Epidemiological studies show that natural killer (NK) cells have anti-HIV activity: they are able to reduce the risk of HIV infection and/or slow HIV disease progression. How NK cells contribute to these outcomes is not fully characterized. We used primary NK cells and autologous HIV-infected cells to examine the role of education through four inhibitory killer immunoglobulin-like receptors (iKIRs) from persons with HLA types that are able to educate NK cells bearing one of these iKIRs. HIV-infected cells activated NK cells through missing-self mechanisms due to the downmodulation of cell surface HLA expression mediated by HIV Nef and Vpu. A higher frequency of educated than uneducated NK cells expressing each of these iKIRs responded to autologous HIV-infected cells by producing CCL4, IFN-γ, and CD107a. Since NK cells were from non-HIV-infected individuals, they model the consequences of healthy NK cell–HIV-infected cell interactions occurring in the HIV eclipse phase, when new infections are susceptible to extinction.
KEYWORDS: HIV, KIR, NK cell education
ABSTRACT
Several studies support a role for specific killer immunoglobulin-like receptor (KIR)–HLA combinations in protection from HIV infection and slower progression to AIDS. Natural killer (NK) cells acquire effector functions through education, a process that requires the interaction of inhibitory NK cell receptors with their major histocompatibility complex (MHC) class I (or HLA class I [HLA-I]) ligands. HLA-C allotypes are ligands for the inhibitory KIRs (iKIRs) KIR2DL1, KIR2DL2, and KIR2DL3, whereas the ligand for KIR3DL1 is HLA-Bw4. HIV infection reduces the expression of HLA-A, -B, and -C on the surfaces of infected CD4 (iCD4) T cells. Here we investigated whether education through iKIR-HLA interactions influenced NK cell responses to autologous iCD4 cells. Enriched NK cells were stimulated with autologous iCD4 cells or with uninfected CD4 cells as controls. The capacities of single-positive (sp) KIR2DL1, KIR2DL2, KIR2DL3, and KIR3DL1 NK cells to produce CCL4, gamma interferon (IFN-γ), and/or CD107a were assessed by flow cytometry. Overall, we observed that the potency of NK cell education was directly related to the frequency of each spiKIR+ NK cell’s ability to respond to the reduction of its cognate HLA ligand on autologous iCD4 cells, as measured by the frequency of production by spiKIR+ NK cells of CCL4, IFN-γ, and/or CD107a. Both NK cell education and HIV-mediated changes in HLA expression influenced NK cell responses to iCD4 cells.
IMPORTANCE Epidemiological studies show that natural killer (NK) cells have anti-HIV activity: they are able to reduce the risk of HIV infection and/or slow HIV disease progression. How NK cells contribute to these outcomes is not fully characterized. We used primary NK cells and autologous HIV-infected cells to examine the role of education through four inhibitory killer immunoglobulin-like receptors (iKIRs) from persons with HLA types that are able to educate NK cells bearing one of these iKIRs. HIV-infected cells activated NK cells through missing-self mechanisms due to the downmodulation of cell surface HLA expression mediated by HIV Nef and Vpu. A higher frequency of educated than uneducated NK cells expressing each of these iKIRs responded to autologous HIV-infected cells by producing CCL4, IFN-γ, and CD107a. Since NK cells were from non-HIV-infected individuals, they model the consequences of healthy NK cell–HIV-infected cell interactions occurring in the HIV eclipse phase, when new infections are susceptible to extinction.
INTRODUCTION
Natural killer (NK) cells are components of the innate immune system with important roles in antiviral and antitumor responses (1). They respond to viruses at early stages of infection by secreting cytokines and chemokines and by degranulating, which results in target cell killing and viral control (1–4). NK cell function depends on the integration of signals received from inhibitory and activating NK cell receptors (NKRs) (5). Among NKRs, killer immunoglobulin-like receptors (KIRs) are a large family of receptors expressed on NK cells and other lymphocyte subsets (6). KIR ligands include major histocompatibility complex (MHC) class I or HLA antigens, expressed on all nucleated cells. The binding of inhibitory KIRs (iKIRs) to their HLA ligands prevents NK cells from responding to healthy HLA-expressing self-cells. NK cells acquire their effector functions through a process called education. NK cell education occurs when iKIR+ NK cells interact with self HLA+ cells during development. Educated NK cells are primed for activation when they encounter target cells with reduced levels of HLA ligands due to virus infection or tumor transformation. The iKIR+ NK cell subsets that are unable to interact with self HLA during development remain uneducated and are hyporesponsive to target cell stimulation (7–12).
KIR and HLA genes are highly polymorphic and map to different chromosomes. Thus, educated and uneducated NK cells can coexist, as can NK cells with various levels of education and responses to stimulation, due to KIR and HLA allelic variation (13–16).
KIR2DL1, KIR2DL2, and KIR2DL3 recognize HLA-C allotypes, which can be classified into C1 and C2 groups. C1 allotypes have an asparagine at position 80 of the heavy chain and are ligands for KIR2DL3. C2 group allotypes have a lysine at this position and are ligands for KIR2DL1 and KIR2DS1 (14, 17, 18). KIR2DL2 is an intermediate receptor that also binds C1 allotypes. However, based on assays measuring the binding of KIR-Fc fusion molecules to HLA class I (HLA-I)-coated microbeads, KIR2DL2 recognizes some C2 allotypes (14, 19).
KIR haplotypes can be divided into A and B, based on KIR gene content (5, 20, 21). KIR haplotype A includes framework KIR genes and genes encoding mostly iKIRs. KIR B haplotypes include various numbers of genes encoding activating KIRs (aKIRs) in addition to the genes present in KIR haplotype A. The genes encoding KIR2DL2 and KIR2DL3 are alleles at the same locus. KIR2DL2 allotypes encoded by centromeric KIR haplotype B (Cen B) alleles have a greater avidity for C1 group HLA than those encoded by centromeric KIR haplotype A (Cen A) alleles. KIR2DL3 variants encoded by Cen A alleles have a higher avidity for C1 allotypes than those encoded by Cen B alleles (13). KIR2DL1 receptors are encoded by a separate KIR locus present in both Cen A and Cen B. The Cen A allele-encoded KIR2DL1 receptors bind C2 group antigens more avidly than the Cen B allele-encoded allotypes (13). In general, KIR2DL1+ NK cells are educated through the binding of KIR2DL1 to C2, whereas KIR2DL2+ and KIR2DL3+ NK cells are educated through the binding of these receptors to C1 ligands.
The iKIR KIR3DL1 interacts with a subset of HLA-A and -B antigens containing Bw4 motifs (22–24). Allotypes belonging to the Bw4 group differ from HLA-Bw6 variants at amino acids 77 to 83 of the HLA heavy chain (23, 25). Bw4 allotypes can have an isoleucine (80I) or a threonine (80T) at position 80 of the HLA heavy chain, which influences the avidity of KIR3DL1 binding to its ligands. In general, Bw4*80I allotypes have a higher avidity than Bw4*80T subtypes for KIR3DL1, leading to more-potent education and responsiveness (15, 16, 26–28). Bw6 allotypes do not interact with KIR3DL1, and KIR3DL1+ NK cells from Bw6 homozygotes (hmzs) remain uneducated. The high level of polymorphism of KIR genes influences their gene products’ cell surface expression, avidity, and specificity for HLA ligands. Boudreau et al. have shown that the expression levels of both KIR3DL1 and Bw4 and the strength of receptor-ligand binding predict NK education and responses to HLA-null cells and autologous HIV-infected CD4 (iCD4) T cells (26). So far, 77 allotypes have been identified for KIR3DL1 and are categorized into four groups based on their surface expression. These include KIR3DL1-null allotypes, with no detectable cell surface expression, KIR3DS1, KIR3DL1-low, and KIR3DL1-high allotype groups (29–31). The presence of the homozygous KIR3DL1 genotype encoding at least one KIR3DL1-high allotype (KIR3DL1*h/*y) cocarried with HLA-B*57, which encodes a Bw4*80I isoform (*h/*y+B*57), is associated with slower progression to AIDS and lower HIV loads in infected individuals, and with more protection from HIV infection, than observed for carriers of the receptor or ligand alone, or neither (32, 33). NK cells expressing combinations of KIR3DL1-high allotypes with Bw4*80I allotypes, such as HLA-B*51, -B*52, and -B*57, have a higher functional capacity to produce gamma interferon (IFN-γ) when stimulated with HLA-null cells (34–38).
