Abstract
Urine cell-free DNA (cfDNA) is a valuable noninvasive biomarker for cancer mutation detection, infectious disease diagnosis (eg, tuberculosis), organ transplantation monitoring, and prenatal screening. Conventional silica DNA extraction does not efficiently capture urine cfDNA, which is dilute (ng/mL) and highly fragmented [30 to 100 nucleotides (nt)]. The clinical sensitivity of urine cfDNA detection increases with decreasing target length, motivating use of sample preparation methods designed for short fragments. We compared the analytical performance of two published protocols (Wizard resin/guanidinium thiocyanate and Q Sepharose), three commercial kits (Norgen, QIAamp, and MagMAX), and an in-house sequence-specific hybridization capture technique. Dependence on fragment length (25 to 150 nt), performance at low concentrations (10 copies/mL), tolerance to variable urine conditions, and susceptibility to PCR inhibition were characterized. Hybridization capture and Q Sepharose performed best overall (60% to 90% recovery), although Q Sepharose had reduced recovery (<10%) of the shortest 25-nt fragment. Wizard resin/guanidinium thiocyanate recovery was dependent on pH and background DNA concentration and was limited to <35%, even under optimal conditions. The Norgen kit led to consistent PCR inhibition but had high recovery of short fragments. The QIAamp and MagMAX kits had minimal recovery of fragments <150 and <80 nt, respectively. Urine cfDNA extraction methods differ widely in ability to capture short, dilute cfDNA in urine; using suboptimal methods may profoundly impair clinical results.
CME Accreditation Statement: This activity (“JMD 2019 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the accreditation requirements and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2019 CME Program in Molecular Diagnostics”) for a maximum of 18.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Urine cell-free DNA (cfDNA) is an emerging noninvasive biomarker for cancer mutation detection,1, 2, 3, 4 infectious disease diagnosis,5, 6, 7 organ transplantation monitoring,8, 9 and prenatal screening.1, 10, 11 As cells die throughout the body, cfDNA is released into the bloodstream. A fraction of circulating cfDNA, composed largely of short fragments, crosses the kidney barrier, is excreted in urine, and can be analyzed by PCR or sequencing.1 This subset of urine cfDNA, derived from circulating cfDNA, is known as transrenal DNA, but cfDNA can also be generated directly in urine from cells shed along the urinary tract.
To maximize the clinical sensitivity and reproducibility of urine cfDNA analysis, extraction methods capable of efficiently capturing short, dilute DNA fragments are essential. Although plasma cfDNA is primarily nucleosomal, with a peak length of 160 to 167 nucleotides (nt),12, 13, 14 urine cfDNA is more fragmented. The upper length limit of the transrenal fraction of urine cfDNA is defined by glomerular filtration, and all urine cfDNA fragments are quickly degraded further in urine.15 Determining the true length distribution of urine cfDNA is challenging because both extraction and library preparation methods may underestimate the presence of shorter fragments, but most fragments are expected to be <100 nt.11, 14, 16 Peak fragment length varies across patients, but may be as low as 30 to 60 nt.11, 14, 16
Because of the extensive fragmentation of urine cfDNA, the diagnostic clinical sensitivity of urine cfDNA detection increases with decreasing target length. Maximizing sensitivity by targeting shorter fragments is especially critical because urine cfDNA is also dilute, with total concentrations ranging from <1 to 200 ng/mL1, 17, 18 and copy numbers of specific targets much lower. In a study detecting fetal cfDNA in maternal urine, decreasing PCR amplicon length from 65 to 39 nt increased clinical sensitivity from 25% to 75%. A further decrease to 25 nt was required before achieving 100% detection.10 This effect may be even more pronounced for bacterial, viral, and mitochondrial cfDNA, which are not protected by histones and are, therefore, more degraded than human genomic cfDNA.12, 15, 16 For tuberculosis urine cfDNA, a modest 10-nt decrease in amplicon length (49 to 39 nt) led to 5- to 10-fold improvement in detected concentration.19 Critically, the ability to target shorter cfDNA fragments lies not only in decreasing amplicon length, but also in design and selection of sample preparation methods capable of capturing and concentrating the short, dilute fragments that constitute the bulk of urine cfDNA.
Unfortunately, conventional extraction methods for cell-associated DNA or even plasma cfDNA are not suitable for urine cfDNA because they are not designed for short fragments. The Boom method, commonly used for both research and clinical work, adsorbs DNA to silica under chaotropic conditions.20 The key driving forces of silica adsorption are hydrophobic interactions due to dehydration of silica and DNA surfaces and hydrogen bonding between silica and the DNA backbone, both of which depend on DNA length.21 Consequently, silica adsorption is less effective at purifying short fragments, with recovery generally decreasing below 50 to 100 nt. Silica adsorption also requires relatively high DNA concentrations for optimal performance because a fraction of DNA may remain irretrievably bound to the silica surface.22, 23 This loss is trivial in most samples, but for low-concentration samples, like urine cfDNA, it may make up a significant portion of the input.
With these limitations in mind, an ideal urine cfDNA extraction method would enable high recovery of short DNA from dilute solutions. Despite the great clinical promise of urine cfDNA as an easy-to-access sample, there has been little quantitative comparison of approaches taken to improve recovery of urine cfDNA. A recent review emphasized the lack of standardization in sample preparation methods, including DNA extraction, as a key limitation in the development of urine cfDNA assays.24 Previous studies have compared clinical detection rates10, 19 and total cfDNA recovery18, 25 of a limited set of extraction methods, but no studies have investigated analytical performance using spiked samples.
Herein, two published urine cfDNA extraction protocols [Wizard resin/guanidinium thiocyanate (Wizard/GuSCN) and Q Sepharose], three commercial kits (Norgen, QIAamp, and MagMAX), and a sequence-specific hybridization capture technique, developed in our laboratory, were analytically compared. The Wizard/GuSCN method uses high concentrations (>3 mol/L) of chaotropic GuSCN to adsorb DNA to Wizard silica resin. This approach was used to originally demonstrate the presence of cfDNA in urine1 and has since been widely applied, most frequently for detecting tuberculosis26 and fetal11 cfDNA. The Q Sepharose method uses a quaternary ammonium anion exchange resin to preconcentrate DNA before desalting on a silica spin column. It improves recovery of short urine cfDNA fragments compared with Wizard/GuSCN10 and has often been used to detect tumor cfDNA mutations for cancer diagnosis, monitoring, and prognosis.2, 3 The Norgen Biotek (Thorold, ON, Canada) Urine Cell-Free Circulating DNA Purification Kit uses a hybrid silica/silicon carbide spin column, where addition of silicon carbide reportedly improves yield of short DNA compared with silica alone (US patent 9,422,596). The Qiagen (Hilden, Germany) QIAamp Circulating Nucleic Acid Kit uses a silica vacuum column and reportedly improves recovery of fragmented DNA compared with other Qiagen kits. It is one of the most widely used commercial kits for plasma cfDNA extraction27 but is not commonly used for urine cfDNA. The Thermo Fisher Scientific (Waltham, MA) MagMAX Cell-Free DNA Isolation Kit uses Dynabeads MyOne Silane to maximize binding kinetics and capacity but is intended primarily for plasma cfDNA. It was included as a reference method to represent best-case silica adsorption without modifications specifically for urine cfDNA, although it has been used previously in urine.28
To enable high-efficiency purification of short fragments, our laboratory has developed a hybridization capture method for urine cfDNA using a biotinylated sequence-specific probe and streptavidin-coated magnetic beads. Hybridization is commonly used for targeted enrichment of sequencing libraries but has been less frequently used as a sample preparation method for capturing target sequences directly from raw samples. Hybridization capture with magnetic beads has been used previously to enrich pathogen DNA and mRNA directly from sputum,29, 30 blood,31 feces,31, 32 vaginal/anal swabs,33 and cell lysates,34, 35 with detection down to 5 to 10 copies/mL30 and recovery up to 60% to 80%.36 Hybridization has also been used in microfluidic37 and lateral flow38 formats. In previous implementations, hybridization capture was used primarily to remove excess nontarget DNA, which can inhibit amplification.30, 38 In the case of urine cfDNA, hybridization's ability to sensitively capture short fragments, regardless of length and concentration, was instead leveraged. To our knowledge, hybridization capture has not been used previously to target urine cfDNA.
For each extraction method, the dependence on DNA fragment length, performance at low DNA concentrations, tolerance to variable urine conditions, and susceptibility to PCR inhibition were characterized. The results of this work will help guide selection and optimization of DNA extraction methods for urine cfDNA analysis. Careful design of sample preparation methods should lead to increased clinical sensitivity and reproducibility of urine cfDNA diagnostics.
