West Nile virus (WNV) is an encephalitic flavivirus that remains endemic in the United States. Previous studies have found dysfunctional T cell responses correlate to severe disease outcomes during human WNV infection. Here, we sought to better understand the ability of WNV to program human dendritic cells (DCs) to prime WNV-specific T cell responses. While productive infection of monocyte-derived DCs activated antiviral and type I interferon responses, molecules associated with inflammation and programming of T cells were minimally induced. Functionally, WNV-infected DCs dampened T cell activation and proliferation during an allogeneic response. Combined, our data support a model whereby WNV infection of human DCs compromises WNV-specific T cell immunity.
KEYWORDS: dendritic cells, RIG-I-like receptors, type I interferon, West Nile virus
ABSTRACT
West Nile virus (WNV) is a neurotropic flavivirus and the leading cause of mosquito-borne encephalitis in the United States. Recent studies in humans have found that dysfunctional T cell responses strongly correlate with development of severe WNV neuroinvasive disease. However, the contributions of human dendritic cells (DCs) in priming WNV-specific T cell immunity remains poorly understood. Here, we demonstrate that human monocyte derived DCs (moDCs) support productive viral replication following infection with a pathogenic strain of WNV. Antiviral effector gene transcription was strongly induced during the log phase of viral growth, while secretion of type I interferons (IFN) occurred with delayed kinetics. Activation of RIG-I like receptor (RLR) or type I IFN signaling prior to log phase viral growth significantly diminished viral replication, suggesting that early activation of antiviral programs can block WNV infection. In contrast to the induction of antiviral responses, WNV infection did not promote transcription or secretion of proinflammatory (interleukin-6 [IL-6], granulocyte-macrophage colony-stimulating factor [GM-CSF], CCL3, CCL5, and CXCL9) or T cell modulatory (IL-4, IL-12, and IL-15) cytokines. There was also minimal induction of molecules associated with antigen presentation and T cell priming, including the costimulatory molecules CD80, CD86, and CD40. Functionally, WNV-infected moDCs dampened allogenic CD4 and CD8 T cell activation and proliferation. Combining these observations, we propose a model whereby WNV subverts human DC activation to compromise priming of WNV-specific T cell immunity.
IMPORTANCE West Nile virus (WNV) is an encephalitic flavivirus that remains endemic in the United States. Previous studies have found dysfunctional T cell responses correlate to severe disease outcomes during human WNV infection. Here, we sought to better understand the ability of WNV to program human dendritic cells (DCs) to prime WNV-specific T cell responses. While productive infection of monocyte-derived DCs activated antiviral and type I interferon responses, molecules associated with inflammation and programming of T cells were minimally induced. Functionally, WNV-infected DCs dampened T cell activation and proliferation during an allogeneic response. Combined, our data support a model whereby WNV infection of human DCs compromises WNV-specific T cell immunity.
INTRODUCTION
West Nile virus (WNV) is a neurotropic flavivirus that remains the leading cause of mosquito-borne encephalitis in the United States (1). It is estimated that upwards of 6 million people have been infected by WNV in the United States since its introduction in 1999, leading to over 1,000 cases of neuroinvasive disease and nearly 100 deaths each year (2). Following the bite of an infected mosquito, approximately 20% of individuals present with clinical outcomes ranging from mild febrile illness to severe neuroinvasive disease. Neuroinvasion is a serious complication, with long-term sequelae that include ocular involvement, cognitive impairment, muscle weakness, and flaccid paralysis (3). The continued public health threat and lack of FDA-approved vaccines or specific therapeutics against WNV underpin the need to better understand the mechanisms of protective immunity during human infection.
The pathogenesis of human WNV infection is incompletely understood although excellent mouse models have illuminated mechanisms of virus-induced encephalitis and critical features of immune control (4). The bite of an infected mosquito delivers high doses of WNV into the skin where keratinocytes, Langerhans cells, and dermal dendritic cells (DCs) are believed to be initial target cells of infection (5, 6). Over the next 24 h, WNV migrates to the skin-draining lymph nodes and replicates within resident DCs. Subsequent viremia promotes peripheral seeding of virus into permissive tissues such as the spleen, where DCs are targeted for infection (7). WNV then crosses the blood-brain barrier and infects neurons within the central nervous system (CNS), leading to viral encephalitis. Restriction of viral replication by DCs during the early, peripheral phases of viral replication has been shown to be critical for limiting neuroinvasion and mitigating viral encephalitis (7, 8).
Within murine DCs, detection of WNV occurs primarily through the concerted efforts of retinoic acid inducible gene I (RIG-I) and MDA5 (9, 10), members of the RIG-I-like receptor (RLR) family of cytosolic pattern recognition receptors. Signal transduction through the adaptor protein mitochondrial antiviral signaling (MAVS) triggers the nuclear factor-κB (NF-κB) and interferon regulatory factor 3 (IRF-3)-, IRF-5-, and IRF-7-dependent induction of type I interferon (IFN) and antiviral effector gene transcription (8). Following the MAVS-dependent secretion of type I IFN (10), signaling through the type I IFN receptor on DCs is required for early virus restriction and host survival (7).
In addition to direct restriction of viral replication, DCs are critical for the programming of antiviral CD8+ T cell responses that are required for clearance of WNV from the peripheral tissues and CNS (11). In humans, analysis of CD4+ and CD8+ T cells from the blood of WNV-infected patients has found that dysfunctional T cell responses correlate with symptomatic disease outcome (12, 13). Decreased frequencies of CD4+ regulatory T (Treg) cells also correlates with symptomatic WNV infection, highlighting the importance of a balanced T cell response (14). However, the contributions of human DCs in programming T cell immunity during human WNV infection remains poorly understood.
Here, we utilized primary human cells to demonstrate that WNV productively replicates within monocyte-derived DCs (moDCs). Log phase viral replication corresponded with induction of type I IFN and antiviral effector genes, with more delayed secretion of IFN-α and IFN-β proteins. Activation of RLR or type I IFN signaling restricted viral replication, with RLR signaling remaining effective even after blockade of signaling through the type I IFN receptor. In contrast, WNV infection failed to upregulate molecules involved in promoting inflammatory responses and priming of T cell immunity. Functionally, impaired DC activation resulted in diminished T cell proliferation by WNV-infected moDCs during an allogeneic response. Combined, our data show that WNV infection of human DCs activated antiviral responses while failing to program DCs to effectively prime WNV-specific T cell immunity.
(This article was submitted to an online preprint archive [15].)
RESULTS
WNV productively infects human DCs.
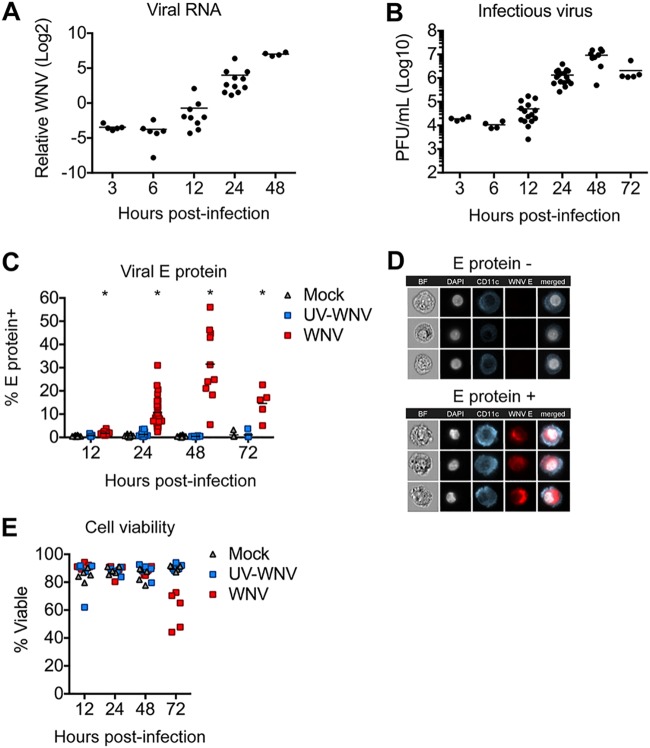
While DCs are an important cell type during infection with multiple flaviviruses, their contributions during human WNV infection remains limited. To model viral replication in human DCs, monocyte-derived DCs (moDCs) were generated from peripheral blood CD14+ monocytes and infected with a pathogenic strain of WNV (16). Viral replication was first detected at 12 h postinfection (hpi), as noted by increased viral RNA synthesis (Fig. 1A). Viral RNA levels continued to increase exponentially over the next 36 h. Consistent with genome replication kinetics, release of infectious virus increased exponentially between 12 and 24 hpi and plateaued at 48 hpi (Fig. 1B). Next, infected populations of moDCs were stained for intracellular expression of a structural protein found within the virus envelope (viral E protein) (17). Corresponding with log phase viral growth, the percentage of infected cells increased exponentially between 12 and 24 hpi (Fig. 1C). Infection plateaued between 24 and 48 hpi, reaching upwards of 50% of cells positive for viral E protein. E protein expression was not observed in mock-infected controls or controls infected with UV-inactivated virus (UV-WNV). Despite the modest increase in WNV titers at 12 hpi, the rise in viral E protein-positive (protein+) cells between 12 and 24 hpi is consistent with earlier work in moDCs where ample detection of flaviviral E protein by immunofluorescence was shown not to occur until 24 hpi (18, 19). ImageStream analysis revealed that WNV E protein was localized predominantly within the cytoplasm and did not colocalize with the cell surface marker CD11c or the nucleus (Fig. 1D). Declining percent infection at 72 hpi corresponded with significant loss of cell viability (Fig. 1E). Combined, three complementary measures of viral replication (viral RNA, infectious virus release, and viral E protein staining) confirm that human moDCs are productively infected by WNV, with log phase viral growth beginning between 12 and 24 hpi.
