10.1056/NEJM199505183322003. [DOI] [PubMed] [Google Scholar]
- 2.Girolami B, Prandoni P, Stefani PM, et al. The incidence of heparin-induced thrombocytopenia in hospitalized medical patients treated with subcutaneous unfractionated heparin: a prospective cohort study. Blood. 2003;101(8):2955–2959. doi: 10.1182/blood-2002-07-2201. [DOI] [PubMed] [Google Scholar]
- 3.Greinacher A, Farner B, Kroll H, Kohlmann T, Warkentin TE, Eichler P. Clinical features of heparin-induced thrombocytopenia including risk factors for thrombosis. a retrospective analysis of 408 patients. Thromb. Haemost. 2005;94(1):132–135. doi: 10.1160/TH04-12-0825. [DOI] [PubMed] [Google Scholar]
- 4.Franchini M. Heparin-induced thrombocytopenia: an update. Thromb. J. 2005;3:14. doi: 10.1186/1477-9560-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martel N, Lee J, Wells PS. Risk for heparin-induced thrombocytopenia with unfractionated and low-molecular-weight heparin thromboprophylaxis: a meta-analysis. Blood. 2005;106(8):2710–2715. doi: 10.1182/blood-2005-04-1546. [DOI] [PubMed] [Google Scholar]
- 6.Linkins LA, Dans AL, Moores LK, et al. Treatment and prevention of heparin-induced thrombocytopenia: Antithrombotic Therapy and Prevention of Thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. 2012;141(2 Suppl.):e495S–e530S. doi: 10.1378/chest.11-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The most recent clinical guidelines for prevention and treatment of heparin-induced thrombocytopenia (HIT).
- 7.Arepally GM. Heparin-induced thrombocytopenia. Blood. 2017;129(21):2864–2872. doi: 10.1182/blood-2016-11-709873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowther MA, Cook DJ, Albert M, et al. The 4Ts scoring system for heparin-induced thrombocytopenia in medical-surgical intensive care unit patients. J. Crit. Care. 2010;25(2):287–293. doi: 10.1016/j.jcrc.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Warkentin TE, Heddle NM. Laboratory diagnosis of immune heparin-induced thrombocytopenia. Curr. Hematol. Rep. 2003;2(2):148–157. [PubMed] [Google Scholar]
- 10.Watson H, Davidson S, Keeling D. Guidelines on the diagnosis and management of heparin-induced thrombocytopenia: second edition. Br. J. Haematol. 2012;159(5):528–540. doi: 10.1111/bjh.12059. [DOI] [PubMed] [Google Scholar]
- 11.Chong BH, Ismail F. The mechanism of heparin-induced platelet aggregation. Eur. J. Haematol. 1989;43(3):245–251. doi: 10.1111/j.1600-0609.1989.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 12.Cuker A, Gimotty PA, Crowther MA, Warkentin TE. Predictive value of the 4Ts scoring system for heparin-induced thrombocytopenia: a systematic review and meta-analysis. Blood. 2012;120(20):4160–4167. doi: 10.1182/blood-2012-07-443051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aljabri A, Huckleberry Y, Karnes JH, et al. Cost–effectiveness of anticoagulants for suspected heparin-induced thrombocytopenia in the United States. Blood. 2016;128(26):3043–3051. doi: 10.1182/blood-2016-07-728030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong BH, Ismail F, Cade J, Gallus AS, Gordon S, Chesterman CN. Heparin-induced thrombocytopenia: studies with a new low molecular weight heparinoid, Org 10172. Blood. 1989;73(6):1592–1596. [PubMed] [Google Scholar]
- 15.Al-Eidan FA. Pharmacotherapy of heparin-induced thrombocytopenia: therapeutic options and challenges in the clinical practices. J. Vasc. Nurs. 2015;33(1):10–20. doi: 10.1016/j.jvn.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Tran PN, Tran MH. Emerging role of direct oral anticoagulants in the management of heparin-induced thrombocytopenia. Clin. Appl. Thromb. Hemost. 2018;24(2):201–209. doi: 10.1177/1076029617696582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linkins LA, Warkentin TE, Pai M, et al. Rivaroxaban for treatment of suspected or confirmed heparin-induced thrombocytopenia study. J. Thromb. Haemost. 2016;14(6):1206–1210. doi: 10.1111/jth.13330. [DOI] [PubMed] [Google Scholar]
