Modulation of cellular antiviral signaling is a key determinant of viral pathogenesis. Human cytomegalovirus (HCMV) is a significant source of morbidity in neonates and the immunosuppressed that contains many genes that modulate antiviral signaling, yet how these genes contribute to shaping the host cell’s transcriptional response to infection is largely unclear. Our results indicate that the HCMV UL26 protein is critical in preventing the establishment of a broad cellular proinflammatory transcriptional environment. Further, we find that the host gene IKKβ is an essential determinant governing the host cell’s antiviral transcriptional response. Given their importance to viral pathogenesis, continuing to elucidate the functional interactions between viruses and the cellular innate immune response could enable the development of therapeutic strategies to limit viral infection.
KEYWORDS: BST1, HCMV, IKK, ISG15, NF-κB, cytokines, innate immunity, interferons
ABSTRACT
Viruses must negotiate cellular antiviral responses in order to replicate. Human cytomegalovirus (HCMV) is a prevalent betaherpesvirus that encodes a number of viral gene products that modulate cellular antiviral signaling. The HCMV UL26 gene has previously been found to attenuate cytokine-activated NF-κB signaling, yet the role that UL26 plays in modulating the host cell’s global transcriptional response to infection is not clear. Here, we find that infection with a UL26 deletion virus (ΔUL26) induces a proinflammatory transcriptional environment that includes substantial increases in the expression of cytokine signaling genes relative to wild-type HCMV. These increases include NF-κB-regulated genes as well as interferon-stimulated genes (ISGs), such as ISG15 and bone marrow stromal cell antigen 2 (BST2). The ΔUL26 mutant-mediated induction of ISG15 expression was found to drive increases in global protein ISGylation during ΔUL26 mutant infection. However, short hairpin RNA (shRNA) and CRISPR-mediated targeting of ISG15 indicated that its induction does not restrict HCMV infection. In contrast, shRNA-mediated targeting of BST2 demonstrated that BST2 restricts HCMV cell-to-cell spread. In addition, the increased expression of both of these ISGs and the global enhancement in protein ISGylation were found to be dependent on the activity of the canonical inhibitor of NF-κB kinase beta (IKKβ). Both CRISPR-based and pharmacologically mediated inhibition of IKKβ blocked the induction of ISG15 and BST2. These results suggest significant cross-talk between the NF-κB and interferon signaling pathways and highlight the importance of IKK signaling and the HCMV UL26 protein in shaping the antiviral response to HCMV.
IMPORTANCE Modulation of cellular antiviral signaling is a key determinant of viral pathogenesis. Human cytomegalovirus (HCMV) is a significant source of morbidity in neonates and the immunosuppressed that contains many genes that modulate antiviral signaling, yet how these genes contribute to shaping the host cell’s transcriptional response to infection is largely unclear. Our results indicate that the HCMV UL26 protein is critical in preventing the establishment of a broad cellular proinflammatory transcriptional environment. Further, we find that the host gene IKKβ is an essential determinant governing the host cell’s antiviral transcriptional response. Given their importance to viral pathogenesis, continuing to elucidate the functional interactions between viruses and the cellular innate immune response could enable the development of therapeutic strategies to limit viral infection.
INTRODUCTION
Human cytomegalovirus (HCMV) is a prevalent, opportunistic betaherpesvirus. Primary infection in immunocompetent hosts is typically asymptomatic. However, the virus establishes latent infection and persists in the host indefinitely (1, 2). While infection in healthy individuals is usually resolved without incident, HCMV infection presents a serious threat to the health of AIDS patients, organ transplant recipients, chemotherapy recipients, and other populations with impaired immune function. HCMV infection can also be transmitted from mother to child through the placenta, and congenital HCMV infection occurs in an estimated 1 to 2% of all live births (3). Congenital infection often results in serious complications, including hearing loss, vision loss, and neurological damage (4).
HCMV is the largest human herpesvirus, with an ∼235-kb genome containing over 200 viral genes (5). The HCMV virion is composed of an outer phospholipid envelope and an inner capsid housing the viral genome. A diverse array of proteins, collectively referred to as the tegument layer, is packaged into the virion between the envelope and capsid layers and is delivered into the cytoplasm upon virion fusion to the host cell (1). Tegument proteins perform a variety of functions to facilitate productive infection, including inducing viral gene transcription, mediating virion assembly, and downregulating cellular innate immune signaling (6–9).
Tegument proteins are present throughout the viral life cycle and have multifaceted roles during infection. The UL26 gene encodes a tegument protein that can initiate from either of two in-frame start codons, resulting in both large (27-kDa) and small (21-kDa) isoforms. UL26 is delivered with the tegument upon initial infection and is expressed de novo during the viral life cycle with early expression kinetics (6, 10). Early during infection, UL26 is required for maximal transcriptional activation of the viral major immediate-early promoter and localizes to the nucleus of the host cell (6, 7). As infection progresses, UL26 exits the nucleus and is recruited to cytoplasmic virion assembly centers, where it has been shown to be required for the formation of stable virions with properly phosphorylated tegument constituents (7). Studies utilizing HCMV mutant strains lacking the UL26 open reading frame have shown that the loss of UL26 during infection results in growth defects, including an ∼90% reduction in productive viral replication and significantly reduced cell-to-cell spread (11).
Innate immune signaling is a critical determinant of the success or failure of infection. Immune activation occurs rapidly upon viral entry into the host cell and is triggered by pattern recognition receptors (PRRs), cellular proteins that interact with components of the virion and activate downstream antiviral responses. PRRs capable of sensing and limiting HCMV infection include Toll-like receptors (TLRs), such as TLR2, which senses the HCMV glycoprotein B (gB) and gH at the plasma membrane and stimulates the production of antiviral inflammatory cytokines by activating NF-κB pathway signaling (12, 13). In some cases, these pathways are coopted to support HCMV infection. For example, TLR9 signaling can increase CMV replication and host cell survival (14). Other PRRs, such as cGAS, IFI16, and ZBP1, sense infection by directly binding HCMV double-stranded DNA (dsDNA) in the cytoplasm and nucleus of the host cell and signal through diverse effectors to trigger a suite of antiviral type I interferon responses (15–20). Notably, the timing and context in which these PRRs function can determine their pro- or antiviral contributions. In addition to inducing an antiviral interferon (IFN) response to infection, IFI16 has also been shown to function provirally by binding the viral tegument protein pp65 and transactivating the HCMV major immediate-early promoter to upregulate viral gene transcription (21). Seemingly contradictory findings like these highlight the complex interplay between virus and innate host cell immunity, but what remains clear is that innate immune signaling is a critical determinant of infectious outcomes.
UL26 attenuates antiviral pathways, including NF-κB signaling (8, 9). NF-κB is activated at early times during infection (22–25) but is strongly inhibited at later time points (26–28). We have previously found that UL26 is both necessary and sufficient to inhibit NF-κB signaling induced by tumor necrosis factor alpha (TNF-α) treatment (8, 29). An HCMV ΔUL26 mutant is more sensitive to TNF-α challenge, fails to inhibit TNF-α-stimulated phosphorylation of the inhibitor of NF-κB kinase (IKK) complex during infection, and induces the expression and secretion of IL-6, a canonical NF-κB transcriptional target (8). In addition to blocking canonical NF-κB activation, a ΔUL26 mutant induces the expression and nuclear localization of the noncanonical NF-κB factor RelB, suggesting that UL26 attenuates signaling from multiple arms of the NF-κB pathway (8).
Here, we investigated how UL26 impacts the host cell’s transcriptional response to infection. We find that the absence of UL26 significantly upregulates the expression of innate immune responsive genes, including NF-κB and interferon pathway constituents. Our results indicate that tegument-derived UL26 is sufficient to downregulate ISG15 gene expression and the accumulation of ISGylated species. Further, CRISPR-mediated knockout of IKKβ, a key canonical NF-κB kinase, demonstrates that IKKβ signaling is required for the transcriptional upregulation of ISG15 and bone marrow stromal cell antigen 2 (BST2), interferon-associated genes that are strongly induced during ΔUL26 mutant infection, as well as the attachment of ISG15 to target proteins. Collectively, our data indicate that the absence of UL26 during viral infection results in a proinflammatory transcriptional environment, and that signaling through the IKKβ is required for the establishment of this transcriptional profile.
RESULTS
Infection with a ΔUL26 HCMV mutant induces a proinflammatory, antiviral transcriptional environment.