HIV infection downmodulates the expression of HLA-A, -B, and -C (39–41). Therefore, it is expected that iKIR+ NK cells educated through these HLA molecules will have higher responses to autologous iCD4 cells than their uneducated counterparts. We tested this hypothesis by stimulating NK cells with autologous iCD4 cells and examining the induction of CCL4, IFN-γ, and CD107a functions in educated versus uneducated single-positive (sp) KIR2DL1+, KIR2DL2+, KIR2DL3+, and KIR3DL1+ NK cells. These educated spKIR2DL1+, spKIR2DL2+, and spKIR2DL3+ NK cells exhibited higher responses to iCD4 cells than their uneducated counterparts. The expression levels of both KIR3DL1 and its Bw4 ligand, as well as the avidity of receptor-ligand binding, influenced spKIR3DL1+ NK cell education potency and iCD4 cell responsiveness.
RESULTS
HLA-C1 and HLA-C2 are downmodulated on iCD4 cells.
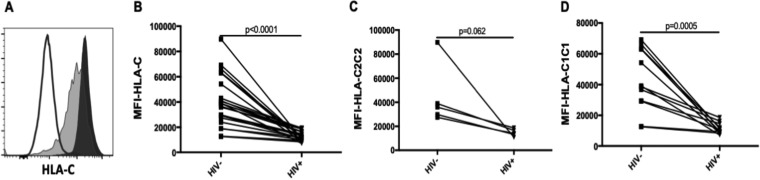
HIV infection downmodulates cell surface HLA-A, -B, and -C expression (39, 41–45). However, HIV-dependent downmodulation of HLA-C is virus strain dependent (41, 43, 46). To investigate whether HIVJR-CSF downmodulated HLA-C expression on iCD4 cells, we stained uninfected CD4 (unCD4) and iCD4 cells with monoclonal antibody (MAb) DT9. Figure 1A shows the staining of unCD4 and iCD4 cells with DT9. The mean fluorescence intensity (MFI) of DT9 staining of iCD4 cells was significantly lower than that of unCD4 cells from 23 subjects (Fig. 1B). HLA-C2 expression trended toward being lower in iCD4 cells than in unCD4 cells (P, 0.062 by the Wilcoxon matched pairs test [Fig. 1C]), while C1 expression was significantly lower on iCD4 cells than on unCD4 cells (P, <0.001 by the Wilcoxon test [Fig. 1D]). Thus, infection of CD4 cells with HIVJR-CSF downmodulated HLA-C expression.
FIG 1.
HLA-C is downmodulated on CD4 cells infected with HIVJR-CSF. (A) MFI of HLA-C expression on uninfected and HIV-infected CD4 cells. Shown is staining for HLA-C on uninfected (filled histogram) and infected (shaded histogram) CD4 cells and on isotype control cells (open histogram). (B) MFI of HLA-C staining on uninfected (HIV–) and HIV-infected (HIV+) CD4 cells from 23 subjects. (C) MFI of HLA-C staining on HIV– and HIV+ CD4 cells from 5 HLA-C2 hmzs. (D) MFI of HLA-C staining on HIV– and HIV+ CD4 cells from 11 HLA-C1 hmzs.
A lower frequency of uneducated than educated spKIR2DL1+, spKIR2DL2+, and spKIR2DL3+ NK cells responded to iCD4 cells.
HLA-C2 group antigens are ligands for KIR2DL1, while HLA-C1 antigens are ligands for KIR2DL2 and KIR2DL3. We questioned whether reduced HLA-C expression on iCD4 cells was enough to abrogate inhibitory signaling through KIR2DL1–HLA-C2, KIR2DL2–HLA-C1, and KIR2DL3–HLA-C1 interactions. The strategy used to gate on spKIR2DL1+, spKIR2DL2+, and spKIR2DL3+ NK cells and their functional profiles following iCD4 cell stimulation are shown in Fig. 2A to D. The frequencies of iCD4 cell-stimulated educated and uneducated spKIR2DL1+, spKIR2DL2+, and spKIR2DL3+ NK cells displaying each of seven possible functional profiles characterized by CCL4, IFN-γ, and CD107a production, total function, total CCL4, total IFN-γ, and total CD107a functions are shown in Fig. 3.
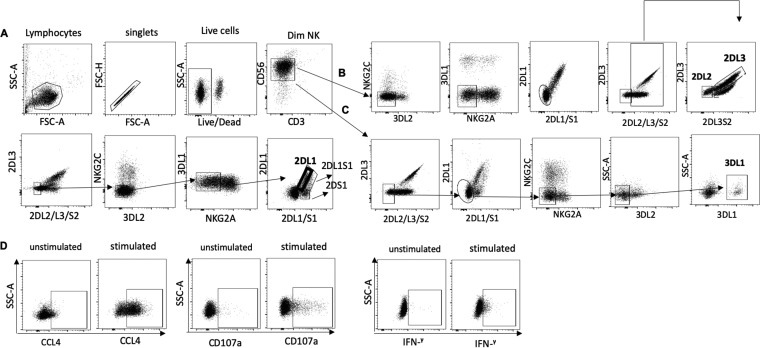
FIG 2.
Strategy used to gate on spKIR2DL1+, spKIR2DL2+, spKIR2DL3+, and spKIR3DL1+ NK cells. (A) Live, singlet lymphocytes were gated on; from these, CD3− CD56dim cells were selected. spKIR2DL1 NK cells were gated from KIR2DL2–, KIR2DL3–, KIR2DS2–, KIR3DL2–, NKG2C–, NKG2A–, KIR3DL1–, and KIR2DS1– NK cells. (B) spKIR2DL2 and spKIR2DL3 NK cells were gated from NKG2C–, KIR3DL2–, KIR3DL1–, NKG2A–, KIR2DL1–, KIR2DS1–, and KIR2DS2– NK cells. (C) spKIR3DL1 NK cells were gated on from KIR2DL2–, KIR2DL3–, KIR2DS2–, KIR2DL1–, KIR2DS1–, NKG2A–, NKG2C–, and KIR3DL2– NK cells. SSC-A, side scatter area; FSC-A, forward scatter area; FSC-H, forward scatter height; 2DL3, KIR2DL3; 2DL2/L3/S2, KIR2DL2/KIR2DL3/KIR2DS2; 3DL2, KIR3DL2; 3DL1, KIR3DL1; 2DL1, KIR2DL1; 2DL1/S1, KIR2DL1/KIR2DS1. (D) spiKIR+ cells were examined for all possible combinations of CCL4, CD107a, and IFN-γ expression by Boolean gating.
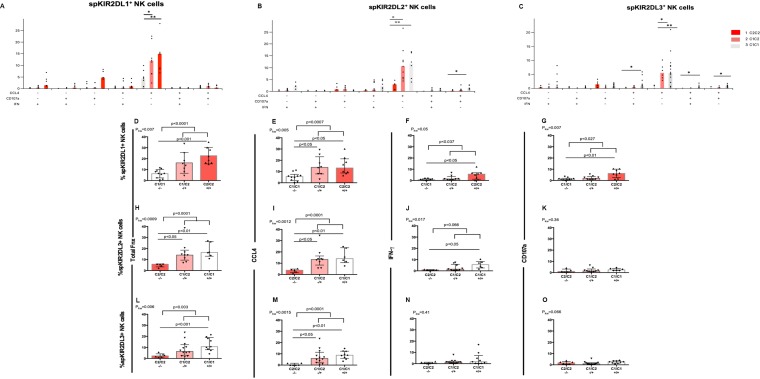
FIG 3.
