Lymphoid leukosis (LL)-like lymphoma is a low-incidence yet costly and poorly understood disease of domestic chickens. The observed unique characteristics of LL-like lymphomas are that the incidence of the disease is chicken line dependent; pathologically, it appeared to mimic avian leukosis but is free of exogenous ALV infection; inoculation of the nonpathogenic ALV-E or MDV-2 (SB-1) boosts the incidence of the disease; and inoculation of both the nonpathogenic ALV-E and SB-1 escalates it to much higher levels. This study was designed to test the impact of two new ALV-E isolates, recently derived from commercial broiler breeder flocks, in combination with the nonpathogenic SB-1 on LL-like lymphoma incidences in both an experimental egg layer line of chickens and a commercial broiler breeder line of chickens under a controlled condition. Data from this study provided an additional piece of experimental evidence on the potency of nonpathogenic ALV-E, MDV-2, and ALV-E plus MDV-2 in boosting the incidence of LL-like lymphomas in susceptible chickens. This study also generated the first piece of genomic evidence that suggests host transcriptomic variation plays an important role in modulating LL-like lymphoma formation.
KEYWORDS: endogenous retrovirus, serotype 2 Marek’s disease virus, spontaneous tumors, genetic resistance, differentially expressed genes, signaling pathways
ABSTRACT
In 2010, sporadic cases of avian leukosis virus (ALV)-like bursal lymphoma, also known as spontaneous lymphoid leukosis (LL)-like tumors, were identified in two commercial broiler breeder flocks in the absence of exogenous ALV infection. Two individual ALV subgroup E (ALV-E) field strains, designated AF227 and AF229, were isolated from two different breeder farms. The role of these ALV-E field isolates in development of and the potential joint impact in conjunction with a Marek’s disease virus (MDV) vaccine (SB-1) were further characterized in chickens of an experimental line and commercial broiler breeders. The experimental line 0.TVB*S1, commonly known as the rapid feathering-susceptible (RFS) line, of chickens lacks all endogenous ALV and is fully susceptible to all subgroups of ALV, including ALV-E. Spontaneous LL-like tumors occurred following infection with AF227, AF229, and a reference ALV-E strain, RAV60, in RFS chickens. Vaccination with serotype 2 MDV, SB-1, in addition to AF227 or AF229 inoculation, significantly enhanced the spontaneous LL-like tumor incidence in the RFS chickens. The spontaneous LL-like tumor incidence jumped from 14% by AF227 alone to 42 to 43% by AF227 in combination with SB-1 in the RFS chickens under controlled conditions. RNA-sequencing analysis of the LL-like lymphomas and nonmalignant bursa tissues of the RFS line of birds identified hundreds of differentially expressed genes that are reportedly involved in key biological processes and pathways, including signaling and signal transduction pathways. The data from this study suggested that both ALV-E and MDV-2 play an important role in enhancement of the spontaneous LL-like tumors in susceptible chickens. The underlying mechanism may be complex and involved in many chicken genes and pathways, including signal transduction pathways and immune system processes, in addition to reported viral genes.
IMPORTANCE Lymphoid leukosis (LL)-like lymphoma is a low-incidence yet costly and poorly understood disease of domestic chickens. The observed unique characteristics of LL-like lymphomas are that the incidence of the disease is chicken line dependent; pathologically, it appeared to mimic avian leukosis but is free of exogenous ALV infection; inoculation of the nonpathogenic ALV-E or MDV-2 (SB-1) boosts the incidence of the disease; and inoculation of both the nonpathogenic ALV-E and SB-1 escalates it to much higher levels. This study was designed to test the impact of two new ALV-E isolates, recently derived from commercial broiler breeder flocks, in combination with the nonpathogenic SB-1 on LL-like lymphoma incidences in both an experimental egg layer line of chickens and a commercial broiler breeder line of chickens under a controlled condition. Data from this study provided an additional piece of experimental evidence on the potency of nonpathogenic ALV-E, MDV-2, and ALV-E plus MDV-2 in boosting the incidence of LL-like lymphomas in susceptible chickens. This study also generated the first piece of genomic evidence that suggests host transcriptomic variation plays an important role in modulating LL-like lymphoma formation.
INTRODUCTION
Lymphoid leukosis (LL) is a B-cell lymphoma of chickens taking place during 4 months of age and older (1, 2). The tumors typically involve liver, spleen, and bursa of Fabricius (3) and are usually composed of aggregates of lymphoblasts of B-cell origin with subsequent production of monoclonal IgM (4).
Commonly, LL is induced by transmissible strains of retroviruses, known as avian leukosis virus (ALV), in susceptible chickens. The specific strains of ALV are defined as exogenous ALVs, since they are transmitted as infectious virus particles. Exogenous ALVs propagate in most tissues and organs of the avian body but only persist in bursal lymphocytes, the target cells of neoplastic transformation (5). Field strains of exogenous ALVs do not harbor any oncogene but instead induce lymphoid leukosis by activation of the cellular myc oncogene (5). Only defective exogenous ALVs harbor oncogenes, such as v-myc, v-src, v-myb, etc., and have been shown to induce acute tumors in susceptible hosts (5).
Spontaneous ALV-like bursal lymphomas, also termed LL-like lymphomas, have been reported in chicken flocks in the absence of exogenous ALV (6, 7). These tumors are of bursal cell origin and are grossly and microscopically similar to exogenous ALV-induced LL but free of detectable ALV infection (7). Some genetic lines of chickens are more susceptible to the development of spontaneous ALV-like tumors, such as line 0 (8, 9) and the transgenic line 0.ALV6 (10), two of the chicken lines maintained by the USDA, Agriculture Research Service, Avian Disease and Oncology Laboratory (ADOL). The 0.ALV6 line, originally derived from line 0, carries a defective subgroup A avian leukosis provirus in its germ line and has been shown to develop spontaneous LL-like tumors similarly to line 0 (11).
There are seven subgroups, A, B, C, D, E, J, and K, of ALVs identified in chickens based upon the viral envelope glycoproteins (12–16). Unlike the other subgroups of exogenous ALVs, the subgroup E viruses are avian retrovirus-like elements that are transmitted genetically in a Mendelian fashion and are termed endogenous viruses (3). The domestic White Leghorn chicken genome carries at least 22 endogenous ALV proviral loci (ev-1 through ev-22) (17–19). Many of the endogenous viruses are genetically defective and incapable of giving rise to infectious virions (20), whereas others may be expressed in an infectious form (21). In the infectious form, endogenous viruses are transmitted similarly to exogenous viruses, although there are chickens or lines of chickens that are genetically resistant to infection of endogenous viruses (3, 22, 23). Rous-associated virus type 0 (RAV-0), a subgroup E endogenous virus, has little or no oncogenic potential (24). However, RAV-60, a subgroup E recombinant of endogenous and exogenous viruses, is highly oncogenic, and infection of RAV-60 can lead to LL (25, 26). Endogenous ALVs also influence the response of birds to infection by exogenous ALV (9, 27–29).
Genetic resistance to avian leukosis occurs at two levels, cellular resistance to virus infection and resistance to tumor formation (30–33). Inheritance of cellular resistance to ALV infection is of a Mendelian type. Independent autosomal loci control the resistance to infection of subgroups ALV-A, -B, -C, and -J and are designated tva (tumor virus A subgroup), tvb, tvc, and tvj, respectively (22, 34–36). Receptors responsible for mediating the cellular resistance and infection for ALV subgroups D and E are also coded by the tvb locus of specific alleles (37).
The TVB receptor complex is coded by a series of three alleles (TVB*S1, TVB*S3, and TVB*R) of tvb. The TVB*S1 allele is dominant and encodes the receptor that mediates infection of ALV subgroups B, D, and E. The TVB*S3 allele is recessive to TVB*S1 but dominant to TVB*R and encodes a receptor that only mediates ALV-B and ALV-D infection, not ALV-E infection. The TVB*R allele encodes a defective (truncated) receptor incapable of facilitating infection by any of the three subgroup ALVs. Resistance to subgroup E ALV is more complex. In addition to the allelic forms of the tvb locus, it is believed that resistance to ALV-E also involves another locus, reportedly known as i (38). Subsequent studies suggested that the i locus is, in fact, a series of competent ALV-E inserts, also known as ev loci, that express high levels of the envelope glycoproteins of ALV-E, which, in turn, competently interfere with the binding processes between the ALV-E receptors and the subgroup ALV-E (23, 26, 39). Therefore, the susceptibility of both homozygous and heterozygous TVB*S1 chickens to subgroup ALV-E might be compromised by the replication-competent ALV-E inserts carried in those chickens’ genome (23).
Vaccination with Marek’s disease virus serotype 2 (MDV-2) has been shown to enhance the development of ALV and reticuloendotheliosis virus-induced bursal lymphomas (40–42) as well as spontaneous bursal lymphomas (11, 43). MDV-2 has been shown to elevate ALV gene expression and ALV replication (44). Furthermore, bursal cells coinfected with ALV and MDV-2 are more likely to be transformed (45).
In 2010, a commercial company observed the development of spontaneous LL-like tumors in broiler breeder flocks raised on more than one farm. The tumors were identified during postmortem examinations of hens between 35 and 45 weeks of age with no significant impact on reproductive performance and overall flock mortality. The incidence of birds with developed spontaneous tumors was sporadic and was found during routine postmortem exams. Lesions consistent with LL-like tumor were found in liver, spleen, kidney, and, less frequently, in other organs. Samples from various tissues received from the commercial broiler breeder chickens tested negative for known exogenous ALV of subgroups A, B, C, D, J, and K, as determined by virus isolation and PCR assays. However, two ALV-E field strains were isolated from two broiler breeder flocks raised on two different farms and were designated AF-227 and AF-229.