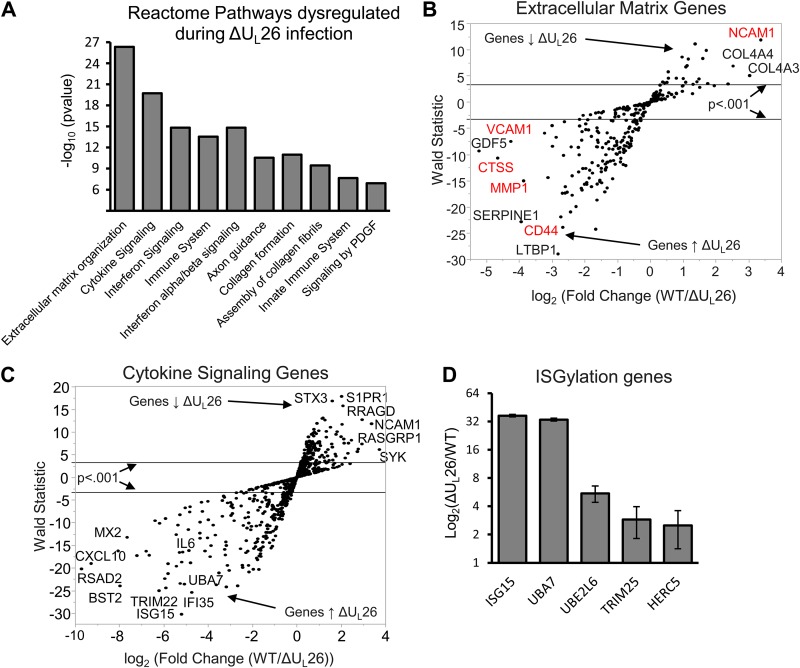
We have previously found that UL26 blocks NF-κB activation in response to various stimuli (8), suggesting that UL26 is a viral antagonist of innate immune signaling. It is unclear how the lack of UL26 globally impacts the cellular transcriptional environment, so we examined the impact of infection with a UL26 deletion mutant on host gene expression. Quiescent, confluent MRC5 fibroblasts were infected with wild-type (WT) HCMV or a ΔUL26 mutant, total cellular RNA was harvested at 48 hours postinfection (hpi), and RNA sequencing (RNA-seq) analysis was performed. Gene ontology analysis of differentially expressed genes using the Reactome database indicated that four of the top five most significantly differentially regulated pathways were involved in immune regulation (Fig. 1A). The most significantly dysregulated pathway ontology during ΔUL26 mutant infection was extracellular matrix organization (Fig. 1A); however, some of the most differentially regulated genes in the extracellular matrix (ECM) pathway ontology were also immune related (Fig. 1B, in red). The second most significantly represented Reactome pathway was cytokine signaling (Fig. 1A and C), followed by various immune and interferon-signaling ontologies. Consistent with our previous findings (8), infection with a ΔUL26 mutant strain significantly induced the expression of interleukin 6 (IL-6) (Fig. 1C), an NF-κB target. Collectively, these data indicate that the UL26 protein is necessary for HCMV-mediated modulation of innate immune gene expression.
FIG 1.
Infection with a ΔUL26 HCMV mutant induces a proinflammatory, antiviral transcriptional environment. MRC5 fibroblasts were infected at an MOI of 3.0 with WT or ΔUL26 HCMV mutant, and total cellular RNA was harvested at 48 hpi for RNA-seq analysis (n = 3). (A) Bar graph showing the Reactome database gene ontology categories that respond most differently to infection in the absence of UL26. PDGF, platelet-derived growth factor. (B) Scatter plot of the genes present in the extracellular matrix. Reactome ontology was plotted according to their average expression fold changes and statistical significance. Genes highlighted in red also belong to the Reactome immune system ontology group. Down arrow indicates downregulation, and up arrow indicates upregulation. (C) Scatter plot of genes present in the cytokine signaling Reactome ontology, plotted according to their fold changes and statistical significance. (D) The increase in mRNA abundance during ΔUL26 mutant infection, relative to WT infection, of specific genes encoding proteins involved in the enzymatic activation, attachment, and cleavage of ISG15.
UL26 is necessary to prevent hyper-ISGylation during HCMV infection.
Our RNA-seq analysis indicated that, relative to WT infection, the single most significantly dysregulated gene during ΔUL26 mutant infection was ISG15. ISG15 is an interferon-inducible ubiquitin-like protein that is conjugated to cellular proteins, in a process termed protein ISGylation, via the activities of an E1-activating enzyme, UBA7, an E2-conjugating enzyme, UBE2L6, and an E3 ligase, either TRIM25 or HERC5 (29, 30). ISG15 possesses antiviral activities (reviewed in reference 31) and has also been found to inhibit HCMV replication (32). Further, expression of UL26 has been reported to attenuate protein ISGylation upon cotransfection with the ISG conjugation machinery (9). In addition to inducing the expression of ISG15, infection with ΔUL26 mutant HCMV strongly induced UBA7 and moderately induced UBE2LS, TRIM25, and HERC5 (Fig. 1D). In total, we find that ΔUL26 infection strongly induces the transcription of the ISGylation machinery in addition to upregulating ISG15 itself.
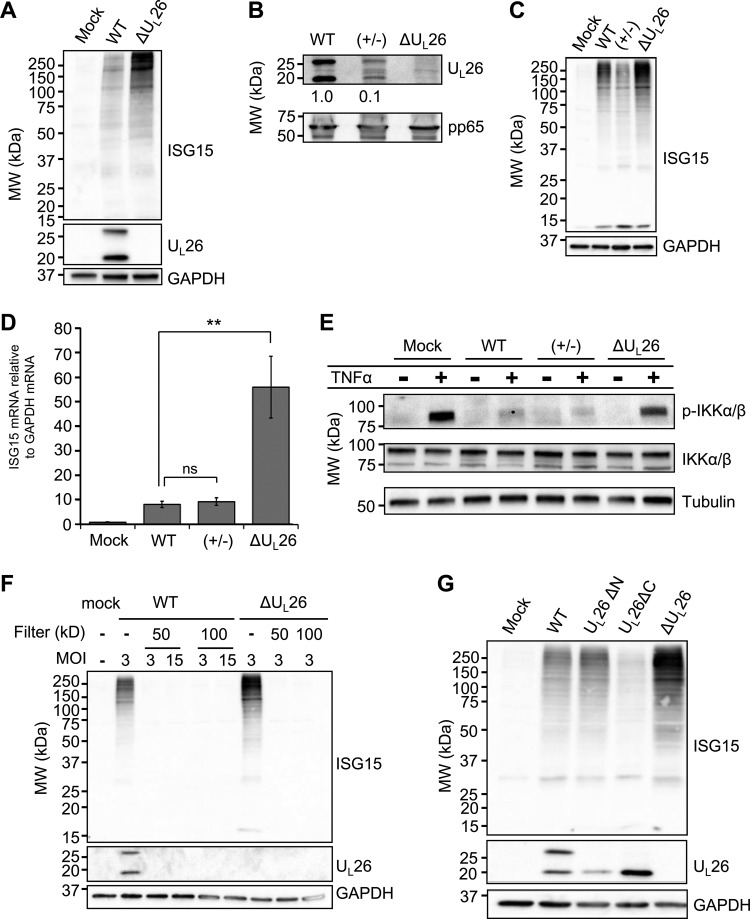
We next examined whether the ΔUL26-associated induction of ISG15-related enzymes correlates with an increase in the attachment of ISG15 to target proteins during infection. While both infection with WT and a ΔUL26 mutant HCMV substantially induced the accumulation of ISGylated proteins over the levels observed in mock-infected cells, ISGylation was significantly more pronounced in ΔUL26 mutant-infected cells than in WT-infected cells (Fig. 2A). In contrast to this observation, a previous report found that infection with a ΔUL26 strain did not change protein ISGylation relative to WT infection, despite the fact that UL26 has been found to inhibit ISGylation in transfected 293T cells (9). When considering possible explanations for this discrepancy, we noted that in other studies, ΔUL26 mutant viral stocks were grown in UL26-expressing cells (9), which helps increase viral titers but presumably enables packaging of UL26 into the virion and delivery to cells upon infection. In contrast, our stocks are propagated in the absence of UL26 and do not contain UL26 in the tegument of the virion. To determine whether this difference could explain the observed ISGylation differences, we grew ΔUL26 mutant virus in UL26-expressing fibroblasts to generate a ΔUL26 mutant viral stock containing UL26 packaged in the virion tegument but lacking the UL26 reading frame in the viral genome and therefore unable to synthesize de novo UL26 during infection, which we refer to as UL26(+/−) virus. Concentrating the virions of UL26(+/−) virus by ultracentrifugation and Western blotting for the UL26 protein confirmed that ectopically expressed UL26 was packaged into the tegument of this UL26(+/−) viral stock, albeit at lower levels than found in WT virions (Fig. 2B). We observed that infection with(+/−) virus containing only tegument-derived UL26 resulted in a decrease in ISGylated species during infection compared to ΔUL26 mutant infection, indicating that tegument-derived UL26 is sufficient to inhibit ISGylation during infection (Fig. 2C). We then assessed ISG15 mRNA levels during infection with WT, UL26(+/−), or ΔUL26 mutant HCMV at 48 hpi and found that UL26(+/−) infection restricted the transcription of ISG15 to low levels similar to those observed during WT infection, suggesting that the presence of tegument-delivered UL26 is sufficient to inhibit the ISG transcriptional response to infection and that de novo production of UL26 is not required to blunt this response (Fig. 2D).
FIG 2.