Materials and Methods
Synthetic DNA Target Design
To study the analytical performance of the urine cfDNA extraction methods, synthetic single-stranded DNA (ssDNA) targets were spiked into pooled urine before extraction and analysis by real-time quantitative PCR (qPCR). The targets were selected from a conserved and specific region of the insertion sequence IS6110 of the Mycobacterium tuberculosis complex (GenBank, https://www.ncbi.nlm.nih.gov/genbank; accession number X17348).39 The targets were 25, 40, 80, and 150 nt in length, as listed in Table 1. The 40-, 80-, and 150-nt targets were designed to be amplified by a shared primer set, with additional bases outside of the primer amplification region added to the 3′ end of the 40-nt target to generate the 80- and 150-nt targets. The 25-nt target was designed to be amplified by a separate set of primers in a two-stage, single-tube PCR for ultrashort targets.10
Table 1.
Target, Primer, and Probe Sequences
| Assay | Oligonucleotide | Sequence |
|---|---|---|
| PCR of 40-, 80-, and 150-nt targets | Forward primer | 5′-CGAACCCTGCCCAGGTCGA-3′ |
| Reverse primer | 5′-GTAGCAGACCTCACCTATGTGT-3′ | |
| 40-nt Target | 5′-CGAACCCTGCCCAGGTCGACACATAGGTGAGGTCTGCTAC-3′ | |
| 80-nt Target | 5′-CGAACCCTGCCCAGGTCGACACATAGGTGAGGTCTGCTACACACCATTCAATTTCATCACTGCCAATACTCCACTCTCAT-3′ | |
| 150-nt Target | 5′-CGAACCCTGCCCAGGTCGACACATAGGTGAGGTCTGCTACACACCATTCAATTTCATCACTGCCAATACTCCACTCTCATCTACACAACCCATTAGTACCTTACCTCGCTTCCTATCCCAATTCACTTAATCTTAAACCGGTCAGGGAAG-3′ | |
| PCR of 25-nt target | 25-nt Target | 5′-CCGGCTGTGGGTAGCAGACCTCACC-3′ |
| First-stage hairpin forward primer | 5′-GCGTAAGAAT/iMe-isodC/AAACGTCGCTCAACTTCCATTCTTACGCCCGGCTGTGG-3′ | |
| Second-stage universal forward primer | 5′-AACGTCGCTCAACTTCCATT-3′ | |
| Reverse primer | 5′-TTAGAGAAGGTGAGGTCTGC-3′ | |
| MGB TaqMan probe | 5′-6FAM/CCGGCTGTGGGTA/MGBNFQ-3′ | |
| Hybridization capture | Biotinylated capture probe for 40-, 80-, and 150-nt targets | 5′-/5BiosG/AGACCTCACCTATGTGTC/3SpC3/-3′ |
| Biotinylated capture probe for 25-nt target | 5′-/5BiotinTEG/GAGGTCTGCTACCCA/3SpC3/-3′ |
Single-stranded synthetic oligonucleotides were used as spike-in targets to study the analytical performance of urine cfDNA extraction methods. The 40-, 80-, and 150-nt targets were designed to be amplified by the same primer set, with the shared primer amplification region boldfaced. The 25-nt target was designed to be amplified by a separate set of primers in a two-stage, single-tube PCR for ultrashort targets. Binding regions for the biotinylated capture probes are underlined. All oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA), except for the MGB TaqMan probe, which was from Thermo Fisher Scientific (Waltham, MA).
DNA Extraction from Pooled Human Urine
Urine from five healthy volunteers was pooled into a representative sample, supplemented with 10 mmol/L EDTA, and stored at −80°C until analysis.
Wizard Resin/Guanidinium Thiocyanate
Urine (5 mL) was mixed with 7.5 mL 6 mol/L GuSCN and 1 mL Wizard Minipreps DNA Purification Resin (Promega, Madison, WI), rotated at room temperature for 2 hours, and vacuum filtered through a syringe fitted with a Wizard minicolumn. The resin was washed twice with 5 mL wash buffer (80 mmol/L KOAc, 8.3 mmol/L Tris-HCl, pH 7.5, 40 μmol/L EDTA, and 55% ethanol). The minicolumn was removed and dried (10,000 × g, 2 minutes). DNA was eluted with 100 μL 60°C nuclease-free water (1-minute incubation, 1 minute at 16,000 × g).
Q Sepharose Anion Exchange Resin
Urine (10 mL) was mixed with 300 μL Q Sepharose Fast Flow (GE Healthcare, Waukesha, WI) and rotated at room temperature for 30 minutes. The resin was pelleted (1800 × g, 5 minutes), resuspended in 1 mL low-salt buffer (0.3 mol/L LiCl and 10 mmol/L NaOAc, pH 5.5), transferred to a Mini Bio-Spin Column (Bio-Rad Laboratories, Hercules, CA), and filtered (800 × g, 1 minute). The resin was washed with 4 × 0.5 mL low-salt buffer (800 × g, 30 seconds). DNA was eluted (800 × g, 3 minutes) using 670 μL high-salt buffer (2 mol/L LiCl and 10 mmol/L NaOAc, pH 5.5). The eluate was mixed with 2 mL 95% ethanol and applied incrementally to a QIAquick column (Qiagen; 800 × g, 30 seconds). The column was washed twice with 0.5 mL 2 mol/L LiCl in 70% ethanol and twice with 0.5 mL 75 mmol/L KOAc, pH 5.5, in 80% ethanol (800 × g, 30 seconds). The column was dried (20,000 × g, 3 minutes) and DNA was eluted (20,000 × g, 2 minutes) in 106 μL elution buffer (Qiagen).
Norgen Urine Cell-Free Circulating DNA Purification Mini Kit
DNA was extracted from 2 mL urine using the manufacturer's protocol and eluted into 50 μL.
Qiagen QIAamp Circulating Nucleic Acid Kit
DNA was extracted from 4 mL urine using the manufacturer's protocol for purification of circulating nucleic acids from urine and eluted into 50 μL. The QIAamp experiments were performed later than those for other methods, so a different urine sample was used.
Thermo Fisher Scientific MagMAX Cell-Free DNA Isolation Kit
DNA was extracted from 1 mL urine using the manufacturer's protocol for manual isolation of cfDNA from urine and eluted into 20 μL.
Hybridization Capture
Urine (1 mL) was mixed with 15 nmol/L biotinylated capture probe (Table 1), 1 mol/L NaCl, and 10 mmol/L Tris-HCl, pH 7.5; denatured (95°C, 10 minutes); and hybridized (45°C, 15 minutes). Hybridized complexes were immobilized on 83.2 μL Dynabeads MyOne Streptavidin C1 (Thermo Fisher Scientific) by 15-minute rotation at room temperature. Beads were washed twice with 1 mL high-salt wash (1 mol/L NaCl and 10 mmol/L Tris-HCl, pH 7.5) and once with 1 mL low-salt wash (15 mmol/L NaCl and 10 mmol/L Tris-HCl, pH 7.5). DNA was eluted (20 μL 20 mmol/L NaOH) and partially neutralized (3.5 μL 100 mmol/L HCl).
Real-Time Quantitative PCR
qPCR of the 40-, 80-, and 150-nt targets was performed in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) with an initial incubation of 94°C for 5 minutes, followed by 45 amplification cycles (94°C for 30 seconds, 58°C for 30 seconds, and 68°C for 1 minute). Each reaction contained 1.25 U OneTaq Hot Start DNA Polymerase [New England Biolabs (NEB), Ipswich, MA], 1× OneTaq GC Reaction Buffer [NEB; 80 mmol/L Tris-SO4, 20 mmol/L (NH4)2SO4, 2 mmol/L MgSO4, 5% glycerol, 5% dimethyl sulfoxide, 0.06% IGEPAL CA-630, and 0.05% Tween 20, pH 9.2], 0.8 mmol/L dNTPs (NEB), 0.4× EvaGreen (Biotium, Fremont, CA), 200 nmol/L forward primer, and 200 nmol/L reverse primer (Table 1). Quantification cycle values were determined using the CFX Manager software version 3.1 (Bio-Rad Laboratories) at a threshold of 500 relative fluorescence units (RFUs), and recovered copies were calculated by a standard curve. Validation of the 40-, 80-, and 150-nt PCR is given in Supplemental Figure S1.
Ultrashort qPCR of the 25-nt target was performed in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) with an initial denaturation phase (94°C for 5 minutes), 10 preamplification cycles to extend the first-stage loop primer (94°C for 30 seconds and 45°C for 1 minute), and 40 amplification cycles (94°C for 30 seconds and 59°C for 1 minute). Each reaction contained 1.25 U Hot Start Taq DNA Polymerase (NEB) and 1× Standard Taq Buffer (NEB; 10 mmol/L Tris-HCl, 50 mmol/L KCl, and 1.5 mmol/L MgCl2, pH 8.3) supplemented with an additional 0.5 mmol/L MgCl2 and 70 mmol/L Tris-HCl, 0.8 mmol/L dNTPs (NEB), 50 nmol/L first-stage hairpin forward primer, 700 nmol/L second-stage universal forward primer, 700 nmol/L reverse primer, and 100 nmol/L MGB TaqMan probe (Table 1). Quantification cycle values were determined using the CFX Manager Software version 3.1 at a threshold of 100 RFUs, and recovered copies were calculated by a standard curve. Validation of the 25-nt PCR is given in Supplemental Figure S2.
qPCR of experimental samples was performed in triplicate in a 50 μL volume containing 5 μL of DNA output, except for hybridization capture experiments, where the entire output (approximately 23 μL) was analyzed in a single PCR well. To control for contamination, no template controls were run not only for PCR (n = 3) but also through the entire DNA extraction procedure for all experiments (n ≥ 3).