Since it was unclear if the two isolate-specific ALV-E field strains were responsible for the incidences of spontaneous LL-like bursa lymphomas observed in the commercial broiler breeder flocks, this study was designed to characterize the two field isolates by infecting birds sampled from a fully susceptible line of chickens, the line 0.TVB*S1, commonly known as the rapid feathering-susceptible (RFS) line, which lacks all endogenous ALV and is fully susceptible to all subgroups of ALV, including ALV-E (46). The affected commercial lines were no longer available at the time of this study; thus, a different line of commercial broiler chickens from the same commercial company was used in the challenge trials of the two field isolates in the presence and absence of vaccination with MDV-2. Complete and partial genome sequence analyses of the two field strains of ALV isolators were conducted for categorization and characterization. Total RNA samples were extracted from fresh LL-like bursa lymphoma tissues of the RFS chickens inoculated with AF227 followed by SB-1 vaccination, normal bursa tissues, and B cells isolated from normal spleen tissues of uninfected RFS chickens, which were subsequently subjected to RNA-sequencing (RNA-Seq) analysis to explore potential genomic variation that may reveal insights into the escalated incidences of spontaneous LL-like lymphomas.
RESULTS
The field isolates AF227 and AF229 are closely related to the ALV subgroup ALV-E.
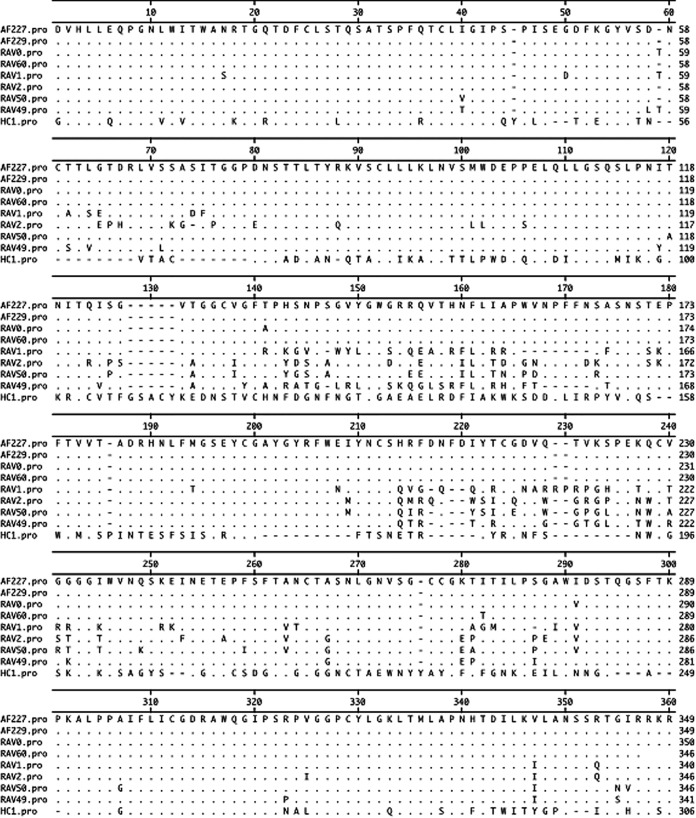
Two avian virus field isolates have been derived from commercial broiler breeder flocks. The genomes of the two field isolates, AF227 and AF229, along with the genomes of the prototype ALV-E strains, RAV-0 and RAV-60, and the prototype ALV-A strain, RAV-1, were sequenced. The complete genomic sequences have been deposited in GenBank. Accession numbers of MF817820 for AF227, MF817821 for AF229, MF817822 for RAV-0, MF817823 for a partial RAV-60 sequence, and MF926337 for RAV-1 were assigned. Sequence analyses of the complete AF227 and AF229 genomes were closely related to those of known endogenous ALV strains, which confirmed our expectation that these two new field isolates were subgroup E ALV, or ALV-E. The sequences of both AF227 and AF229 genomes were more than 99% homologous to the RAV-0 genome sequence. In contrast, the AF227 and AF229 genomes were only 89% homologous to the ALV-A (RAV-1) genome and 79% homologous to the ALV-J (HPRS-103) genome (47) sequences. Further comparison of the gp85 deduced amino acid sequences showed that AF227 and AF229 were 99% to 100% homologous to the sequence of the ALV subgroup E viruses, 81 to 85% homologous to those of ALV subgroup A to D viruses, and 42% homologous to that of ALV subgroup J viruses (Fig. 1). The amino acid sequences of the gp85 envelope protein of the endogenous viruses varied little but obviously differed from that of the exogenous viruses, consistent with the literature (48–52). Furthermore, there was only a 45% nucleotide sequence homology to exogenous long terminal repeats (LTRs), whereas the AF227 and AF229 LTRs showed 96% identity to those in ALV-E strain RAV-0. The endogenous ALV LTRs were 256 nucleotides long and were shorter than the exogenous RAV-1 LTRs, which were 347 nucleotides in length.
FIG 1.
Comparison of gp85 amino acid sequences of avian leukosis virus (ALV) subgroups E (AF227, AF229, RAV0, and RAV60; accession numbers MF817820, MF817821, MF817822, and MF817823, respectively), A (RAV1, accession number MF926337), B (RAV2, accession number M14902.1), C (RAV49, accession number J02342.1), D (RAV50, accession number D10652.1), and J (HC-1, accession number AF247391.1). One-letter amino acid codes are listed. Amino acid residues identical to the majority of codes of AF227 are indicated by dots, dashes indicate amino acid residues missing due to deletions, and one-letter codes represent the differences between AF227 and a subgroup of ALV.
Pathogenicity of the ALV-E field isolates AF227 and AF229.
The pathogenicity of the ALV-E field isolate AF227 observed in the fully susceptible RFS line of chickens is shown in Table 1. The RFS (C/0) chickens inoculated at 7 days of embryonation (DOE) were viremia tolerant for ALV-E, as evidenced by virus isolation at 4 and 32 weeks of age using RFS chicken embryo fibroblasts (CEFs). Cocultivation of buffy coat analysis showed that only the birds inoculated with MDV-2 in this trial were positive for MDV. No significant difference in the incidences of spontaneous LL-like lymphomas was observed following the inoculation of the RFS embryos at embryonation with either AF227 or serotype 2 vaccine (SB-1) alone on the day of hatch, which resulted in 14% and 17% of LL-like lymphomas in the RFS birds, respectively. However, inoculation of the RFS embryos at embryonation with both AF227 and SB-1 on the day of hatch significantly enhanced the incidence of LL-like lymphomas (P < 0.05), which resulted in 42% LL-like lymphomas in the RFS birds (Table 1).
TABLE 1.
Incidences of LL-like lymphomas observed in RFS chickens inoculated with ALV-E AF227, MDV-2 vaccine (SB-1), or both
| Inoculum | No. of chickens at risk | Virus isolation [no. of chickens positive/no. of chickens at risk (%)] at wka
: |
No. of chickens with tumors/no. of chickens at riskb (%) | |
|---|---|---|---|---|
| 4 | 32 | |||
| PBS | 24 | 0/10 (0) | 0/10 (0) | 2/24 (8)a |
| SB-1 | 24 | 0/10 (0) | 0/10 (0) | 4/24 (17)a |
| AF227 | 36 | 9/10 (90) | 9/9 (100) | 5/36 (14)a |
| AF227 + SB-1 | 36 | 10/10 (100) | 10/10 (100) | 15/36 (42)b |
Plasma samples were tested for endogenous ALV on CEFs of the ADOL line RFS (C/E).
The inoculum groups not sharing a lowercase superscript letter differed in tumor incidences with statistical significance based upon chi-square analysis (P < 0.05).
The commercial broiler breeder flocks were typed by single-nucleotide polymorphism as TVB*S1/*S1 homozygous (data not shown) and should be fully susceptible to infection of ALV-E, provided that birds are free of ev gene expression, particularly ev3, ev6, and ev21, by the birds. Only 0 to 5% of the chickens at risk developed LL-like lymphoma following inoculation of SB-1 in combination with either AF229 or AF227. In contrast, 33 to 43% of RFS chickens developed LL-like lymphoma following the same challenges. All virus isolation results on the line RFS CEF were positive, except for those of the SB-1 and the phosphate buffered saline (PBS) inoculum groups of the RFS birds (Table 1). On the other hand, all virus isolation results on the line 0 CEF were negative for both the commercial broilers and the RFS birds at 4 and 32 weeks of age, which evidenced that there was exogenous ALV infection in neither the commercial boilers nor the RFS birds (Table 2).
TABLE 2.
Incidences of LL-like lymphomas observed in commercial broiler breeders and RFS chickens inoculated with SB-1 MDV-2 vaccine, ALV-E isolate AF229, or a combination of SB-1 and AF227 or AF229
| Inoculum | Chicken line | Virus isolation [no. of chickens positive/no. of chickens at risk (%)] at wka
: |
No. of chickens with LL-like lymphoma/no. of chickens at riskb (%) | |||
|---|---|---|---|---|---|---|
| 4 |
52 |
|||||
| C/E | C/0 | C/E | C/0 | |||
| PBS | Broiler | 0/32 (0) | 32/32 (100) | 0/29 (0) | 29/29 (100) | 0/29 (0)a |
| AF229 | Broiler | 0/33 (0) | 33/33 (100) | 0/31 (0) | 31/31 (100) | 0/33 (0)a |
| SB-1 | Broiler | 0/35 (0) | 35/35 (100) | 0/31 (0) | 31/31 (100) | 0/35 (0)a |
| AF229 + SB-1 | Broiler | 0/35 (0) | 35/35 (100) | 0/30 (0) | 30/30 (100) | 0/34 (0)a |
| AF227 + SB-1 | Broiler | 0/27 (0) | 27/27 (100) | 0/14 (0) | 14/14 (100) | 1/19 (5)a |
| PBS | RFS | 0/43 (0) | 0/43 (0) | 0/33 (0) | 0/33 (0) | 2/57 (3.5)a |
| AF229 + SB-1 | RFS | 0/33 (0) | 33/33 (100) | 0/19 (0) | 19/19 (100) | 11/33 (33)b |
| AF227 + SB-1 | RFS | 0/35 (0) | 35/35 (100) | 0/18 (0) | 18/18 (100) | 15/35 (43)b |
Plasma samples were tested for exogenous and endogenous ALVs on CEFs of ADOL lines 0 (C/E) and RFS (C/0).
Tumor incidences between inoculum groups sharing no common lowercase superscript letter differed significantly based upon chi-square analysis (P < 0.05).
Genomic analysis of AF227 by next-generation sequencing.