UL26 is necessary to prevent hyper-ISGylation during HCMV infection. (A) MRC5 fibroblasts were mock infected or infected with WT or ΔUL26 HCMV mutant at an MOI of 3.0. Cell lysates were harvested at 48 hpi and Western blotted with the indicated antibodies. MW, molecular weight. (B) Virions from WT, UL26(+/−), and ΔUL26 mutant viral stocks were concentrated by ultracentrifugation, resuspended in disruption buffer, and Western blotted with the indicated antibodies. The relative intensities of the UL26 bands were quantified using ImageJ, normalized to the band intensity in the WT sample are indicated. (C) MRC5 fibroblasts were mock infected or infected at an MOI of 3.0 with WT, UL26(+/−), or ΔUL26 mutant HCMV. Cell lysates were harvested at 48 hpi and Western blotted with the indicated antibodies. (D) MRC5 cells were infected as in panel C, and the total cellular RNA was harvested at 48 hpi. The abundance of the ISG15 transcript was measured by real-time PCR and normalized to GAPDH. Values are plotted as the means ± standard error of the mean (SEM) (n = 6; **, P < 0.01; ns, not significant). (E) MRC5 cells were infected as in panel C and treated with 10 ng/ml TNF-α for 10 min at 48 hpi before being processed for Western blot analysis with the indicated antibodies. (F) Infectious viral inoculum from either WT or ΔUL26 mutant HCMV was passed through centrifugal filtration units with the indicated MW cutoffs or left unfiltered as a control. These filtrates were then used to infect confluent, serum-starved MRC5 cells at the indicated multiplicities of infection, which were determined by the titer of the viral stock supernatants prior to filtration. Cell lysates were harvested at 48 hpi and Western blotted with the indicated antibodies. (G) MRC5 cells were mock infected or infected with WT or UL26ΔN, UL26ΔC, or ΔUL26 mutant HCMV at an MOI of 3.0. Cellular protein was harvested at 48 hpi and Western blotted with the indicated antibodies.
We have previously observed that infection with ΔUL26 mutant virus induces NF-κB activation and cytokine secretion (8). Given our observation that de novo UL26 is dispensable for HCMV’s inhibition of ISGylation during infection, we next tested whether tegument-delivered UL26 alone was sufficient to mediate another notable UL26 phenotype, inhibition of TNF-α-induced IKK phosphorylation (8). We found that the levels of phosphorylated IKKα/β in TNF-α-treated fibroblasts during(+/−) infection remained near those observed during WT infection, demonstrating that tegument-delivered UL26 is sufficient to inhibit IKK phosphorylation (Fig. 2E). The fact that ΔUL26 mutant infection induces ISG and NF-κB signaling raises the possibility that the inocula of ΔUL26 mutant stocks may contain cytokines that are absent from WT stocks and that these cytokines could be responsible for activating ISGylation irrespective of downstream infection. To determine if paracrine signaling factors present in the conditioned medium of ΔUL26 mutant stocks could be responsible for increasing protein ISGylation, we removed the virions from the infectious inocula via molecular weight cutoff spin filters of sizes ranging from 50 to 100 nm. We find that treatment with the conditioned medium from WT and ΔUL26 mutant infections did not induce the accumulation of ISGylated species even at very high multiplicities of infection (MOIs), suggesting that the increased induction of protein ISGylation observed during ΔUL26 mutant infection is not the result of a secreted small molecule or cytokine but instead dependent on viral infection (Fig. 2F).
We have previously generated a number of UL26 viral mutants that stably express truncated forms of the UL26 protein that lack either the N-terminal 34 amino acids or C-terminal 38 amino acids (11). Notably, the C-terminal UL26 mutant virus displays a 10-fold viral growth defect equal to that observed in UL26-deficient HCMV. These mutants were utilized to assess if the domains of UL26 observed to be important to in vitro viral growth are required to modulate protein ISGylation during infection. We examined the impact of these UL26 truncations on the accumulation of ISGylated species during infection and found that viruses expressing both the N-terminal and the C-terminal UL26 mutants were capable of restricting protein ISGylation to levels at or below those induced by WT infection (Fig. 2G). These data indicate that the UL26 sequences necessary for high-titer replication (i.e., the 38 C-terminal amino acids of UL26) are separable from those necessary for the prevention of ISGylation during infection.
ΔUL26 mutant-mediated enhancement of protein ISGylation is not responsible for inducing IL-6 expression.
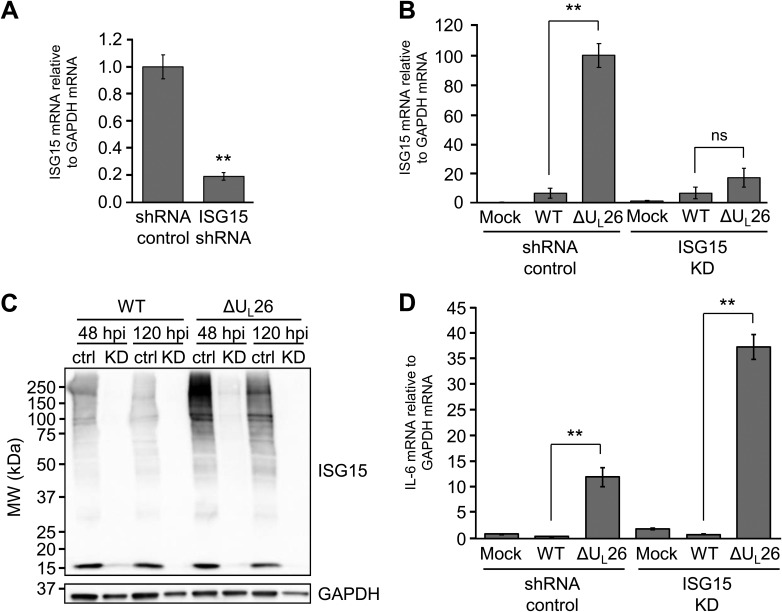
To determine how the enhanced ISGylation associated with ΔUL26 mutant infection impacts viral infection, we targeted ISG15 for shRNA-mediated knockdown (KD). Lentiviral transduction with an ISG15-specific shRNA reduced ISG15 mRNA expression by over 80% compared to a nontargeting shRNA control construct (Fig. 3A), and ISG15 transcription was not observed to be significantly different between WT and ΔUL26 mutant infections in ISG15 KD cells (Fig. 3B). The efficiency of this knockdown was sufficient to block the accumulation of ISGylated species during both WT and ΔUL26 mutant infections at both early (48 hpi) and late (120 hpi) times (Fig. 3C). These data suggest that the ΔUL26 mutant-mediated induction of ISG15 mRNA expression is critical for the induction of protein ISGylation.
FIG 3.
The ΔUL26 mutant-mediated induction of ISG15 transcription is required for the enhancement of protein ISGylation but is dispensable for the induction of IL-6 expression during ΔUL26 mutant infection. (A) HFF cells were transduced with either a control shRNA construct or a construct encoding an ISG15-targeting shRNA. The abundance of ISG15 transcript was measured by real-time PCR and normalized to GAPDH. Values are means ± SEM (n = 5; **, P < 0.01). (B) HFF shRNA control and ISG15 KD shRNA-transduced cells were either mock infected or infected with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cellular RNA was harvested at 48 hpi, and ISG15 mRNA abundance was assessed by real-time PCR and normalized to GAPDH. Values are means ± SEM (n = 5; **, P < 0.01; ns, not significant). (C) HFF shRNA control (ctrl) and ISG15-targeting shRNA-transduced (KD) cells were infected as in panel B. Cellular protein was harvested at the indicated time points and Western blotted with antibodies directed at ISG15 and GAPDH. (D) HFF shRNA control and ISG15 KD cells were infected as in panel B, RNA was harvested at 48 hpi, and IL-6 mRNA abundance was assessed by real-time PCR and normalized to GAPDH. Values are means ± SEM (n = 5; **, P < 0.01).
IL-6 expression and secretion are induced during ΔUL26 mutant infection (Fig. 1C) (8). To determine whether the induction of ISG15 mRNA expression and the enhanced ISGylation observed during ΔUL26 mutant infection contribute to this increase in IL-6 expression, we analyzed how ISG15 knockdown impacts IL-6 expression during infection. ISG15 knockdown did not reduce IL-6 mRNA levels but rather increased IL-6 transcript abundance relative to the controls (Fig. 3D). These results indicate that the enhanced protein ISGylation observed during ΔUL26 mutant infection is not responsible for the ΔUL26 mutant-mediated induction of IL-6 expression. Further, given that the ΔUL26 mutant-mediated induction of IL-6 is dependent on IKKβ/NF-κB signaling (29), this result suggests that the NF-κB-related phenotypes associated with ΔUL26 mutant infection are not mediated via increased ISGylation.