Results
Table 2 summarizes the urine cfDNA extraction methods, including processing time, cost, and volume of urine analyzed.
Table 2.
Overview of Urine cfDNA Extraction Methods
| Urine cfDNA extraction method | Key purification chemistry | Rationale for selection | Processing time (hands-on time), hours∗ | Cost per sample, $† | Effective urine volume analyzed per PCR well, μL |
|---|---|---|---|---|---|
| Hybridization capture | Hybridization to biotinylated probe and capture on streptavidin magnetic beads | Developed in our laboratory specifically for urine cfDNA | 1.75 (1) | 15‡ | 1000§ |
| Wizard/GuSCN | Adsorption to silica resin in presence of high-concentration chaotrope (3–6 mol/L) | Originally used to isolate cfDNA from urine; widely used in the literature | 3 (2.5) | 5 | 472 |
| Q Sepharose | Preconcentration by anion exchange resin, followed by adsorption to silica spin column | Shown to improve recovery of short fragments compared with Wizard/GuSCN method | 3 (1) | 5 | 250 |
| Norgen Urine Cell-Free Circulating DNA Purification Kit | Adsorption to silica/silicon carbide hybrid spin column in presence of chaotrope | Commercial kit designed specifically for urine cfDNA | 1.5 (1.25) | 5 | 200 |
| Qiagen QIAamp Circulating Nucleic Acid Kit | Adsorption to silica vacuum column in presence of chaotrope | Commercial kit commonly used for plasma cfDNA | 2 (1.25) | 25 | 400 |
| Thermo Fisher Scientific MagMAX Cell-Free DNA Isolation Kit | Adsorption to Dynabeads MyOne Silane in presence of chaotrope | Representative commercial silica kit; best-case scenario without specific designs for urine cfDNA | 2.5 (2) | 18 | 250 |
Wizard/GuSCN, Wizard resin/guanidinium thiocyanate.
For 12 samples; sample preparation time only, not including qPCR.
Sample preparation cost only, not including qPCR.
Cost listed for capture of a single target. Cost is due almost exclusively to the magnetic beads and is, thus, not expected to scale up significantly for multiplexed capture (estimated $0.10 to $0.15 per capture probe per sample).
All real-time quantitative PCRs were performed using 5 μL of sample per well, except for hybridization, where the entire approximately 23 μL output was analyzed in a single PCR well.
Effect of DNA Fragment Length on Recovery
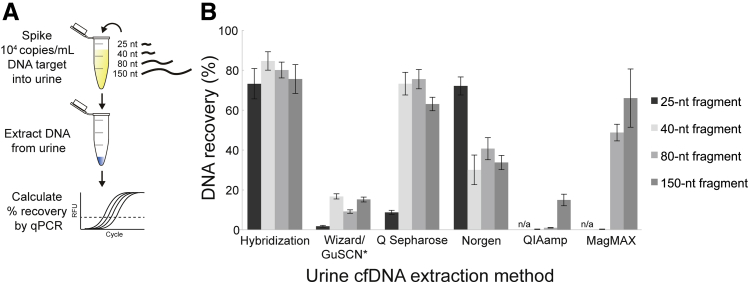
To evaluate the dependence of urine cfDNA extraction methods on fragment length, DNA was extracted from urine spiked with 104 copies/mL of synthetic DNA target of length 25, 40, 80, or 150 nt (Figure 1A). Figure 1B shows the percentage recovery of each extraction method across fragment lengths. Hybridization capture was the only method that maintained high recovery (73% to 84%) across all fragment lengths from 25 to 150 nt. Q Sepharose had similar, high recovery (63% to 75%) of 40- to 150-nt fragments, but reduced recovery (9%) of the shortest 25-nt fragment. Wizard/GuSCN recovery was initially low (<5%) across all fragments. Later experiments showed that Wizard/GuSCN was dependent on urine composition, particularly pH and background DNA. Even after adjusting urine to optimal conditions (pH 6; 1000 ng/mL sheared salmon sperm DNA; Thermo Fisher Scientific), Wizard/GuSCN recovery was still low (9% to 17%) across 40- to 150-nt fragments and further reduced (2%) for the 25-nt fragment. The Norgen kit had moderate recovery (30% to 41%) across 40- to 150-nt fragments and improved recovery (72%) of the 25-nt fragment. Recovery using the QIAamp kit was limited (18%) for the longest 150-nt fragment and was low (1% and 0.2%) for the shorter 80- and 40-nt fragments, respectively. The MagMAX kit recovery was high (66%) for the 150-nt fragment, but quickly diminished with decreasing fragment length and was practically nonexistent (0.2%) for the 40-nt fragment. No template controls were run through the entire DNA extraction procedure for each method (Supplemental Table S1).
Figure 1.
DNA recovery from urine is dependent on both extraction method and target fragment length. A: To evaluate the length dependence of urine cfDNA extraction methods, synthetic targets of various lengths (25, 40, 80, and 150 nt) were spiked into urine at 104 copies/mL before extraction. The dashed line indicates an example relative fluorescence unit (RFU) threshold for determination of the PCR quantification cycle. B: Percentage recoveries are given. Results of extraction no template controls are given in Supplemental Table S1. *Wizard resin/guanidinium thiocyanate (Wizard/GuSCN) samples were adjusted to pH 6 and spiked with 1000 ng/mL background DNA. Data are expressed as means ± SD (B). n = 3 (B). n/a, indicates 25-nt fragment not tested for QIAamp and MagMAX; qPCR, real-time quantitative PCR.
Ability to Detect Low Concentrations of Short DNA Fragments
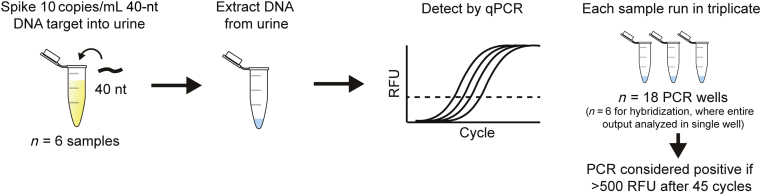
To determine each method's potential for sensitive capture of short, dilute urine cfDNA, 10 copies/mL of 40-nt target were spiked into urine before extraction (Figure 2). Hybridization capture and Q Sepharose reliably yielded detectable DNA from all low concentration spiked samples (Table 3). The Norgen kit also detected all samples, but only weakly. Wizard/GuSCN weakly detected 83% of samples but was inconsistent, with only 44% positive PCR wells. The QIAamp and MagMAX kits did not allow confident detection of any positive samples. Full results of the low-concentration extraction experiments, including no template controls, are given in Supplemental Table S2.
Figure 2.
Design of experiment to test extraction methods' abilities to detect low concentrations of short DNA fragments in urine. Ten copies per milliliter of 40-nt target were spiked into urine, extracted, and detected by real-time quantitative PCR. PCR was performed in triplicate, except for hybridization, where the entire output was analyzed in a single PCR well. PCR was considered positive if >500 relative fluorescence units (RFUs) after 45 amplification cycles. The dashed line indicates an example RFU threshold for determination of the PCR quantification cycle. n = 6.
Table 3.
Ability to Detect Low Concentrations of Short DNA
| Urine cfDNA extraction method | Samples with ≥1 positive PCR well, % (number/total) | Positive PCR wells, % (number/total) | Detected copies/well, mean ± SD | Expected copies/well if theoretical 100% recovery∗ |
|---|---|---|---|---|
| Hybridization capture | 100 (6/6) | 100 (6/6) | 7.8 ± 5.1 | 10 |
| Wizard/GuSCN | 83 (5/6) | 44 (8/18) | 1.6 ± 0.87 | 2.5 |
| Q Sepharose | 100 (6/6) | 89 (16/18) | 5.0 ± 1.5 | 4.7 |
| Norgen Urine Cell-Free Circulating DNA Purification Kit | 100 (6/6) | 89 (16/18) | 1.42 ± 0.67 | 2 |
| Qiagen QIAamp Circulating Nucleic Acid Kit | 17 (1/6) | 6 (1/18) | 0.05 ± 0.12 | 4 |
| Thermo Fisher Scientific MagMAX Cell-Free DNA Isolation Kit | 33 (2/6) | 11 (2/18) | 0.77 ± 0.31 | 2.5 |
Hybridization, Q Sepharose, and Norgen methods can detect low concentrations of short cfDNA fragments; Wizard/GuSCN, QIAamp, and MagMAX methods are not expected to perform well under these conditions. The ability of each urine cfDNA extraction method to detect low-concentration samples was tested using the experiment design shown in Figure 2. Full results, including no template controls (n = 6), are given in Supplemental Table S2.
Wizard/GuSCN, Wizard resin/guanidinium thiocyanate.
