The RIG-I-like receptors (RLRs) are double-stranded RNA-binding proteins that play a role in initiating and modulating cell intrinsic immunity through the recognition of RNA features typically absent from the host transcriptome. While they are initially characterized in the context of RNA virus infection, evidence has now accumulated establishing the role of RLRs in DNA virus infection.
Keywords: RIG-I-like receptors, RIG-I, MDA5, DNA virus, innate immunity, herpesvirus
ABSTRACT
The RIG-I-like receptors (RLRs) are double-stranded RNA-binding proteins that play a role in initiating and modulating cell intrinsic immunity through the recognition of RNA features typically absent from the host transcriptome. While they are initially characterized in the context of RNA virus infection, evidence has now accumulated establishing the role of RLRs in DNA virus infection. Here, we review recent advances in the RLR-mediated restriction of DNA virus infection with an emphasis on the RLR ligands sensed.
INTRODUCTION
The cell intrinsic innate immune system represents one of the first lines of defense from pathogens. Through a collection of pattern-recognition receptors (PRRs), pathogens are detected via conserved molecular structures, known as pathogen-associated molecular patterns (PAMPs), which are essential for their life cycle (1). Nucleic acids, including both DNA and RNA, are essential genetic information carriers for all living organisms, including bacterial and eukaryotic pathogens, as well as viruses, and are, thus, major structures detected by the innate immune PRRs (2). Activation of PRRs by PAMPS results in the production of numerous host defense molecules, including type I and type III interferons (IFNs), proinflammatory cytokines and chemokines, as well as the expression of genes that promote an intracellular antimicrobial state (3).
The RIG-I-like receptors (RLRs), which include RIG-I (retinoic acid-inducible gene-I; DDX58), MDA5 (melanoma-differentiation-associated gene 5), and LGP2 (laboratory of genetics and physiology 2) (4, 5), are prominent intracellular PRRs that sense double-stranded RNA (dsRNA) and discriminate self versus nonself RNA. All three RLRs are part of the large and diverse superfamily 2 (SF2) of nucleic acid-dependent NTPases and share similar domain structures (6). For example, a central DExD/H-box RNA helicase core consisting of two RecA-like helicase domains promotes dsRNA recognition. The helicase domain is attached via a pincer-shaped linker to the zinc-binding C-terminal domain (CTD), which also contributes to dsRNA binding (7–13). In addition, within the N terminus of RIG-I and MDA5 are tandem caspase activation and recruitment domains (CARDs) that facilitate interactions with downstream adapter proteins and confer signaling capabilities (5). While LGP2 retains the ability to bind dsRNA with high affinity, it lacks the N-terminal CARDs and, thus, functions to regulate signaling by RIG-I and MDA5 (14–18). Given the central role of RIG-I and MDA5 in discriminating self from nonself, the remainder of this Gem will focus on these two sensors.
Despite their structural similarity, RIG-I and MDA5 recognize distinct chemical and structural features of RNA and, thus, restrict distinct subsets of viruses. RIG-I preferentially binds short (<300 bp) dsRNAs bearing a 5′ triphosphate (5′-ppp) moiety (13, 19–22). In addition to 5′-ppp RNA, biochemical studies have demonstrated RIG-I binding to dsRNA with 5 diphosphate (5′-pp) ends as well as circular RNA (23–25). The molecular feature of circular RNAs that are responsible for RIG-I sensing is not known, as they lack the necessary phosphate moieties recognized by RIG-I (24). In contrast to RIG-I, the molecular patterns that confer MDA5 discrimination are less well characterized. However, it is clear that MDA5 preferentially binds to long dsRNA (>1,000 bp) with no end specificity (26–29).
Upon the recognition of ligand, RNA signaling through MDA5 and RIG-I is regulated by a series of conformational changes as well as posttranslational modifications. For example, RIG-I K63-linked ubiquitination by tripartite motif protein 25 (TRIM25) and Riplet (RNF135) and MDA5 K63-linked ubiquitination by TRIM65 facilitate RLR oligomerization (30–36). Oligomerization of RIG-I and MDA5 drive their association with their common adapter mitochondrial antiviral signaling protein (MAVS), which initiates the activation of nuclear factor κB (NF-κB), interferon regulatory factor 3 (IRF3), IRF7, and an antimicrobial gene expression response (37, 38).
Given that RIG-I and MDA5 sense RNA, it is perhaps not surprising they were first discovered as restriction factors for RNA viruses (4). Moreover, studies that have sought to define the PAMPS recognized by RIG-I and MDA5 during RNA virus infection have consistently identified RNA viral genomes and replicative intermediates. These studies suggest that the RNA viral pathogens themselves are the predominant PAMPs recognized. Interestingly, however, several DNA viruses have also been reported to activate the RLR pathway as well as encode mechanisms to antagonize it (39–44). Given that the genomes of DNA viruses do not contain the requisite features required for RLR activation, this raises the intriguing question as to what are the PAMPs recognized during DNA virus infection. While investigations into the mechanisms of DNA virus restriction by the RLR pathway have lagged behind those of the RNA virus field, recent studies have shed significant light on the mechanism of RLR activation during DNA virus infection. Here, we discuss these recent advances and focus on findings that elucidate the species and characteristics of RNA ligands sensed during viral infection. Moreover, we emphasize the emerging concept that host-derived RNAs, rather than the viral RNAs, are prominent PAMPs that activate the RLR pathway and contribute to antiviral defense.
