Mice are commonly used as a model to study the growth and virulence of influenza A viruses in mammals but are not a natural host and have distinct sialic acid receptor profiles compared to humans. Using experimental infections with different subtypes of influenza A virus derived from different hosts, we found that evolution of influenza A virus in mice did not necessarily proceed through the linear accumulation of host-adaptive mutations, that there was variation in the patterns of mutations detected in each repetition, and that the mutation dynamics depended on the virus examined. In addition, variation in the viral receptor, sialic acid, did not affect influenza virus evolution in this model. Overall, our results show that while mice provide a useful animal model for influenza virus pathology, host passage evolution will vary depending on the specific virus tested.
KEYWORDS: influenza, host range, animal models, evolution, sialic acid
ABSTRACT
Influenza A viruses have regularly jumped to new host species to cause epidemics or pandemics, an evolutionary process that involves variation in the viral traits necessary to overcome host barriers and facilitate transmission. Mice are not a natural host for influenza virus but are frequently used as models in studies of pathogenesis, often after multiple passages to achieve higher viral titers that result in clinical disease such as weight loss or death. Here, we examine the processes of influenza A virus infection and evolution in mice by comparing single nucleotide variations of a human H1N1 pandemic virus, a seasonal H3N2 virus, and an H3N2 canine influenza virus during experimental passage. We also compared replication and sequence variation in wild-type mice expressing N-glycolylneuraminic acid (Neu5Gc) with those seen in mice expressing only N-acetylneuraminic acid (Neu5Ac). Viruses derived from plasmids were propagated in MDCK cells and then passaged in mice up to four times. Full-genome deep sequencing of the plasmids, cultured viruses, and viruses from mice at various passages revealed only small numbers of mutational changes. The H3N2 canine influenza virus showed increases in frequency of sporadic mutations in the PB2, PA, and NA segments. The H1N1 pandemic virus grew well in mice, and while it exhibited the maintenance of some minority mutations, there was no clear evidence for adaptive evolution. The H3N2 seasonal virus did not establish in the mice. Finally, there were no clear sequence differences associated with the presence or absence of Neu5Gc.
IMPORTANCE Mice are commonly used as a model to study the growth and virulence of influenza A viruses in mammals but are not a natural host and have distinct sialic acid receptor profiles compared to humans. Using experimental infections with different subtypes of influenza A virus derived from different hosts, we found that evolution of influenza A virus in mice did not necessarily proceed through the linear accumulation of host-adaptive mutations, that there was variation in the patterns of mutations detected in each repetition, and that the mutation dynamics depended on the virus examined. In addition, variation in the viral receptor, sialic acid, did not affect influenza virus evolution in this model. Overall, our results show that while mice provide a useful animal model for influenza virus pathology, host passage evolution will vary depending on the specific virus tested.
INTRODUCTION
Many animal viruses naturally infect and spread among a range of host species and can often be experimentally inoculated into alternative hosts to cause infections and disease, and where onward transmission may result in epidemics. Because the spread of a virus in a new host may involve adaptive evolution, with ongoing selection as the virus replicates in the host cells, tissues, and populations, or responds to host immunity (1–3), determining the underlying processes of viral evolution is central to understanding and controlling the spread of viruses in humans and other animal populations.
Here, we combine the infection by influenza A virus (IAV) in mice with complete-genome deep sequencing to examine the underlying processes and dynamics of viral growth in a new host. Most IAVs are naturally maintained as intestinal infections of various bird species in freshwater and saltwater environments (4). Occasionally, IAV will spill over to infect mammalian hosts and more rarely will go on to cause epidemics and pandemics. Natural outbreaks by IAV in mammals have been observed in humans, swine, mink, seals, horses, cats, and dogs (5, 6). We have the greatest knowledge of outbreaks that occurred during the past 100 or so years, which in humans include the H1N1, H2N2, and H3N2 human seasonal viruses that were first recognized in 1918, 1957, and 1968, respectively, as well as of the second human H1N1 pandemic strain that spread worldwide in 2009, replacing the circulating seasonal H1N1 clade (7, 8). The emergence of IAV in new hosts involves the natural selection of mutations involved in host adaptation, which may alter binding to the sialic acid (Sia) receptor by the hemagglutinin (HA), cleavage of the Sia by the neuraminidase (NA), host-specific nuclear transport and replication processes that involve the polymerase subunits, and evasion of the host immune responses (reviewed in references 2 and 9, to ,11). Some of these processes and mutations that impact mammalian transmission and disease have also been identified during experimental host passages, in which IAVs are passaged in new hosts such as ferrets (12–14).
Mice are not a natural host for IAVs but have long been used as an animal model to study the replication, pathogenesis, and immune responses of many different viruses from avian and mammalian hosts (15–18). While some IAV strains appear to infect and replicate to high levels in the lungs or other respiratory tissues of mice, many show relatively limited replication, most do not spread naturally from mouse to mouse, and many cause little disease unless they are adapted by serial passage (19–21). The sequences of mouse-adapted IAVs often contain mutations in various genomic segments, including HA, NA, and NS, as well as the polymerase gene segments (PB2, PB1, and PA) (reviewed in reference 22). The considerable number of mouse adaptation studies of IAV vary in repetition, reproducibility, and specifics of the methodologies and experimental variables, all of which could impact virus evolution. In addition, analyses have frequently relied on laboratory tissue culture isolation and the measurement of population consensus mutations or polymorphisms that rose to high levels, often assuming that these were mouse-adaptive mutations fixed by positive selection. However, aside from a direct fitness advantage, some of the mutations observed in mice might have attained higher frequencies due to founder effects associated with population bottlenecks and/or by hitchhiking with beneficial mutations. The acquisition of increased fitness in a new host may also involve complex epistatic mutations and can depend on the specific sequences of the genomes in which they arise (genetic contingency) (23–25). Together, these issues indicate that endpoint analysis of genome variation in passaged populations may not distinguish functionally adaptive mutations from other, nonadaptive variations (26). Understanding the details of how mutations arise in viruses during passage in mice would therefore provide a better understanding of the intricate evolutionary processes involved and facilitate a comparison to viral emergence seen in other natural hosts.
Viral receptors are often involved in host adaptation, and influenza viruses use Sia-terminated glycans on cell glycoproteins or glycolipids as primary receptors of infection while also interacting with and evading or removing Sia in the mucus of the respiratory tract (27, 28). Sias are a family of glycans that include N-acetylneuraminic acid (Neu5Ac) as well as other modified forms and are primarily connected to the underlying glycan through α2,3 or α2,6 linkages (α2,8 linkages are primarily found in polysialic acids) (29). Modifications of Sia may include the hydroxylation of the 5-acetyl group to glycolyl by the CMP-N-acetylneuraminic acid hydroxylase (CMAH), creating N-glycolylneuraminic acid (Neu5Gc) (30). Humans and ferrets lack a functional CMAH, so that Neu5Gc is not displayed in cells of those hosts, while CMAH is active in other natural influenza virus hosts such as swine and horses, and Neu5Gc is also displayed at high levels in many tissues of mice (31, 32). Other chemical modifications of Sia that have the potential to impact virus-Sia interactions are also abundant and diverse across vertebrate species and include the addition of acetyl groups on the 4, 7, 8, and/or 9 positions, as well as potential lactyl or sulfate modifications of the glycerol side chain (33–35). IAV emergence in new hosts frequently involves adaptation to the specific linkage of the Sia in the respiratory tissues of that host (36). In one common process, the HAs of avian IAVs bind with higher affinity to α2,3-linked Sia receptors, which are more abundant in the avian gastrointestinal tract (37). In contrast, human IAV HAs preferentially bind the α2,6-linked Sias that predominate in the human upper respiratory tract (URT), and adaptation to humans involves adaptive mutation of the HA receptor binding site (36, 38, 39).