Calculated on the basis of the initial 10 copies/mL target concentration and adjusted for the urine input and elution volume of each method.
Tolerance to Varied Urine Conditions
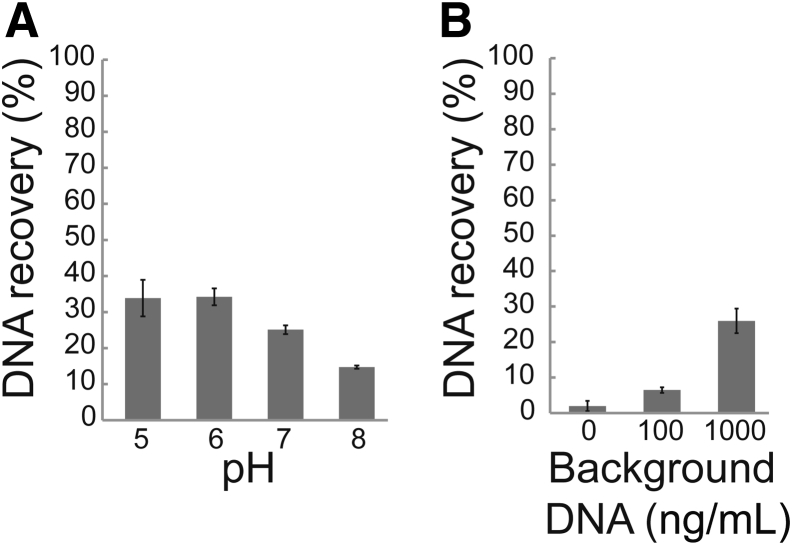
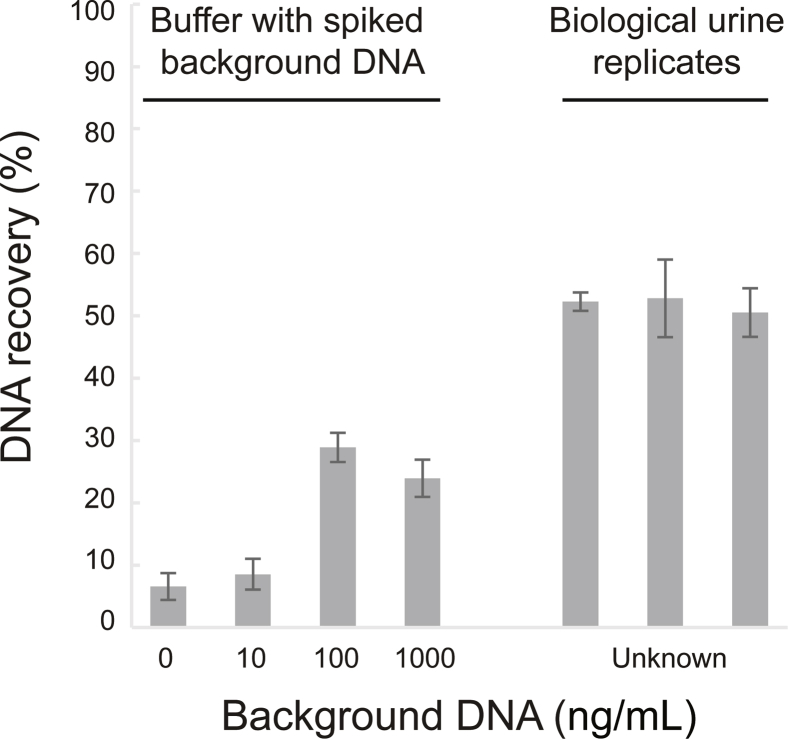
To test the methods' tolerance to varied conditions expected in urine, 104 copies/mL of 150-nt target were extracted from buffer (phosphate-buffered saline or tris-buffered saline) with a range of pH (pH 5, 6, 7, and 8), background DNA (0, 100, and 1000 ng/mL sheared salmon sperm DNA; Thermo Fisher Scientific), and salt (13.7, 137, and 500 mmol/L NaCl) conditions. The Wizard/GuSCN method was highly dependent on urine composition, specifically pH and background DNA. Recovery decreased as pH increased above pH 6 (Figure 3A). Spiking in background DNA (1 μg/mL) improved recovery, but maximum recovery was still well below that of the other methods (Figure 3B). Variation in salt from 13.7 to 500 mmol/L NaCl had no effect on recovery (Supplemental Table S3). The remaining methods were all relatively tolerant to variations in pH, background DNA, and salt (Supplemental Table S3). Recovery by the Q Sepharose method was moderately reduced with especially low background DNA (≤10 ng/mL) but was consistent across biological urine replicates (Supplemental Figure S3).
Figure 3.
The Wizard resin/guanidinium thiocyanate (Wizard/GuSCN) method is dependent on urine pH and background DNA concentration. A: Recovery decreases with increasing pH. Before extraction by the Wizard/GuSCN method, 104 copies/mL of 150-nt target were spiked into phosphate-buffered saline (PBS) with 1000 ng/mL background DNA. B: Recovery increases with the addition of background DNA. Before extraction by the Wizard/GuSCN method, 104 copies/mL of 150-nt target were spiked into PBS, pH 6. Data are expressed as means ± SD (A and B). n = 3 (A and B).
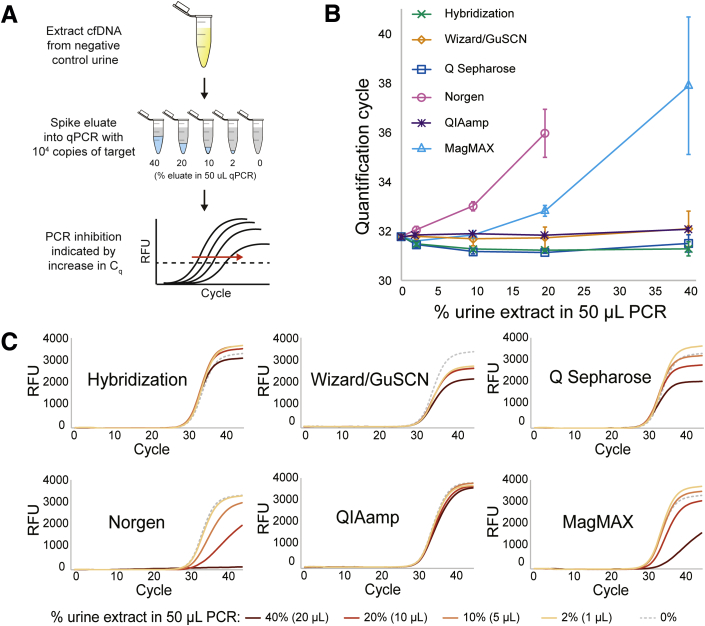
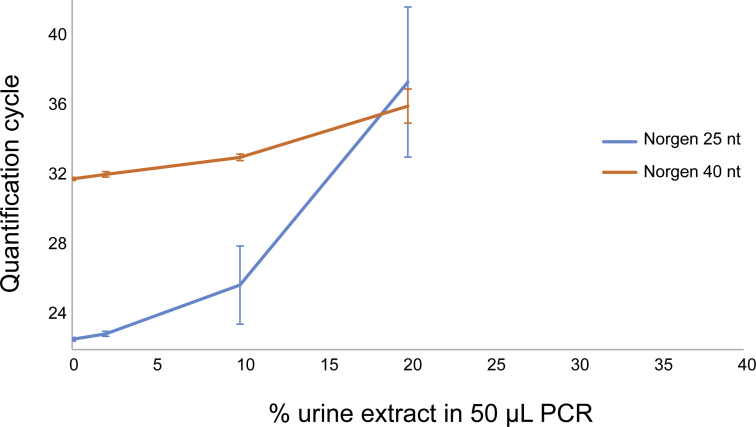
Susceptibility to PCR Inhibition
To test for PCR inhibition resulting from each method, cfDNA was extracted from negative control urine, without added target, and the resulting eluate (0, 1, 5, 10, or 20 μL) was spiked into 50 μL PCR containing 1000 copies of 40-nt target (Figure 4A). PCR inhibition was indicated by an increase in quantification cycle. Hybridization capture, Wizard/GuSCN, Q Sepharose, and QIAamp were resistant to inhibition for up to 40% eluate (Figure 4B). Although not accompanied by an increase in quantification cycle, increasing the fraction of Wizard/GuSCN and Q Sepharose eluate reduced plateau RFU (Figure 4C). MagMAX led to slight inhibition at 20% eluate and severe inhibition at 40% eluate. Norgen led to inhibition at all conditions tested and no amplification at 40% eluate.
Figure 4.
Urine cfDNA extraction methods have varying susceptibility to cause PCR inhibition. A: Eluate extracted from negative control urine (no added target) was spiked into PCR containing a constant target concentration (0-, 1-, 5-, 10-, or 20-μL eluate in 50 μL PCR, for final 0%, 2%, 10%, 20%, or 40% eluate, respectively). The dashed line indicates an example relative fluorescence unit (RFU) threshold for determination of the PCR quantification cycle. B: An increase in quantification cycle (Cq) indicates PCR inhibition. C: Representative PCR curves are shown. Data are expressed as means ± SD (B). n = 3 (B). Wizard/GuSCN, Wizard resin/guanidinium thiocyanate.