The frequency of spKIR2DL1+, spKIR2DL2+, or spKIR2DL3+ NK cells responding to autologous HIV-infected CD4 cells increases with increasing ligand copy number. (A to C) The y axes show the frequencies of functional spKIR2DL1+ (A), spKIR2DL2+ (B), and spKIR2DL3+ (C) cells characterized by the seven possible combinations of CCL4 secretion, IFN-γ secretion, and CD107a expression. The presence of each of these functions in the seven functional subsets studied is indicated by a plus sign under the graph. Each point represents a separate individual. Bar heights show median values for the data sets. P values indicated above the bars linking the data sets being compared were generated using Dunn’s posttests. *, P < 0.05; **, P < 0.01. (D to O) The y axes show the frequencies of functional spKIR2DL1+ (D to G), spKIR2DL2+ (H to K), and spKIR2DL3+ (L to O) cells exhibiting the sum of all functions tested (D, H, and L), total CCL4 secretion (E, I, and M), total IFN-γ secretion (F, J, and N), and total CD107a expression (G, K, and O). For spKIR2DL1 results, cells from 26 donors were included; 10 were from subjects not educated through KIR2DL1, and 8 each were from subjects educated through KIR2DL1 by 1 or 2 HLA-C2 ligands, respectively. For spKIR2DL2 results, cells from 20 donors were included; 5 were from subjects not educated through KIR2DL2, and 9 and 6 were from subjects educated through KIR2DL2 by 1 or 2 HLA-C1 ligands, respectively. For spKIR2DL3 results, cells from 27 donors were included; 6 were from subjects not educated through KIR2DL3, while 12 and 9 were from subjects educated through KIR2DL3 by 1 or 2 ligands, respectively. All results from C1/C1 donors are represented by white bars, from C1/C2 donors by light pink bars, and from C2/C2 donors by dark pink bars. Each point represents results for a single individual. For each data set, the bar height and error bars show the median and interquartile range, respectively. P values given above the bars linking the data sets being compared were generated using Mann-Whitney tests for comparison of results from uneducated versus educated subjects and by Dunn’s posttests for comparison of results from carriers of C1/C1, C1/C2, and C2/C2 genotypes. Fnx, function; –/–, uneducated NK cell condition; –/+, educated by one ligand; +/+, educated by two ligands; Pkw, P value generated by a Kruskal-Wallis test.
In general, as the number of HLA-C ligands available to educate these three types of spiKIR+ NK cells increased, so did the frequency of functional spiKIR+ NK cells. The results were significant after correction for multiple comparisons for spKIR2DL1+ functional subsets characterized by CCL4 alone, total CCL4 secretion, and total function (P, <0.001 for all by Spearman correlation tests [Fig. 3A, D, and E]). The frequency of spKIR2DL2+ NK cell functional subsets also increased significantly with increasing ligand copy number following iCD4 cell stimulation. Results were significant for functional subsets characterized by secretion of CCL4 alone and IFN-γ alone; production of CCL4 with CD107a, CCL4 with IFN-γ and CD107a, total CCL4, and total IFN-γ; and the sum of all functions tested (P, <0.02 for all by the Spearman test [Fig. 3B and H to J]). Among iCD4 cell-stimulated spKIR3DL3+ NK cells, all the functional subsets that included CCL4 production, i.e., those that were trifunctional, CCL4+ IFN-γ+, CCL4+ CD107a+, positive for CCL4 only, total CCL4, and the sum of all functions as well as IFN-γ secretion only, increased in frequency with increasing ligand copy number (P, <0.03 for all by Spearman tests [Fig. 3C, L, and M]).
We next compared the frequencies of functional NK cells bearing each of these spiKIRs based on whether they were educated or not (i.e., by either one or two educating ligands). A higher frequency of educated than uneducated spKIR2DL1+ NK cells produced CCL4 alone, the sum of all functions, total CCL4, total IFN-γ, and total CD107a (P, <0.04 for all by Mann-Whitney tests [Fig. 3A and D to G]). More iCD4 cell-induced educated than uneducated spKIR2DL2+ NK cells produced CCL4 only, the sum of all functions tested, and total CCL4 (P, <0,006 for all by Mann-Whitney tests [Fig. 3B, H, and I]). A higher percentage of educated than uneducated KIR2DL3+ NK cells responded to autologous iCD4 cells by secreting CCL4 only and by producing CCL4, IFN-γ, and CD107a, total CCL4, and the sum of all functions tested (P, <0.04 for all by Mann-Whitney tests [Fig. 3C, L and M]).
Next, comparisons were made between the frequencies of functional cells induced by iCD4 cell stimulation of spiKIR+ NK cells that were not educated and of those that were educated by one or two ligands. Kruskal-Wallis tests were used to determine whether there were significant differences between cells from carriers of the three possible HLA-C ligand genotypes in terms of the ability of spiKIR+ cells to respond to iCD4 cell stimulation. Dunn’s posttests were performed when Kruskal-Wallis tests were significant. The frequencies of iCD4 cell-stimulated spKIR2DL1+ NK cells from subjects producing CCL4 alone, total function, total CCL4, total IFN-γ, and total CD107a were significantly lower in cells from uneducated C1/C1 subjects than in cells from educated C2/C2 subjects (P, <0.05 for all by Dunn’s posttests [Fig. 3A and D to F]). As well, fewer iCD4 cell-induced spKIR2DL1+ NK cells from C1/C1 than from C1/C2 subjects were positive for CCL4 alone and total CCL4 production (P, <0.5 for all by Dunn’s posttests [Fig. 3A and E]).
A higher frequency of iCD4 cell-activated spKIR2DL2+ NK cells from educated C1/C1 carriers than from uneducated C2/C2 carriers produced CCL4 alone, IFN-γ alone, the sum of all functions, total CCL4, and total IFN-γ (P, < 0.05 for all by Dunn’s posttests [Fig. 3B and H to J]). This was also the case for spKIR2DL3+ NK cells secreting CCL4 only or both CCL4 and IFN-γ, and for those producing CCL4 with CD107a, total function, and total CCL4 (P, <0.05 for all by Dunn’s posttests [Fig. 3C, L, and M]). More activated spKIR2DL2+ and spKIR2DL3+ NK cells from educated C1/C2 subjects than from uneducated C2/C2 subjects produced CCL4 alone (P, <0.05 by Dunn’s posttest [Fig. 3B]).
In summary, downmodulation of both C1 and C2 antigens on iCD4 cells was enough to interrupt inhibitory signals mediated by KIR2DL1–C2, KIR2DL2–C1, and KIR2DL3–C1 interactions and to reverse inhibition by educated spiKIR+ NK cells. Furthermore, increasing the C2 and C1 copy numbers was associated with augmented education potency, which translated into an increased frequency of functional spiKIR+ cells responding to autologous iCD4 cells.
Comparisons of the iCD4 cell responsiveness of within-individual spiKIR2DL+ NK cells.
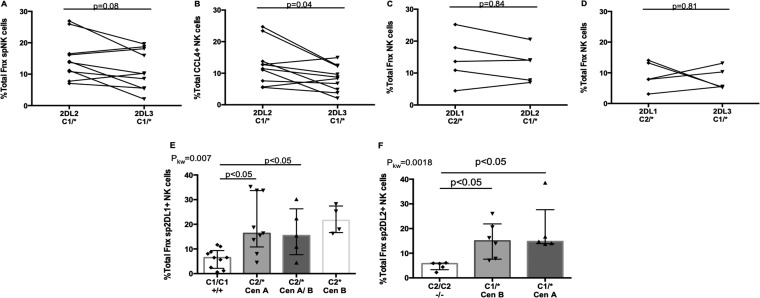
KIR2DL2 is proposed to have a higher avidity for C1 than KIR2DL3 (13). We explored whether differences in spiKIR receptor avidity for HLA-C would lead to variances in the frequency of functional cells responding to autologous iCD4 cells. For this analysis, we used NK cells from KIR2DL2/KIR2DL3 heterozygotes from C1 carriers to examine within-subject differences in the frequency of iCD4 cell-stimulated functional spKIR2DL2+ and spKIR2DL3+ NK cells. The frequency of iCD4 cell-stimulated spKIR2DL2+ NK cells was higher than that of spKIR2DS3+ NK cells for the sum of all functions and CCL4 secretion, although only the differences in CCL4 secretion achieved statistical significance (P, 0.04 by Wilcoxon tests [Fig. 4A and B]). This finding is consistent with the conclusions arrived at by others using KIR-Fc fusion proteins based on wild-type KIR2DL2 and KIR2DL3 and single-amino-acid variants for HLA-C allotype-coated microbeads, as well as inhibition-of-cytotoxicity assays using HLA-null 721.221 cells and panels of 721.221 cells transduced with single HLA-C variants (13, 47). On the other hand, comparison of the frequencies of iCD4 cell-induced functional, educated spKIR2DL1+ NK cells with those of spKIR2DL2+ or spKIR2DL3+ NK cells originating from the same person showed that they did not differ significantly from each other (Fig. 4C and D).
FIG 4.