RLR RECOGNITION OF DNA VIRUS-ENCODED RNAS
The genomes of DNA viruses do not present the necessary biochemical features required for RLR engagement and activation. However, they still produce coding and noncoding RNAs that may adopt structures or contain the chemical moieties recognized by the RLRs. The first demonstration that DNA virus-encoded RNAs are surveyed by the RLR pathway came from studies on the ubiquitous herpesvirus Epstein-Barr virus (EBV) (40). EBV, which is a human gammaherpesvirus, is the causative agent of infectious mononucleosis and is associated with several malignancies, including Burkitt’s lymphoma (BL). Interestingly, it was observed that the exogenous expression of RIG-I in cells latently infected with EBV resulted in the expression of type I interferons (IFNs), suggesting that perhaps latent viral transcripts are sensed. During latent infection, EBV gene expression is restricted to a subset of viral transcripts, with the most abundant being the EBV‐encoded small RNAs (EBERs). The EBERs, consisting of EBER1 and EBER2, are nonpolyadenylated, untranslated RNAs of 167 or 172 nucleotides in length, respectively (45). Both RNAs are transcribed by cellular RNA polymerase III (RNAPIII) and adopt secondary structures containing multiple intramolecular short stem-loops (45, 46). Moreover, RNase fingerprinting experiments determined that the 5′-ends of the EBERs are triphosphorylated (46). In line with the EBERs possessing the necessary structural and chemical moieties for RIG-I recognition, in vivo UV cross-linking followed by immunoprecipitation and reverse-transcription PCR demonstrated that RIG-I directly interacts with both EBERs (40). Furthermore, plasmid-borne EBER expression was sufficient to induce type I IFN and interferon-stimulated gene (ISG) expression in RIG-I-expressing, EBV-negative BL cells (40).
Expression of the EBERs induces interleukin 10 (IL-10) expression which further enhances the growth of BL cells (47). However, the molecular mechanisms responsible for EBER-induced IL-10 expression was unknown. A role for the RLR pathway in EBER-induced IL-10 was discovered when it was demonstrated that knock down of RIG-I or overexpression of a dominant-negative mutant of RIG-I downregulated IL-10 production in EBER-positive cells (48). Moreover, the downstream IRF3 pathway, but not the NF-κB pathway, mediated IL-10 production (48). Thus, the recognition of EBERs by RIG-I provides an exquisite model of viral co-option of cell intrinsic defense mechanisms.
EBER1 is also detected in culture supernatants of EBV-infected cells and sera from patients with active EBV infection, and the transfer of extracellular EBER1 to uninfected cells induces an innate immune response (49, 50). Depending on the mechanism from which the extracellular EBER1 is derived, Toll-like receptor 3 (TLR3) or RIG-I are capable of recognizing the RNA. EBER1 that is released by the active secretion of lupus erythematosus-associated antigen (La)-bound RNAs is detected by TLR3 (49). In contrast, EBER1 is present within exosomes, and the uptake of EBER1-containing exosomes results in a robust IFN response that is mediated by RIG-I in a 5′-ppp-dependent manner (50).
The RLR pathway has also been implicated in the sensing of adenoviruses (Ads), which are nonenveloped, double-stranded DNA (dsDNA) viruses of the Adenoviridae family (42). Interestingly, Ads encode small noncoding RNAs that are highly similar to the EBV-encoded EBERs (45). These RNAs, termed virus-associated RNA I and II (VAI and VAII, VA-RNAs), are also highly expressed RNAPIII-derived transcripts that form dsRNA structures (51). Moreover, the VA-RNAs are also immunostimulatory. For example, transfection of in vitro-transcribed VA-RNAs or stable expression of VA-RNAs in human gastric carcinoma-derived NU-GC-3 cells induces IFN-β and ISG expression in a RIG-I-IRF3-dependent manner (42). Moreover, Ad infection triggers a biphasic induction of IFN-β and ISGs at both 12 to 24 hours postinfection (hpi) and 48 to 60 hpi, and the later induction coincides with VA-RNA expression. Importantly, RIG-I silencing or UV inactivation inhibits IFN-β and ISG expression at the later time points (42).
