Bacteriophages are in a constant evolutionary struggle to overcome their microbial hosts’ defenses and must adapt in unconventional ways to remain viable as infectious agents. One mode of adaptation is modifying the viral genome to contain noncanonical nucleotides. Genome modification in phages is becoming more commonly reported as analytical techniques improve, but guanosine modifications have been underreported. To date, two genomic guanosine modifications have been observed in phage genomes, and both are low in genomic abundance. The significance of our research is in the identification of two novel DNA modification systems in Campylobacter-infecting phages, which replace all guanosine bases in the genome in a genus-specific manner.
KEYWORDS: Campylobacter jejuni, DNA modification, bacteriophages, deazaguanosine, deoxyinosine
ABSTRACT
Several reports have demonstrated that Campylobacter bacteriophage DNA is refractory to manipulation, suggesting that these phages encode modified DNA. The characterized Campylobacter jejuni phages fall into two phylogenetic groups within the Myoviridae: the genera Firehammervirus and Fletchervirus. Analysis of genomic nucleosides from several of these phages by high-pressure liquid chromatography-mass spectrometry confirmed that 100% of the 2′-deoxyguanosine (dG) residues are replaced by modified bases. Fletcherviruses replace dG with 2′-deoxyinosine, while the firehammerviruses replace dG with 2′-deoxy-7-amido-7-deazaguanosine (dADG), noncanonical nucleotides previously described, but a 100% base substitution has never been observed to have been made in a virus. We analyzed the genome sequences of all available phages representing both groups to elucidate the biosynthetic pathway of these noncanonical bases. Putative ADG biosynthetic genes are encoded by the Firehammervirus phages and functionally complement mutants in the Escherichia coli queuosine pathway, of which ADG is an intermediate. To investigate the mechanism of DNA modification, we isolated nucleotide pools and identified dITP after phage infection, suggesting that this modification is made before nucleotides are incorporated into the phage genome. However, we were unable to observe any form of dADG phosphate, implying a novel mechanism of ADG incorporation into an existing DNA strand. Our results imply that Fletchervirus and Firehammervirus phages have evolved distinct mechanisms to express dG-free DNA.
IMPORTANCE Bacteriophages are in a constant evolutionary struggle to overcome their microbial hosts’ defenses and must adapt in unconventional ways to remain viable as infectious agents. One mode of adaptation is modifying the viral genome to contain noncanonical nucleotides. Genome modification in phages is becoming more commonly reported as analytical techniques improve, but guanosine modifications have been underreported. To date, two genomic guanosine modifications have been observed in phage genomes, and both are low in genomic abundance. The significance of our research is in the identification of two novel DNA modification systems in Campylobacter-infecting phages, which replace all guanosine bases in the genome in a genus-specific manner.
INTRODUCTION
Lytic viruses known as bacteriophages (phages) are destructive to their bacterial hosts, and bacteria in turn have evolved mechanisms of defending themselves from phages. Some of these defense mechanisms involve innate immune-like systems for phage DNA degradation, such as CRISPR-Cas and restriction-modification systems (1, 2). Presumably as a result of these pressures, several phages have evolved mechanisms to express hypermodified DNA. Some phages synthesize and incorporate modified nucleotides during replication, while others modify nucleotides after replication. Both mechanisms of DNA modification can involve either complete or partial replacement of a canonical base with a modified base.
One of the best-described examples of phage DNA hypermodification is that of Escherichia coli phage T4, which incorporates 5-hydroxymethylcytosine (5-hmdC) in place of all deoxycytidine (dC) residues during DNA replication and further glucosylates the 5-hmdC with 100% efficiency (3). In the case of Bacillus phages SP10 and SP15, deoxythymidine (dT) is partially replaced by α-glutamyldeoxythymidine and 5-dihydroxypentyldeoxyuridine. Bacillus phage PBS2 replaces dT with deoxyuridine (dU) across 100% of the genome, and Bacillus phages SP8, ϕe, 2C, H1, SP82, and SP01 replace dT with a further modified dU derivative, 5-hydroxymethyl-dU (4). Recently, the Salmonella phage ViI was found to replace 40% of its dT nucleobases with 5-(2-aminoethoxy)methyluridine and the Pseudomonas phage M6 was found to replace 30% of its dT residues with 5-(2-aminoethyl)uridine (5). Purine modifications have been less commonly reported than pyrimidine modifications, but examples include the complete replacement of deoxyadenosine (dA) with 2-aminoadenosine in the cyanophage S-2L and the ∼15% replacement of dA with N6-carbamoyl-methyladenosine in the Escherichia coli phage Mu (6). Recently, the replacement of ∼25% of deoxyguanosine (dG) residues by deoxyarchaeosine (dG+) was described in the E. coli phage 9g (7). To date, phage DNA hypermodification has not been examined in Campylobacter jejuni.
C. jejuni is a ubiquitous gastrointestinal pathogen, accounting for the majority of bacterial diarrhea cases worldwide (8). Most instances of C. jejuni infection occur following mishandling or consumption of contaminated poultry products, since C. jejuni naturally colonizes the gastrointestinal tract of chickens. Campylobacter phages have the potential to be used in C. jejuni biocontrol as alternatives to antibiotics, but further understanding of C. jejuni interactions with phages is important for the success of this strategy (9, 10). Most characterized C. jejuni phages fall within the Eucampyvirinae of the Myoviridae, which is divided into the Firehammervirus and Fletchervirus genera (9, 11–13). C. jejuni phage DNA has previously been regarded as refractory to genetic manipulation, resisting restriction digestion, PCR, and sequencing (13). These observations suggest that Campylobacter phage DNA could be modified through either nucleotide modification or association with histone-like proteins (14).
Here we report the identification of hypermodified nucleotides in the DNA of five lytic Campylobacter phages: the fletcherviruses NCTC 12673 and NCTC 12669, the firehammerviruses CP220 and CPt10, and the unclassified Fletchervirus-like phage F336 (15–18). We observed that each phage completely replaced dG in its DNA in a genus-specific manner. Phages NCTC 12673, NCTC 12669, and F336 replaced dG with 2′-deoxyinosine (dI), while phages CP220 and CPt10 replace dG with 2′-deoxy-7-amido-7-deazaguanosine (dADG). In addition, we found that the dADG-incorporating phages encode functional components of the queuosine (Q) biosynthesis pathway, which, we hypothesize, is diverted to complete the synthesis of the ADG base. We also observed the formation of modified cytoplasmic nucleotide pools after infection with representative Fletchervirus and Firehammervirus phages, which indicates two novel systems of noncanonical nucleotide biosynthesis in these two phage groups resulting in the incorporation of dI and dADG into the respective phage genomes.
RESULTS
Fletchervirus phage DNA displays complete replacement of dG with dI.