Comparison of the responses to autologous iCD4 cells of spKIR2DL1+, spKIR2DL2+, and spKIR2DL3+ NK cells from the same donor and the influence of their allotypes on responses to iCD4 cells. (A to D) The y axes show the frequencies of functional cells characterized by the sum of all functions (A, C, and D) or CCL4 secretion (B) for 10 pairs of educated spKIR2DL2+ and spKIR2DL3+ NK cells (A and B), 5 pairs of educated spKIR2DL1+ and spKIR2DL2+ NK cells (C), and 5 pairs of educated spKIR2DL1+ and spKIR2DL3+ NK cells (D) from the same donors. The significance of within-individual differences was assessed using Wilcoxon matched pairs tests. (E) Frequencies of iCD4 cell-stimulated functional spKIR2DL1+ NK cells generated by donors cocarrying a noneducating C1/C1 genotype or educating C2/* genotypes. The frequency of functional cells was compared for donors cocarrying C2/* with KIR2DL1 genotypes where both alleles mapped to the centromeric KIR haplotype A (Cen A), one allele mapped to Cen A and the other to the centromeric KIR haplotype B (Cen B), or both alleles mapped to Cen B. (F) Frequencies of iCD4 cell-stimulated functional spKIR2DL2+ NK cells generated when donors cocarried a noneducating C2/C2 genotype or an educating C1/* genotype. The frequency of functional cells was compared for donors cocarrying C2/* with KIR2DL2 genotypes where both alleles mapped to Cen A or Cen B. Each point represents results for a single individual. For each data set, the bar height and error bars show the median and interquartile range, respectively. P values are given above the bars linking the data sets being compared. 2DL1, spKIR2DL1+; 2DL2, spKIR2DL2+; 2DL3, spKIR2DL3+; C1/*, either C1/C1 or C1/C2; C2/*, either C1/C2 or C2/C2.
KIR2DL1 allotypes mapping to Cen A have a higher avidity for C2 antigens than those mapping to Cen B, while KIR2DL2 allotypes mapping to Cen B bind C1 antigens with a higher avidity than those mapping to Cen A (13). Since iKIR receptor–HLA ligand avidity affects education potency, we asked whether responsiveness to iCD4 cells differed on the basis of the spKIR2DL1, -L2, or -L3 allotype. Similar frequencies of functional cells from Cen A KIR2DL1 hmzs, Cen B KIR2DL1 hmzs, and Cen A/Cen B KIR2DL1 heterozygotes responded to iCD4 cells (Fig. 4E). So, too, similar frequencies of spKIR2DL2 NK cells from Cen A and Cen B hmzs responded to iCD4 cells (Fig. 4F). Since all the allotypes for spKIR2DL3+ NK cells mapped to Cen A, we were unable to evaluate the influence of the KIR2DL3 allotype on the frequency of functional cells responding to iCD4 cells.
Expression of HLA-Bw4 and KIR3DL1 allotypes on the surfaces of CD4 and NK cells.
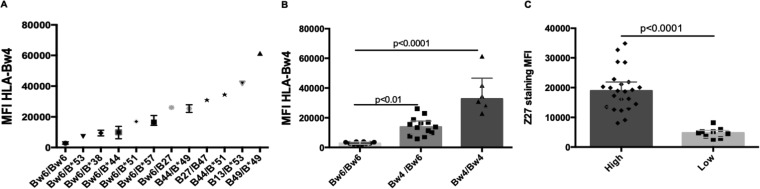
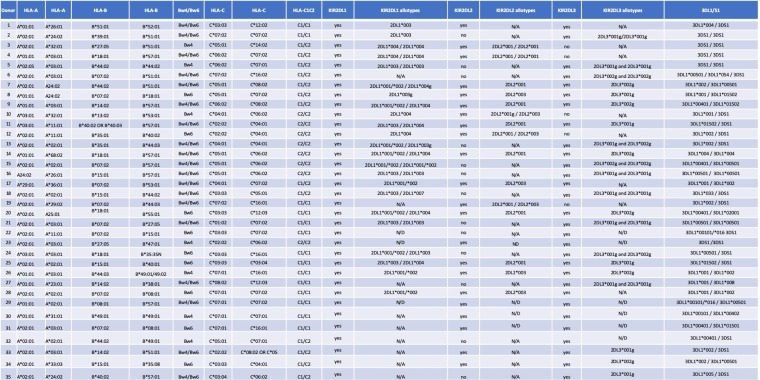
Bw4 subtypes have variable cell surface expression densities (26). We confirmed this by staining donor CD4 cells with a MAb specific for HLA-Bw4 (Fig. 5A). Cells from Bw6 hmzs had background levels of staining, which were significantly lower than those on cells from Bw4/Bw6 heterozygotes expressing only one Bw4 allotype and on cells from Bw4 hmzs (Fig. 5B). KIR3DL1 allotypes are expressed at different intensities on the surfaces of NK cells (16, 31). Table 1 shows the KIR3DL1/KIR3DS1 (KIR3DL1/S1) genotypes and allotypes of the study population. The MFI of binding by MAb Z27 to KIR3DS1 allotypes on NK cells is lower than, and distinct from, that to KIR3DL1-low and KIR3DL1-high allotypes on NK cells. Using MAb Z27 to stain NK cells from KIR3DL1/S1 heterozygotes and KIR3DL1 hmzs, we observed that KIR3DL1-high allotypes had a significantly higher MFI than KIR3DL1-low allotypes on NK cells (Fig. 5C).
FIG 5.
HLA and KIR3DL1 allotypes are expressed at distinct intensities on CD4 T cells and NK cells, respectively. (A) The y axis shows the mean fluorescence intensity (MFI) of expression of HLA-Bw4 antigens from individuals carrying distinct HLA genotypes by use of a MAb specific for HLA-Bw4. (B) The y axis shows the MFI generated by staining CD4 T cells from Bw6 hmzs, Bw4/Bw6 heterozygotes, and Bw4 hmzs with the HLA-Bw4-specific MAb. (C) The y axis shows the MFI of expression of KIR3DL1-high allotypes versus KIR3DL1-low allotypes on spKIR3DL1+ NK cells. Each point indicates results for a single individual. For each data set, the bar height and error bars show the median and interquartile range, respectively.
TABLE 1.
KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1/S1, and HLA genotypes and allotypes of the study populationa
2DL1, KIR2DL1; 2DL2, KIR2DL2; 2DL3, KIR2DL3; 3DL1/S1, KIR3DL1/KIR3DS1.
Extent of HLA-Bw4 downmodulation on iCD4 cells compared to unCD4 cells.
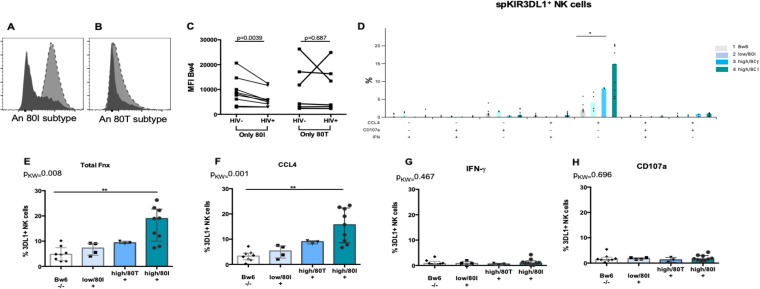
HIV Nef downmodulates HLA-A and -B from the surfaces of infected cells (39, 44, 45). We questioned whether there was a difference between Bw4*80I and Bw4*80T allotypes in terms of the extent to which they were downmodulated on iCD4 cells compared to unCD4 cells. For this analysis, we included donors expressing either Bw4*80I or Bw4*80T isoforms. HIV infection reduced the expression of Bw4*80I to a greater extent than that of Bw4*80T (Fig. 6A to C).
FIG 6.
The extent of HIV-mediated downmodulation of HLA-B predicts the frequency of KIR3DL1+ NK cells stimulated by autologous iCD4 cells. (A to C) HIV infection of CD4 cells downmodulates HLA-Bw4*80I to a greater extent than HLA-Bw4*80T. (A and B) A Bw4-specific MAb was used to stain iCD4 (shaded histograms) and unCD4 (filled histograms) T cells from a subject who expressed HLA-Bw4*80I (A) or HLA-Bw4*80T (B). (C) The y axis shows the MFI of HLA-Bw4*80I expression (left) and HLA-Bw4*80T expression (right) on uninfected (HIV–) and HIV-infected (HIV+) CD4 cells. (D to H) The y axes show the frequencies of spKIR3DL1+ NK cells responding to autologous iCD4 cells by expressing the seven possible functions characterized by secretion of CCL4, secretion of IFN-γ, and expression of CD107a (D), the sum of all functions tested (E), total CCL4 secretion (F), total IFN-γ secretion (G), and total CD107a expression (H). Each point represents a separate individual. For each data set, the bar height and error bars represent the median and interquartile range, respectively. P values indicated above the bars linking the data sets being compared were generated using Kruskal-Wallis tests with Dunn’s posttests (*, P < 0.05; **, P < 0.01). 80I, a KIR3DL1 allotype with an isoleucine at position 80; 80T, a KIR3DL1 allotype with a threonine at position 80; MFI, mean fluorescence intensity; Fxn, function; 3DL1+, spKIR3DL1+ NK cells; Pkw, P value generated by a Kruskal-Wallis test.