- 18.Trossaert M, Gaillard A, Commin PL, Amiral J, Vissac AM, Fressinaud E. High incidence of anti-heparin/platelet factor 4 antibodies after cardiopulmonary bypass surgery. Br. J. Haematol. 1998;101(4):653–655. doi: 10.1046/j.1365-2141.1998.00750.x. [DOI] [PubMed] [Google Scholar]
- 19.Pouplard C, May MA, Iochmann S, et al. Antibodies to platelet factor 4-heparin after cardiopulmonary bypass in patients anticoagulated with unfractionated heparin or a low-molecular-weight heparin: clinical implications for heparin-induced thrombocytopenia. Circulation. 1999;99(19):2530–2536. doi: 10.1161/01.cir.99.19.2530. [DOI] [PubMed] [Google Scholar]
- 20.Greinacher A, Ittermann T, Bagemuhl J, et al. Heparin-induced thrombocytopenia: towards standardization of platelet factor 4/heparin antigen tests. J. Thromb. Haemost. 2010;8(9):2025–2031. doi: 10.1111/j.1538-7836.2010.03974.x. [DOI] [PubMed] [Google Scholar]
- 21.Zucker MB, Katz IR. Platelet factor 4: production, structure, and physiologic and immunologic action. Proc. Soc. Exp. Biol. Med. 1991;198(2):693–702. doi: 10.3181/00379727-198-43309. [DOI] [PubMed] [Google Scholar]
- 22.Kelton JG, Sheridan D, Santos A, et al. Heparin-induced thrombocytopenia: laboratory studies. Blood. 1988;72(3):925–930. [PubMed] [Google Scholar]
- 23.Brandt S, Krauel K, Gottschalk KE, et al. Characterisation of the conformational changes in platelet factor 4 induced by polyanions: towards in vitro prediction of antigenicity. Thromb. Haemost. 2014;112(1):53–64. doi: 10.1160/TH13-08-0634. [DOI] [PubMed] [Google Scholar]
- 24.Greinacher A. Clinical practice. Heparin-induced thrombocytopenia. N. Engl. J. Med. 2015;373(3):252–261. doi: 10.1056/NEJMcp1411910. [DOI] [PubMed] [Google Scholar]
- 25.Rauova L, Poncz M, McKenzie SE, et al. Ultralarge complexes of PF4 and heparin are central to the pathogenesis of heparin-induced thrombocytopenia. Blood. 2005;105(1):131–138. doi: 10.1182/blood-2004-04-1544. [DOI] [PubMed] [Google Scholar]
- 26.Suvarna S, Espinasse B, Qi R, et al. Determinants of PF4/heparin immunogenicity. Blood. 2007;110(13):4253–4260. doi: 10.1182/blood-2007-08-105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warkentin TE, Arnold DM, Nazi I, Kelton JG. The platelet serotonin-release assay. Am. J. Hematol. 2015;90(6):564–572. doi: 10.1002/ajh.24006. [DOI] [PubMed] [Google Scholar]
- 28.Reilly MP, Taylor SM, Hartman NK, et al. Heparin-induced thrombocytopenia/thrombosis in a transgenic mouse model requires human platelet factor 4 and platelet activation through FcγRIIA. Blood. 2001;98(8):2442–2447. doi: 10.1182/blood.v98.8.2442. [DOI] [PubMed] [Google Scholar]
- 29.Staibano P, Arnold DM, Bowdish DM, Nazy I. The unique immunological features of heparin-induced thrombocytopenia. Br. J. Haematol. 2017;177(2):198–207. doi: 10.1111/bjh.14603. [DOI] [PubMed] [Google Scholar]
- 30.Warkentin TE, Kelton JG. Temporal aspects of heparin-induced thrombocytopenia. N. Engl. J. Med. 2001;344(17):1286–1292. doi: 10.1056/NEJM200104263441704. [DOI] [PubMed] [Google Scholar]
- 31.Khandelwal S, Lee GM, Hester CG, et al. The antigenic complex in HIT binds to B cells via complement and complement receptor 2 (CD21) Blood. 2016;128(14):1789–1799. doi: 10.1182/blood-2016-04-709634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greinacher A, Holtfreter B, Krauel K, et al. Association of natural anti-platelet factor 4/heparin antibodies with periodontal disease. Blood. 2011;118(5):1395–1401. doi: 10.1182/blood-2011-03-342857. [DOI] [PubMed] [Google Scholar]; • Identifies bacterial infection associated with periodontal disease as a risk factor for platelet factor 4/heparin antibodies, informing models of secondary immunization with HIT.