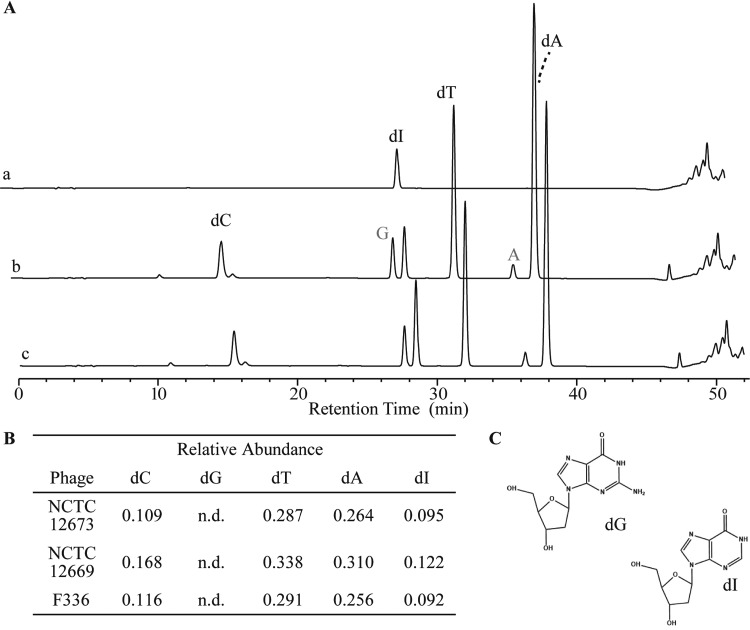
To determine if the Fletchervirus phages incorporate modified nucleotides, we used nucleoside high-performance liquid chromatography (HPLC)-mass spectroscopy (MS) to separate and identify the substituents of NCTC 12673, F336, and NCTC 12669 genomic DNA. Upon analysis of NCTC 12673 DNA, we identified peaks corresponding to three of the canonical nucleosides, dA, dC, and dT, and an unknown peak at a retention time of 28.5 min (Fig. 1A). We did not observe a peak corresponding to dG. Further analysis of the unidentified nucleoside using UV-visible (UV-Vis) spectroscopy (see Fig. S1C in the supplemental material) showed a spectrum consistent with that of deoxyinosine (dI). Following these experiments, we examined the nucleoside content of DNA extracted from two more Campylobacter phages, F336 and NCTC 12669, and observed a similar replacement of dG with dI (Fig. S1A and B). We further calculated the relative abundance of each nucleoside observed and found that it roughly adhered to Chargaff’s rules in each phage genome (Fig. 1B).
FIG 1.
HPLC-MS analyses of acid-hydrolyzed DNA from a representative Fletchervirus member. (A) HPLC-MS spectrum of acid-hydrolyzed genomic DNA from NCTC 12673, a representative Fletchervirus. Trace a, the dI standard; trace b, nucleosides from the NCTC 12673 nucleoside digest; trace c, coinjection of the dI standard with the nucleoside digest from trace b. Gray lettering indicates ribonucleotides. (B) Relative quantities of the nucleosides for phages displaying the deoxyinosine (dI)-containing spectra represented in panel A and in Fig. S2A and S2B in the supplemental material. n.d., not determined. (C) Structures of deoxyguanosine (dG) and deoxyinosine (dI).
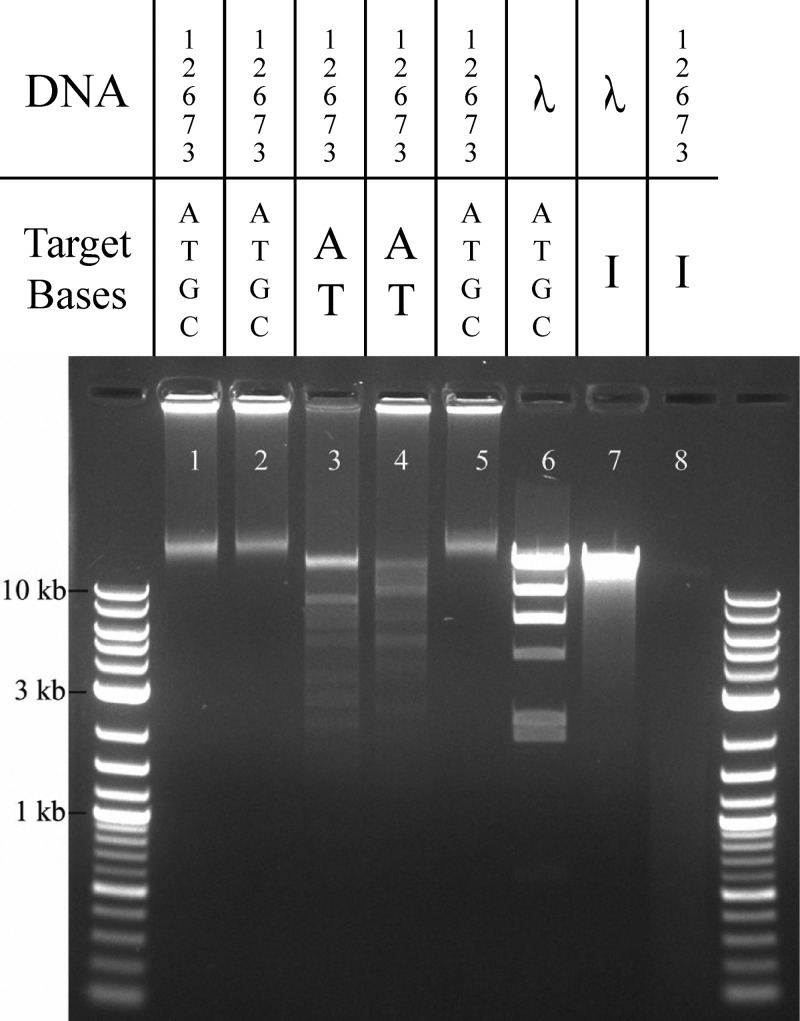
To further confirm that the unknown nucleoside in NCTC 12673 DNA is dI, we tested the effect of endonuclease V (EndoV), a DNA repair enzyme that excises dI from DNA. EndoV completely degraded NCTC 12673 DNA but did not degrade λ phage DNA (Fig. 2). Consistent with previous observations, restriction enzymes recognizing dG-containing binding motifs (EcoRI, HindIII, and XbaI) did not degrade the DNA, even when up to 38 cut sites were predicted in the genome (Fig. 2). Conversely, the enzymes recognizing AT-only sequences (PacI and SwaI) effectively cleaved NCTC 12673 DNA. Overall, these results indicate that dG is absent from NCTC 12673 genomic DNA, while dI is present.
FIG 2.
Restriction digests of Campylobacter phage DNA to confirm base substitution. Agarose gel electrophoresis of NCTC 12673 DNA following incubation with EcoRI (lane 1), HindIII (lane 2), PacI (lane 3), SwaI (lane 4), XbaI (lane 5), and EndoV (lane 8) compared with that of lambda phage DNA incubated with HindIII (lane 6) and EndoV (lane 7).
Detection of dITP pools in NCTC 12673-infected culture.
We wanted to determine if the mechanism by which these nucleobase modifications are made is prereplicative or postreplicative. Due to the 100% efficiency of the substitution, we hypothesized that the modification would be made prereplicatively, with a new pool of modified dITP being formed during infection. To test this, cultures of C. jejuni 11168 MP21 were infected with NCTC 12673 and nucleotide pools were isolated from those infected cultures and analyzed by high-performance anion-exchange chromatography (HPAEC) and UV detection (the results for the deoxynucleoside triphosphate [TP] standards are shown in Fig. S2). Using this method, we observed the formation of a dITP pool at a retention time of ∼48 min in the NCTC 12673-infected culture of C. jejuni 11168 MP21 (Fig. S3A). Coinjection of the dITP standard with the sample increased the peak height at the same retention time. These results were replicated under identical conditions with C. jejuni 11168 MP21 and also with the C. jejuni 12661 propagating strain (Fig. S3B and C).
