Abstract
Diabetic retinopathy is a major complication of diabetes. Increasing evidence has indicated that microRNAs (miRs) serves an important role in diabetic retinopathy. However, the expression and mechanism of miR-217 in high glucose-induced human retinal pigment epithelial cells ARPE-19 is still unclear. Therefore, the aim of this study was to investigate the role of miR-217 in high glucose-induced retinal epithelial cell damage, and further to explore the molecular mechanisms. In our study, we found that compared with control group, miR-217 was upregulated in high glucose-induced ARPE-19 cells. In addition, TargetScan and a dual-luciferase reporter gene assay showed that Sirtuin 1 (SIRT1) was a direct target of miR-217. Then, we performed reverse transcription-quantitative polymerase chain reaction assay and western blot assay to explore the expression of SIRT1 in high glucose-induced ARPE-19 cells. Our results demonstrated that SIRT1 was downregulated at the mRNA and protein levels in high glucose-induced ARPE-19 cells. Then, ARPE-19 cells were transfected with inhibitor control, miR-217 inhibitor or miR-217 inhibitor + SIRT1-small interfering RNA for 6 h, and then the cells were treated with 50 mM D-glucose for 24 h. We then investigated the effects of miR-217 inhibitor on ARPE-19 cell viability and apoptosis. An MTT assay revealed that miR-217 inhibitor significantly increased the viability and decreased the apoptosis of high glucose-induced ARPE-19 cells. ELISA indicated that miR-217 inhibitor significantly reduced the expression of inflammatory factors, such as interleukin (IL)-1β, tumor necrosis factor-α, and IL-6 in high glucose-treated ARPE-19 cells. Additionally, a western blot assay demonstrated that miR-217 inhibitor suppressed the expression of p-p65. The effects of miR-217 inhibitor on high glucose-treated ARPE-19 cells were significantly reversed by the silencing the SIRT1 gene. Therefore, our findings suggested that miR-217 inhibitor protected against retinal epithelial cell damage caused by high glucose via targeting SIRT1, thereby playing a protective role in diabetic retinopathy. Targeting miR-217 may have therapeutic potential in the treatment of diabetic retinopathy.
Keywords: microRNA-217, diabetic retinopathy, Sirtuin 1, ARPE-19 cells, inflammation, apoptosis
Introduction
Diabetic retinopathy (DR) is a major complication of diabetes in the working-age population, and causes serious loss of vision or even blindness (1–3). At present, hyperglycemia is the main cause of the development of this disease, which can cause pathological metabolism and biochemical changes that damage retinal cells (4). During the development of DR, retinal endothelial cell (REC) dysfunction is a crucial initiator of multifactorial pathology, which depends on metabolic abnormalities and inflammation (5–9). There is evidence that high glucose (HG)-induced aseptic inflammation is associated with the development and progression of REC injury (10–13). Retinal pigment epithelial (RPE) cells are important component of the external blood-retinal barrier that selectively regulates the flow of molecules into and out of the retina (14). In the development of DR, retinal micro-vascular dysfunction involves the loss of endothelial cells and pericytes, capillary occlusion and vascular barrier destruction, and endothelial cell hypertrophy and degeneration, leading to capillary perfusion and hypoxia (15,16).
MicroRNAs (miRNAs/miRs) are a group of endogenous, single stranded, small non-coding RNAs (21–25 nucleotides in length), which can post-transcriptionally regulate target gene expression by complementarily binding to their 3′-untranslated region (3′-UTR) (17–20). It has been demonstrated that miRNAs are involved in many cellular biological processes in normal physiology and pathogenesis, such as differentiation, cell growth, apoptosis and inflammation (21). Previously, studies have reported that miR-217 participated in the regulation of a variety of biological processes, such as cell growth, apoptosis, differentiation and metastasis in various types of cells (22,23). In addition, miR-217 was determined to be abnormally expressed in a variety of tumor cells, including pancreatic adenocarcinoma, osteosarcoma, cervical carcinoma and lung cancer (24–27). miR-217 inhibition has been found to upregulate the hypoxia inducible factor (HIF)-1α/vascular endothelial growth factor pathway to promote angiogenesis and ameliorate inflammation of diabetic foot ulcer rats (28). Sun et al (29) reported that miR-217 inhibition can protectively antagonize HG-induced podocyte damage and insulin resistance by restoring the defective autophagy pathway via targeting phosphatase and tensin homolog, indicating that miR-217 was a promising therapeutic target for diabetic nephropathy. Shao et al (30) suggested that miR-217 promotes inflammation and fibrosis in HG-cultured rat glomerular mesangial cells via the Sirtuin 1 (Sirt1)/HIF-1α signaling pathway. Additionally, miR-217 has been reported to be related to the development of proteinuria in type 2 diabetes patients; serum miR-217 may be involved in the development of diabetic kidney disease through promoting chronic inflammation, renal fibrosis and angiogenesis (31). These results indicated that miR-217 plays an important role in diabetes and its complications; however, the role of miR-217 in HG-induced retinal epithelial cell damage remains unclear.
Therefore, in the present study, we aimed to investigate the role of miR-217 in HG-induced retinal epithelial cell damage and its molecular mechanisms to determine the role of miR-217 in diabetic retinopathy.