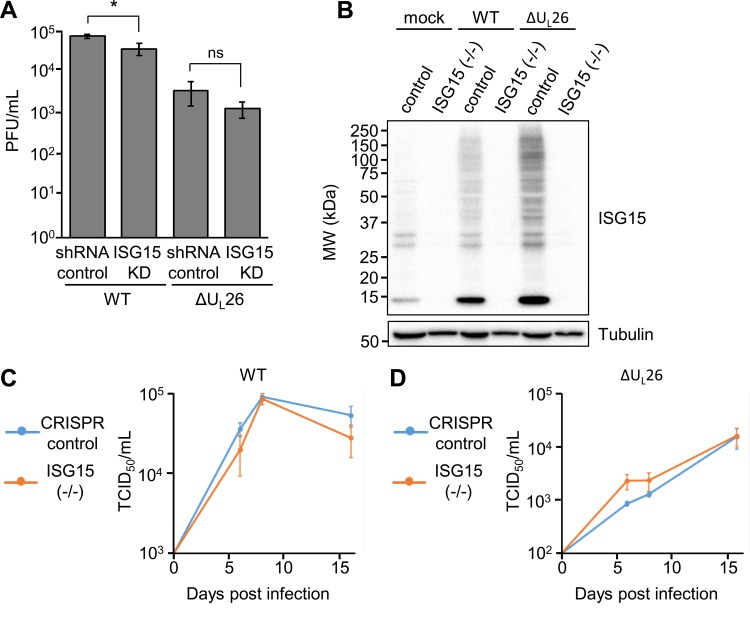
As ISG15 has been previously found to antagonize HCMV infection (33), we tested whether ISG15 knockdown affected HCMV replication. We found that knockdown of ISG15 did not increase the production of HCMV progeny but instead slightly reduced viral growth (Fig. 4A). To corroborate these data, we targeted genomic ISG15 via CRISPR-Cas9, creating an ISG15 knockout cell line that fails to express ISG15 during both mock and ΔUL26 mutant infection (Fig. 4B). To assess the role of ISG15 on viral growth, we performed low-MOI growth curves (MOI, 0.1) in CRISPR control and ISG15(−/−) cell lines (Fig. 4C and D). No statistically significant growth defect was observed in the absence of ISG15 expression. Collectively, these data suggest that increased ISG15 mRNA expression and enhanced protein ISGylation are not responsible for the in vitro growth defect associated with ΔUL26 mutant infection. Further, our data suggest that in the current in vitro context, ISG15 does not restrict HCMV infection.
FIG 4.
ΔUL26 mutant-mediated enhancement of protein ISGylation is independent of its in vitro replication defect. (A) HFF shRNA control and ISG15 KD cells were infected with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Viral supernatants were harvested at 96 hpi, and viral replication was assessed via TCID50. Values are means ± SEM (n = 4; *, P < 0.05; ns, not significant). (B) BJ/hTert CRISPR control and ISG15(−/−) cells were either mock infected or infected with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cell lysates were harvested at 48 hpi and Western blotted with the indicated antibodies. (C and D) CRISPR control and ISG15(−/−) cells were infected with WT HCMV (C) or ΔUL26 mutant HCMV (D) at an MOI of 0.1. Viral supernatants were harvested at days 6, 8, and 16 postinfection and quantified via TCID50 (n = 4).
BST2 contributions to viral infection.
Bone marrow stromal cell antigen 2 (BST2) is another IFN-associated gene that is strongly upregulated during infection in the absence of UL26 (Fig. 1C). BST2 is an antiviral host cell factor with an unusual structural arrangement consisting of a transmembrane region, an internal coiled-coil domain, and a C-terminal glycosylphosphatidylinositol (GPI) anchor (34). The combination of these domains enables antiviral function via the incorporation of the BST2 GPI domain into assembled virions, while the transmembrane domain adheres to lipid rafts in the plasma membrane, thus tethering virions to the cell and preventing egress (35, 36). BST2 is capable of interfering with the release of a diverse array of enveloped viruses, including respiratory syncytial virus (RSV), Mason-Pfizer monkey virus (MPMV), human T-cell leukemia virus type 1 (HTLV-1), Ebola virus, herpes simplex virus 1 (HSV-1), and Japanese encephalitis virus (JEV), among many others (37–41). In response, viruses have evolved anti-BST2 countermeasures, including the HIV accessory protein Vpu, the HIV-2 envelope (Env) protein, and the K5 ubiquitin ligase encoded by Kaposi’s sarcoma-associated herpesvirus (KSHV), which each promote the degradation, sequestration, or otherwise inhibition of BST2 to rescue viral titers (reviewed in reference 41).
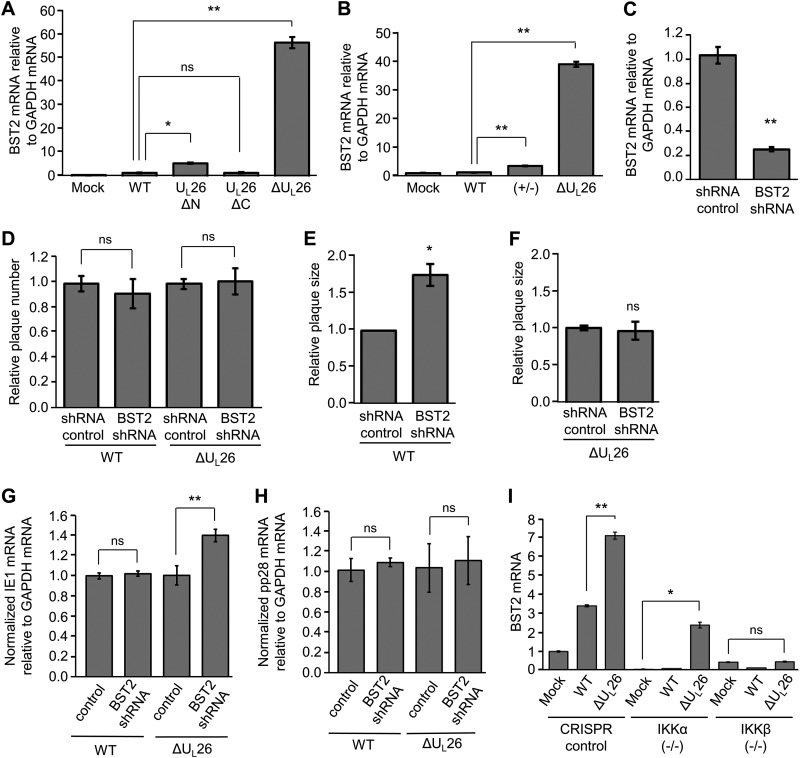
In the context of HCMV infection, it has been reported that BST2 is packaged in virions and enhances viral entry into BST2-expressing cell populations (42). To further examine how UL26 shapes the interferon-associated immune response to infection, BST2 mRNA expression was assessed during infection with both WT and ΔUL26 mutant HCMV, as well as with viruses encoding the UL26ΔN and UL26ΔC mutant isoforms (Fig. 5A). The inhibition of BST2 expression was maintained during both UL26ΔN and UL26ΔC infection, indicating that the ΔUL26 mutant growth defect is likely associated with a separate UL26 function not linked with changes in BST2 expression. Given our finding that de novo production of UL26 during infection is dispensable for inhibition of both protein ISGylation (Fig. 2C) and inhibition of TNF-α-induced IKK phosphorylation in response to infection (Fig. 2E), we measured BST2 mRNA accumulation during infection with UL26(+/−) virus to determine if tegument-derived UL26 was sufficient to keep BST2 transcription inhibited (Fig. 5B). We find that the UL26(+/−) virus restores BST2 mRNA levels to WT levels during infection, suggesting that BST2 is being regulated by tegument-derived UL26 in a manner similar to NF-κB and ISG15 signaling (Fig. 5B).
FIG 5.
Increased BST2 transcription during infection restricts plaque size and is dependent on IKK signaling. (A) MRC5 cells were mock infected or infected with WT or UL26ΔN, UL26ΔC, or ΔUL26 mutant HCMV at an MOI of 3.0. The abundance of BST2 mRNA at 48 hpi was measured by real-time PCR and normalized to GAPDH. Values are means ± SEM (n = 6; *, P < 0.05; **, P < 0.01; ns, not significant). (B) MRC5 cells were mock infected or infected with WT, UL26(+/−), or ΔUL26 mutant HCMV. The abundance of BST2 mRNA at 48 hpi was measured by real-time PCR and normalized to GAPDH. Values are means ± SEM (n = 3; **, P < 0.01; ns, not significant). (C) HFF cells were lentivirally transduced with either a nontargeting control shRNA construct or a BST2-targeting shRNA construct. The abundance of BST2 transcript was measured by real-time PCR and normalized to GAPDH. Values are means ± SEM (n = 5; **, P < 0.01) (D) Nontargeting shRNA control and BST2 KD cells were infected with 40 PFU of either WT or ΔUL26 mutant HCMV. The relative efficiencies of plaque formation were quantified at 11 days postinfection (dpi). The averages are plotted after normalizing to the number of plaques for each virus counted on the infected shRNA controls. Values are means ± SEM (n = 8; ns, not significant). (E and F) shRNA control and BST2 KD cells were infected as in panel C with 40 PFU of either WT (E) or ΔUL26 mutant (F) HCMV. Individual plaque sizes were quantified using the ImageJ software, averaged, and normalized to the average plaque size for each virus in shRNA control cells. Values are means ± SEM (n = 40; *, P < 0.05; ns, not significant). (G and H) Nontargeting shRNA control and BST2 KD cells were infected with either mock infected or infected with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cellular RNA was harvested at 48 hpi, and IE1, pp28, and GAPDH mRNA levels were measured by quantitative PCR (qPCR). The ratio of IE1 mRNA to GAPDH mRNA (G) or pp28 mRNA to GAPDH mRNA (H) for each virus was normalized to the value measured in the shRNA control cell line and plotted as the means ± SEM (n = 4; **, P < 0.01; ns, not significant). (I) CRISPR control, IKKα(−/−), and IKKβ(−/−) BJ/hTert cells were either mock infected or infected with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cellular RNA was harvested at 48 hpi, and the abundance of the BST2 transcript was measured by real-time PCR and normalized to GAPDH. Values are plotted as the means ± SEM (n = 6; *, P < 0.05; **, P < 0.01; ns, not significant).