Mice differ in a number of properties compared to the natural hosts of IAV, which include birds, humans, dogs, swine, horses, seals, and mink. Their use in laboratory studies of pathogenesis and immunology is often in contrast to ferrets and guinea pigs that are commonly used as experimental models of virus contact and airborne transmission (40, 41). Mice also often differ in the forms of the natural Sia receptor, as they display lower proportions of α2,6-linked Sia in their URT relative to humans and some other hosts, and they display both Neu5Ac and Neu5Gc (42, 43). The α2,3- and α2,6-linked Sias and other differences can result in selection in both the HA1 and HA2 domains (44). The HA and NA of some influenza virus strains may distinguish between Neu5Ac and Neu5Gc, generally through lower binding of the HA and lower NA activity on Neu5Gc than on Neu5Ac (45, 46). Mice that lack Neu5Gc have been generated by knocking out the CMAH gene, which was then crossbred into a C57BL/6 background (31), allowing the specific biology of that variation to be assayed, as well as the effects on the Sia specificity of pathogenic bacteria (47, 48).
Here, we use three different IAVs—human viruses A/California/04/2009 H1N1 and A/Wyoming/3/2003 H3N2 and canine virus A/Canine/IL/11613/2015 H3N2—to define, in detail, the sequence variation that arises during infection and passage in mice. Each virus was derived from a genetically homogeneous starting point in reverse genetics plasmids, propagated for only a limited number of passages in cells to prepare an infectious virus stock, and then passed in wild-type C57BL/6 mice and in CMAH−/− mice that lack Neu5Gc. We used full-genome deep sequencing to define the experimental variation of the viruses and the role of potential Neu5Gc receptors in that process.
(This article was submitted to an online preprint archive [49].)
RESULTS
The IAVs tested were strain specific for replication and phenotypic pathology in mice.
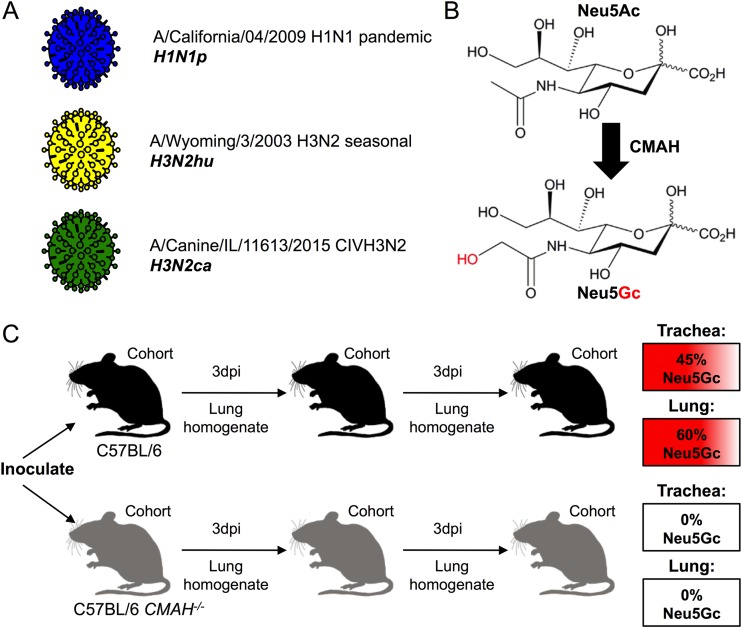
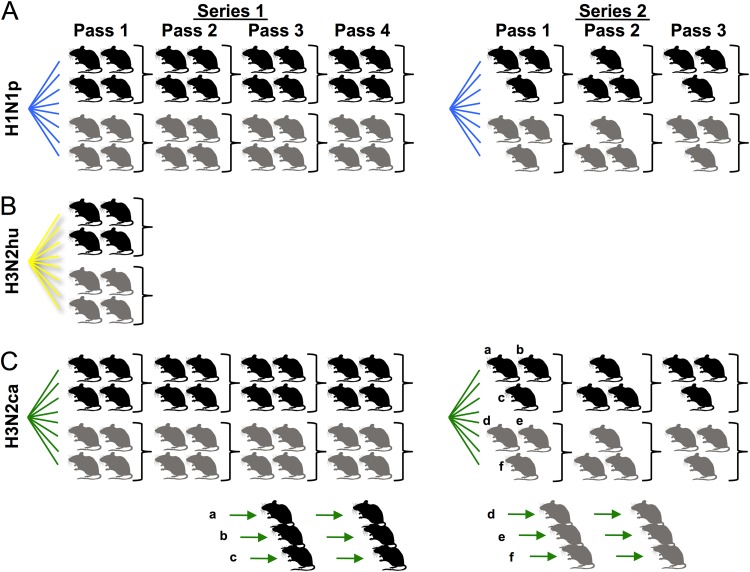
The A/California/04/2009 H1N1 CA04 (H1N1p), A/Wyoming/3/2003 H3N2 WY03 (H3N2hu), and A/Canine/IL/11613/2015 H3N2 IL-15 (H3N2ca) influenza viruses were each derived from plasmid clones (Fig. 1A). The H1N1p and H3N2ca viruses were recovered and passaged in MDCK cells, while the H3N2hu virus was recovered and grown in MDCK-SIAT cells expressing higher levels of α2,6-linked Sia. Analysis of wild-type C57BL/6 mice revealed that Neu5Gc comprised between 45% and 60% of the total Sia present in the trachea and lungs (Table 1). The CMAH−/− mice contained no detectable levels of Neu5Gc within the same tissue samples (Table 1; Fig. 1B). Viruses were intranasally inoculated into wild-type C57BL/6 mice or CMAH−/− mice and then transferred by lung-to-intranasal passages with a fixed homogenate volume of 50 μl (Fig. 1C). The inoculations with H1N1p and H3N2ca were transferred through four groups of C57BL/6 and CMAH−/− mice in the first series of passages and through three groups in the second series of passages (Fig. 2).
FIG 1.
Outline of the experimental design. (A) Influenza virus stocks were generated from reverse genetic clones: H1N1p (blue), H3N2hu (yellow), and H3N2ca (green). (B) The enzyme CMAH catalyzes the enzymatic conversion of Neu5Ac to Neu5Gc. Inactivation of the cmah gene in a C57BL/6 mouse background generates a Neu5Ac-only mouse. (C) Experimental passages of viruses were performed by nasal inoculation of culture-derived virus or lung homogenates into control C57BL/6 (black) or CMAH−/− (gray) mouse cohorts, 3-day incubation, and harvest of lung homogenates. C57BL/6 mice display Neu5Gc, as a proportion of total Sia, at 45% in trachea and 60% in the lungs. CMAH−/− mice contain no Neu5Gc in their trachea or lungs.
TABLE 1.
Relative proportions of Neu5Ac and Neu5Gc identified in mouse respiratory tissues
| Respiratory tissue | % of total Sia in mouse straina
: |
|||
|---|---|---|---|---|
| C57BL/6 |
CMAH−/− |
|||
| Neu5Ac | Neu5Gc | Neu5Ac | Neu5Gc | |
| Trachea | 50 | 45 | 92 | ND |
| Lung | 40 | 60 | 96 | ND |
Determined by HPLC analysis. ND, not detected.
FIG 2.
Diagram of experimental mouse passages performed in this study with H1N1p (A), H3N2hu (B), and H3N2ca (C) or with H3N2ca in mouse-to-mouse lineages (C). Individual mice are displayed as control C57BL/6 (black) or CMAH−/− (gray) mice within cohorts. The experiment was performed in two different iterations: series 1 proceeded for four passages among cohorts of four mice (two male, two female), while series 2 proceeded for three passages among cohorts of three mice (an alternating 2:1 sex ratio). H3N2ca mouse lung homogenates from passage 1 of series 2 were also used to initiate a series of mouse-to-mouse lineages in C57BL/6 (a, b, and c) or CMAH−/− (d, e, and f) individual mice.
Mice were initially inoculated with a fixed number of 50% tissue culture infective dose (TCID50) units of virus as measured in MDCK or MDCK-SIAT cells, followed by a fixed volume of lung homogenates. Subsequent genomic viral RNA (vRNA) quantitation of all inoculation samples revealed that doses varied between 106 and 108 genome copies per inoculum (Fig. 3). The H1N1p virus replicated to give robust genome copy numbers maintained in lungs per passage (generally 2 to 3 log10 units more than the initial dose), and mice displayed moderate to severe signs of disease, including weight loss, observed lethargy, and the presence of lesions on the lungs (Fig. 3A). In contrast, mice inoculated with seasonal human H3N2hu virus (Fig. 3B), even at 108 genome copies, showed few genome copies in their lungs upon the first passage and exhibited no observable signs of infection, including weight loss. No virus was recovered after transfer of materials of the lungs of the first-passage mice to a second series of mice (data not shown). The H3N2ca virus (Fig. 3C) replicated well in the lungs of mice during the experiments, although with 2- or 3 log10-lower titers present in lungs after the initial passage. The H3N2ca virus-infected mice showed occasional behavioral signs of infection during the 3-day incubation (lethargy) but exhibited no measured weight loss and no notable anatomical abnormality of the respiratory tissues.