Materials and methods
Cell culture and HG treatment
The RPE cell line ARPE-19 was acquired from American Type Culture Collection (ATCC; cat. no. ATCC® CRL-2302) and cultured in Dulbecco's Modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Beyotime Institute of Biotechnology) at 37°C in a humidified incubator with 5% CO2.
For HG treatment, ARPE-19 cells were treated with 50 mM D-glucose (Beyotime Institute of Biotechnology) at 37°C for 24 h. Cells cultured in DMEM without glucose served as the control. The cultures were conducted in triplicate.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as per the manufacturer's protocol. RNA concentration was measured using a NanoDrop™ 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). RT was conducted with 1 µg total RNA via a PrimeScript reverse transcription reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocols. RT conditions were as follows: 42°C for 60 min and 75°C for 5 min. Then, qPCR was performed using the Fast SYBR™ Green Master Mix (Thermo Fisher Scientific, Inc.) using the CFX Connect Real-Time System (Bio-Rad Laboratories, Inc.). The thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min and 40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 10 sec, and extension at 72°C for 30 sec. U6 for miRNA and GAPDH for mRNA were used as internal controls. The primer sequences for qPCR were as follows: U6, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′; reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′; GAPDH, forward 5′-CTTTGGTATCGTGGAAGGACTC-3′; miR-217, forward 5′-TACTGCATCAGGAACTGACTGGA-3′; reverse 5′-GTGCAGGGTCCGAGGT-3′; SIRT1, forward 5′-AATCCAGTCATTAAAGGTCTACAA-3′; reverse 5′-TAGGACCATTACTGCCAGAGG-3′; reverse 5′-GTAGAGGCAGGGATGATGTTCT-3′. The 2−ΔΔCq method (32) was used to quantify the relative expression of genes.
Dual-luciferase reporter assay
Bioinformatics software (TargetScan 7.2, http://www.targetscan.org/vert_72/) was used to predict target gene of miR-217. The results revealed the binding sites between the 3′-UTR of SIRT1 and miR-217. To verify whether miR-217 could target SIRT1, we performed a dual-luciferase reporter assay. The wild type (WT-SIRT1) and mutant (MUT-SIRT1) 3′-UTR of SIRT1 were respectively cloned into a pmiR-RB-Report™ dual luciferase reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's instructions. To point-mutate the miR-217 binding domain on the 3′UTR of SIRT1, a QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) was performed following the manufacturer's instructions. Then, 50 nM miR-217 mimic (agomir; 5′-UACUGCAUCAGGAACUGAUUGGA-3′; Shanghai GenePharma Co., Ltd.) or 50 nM mimic control (5′-UUUGUACUACACAAAAGUACUG-3′; Shanghai GenePharma Co., Ltd.), and the WT or MUT 3′-UTR of SIRT1 were co-transfected into ARPE-19 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h. Subsequently, relative luciferase activity was detected using a Dual Luciferase Assay System (Promega Corporation) with a microplate reader (Molecular Devices, LLC). Renilla luciferase was used for the normalization of firefly luciferase activity. Each experiment was performed three times.
Western blotting assay
Total protein from ARPE-19 cells was extracted using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology). We used Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific, Inc.) to measure protein concentration. A total of 20 µg of protein from each sample was subjected to 12% SDS-PAGE electrophoresis and transferred to polyvinylidene difluoride membranes (Pierce; Thermo Fisher Scientific, Inc.). Then, the membrane was blocked with 5% skim milk at room temperature for 1.5 h. After washing with PBST three times, the membrane was probed with primary antibodies against SIRT1 (1:1,000; cat. no. 9475; Cell Signaling Technology, Inc.), phosphorylated (p)-NF-κB p65 (p-p65; 1:1,000; cat. no. 3033; Cell Signaling Technology, Inc.), p65 (1:1,000; cat. no. 8242; Cell Signaling Technology, Inc.), or β-actin (45 kDa; 1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.), at 4°C overnight. The next day, the membrane was washed three times with PBST buffer and then incubated with HRP-conjugated secondary antibody (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 2 h. The residue antibody solution was completely washed off with PBST, and protein band was visualized by enhanced chemiluminescence method (EMD Millipore). β-actin was served as loading control for normalization using AlphaView 3.4.0 software (ProteinSimple).
Cell transfection
ARPE-19 cells were plated into 6-well plates and incubated for 24 h before transfection. Cells were transfected with 100 nM inhibitor control (5′-GCCUCCGGCUUCGCACCUCU-3′; Shanghai GenePharma Co., Ltd., Shanghai, China), 100 nM miR-217 inhibitor (antagomir; 5′-UACUGCAUCAGGAACUGAUUGGA-3′; Shanghai GenePharma Co., Ltd.), 0.2 µM control-small interfering (si)RNA (cat. no. sc-36869; Santa Cruz Biotechnology, Inc.), 0.2 µM SIRT1-siRNA (cat. no. sc-40986; Santa Cruz Biotechnology, Inc.), or 100 nM miR-217 inhibitor + 0.2 µM SIRT1-siRNA using Lipofectamine 2000 reagent for 24 h. Then, we performed RT-qPCR assay to detect the transfection efficiency.
