Abstract
Pharmacogenetic testing increasingly is available from clinical and research laboratories. However, only a limited number of quality control and other reference materials currently are available for the complex rearrangements and rare variants that occur in the CYP2D6 gene. To address this need, the Division of Laboratory Systems, CDC-based Genetic Testing Reference Material Coordination Program, in collaboration with members of the pharmacogenetic testing and research communities and the Coriell Cell Repositories (Camden, NJ), has characterized 179 DNA samples derived from Coriell cell lines. Testing included the recharacterization of 137 genomic DNAs that were genotyped in previous Genetic Testing Reference Material Coordination Program studies and 42 additional samples that had not been characterized previously. DNA samples were distributed to volunteer testing laboratories for genotyping using a variety of commercially available and laboratory-developed tests. These publicly available samples will support the quality-assurance and quality-control programs of clinical laboratories performing CYP2D6 testing.
CYP2D6 is part of the cytochrome P450 gene family, a group of phase I metabolism enzymes that are responsible for the elimination or bioactivation of numerous drugs.1, 2 CYP2D6 substrates belong to different drug classes, including antidepressants (eg, amitriptyline, doxepin, fluvoxamine, nortriptyline, fluoxetine, paroxetine), antipsychotics (eg, chlorpromazine, clozapine, haloperidol, risperidone), antiarrhythmics (eg, flecainide, propafenone), beta-blockers (eg, carvedilol, metoprolol), opioid analgesics (eg, codeine, tramadol), anticancer agents (eg, gefitinib, tamoxifen), and other drugs (eg, atomoxetine, dextromethorphan, ondansetron; Pharmacogenomics Knowledge Base, https://www.pharmgkb.org/vip/PA166170264, last accessed January 29, 2019).
The highly polymorphic CYP2D6 gene is located on chromosome 22q13.2,3 and, to date, more than 100 CYP2D6 star (*) allele haplotypes (not counting suballelic variants) currently are cataloged by the Pharmacogene Variation (PharmVar) Consortium (https://www.pharmvar.org/gene/CYP2D6, last accessed August 29, 2019). Genetic variation is an important factor explaining the wide range of enzymatic activity of drug-metabolizing enzymes that can be observed within and among populations.4
Based on the identified genotypes in a patient, a diplotype is assigned, which then is translated into a phenotype (ie, metabolizer status for drug-metabolizing enzymes and function status for drug transporters).5 For CYP2D6, there currently are four commonly inferred phenotype groups: poor metabolizers, intermediate metabolizers, normal metabolizers, and ultrarapid metabolizers. Normal and intermediate metabolizers generally are expected to tolerate and respond to standard doses of most medications metabolized by CYP2D6. Patients with no CYP2D6 activity (poor metabolizers) may experience adverse events because a drug either is not metabolized or is metabolized inefficiently, which causes levels above the therapeutic range for active drugs. Therapeutic failure also may be observed in patients with a poor metabolizer phenotype if the drug depends on CYP2D6 for bioactivation.6 Conversely, patients with diplotypes leading to higher than normal activity (ultrarapid metabolizers) may experience therapeutic failure because an active drug is metabolized too fast, causing drug levels that are below the therapeutic range.
Alleles (or haplotypes) are named using the star (*) nomenclature referring to a particular allele or haplotype as *allele. An allele (or haplotype) defines variant(s) that are in cis and the combination of the two haplotypes is referred to as a diplotype. The majority of star (*) alleles are defined by single-nucleotide variants or insertions/deletions of one or a few nucleotides. However, the presence of copy number variations (deletions, duplications, and multiplications) and gene rearrangements with the CYP2D7 pseudogene, leading to CYP2D6–CYP2D7 and CYP2D7–CYP2D6 hybrid genes, pose additional challenges for genotype analysis.3, 7, 8 Sequence variations and the structural variants can occur by themselves or in different combinations. Additional information regarding nomenclature can be found at https://www.pharmvar.org; last accessed August 29, 2019. PharmVar maintains nomenclature for most CYP450 genes, including CYP2D6, as well as other selected genes involved in drug metabolism.9, 10 For simplicity, the term allele will be used from here on forward.
CYP2D6 genotyping assays are difficult to design and validate analytically because of the large number of star (*) alleles and variant types [there are 131 major star alleles (excluding suballeles) listed by PharmVar, https://www.pharmvar.org/gene/CYP2D6, accessed August 28, 2019; note that eight have been retired, making the actual count less than the highest numbered allele: *139]. Reference materials containing a wide variety of sequence and structural variants, including copy number variants and hybrid genes, are invaluable for developing and validating assays and test platforms that can accurately detect all of the variants that the assay or platform is designed to detect and ensure that sequence variations found on other alleles are not affecting assay or platform results. Laboratories developing test panels and testing patients for CYP2D6 need a large and diverse set of reference materials, ideally representing all defined alleles.11 These materials also are important for quality control and proficiency testing.
Although many clinical laboratories offer CYP2D6 testing, reference materials are not publicly available for many of the complex structural or low-frequency variants. The CDC Genetic Testing Reference Material Program (GeT-RM) has conducted two previous pharmacogenetic (PGx) studies,12, 13 referred to here as PGX1 and PGX2, with the goal of creating publicly available and well-characterized genomic DNA reference materials for pharmacogenes, including CYP2D6. Together, these studies identified cell line–based genomic DNA samples containing many, but not all, of the star (*) alleles that are included in most clinical CYP2D6 genotyping tests. For a significant number of the genomic DNA samples, results were ambiguous or genotype could not be determined with certainty (eg, gene copy number remained unknown or structural variants were only partially characterized). In addition, no reference materials were identified for many of the rare star (*) alleles, including many of the structural variants. This was because of the low population frequency of these variants as well as the design of the test platforms used to characterize the samples. To address the need for a more comprehensive set of reference materials for CYP2D6 testing, the GeT-RM program, in collaboration with the genetic testing community, have further characterized selected samples previously characterized by Get-RM for CYP2D6 as well as additional samples that harbor rare and/or complex alleles or allele combinations.
Materials and Methods
Cell-Line DNA and Participating Laboratories
DNA from 179 cell lines was selected from the National Institute of General Medical Sciences and the National Human Genome Research Institute Repositories at the Coriell Cell Repositories (Camden, NJ) for this study. Of these, 135 were characterized in the most recent GeT-RM study (PGX2),13 and two (NA17222 and NA17287) were characterized in PGX1.12 Forty-two additional DNA samples carrying rare variants that were not represented in the PGX2 reference material panel13 were selected based on data supplied by the authors of this article or identified by searching the NCBI 1000 Genomes Project (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes, last accessed August 13, 2018). The five laboratories that participated in this follow-up study, Agena Bioscience, ARUP Laboratories, Children's Mercy Kansas City, Medical College of Wisconsin/RPRD (Right Patient Right Drug) Diagnostics, and Icahn School of Medicine at Mount Sinai, used a variety of genotyping and sequencing platforms.
DNA Preparation
DNA was prepared from each of the selected cell lines by the Coriell Cell Repositories using Gentra/Qiagen Autopure (Valencia, CA) per the manufacturer's instructions.
Characterization Protocol
Each of the testing laboratories received one 10-μg aliquot of DNA from each of the cell lines that they volunteered to test. Each laboratory tested the samples using their standard methods and/or additional methods needed to resolve inconclusive genotype calls. The test platforms and genotyping assays used in the study, and the alleles detected by each method, are shown in Supplemental Table S1 and are described in the following sections. The results were submitted to two researchers (A.G. and V.M.P.), who examined the data for quality and discordances, and determined the consensus genotype. If discordances were noted, the participating laboratories were asked to re-evaluate their data for the sample(s) in question to determine the cause of the inconsistency. Supplementary testing was performed if required.
PharmacoScan Array
Following the manufacturer's instructions, genomic DNA first was amplified (DNA amplification and multiplex PCR). The amplified products were pooled, purified, fragmented, labeled, and hybridized to the PharmacoScan Array (Thermo Fisher Scientific, Waltham, MA) per the manufacturer's recommendations. Arrays were stained with a fluorescent antibody and scanned on the GeneTitan Multi-Channel Instrument (Thermo Fisher Scientific). Data were analyzed using the Axiom Analysis Suite 3.1 (Thermo Fisher Scientific). Analysis was performed using either the commercially released allele translation table (version r6; Thermo Fisher Scientific) or a custom translation table that includes additional CYP2D6 star alleles for translation (version v.r6+20180103).
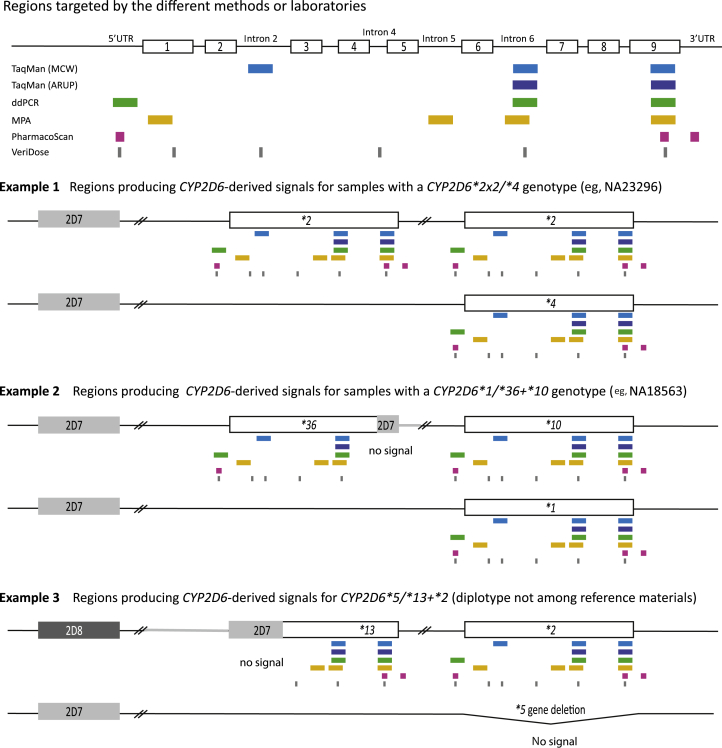
The PharmacoScan Array detects nucleotide variants and copy number state (0, 1, 2, ≥3) on a subset of genes. For CYP2D6, three gene regions (5′ flanking, 3′ flanking, and exon 9) are interrogate by the array (Figure 1 and Table 1).
Figure 1.
Analysis of CYP2D6 copy number and structural variation. The figure provides an overview of the regions targeted for quantitative copy number determination by method and laboratory. The top panel shows the target regions of the TaqMan copy number assays used in the study. The Medical College of Wisconsin/RPRD Diagnostics (MCW) test site interrogated three gene regions whereas ARUP Laboratories (ARUP) tested for two gene regions. TaqMan assays were also used for droplet digital PCR (ddPCR) whereas a quantitative multiplex PCR method (MPA) developed at Children's Mercy Kansas City targets four regions. The PharmacoScan and VeriDose platforms target three and six regions, respectively. CYP2D6 exons are represented by numbered boxes and the 5′ untranslated region (5′UTR) and introns 2, 4, 5, and 6 also are shown. Example 1 shows a diplotype that produces the same copy number calls (three copies) across all targeted regions. Example 2 shows a diplotype containing an allele with a CYP2D7-derived exon 9 conversion; this region does not amplify and therefore produces two-copy number calls, whereas all other regions produce three-copy number calls. Example 3 shows a genotype with a CYP2D6*5 gene deletion on one chromosome and a tandem harboring a CYP2D7–CYP2D6 hybrid on the other chromosome. As illustrated, the 5′UTR, exon 1, and intron 2 target regions corresponding to CYP2D7 do not amplify and produce one-copy number calls, whereas the intron 5 and 6 and exon 9 regions amplify from the CYP2D6 portion of the hybrid as well as the CYP2D6*2 in the tandem producing two-copy number calls. Table 1 summarizes the copy number calls for each example for each of the methods/platforms used.