FIG 3.
General influenza virus dynamics in mouse cohorts inoculated with H1N1p (A), H3N2hu (B), or H3N2ca (C). Control C57BL/6 mouse samples are denoted as squares while CMAH−/− mouse samples are shown as triangles. Top, quantitation of influenza virus genome copies (per RT-qPCR of M segment) for each virus was measured for stock virus and pooled lung homogenate (normalized to inoculum volume) to measure genomic bottleneck size at each passage. H3N2hu-inoculated mice lacked measurable genome copies in their lungs at first passage. Bottom, mouse weights were recorded during the course of infection. Figures are from the first passage of series 1 as a representative example. All virus-specific weight-loss phenotypes persisted during the course of each experimental passage and between repeat passage series. Only mice inoculated with H1N1p showed weight loss during the course of infection, with no significant variation between experimental groups.
We examined the respiratory tissues of C57BL/6 and CMAH−/− mice to determine whether alterations in Neu5Gc expression would impact the qualitative display of Sia receptor linkages. Histochemistry with the lectins MAH and SNA to detect either α2,3- or α2,6-linked Sias showed no obvious differences in the staining of those tissues (Fig. 4A). Immunohistochemistry of the mouse lungs confirmed the presence of IAV antigen (NP) in H1N1p- and H3N2ca-inoculated mice but not in H3N2hu-inoculated mice (Fig. 4B).
FIG 4.
(A) Expression of the α2,3- and α2,6-linked Sias in the trachea and lungs of wild-type C57BL/6 mice, which express ∼45 to 60% Neu5c, or CMAH−/− mice, which lack Neu5Gc. (B) Examples showing the viral infection in the lungs of experimentally passaged mice by immunohistochemistry for IAV antigen (NP), stained in red. Bar, 100 μm.
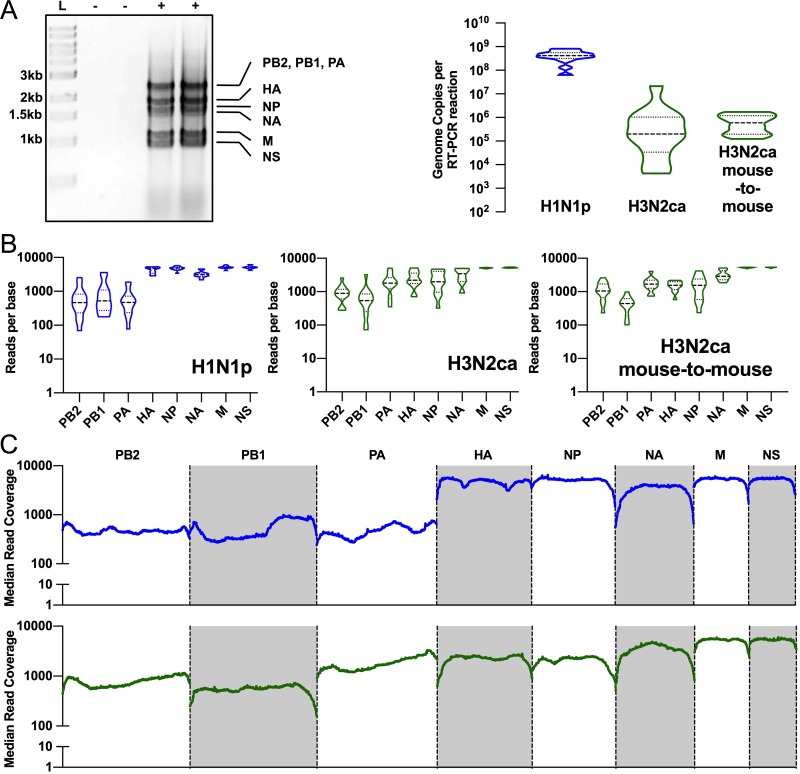
Viral RNA (vRNA) was amplified by IAV whole-genome reverse transcription-PCR (RT-PCR) from pooled and individual experimental lungs (as outlined in Fig. 2), and the cDNA was used to generate libraries for Illumina sequencing. Quantitative reverse transcriptase PCR (RT-qPCR) of the M segment confirmed that genome copies were generally >100,000 RNA copies per reaction, allowing more confident analysis of low-percentage variants (50). The median copy number per reaction of H1N1p samples was 4.15 × 108 (±[2.15 × 108]), while samples of H3N2ca showed greater variation in amounts, although with a median number of copies of 1.99 × 105 (±[2.15 × 106]) per reaction, although even the least abundant sample was still in excess of 4,000 RNA copies (Fig. 5A). Amplification and read coverage varied among segments despite robust quality vRNA in all reactions, and the PB2/PB1/PA segments frequently showed severalfold-lower mean coverage per base than other segments for the same virus strain (Fig. 5B). This may be driven by inconsistent coverage across the length of PB2/PB1/PA segments (Fig. 5C). We set stringency cutoffs for single nucleotide variant (SNV) calling at 1.0% with minimum coverage of 500 reads.
FIG 5.
Whole-genome deep sequencing metrics and quality control. (A) Gel migration of products of IAV whole-genome RT-PCR shows specific bands of 8 genome segments. Products were present only in infected lung homogenates (+) and absent in uninfected controls (−). RT-PCR mixtures generally contained >100,000 input genome copies. (B) Reads across genome segments were not equal, with frequent bias toward smaller segments. (C) Median reads across the genome segments show some variation in coverage across PB2/PB1/PA segments. Blue line, H1N1p; green line, H3N2ca.
Passage of H1N1p in mice resulted in only a small number of polymorphic sites.
Analysis of the H1N1p viruses at each passage in mice showed only very low levels of sequence variation (Fig. 6). The plasmid sequence showed no single nucleotide variants (SNVs) above 0.2% at any position, confirming the low errors associated with library preparation and Illumina sequencing. After three passages in the MDCK cells, the H1N1p viruses contained just 9 SNVs at >1% of reads in the PB1, PA, HA, and NA segments, and two (in HA) were present at ∼5 to 7% (Fig. 6). We deep sequenced the genomes of the virus in the lungs pooled from each group of mice at each passage in each series and found 22 unique SNVs (>1%) across the genome in the final passage of each group. Although mutations were present in multiple genome segments, most were found in the PB1 and HA genes. However, all but two of the SNVs in these genes represented <5% of the sequences at these positions, and none increased substantially during either of the repeated passage series conducted (Fig. 6). The two positions that showed an SNV frequency of >5% were PB1-Val709 and HA-Asp222 (Fig. 6, annotated). The PB1-Val709 locus showed a nonsynonymous mutation to Ile (by G2149A) in ∼12% of reads in the first mouse infection of one iteration of the experiment but then fell to ∼4% by passage 3. The HA-Asp222 locus showed variation in only one passage series and increased in frequency during each passage. In wild-type mice, this locus contained a mixture of two SNVs (nucleotides G747A to code for 222Asn and A748G to code for 222Gly), while in CMAH−/− mice only the Asp222Gly variant was present. These major SNVs were not present above threshold in the stock H1N1p virus and were likely not present, as no PB1-Val709Ile reads were found in the library and HA-Asp222Gly was present in only a single read (unknown if RT-PCR error or virus derived). Besides these changes, other low-level mutations included both synonymous and nonsynonymous changes (Fig. 6) but did not include any positions known to affect viral functions.
FIG 6.
Mutational frequency during H1N1p experimental passage in pooled mouse groups. Single nucleotide variants (SNVs) are represented along the genome segments for the plasmid and virus stocks (black stars), C57BL/6 mice of series 1 (blue circles), CMAH−/− mice of series 1 (blue squares), C57BL/6 mice of series 2 (orange triangles), and CMAH−/− mice of series 2 (orange diamonds). SNVs present at >20% are annotated for their resulting protein sequence changes.