MTT assay
ARPE-19 cells were first transfected with inhibitor control, miR-217 inhibitor, or miR-217 inhibitor + SIRT1-siRNA for 6 h, and then the cells were treated with 50 mM D-glucose at 37°C for 24 h, and cell viability was determined by using MTT assay. In brief, 5 mg/ml MTT (Sigma-Aldrich; Merck KGaA) was added to cell culture medium and incubated for 4 h at 37°C. Subsequently, dimethyl sulfoxide was added to each well to dissolve the formazan crystals. The absorbance was measured at 490 nm using a Synergy™ 2 Multi-function Microplate Reader (Bio-Tek Instruments). Experiments were repeated at least three times.
Flow cytometry assay
We used specific a Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit I (BD Bioscience) to detect cell apoptosis according to the manufacturer's introduction. In brief, cells were washed twice with 1X PBS. Then, cells (1×106) were collected, centrifuged (1,000 × g, 5 min, 4°C), and re-suspended in 100 µl of FITC-binding buffer. Subsequently, the buffer was added with ~5 µl ready-to-use annexin V-FITC (BD Bioscience) and 5 µl propidium iodide (PI). In the dark, cells were incubated for 30 min at room temperature. Annexin V-FITC and PI fluorescence was assessed by BD FACSCalibur flow cytometer (BD Technologies) with FlowJo software (version 7.6.1; FlowJo LLC). The apoptotic rate (early apoptotic rate + late apoptotic rate) was calculated and presented.
ELISA
After treatment, cell supernatant was collected through centrifugation (500 × g, 5 min, 4°C). The levels of (IL)-1β (cat. no. PT305; Beyotime Institute of Biotechnology), tumor necrosis factor α (TNF-α; cat. no. PT518; Beyotime Institute of Biotechnology), and IL-6 (cat. no. PT330; Beyotime Institute of Biotechnology) in the supernatant (100 µl) of ARPE-19 cells were detected using sandwich ELISA kits from according to the manufacturer's protocol of each kit.
Statistical analysis
All experiments were carried out three times. Data were displayed from three independent experiments in triplicate. We conducted analyses with a Student's t-test or one-way ANOVA followed by Tukey's test for biostatistical analysis. Analyses were performed using SPSS 19.0 statistical software (IMB Corp.). The data were presented as the mean ± SD, and P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-217 in HG-induced ARPE-19 cells
To explore the role of miR-271 in HG-induced ARPE-19 cells, we firstly performed RT-qPCR assay to detect the expression of miR-217 in HG-induced ARPE-19 cells. RT-qPCR assay indicated that compared with the control group, the expression of miR-217 was significantly increased in the HG treatment group (Fig. 1).
Figure 1.
miR-217 is upregulated in induced ARPE-19 cells. Reverse transcription-quantitative polymerase chain reaction analysis of miR-217 in control group and HG-induced ARPE-19 cells. Experiments were repeated three times. The data were shown as the mean ± SD. **P<0.01 vs. Control. HG, high glucose; miR, microRNA.
SIRT1 is the direct target gene of miR-217
To further investigate the molecular mechanism of miR-217 in HG-induced ARPE-19 cells, we performed TargetScan to predict the target gene of miR-217. The results showed the binding sites between 3′-UTR of SIRT1 and miR-217 (Fig. 2A). Next, to verify the binding sites between miR-217 and SIRT1, we performed a dual-luciferase reporter assay. First, we confirmed that miR-217 mimic significantly enhanced the level of miR-217 in ARPE-19 cells (Fig. 2B). The dual-luciferase reporter assay showed that miR-217 mimic inhibited the luciferase activity of ARPE-19 cells co-transfected with the SIRT1-WT and miR-217 mimic. However, no significant change was observed in the luciferase activity of ARPE-19 cells co-transfected with the SIRT1-MUT and miR-217 mimic (Fig. 2C). Taken together, SIRT1 was suggested to be a direct target gene of miR-217.
Figure 2.
SIRT1 is the direct target gene of miR-217. (A) The predicted binging sites of miR-217 in SIRT1 mRNA 3′UTR. (B) The level of miR-217 in ARPE-19 cells transfected with mimic control or miR-217 mimic for 48 h was detected using reverse transcription-quantitative polymerase chain reaction. (C) Dual-luciferase reporter assay was performed to confirm the binding sites between miR-217 and SIRT1. Experiments were repeated three times. The data were shown as the mean ± SD. ##P<0.01 vs. Control; **P<0.01 vs. mimic control. miR, microRNA; mut, mutant; SIRT1, Sirtuin 1; 3′UTR, 3′-untranslated region; WT, wild type.
Expression of SIRT1 in HG-induced ARPE-19 cells
To detect the expression of SIRT1 in HG-induced ARPE-19 cells, we performed western blotting and RT-qPCR. Western blotting indicated that the expression of SIRT1 was notably decreased at the protein level in the HG induced-ARPE-19 cells (Fig. 3A). RT-qPCR revealed that compared with the control group, SIRT1 expression was significantly decreased at mRNA level in the HG-induced ARPE-19 cells (Fig. 3B).
Figure 3.