The sensing of VA-RNA by the RLR pathway appears to be cell type specific. Infection of mouse embryonic fibroblasts (MEFs) and granulocyte-macrophage colony stimulating factor-generated bone marrow-derived dendritic cells (GM-DCs) with VA-RNA-deleted Ad results in significantly less IFN-β expression than wild type (WT) Ad (52). Interestingly, however, while the IFN-β expression was MAVS-dependent, it was neither RIG-I- nor MDA5-dependent (52). To date, the receptor that senses VA-RNAs in MEFs and GM-DCs has not been identified.
Ads have gained significant appreciation because of their utility in gene therapy. In fact, replication-incompetent recombinant Ad vectors are the most widely used gene therapy system due to their high efficiency of gene delivery into both dividing and nondividing cells (53). However, administration of Ad vectors can induce a robust anti-Ad immune response, which has limited the application of Ad vector-mediated gene therapy (52). Given the immunostimulatory nature of the VA-RNAs, the development of Ad vectors lacking VA-RNAs may decrease the stimulation of innate immune response and yield better vectors for gene therapy (54, 55).
RNAPIII DETECTION OF CYTOPLASMIC DNA DRIVES RIG-I LIGAND SYNTHESIS
Similar to RNA, the accumulation of foreign or self-DNA in the cytosol also triggers a potent innate immune response. While much of this response is now known to be dependent on the DNA sensor cyclic GMP-AMP synthase (cGAS) (56, 57), interestingly, in some cases this response is dependent on functional RIG-I/MAVS signaling (58, 59). In 2009, the Chen and Hornung groups resolved this mystery when they discovered that cytoplasmic localized RNAPIII is able to recognize and bind AT-rich DNA and initiate promoter-independent transcription to generate 5′-ppp RNA species, which activates the RIG-I/MAVS/IFN pathway (60, 61).
Investigations into the role of RNAPIII in pathogen restriction are still limited. However, cell culture studies coupled with the identification of pediatric patients with mutations in the RNAPIII machinery that present with severe varicella zoster virus (VZV) infection support the role of RNAPIII in detecting DNA virus infection (62). Along this line, the Chen group observed that IFN-β expression induced by Ad and herpes simplex virus 1 (HSV-1; a DNA alphaherpesvirus) infection of Raw264.7 cells was attenuated by pretreatment with the RNAPIII inhibitor ML-60218 (60). Although it is possible that the effect of RNAPIII inhibition on IFN-β expression induced by Ad infection is a result of reduced VA-RNA expression (42), HSV-1 is not known to express RNAPIII-dependent transcripts. The RIG-I-RNAPIII axis in sensing HSV-1 infection was further confirmed in primary astrocytes and microglia where RIG-I and RNAPIII contribute to the restriction of HSV-1 infection (63).
In recent years, additional evidence supporting a role for RNAPIII-mediated DNA sensing has come from investigations into single-gene inborn errors of innate or cell-intrinsic immunity. In particular, the Mogenson group identified and characterized loss-of-function mutations within genes encoding subunits of RNAPIII in four children with severe VZV infection (62). VZV is a neurotropic human alphaherpesvirus that causes varicella (chicken pox) upon primary infection and herpes zoster (shingles) following reactivation. However, these children suffered from severe VZV infection in the central nervous system (CNS) or lungs and experienced VZV encephalitis, cerebellitis, or pneumonia with acute respiratory distress syndrome. Through a whole-exome sequencing approach, rare mutations in the POLR3A and/or POLR3C genes were identified. Importantly, while patient peripheral blood mononuclear cells (PBMCs) had reduced type I and III IFN in response to the RNAPIII ligand poly(dA:dT) as well as VZV infection, the IFN responses were restored when WT POLR3A and/or POLR3C genes were introduced into patient PBMCs. In addition to the initial pediatric patients, subsequent work has identified adult patients with mutations in additional RNAPIII subunits that are unable to properly control VZV infection (64, 65).
Patients with RNAPIII mutations mounted proper IFN responses to HSV-1 (62, 64). Thus, an interesting question that emerges is why RNAPIII mutations appear to selectively reduce VZV intrinsic sensing. One possible explanation has to do with the fact that the VZV genome is unique among several human herpesviruses (i.e., HSV-1, HSV-2, and EBV) in having islands of genomic sequence that are 70% to 80% AT rich (62). Given that the current model of RNAPIII-based immunity is mediated through promoter-independent transcription of AT-rich DNA, the other viruses may just lack the required sequences to initiate transcription. Along this line, it is interesting to note that the cytomegalovirus (CMV) genome also contains regions of high AT content, and moreover, a father of one of the pediatric patients was heterozygous for RNAPIII mutations and suffered from CMV encephalitis in his youth (62).