The entire nucleotide sequence of AF227 was recovered and assembled from RNA sequencing (RNA-Seq) data obtained from the LL-like bursal lymphomas of the AF227- and MDV-2-infected RFS chickens. The recovered AF227 nucleotide sequence was identical to that of the AF227 nucleotide sequence derived from cloned AF227 virus. In addition, the MDV-2 gene, R-LORF1, was detected with significantly high expression levels, while the SORF1 and SORF2 genes were expressed at a relatively low level in the LL-like lymphoma tissues.
The malignant and nonmalignant bursa groups were more comparable on overall expression levels, in contrast to the splenic B-cell group.
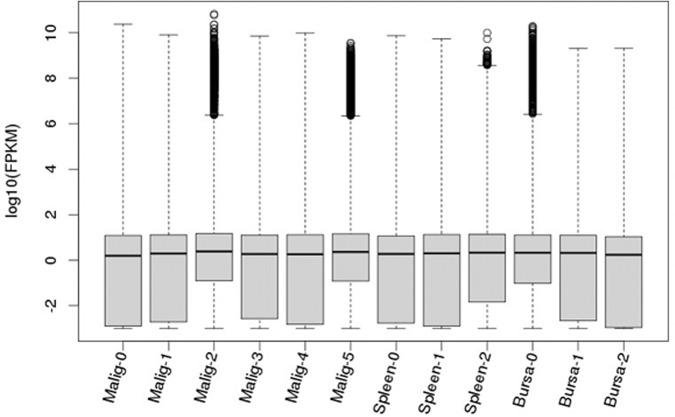
Next-generation sequencing (NGS) read files of the individual RNA samples for the LL-like lymphomas, the control groups of normal bursal tissues, and splenic B cells were deposited in the NCBI SRA database under assigned project accession number PRJNA543277 (https://www.ncbi.nlm.nih.gov/sra/ PRJNA543277). The read data postnormalization are shown in Fig. 2, illustrating the read distributions and variability among the samples. No obvious difference was observable between the lymphoma group (Malig-0 to Malig-5) and either of the control groups (Spleen-0 to Spleen-2 and Bursa-0 to Bursa-2) in distribution, variability, and median values. However, Ward hierarchical clustering analysis was further conducted using Euclidean distance to generate a distance matrix. New clustering analyses showed low biological variability among samples within each of the three biological sample groups, but the splenic B cell group was clearly more distant from the malignant and nonmalignant bursa groups (Fig. 3). Thus, the subsequent comparisons were made between the malignant and nonmalignant bursa tissue groups only, except for the overall gene expression profiling of the LL-like lymphomas and the two control groups of normal bursa tissues and splenic B cells.
FIG 2.
Box-and-whisker plots of gene expression profiles for each of the malignant samples (Malig-0 to Malig-5) and nonmalignant samples, including the control groups of bursa tissues (Bursa-0 to Bursa-2) and splenic B cells (Spleen-0 to Spleen-2), illustrating the RNA-Seq read distribution and variability of the RNA-Seq samples. FPKM, fragments per kilobase per million.
FIG 3.
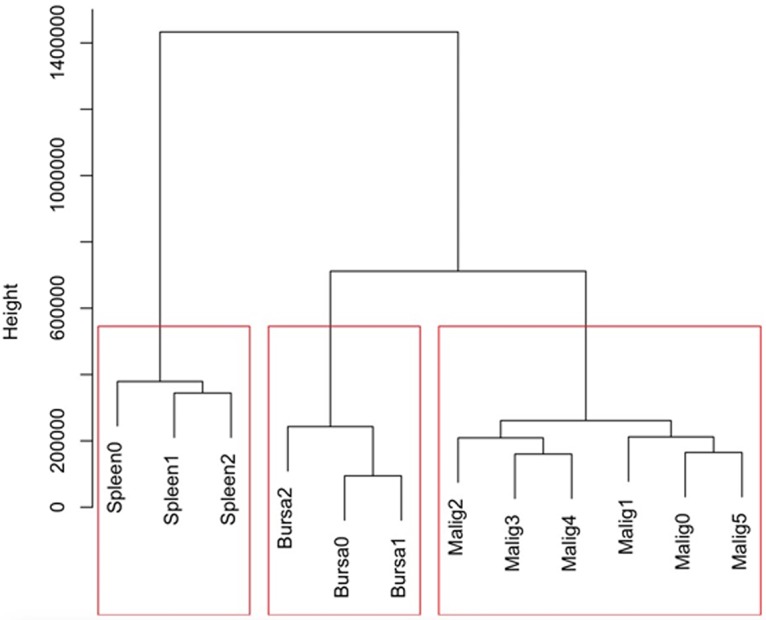
Ward hierarchical clustering by Euclidean distance between the malignant bursa samples (Malig-0 to Malig-5) and the control groups of nonmalignant bursa and splenic B-cell samples (Bursa-0 to Bursa-2 and Spleen-0 to Spleen-2). The result graphically showed that the malignant group was relatively more comparable to the nonmalignant bursa group, in contrast to the splenic B-cell group.
Gene expression profile of the LL-like lymphoma samples.
A total of 10,539 genes/transcripts were identified with a minimum pass filter read count of 10 or more in at least four of the twelve sequenced samples simultaneously. Of those, 521 and 260 expressed genes were exclusively observed in the LL-like lymphoma (tumor) samples and the normal control samples (both normal bursa tissues and splenic B cells), respectively (Fig. 4).
FIG 4.
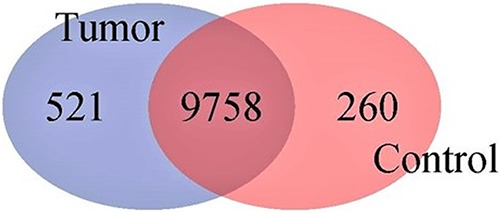
Venn diagram graphically illustrating profile statistics of genes identified by RNA-Seq between the spontaneous lymphoid lymphoma samples and the combined control samples (bursa and splenic B cells). Although most of the genes were expressed in both groups, hundreds of genes were exclusively expressed either in the tumor (lymphoid leukosis-like lymphomas) or the normal control samples.
Identification of differentially expressed genes between the LL-like lymphoma tissues and the normal control of nonmalignant bursa tissues.
The differential expression analysis was conducted between the malignant (LL-like lymphomas) and the nonmalignant bursa groups due to the relatively close compatibility between the LL-like lymphomas and this nonmalignant bursa control group (Fig. 3). A total of 923 genes/transcripts was identified differentially expressed between the LL-like lymphomas and normal bursa control group (0.5 < log2 fold change < −0.5 and P < 0.001). Of those, 196 genes were significantly more highly expressed and 727 genes were significantly less expressed in the LL-like lymphomas tissues than those in the control group of normal bursa tissues (see Table S1 in the supplemental material). A subset of the top genes significantly upregulated and downregulated was further closely examined and classified into categories of known functions, which include oncogenes, tumor suppressor genes, virus-associated genes, and immune response and cytokine genes. The oncogene category includes upregulated T-cell lymphoma invasion and metastasis 2 (TIAM2) and B-cell CLL/lymphoma 9-like (BCL9L) genes along with 11 other up- or downregulated oncogenes; the key tumor suppressor genes include downregulated tumor protein p63 (TP63) and suppression of tumorigenicity 14 (ST14) genes, along with 4 other genes. The key immune response cytokine genes include upregulated interleukin-18 (IL-18), class I histocompatibility antigen, F10 alpha chain-like (HA1F), and CD247 molecule (CD247), along with 15 other up- or downregulated genes (Table 3).
TABLE 3.