Discussion
The highly fragmented and dilute nature of urine cfDNA (≤30 to 100 nt, <1 to 200 ng/mL)1, 11, 14, 16, 17, 18 motivates the use of sample preparation methods capable of recovering short DNA with high efficiency. Our goal was to generate a representative data set to aid in selection and optimization of extraction methods to ensure high-quality results from urine cfDNA studies. The analytical dependence of six methods on a key set of variables was characterized to gain insight into how the methods may perform in clinical samples and to identify any critical pitfalls. In the subsections below, the strengths and limitations of each method are discussed, and the final conclusions regarding choice of urine cfDNA extraction method are stated. Apart from hybridization capture, which was developed in house, published protocols were followed as closely as possible. Further optimizations, including adjusting urine pH, spiking in background DNA, improving elution efficiency, and tailoring sample, elution, and PCR volumes, may improve outcomes.
Wizard Resin/Guanidinium Thiocyanate
Despite its use in the first study isolating urine cfDNA,1 the Wizard/GuSCN method has low and variable recovery. After observing low recovery (<5%) and significant variation across urine samples in preliminary work, the recovery was found to be highly dependent on pH and background DNA concentration, both of which fluctuate widely across clinical urine samples. Urine pH ranges from 5 to 8 (mean, 5.99 to 6.43),40 but Wizard/GuSCN had reduced recovery as pH increased above 6. As silanol groups become deprotonated at higher pH, increased electrostatic repulsion between DNA and silica diminishes adsorption.21, 22, 23 Recovery by Wizard/GuSCN also improved as background DNA increased up to 1 μg/mL. Concentrations >1 μg/mL, already well above the expected biological range of <1 to 200 ng/mL, were not tested.1, 17, 18 Again, this limitation is not surprising for silica. Supplementation with carrier nucleic acids improves silica extraction yields, particularly for dilute samples, and is often implemented in commercial purification kits.41 Limited recovery of dilute DNA may be due to ineffective elution rather than inefficient adsorption. After adsorption in the presence of high-concentration chaotrope, like in the Wizard/GuSCN method, a fraction of DNA may remain irretrievably bound because of strong hydrophobic interactions with silica.22, 23 The resulting DNA loss is particularly detrimental for dilute samples, in which the irretrievable fraction represents a significant portion of the input.22
The dependence of Wizard/GuSCN on urine composition and, thus, its likely failure in a portion of patient samples may partially explain the low, variable clinical sensitivities previously reported when using it as a sample preparation method for urine cfDNA.6, 7, 26 Even under ideal conditions, maximum recovery was limited to <20% from urine and 30% to 35% from buffer. Although Wizard/GuSCN showed improved recovery of moderately short targets (40 nt) compared with conventional silica adsorption (ie, MagMAX), it was still unable to recover the shortest 25-nt fragment. On the basis of this analytical characterization, Wizard/GuSCN is not recommended for use in clinical samples (particularly for low-concentration targets) without further optimization. If used, adjusting urine samples to pH 5 to 6, spiking in ≥1 μg/mL carrier nucleic acid, and using elevated temperature and incubation time to increase elution yield are suggested.23
Q Sepharose Anion Exchange Resin
The Q Sepharose method improves on Wizard/GuSCN in both recovery of short DNA fragments and overall yield. Q Sepharose had high recovery (63% to 75%) of fragments down to 40 nt. Previous comparison showed that Q Sepharose increased clinical detection of fetal cfDNA in maternal urine compared with Wizard/GuSCN.10 The results of this study support this conclusion and suggest that preconcentration of urine cfDNA using anion exchange resin helps compensate for the length and concentration dependence of silica adsorption. Q Sepharose did not, however, completely overcome fragment length dependence, with <10% recovery of the shortest 25-nt fragment. In addition to concentrating cfDNA, Q Sepharose eliminates urine variabilities, like pH, that might otherwise affect silica adsorption. Q Sepharose is expected to perform well in clinical samples, as supported by its successful previous implementation for liquid biopsies.2, 3
Q Sepharose is recommended as an established, ready-to-go protocol that would be sufficient for most applications. It would be well suited for next-generation sequencing, where bulk, not sequence-specific, purification of cfDNA is necessary. It should ideally be paired with single-stranded library preparation, which has been shown to improve sequencing yield of <100-nt cfDNA fragments.12 For amplification applications, its resistance to PCR inhibition suggests that larger effective volumes could be amplified per reaction to increase analytical sensitivity. When extreme sensitivity and retention of the shortest fragments are required, using hybridization capture is recommended instead.
Norgen Urine Cell-Free Circulating DNA Purification Kit
The Norgen kit had moderate recovery (30% to 41%) of fragments 40 to 150 nt, but higher recovery (72%) of the 25-nt fragment. It was the only silica-based method to efficiently capture the shortest fragment, demonstrating that hybrid silica/silicon carbide spin columns improve capture of ultrashort fragments relative to silica alone, as claimed by the manufacturer. Unfortunately, the Norgen kit also led to consistent PCR inhibition, even when using only a small volume of eluate in PCR. Consequently, quantification using the Norgen kit is unreliable because each individual PCR assay will be uniquely affected by inhibition.42 As an example, the differential inhibition of the 40- and 25-nt qPCR assays is given in Supplemental Figure S4. The Norgen kit was weakly capable of detecting low concentrations of DNA, but its analytical sensitivity is limited by a relatively small urine input (2 mL) combined with inhibition-restricted PCR volume. Although not ideal for precise quantification or sensitive detection of dilute targets, the Norgen kit is a commercially available, user-friendly option. It could be used for quick, preliminary urine cfDNA analyses in which qualitative or semiquantitative detection is adequate.
Qiagen QIAamp Circulating Nucleic Acid Kit
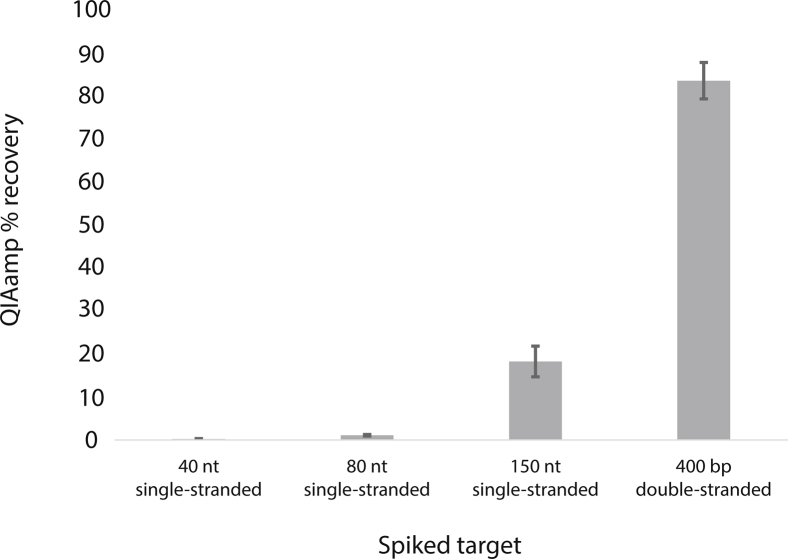
The QIAamp kit had limited recovery (18%), even for the longest 150-nt fragment. It also showed a clear dependence on fragment length, with significantly reduced recovery of the 80- and 40-nt fragments (1% and 0.2%, respectively). To confirm that the low observed recovery was not due to the urine sample used or errors in the extraction procedure, a long 400-bp double-stranded DNA (dsDNA) target was also tested. It had higher recovery (83% ± 4%) (Supplemental Figure S5), indicating that the low recovery using the QIAamp kit was due to the short length and/or single-stranded nature of the spiked target.
The poor performance of the QIAamp kit for short fragments in urine is surprising given its widespread successful use in plasma. Several comparative studies have identified the QIAamp kit as one of the best-performing commercial options for plasma cfDNA.43, 44, 45, 46 Although the kit has high overall yields from plasma, its recovery has been previously shown to decrease as fragment length decreases.44, 46 For spiked dsDNA >100 bp, the QIAamp kit had >80% recovery from plasma,44, 46 but for dsDNA ≤100 bp, the recovery was reduced, with no recovery of a 25-bp fragment.46 This trend is in line with the manufacturer's product information, which claims efficient recovery of fragments down to 75 bp only. Regardless of sample type, both our results and others suggest that the QIAamp kit is inadequate for capturing short DNA fragments. It is unclear what caused the overall recovery from urine seen herein to be lower than that of previous reports from plasma, but it may be at least partly due to the strandedness of the spiked target. Previous plasma studies used dsDNA,44, 46 whereas this study used ssDNA, which interacts differently with silica surfaces on a molecular level.47 Although ssDNA has been reported to bind more strongly to silica at low pH than dsDNA,47 the relative recovery of ssDNA and dsDNA can be tuned by using chaotropic binding buffers of different compositions (ie, higher pH).48 The specific buffer conditions of the QIAamp kit may be better suited for dsDNA than for ssDNA, exacerbating the existing length dependence when using an ssDNA target.