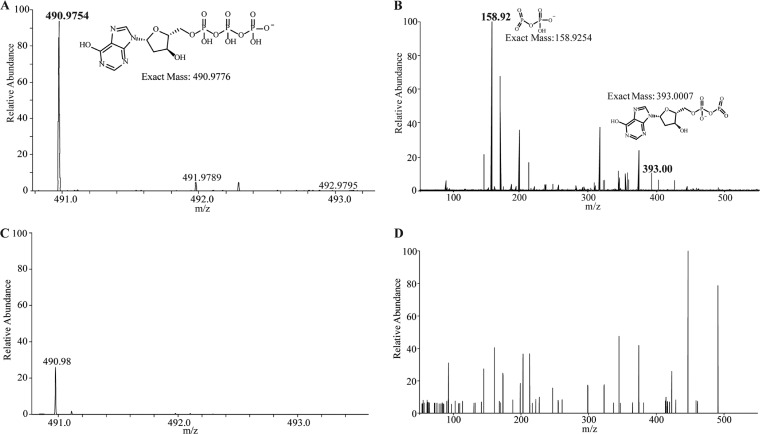
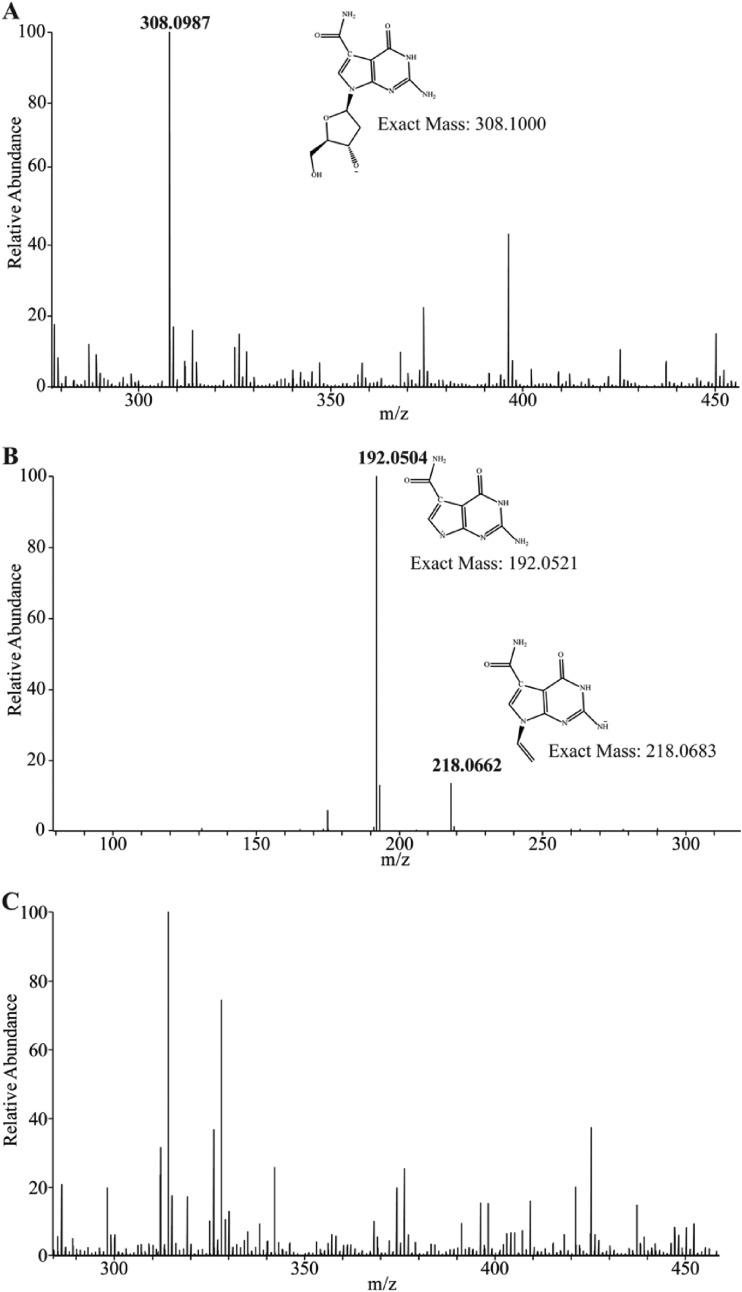
Mass spectrometry (MS) and tandem MS (MS/MS) analyses further confirmed the identity of the dITP in the sample (Fig. 3A and B; the results for the standards are shown in Fig. S5A and B). The MS spectrum of the uninfected sample showed a peak similar to the mass of dITP (Fig. 3C), but further fragmentation by MS/MS did not detect the diagnostic ions of the breakdown product (Fig. 3D). Together, these results demonstrate that a dITP pool is formed in a phage-infected cell.
FIG 3.
Mass spectrometric analyses of C. jejuni 11168 MP21 nucleotide pools. (A) MS spectrum confirming that dITP is present after NCTC 12673 phage infection, consistent with the MS spectrum of the dITP standard shown in Fig. S5A. (B) MS/MS fragmentation spectrum of the dITP peak observed in panel A, consistent with the MS/MS spectrum of the standard shown in Fig. S5B. (C) MS spectrum of the uninfected C. jejuni 11168 MP21 nucleotide pool. (D) MS/MS fragmentation of the peak at m/z 490.98 observed in panel C shows no corresponding dITP fragments.
Firehammervirus phage DNA shows complete replacement of dG with dADG.
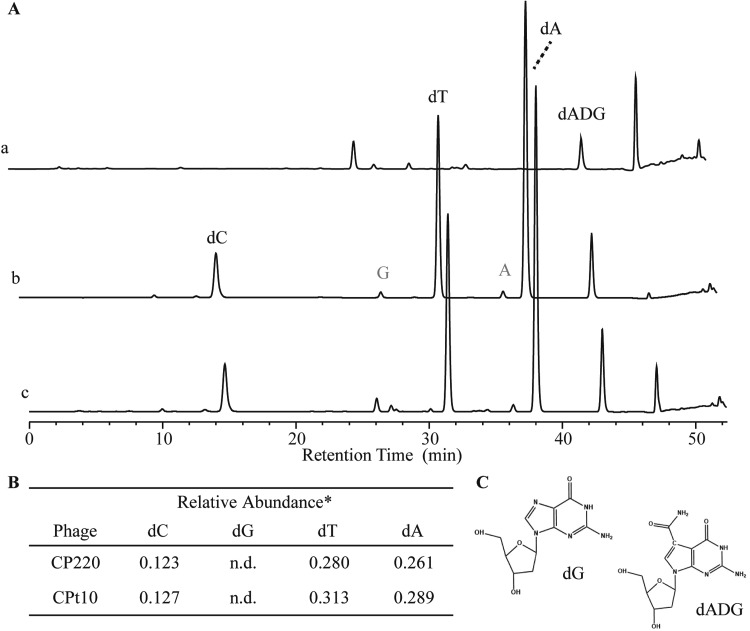
Following the identification of dI as the Fletchervirus phage dG substitution, we wanted to determine whether a similar substitution occurs in the related Firehammervirus genus. Again, nucleoside analysis by HPLC-MS revealed that the canonical nucleotides dA, dT, and dC were found in the representative CP220 and CPt10 genomes, while dG was similarly absent. However, a peak corresponding to the retention time of dI was also not observed; instead, an unknown peak with a retention time of ∼43 min was observed (Fig. 4A). UV-Vis spectroscopy of the unknown peak showed a spectrum that did not correspond to any known nucleotide (Fig. S1). To identify the chemical composition of the nucleotide corresponding to the unknown peak, we used electrospray ionization (ESI)-MS on the isolated nucleosides. From the fragmentation pattern observed, we identified the unknown peak to be 2′-deoxy-7-amido-7-deazaguanosine (dADG) (Fig. S4). The same observation was made after CPt10 HPLC nucleoside analysis (Fig. S1D). Together, these results show that firehammerviruses replace genomic G with the hypermodified base ADG.