SIRT1 expression is downregulated in HG-induced ARPE-19 cells. (A) Western blot analysis of SIRT1 in the control group or HG-induced ARPE-19 cells. (B) Reverse transcription-quantitative polymerase chain reaction analysis of SIRT1 in control group or HG-induced ARPE-19 cells. Experiment was repeated for three times. The data were shown as the mean ± SD. **P<0.01 vs. Control. HG, high glucose; SIRT1, Sirtuin 1.
miR-217 negatively regulates SIRT1 expression in ARPE-19 cells
ARPE-19 cells were transfected with inhibitor control, miR-217 inhibitor, control-siRNA, SIRT1-siRNA, or miR-217 inhibitor + SIRT1-siRNA using Lipofectamine 2000 for 24 h. We performed RT-qPCR to detect the transfection efficiency. RT-qPCR assay indicated that miR-217 inhibitor significantly reduced the expression of miR-217 in ARPE-19 cells compared with the control (Fig. 4A). SIRT1-siRNA significantly decreased SIRT1 mRNA expression in ARPE-19 cells compared with the control (Fig. 4B). miR-217 inhibitor significantly increased the expression of SIRT1 in ARPE-19 cells, but this increase was abolished by SIRT1-siRNA (Fig. 4C and D). These results indicated that SIRT1 was negatively regulated by miR-217 in ARPE-19 cells.
Figure 4.
Transfection efficiency of miR-217 inhibitor or SIRT1-siRNA in ARPE-19 cells. (A) RT-qPCR analysis of the expression of miR-217 in ARPE-19 cells transfected with inhibitor control or miR-217 inhibitor for 24 h. (B) RT-qPCR analysis of the mRNA expression of SIRT1 in ARPE-19 cells transfected with control-siRNA or SIRT1-siRNA for 24 h. (C) Western blot assay was performed to detect the protein expression of SIRT1 in ARPE-19 cells transfected with inhibitor control, miR-217 inhibitor, or miR-217 inhibitor+SIRT1-siRNA for 24 h. (D) RT-qPCR assay was performed to detect the mRNA expression of SIRT1 in ARPE-19 cells transfected with inhibitor control, miR-217 inhibitor, or miR-217 inhibitor + SIRT1-siRNA for 24 h. Experiments were repeated three times. The data were shown as the mean ± SD. **P<0.01 vs. Control; ##P<0.01 vs. inhibitor. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; SIRT1, Sirtuin 1.
Effects of miR-217 inhibitor on the viability and apoptosis of ARPE-19 cells treated with HG
To explore the effects of miR-217 on HG treated ARPE-19 cells, we performed an MTT assay and flow cytometry. The MTT assay indicated that compared with the control group, the cell viability was significantly reduced in the HG group; compared with the HG group, miR-217 inhibitor significantly increased the viability of ARPE-19 cells, which was significantly reversed by the silencing of the SIRT1 gene (Fig. 5A). Flow cytometry assay revealed that compared with the control group, cell apoptosis was significantly increased in the HG group; compared with the HG group, miR-217 inhibitor significantly decreased the apoptosis of ARPE-19 cells, which was significantly reversed by the silencing of the SIRT1 gene (Fig. 5B and C). Taken together, miR-217 inhibitor could enhance cell viability and reduce cell apoptosis in HG-treated ARPE-19 cells.
Figure 5.
miR-217 inhibitor increases ARPE-19 cell viability and inhibits the apoptosis of HG-induced ARPE-19 cells. ARPE-19 cells were transfected with inhibitor control, miR-217 inhibitor, or miR-217 inhibitor + Sirtuin 1-siRNA for 6 h, then the cells were treated with 50 mM D-glucose for 24 h. Then, (A) an MTT assay was conducted to detect cell viability and (B) flow cytometry was performed to detect cell apoptosis. (C) The apoptotic rate was calculated and presented. Experiment was repeated for three times. The data were shown as the mean ± SD. **P<0.01 vs. Control; ##P<0.01 vs. HG; &&P<0.01 vs. inhibitor. FITC, fluorescein isothiocyanate; HG, high glucose; PI, propidium iodide; siRNA, small interfering RNA.
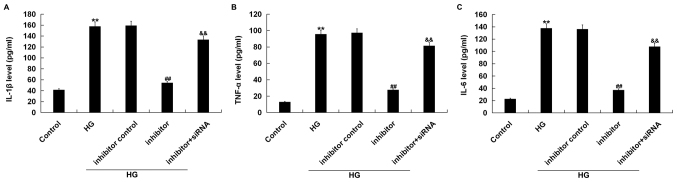
Effects of miR-217 inhibitor on the expression of inflammatory cytokines in HG-induced ARPE-19 cells
In order to determine the expression of inflammatory cytokines, we performed ELISA to detect the expression of IL-1β, TNF-α and IL-6. ELISA showed that compared with the control group, the expression of IL-1β, TNF-α and IL-6 was significantly increased in the HG group. Compared with the HG group, miR-217 inhibitor decreased the expression of inflammatory factors in ARPE-19 cells. This reduction was significantly reversed by the silencing of the SIRT1 gene (Fig. 6).