RNAPIII plays a central role in the expression of many housekeeping noncoding RNAs, including 5S rRNA, tRNA, and several small nuclear noncoding RNAs (snRNAs) (e.g., 7SK snRNA and U6 snRNA). Interestingly, host 5S rRNA expression was not affected in any of the patients with RNAPIII mutations (62, 64). Future studies are needed to determine whether other RNAPIII transcripts are expressed properly. In addition, more biochemical and structural studies are likely required to determine the molecular basis by which RNAPIII mutations disrupt promoter-independent transcription from AT-rich DNA but do not affect promoter-dependent transcription within the nucleus. We anticipate that these studies will shed additional light on the mechanisms of DNA sensing by RLRs but also provide insight into RNAPIII transcriptional control mechanisms.
RLR-SENSING OF HOST RNAS
Until recently, most studies on the RLR-dependent recognition of viruses has focused on viral-encoded RNAs or, in the case for RNAPIII sensing, the transcription of immunostimulatory noncoding RNAs from AT-rich DNA in a promoter-independent manner. However, a new paradigm is emerging in which the RLRs are activated by host RNAs during virus infection. The first implication of host RNAs activating the RLR pathway during DNA virus infection came from studies investigating retrotransposon expression during murine gammaherpesvirus 68 (MHV68) infection (66). During MHV68 de novo infection, the expression of multiple families of short interspersed elements (SINEs), including B1 and B2 SINEs, is induced. SINEs are small nonautonomous noncoding retrotransposons evolutionarily derived from RNAPIII transcripts. It was discovered that MHV68-induced B2 SINEs are robust activators of the NF-κΒ pathway and that this occurs in a MAVS-dependent manner. Although the precise receptor was not identified, B2 RNA activation of NF-κΒ via MAVS suggests that either RIG-I or MDA5 is involved.
RNAPIII is responsible for the expression of B2 RNAs, and thus, given the precedence for RIG-I to detect other RNAPIII transcripts via their 5′-ppp moieties, it is perhaps reasonable to hypothesize that RIG-I senses B2 RNAs. However, expressed B2 RNAs can also form intermolecular interactions with RNAs, generating regions of long dsRNA which may favor recognition via MDA5 (67). Regardless of their mechanism of sensing, expressed retrotransposons are emerging as prominent drivers of cell intrinsic immune responses. Moreover, remarkably, in some cases, such as MHV68 infection, the retrotransposon-induced responses are co-opted to serve proviral functions (66).
In addition to retrotransposon-derived RNAs, expressed pseudogenes have also been identified as potent RIG-I ligands. Pseudogenes are sequences that resemble in sequence a functional intact gene and can arise through either retrotransposition-based mechanisms or gene duplication events. The ability of pseudogene RNA to be recognized by an RLR was first described in studies focused on HSV-1 (68). Immunoprecipitation of RIG-I from HSV-1-infected HEK293T cells and sequencing of the bound RNAs identified many host RNAs as RIG-I ligands. Notably, the top-enriched RNAs were a group of 5S rRNA pseudogenes transcripts, with RNA5SP141 being the most enriched. 5S rRNA adopts a highly compact secondary structure consisting of multiple short stem-loop structures, and RNA secondary structure prediction analyses of the RNA5SP141 sequence suggests a similar structure. Moreover, 5S rRNA is predicted to possess a 5′-ppp moiety. Consistent with RNA5SP141 being a RIG-I ligand, transfection of in vitro-transcribed as well as plasmid-expressed RNA5SP141 stimulated IFN-β expression as well as IRF3 dimerization in a RIG-I-dependent manner. Moreover, an in vitro ATP hydrolysis assay demonstrated that RNA5SP141 activates the ATPase activity of RIG-I, indicating RNA5SP141 is a direct agonist of RIG-I. Further supporting the functional significance of the RIG-I dependent sensing of RNA5SP141, depletion of the RNA via siRNA or locked nucleic acid gapmers dampens IFN expression in response to HSV-1. Remarkably, the RIG-I dependent sensing of RNA5SP141 was found to be important during EBV as well as the RNA virus influenza A virus (IAV) infection.
RNA5SP141 RNA is primarily localized within the nucleus in uninfected cells. Similar to canonical 5S rRNA, in vitro-transcribed RNA5SP141 can interact with ribosomal protein L5 (RPL5), mitochondrial ribosomal protein L18 (MRPL18), and thiosulfate sulfurtransferase (TST) (69–71). Interestingly, upon HSV-1 infection, RNA5SP141 relocalizes to the cytoplasm in HSV-1-infected cells where it is engaged by RIG-I. While the mechanism driving RNA5SP141 relocalization is unknown, it was hypothesized that HSV-1 reduces the expression of RNA5SP141-binding proteins, resulting in its unmasking in the cytoplasm. In support of this hypothesis, infection with HSV-1 defective for the viral mRNA endonuclease VHS does not result in reduced MRPL18 and TST protein levels, and RNA5SP141 is no longer efficiently immunoprecipitated by RIG-I. However, regardless of the mechanism driving RNA5SP141 relocalization, this is an elegant study describing expressed pseudogenes as RLR ligands and prompts several interesting questions. For example, why is that only 5S rRNA pseudogenes, and not also the functional 5S rRNAs, are recognized by RIG-I? In addition, what are the mechanisms driving selective retention of 5S rRNA pseudogene transcripts in the nucleus? While the answers to these questions are unknown, we hypothesize that they are, in fact, linked and are dependent on additional protein components associated with the functional 5S and/or 5S pseudogene RNAs.