Table 1.
Copy Number Variation Calls from Different Platforms for Examples Provided in Figure 1
| Example 1 | 5′UTR | Exon 1 | Intron 2 | Intron 4 | Intron 5 | Intron 6 | Exon 9 | 3′UTR |
|---|---|---|---|---|---|---|---|---|
| TaqMan | – | – | 3 | – | – | 3 | 3 | – |
| TaqMan | – | – | – | – | – | 3 | 3 | – |
| ddPCR | 3 | – | – | – | – | 3 | 3 | – |
| MPA | – | 3 | – | – | 3 | 3 | 3 | – |
| PharmacoScan | 3 | – | – | – | – | – | 3 | 3 |
| VeriDose | 3 | 3 | 3 | 3 | – | 3 | 3 | – |
| Example 2 | 5′UTR | Exon 1 | Intron 2 | Intron 4 | Intron 5 | Intron 6 | Exon 9 | 3′UTR |
|---|---|---|---|---|---|---|---|---|
| TaqMan | – | – | 3 | – | – | 3 | 2 | – |
| TaqMan | – | – | – | – | – | 3 | 2 | – |
| ddPCR | 3 | – | – | – | – | 3 | 2 | – |
| MPA | – | 3 | – | – | 3 | 3 | 2 | – |
| PharmacoScan | 3 | – | – | – | – | – | 2 | 2 |
| VeriDose | 3 | 3 | 3 | 3 | – | 3 | 2 | – |
| Example 3 | 5′UTR | Exon 1 | Intron 2 | Intron 4 | Intron 5 | Intron 6 | Exon 9 | 3′UTR |
|---|---|---|---|---|---|---|---|---|
| TaqMan | – | – | 1 | – | – | 2 | 2 | – |
| TaqMan | – | – | – | – | – | 2 | 2 | – |
| ddPCR | 1 | – | – | – | – | 2 | 2 | – |
| MPA | – | 1 | – | – | 2 | 2 | 2 | – |
| PharmacoScan | 1 | – | – | – | – | – | 2 | 2 |
| VeriDose | 1 | 1 | 1 | 2 | – | 2 | 2 | – |
Different platforms or methods may target different gene regions [5′ or 3′ untranslated regions (UTR), introns, or exons]. Examples 1, 2, and 3 provide the calls for those shown graphically in Figure 1. Example 1 has three-copy calls across all regions tested. Example 2 has a CYP2D6*36 gene copy that generates three-copy calls for the 5′UTR through intron 6 regions and two-copy calls for exon 9. Example 3 shows a sample with a CYP2D7–CYP2D6 hybrid that generates one-copy calls for the 5′UTR through intron 2 regions and two-copy calls for the intron 4 through exon 9 regions.
–, indicated genomic region was not amplified by the method used; ddPCR, droplet digital PCR; MPA, multiplex PCR amplification.
Copy Number Assay Using TaqMan Real-Time PCR
Samples Analyzed by the Medical College of Wisconsin/RPRD Diagnostics
For a subset of samples analyzed with the PharmacoScan Array (HG00463, HG01190, HG02373, NA18545, and 96 Tier 1 samples from the PGX2 study13), TaqMan copy number assays were run as per the manufacturer's instructions on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) to interrogate copy number state in intron 2, intron 6, and exon 9 (assay IDs: Hs04083572_cn, Hs04502391_cn, and Hs00010001_cn; Thermo Fisher Scientific) (Figure 1 and Table 1). The copy number data were analyzed using CopyCaller software version 2.0 (Thermo Fisher Scientific). Seven two-copy control samples were run on each plate; the sample that showed the median ΔCT within the two-copy samples was selected as the calibrator. In addition, a one-copy and a three-copy sample were included on each plate. The manual CT was 0.2 and the baseline was set on automatic. A confidence level of 95% and z-score value of less than 1.75 was applied to call the copy number.
Samples Analyzed by ARUP Laboratories
Two TaqMan real-time PCR assays targeting intron 6 and exon 9 were performed (assay IDs: Hs04502391_cn and Hs00010001_cn) (Figure 1 and Table 1). All assays were performed on a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific) using 96-well plates, in four replicates along with RNase P (assay ID 4403326; Thermo Fisher Scientific) as a reference gene, according to the manufacturer's protocol. A two-gene copy calibrator, a sample with one gene copy, and a sample with three gene copies were included with each run. Relative quantification of the CYP2D6 copy number was performed using CopyCaller (Thermo Fisher Scientific). If the calculated copy number value for a given probe was N ± 0.4 gene copies (where N is a whole number), it was predicted to be that whole number; however, if it was greater than ±0.4 gene copies the assay was repeated. A confidence level of 95% and z-score value of less than 1.75 was applied to call the copy number.
iPLEX CYP2D6 v1.1 and VeriDose CYP2D6 Copy Number Variation Panel
By using the CYP2D6 panel, specific DNA fragments were amplified from genomic DNA in three PCR reactions and 35 of the most clinically relevant variants subsequently were interrogated using iPLEX PRO (Agena Bioscience, San Diego, CA) single-base extension biochemistry. Genotypes were detected using a MassARRAY Analyzer 4 system and haplotypes were assigned using an ADME CYP2D6 Reporter plugin for the Typer Analyzer software version 4.1.183 (Agena Bioscience). A set of 13 additional variant genotyping assays was designed and run using the same single-base extension protocol to distinguish rare haplotypes. These were interpreted manually in addition to the standard analysis.
By using the VeriDose (Agena Bioscience) CYP2D6 copy number variation (CNV) panel for copy number determination, 11 assays in six target regions within the CYP2D6 gene (5′ untranslated region, exon 1, intron 2, intron 4, intron 6, and exon 9) (Figure 1 and Table 1) were amplified from genomic DNA in one PCR reaction together with a control region in either CYP2D7 or CYP2D8. The 11 assays interrogating single-base differences subsequently were amplified using single-base extension. Data for these assays were analyzed using a MassARRAY Analyzer 4 system and the gene copy number was calculated using a CYP2D6 PGx Reporter plugin for the Typer Analyzer software (Agena Bioscience). The CNV software (PGx Reporter software version 3.51; Agena Bioscience) calculates the copy number for the CYP2D6 gene as well as indicates the presence of CYP2D6*13 (CYP2D7–CYP2D6 hybrid genes), CYP2D6*68 (CYP2D6–CYP2D7 hybrid genes), or genes carrying a CYP2D7-derived exon 9 conversion including *36 and *4N. An aggregate CYP2D6 copy number is reported in the main output, but individual calls per assay are reported in a separate CNV output file.
TaqMan-Based Genotyping
Of the samples tested with the TaqMan-based genotyping assays (Thermo Fisher Scientific) at Children's Mercy Kansas City, 72 were run on the OpenArray platform (Thermo Fisher Scientific) using a custom-designed array panel. OpenArrays were performed as prescribed by the manufacturer. The remaining samples were genotyped using single-tube TaqMan genotyping assays in a 96-well reaction format. OpenArray no-calls or uncertain calls were repeated using the single-tube assay as follows. Each 8-μL reaction contained 0.8 μL DNA (15 ng/μL) and 1× Probe Force qPCR Master Mix (KAPA Biosystems, Wilmington, MA). The initial denaturation was 95°C as recommended for the KAPA mastermix, and cycling parameters were as recommended by the provided TaqMan assay protocols. All assays and OpenArrays were run on QuantStudio 12K Flex Real-Time PCR System version 1.2.2 (Thermo Fisher Scientific). Data were analyzed with the TaqMan Genotyper software version 1.4.0 (Thermo Fisher Scientific); each call was inspected manually.
Characterization of Structural Variants with XL-PCR
Children's Mercy Kansas City also tested the samples using long-range (XL) PCR as previously described.14, 15, 16 Briefly, a series of XL-PCR fragments was amplified to show or validate the presence of a CNV event or the presence of hybrid gene copies (Table 2). For example, the presence of a CYP2D6*2×2 duplication on one of the chromosomes generates fragment B and an approximately 8-Kb long fragment D, whereas the presence of a CYP2D6*36+*10 tandem generates an approximately 10-Kb long fragment D (the longer size is because of a CYP2D7-derived region downstream of *36), but not fragment B. An approximately 10-Kb long fragment D also is generated from CYP2D6*68+*4 (*68 is a CYP2D6–CYP2D7 hybrid gene), and CYP2D7–CYP2D6 hybrid genes amplify an approximately 5-Kb long fragment H. To determine which CYP2D6 alleles are present in a gene duplication (two identical or near-identical gene copies such as CYP2D6*1×2, *2×2, or *4×2), fragment D was genotyped for one or more single-nucleotide polymorphisms (SNPs) to discriminate (eg, a CYP2D6*1×2 from a CYP2D6*4×2). For example, if fragment D was generated from a duplicated CYP2D6*4×2 allele, the genotype calls for rs1065852 (c.100C>T) and rs1065852 (c.1847G>A) (positions according to the CYP2D6 RefSeq NG_008376.3) appeared homozygous; if the duplication is on the CYP2D6*1×2 allele, fragment D is negative for these SNPs. The presence of the CYP2D6*5 gene deletion was detected by XL-PCR, as previously described.14
Table 2.