Figure 6.
miR-217 inhibitor decreases the expression of IL-1β, TNF-α and IL-6 in HG induced ARPE-19 cells. ARPE-19 cells were transfected with inhibitor control, miR-217 inhibitor, or miR-217 inhibitor + SIRT1-siRNA for 6 h, then the cells were treated with 50 mM D-glucose for 24 h. Then, ELISA assay was used to measure the expression of (A) IL-1β, (B) TNF-α and (C) IL-6. Experiments were repeated three times. The data were shown as the mean ± SD. **P<0.01 vs. Control; ##P<0.01 vs. HG; &&P<0.01 vs. inhibitor. HG, high glucose; IL, interleukin; siRNA, small interfering RNA; SIRT1, Sirtuin 1; TNF-α, tumor necrosis factor-α.
Effects of miR-217 on the expression of SIRT1 and p-p65 in HG-induced ARPE-19 cells
In order to explore the specific mechanism of miR-217 in HG-induced ARPE-19 cells, we performed western blotting to detect the expression of SIRT1 and p-p65. The results showed that compared with the control group, the protein expression of SIRT1 was decreased and p-p65 expression was significantly increased in the HG group. Compared with the HG group, miR-217 inhibitor increased the protein expression of SIRT1 (Fig. 7A) and significantly decreased p-p65 expression (Fig. 7A and C) in ARPE-19 cells; this change was significantly reversed by the silencing of the SIRT1 gene. Moreover, the data indicated that HG-induced downregulation of SIRT1 mRNA in ARPE-19 cells was significantly reversed by miR-217 inhibitor, and this upregulation was significantly abolished by SIRT1-siRNA (Fig. 7B).
Figure 7.
miR-217 inhibitor increases the expression of SIRT1 and decreased the expression of p-p65 in HG induced ARPE-19 cells. ARPE-19 cells were transfected with inhibitor control, miR-217 inhibitor, or miR-217 inhibitor+SIRT1-siRNA for 6 h, then the cells were treated with 50 mM D-glucose for 24 h. Then, western blotting and reverse transcription-quantitative polymerase chain reaction were performed to detect the (A) protein and (B) mRNA expression of SIRT1 in ARPE-19 cells. Western blotting was also performed to detect the protein level of (A) p-p65 in ARPE-19 cells (A); (C) densitometry analysis of relative p-p65 expression. Experiments were repeated three times. The data were shown as the mean ± SD. **P<0.01 vs. Control; ##P<0.01 vs. HG; &&P<0.01 vs. inhibitor. HG, high glucose; miR, microRNA; p, phosphorylated; siRNA, small interfering RNA; SIRT1, Sirtuin 1.
Discussion
Diabetic retinopathy is a major complication of diabetes, and retinal pigment epithelial cell damage is involved in the development and progression of diabetic retinopathy (1,10–13). It has been reported that chronic hyperglycemia can induce retinal vascular endothelial cell death (4). Leal et al (33) researched that high glucose was found to reduce the rat retinal endothelial cells viability that were exposed to 30 mM glucose (HG) for 7 days (long-term exposure). Fan et al (34) reported that rat retinal capillary endothelial cells stimulated with 30 mM glucose for 48 and 72 h exhibited significantly reduced cell viability. In this study, we found that the expression of miR-217 was significantly increased in the HG-induced ARPE-19 cells.
At present, several studies have demonstrated that miRNAs, as bio-markers and pharmacological targets, play an important role in ocular diseases, including diabetic retinopathy (35–38). For example, it has been revealed that miR-9, miR-23a, miR-27a, miR-34a, miR-146a, and miR-155 represent potential bio-markers, and novel pharmacological targets for age related macular degeneration (36). miR-383 was highly expressed in HG-induced ARPE-19 cells, and overexpression of miR-383 inhibited cell viability and promoted apoptosis and reactive oxygen species formation in ARPE-19 cells; thus, targeting miR-383 may have therapeutic potential in the treatment of diabetic retinopathy (39). miR-145 decreased HG-induced oxidative stress and inflammation in retinal endothelial cells by modulating the Toll-like receptor 4/nuclear factor-κB signaling (40). miR-217, an important miRNA, serves as a tumor suppressor or an oncogene that depended on the cancer type. For example, Zhao et al (41) showed that miR-217 regulated either V-Ki-Ras2 KRAS or SIRT1 in pancreatic cancer. It has been reported that miRNA-217 acts as a anti-oncogene and is related to the drug-resistance of lung cancer (27). Yin et al (26) revealed that miR-217 was downregulated in clinical cervical cancer tissues. A recent study indicated that miR-217 was upregulated in the hearts of CHF patients (42). However, the expression and mechanism of miR-217 in HG-induced retinal epithelial cells remains unclear. In this study, we investigated the role of miR-217 in HG-induced ARPE-19 cells.
To explore the specific mechanism underlying the role of miR-217 in HG-induced ARPE-19 cells, we researched the target gene of miR-217. TargetScan and a dual-luciferase reporter gene assay suggested that SIRT1 was the direct target gene of miR-217. SIRT1 is a cellular histone deacetylase that is highly expressed in many cancers (43). Lu et al (44) reported that SIRT1 was the target gene of miR-138. Sun et al (45) indicated that SIRT1 was the target gene of miR-29b and miR-29b reversibly modulated the expression of SIRT1. Borji et al (46) showed that SIRT1 was target gene of miR-23b and miR-23b mimic suppressed the expression of SIRT1. A recent study indicated that SIRT1 regulates apoptosis, inflammation and oxidative stress, alleviating diabetic retinopathy (47). Improving SIRT1 inhibition is a potentially effective strategy for treating diabetic retinopathy (48). In our study, we found that the expression of SIRT1 was downregulated at the mRNA and protein levels in HG-induced ARPE-19 cells. Furthermore, we reported that miR-217 negatively regulated SIRT1 expression in ARPE-19 cells. The data suggested that miR-217 might serve a role in HG-induced ARPE-19 cells by regulating the expression of SIRT1.