To examine the contributions of BST2 to UL26-associated growth phenotypes, we knocked down BST2 using shRNA and achieved an 80% reduction in BST2 expression (Fig. 5C). As BST2 has been implicated in assisting the entry of virions into host cells, we used these knockdown cells to examine the effect of BST2 inhibition on viral plaque phenotypes. We were unable to detect any defect in the ability of either WT or ΔUL26 mutant HCMV to initiate infection (Fig. 5D), but the loss of BST2 was noted to preferentially increase WT plaque size, while ΔUL26 mutant plaque size remained unaffected (Fig. 5E and F). The observation that BST2 loss increases viral plaque size only in the presence of UL26 potentially indicates that the virus requires UL26 to exploit the absence of BST2 during infection. To more closely examine the enhanced viral replication in the absence of BST2, we measured immediate-early 1 (IE1) and pp28 mRNA levels at 48 hpi in shRNA control and BST2 KD cells infected with either WT or ΔUL26 mutant viruses (Fig. 5G and H). The expression of IE1 was unaffected by the presence or absence of BST2 during WT infection (Fig. 5G). During ΔUL26 mutant infection, the knockdown of BST2 induced a small but statistically significant increase in IE1 mRNA abundance (Fig. 5G). The expression of the viral late protein pp28, which requires successful viral DNA replication, was unaffected by the loss of BST2 during either WT or ΔUL26 mutant infection (Fig. 5H). These results suggest that the observed increase in viral plaque size observed in BST2 KD cells during WT infection is not due to a substantial increase in viral gene expression.
Last, we utilized CRISPR cell lines with either IKKα or IKKβ knocked out to determine if these critical NF-κB regulatory kinases were required for BST2 upregulation. Individual knockouts of IKKα and IKKβ reduced the overall transcription of BST2 relative to CRISPR control cells (Fig. 5I). Notably, while BST2 transcription was still strongly upregulated during ΔUL26 mutant infection (relative to mock infection) in IKKα(−/−) cells, the loss of IKKβ prevented ΔUL26 mutant-mediated induction of BST2 mRNA (Fig. 5I). These data suggest that the IKKs, particularly IKKβ, are playing an important role in shaping the transcriptional response to ΔUL26 mutant infection and may provide clues towards the mechanism through which UL26 downregulates a diverse array of antiviral transcripts.
IKKβ is necessary for hyper-ISGylation during ΔUL26 mutant infection.
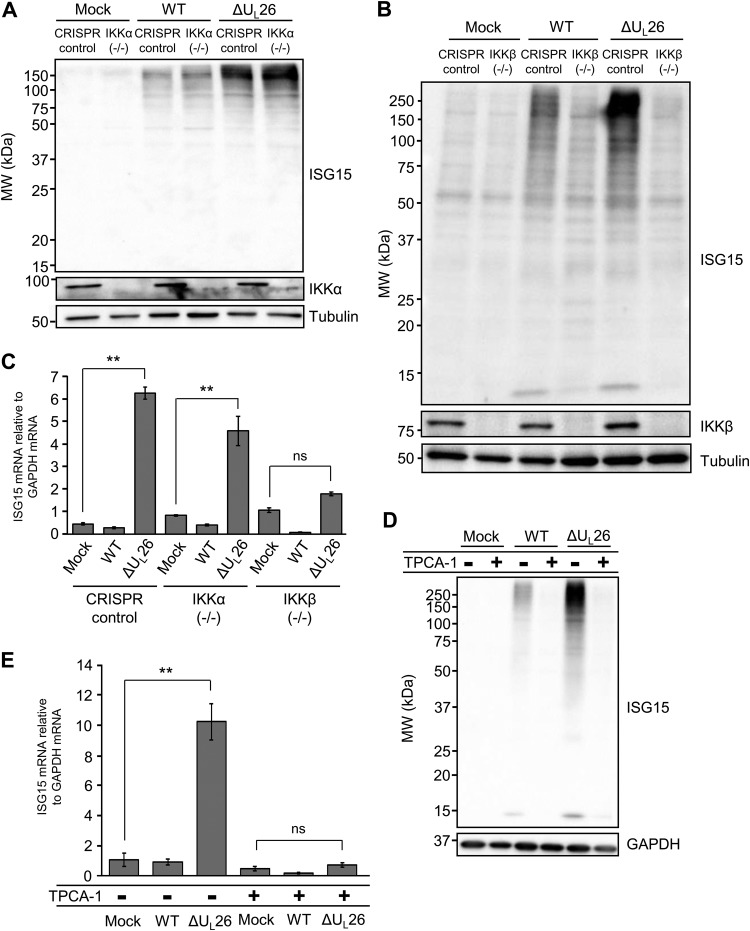
Given that we find IKKβ to be necessary for ΔUL26 mutant-mediated induction of BST2, we wanted to assess whether the IKKs are important for the induction of ISG15 and protein ISGylation observed during ΔUL26 mutant infection. Towards this end, we utilized the aforementioned IKKα and IKKβ knockout (KO) cell lines (29). The presence or absence of IKKα did not affect the increased protein ISGylation associated with ΔUL26 mutant infection (Fig. 6A). In contrast, the IKKβ knockout cells exhibited significantly reduced accumulation of ISGylated species, relative to CRISPR control cells, during both WT and ΔUL26 mutant infections (Fig. 6B). To determine the extent to which the enhanced protein ISGylation correlates with increased ISG15 mRNA expression, we analyzed the abundance of ISG15 mRNA during infection in CRISPR control, IKKα(−/−), and IKKβ(−/−) cells. The ΔUL26 mutant-associated induction of ISG15 mRNA was unchanged in IKKα-deficient cells relative to the CRISPR control, whereas a loss of IKKβ during infection reduced ISG15 transcription to levels indistinguishable from mock infection (Fig. 6C). To further test this dependence, we used the IKK inhibitor 2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide 1 (TPCA-1) (43). Pharmacological inhibition of IKK signaling via TPCA-1 prior to and during infection strongly reduced both the accumulation of ISGylated species as well as the transcription of ISG15 mRNA during WT and ΔUL26 mutant infections (Fig. 6D and E), indicating that host cell activation of ISG15 signaling in response to viral infection depends on the activity of the IKKs. These results suggest that the induction of protein ISGylation during HCMV infection is dependent on IKK signaling, particularly IKKβ.
FIG 6.
Hyper-ISGylation during ΔUL26 mutant infection requires IKKβ. (A) CRISPR control and IKKα(−/−) BJ/hTert cells were either mock infected or infected with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cell lysates were harvested at 48 hpi and Western blotted with the indicated antibodies. (B) CRISPR control and IKKβ(−/−) BJ/hTert cells were infected as in panel A, and cell lysates were harvested at 48 hpi for Western blotting with the indicated antibodies. (C) Total cellular RNA was harvested at 48 hpi from CRISPR control, monoclonal IKKα(−/−), and a pooled polyclonal population of IKKβ(−/−) BJ/hTert cells infected as in panel A. The abundance of ISG15 mRNA was measured by real-time PCR and normalized to GAPDH. Values are the means ± SEM (n = 8; **, P < 0.01; ns, not significant). (D) MRC5 fibroblasts were pretreated with 10 μM TPCA-1 for 4 h prior to either mock infection or infection with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cell lysates were collected at 48 hpi for Western blotting with the indicated antibodies. (E) MRC5 cells were pretreated with TPCA-1 and infected as in panel D. Cellular RNA was harvested at 48 hpi, and the accumulation of ISG15 mRNA was measured by real-time PCR and normalized to GAPDH. Values are plotted as the means ± SEM (n = 5; **, P < 0.01; ns, not significant).
DISCUSSION
Successful viral infection and population persistence require the negotiation of innate and adaptive immune responses. The UL26 protein has emerged as an important viral determinant that modulates antiviral signaling pathways (8, 9). Here, we find that the UL26 protein is necessary to prevent a global inflammatory remodeling of host gene expression during infection. The ΔUL26 mutant-induced transcriptomic inflammatory remodeling includes extensive activation of both NF-κB and interferon signaling genes. ISG15 is one of the most highly upregulated host cell genes during ΔUL26 mutant infection (Fig. 1C and D). Concomitant with increased ISG15 transcription, ΔUL26 mutant infection strongly induces ISGylated species accumulation. Previously, UL26 was found to modulate protein ISGylation (9). However, Kim et al. report that the deletion of UL26 alone was not sufficient to increase protein ISGylation (9). Here, we find that the deletion of UL26 by itself is sufficient to increase protein ISGylation (Fig. 2A). This apparent discrepancy is resolved by our finding that tegument-derived UL26 is sufficient to block increased ISG15 transcription and protein ISGylation during infection. Specifically, the previous report studied ΔUL26 mutant viral stocks produced in the presence of UL26 expressed in trans (9), which supplies UL26 for tegumentation and subsequent delivery to the host cell. We find that the presence of tegument-derived UL26 is sufficient to inhibit protein ISGylation (Fig. 2C).