The H3N2hu virus did not establish in mice.
Recent human seasonal H3N2 viruses have been reported to replicate poorly in many standard cells in culture and in mice (51, 52), due at least in part to a need for multivalent binding to the Sia receptors to allow infection (53, 54). We recovered the H3N2hu virus on MDCK-SIAT cells, which have both higher levels of α2,6-linked Sia and higher densities of Sia overall (55), although growth in wild-type MDCK cells was also successful and the virus produced was comparable in titer and viral genome sequence (data not shown). However, after inoculation into C57BL/6 or CMAH−/− mice, we detected only low levels of viral RNA by RT-qPCR at 3 days and did not see viral antigen in lungs of inoculated mice (Fig. 4B). The H3N2hu virus was clearly propagating weakly in the mice and did not establish sustained infections after inoculation, and no signs of infection of the second-passage mice were detected (data not shown).
Passage of H3N2ca in mice resulted in more polymorphisms and rapid appearance of a small number of mutations.
The H3N2ca virus prepared in MDCK cells after plasmid transfection showed one nonsynonymous mutation in NA (nucleotide G98A, Ala27Thr) that represented ∼15% of the sequences, along with only 7 other minor variants at >1% of reads (Fig. 7). This virus replicated well in mice, and several mutations arose to relatively high levels during both passage series in groups of mice (Fig. 7). SNVs were seen across all genome segments during both passage series and in both mouse backgrounds. Final numbers of unique SNVs (>1%) in H3N2ca groups ranged from 29 to 41, although diversity consistently collapsed during passage series from as many as three times the number of unique SNVs early in the series compared to the last pass. In particular, segments PB2, PA, HA, and NA were most likely to have variants that reached over 10% or that approached fixation. The NA-Ala27Thr change present in the inoculum appeared during both passage series in mice. Several other specific mutations arose during the different passage series of mouse genetics cohorts in the PB2, PA, HA, and NA segments (Fig. 7, annotated). Three of these mutations rose to high frequency, or near fixation, during passage: PB2-Ser286Gly, PA-Tyr112Cys, and the culture-derived NA-Ala27Thr. Other mutations that arose during passage in mice did not reach 50% and occasionally fell off during the later passages, possibly due to competition with the mutation-containing genomes that reached higher levels. To examine whether the midlevel mutants were arising in single mice among the groups of mice where we had examined pooled samples, we isolated individual lung homogenates from the three P1 mice in series 2 and passaged each in an additional series of three single mouse-to-mouse lineages (Fig. 2C and Fig. 8). This revealed that individual mouse-passaged viral lineages acquired mutations in the same genome segments (PB2, PA, HA, and NA) as were seen in the pooled mouse samples. The NA-Ala27Thr mutation was again rapidly selected in all lineages, but there was no obvious convergent evolution of the other specific mutations. Individual lineages also contained mutants in the NS segments (lineages a and d) that were not observed in group passage series, and some mutants in HA also rose to higher frequency in individual lineages (b and f) than in the group series (Fig. 8, annotated).
FIG 7.
Mutational frequencies during H3N2ca experimental passage in pooled mouse groups. Single nucleotide variants (SNVs) are represented along the genome segments for the plasmid and virus stocks (black stars), C57BL/6 mice of series 1 (green circles), CMAH−/− mice of series 1 (green squares), C57BL/6 mice of series 2 (black triangles), and CMAH−/− mice of series 2 (black diamonds). SNVs present at >20% are annotated for their resulting protein sequence changes.
FIG 8.
Mutational frequency at the conclusion (passage 3) of H3N2ca mouse-to-mouse experimental passages in C57BL/6 or CMAH−/− mice. Single nucleotide variants (SNVs) are represented along the genome segments for C57BL/6 lineages a (green circles), b (green squares), and c (green hexagons) and CMAH−/− lineages d (black triangles), e (black diamonds), and f (black inverted triangles). SNVs present at >20% are annotated for their resulting protein sequence changes. FS, frameshift.
DISCUSSION
We sought to better understand the host-specific variation and evolution of two human- and one canine-adapted IAV during infection and passage in mice, providing information central to understanding the evolution and potential adaptation of different influenza viruses in this commonly used experimental host. While mouse passaging of many different mammalian and avian IAVs has been reported, this work is novel in the use of complete-genome deep sequencing to directly compare the variations of different viruses and for testing the specific role of the modified Sia (Neu5Gc) present in mouse tissues at high levels (but which is absent from humans, ferrets, and most dog breeds). Each virus examined was already adapted to mammals, so the main selection pressures would likely be related to host-specific differences of the mouse strain tested, as well as to the experimental transmission route. To allow us to track newly emerged variations in the mice, each virus was initiated from reverse genetics plasmids that were also deep sequenced, providing a baseline for comparison. Viral stocks used to initiate the passage series in the mice were prepared by three passages in either MDCK cells (H1N1p and H3N2ca) or MDCK-SIAT cells which expressed higher levels of α2,6-linked Sia (H3N2hu). To specifically examine the sources of virus variation, passage conditions and combinations (including mouse-to-mouse lineages) were designed based on the preliminary data obtained.
The presence of the nonhuman modified Neu5Gc Sia did not alter the IAV variation in mice.
About 50% of the Sia present in the trachea and lungs of wild-type C57BL/6 mice was Neu5Gc, while CMAH−/− mice expressed 100% Neu5Ac, similar to what is seen in humans, ferrets, and Western dogs (31, 56–58). The variant Sias measured here are most likely components of the mucus present in those tissues, as well as on the cell surfaces, where they act as receptors for infection. We found no significant effect of the presence of Neu5Gc in mouse respiratory tissue in this study. It is likely that the Neu5Gc Sia does not bind the HA of the viruses tested, so that those do not play a role in IAV infection. In the small number of cases where this has been examined, it appears that circulating IAVs preferentially bind the Neu5Ac receptor and largely ignore the abundant Neu5Gc present in some hosts—as was seen in swine H3N2 strains (59). In Neu5Ac binding strains where HA is mutated (at position 155) to allow Neu5Gc binding, their NA cleavage is generally inefficient on that Sia, disrupting HA-NA balance and blocking productive infection (60). One hypothesis stemming from our data is that the HA binding properties of the viruses tested here are under weak or no selection by Neu5Gc in wild-type mice under these experimental conditions. The natural ecology of IAV may involve only Neu5Gc binding by viruses infecting hosts that express high levels of that Sia (61–63). Recent work shows that only a small number of avian variants or equine strains demonstrate true Neu5Gc binding by their HA, yet with 2- to 4-fold-lower NA specificity for Neu5Gc, including avian N1 and N9 and human N1 (45). Human IAV may evolve to avoid HA binding of inhibitory Sia variants found in other hosts but that appear to be missing from humans, such as the case of 4-O-acetyl-modified Sia present in horse serum (64, 65).
Deep sequencing enables a dynamic examination of population variation during evolution.
The plasmid and stock virus sequences suggest that the library preparation and Illumina sequencing contributed no significant variation to the data obtained under our threshold cutoffs. Sequence data from the MDCK-passaged virus stocks showed very few polymorphic positions above 1 or 2% of the total, apart from the NA-Ala27Thr substitution of H3N2ca. The sequences of H3N2ca viruses directly recovered from infected dogs show that Ala is the most common wild-type residue at that position, none show Thr, and a single virus from South Korea that had been passaged in culture showed Gly (GenBank sequence accession no. AFN06542) (66). This mutation lies within the transmembrane domain of NA and does not have a described function. None of the other observed major SNVs (being at >20%) were found in natural isolates of canine H3N2.
Patterns of evolution of IAVs during mouse passage are strain/subtype contingent.