Additionally, we investigated the effects of miR-217 on HG-treated ARPE-19 cell viability, apoptosis, and inflammatory factors release. ARPE-19 cells were first transfected with inhibitor control, miR-217 inhibitor, or miR-217 inhibitor + SIRT1-siRNA for 6 h, and then the cells were treated with 50 mM D-glucose for 24 h. Our data demonstrated that miR-217 inhibitor promoted cell viability and suppressed cell apoptosis. However, only ARPE-19 cell viability at 24 h was investigated after HG treatment, which poses as a limitation of our study. Thus, the cell viability at different time points (such as 24, 48 and 72 h) after HG treatment should be determined to verify our findings. In addition, miR-217 inhibitor could significantly reduce the expression of inflammatory factors, such as IL-1β, TNF-α and IL-6 in ARPE-19 cells. Studies have confirmed that the NF-κB signaling pathway was activated during diabetic retinopathy (49–51). Thus, in this study, we explored the whether miR-217 could affect the activation of NF-κB signaling pathway through measuring the protein levels of p-p65 (52). The results indicated that miR-217 inhibitor inhibited the expression of p-p65, suggesting suppression of the NF-κB signaling pathway. Of note, the effects of miR-217 inhibitor on HG-treated ARPE-19 cells were significantly reversed by SIRT1 silencing. These results indicated that miR-217 regulated the expression of inflammatory cytokines and NF-κB activity in HG-treated ARPE-19 cells by regulating the expression of SIRT1.
In conclusion, miR-217 inhibitor protected against retinal epithelial cell damage caused by HG by targeting SIRT1, thereby playing a protective role in diabetic retinopathy. However, this study was only a preliminary study of the role of miR-217 in diabetic retinopathy in vitro. There were some limitations of the present study, for example, an in vivo study into the role of miR-217 in diabetic retinopathy was not performed. In addition, the expression of miR-217 in patients with diabetic retinopathy was not determined. Moreover, the relationship between miR-217 expression and the clinical features of patients with diabetic retinopathy was not explored. We will further investigate these issues in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed datasets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HX designed the current study, collected and analyzed the data, performed statistical analysis, and prepared the manuscript. ZL contributed to data collection and data interpretation, and searched the literature. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stewart MW. Treatment of diabetic retinopathy: Recent advances and unresolved challenges. World J Diabetes. 2016;7:333–341. doi: 10.4239/wjd.v7.i16.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein BE. Overview of epidemiologic studies of diabetic retinopathy. Ophthalmic Epidemiol. 2007;14:179–183. doi: 10.1080/09286580701396720. [DOI] [PubMed] [Google Scholar]
- 3.Yau JW, Rogers SL, Kawasaki R, Lamoureux EL, Kowalski JW, Bek T, Chen SJ, Dekker JM, Fletcher A, Grauslund J, et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012;35:556–564. doi: 10.2337/dc11-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang CF, Yuan JR, Qin D, Gu JF, Zhao BJ, Zhang L, Zhao D, Chen J, Hou XF, Yang N, et al. Protection of tauroursodeoxycholic acid on high glucose-induced human retinal microvascular endothelial cells dysfunction and streptozotocin-induced diabetic retinopathy rats. J Ethnopharmacol. 2016;185:162–170. doi: 10.1016/j.jep.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Siasos G, Gouliopoulos N, Moschos MM, Oikonomou E, Kollia C, Konsola T, Athanasiou D, Siasou G, Mourouzis K, Zisimos K, et al. Role of endothelial dysfunction and arterial stiffness in the development of diabetic retinopathy. Diabetes Care. 2015;38:e9–e10. doi: 10.2337/dc14-1741. [DOI] [PubMed] [Google Scholar]
- 6.Frank RN. Diabetic retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 7.Mizutani M, Kern TS, Lorenzi M. Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. J Clin Invest. 1996;97:2883–2890. doi: 10.1172/JCI118746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: A sensor for metabolic danger? Science. 2010;327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 9.Roy S, Kern TS, Song B, Stuebe C. Mechanistic insights into pathological changes in the diabetic retina: Implications for targeting diabetic retinopathy. Am J Pathol. 2017;187:9–19. doi: 10.1016/j.ajpath.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis TM, Gardiner TA, Stitt AW. Microvascular lesions of diabetic retinopathy: Clues towards understanding pathogenesis? Eye (Lond) 2009;23:1496–1508. doi: 10.1038/eye.2009.108. [DOI] [PubMed] [Google Scholar]