Because of the QIAamp kit's inefficient recovery of short fragments, and particularly those that are single stranded, its use is not recommended for urine, where cfDNA is more fragmented than in plasma. Although the kit's performance may improve for dsDNA, an ideal urine cfDNA kit would be able to efficiently capture the full diversity of degraded urine cfDNA, which is likely to be a heterogeneous mixture of short ssDNA, dsDNA, and nicked DNA.
Thermo Fisher Scientific MagMAX Cell-Free DNA Isolation Kit
The MagMAX kit was extremely dependent on fragment length, as expected for a silica-based method. It had high recovery of longer fragments but no detectable recovery of the 40-nt fragment. Its use in urine samples, where most cfDNA fragments are too short to be recovered efficiently, is not recommended. Other silica-based plasma cfDNA extraction kits may also experience length-based limitations, like the QIAamp and MagMAX kits. Plasma cfDNA kits should not be used for urine cfDNA extraction without experimentally verifying their ability to capture short DNA fragments.
Hybridization Capture
Our laboratory identified hybridization capture as a sample preparation method likely to perform well for short, dilute urine cfDNA. Unlike silica adsorption, hybridization should be agnostic to both fragment length and concentration and robust against variations in clinical urine samples. Our results confirmed that hybridization capture was the only method to maintain high recovery (73% to 84%) across all fragment lengths tested, even down to the shortest 25-nt fragment. Hybridization capture was capable of reliably detecting low DNA concentrations (down to 10 copies/mL) and was tolerant to changes in urine pH, salt, and background DNA, suggesting that it will be effective in clinical samples. The small elution volume (20 μL) and complete removal of PCR inhibition enable the entire output from 1 mL urine to be analyzed in a single PCR well.
Hybridization capture is recommended for urine cfDNA applications where maximum sensitivity is required. Its improvement over alternate methods will be most apparent when paired with an ultrashort PCR target (eg, 25 nt). Hybridization capture may be particularly beneficial for highly fragmented cfDNA, such as bacterial, viral, or mitochondrial cfDNA. It may also offer the advantage of increased specificity by removing nontarget background DNA, although this was not directly tested in this study.
A key limitation of hybridization capture is that, unlike silica-based methods, it will only isolate specific targeted sequences. Although it is ideal for extraction of a specific diagnostic target, and can be multiplexed to extract multiple targets, it is not suitable for sequencing or other applications requiring broader pull-down of all cfDNA regardless of sequence. Development of capture probes for new targets is straightforward, in our case simply using a truncated version of one of the PCR primers. Cost is currently also a limitation for hybridization capture and is due primarily to the magnetic beads. We are now transitioning to a direct capture approach (probes preimmobilized on beads), which puts the cost of hybridization capture on par with existing published protocols while scaling up the analysis volume to 10 mL.
Summary of the Analytical Performance of Urine cfDNA Extraction Methods
Table 4 summarizes the analytical performance of the urine cfDNA extraction methods. Our results reveal that extraction methods vary widely in their ability to capture the short, dilute cfDNA present in urine. Using suboptimal methods may profoundly compromise clinical results because of low recovery, dependence on urine composition, or PCR inhibition. Overall, hybridization capture and Q Sepharose performed best, with high recovery of short fragments (down to 25 and 40 nt, respectively), sensitive detection of dilute fragments, tolerance to varied urine conditions, and resistance to PCR inhibition. As such, these are the two methods we expect to perform well in clinical samples and, thus, recommend for extraction of urine cfDNA. The results of this work will help inform selection of optimal urine cfDNA extraction methods, which, paired with short PCR amplicons, should lead to improved clinical sensitivity and reproducibility of urine cfDNA diagnostics.
Table 4.
Summary of Analytical Performance of Urine cfDNA Extraction Methods
| Urine cfDNA extraction method | Recovery, % | Minimum target length efficiently recovered, nt∗ | Ability to recover low concentrations of short target | Tolerance to varied urine conditions | Resistance to PCR inhibition |
|---|---|---|---|---|---|
| Hybridization capture | 73–84 | 25 | Good | Good | Good |
| Wizard/GuSCN | 1.6–17 | 40 | Poor | Poor | Good |
| Q Sepharose | 8.6–75 | 40 | Good | Good | Good |
| Norgen Urine Cell-Free Circulating DNA Purification Kit | 30–72 | 25 | Moderate | Good | Poor |
| Qiagen QIAamp Circulating Nucleic Acid Kit | 0.20–18† | 150 | Poor | Good | Good |
| Thermo Fisher Scientific MagMAX Cell-Free DNA Isolation Kit | 0.20–66† | 80 | Poor | Good | Moderate |
Wizard/GuSCN, Wizard resin/guanidinium thiocyanate.
Efficient recovery defined as >50% of the maximum recovery observed across all lengths for that method.
The 25-nt fragment was not tested for QIAamp and MagMAX kits.
Acknowledgments
We thank the University of Washington Coulter Foundation and Global Center for Integrated Health of Women, Adolescents, and Children for seed funding; Diana Marangu for contributing to development of the concept to target tuberculosis urine cell-free DNA using improved sample preparation methods and for contributing to development of the collaboration and seed proposal that initiated this work; and our collaborator David Horne for input on clinical application to tuberculosis detection outside the work presented herein.
Footnotes
Supported by University of Washington Coulter Foundation and Global Center for Integrated Health of Women, Adolescents and Children funding (B.R.L. and J.J.L.); the National Institute of Allergy and Infectious Diseases of the NIH under award R21AI125975 (B.R.L. and J.J.L.); and the National Science Foundation Graduate Research Fellowship program (A.O.).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.07.002.
Supplemental Data
Supplemental Figure S1.
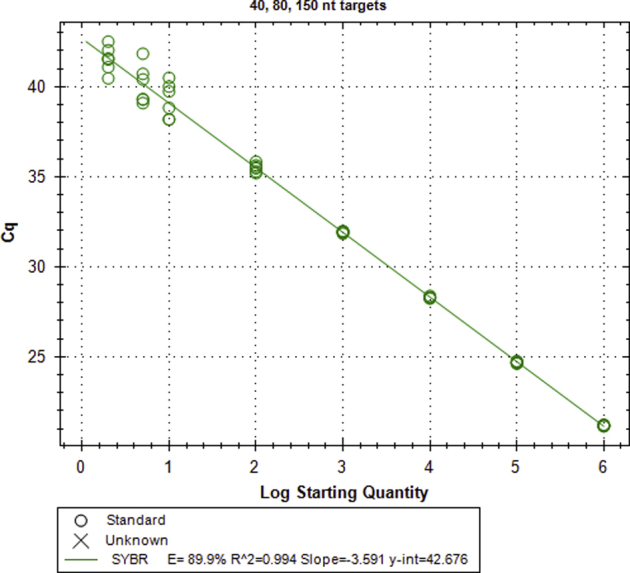
Validation of 40-, 80-, and 150-nt target PCR. A representative calibration curve is given for amplification of the 40-, 80-, and 150-nt targets, but individual calibration curves were run for each experiment to account for any minor variances in template dilution. Amplification of the 40-, 80-, and 150-nt targets had an efficiency of 89.9% (R2 = 0.994) and a linear dynamic range of at least 10 to 106 copies. The limit of detection was less than five copies. PCR specificity for the 40-, 80-, and 150-nt targets was difficult to confirm by melt or gel analysis because the amplicon was only 40 nt, similar to that expected for nonspecific amplification of a primer dimer. To keep the amplicon as short as possible, we instead relied on careful in silico analysis and primer design. Primers were designed against a region of the IS6110 insertion sequence known to be conserved and specific to Mycobacterium tuberculosis complex.39 Candidate primers were screened using National Center for Biotechnology Information Primer BLAST to ensure no amplifiable products outside of the M. tuberculosis complex (including human genomic DNA). After testing the candidate primers, the pair with the most resistance to off-target amplification was selected, despite the trade-off of a slightly reduced amplification efficiency. The final primer set and optimized real-time quantitative PCR conditions reliably result in no amplification of the no template control up to at least 50 cycles. n = 6 per standard concentration. Cq, quantification cycle; y-int, y-intercept.
Supplemental Figure S2.