The three viruses showed distinct patterns of evolution when passaged in mice. This is consistent with the ideas around historical contingency, in which related populations experience distinct evolutionary trajectories despite following the same challenge—in this case mouse host adaptation (67, 68). The failure of H3N2hu (A/Wyoming/03/2003) to replicate appears to be consistent with multiple observations that mice are difficult to infect with more recent seasonal H3N2 strains from humans (52). Recent H3N2 strains show significant adaption to human respiratory Sia receptors that are highly branched or multiantennary N-glycans, likely due to modulated HA avidity in balance with antigenic selection on the epitopes near the top of the HA trimer, close to the Sia binding site (54). In these studies, we saw no differences in the sequences of H3N2hu viruses produced in the MDCK-SIAT versus wild-type cells. Hence, these results highlight the important role of strain and of subtype sequences in the infection of the same animal and in the evolution of host adaptation.
Passage of H1N1p repeatedly in either mouse strain revealed only a few positions with low levels of SNV polymorphism, most frequently in the PB1 and HA gene segments. Most polymorphisms were below 5%, and the two mutations present above 5% were seen in different iterations of the experiment. The mutations detected included both synonymous and nonsynonymous changes. In previous studies of H1N1p, various mutations were reported to be present after mouse passage, including in PB2, PA, HA, and NP (69, 70). Only the HA-Asp222Gly/Asn variant corresponded with mutations previously seen in mouse adaptation studies or selected in other hosts. Changes in that position have been seen in human clinical samples and associated with varied pathogenic outcomes in both humans and mice due to changes in receptor specificity by the Sia α2,3 or α2,6 linkages (71, 72). Our observation of little change in mutational frequencies, along with minimal convergence of sequences in repeated studies, suggests that the changes seen in mouse passage of H1N1p may be random variants that do not directly impact fitness. It is possible that differences between our results and those of previous studies reflect different viral sources, different passaging schemes, or the mouse strains used (BALB/c or DBA in some previous studies versus C57BL/6) (73, 74). Interestingly, the deep sequencing of H1N1p passages in C57BL/6 mice in unrelated studies also revealed unique variants and minimal signatures of selective fixation (R. Honce and S. Schultz-Cherry, personal communication).
The speed of adaptation in novel environments is impacted by the fitness effects of individual mutations (75). The H1N1p virus may already be relatively well adapted to mice, so that most mutations do not provide much fitness benefit. Other things being equal, variants with relatively small effect sizes will fix more slowly even if they are common (76, 77). The mutation rate of IAVs is similar to other RNA polymerases, with a mean estimated rate of around 2.5 × 10−5 substitutions per nucleotide per cell infection (s/n/c) (78), although most of the mutations that arise appear to be removed by purifying selection or are lost during bottlenecks that occur during cell-to-cell infection or host-to-host transmission. The experimental passages of the virus in our studies did not have tight bottlenecks, which led us to expect robust adaptation if natural selection on mutations of different fitness was present. While many of the variants in the H1N1p viruses did not rise in frequency, the polymorphisms that were observed had arisen de novo during the growth of the plasmid-derived viruses in culture and mouse passage, and their maintenance in each virus population was consistent with random sampling effects and perhaps genetic hitchhiking.
H3N2ca inoculation in mice gave rise to considerably more SNV diversity that H1N1p, and the emergence of polymorphisms in PB2, PA, HA, and NA, including near-fixation of PB2-Ser286Gly, PA-Tyr112Cys, and NA-Ala27Thr. Some other mutations that rose in frequency, often to >40% of reads in the population, suggest that they could be on a path of selective fixation in a new background. Unlike past work in the H3 background (human A/HongKong/1/1968/H3N2), we did not see frequent mutations in HA, nor the PB2-Asp701Asn mutation that was strongly associated with mouse pathogenesis and mammalian adaptation by importin-α interaction and nuclear import of viral RNPs (vRNPs) (79–81). Other H3N2ca virus mutations that arose in our experiment that parallel past H3 mouse passage observations included PB2-Asp740Asn, PA-Gln556Arg, and M-Asp232Asn, but these were mostly seen in single mouse-to-mouse lineages (82). It is possible that the NA-Ala27Thr mutation (of apparent strong fitness advantage but unknown function) influenced the evolutionary path of some of our H3N2ca lineages.
Populations evolving with a larger mutational load may be subject to stochastic outcomes without a strong directed-selection coefficient (83). This may explain our results with H3N2ca, in which mutations frequently arose and also increased in frequency, but with little to no convergence of specific mutations between several repeat iterations of the same experiment (apart from the NA-Ala27Thr culture-selected mutation already present in all passage series). It is possible that multiple different mutations can give rise to similar fitness gains or effectively similar phenotypes. Given sufficiently large population sizes and turnover of the virus, evolving populations will rapidly fix highly beneficial mutations, which could converge if related to a strong and focused selective pressure (84); that this does not occur here again suggests that there is likely no central selective force/constraint of mouse host adaptation.
Overall, this study shows that IAV host adaptation in mice is highly contingent on the specific virus used in the experiment and may exhibit only weak signatures of natural selection with little convergent enrichment of polymorphisms toward population fixation. Greater comparison of mouse passage experiments between different laboratories, along with analysis of key population dynamic variables and deep sequencing, will be useful in explaining the host-specific growth of different viruses in mice.
MATERIALS AND METHODS
Cells and viruses.
MDCK cells were obtained from the American Type Culture Collection (ATCC; CCL-34). Variants of those MDCK cells with increased levels of α2,6-linked Sia cells were prepared in the laboratory by transfection with the ST6Gal1 gene in a plasmid under the control of the cytomegalovirus (CMV) promoter (pcDNA3.1; Invitrogen). Cell clones with increased levels of α2,6-linked Sia were identified by staining with the Sambucus nigra (SNA) lectin and termed MDCK-SIAT cells. HEK293T cells were obtained from the ATCC (CRL-3216). All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum and 50 μg/ml gentamicin.
Three IAV strains were derived from reverse genetics plasmids, comprising (i) human H1N1 pandemic IAV (A/California/04/2009, H1N1p) in plasmid pDP2002, (ii) human H3N2 seasonal IAV (A/Wyoming/3/2003, H3N2hu) in plasmid pDZ, and (iii) a canine H3N2 IAV (A/Canine/IL/11613/2015, H3N2ca) in pDZ. The plasmid encoding each viral segment was prepared from a single bacterial colony, and an 8-plasmid mixture for each virus was prepared and used for transfection of a 3:1 coculture of HEK293T cells and MDCK cells (MDCK-SIAT cells for H3N2hu). Each virus was passaged two additional times in the same MDCK, or MDCK-SIAT variant, cells to generate a passage 3 stock, which was tested for infectivity by TCID50 assay and for viral RNA titer by quantitative reverse transcriptase PCR (RT-qPCR). Each plasmid mixture (as DNA) and the resulting virus stocks (from RNA) were then used to generate libraries for Illumina sequencing, as described below, revealing the original sequences and any baseline variation of the viruses used to start the mouse inoculations.
Mouse inoculation and serial passage.
All mouse studies followed protocols approved by the Cornell University Institutional Animal Care and Use Committee. The passage series and analysis of each virus are diagrammed in Fig. 2. Control wild-type (C57BL/6) and CMAH−/− (B6.129 × 1-Cmahtm1Avrk/J) mice were obtained from Jackson Laboratories and housed and/or bred on site. Mice (aged 6 to 10 weeks) were anesthetized using isoflurane gas and inoculated intranasally with 50 μl of 104 TCID50 units (MDCK or MDCK-SIAT) of each virus in PBS. Mice were observed and weighed each day and euthanized 3 days postinfection (dpi), and tissues were harvested. Single lung lobes of each mouse averaged ∼100 mg and were homogenized with 0.5 ml of added sterile PBS (∼20% [wt/vol]). Homogenates were clarified by centrifugation at 1,000 × g for 10 min. Individual mouse sample lung homogenates were stored at −80°C. In some studies, lung homogenate supernatants (n = 4 or 3 of like virus strain and mouse genetics) were also pooled and aliquoted, and 50 μl was used to inoculate each mouse in the next passage.
As the H3N2ca-inoculated mouse samples showed the highest number and level of mutations, and to understand the dynamics and variation within individual mice, for the second passage series lungs of individual mice were passed on to additional individual mice to reveal any differences in the dynamics of single mouse-to-mouse lineages (Fig. 2C).
vRNA extraction and virus copy number quantitation by qPCR.