FIG 4.
HPLC-MS analyses of acid-hydrolyzed DNA from a representative Firehammervirus member. (A) HPLC-MS spectrum of acid-hydrolyzed DNA from CP220, a representative Firehammervirus. Trace a, the dADG standard; trace b, nucleosides from the CP220 nucleoside digest; trace c, coinjection of the dADG standard with the nucleoside digest from trace b. Gray lettering indicates ribonucleotides. (B) Relative quantities of nucleosides for phages displaying 7-amido-7-deaza-deoxyguanosine (dADG)-containing spectra represented by trace b. *, no reliable extinction coefficient number is available for dADG. Therefore, dADG normalization was not performed. (C) Structures of dG and dADG.
CP220 phage-encoded PreQ0 biosynthesis genes folE, queD, and queE can functionally complement E. coli homologs.
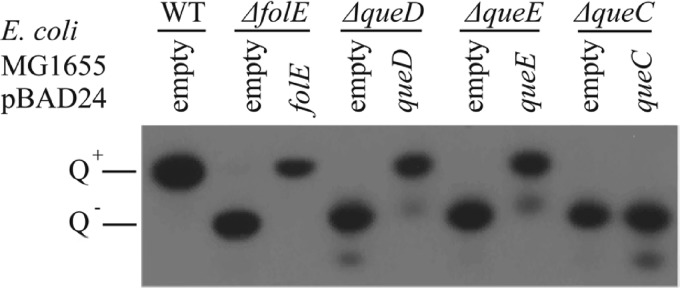
We also wanted to understand how dADG is synthesized by CP220 and CPt10 phages. dADG has already been detected in bacterial DNA as part of a novel restriction-modification system and is synthesized from the precursor base 7-cyano-deazaguanine (PreQ0) (19). The Firehammervirus genomes harbor homologs of folE, queD, queE, and queC, encoding proteins that metabolize PreQ0 from GTP. Plasmids containing Campylobacter phage CP220 queuosine synthesis genes where transformed in the appropriate E. coli MG1655 mutant to test queuosine synthesis complementation. The empty plasmid pBAD24 was transformed into the same strains as a negative control for the complementation and in the E. coli MG1655 wild-type (WT) strain as a positive control. We then extracted the tRNA from overnight cultures of these strains and assayed their Q content using acrylamide gel electrophoresis in which the gel was supplemented with 3-(acrylamido)-phenylboronic acid (APB), a cross-linking reagent that delays the migration of the Q-modified tRNA. We found that queD, queE, and folE functionally complemented the corresponding E. coli mutants, while queC did not (Fig. 5).
FIG 5.
Northern blot of acrylamide-APB gel electrophoresis of tRNA extracted from different strains of E. coli MG1655 (WT, ΔfolE, ΔqueD, ΔqueE, and ΔqueC strains) transformed with pBAD24 (empty vector control) or derivatives expressing the respective homologous genes from the CP220 phage. tRNA lacking Q (Q−) migrates faster than tRNA containing Q (Q+).
Detection of dADG pools in CP220-infected culture.
Nucleotide pools were isolated from a CP220-infected culture of C. jejuni 12661, since CP220 does not infect C. jejuni 11168 MP21. HPAEC did not reveal an obvious dADG triphosphate nucleotide pool after infection, and the synthesized dADG nucleoside standard that we obtained for analysis was not in the triphosphate form and was thus not detectable by HPAEC, so we used MS/MS to observe this pool (Fig. 6).
FIG 6.
Mass spectrometric analyses of C. jejuni 12661 nucleotide pools after CP220 phage infection. (A) MS spectrum showing that dADG is produced in a CP220-infected culture of C. jejuni 12661. (B) MS/MS fragmentation spectrum of the dADG peak observed in panel A. (C) The MS spectrum of the C. jejuni 12661 nucleotide pool shows no mass corresponding to dADG.
Analysis of the nucleotide extract showed a pool of the dADG nucleoside consistent with the fragmentation pattern of the standard (Fig. 6A and B; the results for the standards are shown in Fig. S5C and D). However, consistent with the HPAEC-UV results, no phosphorylated version of the nucleoside was observed, with 1, 2, or 3 phosphate groups being found in the sample. Interestingly, approximately 3% of the nucleoside pool contained dADG further modified with a mass of 163.0499 Da (Fig. S6), possibly corresponding to C5H9NO5, although this hypermodified base was not observed in the original HPLC (Fig. 4). Masses corresponding to neither of these dADG nucleobases were detected in the MS and MS/MS experiments of uninfected C. jejuni 12661 (Fig. 6C). In an effort to find evidence of breakdown products of dADG triphosphate (TP), we searched for masses consistent with Pi, PPi, and inorganic triphosphate (PPPi) in the CP220-infected and uninfected sample. We were able to find evidence of only diphosphate in each extract, and interestingly, the abundance was much greater in the phage-infected sample (Fig. S7). These results indicate that a cytoplasmic pool of the dADG nucleobase (and a further modified form) is formed in C. jejuni 12661 after CP220 infection.
DISCUSSION
The extreme selective pressures encountered during phage and bacterium-host evolution yield unexpected mechanisms for adaptation. Here, we examined the two major classes of C. jejuni-infecting phages and show they have both evolved to replace genomic dG with one of two different noncanonical 2′-deoxypurine bases. Both noncanonical purines, inosine and 7-deazaguanosine derivatives, are found in nature as components of tRNA, and derivatives of their synthetic pathways have been observed in many bacterial genera, indicating that these phage subfamilies may have co-opted existing pathways for their own use in a novel way.
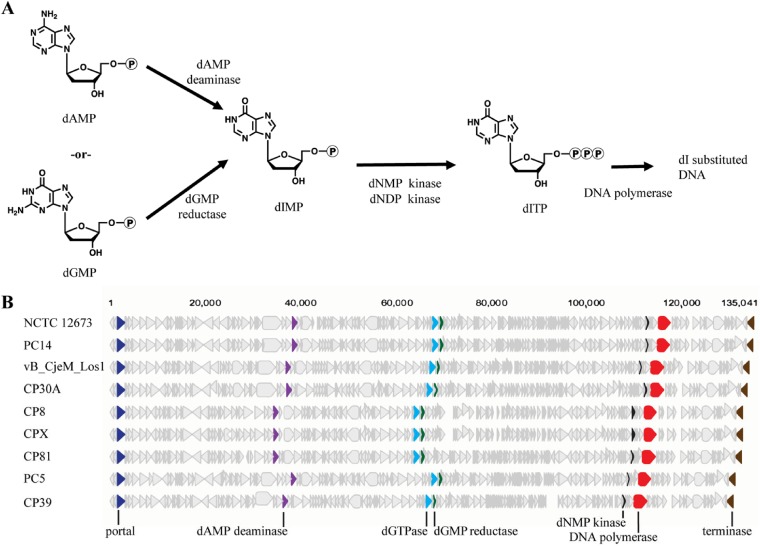
The fletcherviruses NCTC 12673 and NCTC 12669 and the unclassified phage F336 replace all genomic dG residues with dI. We also noted that Chargaff’s rule was observed with these phages, and the proportions of dI and dC together are approximately the predicted G/C content of NCTC 12673, the only phage for which the GC content is available (16). The identification of dI in the NCTC 12673 genomic DNA was confirmed by UV-visible spectroscopy and endonuclease V digestion. These findings explain previous reports describing the refractory nature of C. jejuni phage DNA to genetic manipulation with commercially available restriction endonucleases (13, 16). To explain the mechanism behind this remarkable finding, we hypothesize that the phage utilizes both phage and host-derived purine biosynthesis machinery to accomplish this. dIMP can be created through the action of either a dAMP deaminase or a dGTP reductase, and both intermediates would be present in the C. jejuni cell. A phage-encoded deoxynucleoside monophosphate kinase can then perform the action of phosphorylating the dIMP to dIDP. The dIDP can then be phosphorylated by a general host deoxynucleoside diphosphate kinase, yielding dITP, which can be incorporated into the replicating genome (Fig. 3). Through bioinformatic analyses, we have identified putative enzymes in the proposed pathway, shown in Fig. 7, with the accession numbers for these homologs being found in Table 1. This hypothesis is also supported by the pool of dITP formed in NCTC 12673-infected C. jejuni cells, which is absent in uninfected cells. We expect that further studies of the phage-encoded polymerase will yield insights into the mechanism by which dITP is selectively incorporated over dGTP.