Selected differentially expressed genes with high statistical significance and known functions between the LL-like lymphomas and the control group of normal bursa tissues
| Ensemble IDa | Gene name | Gene description | Log2 fold change (M/N)b | P value | Fold change direction |
|---|---|---|---|---|---|
| Oncogenes | |||||
| 16391 | CNKSR2 | Connector enhancer kinase suppressor of Ras 2 | 2.7 | 1.67E−06 | Up |
| 36005 | TIAM2 | T-cell lymphoma invasion and metastasis 2 | 2.5 | 3.54E−20 | Up |
| 27789 | RAB37 | Ras-related protein Rab-27-like | 1.5 | 1.23E−11 | Up |
| 16817 | RASA3 | RAS p21 protein activator 3 | 1.3 | 5.66E−13 | Up |
| 14277 | RHOH | Ras homolog family member H | 1.3 | 2.37E−07 | Up |
| 07685 | BCL9L | B-cell CLL/lymphoma 9-like | 1.2 | 6.48E−05 | Up |
| 14913 | ROS1 | ROS proto-oncogene 1, receptor tyrosine kinase | −8.4 | 4.19E−69 | Down |
| 11664 | RASSF6 | Ras association domain family member 6 | −7.9 | 4.76E−53 | Down |
| 06076 | RASGEF1C | RasGEF domain family, member 1C | −3.6 | 2.88E−18 | Down |
| 36883 | MET | Proto-oncogene, receptor tyrosine kinase | −3.5 | 2.12E−26 | Down |
| 03129 | RAB11FIP1 | RAB11 family interacting protein 1 (class 1) | −3.5 | 6.97E−15 | Down |
| 00769 | RAB7B | RAB7B, member RAS oncogene family | −2.5 | 9.11E−14 | Down |
| 03503 | MYBL2 | MYB proto-oncogene like 2 | −1.0 | 1.92E−05 | Down |
| Tumor suppressor genes | |||||
| 11715 | HSPA2 | Heat shock 70-kDa protein 2 | 3.6 | 5.09E−15 | Up |
| 17071 | HSPH1 | Heat shock 105-kDa/110-kDa protein 1 | 1.2 | 4.57E−05 | Up |
| 32832 | DNAJC16 | DNAJ heat shock protein family member C16 | 1.0 | 4.77E−06 | Up |
| 07324 | TP63 | Tumor protein p63 | −2.2 | 2.02E−06 | Down |
| 01331 | ST14 | Suppression of tumorigenicity 14 | −1.7 | 3.83E−09 | Down |
| 03855 | SRC | v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog | −1.6 | 6.14E−09 | Down |
| Virus-associated genes | |||||
| 30025 | FABP4 | Fatty acid binding protein 4 | 3.4 | 4.79E−08 | Up |
| 14860 | YES1 | y-Yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | −2.8 | 1.81E−31 | Down |
| 16059 | ETS2 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 | −2.5 | 4.21E−22 | Down |
| 31529 | ERBB2 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 | −2.1 | 8.95E−07 | Down |
| 38154 | YAP1 | Yes-associated protein 1 | −1.3 | 2.51E−05 | Down |
| 03670 | MAFB | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B | −1.2 | 9.92E−09 | Down |
| Immune response genes and cytokines | |||||
| 14585 | ENSGALG14585 | Chemokine | 2.8 | 3.20E−17 | Up |
| 30270 | CD1C | CD1c molecule | 2.7 | 1.02E−15 | Up |
| 12545 | CYTIP | Cytohesin 1 interacting protein | 2.4 | 5.55E−11 | Up |
| 12292 | BANK1 | B-cell scaffold protein with ankyrin repeats 1 | 2.0 | 9.65E−08 | Up |
| 07418 | CD3D | CD3d molecule, delta | 1.9 | 5.22E−05 | Up |
| 07874 | IL18 | Interleukin-18 | 1.9 | 6.25E−06 | Up |
| 26466 | HA1F | Class I histocompatibility antigen, F10 alpha chain-like | 1.6 | 5.90E−07 | Up |
| 15441 | CD247 | CD247 molecule | 1.6 | 2.27E−05 | Up |
| 19322 | TNFRSF13C | Tumor necrosis factor receptor superfamily, member 13C | 1.4 | 9.25E−05 | Up |
| 28496 | NFKBID | NF-kappa-B-inhibitor delta-like | 1.2 | 1.60E−06 | Up |
| 09963 | LYZ | Lysozyme (renal amyloidosis) | −7.6 | 2.60E−62 | Down |
| 08552 | MAL | Mal, T-cell differentiation protein | −7.2 | 8.52E−53 | Down |
| 15348 | ALCAM | Activated leukocyte cell adhesion molecule | −5.5 | 3.02E−62 | Down |
| 11668 | IL8L1 | Interleukin 8-like 1 | −4.0 | 6.37E−18 | Down |
| 08554 | IL17REL | Interleukin 17 receptor E-like | −4.0 | 3.89E−12 | Down |
| 37851 | KK34 | Interleukin-like | −3.6 | 4.81E−09 | Down |
| 26663 | CX3CL1 | Chemokine (C-X3-C motif) ligand 1 | −3.3 | 1.09E−12 | Down |
| 06346 | CXCL14 | C-X-C motif chemokine ligand 14 | −3.2 | 2.59E−07 | Down |
| 38000 | CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | −3.2 | 3.96E−12 | Down |
| 01405 | IRF6 | Interferon regulatory factor 6 | −3.1 | 3.98E−08 | Down |
| 11418 | CCR6 | C-C motif chemokine receptor 6 | −3.0 | 5.32E−07 | Down |
| 21627 | IFI27L2 | Interferon, alpha-inducible protein 27-like 2 | −3.0 | 5.71E−15 | Down |
| 09392 | TLR5 | Toll-like receptor 5 | −2.8 | 4.70E−19 | Down |
| 07174 | TNFSF15 | Tumor necrosis factor superfamily member 15 | −2.8 | 3.31E−09 | Down |
| 29940 | IL-1beta | Interleukin-1β | −2.6 | 1.03E−08 | Down |
| 28466 | IL34 | Interleukin-34 | −2.6 | 1.77E−08 | Down |
| 25599 | CD24 | CD24 molecule | −2.6 | 7.91E−05 | Down |
| 43044 | IL1R1 | Interleukin-1 receptor, type 1 | −2.6 | 9.73E−19 | Down |
| 16785 | IL1RL1 | Interleukin-1 receptor-like 1 | −2.4 | 1.70E−07 | Down |
| 26098 | IL8L2 | Interleukin-8-like 2 | −2.4 | 1.89E−05 | Down |
| 05305 | ACKR2 | Atypical chemokine receptor 2 | −2.3 | 3.30E−12 | Down |
| 03733 | LIFR | Leukemia inhibitory factor receptor alpha | −2.2 | 6.10E−06 | Down |
| 11295 | SOCS2 | Suppressor of cytokine signaling 2 | −2.1 | 6.21E−06 | Down |
| 37413 | IL7 | Interleukin-7 | −1.9 | 1.40E−05 | Down |
| 00884 | CXXC5 | CXXC finger protein 5 | −1.9 | 1.22E−08 | Down |
| 17119 | TNFSF19 | Tumor necrosis factor receptor superfamily member 19 | −1.8 | 3.95E−06 | Down |
| 09612 | TGFB2 | Transforming growth factor, beta 2 | −1.7 | 5.49E−10 | Down |
| 03136 | IKZF2 | IKAROS family zinc finger 2 | −1.7 | 9.74E−05 | Down |
| 41621 | LY6E | Lymphocyte antigen 6 complex, locus E | −1.6 | 3.39E−09 | Down |
| 09179 | TNSF10 | Tumor necrosis factor superfamily member 10 | −1.5 | 3.23E−05 | Down |
| 11446 | TNFAIP2 | TNF-α-induced protein 2 | −1.5 | 1.50E−05 | Down |
| 06407 | TNFRSF23 | Death domain-containing tumor necrosis factor receptor superfamily member 23 | −1.5 | 3.42E−05 | Down |
| 30005 | IGSF1 | Immunoglobulin superfamily, member 1 | −1.2 | 5.69E−06 | Down |
Gene identifiers (ID) are truncated forms of their designations in the ENSEMBL database, e.g., 16391 is ENSGALG0000016391, 36005 is ENSGALG0000036005, etc.
M refers to malignant group, that is, the LL-like lymphoma group; N stands for nonmalignant group, that is, the control group of normal bursa tissues.
GO term and pathway enrichment by lists of differentially expressed genes between the LL-like tumors and the normal bursal controls.
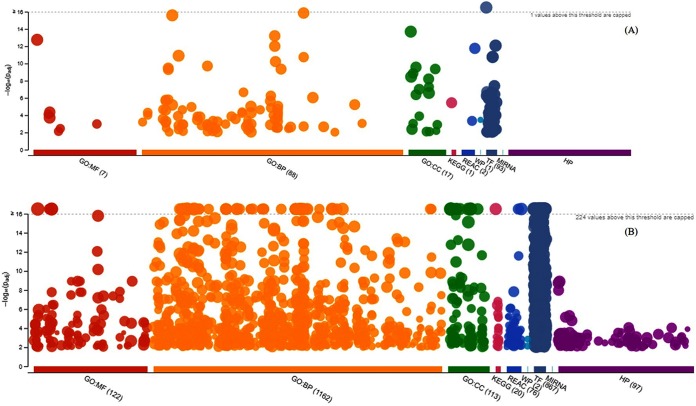
The upregulated genes were enriched in more than 100 Gene Ontology (GO) terms and a few pathways, while the downregulated genes were enriched in more than 1,000 GO terms and more than 100 pathways (Fig. 5). Both lists of differentially expressed genes were enriched across key molecular function, biological process, and cellular component terms (a complete list of GO terms and pathway enrichment outputs for the upregulated and downregulated genes is given in Table S2). Some of the identified most differentially expressed genes of known functions are involved in key GO terms and pathways, which include signal transduction (top oncogene category), apoptotic signaling pathway and regulation of apoptotic signaling pathway (tumor suppressor category), regulation of RNA biosynthetic process and regulation of nucleic acid-templated transcription (viral associated genes), and Toll-like receptor signaling pathways (immune response and cytokine gene category). A complete list of GO terms and pathways that are involved with some of the most differentially expressed genes is detailed in Table 4.
FIG 5.
Manhattan plot illustrating the differentially expressed gene-enriched GO terms (MF, molecular function; BP, biological process; and CC, cellular component) and KEGG pathways across reactome pathways (REAC), WiKi-Pathways (WP), transcription factor (TF), microRNA target base (MIRNA), and human phenotype ontology (HP) term categories. (A) Upregulated genes enriched in GO terms and pathways. (B) Downregulated genes enriched in GO terms and pathways. −log10(Padj), enrichment score calculated using the formula −log10(false discovery rate).
TABLE 4.