- 33.Krauel K, Potschke C, Weber C, et al. Platelet factor 4 binds to bacteria, [corrected] inducing antibodies cross-reacting with the major antigen in heparin-induced thrombocytopenia. Blood. 2011;117(4):1370–1378. doi: 10.1182/blood-2010-08-301424. [DOI] [PubMed] [Google Scholar]
- 34.Arman M, Krauel K, Tilley DO, et al. Amplification of bacteria-induced platelet activation is triggered by FcγRIIA, integrin αIIbβ3, and platelet factor 4. Blood. 2014;123(20):3166–3174. doi: 10.1182/blood-2013-11-540526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greinacher A, Selleng K, Warkentin TE. Autoimmune heparin-induced thrombocytopenia. J. Thromb. Haemost. 2017;15(11):2099–2114. doi: 10.1111/jth.13813. [DOI] [PubMed] [Google Scholar]
- 36.Jaax ME, Krauel K, Marschall T, et al. Complex formation with nucleic acids and aptamers alters the antigenic properties of platelet factor 4. Blood. 2013;122(2):272–281. doi: 10.1182/blood-2013-01-478966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandt S, Krauel K, Jaax M, et al. Polyphosphates form antigenic complexes with platelet factor 4 (PF4) and enhance PF4-binding to bacteria. Thromb. Haemost. 2015;114(6):1189–1198. doi: 10.1160/TH15-01-0062. [DOI] [PubMed] [Google Scholar]
- 38.Khandelwal S, Arepally GM. Immune pathogenesis of heparin-induced thrombocytopenia. Thromb. Haemost. 2016;116(5):792–798. doi: 10.1160/TH16-01-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witten A, Bolbrinker J, Barysenka A, et al. Targeted resequencing of a locus for heparin-induced thrombocytopenia on chromosome 5 identified in a genome-wide association study. J. Mol. Med. (Berl) 2018 doi: 10.1007/s00109-018-1661-6. Epub ahead of print. [DOI] [PubMed] [Google Scholar]; •• The most recent genome-wide association study for HIT and includes both functional confirmation of the majority of HIT cases and a replication cohort.
- 40.Karnes JH, Cronin RM, Rollin J, et al. A genome-wide association study of heparin-induced thrombocytopenia using an electronic medical record. Thromb. Haemost. 2015;113(4):772–781. doi: 10.1160/TH14-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first genome-wide association study for HIT and includes three cohorts of HIT patients. Several genes are implicated in HIT pathology such as HLA-DR and TDAG8.