FIG 7.
Genome analyses reveal proposed pathway enzymes for dITP synthesis. (A) Proposed pathway for the synthesis of dITP in fletcherviruses. (B) Genome maps of available fletcherviruses with proposed genes encoding enzymes associated with dITP synthesis.
TABLE 1.
Accession numbers for Fletchervirus phage genes identified in proposed synthesis pathways
| Campylobacter Fletchervirus phage | GenBank accession no. |
||||
|---|---|---|---|---|---|
| Genome | Predicted dIMP kinase | Predicted dGTPase | Amidinotransferase | Possible GMP reductase | |
| NCTC 12673 | GU296433 | YP_004421657 | YP_004421562 | YP_004421599 | YP_004421564 |
| PC14 | NC_031909 | YP_006908165 | YP_009321662 | YP_009321699 | YP_009321664 |
| vB_CjeM_Los1 | KX879627 | AOT25873 | AOT25943 | AOT25980 | AOT25945 |
| CP30A | NC_018861 | YP_006908165 | YP_006908072 | YP_006908109 | YP_006908074 |
| CP8 | KF148616 | AGS81219 | AGS81277 | AGS81311 | AGS81279 |
| CPX | NC_016562 | YP_004956914 | YP_004956973 | YP_004957007 | YP_004956975 |
| CP81 | FR823450 | CBZ42288 | CBZ42208 | CBZ42172 | CBZ42206 |
| PC5 | KX229736 | ANH51205 | ANH51137 | ANH51273 | ANH51135 |
| CP39 | MH107028 | AVR55800 | AVR55701 | AVR55738 | AVR55703 |
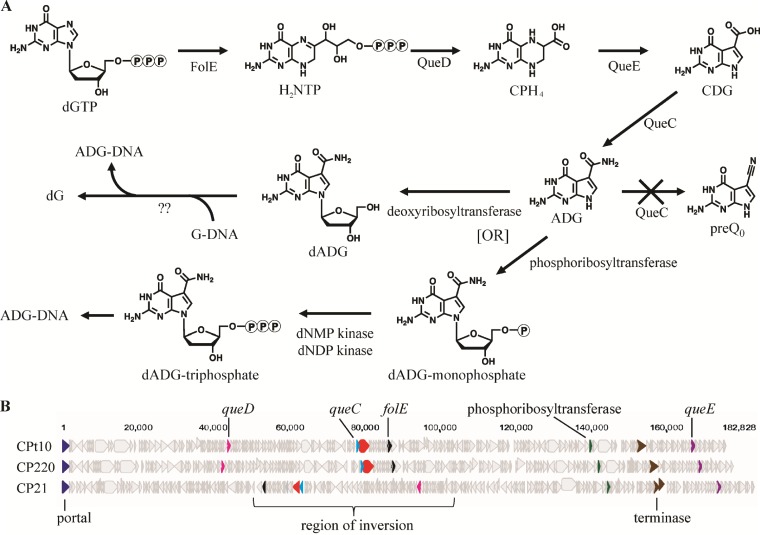
Similar to the fletcherviruses, the firehammerviruses CP220 and CPt10 both demonstrate the complete replacement of genomic dG but did so for a queuosine precursor, dADG. The identity of dADG was confirmed by coinjection of a synthesized nucleoside standard in the HPLC-MS experiments. We hypothesize that the hypermodified nucleobase is synthesized by phage-encoded enzymes. Upon examination of the CP220 genome, homologs of folE, queD, queE, and queC were identified (Table 2). These genes encode proteins that metabolize PreQ0 from GTP in E. coli. Through recombinant expression, we were able to complement the activity of the corresponding E. coli enzymes, with the exception of the bifunctional QueC. In E. coli, QueC catalyzes two reactions, the reaction from CDG to ADG and the reaction from ADG to PreQ0 (7), while the phage-encoded enzyme could not complement the ability to form PreQ0. Since both QueC reactions are required for our reporter assay to indicate complementation, we propose that the CP220 phage homolog may lack the ability to perform the second reaction but generates ADG during phage infection (Fig. 8A). This is consistent with our nucleotide pool analyses by MS, where we observed an abundance of dADG accumulating only during phage infection. We are currently investigating the active-site residues involved in both these reactions to determine if this hypothesis is valid.
TABLE 2.
Accession numbers for Firehammervirus phage genes identified in proposed synthesis pathways
| Campylobacter Firehammervirus phage | GenBank accession no. |
||||
|---|---|---|---|---|---|
| queC | queD | queE | folE | Phosphoribosyltransferase | |
| CPt10 | YP_009169054 | YP_009169009 | YP_009168954 | YP_009169065 | YP_009169122 |
| CP220 | YP_009169248 | YP_009169203 | YP_009169151 | YP_009169258 | YP_009169316 |
| CP21 | YP_007005322 | YP_007005116 | YP_007005215 | YP_007005301 | YP_007005178 |
FIG 8.
Genome analyses reveal enzymes in the proposed pathway for dADG synthesis. (A) Proposed pathway for the synthesis of dADG in firehammerviruses. dNMP, deoxynucleoside monophosphate; dNDP, deoxynucleoside diphosphate. (B) Genome maps of sequenced firehammerviruses with proposed genes encoding enzymes associated with dADG synthesis.
As mentioned above, we also looked for the dADG-TP nucleotide pool in a CP220-infected culture of C. jejuni cells. The cell extract showed an abundance of the dADG nucleoside, but we did not observe phosphorylation on the 5′ carbon of the deoxyribose after 2, 6, or 20 h of infection. With these data, we hypothesize that the phage genome is replicated with canonical nucleotides and that the dG nucleoside is excised and replaced through an unknown mechanism. However, we cannot discount the possibility that the dADG nucleoside is phosphorylated and that the pool is below our limit of detection. This is reflected in the proposed pathway found in Fig. 8A. We are currently looking for a tgt-like gene within the CP220 genome encoding a transglycosyltransferase that would catalyze the replacement of the G nucleobase with dADG. In this nucleoside replacement model, the enzyme would replace all of the dG incorporated into the new genome, which amounts to approximately 24,322 replacements per CP220 genome, with a high efficiency and fidelity.