Summary of Long-Range (XL)-PCR Products Generated for Gene Locus Characterization and/or Sequencing
| Fragment ID | Primer sequences | CYP2D6 XL-PCR amplicons | Product size, Kb | Samples∗ |
|---|---|---|---|---|
| 1 (A)† | F: 5′-GTCCCACACCAGGCACCTGTACT-3′ | Nonduplicated gene and gene deletion | 15.6 (nonduplicated) 3.5 (deletion) | HG00337, NA18632, NA18642 |
| R: 5′-GAATTAGTGGTGGTGGGTGTTTG-3′ | ||||
| 2 (B)† | F: 5′-TCACCCCCAGCGGACTTATCA-3′ | 5′ CYP2D6 gene in duplication | 12 | HG00337, NA18642 |
| R: 5′-CCACAGCCCTCAATAAGTGAA-3′ | ||||
| 3 (C)† | F: 5′-CCCTGGGAAGGCCCCATGGAAG-3′ | 3′ CYP2D6 gene in duplication | 12 | HG00337, NA18642 |
| R: 5′-TAGGTAGCCCTGGCCTATAGCTCCCTGACGCC-3′ | ||||
| 4 (E)† | F: 5′-TCACCCCCAGCGGACTTATCA-3′ | CYP2D6–CYP2D7 hybrid gene | 6.7 | HG00337, NA18632, NA18642 |
| R: 5′-TACGGTGGGCTCCCTGCGAG-3′ | ||||
| 5 (D)† | F: 5′-TTGCCACATTATCGCCCGTGAAA-3′ | Full-length CYP2D6 in any arrangement | 8.4 | HG00337, HG04090, HG04206, NA18632, NA18642, NA18973, NA19908, NA20803, NA20875, NA21105 |
| R: 5′-TAGGTAGCCCTGGCCTATAGCTCCCTGACGCC-3′ | ||||
| 6 (A)‡ | F: 5′-TCACCCCCAGCGGACTTATCAACC-3′ | Full-length CYP2D6 in any arrangement | 6.7 | HG00373, HG03225, HG03246, HG03259, HG03780, NA17128, NA18632, NA19777 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)‡ | F: 5′-TGGAGAGAGGCCACCTGAGGTAGTC-3′ | ASXL-PCR of entire CYP2D6 gene (-2609C) | 7.4 | NA19180 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)‡ | F: 5′-CGTCAAGCTTTCCGACATACACG-3′ | ASXL-PCR of entire CYP2D6 gene (-2532G) | 7.3 | NA23878 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)‡ | F: 5′-CCTCCCAAATCTGATGAAAAATATTAATCC-3′ | ASXL-PCR of entire CYP2D6 gene (-2421C) | 7.2 | NA19917 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)§ | F: 5′-GAGGCAACCTGCTCGGG-3′ | ASXL-PCR of entire CYP2D6 gene (-2178G) | 6.8 | NA07439, NA12154, NA17137, NA18642, NA19174 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)§ | F: 5′-CTGTCCTCAGTGGATGATCCCG-3′ | ASXL-PCR of entire CYP2D6 gene (-1770G) | 6.5 | NA23877 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)‡ | F: 5′-CCTGGACAACTTGGAAGAACCG-3′ | ASXL-PCR of entire CYP2D6 gene (-1584G) | 6.4 | HG00337, HG00436, HG00589, HG01086, HG01094, HG02373, HG03781, NA17448, NA18552, NA18973, NA20289, NA21105 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)‡ | F: 5′-CCTGGACAACTTGGAAGAACCC-3′ | ASXL-PCR of entire CYP2D6 gene (-1584C) | 6.4 | HG00337, HG01806, HG03619, HG03781, NA20289 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)§ | F: 5′-CATGGTGAAACCCTATCTCTACTGAAAATAC-3′ | ASXL-PCR of entire CYP2D6 gene (-1426C) | 6.2 | NA17185 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)‡ | F: 5′-TGTGTGTGAGAGAGAATGTGTGCC-3′ | ASXL-PCR of entire CYP2D6 gene (-740C) | 5.5 | NA19239, NA23877 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| 6 (AS-A)§ | F: 5′-TCACCCCCAGCGGACTTATCAACC-3′ | ASXL-PCR of entire CYP2D6 gene (1023C) | 2.9 | NA19239 |
| R: 5′-CCCGAAACCCAGGATCTGGG-3′ | ||||
| 6 (AS-A)§ | F: 5′-TCACCCCCAGCGGACTTATCAACC-3′ | ASXL-PCR of entire CYP2D6 gene (4401C) | 6.3 | NA17185 |
| R: 5′-GACATCTGCTCAGCCTCAACG-3′ | ||||
| 6 (AS-A)§ | F: 5′-TCACCCCCAGCGGACTTATCAACC-3′ | ASXL-PCR of entire CYP2D6 gene (4723G) | 6.6 | NA17176, NA19908 |
| R: 5′-CTGGGAGGTAGGTAGCCCTGACC-3′ | ||||
| 7 (D)¶ | F: 5′-CCAGAAGGCTTTGCAGGCTTCAG-3′ | CYP2D6–CYP2D7 hybrid gene | 8.6 or 10.2‖ | NA18632, NA18642 |
| R: 5′-CGGCAGTGGTCAGCTAATGAC-3′ | ||||
| 8 (B)¶ | F: 5′-CCATGGAAGCCCAGGACTGAGC-3′ | 5′ CYP2D6 intergenic region in duplication | 3.5 | N/A |
| R: 5′-CGGCAGTGGTCAGCTAATGAC-3′ | ||||
| 9¶ | F: 5′-TCACCCCCAGCGGACTTATCAACC-3′ | CYP2D6–CYP2D7 hybrid genes including *36 | 6.1 | NA17287, NA18632 |
| R: 5′-CACCAGAAAGCTGACGACACGAGA-3′ | ||||
| 10 (H)¶ | F: 5′-TCCGACCAGGCCTTTCTACCAC | CYP2D7–CYP2D6 hybrid genes | 5 | NA19785, NA19790 |
| R: 5′-CGACTGAGCCCTGGGAGGTAGGTAG-3′ | ||||
| N/A∗∗ | F: 5′-CCAGAAGGCTTTGCAGGCTTCAG-3′ (1st PCR) F: 5′-GAACCTCTGGAGCAGCCCATACCC-3′ (nested) |
CYP2D6 genes in downstream position | 8.1 (first PCR) 5.0 (nested) | HG00373, NA17222, NA17287 |
| R: 5′-CGGCAGTGGTCAGCTAATGAC-3′ (first PCR) R: 5′-ACTGAGCCCTGGGAGGTAGGTAG-3′ (nested) |
Fragment IDs match those used in Figure 2 to visualize which region(s) each amplicon represents. Forward and reverse primer sequences are as listed (5′ to 3′). A brief description of the region amplified, purpose of the amplification, and fragment lengths also are provided. Samples used to generate respective XL-PCR products are listed in the right-hand column. The position of the single-nucleotide polymorphism that discriminates among alleles is shown in parentheses. The allele-specific nucleotides in primers are underlined.
AS, allele-specific; ASXL-PCR, allele-specific long-range PCR; F, forward; N/A, not applicable; R, reverse.
Samples listed only when sequenced; some XL-PCRs were also run on other samples to determine which allele is duplicated or to confirm structural variation.
PCR as described.25
XL-PCR performed with primers as described.18
XL-PCR performed as described18 with newly designed primer.
This fragment is 8.6 Kb when the duplicated gene has a CYP2D6 (REP6) downstream region and 10.2 Kb if the duplicated gene has a CYP2D7 (REP7) downstream region (see Figure 2).
XL-PCR performed as described.22
CNV and Structural Variant Detection with Quantitative Multiplex PCR
Quantitative multiplex PCR amplification (MPA) was performed at Children's Mercy Kansas City as previously published,17 with the following modification: 2G Fast HotStart DNA Polymerase (KAPA Biosystems) was used with the supplied buffer at either 1.5× or 1× final concentration. PCR reactions were performed using an Eppendorf MasterCycler ep Gradient S or an Eppendorf MasterCycler Pro Gradient S instrument (Eppendorf, Hamburg, Germany). Four regions were targeted (exon 1, intron 5, intron 6, and exon 9) to determine the number of copies for each region (Table 1 and Figure 1). Each MPA run included samples with 0, 1, 2, 3, and 4 gene copies as positive controls. Calls were within the confidence levels of 95% for each cluster as described previously.17
CNV and Structural Variant Detection with ddPCR
The Children's Mercy Kansas City laboratory interrogated a subset of samples by droplet digital PCR (ddPCR) to confirm selected CNV results. TaqMan copy number assays targeting the 5′ promoter (Hs04078252_cn), intron 6 (Hs04502391_cn), and exon 9 (Hs00010001_cn) (Figure 1 and Table 1) were used and signals were normalized against the RPP30 gene (assay ID: dHsaCP1000485; Bio-Rad) or TERT gene (catalog number 4403316; Thermo Fisher Scientific). Analysis was performed on the QX200 Droplet Digital PCR System (Bio-Rad). Genomic DNA (250 ng) was digested with 1.2 U EcoRI-HF (New England BioLabs, Ipswich, MA) restriction enzyme in a 20-μL reaction at 37°C. After a 1.5-hour incubation, the enzyme was inactivated at 65°C for 20 minutes. Subsequently, ddPCR Supermix for Probes (Bio-Rad), CYP2D6 TaqMan assay, and RPP30 reference TaqMan assay (all at 1× final concentrations) were combined with 50 ng of EcoRI-digested genomic DNA in a 22 μL total volume. Droplets were generated with the Auto Droplet Generator and cycled in a C1000 Touch Thermocycler as follows: 10 minutes at 95°C for 1 cycle, 40 cycles of 30 seconds at 94°C and 1 minute at 60°C, and a final cycle of 98°C for 10 minutes. Droplets were analyzed with the QX200 Droplet Reader instrument and data analysis was performed with the Quantasoft software version 1.0 (both from Bio-Rad). Each ddPCR run included samples with two and four gene copies as positive controls. Calls did not deviate more than 0.15 from the determined copy number (eg, for a copy number of two, calls ranged within 1.85 and 2.15).
Sanger Sequencing of Allele-Specific XL-PCR Amplicons
XL-PCR fragments comprising the entire gene were generated at Children's Mercy Kansas City using a forward primer that specifically amplified the allele of interest and a universal reverse primer. For samples with the CYP2D6*5 gene deletion the XL-PCR fragment was generated with a universal forward primer. Resulting fragments were genotyped to confirm that only the allele of interest was amplified. Fragments were purified and subsequently subjected to Sanger sequencing. Allele-specific XL-PCR and Sanger sequencing were performed as previously described.18 A list of the XL-PCR products generated for each sample is listed in Table 2. The alleles in sample HG00373 could not be separated by allele-specific XL-PCR; a heterozygous amplicon thus was generated using the universal forward primer and sequenced. To unequivocally determine each haplotype, the XL-PCR fragment subsequently was subjected to single-molecule real-time sequencing at the Icahn School of Medicine at Mount Sinai as described in Long-Read, Single-Molecule, Real-Time Sequencing.
Next-Generation Sequencing Sequencing of XL-PCR Amplicons
XL-PCR were generated at the Medical College of Wisconsin/RPRD Diagnostics on a subset of 22 samples from the set of samples identified to potentially contain rare variants. Multiple XL-PCR products were generated from samples containing duplications (HG00337), CYP2D6/CYP2D7 hybrids (NA18632), or duplications and hybrid alleles (NA18642) to amplify both gene copies of a duplication, as well as hybrid genes separately. The XL-PCR products generated are listed in Table 2. Libraries were prepared for sequencing using the Ion Xpress Plus Fragment Library Kit (Thermo Fisher Scientific) per the manufacturer's recommendations. Library template preparation and chip loading was performed on the Ion Chef with Ion Hi-Q Chemistry (Thermo Fisher Scientific) and sequenced on the Ion Proton (Thermo Fisher Scientific) as recommended. Sequencing generated 1.1 to 2.1 million reads per sample (average read length, approximately 155 bp), with an average read depth of approximately 30,000 mapped reads per base. The primary alignment to GRCH37 was performed using Torrent Suite version 5.8.0, and variant calling was performed with the Ion Torrent Variant Caller version 5.8.0.19 (both from Thermo Fisher Scientific). The generated BAM files were reviewed manually with the Integrative Genomics Viewer version 2.4.14 (The Broad Institute, Massachusetts Institute of Technology, Cambridge, MA),19 aligning to GRCh37.20, 20, 21
Long-Read, Single-Molecule, Real-Time Sequencing
Single-molecule, real-time (SMRT) sequencing of the full-length CYP2D6 gene was performed at the Icahn School of Medicine at Mount Sinai on selected samples as previously described.22, 23 In brief, XL-PCR amplified both downstream and upstream (duplicated) copies when present, and these products were used as templates for nested PCR and barcoding before multiplexed SMRT sequencing. All PCR amplicons were purified and pooled before SMRT sequencing on the PacBio RS-II using the P6-C4 Pacific Biosciences (Menlo Park, CA) protocol with a movie collection time of 180 minutes. Circular consensus sequencing was used and FASTQ files were demultiplexed, aligned to chr22:42,522,044-42,527,019 (GRCH37), and subjected to CYP2D6 variant calling. Identified genotypes were translated to common star (*) allele nomenclature and haplotype definitions according to PharmVar.9 XL-PCR products used for sequencing are listed in Table 2.