FIG 1.
WNV productively infects human moDCs. moDCs were infected with WNV or UV-inactivated WNV (UV-WNV) at an MOI of 10 (as determined on Vero cells) and analyzed at the indicated hours postinfection. (A) Viral RNA as quantitated in cell lysates by RT-qPCR, with values shown as log2 normalized expression after normalization to the level of GAPDH. Data are shown for each donor with the mean (n = 5 to 11 donors). (B) Infectious virus release into the supernatant as determined by a viral plaque assay on Vero cells. Data are shown for each donor with the mean (n = 4 to 17 donors). (C) Percentage of E protein+ cells as determined by flow cytometry. Data are shown for each donor with the mean (n = 5 to 31 donors). (D) ImageStream analysis of WNV-infected moDCs labeled for viral E protein at 24 hpi. (E) Percent viable cells. Data are shown for each donor with the mean (n = 5 donors).
Innate immune signaling restricts WNV replication.
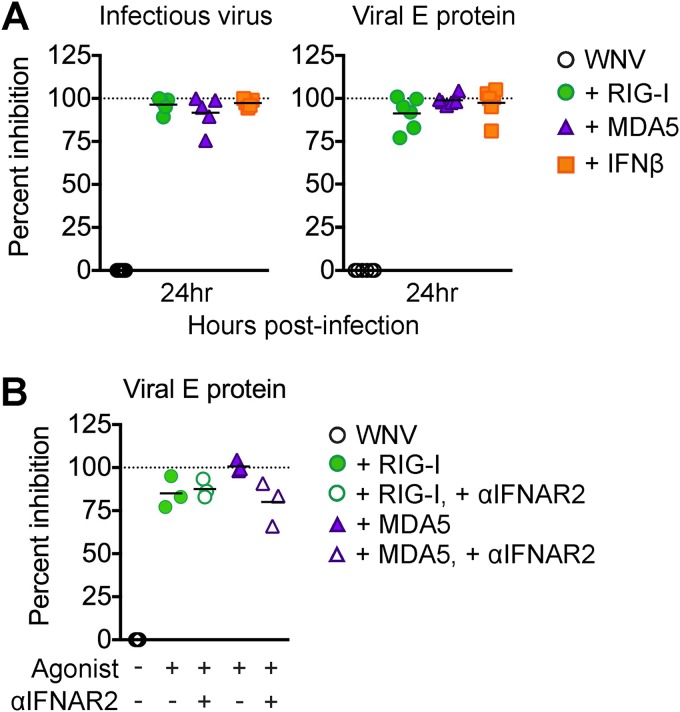
Type I IFN within dendritic cells is critical for mediating protection against lethal infection outcomes and controlling flavivirus replication (20, 21). We next determined the ability of both the RLR and type I IFN signaling pathways to restrict WNV infection of moDCs. We infected moDCs with WNV (multiplicity of infection [MOI] of 10) and treated cells with either a RIG-I agonist, MDA5 agonist, or IFN-β at 1 hpi, and virus replication was measured at 24 hpi. We triggered RIG-I using a previously characterized and highly specific agonist derived from the 3′ untranslated region (UTR) of hepatitis C virus (22) and triggered MDA5 using high-molecular-weight poly(I·C), which preferentially activates MDA5 signaling upon delivery into the cytoplasm (23). Stimulation of RIG-I, MDA5, or IFN-β signaling potently restricted viral replication, with greater than 90% inhibition, as measured by both viral burden in the supernatant and frequency of infected cells (Fig. 2A). To assess whether type I IFN signaling downstream of RLR agonist treatment was responsible for restriction of WNV at 24 hpi, we blocked type I IFN signaling in the presence of either RIG-I or MDA5 agonist in WNV-infected moDCs. We observed reduced viral replication at 24 hpi comparable to that under the RLR treatment conditions alone; however, we did observe a slight reduction in the efficiency of MDA5 signaling to reduce WNV infection in the presence of the anti-IFNAR2 neutralizing antibody (Fig. 2B) (24). Combined, these findings demonstrate that RIG-I, MDA5, and type I IFN signaling can efficiently block WNV replication in human moDCs. In addition, the results may indicate that IFN-independent mechanisms downstream of RLR signaling play an important role in early viral restriction.
FIG 2.
Innate immune signaling restricts WNV replication. (A) moDCs were infected with WNV at an MOI of 10 (as determined on Vero cells) for 1 h and then treated with RIG-I agonist (100 ng/1e6 cells), MDA5 agonist (100 ng/1e6 cells), or IFN-β (1,000 IU/ml) or left untreated (WNV). Supernatants and cells were collected at 24 hpi for flow cytometry and infectious virus release analyses. (B) moDCs were first incubated with or without anti-IFNAR2 (5 μg/ml) for 1 h and then infected and treated with RLR agonists as described for panel A. For the experiments shown in panels A and B, percent inhibition was calculated as follows: (1 − [value for WNV + agonist]/[value for WNV alone]) ×100. Dashed line indicates 100% inhibition, or complete block of viral infection. Data are shown for each donor with the mean (n = 3 donors). *, P < 0.05 (Kruskal-Wallis test).
WNV induces antiviral and type I IFN responses in human DCs.
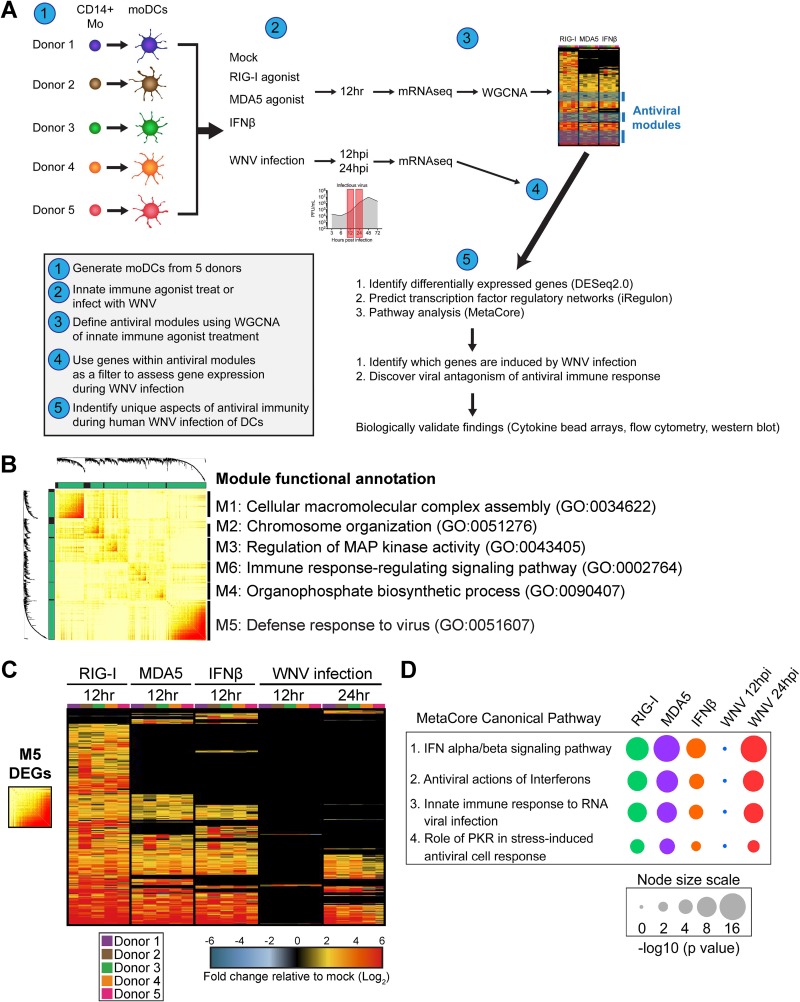
Traditional studies of antiviral responses have predominantly relied on approaches involving genetic ablation, gene knockdown, or gene overexpression methodologies (25). While useful, these approaches remain difficult to perform in primary human cells and may not accurately reflect the role of a given molecule during the normal course of infection. To overcome these limitations, we employed a systems biology approach to assess the antiviral landscape during WNV infection in human DCs (Fig. 3A). We generated moDCs from 5 donors and performed mRNA sequencing following innate immune agonist treatment or infection with WNV. To study the early antiviral response during WNV infection, transcriptional responses were measured preceding (12 hpi) and during (24 hpi) log phase viral replication (Fig. 1). Using weighted gene coexpression network analysis (WGCNA), we defined molecular signatures following stimulation of RIG-I, MDA5, or IFN-β signaling, identifying six clusters of coexpressed genes, or modules (Fig. 3B) (26). The module with the largest gene membership, module 5 (M5), was enriched for genes associated with the biologic process of defense response to virus. Module 6 was also enriched for immune response-related genes, while the remaining four modules were enriched for genes involved in biosynthetic processes, cellular metabolism, and stress responses. Given the large gene number and enrichment for antiviral response pathways, we focused our analyses on the M5 module.