The complete DNA modifications found in these dominant classes of C. jejuni phages pose several tantalizing questions about their impact on the phage life cycle. Prominent among them is, what evolutionary benefit, if any, might these modifications confer to the phage? Previous studies of C. jejuni phages have described the refractory nature of their DNA to restriction by commercially available G/C-recognizing endonucleases (13). We have also demonstrated this, as well as the ability of A/T-recognizing enzymes to cleave the NCTC 12673 genomic DNA in a predictable manner. However, it is unclear whether C. jejuni type I or type II restriction-modification systems are A-T specific or whether they include all nucleotides in their binding motifs (20). In a similar vein, it has been reported that the type II-C CRISPR-Cas systems encoded by C. jejuni possess promiscuous protospacer adjacent motifs (PAM; 5′-NNNNACA-3′) devoid of G residues, which is contrary to the G-rich PAM sequences found in most species (such as 5′-NGG-3′ in Streptococcus pyogenes) (21). The development of binding domains with non-G-containing sequences indicates that the phage G-specific DNA modifications may be widespread in C. jejuni phages and that C. jejuni has potentially adapted a way to attempt to overcome those modifications. However, there has been limited study of restriction-modification and CRISPR-Cas systems in C. jejuni, so we are currently unable to determine the evolutionary effect of the nucleobase modifications observed here.
We have described two abundant genera of C. jejuni phages that demonstrate 100% replacement of genomic dG with dI and dADG in a genus-specific manner. Each hypermodification has evolved an independent biosynthesis pathway with unique polymerases whose sequences have been used to distinguish the phage genera (13). While we do not yet know the impact of DNA hypermodification on the C. jejuni phage life cycle, these observations offer exciting insight into the world of bacterium-phage interactions and the perpetual arms race between the host and virus.
MATERIALS AND METHODS
Growth of bacteria.
C. jejuni strains were grown on 1.5% NZCYM agar plates (Becton, Dickinson and Company) at 37°C under microaerobic conditions (85% N2, 10% CO2, 5% O2). Escherichia coli strains were grown on LB agar supplemented with 50 μg/ml kanamycin or 25 μg/ml chloramphenicol where needed. The bacterial strains used in this study are given in Table 3.
TABLE 3.
Strains and phages used in this study
| Strain or phage (reference) | Description or propagating strain (reference or source) |
|---|---|
| Strains | |
| C. jejuni 12661 | Phage propagation strain (18) |
| C. jejuni 11168 MP21 | Phage propagation strain (22) |
| C. jejuni 12664 | Phage propagation strain (18) |
| E. coli WT MG1655(pBAD24) | Can synthesize queuosine-tRNA (17) |
| E. coli MG1655 ΔqueC(pBAD24) | Cannot synthesize queuosine-tRNA (17) |
| E. coli MG1655 ΔqueD(pBAD24) | Cannot synthesize queuosine-tRNA (27) |
| E. coli MG1655 ΔqueE(pBAD24) | Cannot synthesize queuosine-tRNA (17) |
| E. coli MG1655 ΔfolE(pBAD24) | Cannot synthesize queuosine-tRNA (17) |
| E. coli MG1655 ΔqueC(pBAD24)/queC | Complement expressing the queC homolog gp114 from phage CP220 (this study) |
| E. coli MG1655 ΔqueD(pBAD24)/queD | Complement expressing the queD homolog gp069 from phage CP220 (this study) |
| E. coli MG1655 ΔqueE(pBAD24)/queE | Complement expressing the queE homolog gp017 from phage CP220 (this study) |
| E. coli MG1655 ΔfolE(pBAD24)/folE | Complement expressing the folE homolog gp124 from phage CP220 (this study) |
| Phages | |
| NCTC 12673 (18) | C. jejuni 12661 (18) |
| NCTC 12669 (18) | C. jejuni 12664 (18) |
| F336 (15) | C. jejuni 11168 MP21 (22) |
| CP220 (28) | C. jejuni 12661 (18) |
| CPt10 (28) | C. jejuni 12661 (18) |
Phage propagation, titration, and concentration.
Phages NCTC 12673 and NCTC 12669, along with their propagating strains, C. jejuni 12661 and C. jejuni 12664, respectively (18), were obtained from the National Collection of Type Cultures (NCTC; Salisbury, United Kingdom). Phages CP220 and CPt10 were obtained from Ian and Phillippa Connerton and propagated on C. jejuni 12661. Phage F336 was obtained from Lone Brøndsted and propagated on C. jejuni 11168 MP21 (22). Phage propagation and titration were performed following the methods described previously (23), except that the 4-h pregrowth of the cultures was done with NZCYM broth instead of cation-adjusted brain heart infusion broth (cBHI; Becton, Dickinson and Company) supplemented with 10 mM MgSO4 and 1 mM CaCl2. To increase phage titers following propagation, phage lysates filtered through a 0.22-μm-pore-size filter were ultracentrifuged at 207,870 × g in an SW-28 swinging bucket rotor for 1.5 h at 4°C. Pellets from 25-ml volumes of lysate were resuspended in 1 ml SM buffer containing gelatin (23) and pooled, and the titers were determined. The phages used in this study, along with their propagating strains, are listed in Table 3.
Preparation of bacteriophage genomic DNA.
CsCl-purified bacteriophages in 100 mM Tris, pH 8, 25 mM EDTA, 1% sodium dodecyl sulfate (SDS), and 200 μg/ml proteinase K were incubated at 55°C for 30 min, during which the bluish cast to the solution changed to clear as the virus capsids ruptured. Lysed bacteriophage solution was extracted with an equal volume of a mixture containing phenol, chloroform, and isoamyl alcohol at a ratio of 25:24:1 and neutral pH and agitated by hand for 3 min. The organic and aqueous phases were separated by centrifugation, and the aqueous phase was reextracted two times with equal volumes of chloroform to remove the residual phenol. DNA was precipitated by addition of a 1/10 volume of 3 M sodium acetate and either 2.25 volumes of ice-cold ethanol or an equal volume of isopropanol and incubated on ice for 30 min. The DNA was gently spooled onto a glass rod and successively immersed three times in 70% ethanol to remove the salts. The DNA was dissolved in water over a period of 48 h and dialyzed against water to remove the traces of the alcohols.
Phage genomic DNA digestion to nucleoside and HPLC-MS analysis.
Approximately 5 μg of virion DNA extracted from each of the phages was digested to nucleosides at a 0.1- to 0.2-μg/μl final concentration by treatment overnight at 37°C with 5 μl of the nucleoside digestion mix in 1× nucleoside digestion mix reaction buffer (New England Biolabs [NEB], Ipswich, MA). The resulting DNA nucleoside mixtures were directly subjected to separation and analysis on a reverse-phase HPLC-MS without additional purification.