Allele Designations and Diplotype Reporting
Allele designations are according to those described by the PharmVar Consortium (www.PharmVar.org; last accessed August 28, 2019). Novel allelic variants were submitted to PharmVar for designation.
For the purpose of this report, suballeles are not specified, with the exception of CYP2D6*4N (*4.013), because there are no known functional differences among suballeles. For example, CYP2D6*2A, discriminated from other *2 suballeles by the presence of -1584C>G (rs1080985, NC_000022.11:g.42132375G>C) (now designated *2.001 by PharmVar), are shown as CYP2D6*2. Likewise, although some platforms can discriminate between certain suballeles, for example, CYP2D6*45A and B (now designated *45.001 and *45.002 by PharmVar) are shown as *45. The CYP2D6*4.0013 (formerly *4N) is excepted because this variant contains the CYP2D6-derived exon 9 conversion preventing the generation of signal in CNV assays targeting this region. Of particular note, the decreased function allele originally named CYP2D6*14B is now *14.001 and displayed as *14 in this report whereas the no function allele CYP2D6*14A has been renamed *114.24 Finally, diplotypes determined by the PharmacoScan and iPLEX platforms are shown per respective algorithm outputs without further (manual) interpretation.
Results
Five laboratories participated in this follow-up investigation using a variety of methods, including the matrix-assisted laser desorption/ionization-time of flight–based MassARRAY Analyzer 4 platform (iPLEX panel; Agena Bioscience), the PharmacoScan Array (Medical College of Wisconsin/RPRD Diagnostics), TaqMan-based genotyping (Children's Mercy Kansas City), a number of CNV methods and assays (Medical College of Wisconsin/RPRD Diagnostics, Children's Mercy Kansas City, ARUP Laboratories, and Agena Bioscience), and Sanger sequencing (Children's Mercy Kansas City), next-generation sequencing (Medical College of Wisconsin/RPRD Diagnostics), and SMRT sequencing (Icahn School of Medicine at Mount Sinai). Quantitative CNV analysis was performed by at least one laboratory for a total of 156 samples. In this project, 137 DNA samples from the previously published Get-RM panels (PGX1,12 PGX213) were characterized further using two or more platforms; and an additional 42 DNA samples also were characterized and added to the panel to expand the repertoire of reference materials for CYP2D6. An overview of the test platforms used, including the sequence variations tested, Single Nucleotide Polymorphism database reference SNP cluster ID (rsID) numbers, and positions, is provided in Supplemental Table S1. Selected samples were chosen for sequence and/or CNV analyses to either resolve discordant or inconclusive genotyping results or to unequivocally determine the haplotype of rare alleles. Some samples were analyzed by one or more laboratories before this study and results were kindly provided for this investigation.
Genotyping
Tables 3 and 4 provide the consensus genotypes of all samples for which genotype assignments have changed based on reanalysis as well as samples newly characterized in this investigation, respectively. An extended table (Supplemental Table S2) lists results from each test platform including sequencing and CNV data as well as consensus genotypes for all samples. Consensus CYP2D6 diplotypes were determined by examination of the variant calls made by each platform. Calls were made manually for TaqMan-based genotyping whereas the MassARRAY and PharmacoScan results were obtained with the software tools provided by the manufacturers and reviewed manually. Diplotype assignments across platforms were consistent for the vast majority of samples. A subset of samples, however, showed inconsistent calls that could be explained by the different variant panels tested. For example, HG01680 was called CYP2D6*2/*59 by TaqMan genotyping, *2/*59 or *28/*59 using two different PharmacoScan calling algorithms, and *2/*2 by iPLEX CYP2D6 V1.1. The iPLEX CYP2D6 call was revised to *28/*59 when V1.1 was supplemented with a custom panel. The apparent discordances can be explained by TaqMan not testing for the SNP(s) identifying *28 and iPLEX CYP2D6 V1.1 not testing for the *28 or *59-identifying variant resulting in CYP2D6*2 default assignments. A CYP2D6*28/*59 diplotype ultimately was confirmed by Sanger sequencing. Similarly, a number of other rare alleles were not detected or called by any genotyping platform (eg, CYP2D6*82, *112) or only detected by one (eg, CYP2D6*22) or two platforms (eg, CYP2D6*31), while the others produced default assignments. Similar to CYP2D6*28 described earlier in this paragraph, the current commercial PharmacoScan algorithm did not call a number of alleles because their respective identifying variants, although tested, are not used for haplotype translation. However, an updated algorithm (v.r6+20180103) has been devised to expand the catalog of called alleles including CYP2D6*22 and *35.
Table 3.
Samples from Previous GeT-RM Studies for Which Genotypes Have Been Revised (n = 47)
| Sample ID | Consensus genotype previous study | Consensus genotype this study |
|---|---|---|
| HG00436 | *1/*2×N | *2×2/*71 |
| HG00589 | *1/*2 (*21) | *1/*21 |
| HG01190 | *4/*5 | *68+*4/*5 |
| NA07439 | *4/*41×N | *4×2/*41 |
| NA10855 | *1/*4 | *1/(*68)+*4 |
| NA11832 | *1/*4 | *1/(*68)+*4 |
| NA12878 | *3/*4 | *3/(*68)+*4 |
| NA15245 | *4/*4×N | *4×2/*4 |
| NA17102 | *1/*17 (*40) | *1/*40 |
| NA17222 | *1/*2† | *2/*108 |
| NA17244 | *2/*4×N | *2×2/*4×2 (+hybrid) |
| NA17287 | No consensus† | *1/*83 |
| NA17448 | *1/*2 | *1/*28 |
| NA17454 | *1/*2×N | *1×2/*2×2 |
| NA18524 | *1/*10 (*36) | *1/*36×2+*10 |
| NA18526 | *1/[*10 (*36)] | *1/*36×2+*10 |
| NA18540 | *10/*41 | (*36+)10/*41 |
| NA18544 | *10/*41 | *10/*41 |
| NA18563 | *1/(*36) | *1/*36+*10 |
| NA18564 | *2/[*10 (*36)] | *2A/*36+*10 |
| NA18565 | *10/[*10 (*36)] | *10/*36×2 |
| NA18572 | (*36)/*41 | *36+*10/*41 |
| NA18617 | *10/[*10 (*36)] | *36+*10/*36+*10 |
| NA18959 | *2/[*10 (*36)] | *2/*36+*10 |
| NA18973 | *1/*2 (*21) | *1/*21 |
| NA18980 | *2/[*10 (*36)] | *2/*36+*10 |
| NA19109 | *2×N/*29 | *2×2/*29 |
| NA19143 | *2/*10 | *2 (*45)/*10 |
| NA19174 | (*4/*40) | *4/*40 |
| NA19207 | *2/*10×N | *2×2/*10 |
| NA19226 | *2/*2×N | *2/*2×2 |
| NA19785 | *1/*2×N | *1/*13+*2 |
| NA19819 | *2/*4×N | *2/*4×2 |
| NA19908 | *1/*2 | *1/*46 |
| NA19917 | *1/*17 (*40) | *1/*40 |
| NA19920 | *1/*4×N | *1/*4×2 |
| NA21781 | *2/*4×N | *2×2/*68+*4 |
| NA23090 | *1/(*36) | *1/*36+*10 |
| NA23093 | *1/(*36) | *1/*36+*10 |
| NA23246 | *10/*10×N | *10×2/*36+*10 |
| NA23275 | *1/*17 (*40) | *1/*40 |
| NA23296 | *2/*4×N | *2×2/*4 |
| NA23297 | *10/*17×N | *10×2/*17 |
| NA23313 | *2/*2×N | *2×2/*2 |
| NA23878 | ?/*4 | (*4N)+*4/*39 |
| NA24027 | *2/*6×N | *2×2/*6 |
| NA24217 | *2/*41×N | *2A/*41×3 |
Consensus genotypes from previous Genetic Testing Reference Material Program (GeT-RM) studies are as shown.
?, indicates that one of the alleles could not be determined.
Consensus genotype from the PGX1 study, all other genotypes are from the PGX2 study. Genotype, copy number variation, and sequencing results for these and all other samples for which the genotype did not change are detailed in Supplemental Table S2. Tentative assignments are shown in parentheses.
Table 4.
Consensus Genotypes of Samples that Were Added to the Get-RM Panel (n = 42)
| Sample ID | Consensus genotype in this study |
|---|---|
| HG00111 | *3/*3 |
| HG00156 | *5/*5 |
| HG00337 | *2×2/*22 |
| HG00373 | *2/*2 |
| HG00421 | *2/*10×2 |
| HG00423 | *10/*10×2 |
| HG00463 | *36+*10/*36+*10 |
| HG01086 | *1/*31 |
| HG01094 | *1/*31 |
| HG01108 | *2/*106 |
| HG01680 | *28/*59 |
| HG02373 | *14/*36+*10 |
| HG03225 | *5/*56 |
| HG03246 | *5/*43 |
| HG03259 | *5/*106 |
| HG03619 | *2/*113 |
| HG03643 | *2/*7 |
| HG03703 | *1/*99 |
| HG03780 | *1/*112 |
| HG03781 | *2/*99 |
| HG03882 | *1/*112 |
| HG04206 | *2/*113 |
| NA06989 | *9/*9 |
| NA10860 | *1/*4N+*4 |
| NA12154 | (*68)+*4/*33 |
| NA17113 | *17×2/*45 |
| NA17128 | *1/*43 |
| NA17137 | *29/*45 |
| NA17169 | *17/*56 |
| NA17176 | *3/*45 |
| NA17185 | *4/*58 |
| NA18545 | *5/*36×2+*10×2 |
| NA18632 | *36×2+*10/*52 |
| NA18642 | *36+*10/*1+*90 |
| NA19180 | *1/*58 |
| NA19317 | *5/*5 |
| NA19777 | *1/*82 |
| NA19790 | *1/*13+*2 |
| NA20289 | *6/*11 |
| NA20803 | *2/*22 |
| NA20875 | *1/*111 |
| NA21105 | *3/*111 |
Consensus genotypes of samples characterized only in the current Genetic Testing Reference Material Program (GeT-RM) study and not in past studies. Genotype, copy number variation, and sequencing results for these samples are detailed in Supplemental Table S2.