Viral RNA (vRNA) was isolated from virus-infected lung homogenate supernatant using the QIAamp viral RNA minikit (Qiagen). Influenza virus genome copies were quantified from RNA isolations by reverse transcription-quantitative PCR (RT-qPCR) for the M segment modified from the CDC protocol (85). Products were amplified using Path-ID (Applied Biosystems) with M-specific primers (5′ to 3′, F, GACCRATCCTGTCACCTCTGAC; R, AGGGCATTYTGGACAAAKCGTCTA), probed with 5′-TGCAGTCCTCGCTCACTGGGCACG-3′, and run on a 7500 Fast Real-Time platform against a standard curve.
Library generation and NGS.
Influenza virus whole-genome reverse transcription-PCR (RT-PCR) amplification of cDNA from all 8 viral genome segments was performed using a modification of the method of Zhou et al. (86). Total vRNA was incubated with reaction buffer, Superscript III, and Platinum Taq-HiFi (Invitrogen) in the presence of universal influenza virus-amplifying primers (5′ to 3′, uni12a, GTTACGCGCCAGCAAAAGCAGG; uni12b, GTTACGCGCCAGCGAAAGCAGG; uni13, GTTACGCGCCAGTAGAAACAAGG). RT-PCRs were performed as first-strand synthesis (42°C) for 50 min, followed by 5 cycles at 42°C of annealing and then by 33 cycles at 57°C of annealing. Viral cDNA products were purified with Agencourt AMPure XP magnetic beads (Beckman Coulter), eluted in Tris buffer, and quantified by Qubit (Invitrogen). One-nanogram amounts cDNA were used to prepare next-generation sequencing (NGS) libraries using the Nextera XT DNA library preparation kit (Illumina), with unique index adaptors for each sample. Pooled libraries were run on an Illumina MiSeq v2 for 250-bp paired reads in the Cornell Animal Health and Diagnostic Center Molecular Diagnostics Lab or the Cornell Genomics Facility of the Biotechnology Research Center.
Sequence analysis and variant calling.
Analysis was performed in Geneious v.11.1.5. Read trimming was performed by the BBDuk script (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbduk-guide/), followed by read merging and alignment to the reference genomic plasmid sequence for each virus. For variant calling, we considered those at >1% frequency with minimum 500-read coverage (a threshold that is consistently representative of SNV diversity in all data sets). Recognizing the potential for PCR-derived errors being observed as SNVs, and our use of single library runs per sample, we focused our analysis on those mutations that are maintained passage to passage. Longitudinal sampling of SNVs would allow unique samples to act as proxy duplicates and further suggest biological relevance of those mutations.
HPLC analysis of mouse respiratory tissue variant sialic acids.
The Sia composition of the mouse trachea and lung samples was determined by incubation with 2 M acetic acid at 80°C for 3 h, filtration through a Microcon 10-kDa centrifugal filter (Millipore), and drying in a SpeedVac vacuum concentrator. Released Sias were labeled with 1,2-diamino-4,5-methylenedioxybenzene (DMB; Sigma-Aldrich) for 2.5 h at 50°C (87). High-performance liquid chromatography (HPLC) analysis was performed using a Dionex UltiMate 3000 system with an Acclaim C18 column (ThermoFisher) under isocratic elution in 7% methanol, 7% acetonitrile, and 86% water. Sia standards included bovine submaxillary mucin and commercial standards for Neu5Ac and Neu5Gc (Sigma-Aldrich). Statistical analyses were performed in Prism software (GraphPad; version 8).
Histochemistry of mouse respiratory tissue for sialic acid distribution and experimental lungs for viral antigen.
Expression of α2,3- and α2,6-linked Sias in the trachea and lungs of mice was examined by preparing frozen sections of OCT-embedded tissue. Sections were fixed for 30 min with 10% buffered formalin and then incubated with Maackia amurensis (MAA-II, MAH) or Sambucus nigra (SNA) lectins conjugated with biotin (Vector Laboratories, Burlingame, CA). Sections were then probed by streptavidin-horseradish peroxidase (HRP) followed by incubation with the substrate NovaRED (Vector).
The additional lung lobe of each experimental mouse was stored in 10% buffered formalin. Lungs were paraffin embedded and cut for both hematoxylin and eosin (H&E) staining and immunohistochemistry. The presence of influenza viral antigen in lung tissue was determined by staining with the anti-NP monoclonal antibody (ATCC HB-65, clone H16-L10-4R5), followed by anti-mouse goat IgG conjugated with HRP and then incubation with the substrate NovaRED.
Graphing and analysis.
All figure graphs were generated in GraphPad Prism v.8.1.2.
Data availability.
Sequences have been deposited at the NCBI Sequence Read Archive (SRA) under BioProject no. PRJNA564292 (https://www.ncbi.nlm.nih.gov/bioproject/564292).
ACKNOWLEDGMENTS
Reverse genetics plasmids for A/Wyoming/3/2003/H3N2 were provided by Adolfo García-Sastre at the Icahn School of Medicine, Mount Sinai. Reverse genetics plasmids for A/California/04/2009/H1N1 were provided by Daniel Perez at the University of Georgia. We thank support services at the Cornell College of Veterinary Medicine Animal Health Diagnostic Center (AHDC): the Molecular Diagnostics Lab for support with RT-qPCR analysis as well as Illumina-MiSeq protocol development and sequencing runs, and the Histology lab for support with formalin-fixed, paraffin-embedded (FFPE) tissue embedding and slide preparation.
This work was supported in part by CRIP (Center of Research in Influenza Pathogenesis, an NIAID-funded Center of Excellence in Influenza Research and Surveillance [CEIRS]) contract HHSN272201400008C to C.R.P. and by NIH grant R01 GM080533 to C.R.P. I.E.H.V. was supported by NSF award DGE-1650441. E.C.H. is supported by an ARC Australian Laureate Fellowship (FL170100022).