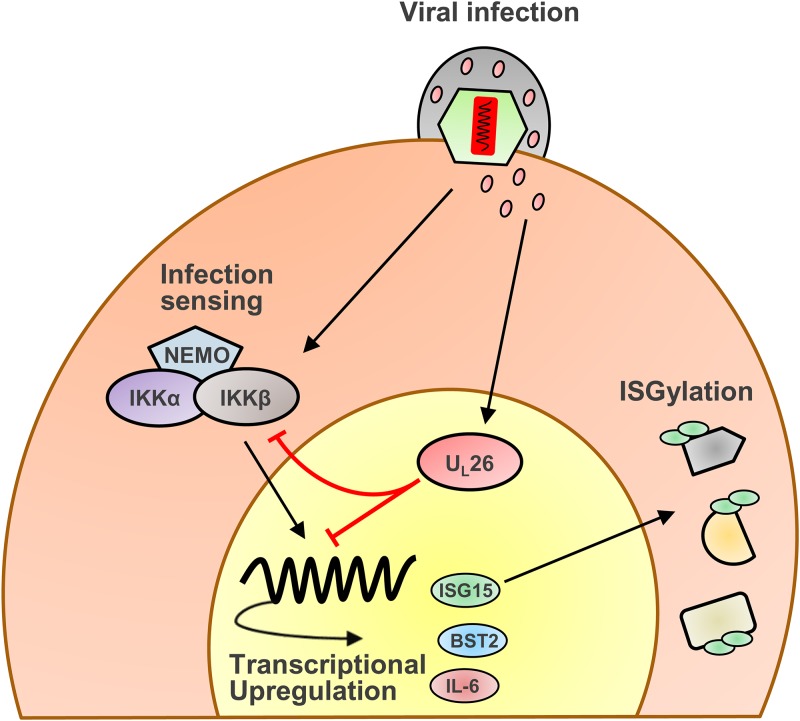
In addition to finding that tegument-derived UL26 is sufficient to inhibit the induction of both ISG15 mRNA expression and the activation of protein ISGylation (Fig. 2C and D), we also find that tegument-derived UL26 is sufficient to block TNF-α-induced IKK phosphorylation (Fig. 2E). These findings provide insight into UL26’s mechanisms of action. Specifically, given that de novo production of UL26 is not required for inhibition of these activities, these results suggest that UL26 is acting relatively early during the viral life cycle to block these pathways. In addition, tegument-derived UL26 primarily localizes to the nucleus (7, 8), suggesting that UL26 may be playing a role in the nucleus to attenuate these antiviral activities (Fig. 7). Interestingly, in one of the first reports describing UL26 function, UL26 was identified as having a strong transcriptional activation domain via an activator trap assay (6). Further experimentation will be required to elucidate potential mechanisms through which nuclear UL26 contributes to the modulation of antiviral signaling.
FIG 7.
Model of UL26 inhibition of IKKβ-dependent induction of antiviral genes. Infection with ΔUL26 mutant HCMV induces the transcription of antiviral genes, including IL-6, BST2, and ISG15, as well as the ISGylation of proteins. Tegument-derived UL26, delivered with the virion, inhibits IKK phosphorylation and prevents both this transcriptional response to ΔUL26 mutant infection as well as downstream ISGylation.
Previously, it was found that UL26 itself is subject to ISGylation at multiple sites (9). It has previously been reported that the UL26 protein is ISGylated at two main lysine residues, K154 and K169. Using a mutated UL26 lacking these lysines in conjunction with a UL26-ISG15 fusion protein, it was found that ISGylation may interfere with the stability of UL26 and impede its ability to downregulate NF-κB signaling (9). The K169 residue is entirely absent in the UL26ΔC mutant. Here, we find that UL26ΔC, which lacks K169, is capable of inhibiting the ISGylation of target proteins (Fig. 2G). Similarly, our previous work has shown that UL26ΔC inhibits TNF-α-induced IKK phosphorylation (29). These results suggest that K169 is dispensable for UL26 stability and modulation of NF-κB signaling and increased protein ISGylation. Further work is required to assess the contribution of K154 and its ISGylation to viral infection and modulation of antiviral signaling.
It is unclear how UL26-mediated inhibition of protein ISGylation contributes to HCMV infection. In general, increased protein ISGylation is considered to have antiviral effects (31). Consistent with this view, increased protein ISGylation during HCMV infection has been reported to be antagonistic to the virus (32). Further, depletion of the ISG15 E3 ligase Herc5 was found to increase viral titers, while depletion of the ISG15-specific protease UBP43 attenuated viral growth (9). We did not observe significant changes in WT or ΔUL26 mutant viral replication during infection in the face of shRNA-mediated knockdown of ISG15 levels and consequent reduced ISGylated species accumulation (Fig. 3 and 4), nor did we see significant changes to viral replication upon CRISPR-mediated knockout of ISG15 (Fig. 4). In considering our data with those of previous reports, we think it likely that the viral interactions with the ISGylation machinery are context dependent and potentially quite complex. One possibility is that HCMV infection might be sensitive to the extent of protein ISGylation activity or the impact of specific ISGylation events. For example, a complete knockdown of ISG15 may erase potential benefits to viral infection that may have resulted from a more modest knockdown. Relatedly, it is known that ISG15 conjugation can affect many different proteins with resulting diverse changes to protein activity (44), which could have differential effects on infection depending on the specific protein being ISGylated. Further clues on the role of ISGylation during HCMV infection will emerge as the functional roles of ISGylation on the activities of target proteins become clearer.
In addition to ISG15, BST2, another interferon-stimulated gene (ISG), is strongly induced by ΔUL26 mutant infection (Fig. 1 and 5). Also known as tetherin, BST2 has been demonstrated to possess significant antiviral activities (45). However, in the context of HCMV infection, it has been reported to enhance viral entry (42). We find that shRNA-mediated knockdown of BST2 increased WT HCMV plaque size (Fig. 5E), suggesting that BST2 limits viral cell-to-cell spread. In contrast to WT HCMV, BST2 knockdown did not impact ΔUL26 mutant plaque size. This suggests that while BST2 limits HCMV’s ability to spread from cell to cell, UL26 is necessary to take advantage of BST2’s absence. The inability of UL26 to capitalize on the loss of BST2 likely reflects the broad antiviral state induced by HCMV infection in the absence of UL26, i.e., the expression of a number of NF-κB target genes and ISGs are increased during infection without UL26.
Our results indicate that IKKβ is required for UL26-mediated induction of two ISGs. Specifically, the expression of both BST2 and ISG15 was dependent on functional IKKβ (Fig. 6). In contrast, knockdown of ISG15 did not reduce the ΔUL26 mutant-induced expression of the NF-κB target gene IL-6, which we have previously found to be dependent on IKKβ activity (29). These data suggest that ΔUL26 mutant-mediated induction of ISG expression and subsequent increased protein ISGylation are a result of the defective NF-κB signaling associated with ΔUL26 mutant infection (Fig. 7). Our findings that the induction of ISG15 expression and protein conjugation is dependent on the presence of functional IKKβ suggest that additional mechanisms of cross-talk between NF-κB and IFN signaling remain to be identified. Given the importance of both of these pathways for controlling a wide variety of infections, elucidating the mechanisms governing their functional interplay should be a high priority.
In summary, our results indicate that UL26 is a major viral determinant that attenuates innate immune signaling during HCMV infection. Tegument-delivered UL26 is sufficient to limit the induction of ISG expression and enhanced protein ISGylation during infection. This UL26-mediated inhibition of protein ISGylation is dependent on IKKβ activity, further emphasizing IKKβ’s role in shaping the host antiviral response to HCMV and, likely, infection more broadly. Viral targeting of cytokine signaling pathways is a critical contributor to successful viral infection, and elucidating the mechanisms through which innate immune signaling limits viral replication, as well as the means by which viruses subvert these mechanisms, will be a key step in identifying ways to tip the balance of this host-pathogen interaction to limit viral pathogenesis.
MATERIALS AND METHODS
Cell culture, viruses, chemicals, and viral infection.
MRC5 fibroblasts (ATCC CCL-171), 293T human embryonic kidney (HEK) cells, human foreskin fibroblast (HFF) cells, and immortalized BJ fibroblasts expressing human telomerase (previously described in reference 46) were cultured in Dulbecco’s modified Eagle medium (DMEM; Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (FBS), 4.5 g/liter glucose, and 1% penicillin-streptomycin (Life Technologies) at 37°C in a 5% (vol/vol) CO2 atmosphere. Unless otherwise indicated, cells were grown to confluence of 3.2 × 104 cells per cm2 in 12-well plates, rendered quiescent via culturing in serum-free medium for 24 h, and exposed to viral inocula for an adsorption period of 1.5 h. Viral inocula were removed after the adsorption period and replaced with fresh, serum-free DMEM. For experiments involving TNF-α-stimulated activation of NF-κB, cells were exposed to 10 ng/ml TNF-α (Sigma) for 10 min at 48 hpi. For experiments using the IKKα/IKKβ inhibitor TPCA-1, cells were pretreated with 10 μM TPCA-1 (Sigma) for 4 h prior to infection as well as the 48 h immediately following infection.