GO terms and pathways involved with some of the top differentially expressed genes between the LL-like lymphomas and normal control group of nonmalignant bursa tissues
| Domaina | GO term | Gene(s) | Upb | Genes | Downc |
|---|---|---|---|---|---|
| Oncogenes | |||||
| BP | Signaling | BCL9L, RHOH, CNKSR2, RASA3 | 4 | RAB7B, RASGEF1C, RASSF6, ROS1, MET | 5 |
| BP | Biological regulation | BCL9L, RHOH, CNKSR2, RASA3, TIAM2 | 5 | RAB7B, RASGEF1C, RASSF6, ROS1, MET, RAB11FIP3 | 6 |
| BP | Regulation of biological process | BCL9L, RHOH, CNKSR2, RASA3, TIAM2 | 5 | RAB7B, RASGEF1C, RASSF6, ROS1, MET, RAB11FIP3 | 6 |
| BP | Regulation of cellular process | BCL9L, RHOH, CNKSR2, RASA3, TIAM2 | 5 | RAB7B, RASGEF1C, RASSF6, ROS1, MET, RAB11FIP3 | 6 |
| BP | Signal transduction | BCL9L, RHOH, CNKSR2, RASA3 | 4 | RAB7B, RASGEF1C, RASSF6, ROS1, MET | 5 |
| Tumor suppressor genes | |||||
| BP | Intracellular estrogen receptor signaling pathway | 0 | SRC, TP63 | 2 | |
| BP | Apoptotic signaling pathway | HSPH1 | 1 | SRC, TP63 | 2 |
| BP | Regulation of apoptotic signaling pathway | HSPH1 | 1 | SRC, TP63 | 2 |
| Virus-associated genes | |||||
| BP | Regulation of gene expression | 0 | MAFB, YES1, ETS2, ERBB2, YAP1 | 5 | |
| BP | Nucleic acid-templated transcription | 0 | MAFB, YES1, ETS2, ERBB2, YAP1 | 5 | |
| BP | Transcription, DNA templated | 0 | MAFB, YES1, ETS2, ERBB2, YAP1 | 5 | |
| BP | Regulation of RNA biosynthetic process | 0 | MAFB, YES1, ETS2, ERBB2, YAP1 | 5 | |
| BP | Regulation of nucleic acid-templated transcription | 0 | MAFB, YES1, ETS2, ERBB2, YAP1 | 5 | |
| BP | Regulation of transcription, DNA templated | 0 | MAFB, YES1, ETS2, ERBB2, YAP1 | 5 | |
| BP | Positive regulation of transcription, DNA templated | 0 | YES1, ETS2, ERBB2, YAP1 | 4 | |
| BP | Positive regulation of gene expression | 0 | YES1, ETS2, ERBB2, YAP1 | 4 | |
| BP | Positive regulation of RNA biosynthetic process | 0 | YES1, ETS2, ERBB2, YAP1 | 4 | |
| BP | Positive regulation of nucleic acid-templated transcription | 0 | YES1, ETS2, ERBB2, YAP1 | 4 | |
| Immune response and cytokine genes | |||||
| BP | Regulation of signaling receptor activity | IL18 | 1 | TNFSF15, TGFB2, IL8L1, IL8L2, IL34, IL-1BETA | 6 |
| BP | Cytokine-mediated signaling pathway | IL18 | 1 | LIFR, ACKR2, IL17REL, CCR6, IL8L1, IL1RL1, IL8L2, IL1R1 | 8 |
| BP | Cell activation | IL18, BANK1 | 2 | CCR6, IL8L1, IL1RL1, IL8L2, IL-1BETA | 5 |
| BP | Leukocyte activation | IL18, BANK1 | 2 | CCR6, IL8L1, IL1RL1, IL8L2, IL-1BETA | 5 |
| BP | Regulation of signaling | IL18, BANK1 | 2 | CXXC5, TNFSF15, TLR5, TGFB2, SOCS2, IL8L1, IL1RL1, IL8L2, IL34, IL-1BETA | 10 |
| BP | Regulation of signal transduction | IL18, BANK1 | 2 | CXXC5, TNFSF15, TLR5, TGFB2, SOCS2, IL8L1, IL1RL1, IL8L2, IL34, IL-1BETA | 10 |
| BP | Immune system process | IL18, BANK1, CD247 | 3 | CXCL14, TNFSF15, TLR5, TGFB2, CCR6, IL8L1, ALCAM, IL1RL1, IL8L2, CX3CL1, IL34, IL-1BETA | 12 |
| BP | Regulation of immune system process | IL18, BANK1, CD247 | 3 | CXCL14, TLR5, TGFB2, CCR6, IL8L1, IL1RL1, IL8L2, IL34, IL-1BETA | 9 |
| MF | Signaling receptor binding | IL18, BANK1, CD247 | 3 | LIFR, CXCL14, TNFSF15, TLR5, TGFB2, SOCS2, IL8L1, IL8L2, CX3CL1, IL34, IL-1BETA | 11 |
| BP | Signaling | IL18, BANK1, CD247, CD3D | 4 | CXXC5, LIFR, ACKR2, CXCL14, TNFSF15, IL17REL, TLR5, TGFB2, SOCS2, CCR6, IL8L1, IL1RL1, IL8L2, IL34, IL-1BETA, IL1R1 | 16 |
| BP | Signal transduction | IL18, BANK1, CD247, CD3D | 4 | CXXC5, LIFR, ACKR2, TNFSF15, IL17REL, TLR5, TGFB2, SOCS2, CCR6, IL8L1, IL1RL1, IL8L2, IL34, IL-1BETA, IL1R1 | 15 |
| BP | Immune response | IL18, CD247 | 2 | CXCL14, TNFSF15, TLR5, TGFB2, CCR6, IL8L1, ALCAM, IL1RL1, IL8L2, CX3CL1, IL-1BETA | 11 |
| BP | Positive regulation of immune system process | IL18, CD247 | 2 | CXCL14, TLR5, IL8L1, IL1RL1, IL8L2, IL34, IL-1BETA | 7 |
| BP | Cell surface receptor signaling pathway | IL18, CD247, CD3D | 3 | LIFR, ACKR2, IL17REL, TGFB2, SOCS2, CCR6, IL8L1, IL1RL1, IL8L2, IL1R1 | 10 |
| keg | KEGG pathways | IL18, CD1C, TNFRSF13C | 3 | LIFR, CXCL14, TNFSF15, TLR5, TGFB2, CCR6, IL8L1, ALCAM, IL8L2, CX3CL1, IL-1BETA, IL7, CX3CR1, IL1R1 | 14 |
| keg | Cytokine-cytokine receptor interaction | IL18, TNFRSF13C | 2 | LIFR, CXCL14, TNFSF15, TGFB2, CCR6, IL8L1, IL8L2, CX3CL1, IL-1BETA, IL7, CX3CR1, IL1R1 | 12 |
| BP | Chemokine-mediated signaling pathway | 0 | ACKR2, CCR6, IL8L1, IL8L2 | 4 | |
| BP | Leukocyte migration | 0 | CXCL14, CCR6, IL8L1, IL8L2, IL-1BETA | 5 | |
| BP | Regulation of leukocyte migration | 0 | CXCL14, CCR6, IL8L1, IL8L2 | 4 | |
| keg | Toll-like receptor signaling pathway | 0 | TLR5, IL8L1, IL8L2, IL-1BETA | 4 |
MF, molecular function; BP, biological process; keg, KEGG pathways.
Up, upregulated.
Down, downregulated.
DISCUSSION
Spontaneous LL-like lymphomas were reported 4 decades ago, having been observed in experimental lines of chickens in the absence of exogenous ALV but with the presence of endogenous ALV (6, 7). Subsequent studies reported that Marek’s disease virus serotype 2, commonly used as one of the bivalent or trivalent Marek’s disease vaccines, augments the incidence of lymphoid leukosis induced by exogenous ALV (41) and the incidence of LL-like lymphomas in chickens free of both exogenous and endogenous ALVs (11, 43).
The putative mechanism had been postulated by researchers encompassing multiple possibilities, whereby MDV-2 could exert influence on bursal cells, which might subsequently modulate the transformation process of B cells alone or in combination with the endogenous, exogenous, or both ALVs. In fact, integration of exogenous herpesviruses into the host genome activates expression of endogenous retroviral genes within the host, as evidenced by Epstein-Barr virus (EBV) (53–55) and Marek’s disease virus (56). Reportedly, MDV-2 only infects and persists in ALV-transformed B cells (45). This supports a model of intracellular cooperation between MDV and ALV resulting in the augmentation of lymphoma development (45). Typically, ALV-E alone demonstrates little to no oncogenicity (24), possibly due to weak promoter activity of the LTR. Coinfection of MDV-2 with ALV results in activation of the retrovirus long terminal repeat by MDV-2. Studies have shown that MDV-2 activates the Rous sarcoma virus LTR (RSV-LTR) promoter 2- to 5-fold more efficiently than serotype 1 MDV (MDV-1) or serotype 3 MDV (MDV-3) (57). Similarly, MDV activates the ALV-LTR promoter, leading to increased expression of ALV RNA, proteins, and infectious viruses in cultured cells (44).
The ALV LTR serves as either a promoter or activator of c-myc (58, 59), c-erbB (60), and c-myb when infection occurs at embryonation (61). The Myc transcription factor induces cell growth and proliferation by influencing the expression of cell cycle regulatory genes (62). ALV induces bursa lymphomas in lymphoma-susceptible strains of chickens after proviral integration within the c-myc gene and subsequent expansion of Myc-overexpressing lymphocytes within transformed follicles (58, 63). Transformed follicles grow much more rapidly than normal follicles following ALV infection. This is not solely due to overexpression of Myc; however, susceptible transformed follicles grow much larger than normal ones because following ALV infection, B-cell differentiation and bursal emigration are blocked at an embryonic stage (64). It is speculated that endogenous ALV LTR genes integrate near c-myc and activate c-myc expression similarly to exogenous ALV LTR. The c-myc transcription levels, however, may be lower following endogenous ALV-LTR integration than exogenous ALV-LTR (65).
This report, in part, characterized the two endogenous ALV field isolates, designated AF227 and AF229, which were isolated from commercial broiler breeder flocks after the spontaneous LL-like lymphomas were observed on the farm. To verify if the new ALV-E isolates alone or in combination with the MDV-2 vaccine are responsible, to an extent, for the incidence of spontaneous LL-like lymphomas, birds from different commercial lines, other than the flocks from which AF227 and AF229 were isolated (those flocks of birds were no longer available at the time of the challenge trials of this study), and a fully susceptible chicken line, developed and maintained on the ADOL farm and known as the RFS line and free of any endogenous ev genes (46), were infected with each of the ALV-E field isolates or inoculated with the MDV-2 vaccine or a combination of the ALV-E isolate and the MDV-2 vaccine. There was no incidence of spontaneous LL-like lymphomas observed in the commercial broiler breeder chickens following infection with either AF229 or AF229 with MDV-2 vaccination in this study (Table 2). No significant difference in LL-like lymphoma incidence was detected between the AF227 isolate group and the MDV-2 vaccination group (P > 0.05) in the RFS birds, as shown in Table 1. However, each ALV-E isolate in combination with MDV-2 vaccine significantly accelerated the incidences of spontaneous LL-like lymphomas in the RFS birds, in contrast to the sole AF227 isolate infection and the sole MDV-2 vaccination groups (Tables 1 and 2; P < 0.05).
Genetic resistance to ALV is multifactorial and depends upon resistance to infection and resistance to tumor development (32, 33, 39). The commercial broiler breeder chickens appear to be resistant to tumor development since they were susceptible to ALV-E infection, as evidenced by the presence of the ALV-E receptor TVB*S1/*S1, and they were viremia tolerant following infection at embryonation. Parghi et al. (66) identified that in ALV-resistant chickens, ALV LTR-enhanced c-myc gene expression is reduced, resulting in normal B-cell differentiation, normal follicle development, and posthatch bursal emigration in resistant transformed bursa follicles. We did not analyze bursa tissue from the commercial broiler breeder chickens to determine the level of gene transcription in transformed bursa follicles. It would, however, be interesting to note the levels of gene expression in transformed bursa follicles between susceptible and resistant chickens.