Copy Number and Structural Variation
In the previous study,13 gene copy number was not assigned [ie, it remained unknown whether a sample had a duplication (eg, CYP2D6*2×2) or more than two gene copies (eg, CYP2D6*2×3)]; such samples were labeled as, for example, CYP2D6*2×N, indicating that the actual number of gene copies is unknown. Furthermore, a number of samples were suspected to carry a CYP2D6*36+*10 tandem, but the presence of this allele was not confirmed. In this follow-up investigation, selected samples were tested further with qualitative (XL-PCR) and other quantitative methods to determine the copy number and characterize the structure of alleles harboring hybrid genes consisting of CYP2D6 and CYP2D7 (sometimes also referred to as fusion genes). Hybrid genes may occur by themselves (as the only gene copy on a chromosome) or in various tandem/duplication arrangements.14, 15, 16, 25, 26
Quantitative copy number testing included two loci (TaqMan-based, ARUP Laboratories), three or four loci [TaqMan-based, ddPCR, or MPA (Children's Mercy Kansas City)], and three loci [TaqMan-based (Medical College of Wisconsin/RPRD Diagnostics)] across CYP2D6 (Figure 1). Equal increased or decreased copy number calls across all loci tested is indicative of a CYP2D6 gene duplication, multiplication, or deletion, for example, a CYP2D6*2×2/*4 genotype generates a 3-copy result for each interrogated region (ie, 3/3/3 or 3/3/3/3 when three and four loci are tested, respectively) and a CYP2D6*1/*5 genotype generates one copy across all targeted regions. In the presence of genes that partially are composed of CYP2D7, PCR primers that target CYP2D6-derived regions will not be able to amplify. Examples are provided in Figure 1, providing CNV calls for different genotypes with structural variation (Table 1). Note that testing for the intron 6 and exon 9 regions only (Figure 1) produces a two-copy call for these regions, which is not consistent with a CYP2D6*5/*13+*2 diplotype, underscoring the importance of targeting regions across the entire CYP2D6 gene for more informative copy number testing. Furthermore, the loss of a CYP2D6 exon 9 copy signal infers the presence of a CYP2D7–exon 9 conversion, which often is interpreted as the presence of CYP2D6*36 in a single or tandem arrangement. However, this may not always be the case. For example, NA10860 contains a CYP2D6*4N in a *4N+*4 tandem structure that is characterized by CYP2D7-derived exon 9 and downstream sequences, which also causes signal loss in the exon 9 CNV assay. Another example is CYP2D6*83 (NA17287). Again, because of the presence of the exon 9 conversion in this allele, the CNV assay does not produce any signal.
A subset of samples with inconclusive MPA results, high copy number, or suspected complex gene structures also were tested using ddPCR, a superior method for absolute quantification that eliminates user and sample quality variability.27 All inconclusive or inconsistent MPA results were resolved (eg, samples HG00337 or HG03781), and those obtained for complex structures were confirmed (eg, samples NA17244 or NA23246) (Supplemental Table S2).
The VeriDose CYP2D6 CNV panel (Agena Bioscience) interrogates copy number at 11 target regions across the gene, detecting gene duplications and hybrid gene structures. VeriDose results are aggregated into each sample's reported genotype and also provided per target region in a separate report. Some inconsistent copy number calls were observed among the iPLEX CYP2D6 V1.1 and iPLEX CYP2D6 V1.1+VeriDose calls. For example, HG00423 was genotyped as CYP2D6*10/*10 by the former, but as *10×2/*10 by the latter. The CNV call obtained with the VeriDose approach was consistent with TaqMan, MPA, and ddPCR CNV results, as well as the formation of a duplication-specific XL-PCR amplicon. It needs to be stressed that the CYP2D6 V1.1 panel was developed for variant genotyping, and CNV detection was focused on detection of the *5 gene deletion. In contrast, the VeriDose CYP2D6 CNV panel was specifically designed and developed to detect CYP2D6 CNVs and structural variants.
The VeriDose CYP2D6 CNV assay provides copy number information based on 11 assays covering six gene regions. By using the data generated, the PGx Reporter plugin for the Typer Analyzer software can detect the number of alleles and detect the presence of gene hybrids such as CYP2D6*4N (*4.0013) (NA10860), *13 (NA19785), *36 (NA18563), and *68 (HG01190), but does not also determine on which allele a duplication is located (NA19207 was called as 3N, *2/*10) (see Supplemental Table S2 for more examples).
The PharmacoScan array reports CYP2D6 CNV events as *5 (0 or 1), CN_Gain (3≥), CN_HybridLoss (partial gene deletion or conversion, eg, a *36 allele), or CN_HybridGain (partial gene duplication). A CN_HybridGain sample includes two full CYP2D6 alleles and a third partial CYP2D6 allele (eg, *2/*10, CN_HybridGain), from which a particular variant may be inferred (eg, CYP2D6*2/*36+*10). For samples that contain duplicated CYP2D6 alleles (either full length or hybrid), the analysis software does not determine which allele is duplicated. For example, samples NA19207 and NA19109 are reported as *2/*10, CN_Gain, and as *2/*29, CN_Gain. For some duplication events, if the duplication did not include the entire 3′ or 5′ flanking regions interrogated by the array, the genotype may be reported as a CN_HybridGain, instead of a CN_Gain. This may lead to discrepancies between test platforms that interrogated alternate regions of the gene and/or flanking sequences.
TaqMan genotyping assays and reaction conditions are not designed to detect CNVs. In some instances, however, the presence of a duplication or multiplication may be inferred by the position of a sample in the TaqMan cluster analysis graph (not shown). For instance, the signal produced from the CYP2D6*2 SNP (rs16947, NM_000106.5:c.886C>T) was positioned off the heterozygous (toward the homozygous) cluster for HG00436, supporting a CYP2D6*1/*2×2 rather than a CYP2D6*1×2/*2 genotype. Such positional shifts may be more or less pronounced and not always apparent, and an absence of a clearly shifted cluster position does not necessarily indicate that a CNV is not present. This same phenomenon holds true for iPLEX chemistry as well as PharmacoScan.
Finally, the presence of duplications and tandem arrangements were verified by specifically amplifying the duplicated or hybrid genes and subsequently genotyping or sequencing these amplification products. For example, NA19109 produced a XL-PCR amplicon encompassing the duplicated gene copy; this amplicon was negative for CYP2D6*29 and, thus, the duplicated gene was attributed to the CYP2D6*2 allele resulting in a CYP2D6*2×2/*29 genotype assignment. XL-PCR also allowed detection of the CYP2D6*36+*10 allele in samples initially genotyped as, for example, CYP2D6*2/*10 (NA18980); the tandem arrangement produced an approximately 10-Kb long XL-PCR amplicon that genotyped positive for rs1065852 (NM_001025161.2:c.100C>T) and the exon 9 conversion. Similarly, XL-PCR confirmed the presence of a CYP2D6*68+*4 tandem in a number of samples that resulted in 2/3/3/3 copy number signals when tested with the MPA assay (eg, NA12878 or HG01190). Because not all samples of the panel were retested systematically, it is possible that genotypes with a CYP2D6*4 or *10 are actually carrying a CYP2D6*68+*4 or *36+*10 allele.
The following gene duplications have been identified among the study samples: CYP2D6*1×2, *2×2, *4×2, *10×2, *17×2, and *36×2 (Tables 3 and 4 and Supplemental Table S2). One sample carried a CYP2D6*41×3 multiplication. Finally, a number of tandem arrangements (ie, structures with two or more hybrid gene copies): CYP2D6*4N+*4, *13+*2, *36+*10, *36×2+*10, *36×2+*10×2, and *68+*4 also were identified. A more detailed description of samples with complex diplotypes containing tandem structures with CYP2D6*52 and *99 are described in Identification of Coriell Samples with Rare Alleles or Genotypes Not Identified in Previous GeT-RM Studies and Samples with Complex Allele Combinations, respectively.
Two samples (NA17244 and NA23246) have inconsistent copy number signals across the interrogated loci. In addition to having a duplication on each chromosome, NA17244 also generated a XL-PCR amplicon signaling the presence of a 2D6-2D7 hybrid gene copy. This sample requires further characterization to fully understand the structural variants causing the MPA 4/5/5/4 and TaqMan 4/5/4 CNV results. NA23246 had a TaqMan 3/4/3 CNV call whereas the MPA and ddPCR calls returned 4/[4]/4/3 and 4/4/3 calls. The three-copy signal for exon 9 in NA23246 is consistent across all CNV methods and the presence of a CYP2D6*36+*10 allele. However, what causes the three-copy signal for the intron 2 region reported by the TaqMan (3/4/3) assay or the tentative four-copy MPA intron 5 call (shown in brackets earlier in this paragraph) remains unknown. It is conceivable that sequence variation(s) in these regions impact assay performance and cause a signal drop-out (three- instead of four-copy signal for TaqMan) or a lower-than-expected signal for the MPA assay. Copy number drop-outs have been observed for the TaqMan intron 2 and intron 6 assays, but have not been investigated systematically. Lower-than-expected signals affecting copy number calls, although consistent among repeated testing, also may be attributed to sample quality and purity.
Identification of Coriell Samples with Rare Alleles or Genotypes Not Identified in Previous GeT-RM Studies
One major goal of this project was to identify rare allelic variants and/or complex genotypes that were not interrogated in the previous GeT-RM studies. In addition, we wanted to identify which allele was duplicated or multiplicated when present. Laboratories volunteered information from their own investigations using Coriell materials to complement the current sample panel and data. Upon the identification of samples of interest, DNA was acquired and provided to the laboratories that participated in testing. In addition, data from the 1000 Genomes Project (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes; last accessed August 13, 2018) was searched for samples harboring rare variants not yet represented in the reference material panel, and candidate samples were tested by the laboratories.
This study identified and characterized a number of rare, novel, and complex CYP2D6 alleles and haplotypes (a complete list of all CYP2D6 alleles is provided in Table 5). Twenty-six CYP2D6 alleles have been added to the panel of available reference materials through the efforts of this study. These include the relatively rare no function CYP2D6*11, *13, *31, *56, and *99 alleles, the CYP2D6*59 decreased function allele, and the normal function CYP2D6*33, *39, *45, and *46 alleles. Function is unknown or uncertain for the remaining 14 newly added alleles. Based on current knowledge, CYP2D6*68, a nonfunctional CYP2D6-2D7 hybrid gene, occurs exclusively in the *68+*4 tandem configuration (up to a quarter of all CYP2D6*4 alleles may indeed have the *68+*4 tandem17). In addition, we have characterized numerous structural arrangements including tandem and hybrid genes.
Table 5.