FIG 3.
Systems biology approach reveals antiviral landscape downstream of RLR signaling and WNV infection. (A) Overview of systems biology approach used in this study. (B) Topologic overlap matrix showing enriched modules defined by WGCNA following a 12-h treatment with RIG-I agonist (100 ng/1e6 cells), MDA5 agonist (100 ng/1e6 cells), or IFN-β (1,000 IU/ml). Functional annotation was performed using DAVID Bioinformatics Resource, version 6.8, with the top enriched biological process shown. GO, Gene Ontology category. (C) Heat map of all module 5 differentially expressed genes with the log2 normalized fold change relative to levels in uninfected, untreated cells shown. Genes that did not reach the significance threshold are depicted in black. (D) Top enriched MetaCore canonical pathways of module 5 differentially expressed genes relative to expression in uninfected and untreated cells (>2-fold change; P < 0.01). Node size corresponds with the pathway enrichment significance score (–log10 P value) for each indicated treatment condition.
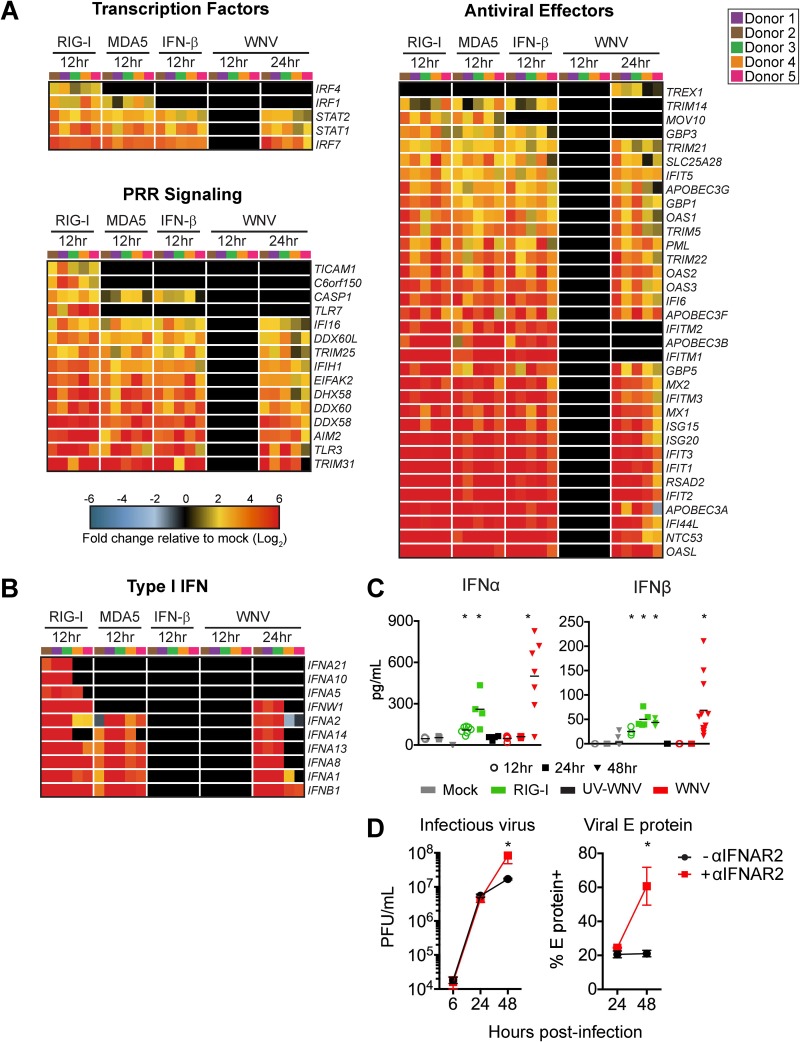
We next identified differentially expressed genes (DEGs) within the M5 module for each treatment condition compared to expression in time-matched untreated and uninfected cells (>2-fold change; significance defined as a P value of <0.01). RIG-I agonist-treated cells induced a greater number of M5-related genes than either MDA5- or IFN-β-treated cells. Gene expression within the M5 module during WNV infection was temporally controlled, showing minimal gene expression at 12 hpi, with more robust gene expression by 24 hpi (Fig. 3C). Despite the overall increase in gene expression between 12 and 24 hpi, a number of genes involved in immune signaling and modulation (IL2RA and SOCS2), DNA binding (ZNF422), DNA repair (POLI) (27), and RNA splicing (TSEN15) (28) were downregulated at 24 hpi. MetaCore pathways enrichment analysis of the M5 DEGs revealed four significantly enriched pathways, including IFN alpha/beta signaling, antiviral activation of interferons, innate immune response to RNA virus infection, and role of PKR in stress-induced antiviral cell response (Fig. 3D). The expression patterns of host defense transcription factors, pattern recognition receptor (PRR) signaling molecules, and antiviral effector genes were largely similar between the RIG-I agonist-, poly(I·C)-, and IFN-β-treated DCs at 12 h poststimulation, suggesting that upregulation of these genes is largely mediated through type I IFN signaling (Fig. 4A). Notably, WNV-infected DCs displayed minimal differentially expressed genes at 12 hpi; however, at 24 hpi numerous antiviral effectors (e.g., IFIT1, IFIT2, IFIT3, RSAD2, and OASL), molecules involved in RNA virus sensing (e.g., DDX58, IFIH1, PKR, and TLR3), and the antiviral transcription factor IRF7 were significantly upregulated. Molecules involved in type I IFN signaling were also not induced at 12 hpi but showed significant enrichment at 24 hpi (Fig. 4B). Despite enrichment of type I IFN genes at 24 hpi, secretion of IFN-α and IFN-β protein was not detected until 48 hpi (Fig. 4C). Given the decrease of WNV replication with RLR agonist treatment (Fig. 2) and the lack of detectable IFN-α or IFN-β protein secretion until 48 hpi in human DCs, we hypothesized that type I IFN secretion is more important in restricting WNV replication at later time points. To confirm the role of type I IFN, we infected moDCs in the presence of an anti-IFNAR2 blocking antibody and observed no effect on viral replication through 24 hpi; however, late viral control was compromised, as shown by a 3-fold increase in the frequency of infected cells and a log-fold increase in viral replication at 48 hpi (Fig. 4D). Combined, our data demonstrate that WNV infection of human DCs induces notable antiviral gene expression and that type I IFN signaling plays a role in late, but not early, restriction of viral replication.
FIG 4.