- 41.Rollin J, Pouplard C, Gratacap MP, et al. Polymorphisms of protein tyrosine phosphatase CD148 influence FcγRIIA-dependent platelet activation and the risk of heparin-induced thrombocytopenia. Blood. 2012;120(6):1309–1316. doi: 10.1182/blood-2012-04-424044. [DOI] [PubMed] [Google Scholar]
- 42.Gruel Y, Pouplard C, Lasne D, Magdelaine-Beuzelin C, Charroing C, Watier H. The homozygous FcγRIIIa-158V genotype is a risk factor for heparin-induced thrombocytopenia in patients with antibodies to heparin-platelet factor 4 complexes. Blood. 2004;104(9):2791–2793. doi: 10.1182/blood-2004-01-0058. [DOI] [PubMed] [Google Scholar]
- 43.Burgess JK, Lindeman R, Chesterman CN, Chong BH. Single amino acid mutation of Fc γ receptor is associated with the development of heparin-induced thrombocytopenia. Br. J. Haematol. 1995;91(3):761–766. doi: 10.1111/j.1365-2141.1995.tb05383.x. [DOI] [PubMed] [Google Scholar]
- 44.Carlsson LE, Santoso S, Baurichter G, et al. Heparin-induced thrombocytopenia: new insights into the impact of the FcγRIIa-R-H131 polymorphism. Blood. 1998;92(5):1526–1531. [PubMed] [Google Scholar]
- 45.Karnes JH, Shaffer CM, Cronin R, et al. Influence of human leukocyte antigen (HLA) alleles and killer cell immunoglobulin-like receptors (KIR) types on heparin-induced thrombocytopenia (HIT) Pharmacotherapy. 2017;37(9):1164–1171. doi: 10.1002/phar.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paparella D, Micelli M, Favoino B, D'Alo M, Fiore T, De Luca Tupputi Schinosa L. Anti-heparin-platelet factor 4 antibodies after cardiopulmonary bypass: role of HLA expression. Haematologica. 2001;86(3):326–327. [PubMed] [Google Scholar]
- 47.Pouplard C, Cornillet-Lefebvre P, Attaoua R, et al. Interleukin-10 promoter microsatellite polymorphisms influence the immune response to heparin and the risk of heparin-induced thrombocytopenia. Thromb. Res. 2012;129(4):465–469. doi: 10.1016/j.thromres.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 48.Rollin J, Pouplard C, Leroux D, May MA, Gruel Y. Impact of polymorphisms affecting the ACP1 gene on levels of antibodies against platelet factor 4-heparin complexes. J. Thromb. Haemost. 2013;11(8):1609–1611. doi: 10.1111/jth.12278. [DOI] [PubMed] [Google Scholar]
- 49.Rollin J, Pouplard C, Sung HC, et al. Increased risk of thrombosis in FcγRIIA 131RR patients with HIT due to defective control of platelet activation by plasma IgG2. Blood. 2015;125(15):2397–2404. doi: 10.1182/blood-2014-09-594515. [DOI] [PubMed] [Google Scholar]; •• Provides some of the most compelling evidence for genetic association with an HIT-related phenotype, supporting the influence of the FCGR2A H131R polymorphism with HIT with thrombosis.
- 50.Harris K, Nguyen P, Van Cott EM. Platelet PlA2 polymorphism and the risk for thrombosis in heparin-induced thrombocytopenia. Am. J. Clin. Pathol. 2008;129(2):282–286. doi: 10.1309/BMW4M8NQBV0TKFRX. [DOI] [PubMed] [Google Scholar]
- 51.Pamela S, Anna Maria L, Elena D, et al. Heparin-induced thrombocytopenia: the role of platelets genetic polymorphisms. Platelets. 2013;24(5):362–368. doi: 10.3109/09537104.2012.701026. [DOI] [PubMed] [Google Scholar]
- 52.Bachelot-Loza C, Saffroy R, Lasne D, Chatellier G, Aiach M, Rendu F. Importance of the FcγRIIa-Arg/His-131 polymorphism in heparin-induced thrombocytopenia diagnosis. Thromb. Haemost. 1998;79(3):523–528. [PubMed] [Google Scholar]
- 53.Arepally G, McKenzie SE, Jiang XM, Poncz M, Cines DB. Fc γ RIIA H/R 131 polymorphism, subclass-specific IgG anti-heparin/platelet factor 4 antibodies and clinical course in patients with heparin-induced thrombocytopenia and thrombosis. Blood. 1997;89(2):370–375. [PubMed] [Google Scholar]
- 54.Brandt JT, Isenhart CE, Osborne JM, Ahmed A, Anderson CL. On the role of platelet Fc γ RIIa phenotype in heparin-induced thrombocytopenia. Thromb. Haemost. 1995;74(6):1564–1572. [PubMed] [Google Scholar]