REFERENCES
- 1.Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM. 2014. The evolution and genetics of virus host shifts. PLoS Pathog 10:e1004395. doi: 10.1371/journal.ppat.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subbarao K. 2019. The critical interspecies transmission barrier at the animal-human interface. Trop Med Infect Dis 4:E72. doi: 10.3390/tropicalmed4020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasik BR, de Wit E, Munster V, Lloyd-Smith JO, Martinez-Sobrido L, Parrish CR. 2019. Onward transmission of viruses: how do viruses emerge to cause epidemics after spillover? Philos Trans R Soc Lond B Biol Sci 374:20190017. doi: 10.1098/rstb.2019.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon S-W, Webby RJ, Webster RG. 2014. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol 385:359–375. doi: 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- 5.Mostafa A, Abdelwhab EM, Mettenleiter TC, Pleschka S. 2018. Zoonotic potential of influenza A viruses: a comprehensive overview. Viruses 10:497. doi: 10.3390/v10090497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parrish CR, Murcia PR, Holmes EC. 2015. Influenza virus reservoirs and intermediate hosts: dogs, horses, and new possibilities for influenza virus exposure of humans. J Virol 89:2990–2994. doi: 10.1128/JVI.03146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horimoto T, Kawaoka Y. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol 3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 8.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altman MO, Angeletti D, Yewdell JW. 2018. Antibody immunodominance: the key to understanding influenza virus antigenic drift. Viral Immunol 31:142–149. doi: 10.1089/vim.2017.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H, Webster RG, Webby RJ. 2018. Influenza virus: dealing with a drifting and shifting pathogen. Viral Immunol 31:174–183. doi: 10.1089/vim.2017.0141. [DOI] [PubMed] [Google Scholar]
- 11.Long JS, Mistry B, Haslam SM, Barclay WS. 2019. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17:67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- 12.Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus A, Fouchier R. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrauwen EJ, Fouchier RA. 2014. Host adaptation and transmission of influenza A viruses in mammals. Emerg Microbes Infect 3:e9. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton TC, Finch C, Shao H, Angel M, Chen H, Capua I, Cattoli G, Monne I, Perez DR. 2014. Airborne transmission of highly pathogenic H7N1 influenza virus in ferrets. J Virol 88:6623–6635. doi: 10.1128/JVI.02765-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hers JF, Mudler J, Masurel N, van der Kuip L, Tyrrell DA. 1962. Studies on the pathogenesis of influenza virus pneumonia in mice. J Pathol Bacteriol 83:207–217. doi: 10.1002/path.1700830123. [DOI] [PubMed] [Google Scholar]
- 16.Hirst GK. 1947. Studies on the mechanism of adaptation of influenza virus to mice. J Exp Med 86:357–366. doi: 10.1084/jem.86.5.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raut S, Hurd J, Blandford G, Heath RB, Cureton RJ. 1975. The pathogenesis of infections of the mouse caused by virulent and avirulent variants of an influenza virus. J Med Microbiol 8:127–136. doi: 10.1099/00222615-8-1-127. [DOI] [PubMed] [Google Scholar]
- 18.Wyde PR, Couch RB, Mackler BF, Cate TR, Levy BM. 1977. Effects of low- and high-passage influenza virus infection in normal and nude mice. Infect Immun 15:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Donnell CD, Subbarao K. 2011. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect 13:502–515. doi: 10.1016/j.micinf.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudneva IA, Kaverin NV, Varich NL, Gitelman AK, Makhov AM, Klimenko SM, Zhdanov VM. 1986. Studies on the genetic determinants of influenza virus pathogenicity for mice with the use of reassortants between mouse-adapted and non-adapted variants of the same virus strain. Arch Virol 90:237–248. doi: 10.1007/bf01317373. [DOI] [PubMed] [Google Scholar]
- 21.Ward AC. 1997. Virulence of influenza A virus for mouse lung. Virus Genes 14:187–194. doi: 10.1023/A:1007979709403. [DOI] [PubMed] [Google Scholar]
- 22.Kamal RP, Katz JM, York IA. 2014. Molecular determinants of influenza virus pathogenesis in mice. Curr Top Microbiol Immunol 385:243–274. doi: 10.1007/82_2014_388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burch CL, Chao L. 2000. Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature 406:625–628. doi: 10.1038/35020564. [DOI] [PubMed] [Google Scholar]
- 24.Elena SF, Solé RV, Sardanyés J. 2010. Simple genomes, complex interactions: epistasis in RNA virus. Chaos 20:026106. doi: 10.1063/1.3449300. [DOI] [PubMed] [Google Scholar]
- 25.Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB. 2011. Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet 7:e1001301. doi: 10.1371/journal.pgen.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrick JE, Lenski RE. 2013. Genome dynamics during experimental evolution. Nat Rev Genet 14:827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matrosovich M, Herrler G, Klenk HD. 2015. Sialic acid receptors of viruses. Top Curr Chem 367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zanin M, Baviskar P, Webster R, Webby R. 2016. The interaction between respiratory pathogens and mucus. Cell Host Microbe 19:159–168. doi: 10.1016/j.chom.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angata T, Varki A. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev 102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 30.Varki A. 2009. Multiple changes in sialic acid biology during human evolution. Glycoconj J 26:231–245. doi: 10.1007/s10719-008-9183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedlund M, Tangvoranuntakul P, Takematsu H, Long JM, Housley GD, Kozutsumi Y, Suzuki A, Wynshaw-Boris A, Ryan AF, Gallo RL, Varki N, Varki A. 2007. N-glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Mol Cell Biol 27:4340–4346. doi: 10.1128/MCB.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peri S, Kulkarni A, Feyertag F, Berninsone PM, Alvarez-Ponce D. 2018. Phylogenetic distribution of CMP-Neu5Ac hydroxylase (CMAH), the enzyme synthetizing the proinflammatory human xenoantigen Neu5Gc. Genome Biol Evol 10:207–219. doi: 10.1093/gbe/evx251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnard KN, Wasik BR, LaClair JR, Weichert WS, Lawrence BK, Parrish CR. 2019. Expression of 9-O- and 7,9-O-acetyl modified sialic acid in cells and their effects on influenza viruses. bioRxiv doi: 10.1101/650341. [DOI] [PMC free article] [PubMed]
- 34.Muñoz-Barroso I, García-Sastre A, Villar E, Manuguerra JC, Hannoun C, Cabezas JA. 1992. Increased influenza A virus sialidase activity with N-acetyl-9-O-acetylneuraminic acid-containing substrates resulting from influenza C virus O-acetylesterase action. Virus Res 25:145–153. doi: 10.1016/0168-1702(92)90106-j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasik BR, Barnard KN, Ossiboff RJ, Khedri Z, Feng KH, Yu H, Chen X, Perez DR, Varki A, Parrish CR. 2017. Distribution of O-acetylated sialic acids among target host tissues for influenza virus. mSphere 2:e00379-16. doi: 10.1128/mSphere.00379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa T, Chaves AJ, Valle R, Darji A, van Riel D, Kuiken T, Majó N, Ramis A. 2012. Distribution patterns of influenza virus receptors and viral attachment patterns in the respiratory and intestinal tracts of seven avian species. Vet Res 43:28. doi: 10.1186/1297-9716-43-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 39.Walther T, Karamanska R, Chan RWY, Chan MCW, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, Haslam SM, Nicholls JM. 2013. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog 9:e1003223. doi: 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belser JA, Barclay W, Barr I, Fouchier RAM, Matsuyama R, Nishiura H, Peiris M, Russell CJ, Subbarao K, Zhu H, Yen H-L. 2018. Ferrets as models for influenza virus transmission studies and pandemic risk assessments. Emerg Infect Dis 24:965–971. doi: 10.3201/eid2406.172114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouvier NM, Lowen AC. 2010. Animal models for influenza virus pathogenesis and transmission. Viruses 2:1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol 80:7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 44.Keleta L, Ibricevic A, Bovin NV, Brody SL, Brown EG. 2008. Experimental Evolution of human influenza virus H3 hemagglutinin in the mouse lung identifies adaptive regions in HA1 and HA2. J Virol 82:11599–11608. doi: 10.1128/JVI.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broszeit F, Tzarum N, Zhu X, Nemanichvili N, Eggink D, Leenders T, Li Z, Liu L, Wolfert MA, Papanikolaou A, Martínez-Romero C, Gagarinov IA, Yu W, García-Sastre A, Wennekes T, Okamatsu M, Verheije MH, Wilson IA, Boons G-J, de Vries RP. 2019. N-glycolylneuraminic acid as a receptor for influenza A viruses. Cell Rep 27:3284–3294.e6. doi: 10.1016/j.celrep.2019.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masuda H, Suzuki T, Sugiyama Y, Horiike G, Murakami K, Miyamoto D, Jwa Hidari KI, Ito T, Kida H, Kiso M, Fukunaga K, Ohuchi M, Toyoda T, Ishihama A, Kawaoka Y, Suzuki Y. 1999. Substitution of amino acid residue in influenza A virus hemagglutinin affects recognition of sialyl-oligosaccharides containing N-glycolylneuraminic acid. FEBS Lett 464:71–74. doi: 10.1016/s0014-5793(99)01575-6. [DOI] [PubMed] [Google Scholar]