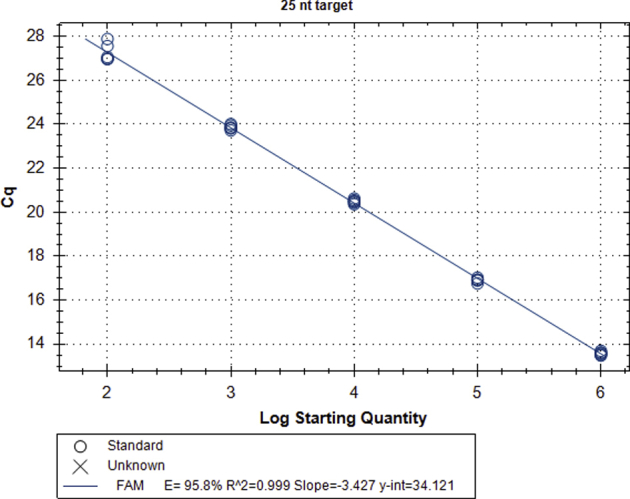
Validation of 25-nt target PCR. A representative calibration curve is given for amplification of the 25-nt target, but individual calibration curves were run for each experiment to account for any minor variances in template dilution. Amplification of the 25-nt target has an efficiency of 95.8% (R2 = 0.999) and a linear dynamic range of 102 to 106 copies. Amplification of 5 to 10 copies was possible, but with reduced efficiency. The quantification cycle (Cq) of the no template control (NTC) for the 25-nt assay is >40 cycles (not including the 10 first-stage preamplification cycles). In the representative calibration curve, four of six NTCs did not amplify above the 100 relative fluorescence unit (RFU) threshold. The other two NTCs had Cqs of 42.5 and 44.8 (effectively, 52.5 and 54.8 cycles, respectively). Specificity of the 25-nt target amplification is ensured by using a sequence-specific probe. n = 6 per standard concentration. y-int, y-intercept.
Supplemental Figure S3.
Dependence of Q Sepharose on background DNA concentration in buffer and performance across biological urine replicates. A total of 104 copies/mL of 150-nt target were spiked into 1× tris-buffered saline (TBS) or urine before extraction. In buffer, the Q Sepharose method had lower recovery at lower background DNA concentrations (0 and 10 ng/μL) than at higher background DNA concentrations (100 and 1000 ng/μL). Unlike the Wizard resin/guanidinium thiocyanate method, however, the 100 ng/mL threshold is within the expected clinical range of urine cfDNA concentrations likely to be found in patient samples. In urine, recovery was consistent across biological urine replicates, despite the moderate dependence on background DNA concentration in buffer. All urine samples had higher recovery than seen in buffer. On the basis of these results, Q Sepharose was classified as tolerant to variable urine conditions, but there is a possibility that unusually dilute samples may have reduced recovery. Data are expressed as means ± SD. n = 3 technical replicates (TBS); n = 3 biological replicates with 3 technical replicates each (urine).
Supplemental Figure S4.
To demonstrate differential PCR inhibition after extraction using the Norgen kit, 104 copies of 40- or 25-nt target were amplified in the presence of 0, 1, 5, 10, or 20 μL eluate from negative control urine extracted using the Norgen kit (0%, 2%, 10%, 20%, or 40% of 50 μL PCR, respectively). An increase in quantification cycle (Cq) indicates PCR inhibition. Amplification of the 25-nt target was more susceptible to PCR inhibition than amplification of the 40-nt target. This result demonstrates how quantification of urine cfDNA after extraction using the Norgen kit would be unreliable because of differential PCR inhibition of different real-time quantitative PCR assays. Calculation of starting quantity based on Cq alone would be biased by the extent of PCR inhibition. Data are expressed as means ± SD. n = 3.
Supplemental Figure S5.
Recovery of QIAamp kit increases for longer double-stranded target. Before extraction using the Qiagen QIAamp Circulating Nucleic Acid Kit, 104 copies/mL of 40-, 80-, or 150-nt (all single-stranded) or 400-bp (double-stranded) targets were spiked into urine. Percentage recoveries are given. Although the recovery of short single-stranded fragments was low (<20%), recovery increased (83%) to match the kit's expected performance for a longer double-stranded target. This indicates that the low recovery of the original 40-, 80-, and 150-nt fragments was because of their short length and/or single-stranded nature, rather than a failure of the QIAamp kit protocol in our laboratory. Data are expressed as means ± SD. n = 3 extractions.
References
- 1.Botezatu I., Serdyuk O., Potapova G., Shelepov V., Alechina R., Molyaka Y., Ananév V., Bazin I., Garin A., Narimanov M., Knysh V., Melkonyan H., Umansky S., Lichtenstein A. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem. 2000;46:1078–1084. [PubMed] [Google Scholar]
- 2.Reckamp K.L., Melnikova V.O., Karlovich C., Sequist L.V., Camidge D.R., Wakelee H., Perol M., Oxnard G.R., Kosco K., Croucher P., Samuelsz E., Vibat C.R., Guerrero S., Geis J., Berz D., Mann E., Matheny S., Rolfe L., Raponi M., Erlander M.G., Gadgeel S. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11:1690–1700. doi: 10.1016/j.jtho.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 3.Fujii T., Barzi A., Sartore-Bianchi A., Cassingena A., Siravegna G., Karp D.D., Piha-Paul S.A., Subbiah V., Tsimberidou A.M., Huang H.J., Veronese S., Di Nicolantonio F., Pingle S., Vibat C.R.T., Hancock S., Berz D., Melnikova V.O., Erlander M.G., Luthra R., Kopetz E.S., Meric-Bernstam F., Siena S., Lenz H.J., Bardelli A., Janku F. Mutation-enrichment next-generation sequencing for quantitative detection of KRAS mutations in urine cell-free DNA from patients with advanced cancers. Clin Cancer Res. 2017;23:3657–3666. doi: 10.1158/1078-0432.CCR-16-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu T., Li J. Clinical applications of urinary cell-free DNA in cancer: current insights and promising future. Am J Cancer Res. 2017;7:2318–2332. [PMC free article] [PubMed] [Google Scholar]
- 5.Green C., Huggett J.F., Talbot E., Mwaba P., Reither K., Zumla A.I. Rapid diagnosis of tuberculosis through the detection of mycobacterial DNA in urine by nucleic acid amplification methods. Lancet Infect Dis. 2009;9:505–511. doi: 10.1016/S1473-3099(09)70149-5. [DOI] [PubMed] [Google Scholar]
- 6.Labugger I., Heyckendorf J., Dees S., Häussinger E., Herzmann C., Kohl T.A., Richter E., Rivera-Milla E., Lange C. Detection of transrenal DNA for the diagnosis of pulmonary tuberculosis and treatment monitoring. Infection. 2017;45:269–276. doi: 10.1007/s15010-016-0955-2. [DOI] [PubMed] [Google Scholar]
- 7.Patel K., Nagel M., Wesolowski M., Dees S., Rivera-Milla E., Geldmacher C., Dheda K., Hoelscher M., Labugger I. Evaluation of a urine-based rapid molecular diagnostic test with potential to be used at point-of-care for pulmonary tuberculosis: Cape Town cohort. J Mol Diagn. 2018;20:215–224. doi: 10.1016/j.jmoldx.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Tong K.L., Li P.K.T., Chan A.Y.W., Yeung C.K., Pang C.C.P., Wong T.Y.H., Lee K.C., Lo Y.M.D. Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: urinary DNA chimerism. Clin Chem. 1999;45:1741–1746. [PubMed] [Google Scholar]
- 9.Gielis E.M., Ledeganck K.J., De Winter B.Y., Del Favero J., Bosmans J.L., Claas F.H., Abramowicz D., Eikmans M. Cell-free DNA: an upcoming biomarker in transplantation. Am J Transplant. 2015;15:2541–2551. doi: 10.1111/ajt.13387. [DOI] [PubMed] [Google Scholar]
- 10.Shekhtman E.M., Anne K., Melkonyan H.S., Robbins D.J., Warsof S.L., Umansky S.R. Optimization of transrenal DNA analysis: detection of fetal DNA in maternal urine. Clin Chem. 2009;55:723–729. doi: 10.1373/clinchem.2008.113050. [DOI] [PubMed] [Google Scholar]
- 11.Tsui N.B.Y., Jiang P., Chow K.C.K., Su X., Leung T.Y., Sun H., Chan K.C.A., Chiu R.W.K., Lo Y.M.D. High resolution size analysis of fetal DNA in the urine of pregnant women by paired-end massively parallel sequencing. PLoS One. 2012;7:e48319. doi: 10.1371/journal.pone.0048319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnham P., Kim M.S., Agbor-Enoh S., Luikart H., Valantine H.A., Khush K.K., De Vlaminck I. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep. 2016;6:27859. doi: 10.1038/srep27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Underhill H.R., Kitzman J.O., Hellwig S., Welker N.C., Daza R., Baker D.N., Gligorich K.M., Rostomily R.C., Bronner M.P., Shendure J. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu S.C.Y., Lee S.W.Y., Jiang P., Leung T.Y., Chan K.C.A., Chiu R.W.K., Lo Y.M.D. High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin Chem. 2013;59:1228–1237. doi: 10.1373/clinchem.2013.203679. [DOI] [PubMed] [Google Scholar]
- 15.Yao W., Mei C., Nan X., Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. Gene. 2016;590:142–148. doi: 10.1016/j.gene.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 16.Burnham P., Dadhania D., Heyang M., Chen F., Westblade L.F., Suthanthiran M., Lee J.R., De Vlaminck I. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat Commun. 2018;9:2412. doi: 10.1038/s41467-018-04745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Y.-H., Wang M., Brenner D.E., Ng A., Melkonyan H., Umansky S., Syngal S., Block T.M. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J Mol Diagn. 2004;6:101–107. doi: 10.1016/S1525-1578(10)60497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Streleckiene G., Reid H.M., Arnold N., Bauerschlag D., Forster M. Quantifying cell free DNA in urine: comparison between commercial kits, impact of gender and inter-individual variation. Biotechniques. 2018;64:225–230. doi: 10.2144/btn-2018-0003. [DOI] [PubMed] [Google Scholar]