KIR3DL1–Bw4 NK cell education potency influences the frequency of iCD4 cell-stimulated functional KIR3DL1+ NK cells.
Bw4*80I molecules have a higher avidity for KIR3DL1 than Bw4*80T subtypes (15, 16, 26–28). Boudreau et al. demonstrated that the expression levels of KIR3DL1 and HLA-Bw4, in addition to their affinity for each other, predicted the frequency of NK cells that express CD107a in response to HLA-null and iCD4 cell stimulation (26). We extended these observations by examining the frequencies of KIR3DL1+ NK cells responding to autologous iCD4 cells by producing all possible combinations of CCL4, IFN-γ, and CD107a, as well as the sum of all functions, total CCL4, total IFN-γ, and total CD107a. We assumed that the greater downmodulation of Bw4*80I than Bw4*80T ligands by HIV Nef would interrupt inhibitory signals through KIR3DL1–Bw4*80I interactions to a greater extent than those from KIR3DL1–Bw4*80T interactions. To verify this, we stratified subjects into four groups: group 1, Bw6 hmzs (regardless of KIR3DL1 allotype) as a negative control for KIR3DL1+ cell education; group 2, the KIR3DL1-low–Bw4*80I combination; group 3, the KIR3DL1-high–Bw4*80T combination; group 4, the KIR3DL1-high–Bw4*80I combination. The frequency of iCD4 cell-induced spKIR3DL1+ NK cells was highest for those originating from group 4 and was significantly different from that of the Bw6 hmz group for CCL4 only, the sum of all functions, and total CCL4 secretion (P, ≤0.05 for all by Dunn’s posttest [Fig. 6D to F]). Although the frequencies of iCD4 cell-induced spKIR3DL1+ functional subsets did not differ significantly among the other groups, there was a significant linear trend for increased function as the avidity and number of Bw4 ligands for KIR3DL1 increased (r, 0.77, 0.81, and 0.76 [P < 0.0001] for CCL4 only, total functions, and CCL4 functions, respectively, by Spearman tests [Fig. 6D to F]).
DISCUSSION
We studied the functional responses of four types of spiKIR+ NK cells to autologous iCD4 cells. Overall, a higher frequency of educated spiKIR+ NK cells than of their uneducated counterparts responded to iCD4 cells by producing CCL4 and, to a lesser extent, IFN-γ and CD107a. The frequency of iCD4 cell-stimulated functional spKIR+ NK cells increased as the ligand copy number increased. For spKIR3DL1+ NK cells, a higher frequency of functional KIR3DL1-high+ cells from HLA-Bw4*80I+ subjects than of those from subjects from the Bw6 hmzs group responded to iCD4 cells. KIR3DL1/HLA-Bw4 genotypes encoding either a KIR3DL1-low receptor, an HLA-Bw4*80T ligand, or both responded to autologous iCD4 cells with a frequency that was intermediate between that of NK cells from carriers of spKIR3DL1-high–HLA-Bw4*80I combinations and those from Bw6 hmzs. Both the potency of NK cell education and HIV-mediated changes in HLA expression levels on iCD4 cells affect NK cell responses to autologous iCD4 cells. These observations illustrate how educated NK cells exert anti-HIV activity.
NK cells acquire their functional potential through education mediated by iNKR interactions with their HLA-I ligands during NK cell development. NK cell education is influenced by the expression levels of iNKRs on the surfaces of NK cells and of HLA-I on stimulatory/target cells, receptor-ligand avidity, and the number of iNKRs to self HLA class I ligands (14, 26, 35, 38, 48). NK cell activation occurs when inhibitory signals mediated through iNKR–HLA-I interactions are interrupted as a result of the downmodulation of self HLA-I ligands on iCD4 target cells.
HIV infection reduced the expression levels of HLA-A, -B, and -C on iCD4 cells. This has been shown by others (39, 41, 44, 45). HLA-C expression on iCD4 cells is downmodulated in a Vpu-dependent manner (41). While HIV strains differ in their abilities to reduce HLA-C expression on iCD4 cells, we confirmed that HIVJR-CSF, the isolate used to infect CD4 cells, decreased the densities of both HLA-C1 and -C2 subtypes on the surfaces of iCD4 cells (41, 43, 46). The reduction in the density of HLA-C2 on the surfaces of iCD4 cells did not achieve statistical significance, likely due to the small number (n = 5) of study subjects tested who were C2/C2 hmzs. C1 expression was significantly lower on the surfaces of iCD4 cells than on those of unCD4 cells.
Functional, educated spKIR2DL1+, -L2+, and -L3+ NK cells from donors carrying two copies of an educating ligand responded to iCD4 cell stimulation with higher frequencies than those of their one-ligand-educated and uneducated counterparts. Thus, C2 and C1 downmodulation was enough to reduce or reverse inhibitory signals mediated by spiKIR–HLA-C interactions and to shift the balance of inhibitory and activating signals received toward activation in response to iCD4 cell stimulation. Thus, a determinant of iCD4 cell-induced spiKIR2DL function is the presence or absence of an educating iKIR–ligand pair. Whether one or two copies of ligands for iKIR are present also influences education potency and NK cell responsiveness to iCD4 target cells with downmodulated ligand expression.
Moesta and colleagues reported that KIR2DL2 and KIR2DL3 are less specific for C1 subtypes than KIR2DL1 is for C2 subtypes (14, 49). So, too, KIR2DL2 is reported to be less specific for C1 ligands than KIR2DL3 (49). We found no evidence that uneducated spKIR2DL2+ NK cells from carriers of promiscuous C2 subtypes previously reported to interact with KIR2DL2 had higher responses to autologous iCD4 cells than those from carriers of nonpromiscuous C2 subtypes. These findings were limited by the small size of the study population available for this analysis. However, our results may differ from those predicted by others due to differences in the experimental conditions used. Moesta et al. measured cytotoxicity using NK cell clones transduced with KIRs cocultured with the HLA-null cell line 721.221 transfected with a panel of HLA-C allotypes (14). They also evaluated the avidity of the binding of KIR-Fc fusion proteins to HLA isotype-coated beads (13, 49). We used primary NK cells, iCD4 cell stimulation of NK cells, and flow cytometry gating strategies to ascertain the frequency of functional spiKIR+ cells. Alter et al. showed that HIV peptides alter KIR–HLA binding and can shift responses from inhibition to activation and vice versa (50). HIV epitopes that affect receptor-ligand binding may be present under our experimental conditions, whereas such peptides would be absent in binding assays and inhibition-of-cytotoxicity assays performed by others (14, 47).
The frequencies of iCD4 cell-induced functional cells from KIR2DL2/KIR2DL3 heterozygotes were higher among spKIR2DL2+ than among spKIR2DL3+ cells. Both these NK cell subsets were educated in the same host by the same HLA-C1 antigens, reducing between-subject differences in educating HLA-I antigens. This is in line with KIR2DL2 having a higher avidity than KIR2DL3 for C1 antigens, due to differences in amino acids in the D1 and D2 domains of these two receptors (13, 18, 47). A comparison of the iCD4 cell-induced responses of educated spKIR2DL1+ NK cells with those of educated spKIR2DL2+ or spKIR2DL3+ NK cells from the same individual failed to detect differences in the frequency of responsive NK cells, even though KIR2DL1–C2 is a stronger binding pair than KIR2DL2–C1 and KIR2DL3–C1 (47, 51).