We did assess the whole-genome transcriptomic levels between the LL-like bursa lymphomas collected from the RFS birds after SB-1 and AF227 inoculation and the normal control group of bursa tissue samples of the same line of birds. Next-generation sequence analysis resulted in hundreds of differentially expressed genes/transcripts for the first time between LL-like lymphoma samples and the normal bursa tissue samples, which include genes of known function in the categories of oncogenes, tumor suppressor genes, virus-associated genes, and immune response and cytokine genes. A minimum of six oncogenes were upregulated and seven oncogenes downregulated in the spontaneous LL-like lymphoma samples, in contrast to the normal control group of nonmalignant bursa tissues and in addition to up- and downregulated genes with categorically known functions of tumor suppression, viral functions, immune responses, and activities of cytokines (Table 3). Further bioinformatics analyses showed that some of those differentiated genes reportedly are involved in key GO terms and pathways, including signaling pathways, signal transduction, immune response, and KEGG pathways.
The upregulated known oncogenes and downregulated tumor suppressor genes represented only a small subset of the 923 genes significantly expressed between the LL-like lymphoma samples and the nonmalignant normal bursa tissues. Sequence analyses of higher depth than those of this study and additional LL-like lymphoma samples for such analyses are warranted and necessary to advance the understanding of the mechanism at genomic levels on what constitutes the LL-like lymphoma susceptibility and how a nonpathogenic subgroup of ALV in conjunction with little or no pathogenic MDV-2 jointly boost the incidence of LL-like lymphomas in susceptible chickens.
In summary, we have isolated two endogenous ALV field isolates capable of inducing spontaneous LL-like lymphomas in susceptible chickens in conjunction with MDV-2. We have characterized the two ALV field isolates at the genomic level, designated the isolates AF227 and AF229. The genomic sequence analyses of the AF227 and AF229 isolates showed these two ALV isolates belong to subgroup E ALV. RNA sequencing analyses of the spontaneous LL-like lymphomas induced by AF227 and SB-1 in an experimental line of birds under controlled conditions and normal bursa tissues of the same genetic line of birds resulted in a total of 923 differentially expressed genes between the two groups. Some of the differentially expressed genes with known functions of oncogenicity and tumor suppression, association with viral functions, immune responses, and cytokine activities are involved with multiple key pathways, including signaling, signal transduction, and KEGG pathways.
MATERIALS AND METHODS
Virus isolation.
Plasma, spleen, and liver samples of eight breeders of a broiler breeder flock were received from a commercial farm at which spontaneous lymphoid leukosis-like bursal lymphoma incidences were observed. The samples were tested for virus growth using chicken embryo fibroblasts (CEF) from ADOL-specific pathogen-free lines of chickens, RFS (C/0) and line 0 (C/E), and followed by PCR using viral subgroup-specific primers. Aliquots of plasma, spleen, and liver homogenates were seeded onto secondary CEF from line 0 (9) and line RFS (46) for virus isolation. CEFs were maintained in Leibovitz L-15 medium plus McCoy 5A medium (1:1), supplemented with 1% bovine serum and antibiotics for 10 to 14 days before harvest of cell-free viruses. Two isolates, designated AF227 and AF229, were obtained and were further characterized molecularly and biologically. Total viral DNA from CEF-infected cells was extracted using standard proteinase K, phenol-chloroform extraction procedures. The viral DNA was analyzed by PCR using ALV subgroup-specific primers as described by Silva et al. (67) to determine which subgroup or subgroups of ALV were present in the viral DNA sample.
Viral DNA sequence analysis.
The proviral DNA samples from the infected CEFs were sequenced at the Research Technology Support Facility, Michigan State University (East Lansing, MI). Contigs were constructed using Sequencer (Gene Codes Corp., Ann Arbor, MI). DNA sequences were aligned using the Clustal W model in the MegAlign program of Lasergene (version 11; DNASTAR, Inc., Madison, WI). Phylogenetic relatedness was also calculated using the MegAlign program. The sequence analyses included the isolates AF227 and AF229, along with the prototype ALV-E virus RAV-0 (24), partial sequence of the recombinant ALV-E strain RAV-60 (68), and ALV subgroup A strain RAV-1 (69).
Lines of chickens used in the challenge trials.
Chickens from the line RFS were used in this study, since they are free of endogenous viruses, TVB*S1/*S1 homozygous, and thus are fully susceptible to infection of all subgroups of ALV viruses, including the ALV-E subgroup (46). Fertile eggs were also obtained from a commercial broiler breeder, which were incubated on the ADOL farm, and the chickens hatched from the eggs were included in one of the experiments. We note that the fertile eggs obtained this time were from the same commercial broiler breeder from which the specimens were received earlier for the virus isolations of AF227 and AF229, but the fertile eggs received this time for the experiment were from a different broiler flock housed on a different farm rather than the original flock and farm from which the spontaneous LL-like tumor incidences were observed or AF227 and AF229 were isolated. Two challenge trials were conducted. One was with chickens only from the ADOL line RFS and the other with chickens from both ADOL line RFS and the commercial broiler breeder eggs hatched on the ADOL farm. All of the birds in each experiment from each line were housed in a biosafety level 2 facility on the ADOL farm. Feed and water were supplied ad libitum.
A challenge trial to test the pathogenicity of ALV isolate AF227 in RFS chickens.
Sampled fertile eggs from the ADOL line RFS chickens were divided into four groups. One group of the embryos, a control group, was inoculated with 100 μl of sterile PBS, and two of the groups were inoculated with 100 μl of ALV-E isolate AF227 at 1,000 50% tissue culture infectious doses/0.1 ml per bird via yolk sac at 7 DOE. At 1 day of age, the other group of chickens that did not receive any treatment and one of the groups that had received AF227 inoculation at 7 DOE was given 500 PFU each of the MDV-2 vaccine SB-1 intraperitoneally. Chickens of the different treatment groups were housed in separate isolators and monitored up to 50 weeks of age. At 2 weeks of age, blood samples were collected from a subset of chickens from each group, and buffy coats were analyzed for the presence of MDV-2-specific plaques as described by Aly et al. (40).
A challenge trial to test the pathogenicity of AF227 and AF229 ALV isolates in commercial breeder broilers and RFS chickens.
The received commercial broiler fertile eggs were divided into five groups in incubation. One group received PBS (control), two groups received AF229, and one group received AF227 at 7 DOE. At 1 day of age, the group of birds pretreated with AF227 and the group of birds that did not receive any treatment at 7 DOE, along with one group of birds pretreated with AF229, were vaccinated with MDV-2 as described above. Fertilized eggs from line RFS were divided into three groups. One group was given PBS (control), and the other two groups were given AF227 or AF229 at 7 DOE. The latter two groups were inoculated with SB-1 vaccine at 1 day of age as described above. All chickens were bled at 4 weeks of age, and all surviving chickens were bled at 52 weeks of age prior to termination. The procedures for handling and sampling of the chickens were preapproved by the ADOL Animal Care and Use Committee (ACUC). The experimental chickens were monitored daily throughout the 52-week experiment period, and all moribund chickens were euthanized humanely by following American Veterinary Medical Association-approved methods.
Virus and antibody assays.
The plasma samples were assayed for infectious exogenous and endogenous ALV as well as the presence of antibody by preestablished procedures described previously by Fadly and Witter (70). The presence or absence of MDV-2 in individual chickens was determined by cocultivation of 1 × 106 buffy coat cells from centrifuged and heparinized blood samples onto duck embryo fibroblasts (DEF), as described previously by Aly et al. (40).
Pathology examinations.
All chickens that died during experiments or were euthanized at the end of the experiments were subjected to individual necropsy. Lymphomas were diagnosed on the basis of visual and histological examinations of tissues with gross tumors or suspicious microtumors, respectively (3).
TVB genotyping by pyrosequencing.
TVB genotypes of all the commercial broilers included in this experiment were determined by pyrosequencing analysis by following procedures described by Zhang et al. (22). Briefly, short PCR amplicons were generated from the broiler DNA samples with a pair of primers, of which one of the primers was biotinylated at the 5′ end. The PCR products then were subjected to binding, shaking, annealing, washing, and denaturation processes. The end products of the PCR amplicons were biotin-labeled and single-stranded DNA, which were then analyzed on a PSQ 96MA pyrosequencer system (Qiagen, Inc., MD) for TVB genotypes. There are six commonly observed TVB genotypes in commercial birds, which are TVB*S1/*S1, TVB*S1/*S3, TVB*S1/*R, TVB*S3/*S3, TVB*S3/*R, and TVB*R/*R.
Tissue samples, B-cell isolation, and total RNA extraction.
LL-like bursal lymphoma tissues were collected from six RFS chickens treated with AF227 at 7 DOE and SB-1 on the day of hatch during postmortem examination between 32 and 43 weeks of age (see Table S3 in the supplemental material). Fresh normal bursal tissues from three 3-week-old noninoculated RFS line chickens were also collected. Splenic B cells from three noninfected and age-matched RFS line chickens were isolated using a magnetically activated cell sorting (MACS) cell separation system by following the manufacturer’s instructions (Miltenyi Biotech, San Diego, CA). Briefly, the spleen tissues were first individually homogenized. Separated cells were stained with a fluorescein isothiocyanate (FITC)-conjugated primary antibody. Subsequently, the cells were magnetically labeled with anti-FITC microbeads. The cell suspension was loaded onto a MACS column, and labeled cells were separated via a MACS separator. Total RNA was extracted from tissue homogenates and the B cells using an RNeasy kit as recommended by the manufacturer (Qiagen, Valencia, CA). The last two groups of samples, the normal bursal tissues and the splenic B cells, served as the normal control groups of the LL-like bursal lymphoma samples in RNA-Seq analysis.
Next-generation sequencing analysis.