- 19.Melkonyan H.S., Feaver W.J., Meyer E., Scheinker V., Shekhtman E.M., Xin Z., Umansky S.R. Transrenal nucleic acids: from proof of principle to clinical tests. Ann N Y Acad Sci. 2008;1137:73–81. doi: 10.1196/annals.1448.015. [DOI] [PubMed] [Google Scholar]
- 20.Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melzak K.A., Sherwood C.S., Turner R.F.B., Haynes C.A. Driving forces for DNA adsorption to silica in perchlorate solutions. J Colloid Interface Sci. 1996;181:635–644. [Google Scholar]
- 22.Katevatis C., Fan A., Klapperich C.M. Low concentration DNA extraction and recovery using a silica solid phase. PLoS One. 2017;12:e0176848. doi: 10.1371/journal.pone.0176848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandeventer P.E., Lin J.S., Zwang T.J., Nadim A., Johal M.S., Niemz A. Multiphasic DNA adsorption to silica surfaces under varying buffer, pH, and ionic strength conditions. J Phys Chem B. 2012;116:5661–5670. doi: 10.1021/jp3017776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Carballo B.L., Broger T., Wyss R., Banaei N., Denkinger C.M. Towards the development of a cfDNA-based in-vitro diagnostic test for infectious diseases: a review of evidence for tuberculosis. Clin Microbiol. 2019;57:e01234-18. doi: 10.1128/JCM.01234-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Bali L., Diman A., Bernard A., Roosens N.H.C., De Keersmaecker S.C.J. Comparative study of seven commercial kits for human DNA extraction from urine samples suitable for DNA biomarker-based public health studies. J Biomol Tech. 2014;25:96–110. doi: 10.7171/jbt.14-2504-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cannas A., Goletti D., Girardi E., Chiacchio T., Calvo L., Cuzzi G., Piacentini M., Melkonyan H., Umansky S.R., Lauria F.N., Ippolito G., Tomei L.D. Mycobacterium tuberculosis DNA detection in soluble fraction of urine from pulmonary tuberculosis patients. Int J Tuberc Lung Dis. 2008;12:146–151. [PubMed] [Google Scholar]
- 27.Trigg R.M., Martinson L.J., Parpart-Li S., Shaw J.A. Factors that influence quality and yield of circulating-free DNA: a systematic review of the methodology literature. Heliyon. 2018;4:e00699. doi: 10.1016/j.heliyon.2018.e00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee D.H., Yoon H., Park S., Kim J.S., Ahn Y.H., Kwon K., Lee D., Kim K.H. Urinary exosomal and cell-free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Sci Rep. 2018;8:14707. doi: 10.1038/s41598-018-32900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangiapan G., Vokurka M., Schouls L., Cadranel J., Lecossier D., Van Embden J., Hance A.J. Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J Clin Microbiol. 1996;34:1209–1215. doi: 10.1128/jcm.34.5.1209-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed J.L., Basu D., Butzler M.A., McFall S.M. XtracTB Assay, a Mycobacterium tuberculosis molecular screening test with sensitivity approaching culture. Sci Rep. 2017;7:3653. doi: 10.1038/s41598-017-03930-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muir P., Nicholson F., Jhetam M., Neogi S., Banatvala J.E. Rapid diagnosis of enterovirus infection by magnetic bead extraction and polymerase chain reaction detection of enterovirus RNA in clinical specimens. J Clin Microbiol. 1993;31:31–38. doi: 10.1128/jcm.31.1.31-38.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millar D.S., Withey S.J., Tizard M.L.V., Ford J.G., Hermontaylor J. Solid-phase hybridization capture of low-abundance target DNA sequences: application to the polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp. silvaticum. Anal Biochem. 1995;226:325–330. doi: 10.1006/abio.1995.1232. [DOI] [PubMed] [Google Scholar]
- 33.Parham N.J., Ois F., Picard J., Peytavi R., Gagnon M., Seyrig G., Gagné P.A., Boissinot M., Bergeron M.G. Specific magnetic bead–based capture of genomic DNA from clinical samples: application to the detection of group B Streptococci in vaginal/anal swabs. Clin Chem. 2007;53:1570–1576. doi: 10.1373/clinchem.2007.091389. [DOI] [PubMed] [Google Scholar]
- 34.Adams N.M., Bordelon H., Wang K-KA., Albert L.E., Wright D.W., Haselton F.R. Comparison of three magnetic bead surface functionalities for RNA extraction and detection. ACS Appl Mater Interfaces. 2015;7:6062–6069. doi: 10.1021/am506374t. [DOI] [PubMed] [Google Scholar]
- 35.Albretsen C., Kalland K.H., Haukanes B.I., Håvarstein L.S., Kleppe K. Applications of magnetic beads with covalently attached oligonucleotides in hybridization: isolation and detection of specific measles virus mRNA from a crude cell lysate. Anal Biochem. 1990;189:40–50. doi: 10.1016/0003-2697(90)90041-7. [DOI] [PubMed] [Google Scholar]
- 36.Peeters S., Stakenborg T., Colle F., Liu C., Lagae L., Van Ranst M. Real-time PCR to study the sequence specific magnetic purification of DNA. Biotechnol Prog. 2010;26:1678–1684. doi: 10.1002/btpr.492. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Morabito K., Tang J.X., Tripathi A. Microfluidic platform for isolating nucleic acid targets using sequence specific hybridization. Biomicrofluidics. 2013;7:44107. doi: 10.1063/1.4816943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohrman B., Richards-Kortum R. Inhibition of recombinase polymerase amplification by background DNA: a lateral flow-based method for enriching target DNA. Anal Chem. 2015;87:1963–1967. doi: 10.1021/ac504365v. [DOI] [PubMed] [Google Scholar]
- 39.Hellyer T.J., Desjardin L.E., Assaf M.K., Bates J.H., Cave M.D., Eisenach K.D. Specificity of IS6110-based amplification assays for Mycobacterium tuberculosis complex. J Clin Microbiol. 1996;34:2843–2846. doi: 10.1128/jcm.34.11.2843-2846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alguacil J., Pfeiffer R.M., Moore L.E., del Fresno M.R., Medina-Lopez R., Kogevinas M., Vermeulen R., Dosemeci M., Silverman D.T., Rothman N., García-Closas M. Measurement of urine pH for epidemiological studies on bladder cancer. Eur J Epidemiol. 2007;22:91–98. doi: 10.1007/s10654-006-9101-2. [DOI] [PubMed] [Google Scholar]
- 41.Kishore R., Reef Hardy W., Anderson V.J., Sanchez N.A., Buoncristiani M.R. Optimization of DNA extraction from low-yield and degraded samples using the BioRobot EZ1 and BioRobot M48. J Forensic Sci. 2006;51:1055–1061. doi: 10.1111/j.1556-4029.2006.00204.x. [DOI] [PubMed] [Google Scholar]
- 42.Huggett J.F., Novak T., Garson J.A., Green C., Morris-Jones S.D., Miller R.F., Zumla A. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes. 2008;1:70. doi: 10.1186/1756-0500-1-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page K., Guttery D.S., Zahra N., Primrose L., Elshaw S.R., Pringle J.H., Blighe K., Marchese S.D., Hilles A., Woodley L., Stebbing J., Coombes R.C., Shaw J.A. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One. 2013;8:e77963. doi: 10.1371/journal.pone.0077963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devonshire A.S., Whale A.S., Gutteridge A., Jones G., Cowen S., Foy C.A., Huggett J.F. Towards standardization of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406:6499–6512. doi: 10.1007/s00216-014-7835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorber L., Zwaenepoel K., Deschoolmeester V., Roeyen G., Lardon F., Rolfo C., Pauwels P. A comparison of cell-free DNA isolation kits: isolation and quantification of cell-free DNA in plasma. J Mol Diagn. 2017;9:162–168. doi: 10.1016/j.jmoldx.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Diefenbach R.J., Lee J.H., Kefford R.F., Rizos H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018;228-229:21–27. doi: 10.1016/j.cancergen.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Shi B., Shin Y.K., Hassanali A.A., Singer S.J. DNA binding to the silica surface. J Phys Chem. 2015;119:11030–11040. doi: 10.1021/acs.jpcb.5b01983. [DOI] [PubMed] [Google Scholar]
- 48.Beld M., Sol C., Goudsmit J., Boom R. Fractionation of nucleic acids into single-stranded and double-stranded forms. Nucleic Acids Res. 1996;24:2618–2619. doi: 10.1093/nar/24.13.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.