The wild-type (WT) HCMV strain used in this work was the green fluorescent protein (GFP)-expressing BADsubUL21.5 (47), and the ΔUL26 mutant HCMV strain was a GFP-expressing BADUL26 transposon insertion mutant (48). The UL26ΔN and UL26ΔC mutants used were derived from the HCMV BADwt clone, as previously described (11). Viral stocks were propagated in MRC5 fibroblasts except in the case of UL26(+/−) virus, which was grown in immortalized BJ/hTert cells lentivirally transduced with a mammalian UL26 expression construct. The titers of the viral stocks were determined using 50% tissue culture infective dose (TCID50) analysis. For experiments involving concentrated virions, an equivalent number of PFU of each viral stock to be analyzed was underlaid with a standard sorbitol cushion and centrifuged for 1.5 h at 26,000 rpm. The virion pellet was resuspended in 1× disruption buffer prior to SDS-PAGE and Western blot analysis (see “Immunoblotting,” below).
RNA-seq and computational analysis.
Quiescent MRC5 fibroblasts in 15-cm dishes were infected at a multiplicity of infection (MOI) of 3.0 with either WT or ΔUL26 mutant HCMV. Total cellular RNA was isolated at 48 hpi using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. Samples were DNase treated using a Turbo DNase kit (Invitrogen) and purified with an RNeasy MinElute cleanup kit (Qiagen). The total RNA concentration was determined with the NanoDrop 1000 spectrophotometer and RNA quality assessed with an Agilent Bioanalyzer. The TruSeq stranded mRNA sample preparation kit (Illumina) was used for next-generation sequencing library construction, per the manufacturer’s protocols. Briefly, mRNA was purified from 200 ng total RNA with oligo(dT) magnetic beads and fragmented. First-strand cDNA synthesis was performed with random hexamer priming, followed by second-strand cDNA synthesis using dUTP incorporation for strand marking. End repair and 3′ adenylation were then performed on the double-stranded cDNA. Illumina adapters were ligated to both ends of the cDNA, which was then purified by gel electrophoresis and amplified with PCR primers specific to the adapter sequences to generate cDNA amplicons ranging in size from 200 to 500 bp. Library preparation and subsequent sequencing were performed at the University of Rochester Genomics Research Center (GRC). Amplified libraries were hybridized to the Illumina single-end flow cell and amplified using the cBot (Illumina). Single-end reads of 100 nucleotides (nt) were generated for each sample using a HiSeq 2500v4 platform (Illumina). Sequence reads were cleaned and adapter trimmed using Trimmomatic-0.36 (49) before mapping each sample individually to the human reference genome (GRCh38.p7, primary assembly + gencode 25 annotation) with STAR2.5.2b (33). Raw read counts were obtained using featurecounts (50) from the subread1.5.0p3 package and gencode 25 human gene annotations using only uniquely aligned reads (default) and including multimapping reads (-M). DESeq2-1.14.1 (51) within R 3.3.2 was used to perform data normalization and differential expression analysis with a false-discovery rate adjusted P value threshold of 0.05 on each set of raw expression measures.
Lentiviral transduction and cell line generation.
Pseudotyped lentivirus was produced in 293T cells seeded in 10-cm dishes at a density of 2 × 106 cells per cm2 and grown for 24 h prior to transfection with 2.6 μg lentiviral vector, 2.4 μg PAX2, and 0.25 μg vesicular stomatitis virus G glycoprotein expression plasmid using FuGENE 6 (Promega). The medium was aspirated after 24 h, and 4 ml fresh medium was added to the plate. After another 24 h, the supernatant was filtered through a 0.45-μm filter and applied to fibroblast cells in the presence of 5 μg/ml Polybrene (Millipore), and the cells were incubated for 24 h. The medium was then refreshed, and the cells were allowed to recover for 72 h prior to selection with 1 μg/ml puromycin (VWR).
UL26-expressing fibroblasts were generated via lentiviral transduction for the production of UL26(+/−) viral stocks. The pCW57.1 lentiviral vector (Addgene no. 41393) was modified by deleting the gateway cloning attR1 and attR2 sites, including the intervening sequences, to generate pCW57.1GA. The UL26 open reading frame was PCR amplified using forward primer 5′-CCGTCAGATCGCCTGGAGAATTGGCTAGATGTACGCCGTTTTCGGCCTCACGAGGTCG-3′ and reverse primer 5′-GGAAAAGGCGCAACCCCAACCCCGGATCTTACGGCAACAGCGCTGATGGCACGTTGC-3′ and inserted downstream of the doxycycline-inducible promoter via Gibson Assembly (52). Prior to infection for the production of UL26(+/−) viral stocks, UL26-expressing cells were treated with 1 μg/ml doxycycline (Sigma) for 24 h. Doxycycline-containing medium was refreshed every 2 days until the stock was harvested.
To achieve targeted knockdown of ISG15 in HFF cells, a pLKO.1-based Mission shRNA construct targeting ISG15 (Sigma/Broad Institute clone number TRCN0000237826) was obtained from the Sigma-Aldrich Mission shRNA library. Knockdown of BST2 in HFF cells was achieved with a BST2-targeting Mission shRNA construct (Sigma/Broad Institute clone number TRCN0000299179). Mission pLKO.1-puro nonmammalian shRNA control construct (Sigma SHC002) was used as a nontargeting control in all shRNA experiments. Plasmid preps and lentiviral transductions were performed according to manufacturer’s instructions.
CRISPR knockout cell lines of ISG15, IKKα, and IKKβ as well as CRISPR control cells were generated as previously described (29). Briefly, ISG15-targeting or IKK-targeting genomic RNAs (gRNAs) were cloned into the LentiCRISPRv2 construct and transduced into immortalized BJ/hTert fibroblasts to create polyclonal knockout cell populations. Monoclonal knockout cell lines were clonally isolated by single-cell dilution into 96-well plates. CRISPR indels were confirmed via TA cloning and tracking of indels by decomposition (TIDE) analysis (29). The gRNA sequence used to target ISG15 was 5′-GCAGCGCCACACCGCTCGGG-3′, and the following primers were used to PCR amplify the ISG15 region for TA cloning: ISG15 forward primer, 5′-CAGCTGGCCTTCTAGTAACGA-3′, and ISG15 reverse primer, 5′-CTGCGTCAGCCGTACCTC-3′. The gRNA and primer sequences used in the generation and analysis of the CRISPR IKK knockout cell lines have been described previously (29). The following frameshift indels were present in the CRISPR knockout cell lines: ISG15(−/−), positions +1 and +1; IKKα(−/−), positions −1 and −37; and IKKβ(−/−), positions −13 and −13. The IKKβ(−/−) pooled cell population was composed of equivalent cell numbers of multiple IKKβ(−/−) clones with the following frameshift deletions: positions −7 and +1, positions +1 and +10, and positions +1 and +1.
Immunoblotting.
Cellular supernatants were scraped in disruption buffer (50 mM Tris [pH 7.0], 2% SDS, 5% 2-mercaptoethanol, and 2.75% sucrose) for Western analysis. Samples were sonicated and boiled for 5 min prior to being briefly centrifuged to pellet cellular debris and separated by 10% SDS-PAGE gels. Samples were transferred to nitrocellulose in Tris-glycine transfer buffer, blocked via incubation in 5% milk in TBST solution (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.1% Tween 20), and reacted with primary and secondary antibodies. Protein bands were visualized using an enhanced chemiluminescence (ECL) kit (Bio-Rad) and a Molecular Imager Gel Doc XR+ system (Bio-Rad). Primary antibodies were specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH; D16H11; Cell Signaling), ISG15 (F-9; Santa Cruz Biotechnology), IKKα (Cell Signaling), IKKβ (Cell Signaling), α-tubulin (11H10; Cell Signaling), BST2 (E-4; Santa Cruz Biotechnology), pp65 (27), and UL26 (7H19 [7, 22]). The secondary antibodies were rabbit polyclonal (Santa Cruz Biotechnology) and mouse monoclonal (Abcam) anti-IgG antibodies.
Real-time PCR.
Total cellular RNA was extracted with TRIzol (Invitrogen) and used to generate cDNA using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Transcript abundance was measured by real-time PCR analysis using Fast SYBR green master mix (Applied Biosystems), a model 7500 Fast real-time PCR system (Applied Biosystems), and the Fast 7500 software (Applied Biosystems). Gene expression equivalent values were determined using the 2−ΔΔCT method and normalized to GAPDH levels. The following primers were used for real-time PCR: ISG15, 5′-CAGCCATGGGCTGGGAC-3′ (forward) and 5′-CTTCAGCTCTGACACCGACA-3′ (reverse); IL-6, 5′-AAATTCGGTACATCCTCGACGGCA-3′ (forward) and 5′-AGTGCCTCTTTGCTGCTTTCACAC-3′ (reverse); BST2, 5′-CACACTGTGATGGCCCTAATG-3′ (forward) and 5′-GTCCGCGATTCTCACGCTT-3′ (reverse); IE1, 5′-TGATTCTATGCCGCACCATGTCCA-3′ (forward) and 5′-AGAGTTGGCCGAAGAATCCCTCAA-3′ (reverse); pp28, 5′-CACCACCATCAGCAAAGCTCCATT-3′ (forward) and 5′-GGTGGGTGGACGTTGTGAAATCTT-3′ (reverse); and GAPDH, 5′-CATGTTCGTCATGGGTGTGAACCA-3′ (forward) and 5′-ATGGCATGGACTGTGGTCATGAGT-3′ (reverse).
Plaque formation and size assays.