Summary of CYP2D6 Alleles Identified in GeT-RM Studies
| A CYP2D6 alleles |
B Function |
C Alleles tested in this study (by at least one platform/method) |
D CYP2D6 alleles identified in GeT-RM studies |
|---|---|---|---|
| *2 | Normal function | Yes | Yes |
| *3 | No function | Yes | Yes |
| *4 | No function | Yes | Yes |
| *5 | No function | Yes† | Yes |
| *6 | No function | Yes | Yes |
| *7 | No function | Yes | Yes |
| *8 | No function | Yes | No |
| *9 | Decreased function | Yes | Yes |
| *10 | Decreased function | Yes | Yes |
| *11 | No function | Yes | Yes (new) |
| *12 | No function | Yes | No |
| *13 | No function | Yes† | Yes (new) |
| *14 | Decreased function | Yes | Yes |
| *15 | No function | Yes | Yes |
| *17 | Decreased function | Yes | Yes |
| *18 | No function | Yes | No |
| *19 | No function | Yes | No |
| *20 | No function | Yes | No |
| *21 | No function | Yes | Yes (new) |
| *22 | Uncertain function | No | Yes (new)‡ |
| *23 | Uncertain function | Yes | No |
| *24 | Uncertain function | No | No |
| *25 | Uncertain function | Yes | No |
| *26 | Uncertain function | No | No |
| *27 | Normal function | Yes | No |
| *28 | Uncertain function | Yes | Yes (new) |
| *29 | Decreased function | Yes | Yes |
| *30 | Uncertain function | Yes | No |
| *31 | No function | Yes | Yes (new) |
| *32 | Unknown function | Yes | No |
| *33 | Normal function | Yes | Yes (new) |
| *34 | Normal function | No | No |
| *35 | Normal function | Yes | Yes |
| *36 | No function | Yes† | Yes |
| *37 | Uncertain function | Yes | No |
| *38 | No function | Yes | No |
| *39 | Normal function | No | Yes (new)‡ |
| *40 | No function | Yes | Yes |
| *41 | Decreased function | Yes | Yes |
| *42 | No function | Yes | No |
| *43 | Uncertain function | Yes | Yes (new) |
| *44 | No function | Yes | No |
| *45 | Normal function | Yes | Yes (new) |
| *46 | Normal function | Yes | Yes (new) |
| *47 | No function | Yes | No |
| *48 | Normal function | Yes | No |
| *49 | Decreased function | Yes | No |
| *50 | Decreased function | Yes | No |
| *51 | No function | Yes | No |
| *52 | Uncertain function | No | Yes (new)‡ |
| *53 | Normal function | No | No |
| *54 | Decreased function | Yes | No |
| *55 | Decreased function | Yes | No |
| *56 | No function | Yes | Yes (new) |
| *57 | No function | Yes | No |
| *58 | Unknown function | Yes | Yes (new) |
| *59 | Decreased function | Yes | Yes (new) |
| *60 | No function | Yes | No |
| *61 | Uncertain function | No | No |
| *62 | No function | Yes | No |
| *63 | Uncertain function | No | No |
| *64 | Uncertain function | No | No |
| *65 | Uncertain function | No | No |
| *68 | No function | Yes† | Yes (new) |
| *69 | No function | No | No |
| *70 | Uncertain function | Yes | No |
| *71 | Uncertain function | Yes | Yes (new) |
| *72 | Decreased function | Yes | No |
| *73 | Unknown function | Yes | No |
| *74 | Unknown function | No | No |
| *75 | Uncertain function | Yes | No |
| *81 | Uncertain function | Yes | No |
| *82 | Unknown function | Yes | Yes (new) |
| *83 | Unknown function | Yes† | Yes (new) |
| *84 | Decreased function | Yes | No |
| *85 | Unknown function | Yes | No |
| *86 | Unknown function | No | No |
| *87 | Uncertain function | No | No |
| *88 | Uncertain function | No | No |
| *89 | Uncertain function | Yes | No |
| *90 | Uncertain function | No | Yes (new) |
| *91 | Uncertain function | No | No |
| *92 | No function | No | No |
| *93 | Uncertain function | No | No |
| *94 | Uncertain function | No | No |
| *94 | Uncertain function | No | No |
| *95 | Uncertain function | Yes | No |
| *96 | No function | No | No |
| *97 | Uncertain function | No | No |
| *98 | Uncertain function | No | No |
| *99 | No function | Yes | Yes (new) |
| *100 | No function | Yes | No |
| *101 | No function | Yes | No |
| *102 | Unknown function | Yes | No |
| *103 | Unknown function | Yes | No |
| *104 | Unknown function | No | No |
| *105 | Unknown function | No | No |
| *106 | Unknown function | No | Yes (new) |
| *107 | Unknown function | No | No |
| *108 | Unknown function | Yes | Yes (new) |
| *109 | Unknown function | No | No |
| *110 | Unknown function | No | No |
| *111 | Unknown function | No | Yes (new) |
| *112 | Unknown function | No | Yes (new) |
| *113 | Unknown function | No | Yes (new) |
| *114 (*14A)† | No function | Yes | No |
| Structural variants (gene deletion, duplication, hybrids, and tandem arrangements) | *1×2, *2×2, *4×2, *4N+*4, *5, *10×2, *13+*2, *17×2, *36×2, *36+*1, *36+*10, *36×2+*10, *36×2+*10×2, *41×3, and *68+*4 |
All defined star alleles as of December 18, 2018, are listed in Column A with the exception of CYP2D6*115–*139, which were added by PharmVar after the experimental portion of this study had concluded. Column B lists the function of each allele per the functionality table listed by the Pharmacogenomics Knowledge Base (https://www.pharmgkb.org/page/cyp2d6RefMaterials). Column C shows whether an allele has been tested by at least one platform or method (“Yes” indicates tested, “No” indicates not tested).
GeT-RM, Genetic Testing Reference Material Program.
Gene deletion or structural variant inferred by quantitative CNV assays. Column D indicates whether an allele has been found in any of the GeT-RM studies. “Yes” indicates that an allele was detected in a number of samples in a previous GeT-RM study (n = 16), and “Yes (new)” denotes those samples newly detected in this study (n = 26). “No” indicates that the allele has not been found in a reference materials sample (n = 63).
An allele that has been identified by sequencing (not tested for by any of the platforms).
Samples with Novel Haplotypes
Novel CYP2D6*2 Suballeles
All platforms identify the CYP2D6*59 decreased function allele via g.2292G>A in intron 4 (rs267608300, NM_000106.5:c.843+44G>A). Decreased function of this allele, however, is conveyed by g.2940G>A (rs79292917, NM_000106.5:c.975G>A) located at the 3′ end of exon 6, which has been shown to cause reduced levels of mRNA expression.28 HG00373 consistently was genotyped heterozygous for rs267608300 (g.2292G>A), causing CYP2D6*59 calls across platforms. Subsequent confirmatory Sanger sequencing of a heterozygous XL-PCR amplicon did not find the functional CYP2D6*59 SNP (g.2940G>A); however, a number of variants that could not be reconciled with known haplotype definitions were identified. Because no variants in the upstream or downstream regions allowed performing allele-specific XL-PCR to sequence alleles separately, SMRT sequencing was performed to determine the phase of each haplotype unequivocally. Both alleles of this sample were submitted to PharmVar and designated as CYP2D6*2 subvariants (*2.0012 and *2.0013; www.pharmvar.org/gene/CYP2D6; last accessed August 28, 2019). The CYP2D6*2.013 suballele, lacking the key variant of CYP2D6*59 (c.2940G>A; rs79292917), causes false-positive CYP2D6*59 calls, which may lead to inaccurate phenotype assignments.
Other Novel Suballeles
Sequencing of allele-specific XL-PCR products showed novel CYP2D6*1, *2, *6, *15, *43, *56, and *71 suballeles (Supplemental Table S2). For example, a novel CYP2D6*15 suballele was found in NA19239 (designated *15.002 by PharmVar), and NA20289 showed a novel *6 suballele (designated *6.006). None of the newly discovered suballeles interfered with the genotyping platforms tested in this investigation.
Samples with Complex Allele Combinations
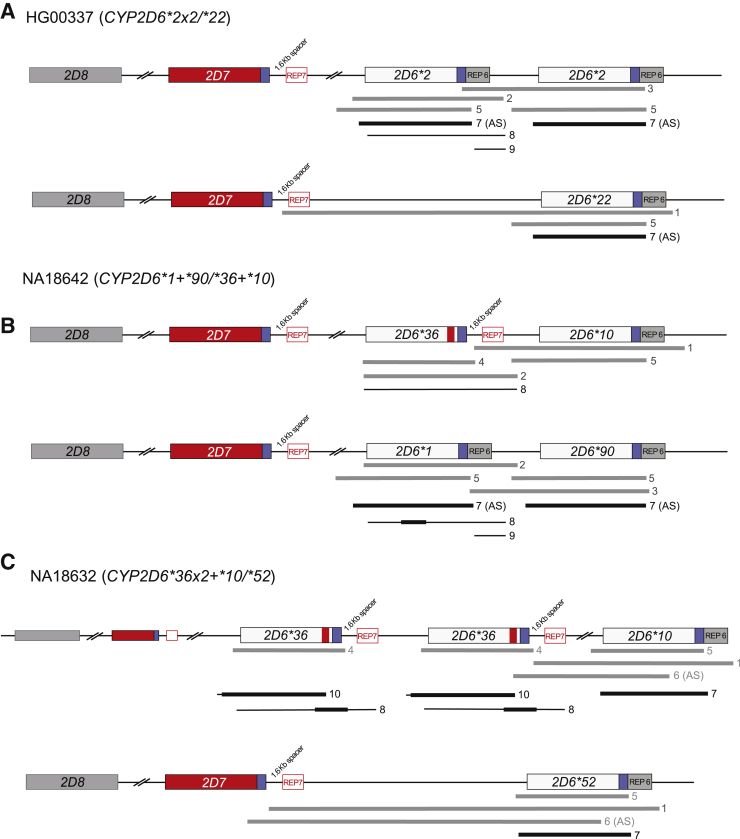
To better understand and describe the complex allele combinations, the Medical College of Wisconsin/RPRD Diagnostics performed multiple XL-PCR reactions on each identified sample to facilitate mapping of the detected variants to their respective haplotypes. For example, sample HG00337 was genotyped as containing a duplication event and variants that suggested the presence of CYP2D6*2 and *22 alleles. To determine which allele was duplicated, each of the three gene copies were amplified separately by XL-PCR (Figure 2A and Table 2). Next-generation sequencing analysis of the XL-PCR amplicons, each containing an entire gene copy, allowed the determination that one chromosome carried two identical copies of CYP2D6*2 and the other chromosome carried the ∗22 gene copy giving rise to a CYP2D6*2×2/*22 genotype. The duplication also was identified to harbor CYP2D6*2 by the Children's Mercy Kansas City group by amplifying and genotyping an XL-PCR product (fragment 7).
Figure 2.
Graphic overview of positional and allele-specific long-range (XL) PCR to generate amplicons to characterize gene copy number variation and resolve complex structures and diplotypes. Gray bars indicate XL-PCR products generated and sequenced by next-generation sequencing (NGS) by the Medical College of Wisconsin/RPRD Diagnostics group, whereas black lines and bars show the XL-PCR products generated by the Children's Mercy Kansas City group and regions sequenced by Sanger, respectively. Some XL-PCR fragments were generated in an allele-specific (AS) manner. Fragments are labeled as listed in Table 2. Blue boxes and boxes labeled REP denote common and repetitive regions downstream of CYP2D6 and CYP2D7, respectively; CYP2D7-derived downstream regions are characterized by the presence of a 1.6-Kb long spacer sequence. A: A sample for which a CYP2D6*2×2/*22 structure was determined. Duplication and AS XL-PCR products showed that a *2 duplication and *22 was shown unequivocally to be the only gene copy on the second chromosome. B: A sample (NA18642) with two different tandem structures on each chromosome. CYP2D6*36+*10 and *1+*90 are the only possible configurations consistent with the XL-PCR products formed and sequences derived from these fragments. The red boxes and blue boxes and the spacer in the CYP2D*36 allele graph indicate that exon 9 and the downstream gene regions are CYP2D7-derived. C: A sample (NA18632) with a rare CYP2D6*36×2+*10 tandem allele, and another rare allele, CYP2D6*52, on the second chromosome. Unequivocal determination of the structure of both alleles required a complex approach of amplifying and sequencing XL-PCR products, some of which were generated in an allele-specific manner.