- 55.Denomme GA, Warkentin TE, Horsewood P, Sheppard JA, Warner MN, Kelton JG. Activation of platelets by sera containing IgG1 heparin-dependent antibodies: an explanation for the predominance of the Fc γRIIa low responder (his131) gene in patients with heparin-induced thrombocytopenia. J. Lab. Clin. Med. 1997;130(3):278–284. doi: 10.1016/s0022-2143(97)90022-6. [DOI] [PubMed] [Google Scholar]
- 56.Trikalinos TA, Karassa FB, Ioannidis JP. Meta-analysis of the association between low-affinity Fcγ receptor gene polymorphisms and hematologic and autoimmune disease. Blood. 2001;98(5):1634–1635. doi: 10.1182/blood.v98.5.1634. [DOI] [PubMed] [Google Scholar]
- 57.Lehrnbecher T, Foster CB, Zhu S, et al. Variant genotypes of the low-affinity Fcγ receptors in two control populations and a review of low-affinity Fcγ receptor polymorphisms in control and disease populations. Blood. 1999;94(12):4220–4232. [PubMed] [Google Scholar]
- 58.Slavik L, Svobodova G, Ulehlova J, et al. Polymorphism of the Fcγ receptor II as a possible predisposing factor for heparin-induced thrombocytopenia. Clin. Lab. 2015;61(8):1027–1032. doi: 10.7754/clin.lab.2015.141207. [DOI] [PubMed] [Google Scholar]
- 59.Karnes JH, Miller MA, White KD, et al. Applications of immunopharmacogenomics: predicting, preventing, and understanding immune-mediated adverse drug reactions. Annu. Rev. Pharmacol. Toxicol. 2018 doi: 10.1146/annurev-pharmtox-010818-021818. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlsson LE, Lubenow N, Blumentritt C, et al. Platelet receptor and clotting factor polymorphisms as genetic risk factors for thromboembolic complications in heparin-induced thrombocytopenia. Pharmacogenetics. 2003;13(5):253–258. doi: 10.1097/00008571-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 61.Lee DH, Warkentin TE, Denomme GA, Lagrotteria DD, Kelton JG. Factor V Leiden and thrombotic complications in heparin-induced thrombocytopenia. Thromb. Haemost. 1998;79(1):50–53. [PubMed] [Google Scholar]
- 62.Haim N, Lanir N, Hoffman R, Haim A, Tsalik M, Brenner B. Acquired activated protein C resistance is common in cancer patients and is associated with venous thromboembolism. Am. J. Med. 2001;110(2):91–96. doi: 10.1016/s0002-9343(00)00691-4. [DOI] [PubMed] [Google Scholar]
- 63.Ryan DH, Crowther MA, Ginsberg JS, Francis CW. Relation of factor V Leiden genotype to risk for acute deep venous thrombosis after joint replacement surgery. Ann. Intern. Med. 1998;128(4):270–276. doi: 10.7326/0003-4819-128-4-199802150-00003. [DOI] [PubMed] [Google Scholar]
- 64.Warkentin TE. Prothrombotic genetic risk factors and heparin-induced thrombocytopenia. Pharmacogenetics. 2003;13(5):245–246. doi: 10.1097/00008571-200305000-00001. [DOI] [PubMed] [Google Scholar]
- 65.Padmanabhan A. Why ‘R’ HIT patients predisposed to thrombosis? Blood. 2015;125(15):2319–2320. doi: 10.1182/blood-2015-02-629782. [DOI] [PubMed] [Google Scholar]
- 66.Rollin J, Pouplard C, Gruel Y. Risk factors for heparin-induced thrombocytopenia:focus on Fcγ receptors. Thromb. Haemost. 2016;116(5):799–805. doi: 10.1160/TH16-02-0109. [DOI] [PubMed] [Google Scholar]; • A comprehensive review of genetic risk factors for HIT and HIT-related thromboses.
- 67.Caudle KE, Rettie AE, Whirl-Carrillo M, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clin. Pharmacol. Ther. 2014;96(5):542–548. doi: 10.1038/clpt.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leckband SG, Kelsoe JR, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and carbamazepine dosing. Clin. Pharmacol. Ther. 2013;94(3):324–328. doi: 10.1038/clpt.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saito Y, Stamp LK, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA-B) genotype and allopurinol dosing: 2015 update. Clin. Pharmacol. Ther. 2016;99(1):36–37. doi: 10.1002/cpt.161. [DOI] [PMC free article] [PubMed] [Google Scholar]