Recent studies investigating the RLR-dependent restriction of the oncogenic herpesvirus Kaposi’s sarcoma-associated herpesvirus (KSHV) have also revealed new insights into RLR ligands. Several groups, including our own, have described a role for the RLRs in restricting KSHV de novo infection as well as lytic reactivation (72–74). However, the RNAs that were sensed have remained largely unexplored. To identify the RNAs responsible for RLR activation, we performed RIG-I and MDA5 formaldehyde cross-linking RNA immunoprecipitation and deep sequencing (fRIP-seq) on primary effusion lymphoma (PEL) cells infected with KSHV (72). Similar to HSV-1, host RNAs were the most significantly enriched. While no obvious similarities emerged regarding what drives MDA5 ligand biogenesis, the top-enriched RIG-I-bound RNAs were all RNAPIII-transcribed small cytosolic noncoding RNAs that adopt highly compact dsRNA structures, including the vault RNAs (vtRNAs), Y RNAs, and retrotransposon Alu RNAs. Interestingly, all of these RNAs were previously described to be substrates for the cellular RNA triphosphatase dual specificity phosphatase 11 (DUSP11) (75), and thus, it was hypothesized that DUSP11 activity was significantly reduced during KSHV lytic infection. Indeed, RNAPII chromatin immunoprecipitation, Western blots, and reverse transcriptase quantitative PCR (RT-qPCR) analyses confirmed the downregulation of DUSP11 expression. Moreover, using a splint-ligation based assay, vtRNAs were found to accumulate as 5′-ppp or 5′-pp species specifically in the lytic cycle. Although experiments testing the contribution of the individual RNAs to the IFN response were not performed, this study highlights how defective or altered RNA processing can generate ligands for RLRs during DNA virus infection. Moreover, given the high abundance of the multiple RIG-I ligands, as well as the fact that MDA5 contributes to KSHV restriction, it is unlikely that depleting a single RNA species would significantly impact the IFN response to lytically replicating KSHV.
It should also be noted that a recent study by the Damania group also identified RIG-I ligands during KSHV infection, with the exception that the RNAs sequenced were isolated from RIG-I immunoprecipitated from the epithelial cell line iSLK.219 (73). While the Damania group also identified host RNAs associated with RIG-I, they identified multiple KSHV genomic regions that give rise to RNA fragments recognized by RIG-I. While the mechanisms responsible for the biogenesis of the immunostimulatory RNAs was not defined, together with our study, a picture emerges in which KSHV is sensed by the RLRs via the detection of both viral and host RNA.
5S rRNA pseudogene RNAs were not bound by RIG-I during KSHV lytic reactivation in PEL cells. Interestingly, however, U6 spliceosomal snRNA pseudogene RNA was identified as a RIG-I ligand. Thus, expressed pseudogene RNAs are also recognized by RIG-I during KSHV infection. Interestingly, U6 snRNA is unique among the spliceosomal snRNAs in that it does not have a cytoplasmic stage involved in its maturation (76). Given that RNA5SP141 is also nuclear localized and relocalizes upon HSV-1 infection, it will be interesting to determine whether U6 snRNA pseudogene RNA exhibits a similar infection-dependent relocalization. Thus, we anticipate that future studies investigating RLR activation by HSV-1 or KSHV will yield fundamental discoveries in both RLR sensing and RNA biology.
CONCLUDING REMARKS
The RLR-dependent restriction of DNA viruses is a rapidly emerging field, and recent literature suggests both viral and host RNAs can serve as RLR ligands (Fig. 1). Here, we have focused on describing the mechanisms of RLR activation during DNA virus infection, with an emphasis on the RNA ligands sensed, including viral RNAs, cytosolic RNAPIII-transcribed RNAs from AT-rich templates, and host RNAs. RLR activation and restriction during a specific DNA virus infection is likely mediated by multiple RNA ligands that are generated through the multiple mechanisms described here. For example, both the EBERs and RNA5SP141 are reported to have roles on the EBV life cycle (40, 48–50, 68). A similar scenario is also apparent when considering the RLR ligands that have been identified during KSHV infection, as both viral and unprocessed host noncoding RNAs are recognized by RIG-I (72, 73). Moreover, although our review has focused on DNA virus infection, the RLR-sensing and restriction of RNA viruses is also mediated through a diverse set of viral and host RNAs. In fact, during IAV infection, the RIG-I sensing of IAV genomes (77), RNAs derived from defective interfering particles (78), RNA5SP141 (68), and endogenous retroviral (ERV) elements (79) have all been reported to be key to RLR-mediated restriction.