In addition, the study identified several samples that contain both rare alleles and a CYP2D6/CYP2D7 hybrid. Samples NA18642 and NA18632 were genotyped by the PharmacoScan array and were shown to have variants signaling the presence of CYP2D6*90 and *52, respectively. Copy number analysis indicated the presence of a duplication allele plus a tandem arrangement containing a hybrid for sample NA18642 and a tandem arrangement containing a hybrid for sample NA18632. Again, a series of XL-PCR amplicons were generated from the different gene regions of interest to sequence them separately. The regions amplified for NA18642 and NA18632 are illustrated in Figure 2, B and C, and detailed in Table 2. The XL-PCR fragments generated for NA18642 were consistent with the presence of a tandem on each chromosome, specifically a CYP2D6*10+*36 and a CYP2D6*1+*90 (note that *1 is upstream of *90 in the duplicated position). The study first reporting on CYP2D6*90 did not investigate structural variation,29 and thus it remains unknown whether *90 exclusively occurs in tandem with a CYP2D6*1 or also by itself as a single-gene copy. This structure was confirmed by the Children's Mercy Kansas City group with Sanger sequencing derived from allele-specific XL-PCR products. In addition to quantitative CNV testing, NA18632 required the generation of a series of XL-PCR products and sequencing to determine a CYP2D6*36×2+*10/*52 diplotype. Sequence analysis of the CYP2D6*36 alleles identified a unique region in exon 9, which was used for designing allele-specific primers (Table 2). This enabled us to show unequivocally that the CYP2D6*10 gene copy is located downstream of the two CYP2D6*36 gene copies and that the CYP2D*52 allele is located on the second chromosome (Figure 2C). Of note, this sample may be called CYP2D6*36+*10/*36+*52, *36×2+*52/*10, or *36×2+*10/*52 if only limited testing is performed. The generation of XL-PCR product 1, however, in combination with all other results, ultimately showed that CYP2D6*52 is not in tandem with *10 or *36, but the sole gene copy on this chromosome. The original study describing CYP2D6*52 did not test for duplication events, hence, it is likely the CNV state was unknown.30 Of note, neither NA18642 nor NA18632 are part of a pedigree facilitating the characterization of these complex diplotypes.
Discussion
Because of the large number of sequence variations, CNVs, and gene rearrangements with the CYP2D7 pseudogene, CYP2D6 is one of the most difficult pharmacogenes to characterize, posing particular challenges for clinical testing laboratories. The first GeT-RM pharmacogenetic reference material study published in 201012 characterized 107 samples and targeted only a limited number of CYP2D6 alleles. The second study published in 201613 was performed on a larger set of samples (n = 137) and covered additional alleles, but many were either not detected by the methods used or alleles were not present in the limited sample panel. As a result, many samples were reported as inconclusive and shown as example CYP2D6 *1/*2 (*21) or *1/*17 (*40), indicating the possibility of the presence of a *21 or *40 in respective samples. In addition, CNV analysis was rather limited and left many samples undetermined with regard to copy number or structural variation, such as the identification of CYP2D6*13-like CYP2D7–CYP2D6 hybrid genes. Another limitation was the lack of discrimination on which an allele duplication or multiplication event was located. Those with unresolved CNVs or tentatively carrying tandem structures were shown, for example, as CYP2D6*2/*4×N (gene copy number unknown and not determined which allele is duplicated/multiplicated) or *2/[*10 (*36)] (tentatively having a *36+*10 tandem). Failure to detect such structures may lead to incorrect phenotype classification.3, 15, 16
Although the previous two studies identified 18 CYP2D6 alleles among the reference materials tested,12, 13 the majority remained elusive. Furthermore, as new test panels and platforms are continually being developed, it is important and timely to complement the existing set of reference materials with additional rare and/or difficult-to-analyze alleles and/or diplotypes to ensure that laboratories can perform comprehensive validation studies to test new assays, platforms, or methods. Comprehensively characterized reference materials also are invaluable for quality-control procedures and proficiency testing.
In this investigation, the commercial PharmacoScan and MassARRAY platforms, which also can detect CNVs and CYP2D6–CYP2D7 hybrid gene structures, were used. Testing was complemented with well-established TaqMan assays because this technology is widely used for clinical testing as well as research. CNVs were tested extensively using three different quantitative approaches as well as XL-PCR. In addition, selected samples also were sequenced to fully characterize the haplotypes present in a sample. Taken together, these efforts resulted in complete characterization of the CYP2D6 gene in numerous samples, as well as the detection of CYP2D6 alleles that were not identified using the methods used in the previous GeT-RM studies. The sample set of the PGX2 study was expanded by 42 DNA samples based on data from the participating laboratories, or information from the 1000 Genomes Project, indicating the presence of variants with rsID numbers that suggested the existence of rare CYP2D6 alleles. Through the current study, we were able to identify reference materials for an additional 26 CYP2D6 alleles that were not found during previous GeT-RM projects (Table 5). Thus, a total of 41 alleles now are represented in publicly available characterized genomic DNA references materials, leaving 69 of the CYP2D6 alleles listed in the PharmVar database before December 17, 2018 (https://www.pharmvar.org/gene/CYP2D6) unidentified during any GeT-RM studies (Table 4). The available reference materials also now contain specimens with rare or unusual diplotypes such as CYP2D6*3/*3, *5/*5, *5/*56, or *28/*59 (Tables 3 and 4 and Supplemental Table S2). Samples homozygous for the CYP2D6*5 gene deletion are particularly valuable for assay development to control for unspecific amplification or assay signals from the CYP2D7 or CYP2D8 pseudogenes. Ideally, laboratories developing test panels and offering clinical testing for CYP2D6 should have a large and diverse set of reference materials representing all alleles included in the test. Although not all CYP2D6 alleles currently defined by PharmVar are represented in the three GeT-RM studies, the addition of these publicly available CYP2D6 reference materials greatly improves the availability of materials to ensure the quality of clinical testing. The GeT-RM program will continue to work with the genetics community to generate additional reference materials for testing as the number of allelic CYP2D6 variants continues to grow.
CNVs still are poorly characterized by many commercial tests for a number of reasons. Although quantitative CNV assays are becoming more standard, the majority are not able to discriminate which of the two alleles has a duplication event. For example, a patient genotyped as CYP2D6*2/*4 and a copy number of three often is reported as (*2/*4) dup or 3N, *2/*4 in lieu of a CYP2D6*2×2/*4 or *2/*4×2 assignment. Another challenge is the interpretation of CNV results with different copy number calls across the gene. The presence of the CYP2D7-derived exon 9 conversion, for example, causes the copy number call to be lower compared with other gene regions. A sample homozygous for the CYP2D6*36+*10 tandem allele therefore will yield copy number calls of two for the exon 9 region, but four for all other regions. Samples with a CYP2D6*68+*4 will have a one-copy gain for the CYP2D6-derived regions of *68 (Supplemental Table S2). These and other reference materials can be used by laboratories to validate their assays within the limitations of their technologies, and to improve the ability of the assays to detect copy number variations.
We have carefully characterized the new reference materials and most of the samples from the PGX2 study were retested using a quantitative CNV assay in at least one laboratory. The panel now includes samples for a variety of CNV structures including the commonly observed CYP2D6*1×2, *2×2, and *4×2 duplications and *36+*10 and *68+*4 tandems, but also some rare structures such as *36×2+*10, *13+*2, and *17×2 (Tables 3 and 4 and Supplemental Table S2).
Consistent test results were observed for NA17244 across platforms and CNV assays, but the nature of the structural variation has not been resolved. It appears that this sample has a hybrid gene in addition to CYP2D6*2 and *4 duplications, but it remains unknown on which chromosome it resides. Regardless, this is the only sample among reference materials yielding a five-copy result for any of the target regions (intron 6). Despite our efforts, a number of other samples also remain inconclusive because not all materials from the PGX2 study were reanalyzed systematically. Furthermore, only a fraction of the samples have been confirmed by sequencing; thus, additional subvariants may be identified in this sample cohort in the future.
In an effort to identify materials with alleles not yet represented by the GeT-RM panel, the 1000 Genomes Project database was searched for samples containing key variants identifying the sought-after alleles. Although samples containing CYP2D6*22, *31, *52, *72, *90, *99, *106, *112, and *113 alleles have been identified successfully by sequence analysis, this approach was not successful for identification of other CYP2D6 alleles. This is because no samples were identified that contained the variant of interest, the unique variant identifying an allele did not have an rs number, or the only signature variant also occurs in other haplotypes. In addition, for some alleles (eg, CYP2D6*74), the samples identified by the search probably do not represent the haplotype of interest; that is, per the 1000 Genomes Project data, the samples appeared to carry additional variants inconsistent with the haplotype of interest. Although these samples may represent haplotypes of potential interest (new and known), they were not pursued owing to limited resources. Other samples identified by the 1000 Genomes Project as potentially carrying alleles of interest could not be confirmed by resequencing, and hence were not included in the current reference materials panel with one exception. HG03781 was predicted by the 1000 Genomes Project to have CYP2D6*99 and *102 alleles. Although the CYP2D6*99 allele was confirmed, the second allele was a common CYP2D6*2.001. The 1000 Genomes Project database is a rich data resource; however, these findings corroborate that CYP2D6 haplotype data should be used with caution and confirmed experimentally. Finally, many of these alleles for which no reference sample has been identified are extremely rare and may have been found in populations that may not be represented, or may be underrepresented, in the 1000 Genomes Project.
Reference materials are used by clinical laboratories to design and validate assays, and to ensure accuracy, specificity, and the quality of genetic test results. In addition, providers of proficiency testing often use well-characterized reference material samples in their surveys. Because characterized genomic DNA, either from cell lines or residual clinical samples containing variants in the gene(s), most closely resemble patient samples, they are the preferred type of reference materials.
Clinical genetic testing laboratories in the United States are required by regulation and guided by professional or best practice standards to use reference materials for assay development and validation, quality control, and proficiency testing31, 32, 33, 34 [American College of Medical Genetics, https://www.acmg.net/PDFLibrary/Standards-Guidelines-Clinical-Molecular-Genetics.pdf, last accessed April 30, 2019; Washington State Legislature, http://app.leg.wa.gov/WAC/default.aspx?cite=246-338-090, last accessed March 25, 2019; College of American Pathologists https://www.cap.org, last accessed March 25, 2019 (registration required); and New York State Clinical Laboratory Evaluation Program, http://www.wadsworth.org/clep, last accessed March 25, 2019]. Despite these requirements, there are a limited number of well-characterized quality-control and other reference material samples for many genetic tests, including pharmacogenetic tests. This lack of reference material samples hinders the ability of laboratories to develop and validate assays, perform necessary quality control, and complicates comparison of assays and assay standardization. The lack of available materials also affects not only the ability of proficiency testing programs to provide challenges with a variety of clinically relevant variants, but also prevents provision of proficiency testing challenges for rare variants.