Liquid chromatography (LC)-MS analysis was performed on an Agilent LC-MS system 1200 series instrument equipped with a G1316A diode array detector and a 6120 single quadrupole mass detector operating in positive and negative electrospray ionization (ESI) modes. LC was carried out on a Waters Atlantis T3 column (4.6 by 150 mm; particle size, 3 μm) with an in-line filter and guard and with mobile phases consisting of 10 mM aqueous ammonium acetate (pH 4.5) and methanol. A linear gradient generated by mixing the two mobile phases from 2% to 25% methanol in 30 min at a 0.5-ml/min flow rate was applied to effectively resolve each nucleoside component after DNA digestion. HPLC data were recorded with a multisignal diode array detector (DAD) as the full UV-Vis spectrum (200 to 400 nm), where the 260-nm absorbance signal was extracted and followed along the HPLC run. MS acquisition was recorded in total ion chromatogram (TIC) mode. For the analysis of the nucleoside composition, the peak area of each nucleoside species resolved in the HPLC trace (recorded as the absorbance at 260 nm) was measured using the integration function of Agilent ChemStation software. The obtained peak area was subsequently divided by the corresponding nucleoside molar extinction coefficient (ε) at 260 nm in order to obtain the normalized molarity for each nucleoside species, in which the extinction coefficients used were 7,100 cm−1 M−1 for dC, 15,066 cm−1 M−1 for dG, 8,560 cm−1 M−1 for dT, 15,060 cm−1 M−1 for dA, and 7,500 cm−1 M−1 for dI (24, 25).
UV-visible spectroscopy.
The UV-Vis spectrum was recorded as part of the data for single HPLC analysis. Each analysis datum was recorded by the multisignal diode array detector as the full UV-Vis spectrum ranging from 200 nm to 400 nm with steps of 2.0 nm.
Restriction endonuclease assay.
The restriction endonuclease assay of the phage DNAs was conducted following the manufacturer’s instructions. Briefly, approximately 250 ng of virion DNA from each of the phages was treated with 1 μl of restriction endonuclease in a 20-μl reaction volume at 37°C for 1 h. The resulting DNA digestion products were directly subjected to agarose gel electrophoresis without further purification. The agarose gel was cast with a supplement of ethidium bromide at a final concentration of 0.2 μg/ml. To visualize the DNA product on the gel, after electrophoresis, the agarose gel was illuminated under UV light, where the DNA bands become visible, and the whole gel was photographed and documented using an AlphaImager gel documentation system.
Nucleotide pool preparation and HPAEC-UV spectroscopy.
Liquid cultures of C. jejuni were inoculated at an optical density at 600 nm of 0.05 and grown in liquid brain heart infusion for 4 h and then infected with phage at a multiplicity of infection of 0.01. The cultures were incubated for an additional 6 h, and then the cells were collected by centrifugation and washed with 1× phosphate-buffered saline. The cells were lysed using an Avestin Emulsiflex C5 homogenizer (ATA Scientific, Australia) in a 50% methanol–50% H2O solution. After lysis, the cell material was pelleted and nucleotide fractions were centrifuged through a 10,000-Da-cutoff column (GE Life Sciences, MA, USA) according to the manufacturer’s instructions and collected. The methanol was evaporated, and the aqueous nucleotide fractions were concentrated with a SpeedVac concentrator and stored at −20°C until analysis.
High-performance anion-exchange chromatography (HPAEC) was done as previously described (26) using HPLC instruments (a GP50 gradient pump and an AD25 absorbance detector) and a CarboPac PA-1 column with a PA-1 guard column (Dionex, Sunnyvale, CA). Eluent 1 was 1 mM sodium hydroxide, and eluent 2 was 1 M sodium acetate in 1 mM sodium hydroxide. The column was equilibrated with a mixture (80:20, vol/vol) of eluents 1 and 2 at a flow rate of 1 ml/min at room temperature. After sample injection, elution was performed with the following gradient: 20% (vol/vol) eluent 2 at 0 min, 55% (vol/vol) at 10 min, 55% (vol/vol) at 25 min, 80% (vol/vol) at 35 min, 100% (vol/vol) at 40 min, and 100% (vol/vol) at 50 min at a flow rate of 1 ml/min at room temperature. Nucleotides were detected by use of the absorbance at 260 nm.
Direct-infusion MS/MS of nucleotides/nucleosides.
The cell extracts with the nucleotides (20 μl) were mixed with 80 μl of ESI-MS infusion buffer (1:1:1 methanol–acetonitrile–1 mM NaOH in water) and filtered through a 0.2-μm-pore-size centrifuge filter. The extracts and standards (0.1 μg/μl) were infused into an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, San Jose, CA, USA) through a nanospray ionization (NSI) source in negative ion mode. The MS1 and MSn spectra collision-induced dissociation (CID) of the nucleotide dITP and nucleoside dADG were acquired at high resolution in an Orbitrap analyzer. The MS1 and MS2 spectra of dITP and dADG from the treated samples were compared with those of the standards.
Cloning of que homologs.
The genes queC, queD, queE, and folE from CP220 were cloned into pBAD24 using restriction digestion and ligation. To generate unmodified DNA that could be amplified by PCR, CP220 genomic DNA was preamplified using phi29 polymerase (GE Life Sciences, MA, USA) according to the manufacturer’s instructions. The amplified DNA was then used as a template for PCR amplification using OneTaq polymerase (NEB, Ipswich, MA) under the manufacturer’s suggested conditions and by using the primers listed in Table 4. The forward primers each encoded their own ribosome binding site. PCR products were digested using appropriate restriction enzymes (Thermo Fisher Scientific, Waltham, MA) and then ligated into equivalently digested pBAD24. The ligation mixtures were transformed into E. coli OneShot TOP10 chemically competent cells (Thermo Fisher Scientific, Waltham, MA), and positive clones were verified by Sanger sequencing.
TABLE 4.
Primers used in this study
| Primer | Organism | Sequence (5′–3′) | RE sitea |
|---|---|---|---|
| queD-F | CP220 | ATGATGTCGACAGGAGGAGATTAATGAAAACATTTAAATGGACTATAGACAAAC | SalI |
| queD-R | CP220 | ATGATCCTGCAGGTTATGAAAATTTAGGGTTTAAAAATCTACAATG | SbfI |
| queC-F | CP220 | ATGATGTCGACAGGAGGAGATTAATGAAAGATACAGTTTTAAGCCTAAGC | SalI |
| queC-R | CP220 | ATGATCCTGCAGGTTAAGAACTAAAATGTTTTAAAACTGTTGTAGC | SbfI |
| queE-F | CP220 | ATGATGTCGACAGGAGGAGATTAATGATTTTCAAAACACGAACTAAAAAAG | SalI |
| queE-R | CP220 | ATGATCCTGCAGGTTAAATACCCTCCTTAGTATCAAAAGC | SbfI |
| folE-F | CP220 | ATGATGTCGACAGGAGGAGATTAATGTTAAAAGAAGATAAATTTATTTGGG | SalI |
| folE-R | CP220 | ATGATCCTGCAGGTTAAACTCGGATTTGAACCTG | SbfI |
| tRNAAsp | E. coli MG1655 | Biotin-CCCTCGGTGACAGGCAGG | NA |
RE, restriction endonuclease; NA, not applicable.
Queuosine detection assay.
Overnight cultures were diluted 1/100-fold into 5 ml of LB supplemented with 0.4% arabinose and 100 μg/ml ampicillin and grown for 2 h at 37°C. Cells were harvested by centrifugation at 16,000 × g for 2 min at 4°C. Cell pellets were immediately resuspended in 1 ml of the TRIzol reagent (Life Technologies, Carlsbad, CA). Small RNAs were extracted using a PureLink microRNA isolation kit from Invitrogen (Carlsbad, CA) according to the manufacturer’s protocol. The purified RNAs were eluted in 50 μl of RNase-free water, and tRNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA). Then, 200 μg of RNA was used in the 3-(acrylamido)-phenylboronic acid (APB) assay as previously described in detail (19) using the probe 5′-biotin-CCCTCGGTGACAGGCAGG-3′, which detects tRNAAsp (GUC), at a final concentration of 0.3 μM.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kazuhiro Aoki and Michael Tiemeyer for their assistance with the HPAEC-UV analysis and Ian and Phillippa Connerton for supplying the phages CP220 and CPt10 used in this study. We also thank Peter Dedon for supplying the dADG standard used for nucleoside analyses. Additionally, we thank Bernadette Beadle for her assistance in sample preparation for HPLC analysis.