- 11.Spijkerman AM, Gall MA, Tarnow L, Twisk JW, Lauritzen E, Lund-Andersen H, Emeis J, Parving HH, Stehouwer CD. Endothelial dysfunction and low-grade inflammation and the progression of retinopathy in type 2 diabetes. Diabet Med. 2007;24:969–976. doi: 10.1111/j.1464-5491.2007.02217.x. [DOI] [PubMed] [Google Scholar]
- 12.Shin ES, Sorenson CM, Sheibani N. Diabetes and retinal vascular dysfunction. J Ophthalmic Vis Res. 2014;9:362–373. doi: 10.4103/2008-322X.143378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toma L, Stancu CS, Botez GM, Sima AV, Simionescu M. Irreversibly glycated LDL induce oxidative and inflammatory state in human endothelial cells; added effect of high glucose. Biochem Biophys Res Commun. 2009;390:877–882. doi: 10.1016/j.bbrc.2009.10.066. [DOI] [PubMed] [Google Scholar]
- 14.Cunha-Vaz J, Bernardes R, Lobo C. Blood-retinal barrier. Eur J Ophthalmol. 2011;21(Suppl 6):S3–S9. doi: 10.5301/EJO.2010.6049. [DOI] [PubMed] [Google Scholar]
- 15.Lorenzi M, Gerhardinger C. Early cellular and molecular changes induced by diabetes in the retina. Diabetologia. 2001;44:791–804. doi: 10.1007/s001250100544. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Zhang Z, Kador PF. Polyol effects on growth factors and MAPK signaling in rat retinal capillary cells. J Ocul Pharmacol Ther. 2014;30:4–11. doi: 10.1089/jop.2013.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specifc microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Liu N, Yang X, Tu L, Zhang X. Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci Rep. 2016;6:35558. doi: 10.1038/srep35558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Mendell J, Olson E. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei R, Deng Z, Su J. miR-217 targeting Wnt5a in osteosarcoma functions as a potential tumor suppressor. Biomed. Pharmacother. 2015;72:158–164. doi: 10.1016/j.biopha.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Yin H, Liang X, Jogasuria A, Davidson NO, You M. miR-217 regulates ethanolinduced hepatic inflammation by disrupting sirtuin 1-lipin-1 signaling. Am J Pathol. 2015;185:1286–1296. doi: 10.1016/j.ajpath.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azam AT, Bahador R, Hesarikia H, Shakeri M, Yeganeh A. Downregulation of microRNA-217 and microRNA-646 acts as potential predictor biomarkers in progression, metastasis, and unfavorable prognosis of human osteosarcoma. Tumour Biol. 2016;37:5769–5773. doi: 10.1007/s13277-015-3821-4. [DOI] [PubMed] [Google Scholar]
- 25.Popov A, Szabo A, Mandys V. Small nucleolar RNA U91 is a new internal control for accurate microRNAs quantifcation in pancreatic cancer. BMC Cancer. 2015;15:774. doi: 10.1186/s12885-015-1785-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Z, Ren W. MicroRNA-217 acts as a tumor suppressor and correlates with the chemoresistance of cervical carcinoma to cisplatin. OncoTargets and Terapy. 2019;12:759–771. doi: 10.2147/OTT.S176618. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Guo J, Feng Z, Huang Z, Wang H, Lu W. MicroRNA-217 functions as a tumour suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Mol Cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin CJ, Lan YM, Ou MQ, Ji LQ, Lin SD. Expression of miR-217 and HIF-1α/VEGF pathway in patients with diabetic foot ulcer and its effect on angiogenesis of diabetic foot ulcer rats. J Endocrinol Invest. 2019 May 11; doi: 10.1007/s40618-019-01053-2. doi: 10.1007/s40618-019-01053-2 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 29.Sun J, Li ZP, Zhang RQ, Zhang HM. Repression of miR-217 protects against high glucose-induced podocyte injury and insulin resistance by restoring PTEN-mediated autophagy pathway. Biochem Biophys Res Commun. 2017;483:318–324. doi: 10.1016/j.bbrc.2016.12.145. [DOI] [PubMed] [Google Scholar]
- 30.Shao Y, Lv C, Wu C, Zhou Y, Wang Q. Mir-217 promotes inflammation and fibrosis in high glucose cultured rat glomerular mesangial cells via Sirt1/HIF-1α signaling pathway. Diabetes Metab Res Rev. 2016;32:534–543. doi: 10.1002/dmrr.2788. [DOI] [PubMed] [Google Scholar]