Total RNAs of the bursal lymphoma tissue, the normal bursal tissues, and the splenic B cells were subjected to NGS analysis. NGS libraries were built using Illumina’s TruSeq stranded mRNA library preparation kit by following the manufacturer’s instructions (Illumina, San Diego, CA). Sequencing was performed on an Illumina HiSeq 2500 machine running in high-output mode in a 2× 100-bp paired-end format using an Illumina TruSeq PE cluster kit (v3) and TruSeq SBS kit (v3). The raw reads were quality assessed using FastQC, version 0.11.2 (http://www.bioinformatics.babraham.ac.uk/), and adaptors were removed using Trimmomatic, version 0.30 (71). Low-quality bases were trimmed using custom Python scripts to remove the first 13 nucleotides, and Sickle v1.33 (72) was used, under a sliding window with an average quality score of 30, for removal of reads with N nucleotides and reads that were under the 50-bp minimum read length threshold. The good-quality reads were mapped to a combined reference sequence containing the chicken (galGal4) genome (73), SB-1 (GenBank accession no. HQ840738.1) (74), and Rous sarcoma virus (NCBI reference sequence NC_001407.1) (75) with Ensembl annotation (76) using TopHat2, version 2.0.8b (77), and Bowtie2, version 2.1.0 (78). This resulted in approximately 33,806,146 minimum reads, 55,558,773 median reads, and 118,577,764 maximum reads in the alignments of the 12 sample libraries. The alignments were subjected to subsequent analysis with CuffDiff2, version 2.2.0 (79), to identify genes that were differentially expressed between the LL-like bursal lymphoma group and the control group. Ensembl gene identifiers for the differentially expressed genes were used to form the upregulated and downregulated gene lists, which were used as the input for Gene Ontology and pathway enrichment analyses using both custom R scripts and the g:Profiler online resources (80).
Reconstruction of AF227 genome from the LL-like lymphoma RNA-Seq data.
The RNA-Seq reads of the LL-like lymphoma samples were aligned to the NCBI viral genomes database (81) using BLAST+, version 2.2.28 (82). All reads that matched to an annotated avian virus were retained, pooled, and then assembled into a reconstructed version of the AF227 genome with Trinity, version 20140413p1 (83). All next-generation sequence data processes were performed at the Michigan State University High-Performance Computing Facility (East Lansing, MI).
Data availability.
The complete genomic sequences have been deposited in GenBank under accession numbers MF817820 for AF227, MF817821 for AF229, MF817822 for RAV-0, MF817823 for a partial RAV-60 sequence, and MF926337 for RAV-1.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of Melanie Flesberg and Bernice Li in conducting the challenge trials, performance of ALV assays, and preparation of total RNA samples for RNA deep sequencing for this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00861-19.
REFERENCES
- 1.Rispens BH, de Boer GF, Hoogerbrugge A, van Vioten J. 1976. A method for the control of lymphoid leukosis in chickens. J Natl Cancer Inst 57:1151–1156. doi: 10.1093/jnci/57.5.1151. [DOI] [PubMed] [Google Scholar]
- 2.de Boer GF, van Vloten J, Groenendal JE, Maas HJ, Borm F, Hoogerbrugge A, Krasselt M. 1979. The control of lymphoid leukosis in a flock White Plymouth Rock chickens. Tijdschr Diergeneeskd 104:23–28. doi: 10.1080/01652176.1979.9693717. [DOI] [PubMed] [Google Scholar]
- 3.Fadly A, Nair V. 2008. Leukosis/sarcoma group, p 514–568. In Saif Y, Fadly A, Glisson JR, McDougald LR, Nolan LK, Swayne DE (ed), Diseases of poultry, 12th ed Blackwell Publishing, Ames, IA. [Google Scholar]
- 4.Payne GS, Bishop JM, Varmus HE. 1982. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 5.Baba TW, Humphries EH. 1986. Selective integration of avian leukosis virus in different hematopoietic tissues. Virology 155:557–566. doi: 10.1016/0042-6822(86)90216-3. [DOI] [PubMed] [Google Scholar]
- 6.Crittenden LB, Witter RL, Fadly AM. 1979. Low incidence of lymphoid tumors in chickens continuously producing endogenous virus. Avian Dis 23:646–653. doi: 10.2307/1589740. [DOI] [PubMed] [Google Scholar]
- 7.Crittenden LB, Witter RL, Okazaki W, Neiman PE. 1979. Lymphoid neoplasms in chicken flocks free of infection with exogenous avian tumor viruses. J Natl Cancer Inst 63:191–200. [PubMed] [Google Scholar]
- 8.Astrin SM, Buss EG, Haywards WS. 1979. Endogenous viral genes are non-essential in the chicken. Nature 282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 9.Crittenden LB, Fadly AM. 1985. Responses of chickens lacking or expressing endogenous avian leukosis virus genes to infection with exogenous virus. Poult Sci 64:454–463. doi: 10.3382/ps.0640454. [DOI] [PubMed] [Google Scholar]
- 10.Salter DW, Crittenden LB. 1989. Artificial insertion of a dominant gene for resistance to avian leukosis virus into the germ line of the chicken. Theor Appl Genet 77:457–461. doi: 10.1007/BF00274263. [DOI] [PubMed] [Google Scholar]
- 11.Salter DW, Payne W, Kung HJ, Robinson D, Ewert D, Olson W, Crittenden LB, Fadly AM. 1999. Enhancement of spontaneous bursal lymphoma frequency by serotype 2 Marek’s disease vaccine, SB-1, in transgenic and non-transgenic line 0 white leghorn chickens. Avian Pathol 28:147–154. doi: 10.1080/03079459994876. [DOI] [PubMed] [Google Scholar]
- 12.Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME. 1991. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol 72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 13.Vogt PK. 1997. Historical introduction to the general properties of retroviruses In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 14.Weiss RA. 1981. Retrovirus receptors In Longberg-Holm K, Philipson L (ed), Virus receptors. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 15.Cui N, Su S, Chen Z, Zhao X, Cui Z. 2014. Genomic sequence analysis and biological characteristics of a rescued clone of avian leukosis virus strain JS11C1, isolated from indigenous chickens. J Gen Virol 95:2512–2522. doi: 10.1099/vir.0.067264-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Zhao P, Cui ZZ. 2012. Identification of a new subgroup of avian leukosis virus isolated from Chinese indigenous chicken breeds. Bing Du Xue Bao 28:609–614. [PubMed] [Google Scholar]
- 17.Rovigatti VG, Astrin SM. 1983. Avian endogenous viral genes. Curr Top Microbiol Immunol 103:1–21. [DOI] [PubMed] [Google Scholar]
- 18.Johnson JA, Heneine W. 2001. Characterization of endogenous avian leukosis viruses in chicken embryonic fibroblast substrates used in production of measles and mumps vaccines. J Virol 75:3605–3612. doi: 10.1128/JVI.75.8.3605-3612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benkel BF. 1998. Locus-specific diagnostic tests for endogenous avian leukosis-type viral loci in chickens. Poult Sci 77:1027–1035. doi: 10.1093/ps/77.7.1027. [DOI] [PubMed] [Google Scholar]
- 20.Crittenden LB, Astrin SM. 1981. Genes, viruses, and avian leukosis. BioScience 31:305–310. doi: 10.2307/1308148. [DOI] [Google Scholar]
- 21.Crittenden LB, Astrin SM, Smith EJ. 1983. Independent segregation of ev 10 and ev 11, genetic loci for spontaneous production of endogenous avian retroviruses. Virology 129:514–516. doi: 10.1016/0042-6822(83)90192-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhang HM, Bacon LD, Heidari M, Muir WM, Groenen MAM, Zhang Y, Wong GKS, Fulton JE, O'Sullivan NP, Albers GAA, Vereijken ALJ, Rattink AP, Okimoto R, McKay JC, McLeod S, Cheng HH. 2007. Genetic variation at the tumour virus B locus in commercial and laboratory chicken populations assessed by a medium-throughput or a high-throughput assay. Avian Pathol 36:283–291. doi: 10.1080/03079450701449248. [DOI] [PubMed] [Google Scholar]
- 23.Hunt H, Fadly A, Silva R, Zhang H. 2008. Survey of endogenous virus and TVB receptor status of commercial chicken stocks supplying specific-pathogen free eggs. Avian Dis 52:433–440. doi: 10.1637/8183-112907-Reg.1. [DOI] [PubMed] [Google Scholar]
- 24.Motta JV, Crittenden LB, Purchase HG, Stone HA, Witter RL. 1975. Low oncogenic potential of avian endogenous RNA tumor virus infection or expression. J Natl Cancer Inst 55:685–689. doi: 10.1093/jnci/55.3.685. [DOI] [PubMed] [Google Scholar]
- 25.Crittenden LB, Hayward WS, Hanafusa H, Fadly AM. 1980. Induction of neoplasms by subgroup E recombinants of exogenous and endogenous avian retroviruses (Rous-associated virus type 60). J Virol 33:915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson HL, Astrin SM, Senior AM, Salazar FH. 1981. Host susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J Virol 40:745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crittenden LB, Fadly AM, Smith EJ. 1982. Effect of endogenous leukosis virus genes on response to infection with avian leukosis and reticuloendotheliosis viruses. Avian Dis 26:279–294. doi: 10.2307/1590097. [DOI] [PubMed] [Google Scholar]
- 28.Crittenden LB, Smith EJ, Fadly AM. 1984. Influence of endogenous viral (ev) gene expression and strain of exogenous avian leukosis virus (ALV) on mortality and ALV infection and shedding in chickens. Avian Dis 28:1037–1056. doi: 10.2307/1590280. [DOI] [PubMed] [Google Scholar]
- 29.Smith EJ, Fadly AM. 1988. Influence of congenital transmission of endogenous virus-21 on the immune response to avian leukosis virus infection and the incidence of tumors in chickens. Poult Sci 67:1674–1679. doi: 10.3382/ps.0671674. [DOI] [PubMed] [Google Scholar]
- 30.Bacon LD. 1987. Influence of the major histocompatibility complex on disease resistance and productivity. Poult Sci 66:802–811. doi: 10.3382/ps.0660802. [DOI] [PubMed] [Google Scholar]
- 31.Barnard RJ, Elleder D, Young JA. 2006. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology 344:25–29. doi: 10.1016/j.virol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 32.Crittenden LB. 1975. Two levels of genetic resistance to lymphoid leukosis. Avian Dis 19:281–292. doi: 10.2307/1588982. [DOI] [PubMed] [Google Scholar]