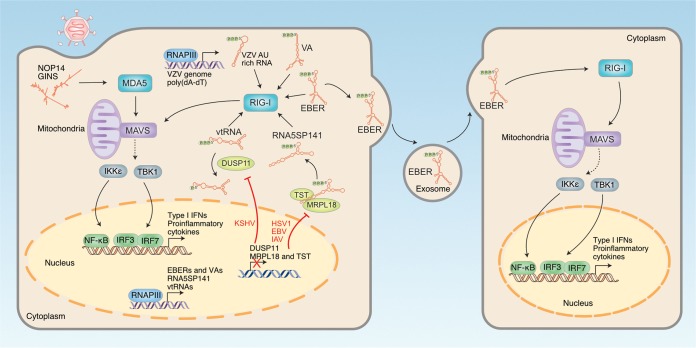
FIG 1.
Schematic of RIG-I-like receptor (RLR) activation by various RNAs during DNA virus infection. Both viral and host RNAs are recognized by RLRs and are capable of activating downstream signaling through interactions with MAVS. Activation of MAVS via the RLRs results in the production of type I IFNs and proinflammatory cytokines. Solid lines indicate direct effects or pathway connections. Dashed lines indicate indirect signaling events and are omitted here because of space. Red lines indicate inhibitory effects. DUSP11, dual specificity phosphatase 11; EBER, Epstein-Barr virus-encoded small RNA; EBV, Epstein-Barr virus; HSV-1, herpes simplex virus type 1; IAV, influenza A virus; IKKε, IκB kinase-ε; IFN, interferon; IRF, IFN regulatory factor; KSHV, Kaposi’s sarcoma-associated herpesvirus; MAVS, mitochondrial antiviral signaling protein; MDA5, melanoma differentiation-associated protein 5; MRPL18, mitochondrial ribosomal protein L18; NF-κB, nuclear factor-κB; NOP14, nucleolar protein 14; P, phosphate; RIG-I, retinoic acid-inducible gene-I; RNA5SP141, 5S ribosomal pseudogene 141 RNA; RNAPIII, RNA polymerase III; TBK-1, TANK-binding kinase 1; TST, thiosulfate sulfurtransferase; VA, viral associated RNA of adenovirus; vtRNA, vault RNA; VZV, varicella-zoster virus.
Despite significant progress over the recent years, there remain many important unknowns. For example, although the RIG-I ligand has been biochemically and structurally defined, there is still limited information regarding the requirements of a MDA5 ligand. Along a similar line, to date only one study has identified MDA5 ligands during DNA virus infection, and thus, the full range of RNA species recognized by MDA5 remains to be determined. Additionally, evidence supporting a role for RNAPIII sensing is still limited, and if its contribution to virus restriction and disease pathogenesis is to be further understood, more molecular and biochemical analyses are required. Given the relatively recent identification of patients with mutations in RNAPIIII subunits that present with serious DNA virus-mediated disease, we anticipate that investigations into RNAPIII sensing will increase. Last, intact RLR-sensing has, in some cases, been found to contribute to effective chemotherapeutic approaches, and mutations in the RLRs are associated with various autoimmune diseases. Thus, given the roles of the RLRs outside the context of virus restriction, a better understanding of the mechanisms of RLR activation, including the ligands recognized, is likely to have a broad impact.
ACKNOWLEDGMENTS
We thank members of the Karijolich laboratory for discussions.
Research in the Karijolich laboratory is funded by institutional startup funds and the Pew Charitable Trusts. J.K. is a Pew Biomedical Scholar. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. 2015. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 3.Tan X, Sun L, Chen J, Chen ZJ. 2018. Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol 72:447–478. doi: 10.1146/annurev-micro-102215-095605. [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr, Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 6.Luo D, Kohlway A, Pyle AM. 2013. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol 10:111–120. doi: 10.4161/rna.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A 104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, Inagaki F. 2009. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem 284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. 2009. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res 37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Lu C, Stewart M, Xu H, Strong RK, Igumenova T, Li P. 2009. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys 488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Kageyama M, Takahasi K, Narita R, Hirai R, Yoneyama M, Kato H, Fujita T. 2011. 55 Amino acid linker between helicase and carboxyl terminal domains of RIG-I functions as a critical repression domain and determines inter-domain conformation. Biochem Biophys Res Commun 415:75–81. doi: 10.1016/j.bbrc.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell 29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 13.Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. 2010. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 15.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol 175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 16.Murali A, Li X, Ranjith-Kumar CT, Bhardwaj K, Holzenburg A, Li P, Kao CC. 2008. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J Biol Chem 283:15825–15833. doi: 10.1074/jbc.M800542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruns AM, Leser GP, Lamb RA, Horvath CM. 2014. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell 55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A 107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, Hartmann G, Patel DJ. 2010. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, Endres S, Rothenfusser S. 2009. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A 106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 22.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 23.Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA, Barchet W, Ludwig J, Dermody TS, Hartmann G, Reis e Sousa C. 2014. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY. 2017. Sensing self and foreign circular RNAs by intron identity. Mol Cell 67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee AK, Shatkin AJ. 1971. Guanosine-5′-diphosphate at the 5′ termini of reovirus RNA: evidence for a segmented genome within the virion. J Mol Biol 61:643–653. doi: 10.1016/0022-2836(71)90069-6. [DOI] [PubMed] [Google Scholar]