The Ab panel and gating strategy we used allowed us to assess the functions of exclusively gated KIR2DL1+, -L2+, and -L3+ primary NK cells stimulated with autologous iCD4 cells. Because there are no KIR2DL2-specific Abs available, we used a MAb combination that included DX27 (KIR2DL2, -L3, and -S2 specific), 180701 (KIR2DL3 specific), and 1F12 (KIR2DL3 and -S2 specific) to distinguish between spKIR2DL2+, spKIR2DL3+, and spKIR2DS2+ NK cells. These were detected using a gating strategy described by us and others (52–54). Work by others using either KIR2DL2-transduced NK clones or inclusively gated KIR2DL2 NK cells coexpressing other NKRs demonstrated that NKRs other than KIR2DL2 can affect the education of KIR2DL2+ NK cells (53). KIR2DS2 is particularly important to consider in this regard. The KIR2DS2 gene is in a close linkage disequilibrium with KIR2DL2, and all carriers of KIR2DL2 also carry KIR2DS2 genes. The gating strategy we used in this report to isolate and study spKIR2DL2+ NK cell responses to iCD4 cells minimized the possibility that these responses were due to coexpression of KIR2DS2 and many other NKRs coexpressed on this subset of NK cells.
Several studies have shown the importance of specific KIR3DL1–HLA-B allotype combinations in slower progression to AIDS, HIV viral load control, inhibition of HIV replication, and a reduced risk of HIV infection in HIV-exposed seronegative subjects (32, 33, 37, 38, 55–57). NK cells respond rapidly to HIV infection without the need for prior priming or a lengthy period of differentiation into activated cells (58). The NK cells we used in this study originated from non-HIV-infected donors. Their interactions with autologous HIV-infected cells reflect the way in which healthy NK cells respond to recently infected self CD4 cells that would be generated soon after HIV infection. If present during the eclipse phase of infection, such NK cells would have the potential to contribute to the extinction of small foci of HIV-infected cells present at the portals of entry where NK cells have been reported to be present (59, 60). Such activity has the potential to prevent the establishment of infection. Our results suggest that not only do KIR3DL1+ cells respond to autologous HIV-infected cells but that KIR2DL1+, -L2+, and -L3+ NK cells do so as well. It is worth noting that each of the spiKIR populations we worked with represents a small subset of the total NK cell population, on the order of <5% of NK cells (34, 61). Although NK cell populations other than spiKIRs likely also contribute to HIV control, investigation of the responses of spiKIRs to autologous iCD4 cells is useful for dissecting the roles of NK cell education and HLA downmodulation by HIV Nef in the activation of these cells for antiviral activity.
Our results for the functional responsiveness of spKIR3DL1+ NK cells to iCD4 cells are in line with those reported by Boudreau et al. (26), with some differences. Among these was the assessment of CCL4 secretion, which made up the bulk of the functional responses assessed. CCL4 secretion is an important anti-HIV function that blocks the infection of new target cells (62). NK cells from carriers of the KIR3DL1-high–Bw4*80I combination responded to iCD4 cells with the highest frequency of functional cells. The functionality of NK cells from Bw6 hmz donors was the lowest, given that Bw6 is not a ligand for KIR3DL1.
Some of the limitations of this study merit noting. The size of the study population limited our ability to detect a broader range of between-group differences. Also, we did not stain for all NK cell receptors. The LSR II Fortessa X-20 instrument that we used for data acquisition detected fluorochromes in as many as 18 channels. It is possible that other NKRs, either activating or inhibitory, were differentially expressed on the NK cell subsets we focused on and could have contributed unequally to NK cell function. Questions to investigate in future studies include the contributions of the activating natural cytotoxicity receptors and NKG2D to anti-HIV activity or to modulation of the anti-HIV activities of KIR2DL1+, -L2+, and -L3+ and KIR3DL1+ NK cells.
In conclusion, we demonstrated the impact of education through iKIR–HLA pairs on NK cell responsiveness to autologous iCD4 cells. The higher frequencies of educated than uneducated NK cells responding to iCD4 cells by secreting CCL4 in all cases (for KIR2DL1+, -L2+, and -L3+ and KIR3DL1+ cells) and by producing IFN-γ and CD107a in a subset of comparisons highlight the potential role of educated NK cells in viral control by inducing the production of CC chemokines, which compete with HIV to bind to the CCR5 coreceptor for HIV entry; by eliciting the secretion of IFN-γ, an important antiviral cytokine; and by inducing the expression of CD107a, an NK cell degranulation marker (37, 62).
MATERIALS AND METHODS
Ethics statement.
This study was conducted in accordance with the principles expressed in the Declaration of Helsinki. It was approved by the Institutional Review Boards of the Comité d’Éthique de la Recherche du Centre Hospitalier de I’Université de Montréal and the Research Ethics Board of the McGill University Health Center. All subjects were healthy adults who provided written informed consent for specimen collection and subsequent analyses.
Study population.
The study population included 35 subjects. All were healthy HIV-negative individuals recruited from the St. Luc cohort or from the Chronic Viral Illness Service at the McGill University Health Centre in Montreal, QC, Canada. Table 1 shows the HLA allotypes of the 35 subjects; whether KIR2DL1, KIR2DL2, and KIR2DL3 genes were present; and if so, the alleles present at these loci as well as at the KIR3DL1/S1 locus.
Genotyping.
Genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) or from Epstein-Barr virus-transformed cells using QIAamp DNA Blood minikits (Qiagen, Mississauga, ON, Canada) according to the manufacturer’s instructions. Typing for HLA class I (HLA-I) was performed by sequencing using commercial reagents (Atria Genetics, Inc., South San Francisco, CA). KIR gene content determination and allele typing were carried out using the ScisGo KIR v3 typing kit (Scisco Genetics Inc., Seattle, WA) according to the manufacturer’s protocol. Briefly, the method, modified from the approach described by Nelson et al. (63), employs an amplicon-based two-stage PCR, followed by sample pooling and sequencing using a MiSeq v2 PE500 system (Illumina, San Diego, CA). Results were assembled using custom software supplied as an adjunct to the ScisGo KIR v3 typing kit.
Cells.
PBMCs were isolated from leukapheresis samples by density gradient centrifugation (Lymphocyte Separation Medium; Wisent Bioproducts, St-Jean-Baptiste, QC, Canada) and were cryopreserved in 10% dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO)–90% fetal bovine serum (FBS; Wisent, Inc.). CD4 and NK cells were isolated from frozen PBMCs by negative selection using EasySep human CD4+ T cell isolation kits and EasySep human NK cell enrichment kits for CD4 and NK cell isolation, respectively (Stemcell Technologies, Inc., Vancouver, BC, Canada). Purity, verified by flow cytometry, was an average of 99.1% for CD4 cells and 96.3% for NK cells.
Preparation of HIV-infected CD4 T cell stimulators.
Isolated CD4 cells were stimulated with CD3/CD28 tetramers (ImmunoCult Human CD3/CD28 T Cell Activator; Stemcell Technologies, Inc.) and 100 IU/ml of recombinant human interleukin-2 (hrIL-2; Cedarlane, Burlington, ON, Canada) in RPMI 1640 medium containing 10% FBS, 2 mM l-glutamine, 100 IU/ml penicillin and 100 mg/ml streptomycin (all from Wisent), and 100 IU hrIL-2 (R10-100) for 4 days at 37°C in a humidified 5% CO2 incubator. On day 4, stimulated CD4+ T cells were infected with HIV-1JR-CSF-VSVG in R10-100 for 4 h. iCD4 cells were washed and were cultured for 3 days in R10-100. On day 3, the median (interquartile range) frequency of HIV-infected cells that were HIV p24+ and CD4− was 12% (9.9%, 15%). unCD4 cells were cultured in parallel with iCD4 cells for 3 days in R10-100. On day 3, both unCD4 and iCD4 cells were used to stimulate NK cells.
Detection of HLA-A, -B, and -C on the surfaces of CD4 cells.