WNV induces robust antiviral and type I IFN responses. mRNA sequencing was performed on moDCs generated from 5 donors after treatment with RIG-I agonist (100 ng/1e6 cells for 12 h), high-molecular-weight poly(I·C), MDA5 agonist (100 ng/1e6 cells), or IFN-β (100 IU/ml) or WNV infection (MOI of 10; 12 and 24 hpi). (A) Heat map of differentially expressed genes (DEGs) corresponding to antiviral transcription factors, innate immune sensors, and antiviral effector genes. Genes that did not reach the significance threshold are depicted in black. (B) Heat map of DEGs corresponding to type I IFN responses. For all heat maps, the log2 normalized fold change in expression relative to expression in uninfected, untreated cells is shown (>2-fold change; significance, P < 0.01). Genes that did not reach the significance threshold are depicted in black. Each column within a treatment condition is marked by a unique color and represents a different donor (n = 5 donors). (C) Secretion of IFN-α and IFN β proteins into the supernatant following RIG-I agonist treatment (100 ng/1e6 cells), infection with UV-inactivated WNV (MOI of 10; UV-WNV), or infection with replication-competent WNV (MOI of 10; WNV). Data are shown for each donor with the mean (n = 4 to 11 donors). *, P < 0.05 (Kruskal-Wallis test). (D) moDCs were first incubated with or without anti-IFNAR2 (5 μg/ml) for 1 h and then infected with WNV at an MOI of 10. Infectious virus release into the supernatant (left panel) or viral E protein staining (right panel) was assessed at 6, 24, or 48 hpi. Data are represented as percent inhibition and shown for each donor with the mean (n = 5 to 6 donors). *, P < 0.05 (Kruskal-Wallis test).
WNV infection fails to promote inflammatory and T cell modulatory cytokine responses.
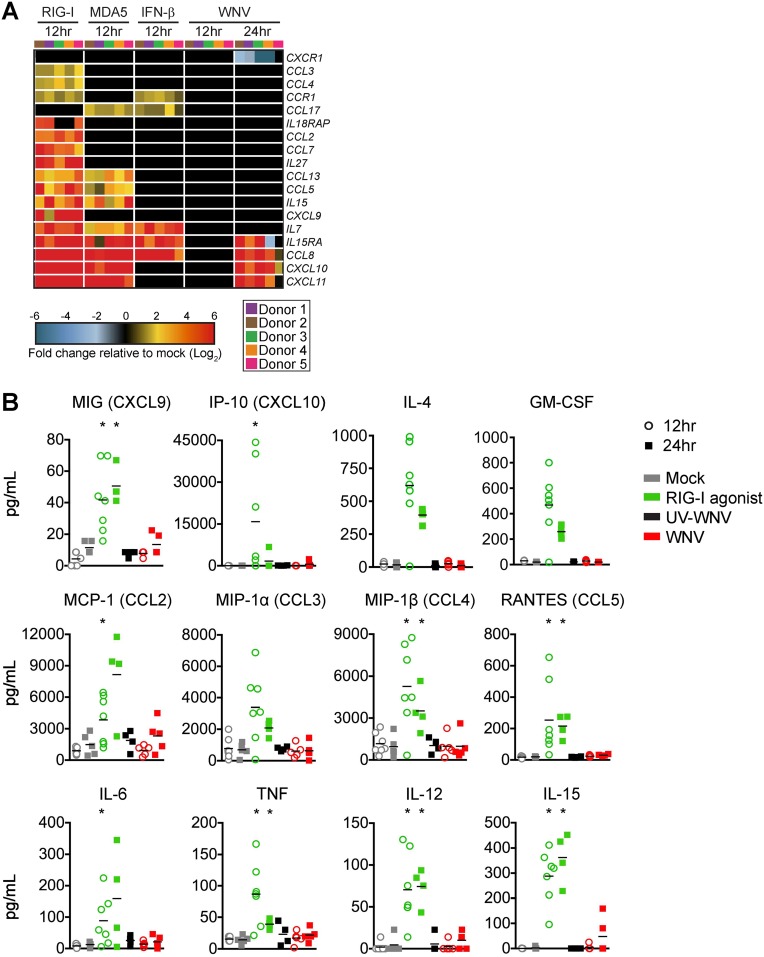
We next assessed the induction of inflammatory cytokine and chemokine responses, an important component of antiviral immunity, DC activation, immune cell recruitment, and T cell priming (29, 30). In contrast to type I IFN and antiviral effector responses, WNV infection promoted minimal transcriptional enrichment of multiple cytokines involved in inflammatory cytokine responses (e.g., interleukin-15 [IL-15], IL-7, and IL-27), and chemotaxis (e.g., CCL2, CCL3, CCL4, CCL5, and CXCL9) (Fig. 5A). CXCR1 transcription was also selectively downregulated during WNV infection. Importantly, RIG-I agonist treatment induced transcriptional expression of multiple inflammatory and T cell modulatory cytokines, confirming the ability of moDCs to mount proinflammatory responses upon innate immune stimulation. RIG-I agonist treatment induced inflammatory cytokines (IL-6 and granulocyte-macrophage colony-stimulating factor [GM-CSF]), T cell promoting cytokines (IL-4, IL-15, and IL-12) and chemokines (CCL3, CCL5, and CXCL9). WNV-infected moDCs displayed little to no induction at the protein level of these cytokines/chemokines at 24 hpi (Fig. 5B). These findings strongly suggest that WNV blocks the induction of proinflammatory cytokines/chemokines during infection of moDCs.
FIG 5.
WNV-infected DCs do not generate robust proinflammatory cytokine and chemokine responses. (A) Heat map of genes involved in inflammatory cytokine responses and chemotaxis. The log2 normalized fold change in expression relative to that in uninfected, untreated cells is shown (>2-fold change; significance, P < 0.01). Genes that did not reach the significance threshold are depicted in black. Each column within a treatment condition is marked by a unique color and represents a different donor (n = 5 donors). (B) Secretion of inflammatory cytokines, T cell modulatory cytokines, and chemokines was assessed by multiplex bead array following RIG-I agonist treatment (100 ng/1e6 cells), infection with UV-inactivated WNV (MOI of 10; UV-WNV), or infection with replication-competent WNV (MOI of 10; WNV). Responses were assessed at 24 h following treatment or infection. Data for each donor is shown with the mean (n = 4 to 7 donors). *, P < 0.05 (Kruskal-Wallis test).
WNV infection does not induce molecules involved in T cell priming.
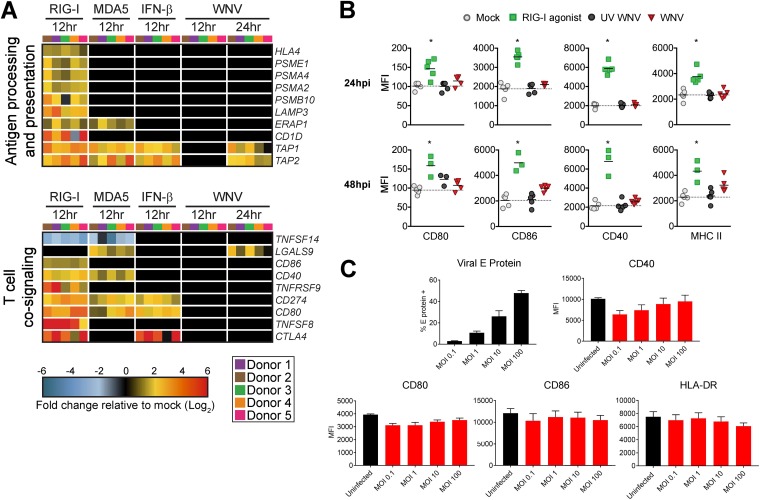
In addition to the secretion of cytokines that modulate T cell responses, engagement of virus-associated molecular patterns increases the surface expression of T cell cosignaling and major histocompatibility complex (MHC) molecules on activated DCs (31). At the transcriptional level, WNV infection failed to induce multiple molecules associated with antigen presentation on MHC (HLA-A and ERAP1), LAMP3 (32), proteasome subunits (PSME1, PSMA2, PSMA4, and PSMB10), and CD1D (33) (Fig. 6A). WNV also failed to significantly upregulate genes involved in T cell cosignaling (e.g., CD80, CD86, and CD40) and selectively upregulated expression of galectin-9 (LGALS9), a ligand for the T cell inhibitory receptor TIM3 (12). These findings were biologically validated by flow cytometry, which showed that WNV infection did not upregulate cell surface levels of CD80, CD86, CD40, or MHC class II (MHC-II) proteins within E protein+ cells at 24 hpi or 48 hpi (Fig. 6B). Notably, high levels of WNV infection (MOI of 100) still failed to induce expression of costimulatory and MHC-II molecules (Fig. 6C). In contrast to WNV infection, RIG-I agonist significantly upregulated transcription of multiple molecules involved in antigen presentation and T cell cosignaling, corresponding with increased cell surface expression of CD80, CD86, CD40, and MHC-II proteins. While WNV infection induces type I IFN and antiviral effector responses, WNV-infected DCs were compromised in their ability to induce inflammatory and chemotactic mediators important for immune activation as well as antigen presentation and costimulatory molecules required for optimal T cell priming.