- 26.Triantafilou K, Vakakis E, Kar S, Richer E, Evans GL, Triantafilou M. 2012. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci 125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 27.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol 83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJ. 2012. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep 2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oshiumi H, Miyashita M, Matsumoto M, Seya T. 2013. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog 9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem 284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 33.Lang X, Tang T, Jin T, Ding C, Zhou R, Jiang W. 2017. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med 214:459–473. doi: 10.1084/jem.20160592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 36.Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A 105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. 2013. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 38.Peisley A, Wu B, Xu H, Chen ZJ, Hur S. 2014. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West JA, Wicks M, Gregory SM, Chugh P, Jacobs SR, Zhang Z, Host KM, Dittmer DP, Damania B. 2014. An important role for mitochondrial antiviral signaling protein in the Kaposi’s sarcoma-associated herpesvirus life cycle. J Virol 88:5778–5787. doi: 10.1128/JVI.03226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J 25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen SB, Jensen SB, Nielsen C, Quartin E, Kato H, Chen ZJ, Silverman RH, Akira S, Paludan SR. 2009. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene- like receptors, which synergize to induce type I interferon production. J Gen Virol 90:74–78. doi: 10.1099/vir.0.005389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minamitani T, Iwakiri D, Takada K. 2011. Adenovirus virus-associated RNAs induce type I interferon expression through a RIG-I-mediated pathway. J Virol 85:4035–4040. doi: 10.1128/JVI.02160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Sastre A. 2017. Ten strategies of interferon evasion by viruses. Cell Host Microbe 22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan YK, Gack MU. 2016. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol 14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosa MD, Gottlieb E, Lerner MR, Steitz JA. 1981. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol 1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glickman JN, Howe JG, Steitz JA. 1988. Structural analyses of EBER1 and EBER2 ribonucleoprotein particles present in Epstein-Barr virus-infected cells. J Virol 62:902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitagawa N, Goto M, Kurozumi K, Maruo S, Fukayama M, Naoe T, Yasukawa M, Hino K, Suzuki T, Todo S, Takada K. 2000. Epstein-Barr virus-encoded poly(A)(-) RNA supports Burkitt’s lymphoma growth through interleukin-10 induction. EMBO J 19:6742–6750. doi: 10.1093/emboj/19.24.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samanta M, Iwakiri D, Takada K. 2008. Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 27:4150–4160. doi: 10.1038/onc.2008.75. [DOI] [PubMed] [Google Scholar]
- 49.Iwakiri D, Zhou L, Samanta M, Matsumoto M, Ebihara T, Seya T, Imai S, Fujieda M, Kawa K, Takada K. 2009. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J Exp Med 206:2091–2099. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baglio SR, van Eijndhoven MAJ, Koppers-Lalic D, Berenguer J, Lougheed SM, Gibbs S, Léveillé N, Rinkel RNPM, Hopmans ES, Swaminathan S, Verkuijlen SAWM, Scheffer GL, van Kuppeveld FJM, de Gruijl TD, Bultink IEM, Jordanova ES, Hackenberg M, Piersma SR, Knol JC, Voskuyl AE, Wurdinger T, Jiménez CR, Middeldorp JM, Pegtel DM. 2016. Sensing of latent EBV infection through exosomal transfer of 5′pppRNA. Proc Natl Acad Sci U S A 113:E587–E596. doi: 10.1073/pnas.1518130113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akusjarvi G, Mathews MB, Andersson P, Vennstrom B, Pettersson U. 1980. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc Natl Acad Sci U S A 77:2424–2428. doi: 10.1073/pnas.77.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi T, Kawabata K, Kouyama E, Ishii KJ, Katayama K, Suzuki T, Kurachi S, Sakurai F, Akira S, Mizuguchi H. 2010. Induction of type I interferon by adenovirus-encoded small RNAs. Proc Natl Acad Sci U S A 107:17286–17291. doi: 10.1073/pnas.1009823107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee CS, Bishop ES, Zhang R, Yu X, Farina EM, Yan S, Zhao C, Zheng Z, Shu Y, Wu X, Lei J, Li Y, Zhang W, Yang C, Wu K, Wu Y, Ho S, Athiviraham A, Lee MJ, Wolf JM, Reid RR, He TC. 2017. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis 4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Machitani M, Sakurai F, Katayama K, Tachibana M, Suzuki T, Matsui H, Yamaguchi T, Mizuguchi H. 2013. Improving adenovirus vector-mediated RNAi efficiency by lacking the expression of virus-associated RNAs. Virus Res 178:357–363. doi: 10.1016/j.virusres.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 55.Machitani M, Katayama K, Sakurai F, Matsui H, Yamaguchi T, Suzuki T, Miyoshi H, Kawabata K, Mizuguchi H. 2011. Development of an adenovirus vector lacking the expression of virus-associated RNAs. J Control Release 154:285–289. doi: 10.1016/j.jconrel.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 56.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol 7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 60.Chiu Y-H, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogunjimi B, Zhang SY, Sorensen KB, Skipper KA, Carter-Timofte M, Kerner G, Luecke S, Prabakaran T, Cai Y, Meester J, Bartholomeus E, Bolar NA, Vandeweyer G, Claes C, Sillis Y, Lorenzo L, Fiorenza RA, Boucherit S, Dielman C, Heynderickx S, Elias G, Kurotova A, Auwera AV, Verstraete L, Lagae L, Verhelst H, Jansen A, Ramet J, Suls A, Smits E, Ceulemans B, Van Laer L, Plat Wilson G, Kreth J, Picard C, Von Bernuth H, Fluss J, Chabrier S, Abel L, Mortier G, Fribourg S, Mikkelsen JG, Casanova JL, Paludan SR, Mogensen TH. 2017. Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. J Clin Invest 127:3543–3556. doi: 10.1172/JCI92280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crill EK, Furr-Rogers SR, Marriott I. 2015. RIG-I is required for VSV-induced cytokine production by murine glia and acts in combination with DAI to initiate responses to HSV-1. Glia 63:2168–2180. doi: 10.1002/glia.22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carter-Timofte ME, Hansen AF, Christiansen M, Paludan SR, Mogensen TH. 2019. Mutations in RNA polymerase III genes and defective DNA sensing in adults with varicella-zoster virus CNS infection. Genes Immun 20:214–223. doi: 10.1038/s41435-018-0027-y. [DOI] [PubMed] [Google Scholar]
- 65.Carter-Timofte ME, Hansen AF, Mardahl M, Fribourg S, Rapaport F, Zhang SY, Casanova JL, Paludan SR, Christiansen M, Larsen CS, Mogensen TH. 2018. Varicella-zoster virus CNS vasculitis and RNA polymerase III gene mutation in identical twins. Neurol Neuroimmunol Neuroinflamm 5:e500. doi: 10.1212/NXI.0000000000000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karijolich J, Abernathy E, Glaunsinger BA. 2015. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-kappaB pathway. PLoS Pathog 11:e1005260. doi: 10.1371/journal.ppat.1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karijolich J, Zhao Y, Alla R, Glaunsinger B. 2017. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res 45:6194–6208. doi: 10.1093/nar/gkx180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiang JJ, Sparrer KMJ, van Gent M, Lassig C, Huang T, Osterrieder N, Hopfner KP, Gack MU. 2018. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol 19:53–62. doi: 10.1038/s41590-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL Jr. 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev 21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smirnov A, Entelis N, Martin RP, Tarassov I. 2011. Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev 25:1289–1305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smirnov A, Comte C, Mager-Heckel AM, Addis V, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I. 2010. Mitochondrial enzyme rhodanese is essential for 5 S ribosomal RNA import into human mitochondria. J Biol Chem 285:30792–30803. doi: 10.1074/jbc.M110.151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y, Ye X, Dunker W, Song Y, Karijolich J. 2018. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat Commun 9:4841. doi: 10.1038/s41467-018-07314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Dittmer DP, Mieczkowski PA, Host KM, Fusco WG, Duncan JA, Damania B. 2018. RIG-I detects Kaposi’s sarcoma-associated herpesvirus transcripts in a RNA polymerase III-independent manner. mBio 9:e00823-18. doi: 10.1128/mBio.00823-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inn K-S, Lee S-H, Rathbun JY, Wong L-Y, Toth Z, Machida K, Ou J-H, Jung JU. 2011. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol 85:10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burke JM, Kincaid RP, Nottingham RM, Lambowitz AM, Sullivan CS. 2016. DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev 30:2076–2092. doi: 10.1101/gad.282616.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kiss T. 2004. Biogenesis of small nuclear RNPs. J Cell Sci 117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 77.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 78.Liu G, Lu Y, Liu Q, Zhou Y. 2019. Inhibition of ongoing influenza A virus replication reveals different mechanisms of RIG-I activation. J Virol 93:e02066-18. doi: 10.1128/JVI.02066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt N, Domingues P, Golebiowski F, Patzina C, Tatham MH, Hay RT, Hale BG. 2019. An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc Natl Acad Sci U S A 116:17399–17408. doi: 10.1073/pnas.1907031116. [DOI] [PMC free article] [PubMed] [Google Scholar]