This work was funded in part by the National Institutes of Health (grant GM70641 to V.D.C.-L. and grant 1S10OD018530 to the Complex Carbohydrate Research Center), the Human Frontier Science Program (grant RGP0024 to V.D.C.-L.), and the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy (grant DE-SC0015662 to P.A.). C.M.S. is an Alberta Innovates Strategic Chair in Bacterial Glycomics.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01111-19.
REFERENCES
- 1.Westra ER, Dowling AJ, Broniewski JM, van Houte S. 2016. Evolution and ecology of CRISPR. Annu Rev Ecol Evol Syst 47:307–331. doi: 10.1146/annurev-ecolsys-121415-032428. [DOI] [Google Scholar]
- 2.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–77. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren R. 1980. Modified bases in bacteriophage DNAs. Annu Rev Microbiol 34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 4.Weigele P, Raleigh EA. 2016. Biosynthesis and function of modified bases in bacteria and their viruses. Chem Rev 116:12655–12687. doi: 10.1021/acs.chemrev.6b00114. [DOI] [PubMed] [Google Scholar]
- 5.Lee YJ, Dai N, Walsh SE, Muller S, Fraser ME, Kauffman KM, Guan C, Correa IR Jr, Weigele PR. 2018. Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc Natl Acad Sci U S A 115:E3116–E3125. doi: 10.1073/pnas.1714812115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swinton D, Hattman S, Crain P, Cheng C, Smith D, McCloskey J. 1983. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-fi-d-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci U S A 80:7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiaville JJ, Kellner SM, Yuan Y, Hutinet G, Thiaville PC, Jumpathong W, Mohapatra S, Brochier-Armanet C, Letarov AV, Hillebrand R, Malik CK, Rizzo CJ, Dedon PC, de Crécy-Lagard V. 2016. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci U S A 113:E1452–E1459. doi: 10.1073/pnas.1518570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The MAL-ED Network Investigators. 2014. The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis 59(Suppl 4):S193–S206. doi: 10.1093/cid/ciu653. [DOI] [PubMed] [Google Scholar]
- 9.Atterbury RJ, Connerton PL, Dodd CER, Rees CED, Connerton IF. 2003. Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl Environ Microbiol 69:4511–4518. doi: 10.1128/AEM.69.8.4511-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connerton PL, Timms AR, Connerton IF. 2011. Campylobacter bacteriophages and bacteriophage therapy. J Appl Microbiol 111:255–265. doi: 10.1111/j.1365-2672.2011.05012.x. [DOI] [PubMed] [Google Scholar]
- 11.El-Shibiny A, Connerton PL, Connerton IF. 2005. Enumeration and diversity of campylobacters and bacteriophages isolated during the rearing cycles of free-range and organic chickens. Appl Environ Microbiol 71:1259–1266. doi: 10.1128/AEM.71.3.1259-1266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firlieyanti AS, Connerton PL, Connerton IF. 2016. Campylobacters and their bacteriophages from chicken liver: the prospect for phage biocontrol. Int J Food Microbiol 237:121–127. doi: 10.1016/j.ijfoodmicro.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javed MA, Ackermann HW, Azerdo J, Carvalho CM, Connerton IF, Evoy S, Hammerl JA, Timms A, Kropinski AM. 2013. A suggested classification for two groups of Campylobacter myoviruses. Arch Virol 159:181–190. doi: 10.1007/s00705-013-1788-2. [DOI] [PubMed] [Google Scholar]
- 14.Arutyunov D, Szymanski CM. 2015. A novel DNA-binding protein from Campylobacter jejuni bacteriophage NCTC12673. FEMS Microbiol Lett 362:fnv160. doi: 10.1093/femsle/fnv160. [DOI] [PubMed] [Google Scholar]
- 15.Hansen VM, Rosenquist H, Baggesen DL, Brown S, Christensen BB. 2007. Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiol 7:90. doi: 10.1186/1471-2180-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kropinski AM, Arutyunov D, Foss M, Cunningham A, Ding W, Singh A, Pavlov AR, Henry M, Evoy S, Kelly J, Szymanski CM. 2011. Genome and proteome of Campylobacter jejuni bacteriophage NCTC 12673. Appl Environ Microbiol 77:8265–8271. doi: 10.1128/AEM.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutinet G, Kot W, Cui L, Hillebrand R, Balamkundu S, Gnanakalai S, Meelakandan R, Carstens AB, Fa LC, Tremblay D, Jacobs-Sera D, Sassanfar M, Weigele P, Moineau S, Hatfull GF, Dedon PC, Hansen LH, de Crecy-Lagard V. Nat Commun, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frost J, Kramer J, Gillanders S. 1999. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol Infect 123:47–55. doi: 10.1017/S095026889900254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Hutinet G, Valera JG, Hu J, Hillebrand R, Gustafson A, Iwata-Reuyl D, Dedon PC, de Crécy-Lagard V. 2018. Identification of the minimal bacterial 2′-deoxy-7-amido-7-deazaguanine synthesis machinery. Mol Microbiol 110:469–483. doi: 10.1111/mmi.14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller WG, Pearson BM, Wells JM, Parker CT, Kapitonov VV, Mandrell RE. 2005. Diversity within the Campylobacter jejuni type I restriction-modification loci. Microbiology 151:337–351. doi: 10.1099/mic.0.27327-0. [DOI] [PubMed] [Google Scholar]
- 21.Yamada M, Watanabe Y, Gootenberg JS, Hirano H, Ran FA, Nakane T, Ishitani R, Zhang F, Nishimasu H, Nureki O. 2017. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol Cell 65:1109–1121.e3. doi: 10.1016/j.molcel.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Sørensen MCH, van Alphen LB, Harboe A, Li J, Christensen BB, Szymanski CM, Brøndsted L. 2011. Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J Bacteriol 193:6742–6749. doi: 10.1128/JB.05276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gencay YE, Birk T, Sorensen MC, Brondsted L. 2017. Methods for isolation, purification, and propagation of bacteriophages of Campylobacter jejuni. Methods Mol Biol 1512:19–28. doi: 10.1007/978-1-4939-6536-6_3. [DOI] [PubMed] [Google Scholar]
- 24.Cavaluzzi MJ, Borer PN. 2004. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res 32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watkins NE Jr, SantaLucia J Jr. 2005. Nearest-neighbor thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res 33:6258–6267. doi: 10.1093/nar/gki918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. 2001. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem 293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 27.Phillips G, Grochowski LL, Bonnett S, Xu H, Bailly M, Blaby-Haas C, El Yacoubi B, Iwata-Reuyl D, White RH, de Crécy-Lagard V. 2012. Functional promiscuity of the COG0720 family. ACS Chem Biol 7:197–209. doi: 10.1021/cb200329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timms AR, Cambray-Young J, Scott AE, Petty NK, Connerton PL, Clarke L, Seeger K, Quail M, Cummings N, Maskell DJ, Thomson NR, Connerton IF. 2010. Evidence for a lineage of virulent bacteriophages that target Campylobacter. BMC Genomics 11:214. doi: 10.1186/1471-2164-11-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.