FIG 6.
WNV-infected DCs fail to increase expression of molecules involved in antigen presentation and T cell costimulation. (A) Heat map of genes involved in antigen processing and presentation or T cell cosignaling. The log2 normalized fold change in expression relative to that in uninfected, untreated cells is shown (>2-fold change; significance, P < 0.01). Genes that did not reach the significance threshold are depicted in black. Each column within a treatment condition is marked by a unique color and represents a different donor (n = 5 donors). (B) Cell surface expression of CD80, CD86, CD40, or MHC-II was quantitated by flow cytometry following RIG-I agonist treatment (100 ng/1e6 cells), infection with UV-inactivated WNV (MOI of 10; UV-WNV), or infection with replication-competent WNV (MOI of 10; WNV). Responses were assessed at 24 h and 48 h following treatment or infection. (C) Cell surface expression of CD80, CD86, CD40, or MHC-II was quantitated by flow cytometry following infection with increasing MOIs of WNV at 24 hpi (MOIs of 0.1, 1, 10, and 100). For the experiments shown in panels B and C, WNV-infected moDCs were labeled for viral E protein, and data are shown for the E protein+ population. Data for each donor are shown as median fluorescence intensity (MFI) with the mean (n = 3 to 5 donors). *, P < 0.05 (Kruskal-Wallis test).
WNV-infected DCs dampen allogeneic T cell proliferation.
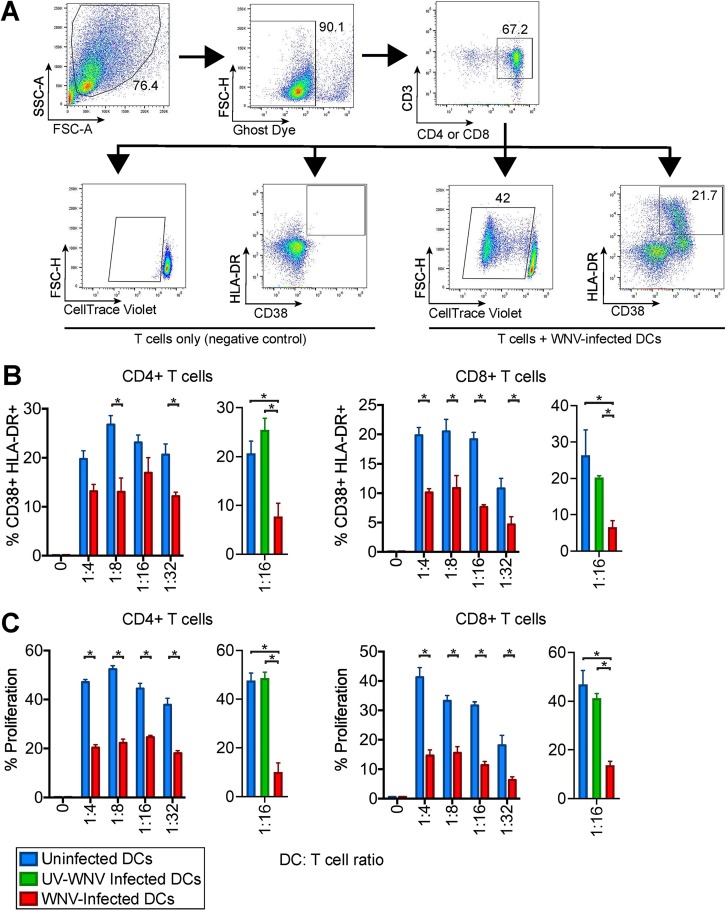
To determine if the minimal DC activation induced during WNV infection impairs T cell proliferation, we assessed the capacity of WNV-infected moDCs to drive an allogeneic T cell response. Uninfected moDCs induced notable activation of donor-mismatched CD4 and CD8 T cells in a DC/T cell ratio-dependent manner, as indicated by increased expression of the human T cell activation markers CD38 and HLA-DR (34) (Fig. 7A). Allogeneic activation of T cells corresponded with proliferation of upwards of 40% of CD4 and CD8 T cells in a DC/T cell ratio-dependent manner. In contrast, WNV-infected moDCs diminished allogeneic CD4 and CD8 T cell activation, corresponding with significantly lower percentages of CD38+ HLA-DR+ and proliferated T cells. UV-WNV-infected moDCs failed to inhibit proliferation and activation of both CD4+ and CD8+ T cells (Fig. 7B and C). Combined, our data suggest that WNV infection induces minimal enrichment of molecules involved in DC activation, resulting in impaired T cell proliferation.
FIG 7.
WNV-infected DCs are compromised in T cell proliferation. moDCs were left uninfected or infected with WNV (MOI of 10) for 24 h. Allogeneic CD4 or CD8 T cells were labeled with CellTrace violet (CTV) and incubated with uninfected or WNV-infected moDCs at the indicated DC/T cell ratios in the presence of an anti-E16 WNV blocking antibody to limit spreading of infection (5 μg/ml) for 6 days. (A) Representative flow cytometry gating strategy for proliferation by CellTrace violet as well as CD38 and HLA-DR positivity in CD4+ and CD8+ T cells. FSC-A, forward scatter area; FSC-H, forward scatter height; SSC-A, side scatter area. (B) The percentage of cells double positive for the T cell activation markers CD38 and HLA-DR on day 6 of allogeneic coculture of mock-, WNV-, or UV-WNV-infected moDCs with T cells. (C) The percentage of cells that had proliferated by day 6 of allogeneic coculture of mock-, WNV-, or UV-WNV-infected moDCs with T cells. The calculation of percent proliferation included any cell that diluted CTV compared to the level in a no-DC, T cell-only control. *, P < 0.05 (one-way and two-way ANOVA).
DISCUSSION
In this study, we combined virologic and immunologic measures with transcriptomic analysis to better understand antiviral responses during WNV infection in primary human DCs. WNV productively infected human moDCs and induced cell death, coinciding with declining viral growth kinetics. RIG-I, MDA5, and IFN-β signaling potently restricted viral replication, corresponding with strong activation of antiviral defense response genes. In contrast, there was minimal upregulation of inflammatory mediators or molecules involved in T cell priming. Functionally, WNV-infected moDCs were impaired in driving allogeneic T cell proliferation and activation.
Studies in mice have found that RLR and type I IFN signaling are critical for viral restriction and host survival during WNV infection; however, the contributions of innate immune signaling during infection of human cells remains limited (7, 35). Here, we demonstrated that RIG-I, MDA5, and IFN-β signaling restricts WNV replication through induction of strong antiviral gene transcription, suggesting that, similar to infection in mice, RLR and type I IFN signaling are important for viral control during human WNV infection. RIG-I and MDA5 agonists also remained efficient in blocking WNV replication independent of type I IFN signaling, consistent with the ability of RLR signaling to induce antiviral gene expression in the absence of the type I IFN receptor in mice (35). Combined, our results confirm the importance of RLR and type I IFN signaling in the induction of antiviral responses and restriction of viral replication within primary human DCs.
These findings are similar to those of previous work, where WNV infection also failed to induce inflammatory cytokine secretion. Infection of moDCs with low-passage-number pathogenic WNV grown in both human and insect cell lines induces IRF3 translocation and ample type I IFN production (19). Despite this, production levels of inflammatory chemokines, including MCP-1, MIP-1α, MIP-1β, and RANTES, was not appreciable over levels in mock-infected samples (19). In addition, infection of moDCs with a nonpathogenic WNV isolate, WNV Kunjin, also induced minimal production of IL-12, despite notable upregulation of both CD86 and CD40 (33). In contrast, another study has shown that WNV-NY99 infection can induce tumor necrosis factor alpha (TNF-α) and IL-6 gene expression at 48 and 72 hpi, respectively (36). In our work, we observed a notable decrease in moDC viability between 48 and 72 hpi, so the upregulation of TNF-α and IL-6 could be attributed to release of pathogen-associated molecular patterns (PAMPs) from dying cells and subsequent stimulation of inflammatory responses in the remaining viable moDC population. Another caveat to the previous study is the lack of inflammatory cytokine translation or secretion data to support its quantitative reverse transcription-PCR (qRT-PCR) results. In our work, we observed upregulation of CXCL10 at 24 hpi in WNV-infected moDCs; however, we did not observe appreciable translation or secretion of this protein in any of our donors. This suggests that upregulation of proinflammatory genes during WNV infection does not always equate to increased cytokine production or release. Altogether, our work, along with that of others, may indicate that an inability to induce inflammatory cytokine responses may be shared among WNV strains, while pathogenic strains have evolved unique mechanisms to subvert antigen presentation and T cell activation.