- 47.Alisson-Silva F, Liu JZ, Diaz SL, Deng L, Gareau MG, Marchelletta R, Chen X, Nizet V, Varki N, Barrett KE, Varki A. 2018. Human evolutionary loss of epithelial Neu5Gc expression and species-specific susceptibility to cholera. PLoS Pathog 14:e1007133. doi: 10.1371/journal.ppat.1007133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galán JE, Varki A. 2014. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wasik BR, Voorhees IEH, Barnard KN, Lawrence BK, Weichert WS, Hood G, Nogales A, Martinez-Sobrido L, Holmes EC, Parrish CR. 2019. Influenza viruses in mice: deep sequencing analysis of serial passage and effects of sialic acid structural variation. bioRxiv 683516. doi: 10.1101/683516. [DOI] [PMC free article] [PubMed]
- 50.McCrone JT, Lauring AS. 2016. Measurements of Intrahost viral diversity are extremely sensitive to systematic errors in variant calling. J Virol 90:6884–6895. doi: 10.1128/JVI.00667-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chambers BS, Li Y, Hodinka RL, Hensley SE. 2014. Recent H3N2 influenza virus clinical isolates rapidly acquire hemagglutinin or neuraminidase mutations when propagated for antigenic analyses. J Virol 88:10986–10989. doi: 10.1128/JVI.01077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Groves HT, McDonald JU, Langat P, Kinnear E, Kellam P, McCauley J, Ellis J, Thompson C, Elderfield R, Parker L, Barclay W, Tregoning JS. 2018. Mouse models of influenza infection with circulating strains to test seasonal vaccine efficacy. Front Immunol 9:126. doi: 10.3389/fimmu.2018.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh DY, Barr IG, Mosse JA, Laurie KL. 2008. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol 46:2189–2194. doi: 10.1128/JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ, Paulson JC. 2017. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe 21:23–34. doi: 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk H-D. 2003. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol 77:8418–8425. doi: 10.1128/jvi.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Löfling J, Lyi SM, Parrish CR, Varki A. 2013. Canine and feline parvoviruses preferentially recognize the non-human cell surface sialic acid N-glycolylneuraminic acid. Virology 440:89–96. doi: 10.1016/j.virol.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng PSK, Böhm R, Hartley-Tassell LE, Steen JA, Wang H, Lukowski SW, Hawthorne PL, Trezise AEO, Coloe PJ, Grimmond SM, Haselhorst T, von Itzstein M, Paton AW, Paton JC, Jennings MP. 2014. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat Commun 5:5750. doi: 10.1038/ncomms6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varki A. 2001. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol Suppl 33:54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bateman AC, Karamanska R, Busch MG, Dell A, Olsen CW, Haslam SM. 2010. Glycan analysis and influenza A virus infection of primary swine respiratory epithelial cells: the importance of NeuAc{alpha}2-6 glycans. J Biol Chem 285:34016–34026. doi: 10.1074/jbc.M110.115998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi T, Takano M, Kurebayashi Y, Masuda M, Kawagishi S, Takaguchi M, Yamanaka T, Minami A, Otsubo T, Ikeda K, Suzuki T. 2014. N-glycolylneuraminic acid on human epithelial cells prevents entry of influenza A viruses that possess N-glycolylneuraminic acid binding ability. J Virol 88:8445–8456. doi: 10.1128/JVI.00716-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higa H, Rogers G, Paulson J. 1985. Influenza virus hemagglutinins differentiate between receptor determinants bearing N-acetyl-, N-glycollyl-, and N,O-diacetylneuraminic acids. Virology 144:279–282. doi: 10.1016/0042-6822(85)90325-3. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki Y, Ito T, Suzuki T, Holland R, Chambers T, Kiso M, Ishida H, Kawaoka Y. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 74:11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M, Tscherne DM, McCullough C, Caffrey M, García-Sastre A, Rong L. 2012. Residue Y161 of influenza virus hemagglutinin is involved in viral recognition of sialylated complexes from different hosts. J Virol 86:4455–4462. doi: 10.1128/JVI.07187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matrosovich M, Gao P, Kawaoka Y. 1998. Molecular mechanisms of serum resistance of human influenza H3N2 virus and their involvement in virus adaptation in a new host. J Virol 72:6373–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogers GN, Pritchett TJ, Lane JL, Paulson JC. 1983. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology 131:394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- 66.Voorhees IEH, Dalziel BD, Glaser A, Dubovi EJ, Murcia PR, Newbury S, Toohey-Kurth K, Su S, Kriti D, Van Bakel H, Goodman LB, Leutenegger C, Holmes EC, Parrish CR. 2018. Multiple incursions and recurrent epidemic fade-out of H3N2 canine influenza A vrus in the United States. J Virol 92:e00323-18. doi: 10.1128/JVI.00323-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blount ZD, Lenski RE, Losos JB. 2018. Contingency and determinism in evolution: replaying life’s tape. Science 362:eaam5979. doi: 10.1126/science.aam5979. [DOI] [PubMed] [Google Scholar]
- 68.Bull JJ, Badgett MR, Springman R, Molineux IJ. 2004. Genome properties and the limits of adaptation in bacteriophages. Evolution 58:692–701. doi: 10.1111/j.0014-3820.2004.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 69.Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J Virol 84:8607–8616. doi: 10.1128/JVI.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye J, Sorrell EM, Cai Y, Shao H, Xu K, Pena L, Hickman D, Song H, Angel M, Medina RA, Manicassamy B, Garcia-Sastre A, Perez DR. 2010. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog 6:e1001145. doi: 10.1371/journal.ppat.1001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJA, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus ADME, Garcia-Sastre A, Perez DR, Fouchier RAM. 2010. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol 84:11802–11813. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vazquez-Perez JA, Isa P, Kobasa D, Ormsby CE, Ramírez-Gonzalez JE, Romero-Rodríguez DP, Ranadheera C, Li Y, Bastien N, Embury-Hyatt C, González-Duran E, Barrera-Badillo G, Ablanedo-Terrazas Y, Sevilla-Reyes EE, Escalera-Zamudio M, Cobián-Güemes AG, Lopez I, Ortiz-Alcántara J, Alpuche-Aranda C, Perez-Padilla JR, Reyes-Terán G. 2013. A (H1N1) pdm09 HA D222 variants associated with severity and mortality in patients during a second wave in Mexico. Virol J 10:41. doi: 10.1186/1743-422X-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otte A, Sauter M, Alleva L, Baumgarte S, Klingel K, Gabriel G. 2011. Differential host determinants contribute to the pathogenesis of 2009 pandemic H1N1 and human H5N1 influenza A viruses in experimental mouse models. Am J Pathol 179:230–239. doi: 10.1016/j.ajpath.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Srivastava B, Błazejewska P, Hessmann M, Bruder D, Geffers R, Mauel S, Gruber AD, Schughart K. 2009. Host genetic background strongly influences the response to influenza A virus infections. PLoS One 4:e4857. doi: 10.1371/journal.pone.0004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crow JF, Kimura M. 1970. An introduction to population genetics theory. Harper & Row, New York, NY. [Google Scholar]
- 76.Burch CL, Chao L. 1999. Evolution by small steps and rugged landscapes in the RNA virus phi6. Genetics 151:921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peck KM, Lauring AS. 2018. Complexities of viral mutation rates. J Virol 92:e01031-17. doi: 10.1128/JVI.01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanjuán R, Domingo-Calap P. 2016. Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gabriel G, Herwig A, Klenk H-D. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog 4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ping J, Dankar SK, Forbes NE, Keleta L, Zhou Y, Tyler S, Brown EG. 2010. PB2 and hemagglutinin mutations are major determinants of host range and virulence in mouse-adapted influenza A virus. J Virol 84:10606–10618. doi: 10.1128/JVI.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sediri H, Schwalm F, Gabriel G, Klenk H-D. 2015. Adaptive mutation PB2 D701N promotes nuclear import of influenza vRNPs in mammalian cells. Eur J Cell Biol 94:368–374. doi: 10.1016/j.ejcb.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 82.Ping J, Keleta L, Forbes NE, Dankar S, Stecho W, Tyler S, Zhou Y, Babiuk L, Weingartl H, Halpin RA, Boyne A, Bera J, Hostetler J, Fedorova NB, Proudfoot K, Katzel DA, Stockwell TB, Ghedin E, Spiro DJ, Brown EG. 2011. Genomic and protein structural maps of adaptive evolution of human influenza A virus to increased virulence in the mouse. PLoS One 6:e21740. doi: 10.1371/journal.pone.0021740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao L, Abbasi AB, Illingworth C. 2019. Mutational load causes stochastic evolutionary outcomes in acute RNA viral infection. Virus Evol 5:vez008. doi: 10.1093/ve/vez008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ. 1997. Exceptional convergent evolution in a virus. Genetics 147:1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shu B, Wu K-H, Emery S, Villanueva J, Johnson R, Guthrie E, Berman L, Warnes C, Barnes N, Klimov A, Lindstrom S. 2011. Design and performance of the CDC real-time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. J Clin Microbiol 49:2614–2619. doi: 10.1128/JCM.02636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J Virol 83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Varki A, Diaz S. 1984. The release and purification of sialic acids from glycoconjugates: methods to minimize the loss and migration of O-acetyl groups. Anal Biochem 137:236–247. doi: 10.1016/0003-2697(84)90377-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences have been deposited at the NCBI Sequence Read Archive (SRA) under BioProject no. PRJNA564292 (https://www.ncbi.nlm.nih.gov/bioproject/564292).