Plaque formation efficiency and plaque size analyses were conducted as previously described (29). Briefly, cells were grown to confluence, rendered quiescent as indicated above, and infected with a known amount of PFU for 1.5 h before being placed under a standard agarose gel overlay. Plaques were allowed to grow for 10 days prior to analysis, at which point an individual plaque was scored if it presented as a locus of GFP-positive cells exhibiting cytopathic effect. Plaque sizes were assessed using the ImageJ software.
Statistical analysis.
Western blot images are representative blots from at least two independent experiments. Statistical significance was assessed using a nonpaired two-tailed homoscedastic Student’s t test, wherein a probability value of <0.05 was considered statistically significant.
Data availability.
The RNA-seq data sets are available in the Gene Expression Omnibus (GEO) database with the accession number GSE137065.
ACKNOWLEDGMENTS
C.M.G. was supported by an NIH training grant from the Training Program in Oral Science (T90-DE021985-08). The work was also supported by an NIH grant AI127370 to J.M. and by a Research Scholar Grant from the American Cancer Society (grant RSG-15-049-01-MPC).
REFERENCES
- 1.Mocarski ES, Shenk T, Pass RF. 2006. Cytomegaloviruses, p 2701–2757. In Knipe DM, Howley PM (ed), Fields virology. Lippincott-Williams & Wilkins, New York, NY. [Google Scholar]
- 2.Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325:63–83. doi: 10.1007/978-3-540-77349-8_4. [DOI] [PubMed] [Google Scholar]
- 3.Andrei G, De Clercq E, Snoeck R. 2008. Novel inhibitors of human CMV. Curr Opin Investig Drugs 9:132–145. [PubMed] [Google Scholar]
- 4.Burny W, Liesnard C, Donner C, Marchant A. 2004. Epidemiology, pathogenesis and prevention of congenital cytomegalovirus infection. Expert Rev Anti Infect Ther 2:881–894. doi: 10.1586/14789072.2.6.881. [DOI] [PubMed] [Google Scholar]
- 5.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, Mann M, Ingolia NT, Weissman JS. 2012. Decoding human cytomegalovirus. Science 338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stamminger T, Gstaiger M, Weinzierl K, Lorz K, Winkler M, Schaffner W. 2002. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J Virol 76:4836–4847. doi: 10.1128/JVI.76.10.4836-4847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munger J, Yu D, Shenk T. 2006. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J Virol 80:3541–3548. doi: 10.1128/JVI.80.7.3541-3548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathers C, Schafer X, Martinez-Sobrido L, Munger J. 2014. The human cytomegalovirus UL26 protein antagonizes NF-kappaB activation. J Virol 88:14289–14300. doi: 10.1128/JVI.02552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YJ, Kim ET, Kim YE, Lee MK, Kwon KM, Kim KI, Stamminger T, Ahn JH. 2016. Consecutive inhibition of ISG15 expression and ISGylation by cytomegalovirus regulators. PLoS Pathog 12:e1005850. doi: 10.1371/journal.ppat.1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abate DA, Watanabe S, Mocarski ES. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol 78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathers C, Spencer CM, Munger J. 2014. Distinct domains within the human cytomegalovirus UL26 protein are important for wildtype viral replication and virion stability. PLoS One 9:e88101. doi: 10.1371/journal.pone.0088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boehme KW, Guerrero M, Compton T. 2006. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol 177:7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- 13.Barbalat R, Lau L, Locksley RM, Barton GM. 2009. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol 10:1200. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iversen A-C, Steinkjer B, Nilsen N, Bohnhorst J, Moen SH, Vik R, Stephens P, Thomas DW, Benedict CA, Espevik T. 2009. A proviral role for CpG in cytomegalovirus infection. J Immunol 182:5672–5681. doi: 10.4049/jimmunol.0801268. [DOI] [PubMed] [Google Scholar]
- 15.Biolatti M, Dell’Oste V, Pautasso S, Gugliesi F, von Einem J, Krapp C, Jakobsen MR, Borgogna C, Gariglio M, De Andrea M, Landolfo S. 2017. Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J Virol 92:e01774-17. doi: 10.1128/JVI.01774-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diner BA, Lum KK, Toettcher JE, Cristea IM. 2016. Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. mBio 7:e01553-16. doi: 10.1128/mBio.01553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 18.DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Früh K. 2010. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol 84:585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Chen J, Cristea IM. 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paijo J, Döring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, Witte G, Messerle M, Hornung V, Kaever V, Kalinke U. 2016. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog 12:e1005546. doi: 10.1371/journal.ppat.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O’Keefe ES, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol 84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caposio P, Dreano M, Garotta G, Gribaudo G, Landolfo S. 2004. Human cytomegalovirus stimulates cellular IKK2 activity and requires the enzyme for productive replication. J Virol 78:3190–3195. doi: 10.1128/jvi.78.6.3190-3195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caposio P, Luganini A, Hahn G, Landolfo S, Gribaudo G. 2007. Activation of the virus-induced IKK/NF-kappaB signalling axis is critical for the replication of human cytomegalovirus in quiescent cells. Cell Microbiol 9:2040–2054. doi: 10.1111/j.1462-5822.2007.00936.x. [DOI] [PubMed] [Google Scholar]
- 24.Caposio P, Musso T, Luganini A, Inoue H, Gariglio M, Landolfo S, Gribaudo G. 2007. Targeting the NF-kappaB pathway through pharmacological inhibition of IKK2 prevents human cytomegalovirus replication and virus-induced inflammatory response in infected endothelial cells. Antiviral Res 73:175–184. doi: 10.1016/j.antiviral.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Yurochko AD, Kowalik TF, Huong SM, Huang ES. 1995. Human cytomegalovirus upregulates NF-kappa B activity by transactivating the NF-kappa B p105/p50 and p65 promoters. J Virol 69:5391–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montag C, Wagner J, Gruska I, Hagemeier C. 2006. Human cytomegalovirus blocks tumor necrosis factor alpha- and interleukin-1β-mediated NF-κB signaling. J Virol 80:11686–11698. doi: 10.1128/JVI.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browne EP, Shenk T. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci U S A 100:11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarvis MA, Borton JA, Keech AM, Wong J, Britt WJ, Magun BE, Nelson JA. 2006. Human cytomegalovirus attenuates interleukin-1beta and tumor necrosis factor alpha proinflammatory signaling by inhibition of NF-kappaB activation. J Virol 80:5588–5598. doi: 10.1128/JVI.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodwin CM, Munger J. 2019. The IkappaB kinases restrict human cytomegalovirus infection. J Virol 93:e02030-18. doi: 10.1128/JVI.02030-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morales DJ, Lenschow DJ. 2013. The antiviral activities of ISG15. J Mol Biol 425:4995–5008. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C, Collins MN, Hsiang TY, Krug RM. 2013. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol 21:181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianco C, Mohr I. 2017. Restriction of human cytomegalovirus replication by ISG15, a host effector regulated by cGAS-STING double-stranded-DNA sensing. J Virol 91:e02483-16. doi: 10.1128/JVI.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. 2003. Bst‐2/HM1.24 is a raft‐associated apical membrane protein with an unusual topology. Traffic 4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 35.Bieniasz PD. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 37.Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol 83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. 2007. An interferon-α-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blondeau C, Pelchen-Matthews A, Mlcochova P, Marsh M, Milne RS, Towers GJ. 2013. Tetherin restricts herpes simplex virus 1 and is antagonized by glycoprotein M. J Virol 87:13124–13133. doi: 10.1128/JVI.02250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Wang P, Zheng Z, Hu K, Zhang M, Guan X, Fu M, Zhang D, Wang W, Xiao G, Hu Q, Liu Y. 2017. Japanese encephalitis virus counteracts BST2 restriction via its envelope protein E. Virology 510:67–75. doi: 10.1016/j.virol.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, Früh K. 2010. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog 6:e1000913. doi: 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viswanathan K, Smith MS, Malouli D, Mansouri M, Nelson JA, Früh K. 2011. BST2/tetherin enhances entry of human cytomegalovirus. PLoS Pathog 7:e1002332. doi: 10.1371/journal.ppat.1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birrell MA, Hardaker E, Wong S, McCluskie K, Catley M, De Alba J, Newton R, Haj-Yahia S, Pun KT, Watts CJ, Shaw RJ, Savage TJ, Belvisi MG. 2005. Ikappa-B kinase-2 inhibitor blocks inflammation in human airway smooth muscle and a rat model of asthma. Am J Respir Crit Care Med 172:962–971. doi: 10.1164/rccm.200412-1647OC. [DOI] [PubMed] [Google Scholar]
- 44.Zhang D, Zhang DE. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res 31:119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neil SJ. 2013. The antiviral activities of tetherin. Curr Top Microbiol Immunol 371:67–104. doi: 10.1007/978-3-642-37765-5_3. [DOI] [PubMed] [Google Scholar]
- 46.Xu S, Schafer X, Munger J. 2016. Expression of oncogenic alleles induces multiple blocks to human cytomegalovirus infection. J Virol 90:4346–4356. doi: 10.1128/JVI.00179-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Bresnahan W, Shenk T. 2004. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc Natl Acad Sci U S A 101:16642–16647. doi: 10.1073/pnas.0407233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq data sets are available in the Gene Expression Omnibus (GEO) database with the accession number GSE137065.