Failure of WNV to activate human moDCs is also similar to our recent work with Zika virus (ZIKV) (18). In contrast to WNV and ZIKV, infection of moDCs with the yellow fever virus vaccine strain 17D (YFV-17D) upregulated multiple inflammatory mediators and surface expression of CD80 and CD86 (31). The ability of YFV-17D to induce strong DC activation may reflect the loss of a viral antagonist during the attenuation process, similar to the ability of WNV Kunjin to induce upregulation of CD86 and CD40 (33). Alternatively, the ability of YFV-17D to induce DC activation may be an inherent property of certain flaviviruses. Indeed, dengue virus (DENV) has also been found to activate inflammatory responses and upregulate costimulatory molecules following infection (37, 38). Altogether, our recent studies may suggest that certain neurotropic flaviviruses are particularly adept at subverting DC-T cell signaling.
Due to the largely subclinical presentation of WNV infection in humans, understanding genetic correlates of susceptibility and viral restriction remains exceedingly difficult. However, modeling WNV infection in mice lacks the genetic variation seen within outbred human populations. To combat this issue, a collaborative cross (CC) mouse model system, which uses recombinant inbred mice containing genetic diversity from eight founder mouse strains, has been recently developed to study host antiviral responses within a genetically diverse population (39). Using the CC mouse system, one group of investigators observed increased regulatory T (Treg) cell infiltration and no immunopathology in the brains of asymptomatic WNV-infected mice (40). This corroborated well with earlier human studies showing that increased levels of Tregs correlated with improved outcomes during WNV infection (14). These studies have focused largely on observing WNV-specific T and B cell responses to in the CC model system; however, the effects of these diverse polymorphisms on DC function during WNV infection remain largely untouched. Similar to our current study in human moDCs, transcriptomic analyses from whole spleens and brains of WNV-infected CC mice have also shown differences in antigen presentation, T cell signaling, and inflammatory cytokine production (41, 42). Altogether, the CC mouse system can be utilized in future studies to recapitulate human disease and understand DC responses during WNV infection.
An important observation of our study was that WNV infection did not trigger DC activation, as determined by upregulation of costimulatory protein expression. Through an allogeneic T cell assay, we found that WNV-infected moDCs were less efficient in inducing CD4+ and CD8+ T cell proliferation than mock-infected moDCs. WNV-specific T cell responses have been detected in both symptomatic and asymptomatic WNV infection in humans (43). However, the quality rather than quantity of the CD4+ and CD8+ T cell responses during WNV infection is an important predictor of symptomatic infection outcome. Dysregulated Th1 CD4+ T cell responses were found to strongly correlate with neuroinvasive disease (13). Additionally, decreased numbers of Tregs have also been implicated in symptomatic and neuroinvasive infection in WNV-infected individuals, suggesting that immunomodulation of WNV-specific T cells responses is essential for avoiding immunopathology (14). Last, a recent study linked expression of the inhibitory T cell receptor Tim-3 on T cells with progression to symptomatic disease outcome (12). Combined, these findings demonstrate that development of an effective T cell response is critical for modulating infection outcome (symptomatic versus asymptomatic) during WNV infection. Our studies have now determined that WNV interferes with DC activation through inhibition of costimulatory molecule expression and proinflammatory cytokine production, which can lead to dysregulated T cell responses, immunopathology, and excessive neuronal injury.
In summary, our systems biology approach defined the antiviral landscape seen during RLR and type I IFN signaling as well as WNV infection in human DCs. Through our study, we observed that WNV can downregulate numerous genes responsible for establishing proper WNV-specific adaptive immune responses in human DCs, negatively affecting proper CD4+ and CD8+ T cell responses. Altogether, our study significantly advances our understanding of how WNV disrupts antiviral immunity during human infection.
MATERIALS AND METHODS
Ethics statement.
Human peripheral blood mononuclear cells (PBMCs) were obtained from deidentified healthy adult blood donors and processed immediately. All individuals who participated in this study provided informed consent in writing in accordance to the protocol approved by the Institutional Review Board of Emory University (IRB no. 00045821), entitled “Phlebotomy of healthy adults for the purpose of evaluation and validation of immune response assays.”
Viruses.
WNV stocks were generated from an infectious clone, WNV isolate TX 2002-HC, and passaged once in Vero cells, as previously described (16). WNV stocks were titrated on Vero cells by plaque assay. moDCs were infected with WNV at an MOI 10 for 1 h at 37°C in complete RPMI [cRPMI] medium (without GM-CSF or IL-4). After 1 h, virus was washed off, and cells were resuspended in fresh cRPMI medium and incubated at 37°C for 3 to 72 h.
Cell lines.
Vero cells (WHO Reference Cell Banks) were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM). Complete DMEM was prepared as follows: DMEM (Corning) supplemented with 10% fetal bovine serum (Optima, Atlanta Biologics), 2 mM l-glutamine (Corning), 1 mM HEPES (Corning), 1 mM sodium pyruvate (Corning), 1× minimal essential medium (MEM) nonessential amino acids (Corning), and 1× antibiotics/antimycotics (Corning). Complete RPMI (cRPMI) medium was prepared as follows: RPMI 1640 medium (Corning) supplemented with 10% fetal bovine serum (Optima, Atlanta Biologics), 2 mM l-glutamine (Corning), 1 mM sodium pyruvate (Corning), 1× MEM nonessential amino acids (Corning), and 1× antibiotics/antimycotics (Corning).
Generation of monocyte derived dendritic cells.
To generate human moDCs, CD14+ monocytes were differentiated in cRPMI medium supplemented with 100 ng/ml GM-CSF and IL-4 for 5 to 6 days, as previously described (18). In brief, freshly isolated PBMCs obtained from healthy donor peripheral blood (lymphocyte separation medium; StemCell Technologies) were subjected to CD14+ magnetic bead positive selection using a MojoSort human CD14 selection kit (BioLegend). Purified CD14+ monocytes were cultured in complete RPMI medium supplemented with 100 ng/ml each of recombinant human IL-4 and GM-CSF (PeproTech) at a cell density of 2e6 cells/ml. After 24 h of culture, medium and nonadherent cells were removed and replaced with fresh medium and cytokines. Suspension cells (moDCs) were harvested after 5 to 6 days of culture and were consistently CD14− CD11c+ HLA-DR+ DC-SIGN+ CD1a+ by flow cytometry. For experimentation, moDCs were maintained in complete RPMI medium without GM-CSF or IL-4.
qRT-PCR.
Total RNA was purified (Quick-RNA MiniPrep kit; Zymo Research), and viral RNA was reverse transcribed (High Capacity cDNA kit; Applied Biosystems) using 1 pmol of a grapevine virus A (GVA)-tagged (underlined) primer (5′-TTTGCTAGCTTTAGGACCTACTATATCTACCTGGGTCAGCACGTTTGTCATTG-3′) directed against the E gene (21, 44). Reverse transcribed viral sequences were detected by quantitative reverse transcription-PCR (qRT-PCR) (TaqMan Gene Expression Master Mix; Applied Biosystems) using 10 pmol of primers (5′-TTTGCTAGCTTTAGGACCTACTATATCTACCT-3′ and 5′-TCAGCGATCTCTCCACCAAAG-3′) and 2.5 pmol of a hydrolysis probe (5′-FAM-TGCCCGACCATGGGAGAAGCTC-3IABkFQ-3′, where FAM is 6-carboxyfluorescein and 3IABkFQ is 3′ Iowa Black fluorescent quencher). All custom primers and probes were obtained from Integrated DNA Technologies. All qRT-PCR products were normalized to the amount of GAPDH (Hs02758991_g1; Applied Biosystems) in each respective sample.
Quantitation of infectious virus.
Infectious virus was quantitated using a plaque assay on Vero cells with a 1% agarose overlay and crystal violet counterstain, as previously described (16).
Innate immune agonists.
To stimulate RIG-I signaling, 100 ng of RIG-I agonist derived from the 3′ UTR of hepatitis C virus (22) was transfected per 1e6 cells using a TransIT-mRNA transfection kit (Mirus). For stimulation of MDA5 signaling, 100 ng of high-molecular-weight poly(I·C) was transfected per 1e6 cells using LyoVec transfection reagent (Invivogen). To stimulate type I IFN signaling, cells were incubated with 100 IU/ml of human recombinant IFN-β. In select experiments, different doses of agonists were used, and this is indicated within the respective figure legend. To inhibit type I IFN signaling, 5 μg/ml anti-human interferon-α/β receptor chain 2 (MMHAR-2; EMD Millipore) blocking monoclonal antibody was used.