Expression levels of HLA-I Bw4 and C were measured on unCD4 and iCD4 cells by use of MAb REA274, specific for HLA-Bw4 (Miltenyi Biotec, Auburn, CA), and MAb DT9, specific for HLA-C (Millipore Sigma, St. Louis, MO). The viabilities of unCD4 and iCD4 cells were determined using UV LIVE/DEAD fixable dead cell stain kits, followed by surface staining using either unconjugated DT9 or fluorochrome-conjugated Abs of the following specificities: CD3–BV785 (OKT3), CD4–BV421 (OKT4), CD8–PC7 (SK1), and HLA-A/B/C–fluorescein isothiocyanate (FITC) (W6/32) (all from BioLegend, San Diego, CA). After surface staining, cells were washed, fixed with 2% paraformaldehyde (PFA), and permeabilized for intracellular staining (ICS) for HIV Gag p24 using a phycoerythrin (PE)-conjugated anti-p24 MAb (KC57; Beckman Coulter, Atlanta GA). DT9 binding was detected by adding a BV605-conjugated goat anti-mouse secondary Ab (BioLegend). Cells were fixed and permeabilized before ICS with PE-conjugated anti-p24; they were then washed before resuspension in 2% PFA until acquisition within 1 h. A total of 5 × 105 to 1 × 106 cells were acquired using an LSRFortessa X-20 flow cytometer (BD Biosciences, Mississauga, ON, Canada). The MFI of Bw4 and HLA-C staining and the frequency of p24+ cells among both unCD4 and iCD4 cells were assessed.
NK cell stimulation and staining.
A total of 1 × 106 overnight-rested NK effector (E) cells were cocultured in duplicate with either unCD4 or iCD4 target (T) cells at an E:T ratio of 1:1 in R10 for 6 h at 37°C in a humidified 5% CO2 incubator. BV711-conjugated anti-CD107a (H4A3; BioLegend) was added at the beginning of the coculture. Brefeldin A (24 μg/ml; Sigma-Aldrich, Oakville, ON, Canada) and monensin (3 μg/ml; BD Biosciences) were added 1 h after coculture initiation. NK cells cultured in R10 alone served as an unstimulated negative control. An aliquot of NK cells from each study subject was tested, in parallel, for their ability to respond to stimulation with 5.8 μg/ml phorbol 12-myristate 13-acetate (PMA) and 1.2 μg/ml ionomycin (Sigma-Aldrich); all responded to this positive-control stimulus. Cells were first stained for viability using UV LIVE/DEAD staining (Thermo Fisher Scientific, St. Laurent, QC, Canada) and were then stained for cell surface markers for 25 min at 4°C using fluorochrome-conjugated Abs: CD3–BV785 (OKT3), CD14–BV785 (M5E2), CD19–BV785 (HIB19), and CD56–BV605 (HDC56) (BioLegend), KIR2DL1–allophycocyanin (APC)–Vio770 (REA284; Miltenyi Biotec), KIR2DL3–FITC (180701; R&D Systems), KIR2DS2/L3–Alexa Fluor 647 (1F12) (54), KIR3DL2–Alexa Fluor 594 (DX31) (64), KIR3DL1/S1–PE (REA168), NKG2C–PE–Vio770 (REA205), and NKG2A-biotin (REA110) (the last three from Miltenyi Biotec). Next, Abs to KIR2DL2/L3/S2–peridinin chlorophyll-A protein (PerCP)–Cy5.5 (DX27; BioLegend) and KIR2DL1/S1–VioBlue (REA1010; Miltenyi Biotec) were added for 15 min at 4°C. After washing, Qdot 655 streptavidin conjugate was added to the cells for 20 min at 4°C; they were then washed, fixed with 2% PFA, permeabilized, and subjected to ICS with anti-CCL4–AF700 (D21-1351) and anti-IFN-γ–BV510 (B27) (both from BD Biosciences). Samples were then prepared for acquisition within 1 h. A list of the Abs and reagents used in this panel is available upon request.
Flow cytometry analysis.
A total of 1.5 × 106 to 1.8 × 106 events were acquired for each sample using an LSRFortessa X-20 instrument. Results were analyzed using FlowJo software v10.3 (Tree Star, Inc., Ashland, OR). Single-stained beads (Comp Beads; BD) were used to calculate compensation. We measured the frequency of NK cells exhibiting the sum of the frequencies of all functions tested (total) and the sum of the frequencies of each functional subset secreting CCL4, secreting IFN-γ, and expressing CD107a, as well as all possible combinations of these functions, i.e., trifunctional cells, three combinations of bifunctional cells, and three kinds of monofunctional cells. These functional subsets were assessed within spKIR2DL1, spKIR2DL2, spKIR2DL3, and spKIR3DL1 NK cell populations. The data presented were background subtracted using results for the matched unCD4 cell control condition.
Statistical analysis.
GraphPad Prism 6 (GraphPad Software, La Jolla, CA) was used for data analysis and graphical presentation. Mann-Whitney and Wilcoxon matched pairs tests were used to assess the statistical significance of differences between two unmatched and matched groups, respectively. Kruskal-Wallis tests with Dunn’s posttests were used to assess the significance of differences among more than two unmatched groups. When multiple comparisons were performed, a Bonferroni correction was employed.
ACKNOWLEDGMENTS
We thank Xiaoyan Ni, Tsoarello Mabanga, and Louise Gilbert for expert technical assistance, Pascale Arlotto for nursing skills in obtaining leukapheresis specimens, and Rachel Bouchard for coordinating leukaphereses from selected study subjects. We also acknowledge and appreciate the contribution of the study participants, without whose generous donation of leukapheresis specimens this study would not have been possible.
This project received support from Canadian Institutes for Health Research grant no. 142494 and from the Fonds de Recherche du Québec-Santé AIDS and Infectious Disease Network.
Z.K. designed the study, performed the experiments and data analysis, and prepared the manuscript draft. F.P.D. contributed to designing and optimizing the multiparametric antibody panel and performed some of the optimization experiments. J.B. and B.L. recruited and followed study subjects and provided cell samples from study subjects. C.R. generated monoclonal antibody 1F12, characterized its specificity, and designed an antibody panel and strategy that allowed the identification of KIR2DL2+ NK cells. D.E.G. performed KIR region gene typing and allotyping. N.F.B. designed the study and participated in data analysis and figure and manuscript preparation. Z.K., F.P.D., J.B., B.L., C.R., D.E.G., and N.F.B. edited and revised the manuscript.
We have no financial conflicts of interest.
REFERENCES
- 1.Trinchieri G. 1989. Biology of natural killer cells. Adv Immunol 47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. 1999. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol 17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 3.Robertson MJ, Ritz J. 1990. Biology and clinical relevance of human natural killer cells. Blood 76:2421–2438. [PubMed] [Google Scholar]
- 4.Cooper MA, Fehniger TA, Caligiuri MA. 2001. The biology of human natural killer-cell subsets. Trends Immunol 22:633–640. doi: 10.1016/S1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 5.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. 2000. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A 97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trowsdale J, Jones DC, Barrow AD, Traherne JA. 2015. Surveillance of cell and tissue perturbation by receptors in the LRC. Immunol Rev 267:117–136. doi: 10.1111/imr.12314. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. 2005. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 8.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Heller G, Chewning J, Kim S, Yokoyama WM, Hsu KC. 2007. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol 179:5977–5989. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
- 10.Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, Modak S, Heller G, Dupont B, Cheung NK, Hsu KC. 2012. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest 122:3260–3270. doi: 10.1172/JCI62749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orr MT, Murphy WJ, Lanier LL. 2010. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol 11:321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. 2005. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, Parham P. 2015. Polymorphic HLA-C receptors balance the functional characteristics of KIR haplotypes. J Immunol 195:3160–3170. doi: 10.4049/jimmunol.1501358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. 2008. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol 180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 15.Carr WH, Pando MJ, Parham P. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol 175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 16.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. 1993. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A 90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. 1998. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol 161:571–577. [PubMed] [Google Scholar]
- 19.Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, Parham P. 2010. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol 185:4233–4237. doi: 10.4049/jimmunol.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhrberg M, Parham P, Wernet P. 2002. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics 54:221–229. doi: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- 21.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. 1997. Human diversity in killer cell inhibitory receptor genes. Immunity 7:753–763. doi: 10.1016/S1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 22.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. 1994. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med 180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. 1995. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med 181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stern M, Ruggeri L, Capanni M, Mancusi A, Velardi A. 2008. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood 112:708–710. doi: 10.1182/blood-2008-02-137521. [DOI] [PubMed] [Google Scholar]