- 31.Shao Y, Ren H, Lv C, Ma X, Wu C, Wang Q. Changes of serum Mir-217 and the correlation with the severity in type 2 diabetes patients with different stages of diabetic kidney disease. Endocrine. 2017;55:130–138. doi: 10.1007/s12020-016-1069-4. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Leal EC, Aveleira CA, Castilho AF, Serra AM, Baptista FI, Hosoya K, Forrester JV, Ambrósio AF. High glucose and oxidative/nitrosative stress conditions induce apoptosis in retinal endothelial cells by a caspase-independent pathway. Exp Eye Res. 2009;88:983–991. doi: 10.1016/j.exer.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Fan Y, Qiao Y, Huang J, Tang M. Protective effects of panax notoginseng saponins against high glucose-induced oxidativeinjury in rat retinal capillary endothelial cells. Evid Based Complement Alternat Med. 2016;2016:5326382. doi: 10.1155/2016/5326382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rassi DM, De Paiva CS, Dias LC, Módulo CM, Adriano L, Fantucci MZ, Rocha EM. Review: MicroRNAS in ocular surface and dry eye diseases. Ocul Surf. 2017;15:660–669. doi: 10.1016/j.jtos.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Romano GL, Platania CBM, Drago F, Salomone S, Ragusa M, Barbagallo C, Di Pietro C, Purrello M, Reibaldi M, Avitabile T, et al. Retinal and circulating miRNAs in age-related macular degeneration: An in vivo animal and human study. Front Pharmacol. 2017;8:168. doi: 10.3389/fphar.2017.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma J, Wang J, Liu Y, Wang C, Duan D, Lu N, Wang K, Zhang L, Gu K, Chen S, et al. Comparisons of serum miRNA expression profiles in patients with diabetic retinopathy and type 2 diabetes mellitus. Clinics (Sao Paulo) 2017;72:111–115. doi: 10.6061/clinics/2017(02)08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong Q, Su G. Roles of miRNAs and long noncoding RNAs in the progression of diabetic retinopathy. Biosci Rep. 2017;37:BSR20171157. doi: 10.1042/BSR20171157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y, Sang Y, Qiu Q. microRNA-383 mediates high glucose-induced oxidative stress and apoptosis in retinal pigment epithelial cells by repressing peroxiredoxin 3. Am J Transl Res. 2017;9:2374–2383. [PMC free article] [PubMed] [Google Scholar]
- 40.Ying H, Yan Y. MicroRNA-145 attenuates high glucose-induced oxidative stress and inflammation in retinal endothelial cells through regulating TLR4/NF-κB signaling. Life Sci. 2018;207:212–218. doi: 10.1016/j.lfs.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Fan J, Yin Z, Wang F, Chen C, Wang DW. Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget. 2016;7:33–45. doi: 10.18632/oncotarget.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lian B, Yang D, Liu Y, Shi G, Li J, Yan X, Jin K, Liu X, Zhao J, Shang W, Zhang R. miR-128 Targets the SIRT1/ROS/DR5 pathway to sensitize colorectal cancer to TRAIL-induced apoptosis. Cell Physiol Biochem. 2018;49:2151–2162. doi: 10.1159/000493818. [DOI] [PubMed] [Google Scholar]
- 44.Lu Y, Tan L, Wang X. Circular HDAC9/microRNA-138/Sirtuin-1 pathway mediates synaptic and amyloid precursor protein processing deficits in Alzheimer's disease. Neurosci Bull. 2019 Mar 18; doi: 10.1007/s12264-019-00361-0. doi: 10.1007/s12264-019-00361-0 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun QR, Zhang X, Fang K. Phenotype of vascular smooth muscle cells (VSMCs) is regulated by miR-29b by targeting Sirtuin 1. Med Sci Monit. 2018;24:6599–6607Yue. doi: 10.12659/MSM.910068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Borji M, Nourbakhsh M, Shafiee SM, Owji AA, Abdolvahabi Z, Hesari Z, Ilbeigi D, Seiri P, Yousefi Z. Down-regulation of SIRT1 expression by mir-23b contributes to lipid accumulation in HepG2 Cells. Biochem Genet. 2019;57:507–521. doi: 10.1007/s10528-019-09905-5. [DOI] [PubMed] [Google Scholar]
- 47.Karbasforooshan H, Karimi G. The role of SIRT1 in diabetic retinopathy. Biomed Pharmacother. 2018;97:190–194. doi: 10.1016/j.biopha.2017.10.075. [DOI] [PubMed] [Google Scholar]
- 48.Mishra M, Duraisamy AJ, Kowluru RA. Sirt1: A guardian of the development of diabetic retinopathy. Diabetes. 2018;67:745–754. doi: 10.2337/db17-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Zhang Y, Jin W, Xing Y, Yang A. Catechin weakens diabetic retinopathy by inhibiting the expression of NF-κB signaling pathway-mediated inflammatory factors. Ann Clin Lab Sci. 2018;48:594–600. [PubMed] [Google Scholar]
- 50.Yin Y, Chen F, Wang W, Wang H, Zhang X. Resolvin D1 inhibits inflammatory response in STZ-induced diabetic retinopathy rats: Possible involvement of NLRP3 inflammasome and NF-κB signaling pathway. Mol Vis. 2017;23:242–250. [PMC free article] [PubMed] [Google Scholar]
- 51.Kim SJ, Yoo WS, Choi M, Chung I, Yoo JM, Choi W. Increased O-GlcNAcylation of NF-κB enhances retinal ganglion cell death in streptozotocin-induced diabetic retinopathy. Curr Eye Res. 2016;41:249–257. doi: 10.3109/02713683.2015.1006372. [DOI] [PubMed] [Google Scholar]
- 52.Tey SK, Tse EYT, Mao X, Ko FCF, Wong AST, Lo RC, Ng IO, Yam JWP. Nuclear Met promotes hepatocellular carcinoma tumorigenesis and metastasis by upregulation of TAK1 and activation of NF-κB pathway. Cancer Lett. 2017;411:150–161. doi: 10.1016/j.canlet.2017.09.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the present study are available from the corresponding author on reasonable request.