RNA sequencing and bioinformatics.
moDCs were generated from five donors and either treated with innate immune agonists for 12 h (RIG-I, MDA5, or IFN-β) or infected with WNV (12 hpi and 24 hpi). Total RNA was purified (Quick-RNA MiniPrep kit; Zymo Research), and mRNA sequencing libraries were prepared for RNA sequencing (Illumina TruSeq chemistry). RNA sequencing was performed on an Illumina HiSeq 2500 System (100-bp single-end reads). Sequencing reads were mapped to the human reference genome 38. Reads were normalized, and differential expression analysis was performed using DESeq2 (45). Differentially expressed genes were determined by 2-fold change and a P value of <0.01. Weighted gene coexpression module analysis was performed on DESeq2 normalized mapped reads (TIBCO Spotfire with Integromics, version 7.0) from RIG-I agonist-, MDA5 agonist-, IFN-β-, and mock-treated samples. First, the data sets were reduced to focus the network analysis on the 5,446 most variable genes (as determined by variation value greater than 1) using the variance function in R. We constructed a signed weighted correlation network by generating a matrix pairwise correlation between all annotated gene pairs. The resulting biweight midcorrelation matrix was transformed into an adjacency matrix using the soft thresholding power (β1) of 12. The adjacency matrix was used to define the topological overlap matrix (TOM) based on a dissimilarity measurement of 1 − TOM. Genes were hierarchically clustered using average linkage, and modules were assigned using the dynamic tree-cutting algorithm (module eigengenes were merged if the pairwise calculation was larger than 0.75). This resulted in the construction of six modules.
Flow cytometry.
Cells were prepared for analysis as previously described (18). In brief, cells were Fc receptor blocked for 10 min, stained for phenotypic and activation markers for 20 min, and viability stained for 20 min (Ghost Dye Violet 510; Tonbo Biosciences). For intracellular staining of WNV E protein, cells were fixed and permeabilized (Transcription Factor Staining Buffer kit; Tonbo Biosciences) and labeled with E16-allophycocyanin (E16-APC) for 20 min at room temperature (17). Flow cytometry data were analyzed using FlowJo, version 10, software. ImageStream data were analyzed using the Amnis IDEAS software. For moDC studies, primary antibodies are listed in Table 1.
TABLE 1.
Primary antibodies used in this study
| Primary antibodya | Source or reference | Catalog no. |
|---|---|---|
| Mouse anti-human CD11c (BV710, clone B-Ly6) | BD Biosciences | 563130 |
| Mouse anti-human HLA-DR (PE-CF594, clone G46-6) | BD Biosciences | 562304 |
| Mouse anti-human CD1a (BV421, clone HI149) | BioLegend | 300128 |
| Mouse anti-human CD209 (PE, clone 9E9A8) | BioLegend | 330106 |
| Mouse anti-human CD14 (PE, clone M5E2) | BioLegend | 301806 |
| Mouse anti-human CD80 (AF488, clone 2D10) | BioLegend | 305214 |
| Mouse anti-human CD86 (BV605, clone IT2.2) | BioLegend | 305430 |
| Mouse anti-human CD40 (PE-Cy7, clone 5C3) | BioLegend | 334322 |
| Mouse anti-human CD3 (BUV496, clone UCHT1) | BD Biosciences | 564809 |
| Mouse anti-human CD4 (FITC, clone A161A1) | BioLegend | 357406 |
| Mouse anti-human CD8 (FITC, clone HIT8a) | BioLegend | 300906 |
| Mouse anti-human HLA-DR (AF700, clone L243) | BioLegend | 307626 |
| Mouse anti-human CD38 (BUV395, clone HIT2) | BD Biosciences | 740294 |
| Unconjugated monoclonal humanized E16 antibody | 17 |
PE, phycoerythrin; FITC, fluorescein isothiocyanate.
T cell proliferation assay.
Freshly isolated PBMCs obtained from healthy donor peripheral blood (lymphocyte separation medium; StemCell Technologies) were subjected to CD4 or CD8 T cell magnetic bead negative selection using a MojoSort human CD4 or CD8 selection kit (BioLegend). Isolated CD4 or CD8 T cells were labeled with a CellTrace violet (CTV) cell proliferation kit (ThermoFisher) per the manufacturer’s instructions. In a 96-well U-bottom plate, CTV-labeled CD4 or CD8 T cells (2e5 ells) were mixed at different ratios with either uninfected moDCs or moDCs infected with WNV for 24 h (1:4, 1:8, 1:16, 1:32, 1:64, and 1:128 DC/T cell ratios). To prevent spreading of infection, we added anti-E16 neutralizing antibody at 5 μg/ml throughout the DC/T cell coculture period (17). After 6 days of coculture, cells were stained for surface expression of CD4 or CD8, CD3, CD38, and HLA-DR. Proliferation, by CTV dilution, and T cell activation (CD38+ HLA-DR+) were assessed by flow cytometry (46). Primary antibodies are listed in Table 1.
Multiplex bead array.
Cytokine analysis was performed on supernatants using a human 25-plex panel (ThermoScientific) and a custom 2-plex panel with human IFN-β and IFN-α simplex kits (eBioscience) as described previously (18). Cytokines analyzed included: IFN-α, IFN-β, GM-CSF, TNF-α, IL-4, IL-6, MIP-1α, IL-8, IL-15, IL-2R, IP-10, MIP-1β, eotaxin, RANTES, MIG, IL-1RA, IL-12 (p40/p70) IL-13, IFN-γ, MCP-1, IL-7, IL-17, IL-10, IL-5, IL-2, and IL-1β.
Statistics.
All statistical analysis was performed using GraphPad Prism, version 8, software. The number of donors varied by experiment and is indicated within the figure legends. Statistical significance was determined as a P value of <0.05 using a Kruskal-Wallis test (when more than two groups lacking paired measurements were compared), a Wilcoxon test (when two groups with paired measurements were compared), or one-way/two-way analysis of variance (ANOVA) (when two groups across multiple independent variables were compared). All comparisons were made between treatment or infection conditions with a time point-matched, uninfected and untreated control.
Data availability.
The raw data of all RNA sequencing in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) under GEO accession number GSE136342.
ACKNOWLEDGMENTS
We thank the Children’s Healthcare of Atlanta and Emory University Pediatric Flow Cytometry Core for providing access to flow cytometry, ImageStream, and Luminex systems and the Yerkes Genomics Core for performing RNA sequencing.
This work was funded in part by National Institutes of Health grants U19AI083019 (M.S.S.), R56AI110516 (M.S.S.), R21AI113485 (M.S.S.), 2U19AI090023 (B.P.), 5R37DK057665 (B.P.), 5R37AI048638 (B.P.), 2U19AI057266 (B.P.), and ORIP/OD P51OD011132 (M.S.S. and B.P.), the Emory University Department of Pediatrics Junior Faculty Focused Award (M.S.S.), Children’s Healthcare of Atlanta, the Emory Vaccine Center, and the Georgia Research Alliance (M.S.S.).