- 25.Wan AM, Ennis P, Parham P, Holmes N. 1986. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol 137:3671–3674. [PubMed] [Google Scholar]
- 26.Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, Hsu KC. 2016. KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. J Immunol 196:3398–3410. doi: 10.4049/jimmunol.1502469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. 2007. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol 178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 28.O’Connor GM, Vivian JP, Widjaja JM, Bridgeman JS, Gostick E, Lafont BA, Anderson SK, Price DA, Brooks AG, Rossjohn J, McVicar DW. 2014. Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J Immunol 192:2875–2884. doi: 10.4049/jimmunol.1303142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. 2007. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol 37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 30.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. 2001. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol 166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 31.Boudreau JE, Le Luduec JB, Hsu KC. 2014. Development of a novel multiplex PCR assay to detect functional subtypes of KIR3DL1 alleles. PLoS One 9:e99543. doi: 10.1371/journal.pone.0099543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulet S, Kleyman M, Kim JY, Kamya P, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, Bernard NF. 2008. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 22:1487–1491. doi: 10.1097/QAD.0b013e3282ffde7e. [DOI] [PubMed] [Google Scholar]
- 34.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. 2008. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood 112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamya P, Boulet S, Tsoukas CM, Routy JP, Thomas R, Cote P, Boulassel MR, Baril JG, Kovacs C, Migueles SA, Connors M, Suscovich TJ, Brander C, Tremblay CL, Bernard N. 2011. Receptor-ligand requirements for increased NK cell polyfunctional potential in slow progressors infected with HIV-1 coexpressing KIR3DL1*h/*y and HLA-B*57. J Virol 85:5949–5960. doi: 10.1128/JVI.02652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamya P, Tallon B, Melendez-Pena C, Parsons MS, Migueles SA, Connors M, Miconiatis S, Song R, Boulet S, Bruneau J, Tremblay CL, Bernard NF. 2012. Inhibitory killer immunoglobulin-like receptors to self HLA-B and HLA-C ligands contribute differentially to natural killer cell functional potential in HIV infected slow progressors. Clin Immunol 143:246–255. doi: 10.1016/j.clim.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Song R, Lisovsky I, Lebouche B, Routy JP, Bruneau J, Bernard NF. 2014. HIV protective KIR3DL1/S1-HLA-B genotypes influence NK cell-mediated inhibition of HIV replication in autologous CD4 targets. PLoS Pathog 10:e1003867. doi: 10.1371/journal.ppat.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM, Bernard NF. 2010. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J Immunol 184:2057–2064. doi: 10.4049/jimmunol.0902621. [DOI] [PubMed] [Google Scholar]
- 39.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671. doi: 10.1016/S1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 40.Bonaparte MI, Barker E. 2004. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood 104:2087–2094. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 41.Apps R, Del Prete GQ, Chatterjee P, Lara A, Brumme ZL, Brockman MA, Neil S, Pickering S, Schneider DK, Piechocka-Trocha A, Walker BD, Thomas R, Shaw GM, Hahn BH, Keele BF, Lifson JD, Carrington M. 2016. HIV-1 Vpu mediates HLA-C downregulation. Cell Host Microbe 19:686–695. doi: 10.1016/j.chom.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ende Z, Deymier MJ, Claiborne DT, Prince JL, Monaco DC, Kilembe W, Allen SA, Hunter E. 2018. HLA class I downregulation by HIV-1 variants from subtype C transmission pairs. J Virol 92:e01633-17. doi: 10.1128/JVI.01633-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korner C, Simoneau CR, Schommers P, Granoff M, Ziegler M, Holzemer A, Lunemann S, Chukwukelu J, Corleis B, Naranbhai V, Kwon DS, Scully EP, Jost S, Kirchhoff F, Carrington M, Altfeld M. 2017. HIV-1-mediated downmodulation of HLA-C impacts target cell recognition and antiviral activity of NK cells. Cell Host Microbe 22:111–119.e4. doi: 10.1016/j.chom.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med 2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 45.Specht A, Telenti A, Martinez R, Fellay J, Bailes E, Evans DT, Carrington M, Hahn BH, Goldstein DB, Kirchhoff F. 2010. Counteraction of HLA-C-mediated immune control of HIV-1 by Nef. J Virol 84:7300–7311. doi: 10.1128/JVI.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachtel ND, Umviligihozo G, Pickering S, Mota TM, Liang H, Del Prete GQ, Chatterjee P, Lee GQ, Thomas R, Brockman MA, Neil S, Carrington M, Bwana B, Bangsberg DR, Martin JN, Kallas EG, Donini CS, Cerqueira NB, O’Doherty UT, Hahn BH, Jones RB, Brumme ZL, Nixon DF, Apps R. 2018. HLA-C downregulation by HIV-1 adapts to host HLA genotype. PLoS Pathog 14:e1007257. doi: 10.1371/journal.ppat.1007257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilton HG, Vago L, Older Aguilar AM, Moesta AK, Graef T, Abi-Rached L, Norman PJ, Guethlein LA, Fleischhauer K, Parham P. 2012. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol 189:1418–1430. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brodin P, Lakshmikanth T, Johansson S, Karre K, Hoglund P. 2009. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood 113:2434–2441. doi: 10.1182/blood-2008-05-156836. [DOI] [PubMed] [Google Scholar]
- 49.Moesta AK, Parham P. 2012. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front Immunol 3:336. doi: 10.3389/fimmu.2012.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, Carrington M, Allen TM, Altfeld M. 2011. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilton HG, Parham P. 2017. Missing or altered self: human NK cell receptors that recognize HLA-C. Immunogenetics 69:567–579. doi: 10.1007/s00251-017-1001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, Sverremark-Ekstrom E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaelsson J, Ljunggren HG, Malmberg KJ. 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.David G, Djaoud Z, Willem C, Legrand N, Rettman P, Gagne K, Cesbron A, Retiere C. 2013. Large spectrum of HLA-C recognition by killer Ig-like receptor (KIR)2DL2 and KIR2DL3 and restricted C1 specificity of KIR2DS2: dominant impact of KIR2DL2/KIR2DS2 on KIR2D NK cell repertoire formation. J Immunol 191:4778–4788. doi: 10.4049/jimmunol.1301580. [DOI] [PubMed] [Google Scholar]
- 54.David G, Morvan M, Gagne K, Kerdudou N, Willem C, Devys A, Bonneville M, Follea G, Bignon JD, Retiere C. 2009. Discrimination between the main activating and inhibitory killer cell immunoglobulin-like receptor positive natural killer cell subsets using newly characterized monoclonal antibodies. Immunology 128:172–184. doi: 10.1111/j.1365-2567.2009.03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carrington M, Alter G. 2012. Innate immune control of HIV. Cold Spring Harb Perspect Med 2:a007070. doi: 10.1101/cshperspect.a007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Habegger de Sorrentino A, Sinchi JL, Marinic K, López R, Iliovich E. 2013. KIR-HLA-A and B alleles of the Bw4 epitope against HIV infection in discordant heterosexual couples in Chaco Argentina. Immunology 140:273–279. doi: 10.1111/imm.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. 2007. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med 204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alter G, Teigen N, Ahern R, Streeck H, Meier A, Rosenberg ES, Altfeld M. 2007. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J Infect Dis 195:1452–1460. doi: 10.1086/513878. [DOI] [PubMed] [Google Scholar]
- 59.Haase AT. 2011. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu Rev Med 62:127–139. doi: 10.1146/annurev-med-080709-124959. [DOI] [PubMed] [Google Scholar]
- 60.Shah SV, Manickam C, Ram DR, Kroll K, Itell H, Permar SR, Barouch DH, Klatt NR, Reeves RK. 2018. CMV primes functional alternative signaling in adaptive Δg NK cells but is subverted by lentivirus infection in rhesus macaques. Cell Rep 25:2766–2774.e3. doi: 10.1016/j.celrep.2018.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lisovsky I, Kant S, Tremblay-McLean A, Isitman G, Kiani Z, Dupuy FP, Gilbert L, Bruneau J, Shoukry NH, Lebouché B, Bernard NF. 2019. Differential contribution of education through KIR2DL1, KIR2DL3, and KIR3DL1 to antibody-dependent (AD) NK cell activation and ADCC. J Leukoc Biol 105:551–563. doi: 10.1002/JLB.4A0617-242RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oliva A, Kinter AL, Vaccarezza M, Rubbert A, Catanzaro A, Moir S, Monaco J, Ehler L, Mizell S, Jackson R, Li Y, Romano JW, Fauci AS. 1998. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J Clin Invest 102:223–231. doi: 10.1172/JCI2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson WC, Pyo CW, Vogan D, Wang R, Pyon YS, Hennessey C, Smith A, Pereira S, Ishitani A, Geraghty DE. 2015. An integrated genotyping approach for HLA and other complex genetic systems. Hum Immunol 76:928–938. doi: 10.1016/j.humimm.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. 2004. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol 34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]