- 33.Pinard-van der Laan MH, Soubieux D, Merat L, Bouret D, Luneau G, Dambrine G, Thoraval P. 2004. Genetic analysis of a divergent selection for resistance to Rous sarcomas in chickens. Genet Sel Evol 36:65–81. doi: 10.1051/gse:2003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crittenden LB. 1991. Retroviral element in the genome of the chickens: implications for poultry genetics and breeding. Crit Rev Poultry Biol 3:73–109. [Google Scholar]
- 35.Plachy J, Reinisova M, Kucerova D, Senigl F, Stepanets V, Hron T, Trejbalova K, Elleder D, Hejnar J. 2017. Identification of New World quails susceptible to infection with avian leukosis virus subgroup J. J Virol 91:e02002-16. doi: 10.1128/JVI.02002-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W, Liu Y, Li H, Chang S, Shu D, Zhang H, Chen F, Xie Q. 2015. Intronic deletions of tva receptor gene decrease the susceptibility to infection by avian sarcoma and leukosis virus subgroup A. Sci Rep 5:9900. doi: 10.1038/srep09900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adkins HB, Brojatsch J, Young J. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J Virol 74:3572–3578. doi: 10.1128/JVI.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne LN, Pani PK, Weiss RA. 1971. A dominant epistatic gene which inhibits cellular susceptibility to RSV(RAV-O). J Gen Virol 13:455–462. doi: 10.1099/0022-1317-13-3-455. [DOI] [PubMed] [Google Scholar]
- 39.Bacon LD, Hunt HD, Cheng HH. 2000. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult Sci 79:1082–1093. doi: 10.1093/ps/79.8.1082. [DOI] [PubMed] [Google Scholar]
- 40.Aly MM, Witter RL, Fadly AM. 1996. Enhancement of reticuloendotheliosis virus-induced bursal lymphomas by serotype 2 Marek’s disease virus. Avian Pathol 25:81–94. doi: 10.1080/03079459608419122. [DOI] [PubMed] [Google Scholar]
- 41.Bacon LD, Witter RL, Fadly AM. 1989. Augmentation of retrovirus-induced lymphoid leukosis by Marek’s disease herpesviruses in White Leghorn chickens. J Virol 63:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fadly AM, Witter RL. 1993. Effects of age at infection with serotype 2 Marek’s disease virus on enhancement of avian leukosis virus-induced lymphomas. Avian Pathol 22:565–576. doi: 10.1080/03079459308418944. [DOI] [PubMed] [Google Scholar]
- 43.Cao W, Mays J, Kulkarni G, Dunn J, Fulton RM, Fadly A. 2015. Further observations on serotype 2 Marek’s disease virus-induced enhancement of spontaneous avian leukosis virus-like bursal lymphomas in ALVA6 transgenic chickens. Avian Pathol 44:23–27. doi: 10.1080/03079457.2014.989195. [DOI] [PubMed] [Google Scholar]
- 44.Pulaski JT, Tieber VL, Coussens PM. 1992. Marek’s disease virus-mediated enhancement of avian leukosis virus gene expression and virus production. Virology 186:113–121. doi: 10.1016/0042-6822(92)90065-W. [DOI] [PubMed] [Google Scholar]
- 45.Fynan E, Block TM, DuHadaway J, Olson W, Ewert DL. 1992. Persistence of Marek’s disease virus in a subpopulation of B cells that is transformed by avian leukosis virus, but not in normal bursal B cells. J Virol 66:5860–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Bacon LD, Fadly AM. 2008. Development of an endogenous virus-free line of chickens susceptible to all subgroups of avian leukosis virus. Avian Dis 52:412–418. doi: 10.1637/8180-112707-Reg. [DOI] [PubMed] [Google Scholar]
- 47.Bai J, Howes K, Payne LN, Skinner MA. 1995. Sequence of host-range determinants in the env gene of a full-length, infectious proviral clone of exogenous avian leukosis virus HPRS-103 confirms that it represents a new subgroup (designated J). J Gen Virol 76:181–187. doi: 10.1099/0022-1317-76-1-181. [DOI] [PubMed] [Google Scholar]
- 48.Dorner AJ, Coffin JM. 1986. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell 45:365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- 49.Dorner AJ, Stoye JP, Coffin JM. 1985. Molecular basis of host range variation in avian retroviruses. J Virol 53:32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva RF, Fadly AM, Hunt HD. 2000. Hypervariability in the envelope genes of subgroup J avian leukosis viruses obtained from different farms in the United States. Virology 272:106–111. doi: 10.1006/viro.2000.0352. [DOI] [PubMed] [Google Scholar]
- 51.Bova CA, Olsen JC, Swanstrom R. 1988. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol 62:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valsesia-Wittmann S, Drynda A, Deleage G, Aumailley M, Heard JM, Danos O, Verdier G, Cosset FL. 1994. Modifications in the binding domain of avian retrovirus envelope protein to redirect the host range of retroviral vectors. J Virol 68:4609–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiao FC, Tai AK, Deglon A, Sutkowski N, Longnecker R, Huber BT. 2009. EBV LMP-2A employs a novel mechanism to transactivate the HERV-K18 superantigen through its ITAM. Virology 385:261–266. doi: 10.1016/j.virol.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 54.Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL. 2002. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res 86:93–100. doi: 10.1016/S0168-1702(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 55.Sutkowski N, Chen G, Calderon G, Huber BT. 2004. Epstein-Barr virus latent membrane protein LMP-2A is sufficient for transactivation of the human endogenous retrovirus HERV-K18 superantigen. J Virol 78:7852–7860. doi: 10.1128/JVI.78.14.7852-7860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu X, Zhu W, Chen S, Liu Y, Sun Z, Geng T, Wang X, Gao B, Song C, Qin A, Cui H. 2016. Expression of the env gene from the avian endogenous retrovirus ALVE and regulation by miR-155. Arch Virol 161:1623–1632. doi: 10.1007/s00705-016-2833-8. [DOI] [PubMed] [Google Scholar]
- 57.Tieber VL, Zalinskis LL, Silva RF, Finkelstein A, Coussens PM. 1990. Transactivation of the Rous sarcoma virus long terminal repeat promoter by Marek’s disease virus. Virology 179:719–727. doi: 10.1016/0042-6822(90)90139-I. [DOI] [PubMed] [Google Scholar]
- 58.Hayward WS, Neel BG, Astrin SM. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 59.Payne LN, Rennie M. 1975. B cell antigen markers on avian lymphoid leukosis tumour cells. Vet Rec 96:454–455. doi: 10.1136/vr.96.20.454. [DOI] [PubMed] [Google Scholar]
- 60.Fung YK, Lewis WG, Crittenden LB, Kung HJ. 1983. Activation of the cellular oncogene c-erbB by LTR insertion: molecular basis for induction of erythroblastosis by avian leukosis virus. Cell 33:357–368. doi: 10.1016/0092-8674(83)90417-8. [DOI] [PubMed] [Google Scholar]
- 61.Pizer E, Humphries EH. 1989. RAV-1 insertional mutagenesis: disruption of the c-myb locus and development of avian B-cell lymphomas. J Virol 63:1630–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grandori C, Cowley SM, James LP, Eisenman RN. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 63.Evert DL, deBoer GF. 1988. Avian lymphoid leukosis: mechanisms of lymphomagenesis. Adv Vet Sci Comp Med 32:37–55. doi: 10.1016/B978-0-12-039232-2.50006-2. [DOI] [PubMed] [Google Scholar]
- 64.Brandvold KA, Ewert DL, Kent SC, Neiman P, Ruddell A. 2001. Blocked B cell differentiation and emigration support the early growth of Myc-induced lymphomas. Oncogene 20:3226–3234. doi: 10.1038/sj.onc.1204431. [DOI] [PubMed] [Google Scholar]
- 65.Hughes SH. 1982. Sequence of the long terminal repeat and adjacent segments of the endogenous avian virus Rous-associated virus 0. J Virol 43:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parghi SS, Brandvold KA, Bowers SJ, Neiman PE, Ruddell A. 2004. Reduced Myc overexpression and normal B-cell differentiation mediate resistance to avian leukosis virus lymphomagenesis. Oncogene 23:4413–4421. doi: 10.1038/sj.onc.1207577. [DOI] [PubMed] [Google Scholar]
- 67.Silva RF, Fadly AM, Taylor SP. 2007. Development of a polymerase chain reaction to differentiate avian leukosis virus (ALV) subgroups: detection of an ALV contaminant in commercial Marek’s disease vaccines. Avian Dis 51:663–667. doi: 10.1637/0005-2086(2007)51[663:DOAPCR]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 68.Hanafusa T, Hanafusa H, Miyamoto T. 1970. Recovery of a new virus from apparently normal chick cells by infection with avian tumor viruses. Proc Natl Acad Sci U S A 67:1797–1803. doi: 10.1073/pnas.67.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogt PK, Ishizaki R. 1965. Reciprocal patterns of genetic resistance to avian tumor viruses in two lines of chickens. Virology 26:664–672. doi: 10.1016/0042-6822(65)90329-6. [DOI] [PubMed] [Google Scholar]
- 70.Fadly A, Witter RL. 1998. Oncornaviruses: leukosis/sarcoma and reticuloendotheliosis, p 185–196. In Glisson JR, Jackwood DJ, Pearson JE, reed WM, Swayne DE (ed), Laboratory manual for the isolation of avian pathogens, 4th ed American Association of Avian Pathologists, Kennett Square, PA. [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files, v1.33. https://github.com/najoshi/sickle.
- 73.Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, Dodgson JB. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 74.Spatz SJ, Schat KA. 2011. Comparative genomic sequence analysis of the Marek’s disease vaccine strain SB-1. Virus Genes 42:331–338. doi: 10.1007/s11262-011-0573-0. [DOI] [PubMed] [Google Scholar]
- 75.Petropoulos CJ. 1997. Appendix 2: retroviral taxonomy, protein structure, sequences, and genetic maps, p 757 In Coffin JM. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY. [Google Scholar]
- 76.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. 2015. Ensembl 2015. Nucleic Acids Res 43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brister JR, Ako-Adjei D, Bao Y, Blinkova O. 2015. NCBI viral genomes resource. Nucleic Acids Res 43:D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genomic sequences have been deposited in GenBank under accession numbers MF817820 for AF227, MF817821 for AF229, MF817822 for RAV-0, MF817823 for a partial RAV-60 sequence, and MF926337 for RAV-1.