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Lindsey NP, Lehman JA, Staples JE, Fischer M. 2015. West Nile virus and other nationally notifiable arboviral diseases—United States, 2014. MMWR Morb Mortal Wkly Rep 64:929–934. doi: 10.15585/mmwr.mm6434a1. [DOI] [PubMed] [Google Scholar]
- 2.Chancey C, Grinev A, Volkova E, Rios M. 2015. The global ecology and epidemiology of West Nile virus. Biomed Res Int 2015:376230. doi: 10.1155/2015/376230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel H, Sander B, Nelder MP. 2015. Long-term sequelae of West Nile virus-related illness: a systematic review. Lancet Infect Dis 15:951–959. doi: 10.1016/S1473-3099(15)00134-6. [DOI] [PubMed] [Google Scholar]
- 4.Suthar MS, Diamond MS, Gale M Jr. 2013. West Nile virus infection and immunity. Nat Rev Microbiol 11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 5.Schmid MA, Harris E. 2014. Monocyte recruitment to the dermis and differentiation to dendritic cells increases the targets for dengue virus replication. PLoS Pathog 10:e1004541. doi: 10.1371/journal.ppat.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim PY, Behr MJ, Chadwick CM, Shi PY, Bernard KA. 2011. Keratinocytes are cell targets of West Nile virus in vivo. J Virol 85:5197–5201. doi: 10.1128/JVI.02692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinto AK, Ramos HJ, Wu X, Aggarwal S, Shrestha B, Gorman M, Kim KY, Suthar MS, Atkinson JP, Gale M Jr, Diamond MS. 2014. Deficient IFN signaling by myeloid cells leads to MAVS-dependent virus-induced sepsis. PLoS Pathog 10:e1004086. doi: 10.1371/journal.ppat.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, Thackray L, Brassil MM, Virgin HW, Nikolich-Zugich J, Moses AV, Gale M, Früh K, Diamond MS. 2013. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog 9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M Jr. 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol 87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, Diamond MS, Gale M Jr. 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog 6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM. 2008. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanteri MC, Diamond MS, Law JP, Chew GM, Wu S, Inglis HC, Wong D, Busch MP, Norris PJ, Ndhlovu LC. 2014. Increased frequency of Tim-3 expressing T cells is associated with symptomatic West Nile virus infection. PLoS One 9:e92134. doi: 10.1371/journal.pone.0092134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James EA, Gates TJ, LaFond RE, Yamamoto S, Ni C, Mai D, Gersuk VH, O’Brien K, Nguyen QA, Zeitner B, Lanteri MC, Norris PJ, Chaussabel D, Malhotra U, Kwok WW. 2016. Neuroinvasive West Nile infection elicits elevated and atypically polarized T cell responses that promote a pathogenic outcome. PLoS Pathog 12:e1005375. doi: 10.1371/journal.ppat.1005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanteri MC, O'Brien KM, Purtha WE, Cameron MJ, Lund JM, Owen RE, Heitman JW, Custer B, Hirschkorn DF, Tobler LH, Kiely N, Prince HE, Ndhlovu LC, Nixon DF, Kamel HT, Kelvin DJ, Busch MP, Rudensky AY, Diamond MS, Norris PJ. 2009. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest 119:3266–3277. doi: 10.1172/JCI39387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerman MG, Bowen JR, McDonald CE, Pulendran B, Suthar MS. 2019. West Nile virus subverts T cell stimulatory capacity of human dendritic cells. bioRxiv 10.1101/602839. [DOI] [PMC free article] [PubMed]
- 16.Suthar MS, Brassil MM, Blahnik G, Gale M Jr. 2012. Infectious clones of novel lineage 1 and lineage 2 West Nile virus strains WNV-TX02 and WNV-Madagascar. J Virol 86:7704–7709. doi: 10.1128/JVI.00401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med 11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowen JR, Quicke KM, Maddur MS, O’Neal JT, McDonald CE, Fedorova NB, Puri V, Shabman RS, Pulendran B, Suthar MS. 2017. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog 13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva MC, Guerrero-Plata A, Gilfoy FD, Garofalo RP, Mason PW. 2007. Differential activation of human monocyte-derived and plasmacytoid dendritic cells by West Nile virus generated in different host cells. J Virol 81:13640–13648. doi: 10.1128/JVI.00857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M Jr. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol 80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuel MA, Diamond MS. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol 79:13350–13361. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YE, Ahn JH. 2015. Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog 11:e1004785. doi: 10.1371/journal.ppat.1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowen JR, Ferris MT, Suthar MS. 2016. Systems biology: a tool for charting the antiviral landscape. Virus Res 218:2–9. doi: 10.1016/j.virusres.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faili A, Aoufouchi S, Flatter E, Gueranger Q, Reynaud CA, Weill JC. 2002. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature 419:944–947. doi: 10.1038/nature01117. [DOI] [PubMed] [Google Scholar]
- 28.Trotta CR, Paushkin SV, Patel M, Li H, Peltz SW. 2006. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441:375–377. doi: 10.1038/nature04741. [DOI] [PubMed] [Google Scholar]
- 29.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. 1992. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med 176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heufler C, Koch F, Stanzl U, Topar G, Wysocka M, Trinchieri G, Enk A, Steinman RM, Romani N, Schuler G. 1996. Interleukin-12 is produced by dendritic cells and mediates T helper 1 development as well as interferon-gamma production by T helper 1 cells. Eur J Immunol 26:659–668. doi: 10.1002/eji.1830260323. [DOI] [PubMed] [Google Scholar]
- 31.Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B. 2006. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med 203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Saint-Vis B, Vincent J, Vandenabeele S, Vanbervliet B, Pin JJ, Aït-Yahia S, Patel S, Mattei MG, Banchereau J, Zurawski S, Davoust J, Caux C, Lebecque S. 1998. A novel lysosome-associated membrane glycoprotein, DC-LAMP, induced upon DC maturation, is transiently expressed in MHC class II compartment. Immunity 9:325–336. doi: 10.1016/S1074-7613(00)80615-9. [DOI] [PubMed] [Google Scholar]
- 33.Kovats S, Turner S, Simmons A, Powe T, Chakravarty E, Alberola-Ila J. 2016. West Nile virus-infected human dendritic cells fail to fully activate invariant natural killer T cells. Clin Exp Immunol 186:214–226. doi: 10.1111/cei.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, Keyserling HL, Ploss A, Rice CM, Orenstein WA, Mulligan MJ, Ahmed R. 2009. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol 183:7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suthar MS, Brassil MM, Blahnik G, McMillan A, Ramos HJ, Proll SC, Belisle SE, Katze MG, Gale M Jr. 2013. A systems biology approach reveals that tissue tropism to West Nile virus is regulated by antiviral genes and innate immune cellular processes. PLoS Pathog 9:e1003168. doi: 10.1371/journal.ppat.1003168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawle DJ, Setoh YX, Edmonds JH, Khromykh AA. 2015. Comparison of attenuated and virulent West Nile virus strains in human monocyte-derived dendritic cells as a model of initial human infection. Virol J 12:46. doi: 10.1186/s12985-015-0279-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Madoz JR, Bernal-Rubio D, Kaminski D, Boyd K, Fernandez-Sesma A. 2010. Dengue virus inhibits the production of type I interferon in primary human dendritic cells. J Virol 84:4845–4850. doi: 10.1128/JVI.02514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olagnier D, Peri S, Steel C, van Montfoort N, Chiang C, Beljanski V, Slifker M, He Z, Nichols CN, Lin R, Balachandran S, Hiscott J. 2014. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 10:e1004566. doi: 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris MT, Aylor DL, Bottomly D, Whitmore AC, Aicher LD, Bell TA, Bradel-Tretheway B, Bryan JT, Buus RJ, Gralinski LE, Haagmans BL, McMillan L, Miller DR, Rosenzweig E, Valdar W, Wang J, Churchill GA, Threadgill DW, McWeeney SK, Katze MG, Pardo-Manuel de Villena F, Baric RS, Heise MT. 2013. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog 9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham JB, Thomas S, Swarts J, McMillan AA, Ferris MT, Suthar MS, Treuting PM, Ireton R, Gale M Jr, Lund JM. 2015. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. mBio 6:e00493-15. doi: 10.1128/mBio.00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green R, Wilkins C, Thomas S, Sekine A, Ireton RC, Ferris MT, Hendrick DM, Voss K, de Villena FP, Baric R, Heise M, Gale M Jr. 2016. Identifying protective host gene expression signatures within the spleen during West Nile virus infection in the collaborative cross model. Genom Data 10:114–117. doi: 10.1016/j.gdata.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green R, Wilkins C, Thomas S, Sekine A, Ireton RC, Ferris MT, Hendrick DM, Voss K, Pardo-Manuel de Villena F, Baric RS, Heise MT, Gale M Jr. 2016. Transcriptional profiles of WNV neurovirulence in a genetically diverse collaborative cross population. Genom Data 10:137–140. doi: 10.1016/j.gdata.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanteri MC, Heitman JW, Owen RE, Busch T, Gefter N, Kiely N, Kamel HT, Tobler LH, Busch MP, Norris PJ. 2008. Comprehensive analysis of West Nile virus-specific T cell responses in humans. J Infect Dis 197:1296–1306. doi: 10.1086/586898. [DOI] [PubMed] [Google Scholar]
- 44.Lim SM, Koraka P, Osterhaus AD, Martina BE. 2013. Development of a strand-specific real-time qRT-PCR for the accurate detection and quantitation of West Nile virus RNA. J Virol Methods 194:146–153. doi: 10.1016/j.jviromet.2013.07.050. [DOI] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McElroy AK, Akondy RS, Davis CW, Ellebedy AH, Mehta AK, Kraft CS, Lyon GM, Ribner BS, Varkey J, Sidney J, Sette A, Campbell S, Stroher U, Damon I, Nichol ST, Spiropoulou CF, Ahmed R. 2015. Human Ebola virus infection results in substantial immune activation. Proc Natl Acad Sci U S A 112:4719–4724. doi: 10.1073/pnas.1502619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data of all RNA sequencing in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) under GEO accession number GSE136342.