The publicly available and renewable CYP2D6 reference materials developed during this study can be used by laboratories to design and validate assays as well as for quality control and proficiency testing. The public availability of these characterized reference materials can help to inform groups such as the Association for Molecular Pathology PGx Working Group and others who may develop recommendations for alleles that should be included in clinical CYP2D6 tests. These and other reference materials characterized by the GeT-RM program are available from the Coriell Institute for Medical Research. More information about the GeT-RM program is available through the GeT-RM website (https://www.cdc.gov/clia/get-rm/index.html, last accessed August 28, 2019). Finally, to facilitate finding reference samples for particular star alleles, PharmVar will cross-reference Coriell IDs to haplotypes in the future for samples that have been fully sequenced and information submitted to PharmVar.
Acknowledgment
We thank the late Dr. Lorraine Toji, Coriell Institute for Medical Research, for her help obtaining the samples.
Footnotes
Supported by NIH grant R24GM123930 to the Pharmacogene Variation Consortium (A.G.); RPRD (Right Patient Right Drug) Diagnostics, an independent clinical laboratory offering pharmacogenetic testing services, including PharmacoScan (A.T. and P.A.); and Implementing Genomics in Practice (IGNITE) project grant U01 HG007762 (V.M.P.).
Disclosures: ARUP Laboratories, the Indiana University School of Medicine Pharmacogenomics Laboratory, RPRD Diagnostics LLC, and Sema4 are fee-for-service clinical laboratories that offer clinical pharmacogenetic testing. U.B. is the CEO of RPRD Diagnostics and holds equity, and R.E.E. is a paid employee of Agena Bioscience and holds company stock options.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry. Use of tradenames and commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention, the Public Health Service, or the US Department of Health and Human Services.
A guest editor acted as Editor-in-Chief for this manuscript. No person at the Centers for Disease Control and Prevention was involved in the peer-review process or final disposition for this article.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.06.007.
Supplemental Data
References
- 1.Zhou S.F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part I. Clin Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Zhou S.F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet. 2009;48:761–804. doi: 10.2165/11318070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Gaedigk A. Complexities of CYP2D6 gene analysis and interpretation. Int Rev Psychiatry. 2013;25:534–553. doi: 10.3109/09540261.2013.825581. [DOI] [PubMed] [Google Scholar]
- 4.Gaedigk A., Sangkuhl K., Whirl-Carrillo M., Klein T., Leeder J.S. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19:69–76. doi: 10.1038/gim.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caudle K.E., Dunnenberger H.M., Freimuth R.R., Peterson J.F., Burlison J.D., Whirl-Carrillo M., Scott S.A., Rehm H.L., Williams M.S., Klein T.E., Relling M.V., Hoffman J.M. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet Med. 2017;19:215–223. doi: 10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y., Botton M.R., Scott E.R., Scott S.A. Sequencing the CYP2D6 gene: from variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics. 2017;18:673–685. doi: 10.2217/pgs-2017-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nofziger C., Paulmichl M. Accurately genotyping CYP2D6: not for the faint of heart. Pharmacogenomics. 2018;19:999–1002. doi: 10.2217/pgs-2018-0105. [DOI] [PubMed] [Google Scholar]
- 9.Gaedigk A., Ingelman-Sundberg M., Miller N.A., Leeder J.S., Whirl-Carrillo M., Klein T.E., PharmVar Steering Committee The Pharmacogene Variation (PharmVar) Consortium: incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther. 2018;103:399–401. doi: 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaedigk A., Sangkuhl K., Whirl-Carrillo M., Twist G.P., Klein T.E., Miller N.A., PharmVar Steering Committee The evolution of PharmVar. Clin Pharmacol Ther. 2019;105:29–32. doi: 10.1002/cpt.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B., O'Connell C.D., Boone D.J., Amos J.A., Beck J.C., Chan M.M. Developing a sustainable process to provide quality control materials for genetic testing. Genet Med. 2005;7:534–549. doi: 10.1097/01.gim.0000183043.94406.81. [DOI] [PubMed] [Google Scholar]
- 12.Pratt V.M., Zehnbauer B., Wilson J.A., Baak R., Babic N., Bettinotti M., Buller A., Butz K., Campbell M., Civalier C., El-Badry A., Farkas D.H., Lyon E., Mandal S., McKinney J., Muralidharan K., Noll L., Sander T., Shabbeer J., Smith C., Telatar M., Toji L., Vairavan A., Vance C., Weck K.E., Wu A.H., Yeo K.T., Zeller M., Kalman L. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J Mol Diagn. 2010;12:835–846. doi: 10.2353/jmoldx.2010.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratt V.M., Everts R.E., Aggarwal P., Beyer B.N., Broeckel U., Epstein-Baak R., Hujsak P., Kornreich R., Liao J., Lorier R., Scott S.A., Smith C.H., Toji L.H., Turner A., Kalman L.V. Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: a GeT-RM collaborative project. J Mol Diagn. 2016;18:109–123. doi: 10.1016/j.jmoldx.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaedigk A., Ndjountche L., Divakaran K., Dianne Bradford L., Zineh I., Oberlander T.F., Brousseau D.C., McCarver D.G., Johnson J.A., Alander S.W., Wayne Riggs K., Steven Leeder J. Cytochrome P4502D6 (CYP2D6) gene locus heterogeneity: characterization of gene duplication events. Clin Pharmacol Ther. 2007;81:242–251. doi: 10.1038/sj.clpt.6100033. [DOI] [PubMed] [Google Scholar]
- 15.Gaedigk A., Fuhr U., Johnson C., Berard L.A., Bradford D., Leeder J.S. CYP2D7-2D6 hybrid tandems: identification of novel CYP2D6 duplication arrangements and implications for phenotype prediction. Pharmacogenomics. 2010;11:43–53. doi: 10.2217/pgs.09.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaedigk A., Jaime L.K., Bertino J.S., Jr., Berard A., Pratt V.M., Bradfordand L.D., Leeder J.S. Identification of novel CYP2D7-2D6 hybrids: non-functional and functional variants. Front Pharmacol. 2010;1:121. doi: 10.3389/fphar.2010.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaedigk A., Twist G.P., Leeder J.S. CYP2D6, SULT1A1 and UGT2B17 copy number variation: quantitative detection by multiplex PCR. Pharmacogenomics. 2012;13:91–111. doi: 10.2217/pgs.11.135. [DOI] [PubMed] [Google Scholar]
- 18.Gaedigk A., Riffel A.K., Leeder J.S. CYP2D6 haplotype determination using long range allele-specific amplification: resolution of a complex genotype and a discordant genotype involving the CYP2D6*59 allele. J Mol Diagn. 2015;17:740–748. doi: 10.1016/j.jmoldx.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunnenberger H.M., Crews K.R., Hoffman J.M., Caudle K.E., Broeckel U., Howard S.C., Hunkler R.J., Klein T.E., Evans W.E., Relling M.V. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao W., Yang Y., Sebra R., Mendiratta G., Gaedigk A., Desnick R.J., Scott S.A. Long-read single molecule real-time full gene sequencing of cytochrome P450-2D6. Hum Mutat. 2016;37:315–323. doi: 10.1002/humu.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y., Sebra R., Pullman B.S., Qiao W., Peter I., Desnick R.J., Geyer C.R., DeCoteau J.F., Scott S.A. Quantitative and multiplexed DNA methylation analysis using long-read single-molecule real-time bisulfite sequencing (SMRT-BS) BMC Genomics. 2015;16:350. doi: 10.1186/s12864-015-1572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claudio-Campos K.I., Gonzalez-Santiago P., Renta J.Y., Rodriguez J., Carrasquillo K., Gaedigk A., Roche A., Duconge J. CYP2C9*61, a rare missense variant identified in a Puerto Rican patient with low warfarin dose requirements. Pharmacogenomics. 2019;20:3–8. doi: 10.2217/pgs-2018-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer W.E., Walker D.L., O'Kane D.J., Mrazek D.A., Fisher P.K., Dukek B.A., Bruflat J.K., Black J.L. CYP2D6: novel genomic structures and alleles. Pharmacogenet Genomics. 2009;19:813–822. doi: 10.1097/FPC.0b013e3283317b95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaedigk A., Bradford L.D., Alander S.W., Leeder J.S. CYP2D6*36 gene arrangements within the cyp2d6 locus: association of CYP2D6*36 with poor metabolizer status. Drug Metab Dispos. 2006;34:563–569. doi: 10.1124/dmd.105.008292. [DOI] [PubMed] [Google Scholar]
- 27.Motoi Y., Watanabe K., Honma H., Tadano Y., Hashimoto H., Kubota T. Digital PCR for determination of cytochrome P450 2D6 and sulfotransferase 1A1 gene copy number variations. Drug Discov Ther. 2017;11:336–341. doi: 10.5582/ddt.2017.01057. [DOI] [PubMed] [Google Scholar]
- 28.Toscano C., Raimundo S., Klein K., Eichelbaum M., Schwab M., Zanger U.M. A silent mutation (2939G>A, exon 6; CYP2D6*59) leading to impaired expression and function of CYP2D6. Pharmacogenet Genomics. 2006;16:767–770. doi: 10.1097/01.fpc.0000236331.03681.24. [DOI] [PubMed] [Google Scholar]
- 29.Qian J.C., Xu X.M., Hu G.X., Dai D.P., Xu R.A., Hu L.M., Li F.H., Zhang X.H., Yang J.F., Cai J.P. Genetic variations of human CYP2D6 in the Chinese Han population. Pharmacogenomics. 2013;14:1731–1743. doi: 10.2217/pgs.13.160. [DOI] [PubMed] [Google Scholar]
- 30.Lee S.J., Lee S.S., Jung H.J., Kim H.S., Park S.J., Yeo C.W., Shin J.G. Discovery of novel functional variants and extensive evaluation of CYP2D6 genetic polymorphisms in Koreans. Drug Metab Dispos. 2009;37:1464–1470. doi: 10.1124/dmd.108.022368. [DOI] [PubMed] [Google Scholar]
- 31.Chen B., Gagnon M., Shahangian S., Anderson N.L., Howerton D.A., Boone J.D., Centers for Disease Control and Prevention (CDC) Good laboratory practices for molecular genetic testing for heritable diseases and conditions. MMWR Recomm Rep. 2009;58:1–37. [PubMed] [Google Scholar]
- 32.Association for Molecular Pathology statement. Recommendations for in-house development and operation of molecular diagnostic tests. Am J Clin Pathol. 1999;111:449–463. doi: 10.1093/ajcp/111.4.449. [DOI] [PubMed] [Google Scholar]
- 33.International Organization for Standardization . International Organization for Standardization; Geneva: 2012. ISO 15189 Medical laboratories: requirements for quality and competence. [Google Scholar]
- 34.Electronic Code of Federal Regulations: The Clinical Laboratory Improvement Amendments (CLIA). Title 42, Chapter IV, Subchapter G, Part 493.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.