Abstract
Transposases are ubiquitous mobile genetic elements responsible for genome development, driving rearrangements, such as insertions, deletions and translocations. Across species evolution, some transposases are tamed by their host and are made part of complex cellular systems. The proliferation of retroviruses is also dependent on transposase related enzymes termed integrases. Recombination-activating gene protein (RAG)1 and metnase are just two examples of transposase domestication and together with retroviral integrases (INs), they belong to the DDE polynucleotidyl transferases superfamily. They share mechanistic and structural features linked to the RNase H-like fold, harboring a DDE(D) metal dependent catalytic motif. Recent antiretroviral compounds target the catalytic domain of integrase, but they also have the potential of inhibiting other related enzymes. In this review, we report the activity of different classes of integrase inhibitors on various DDE transposases. Computational simulations are useful to predict the extent of off-target activity and have been employed to study the interactions between RAG1 recombinase and compounds from three different pharmacologic classes. We demonstrate that strand-transfer inhibitors display a higher affinity towards the RAG1 RNase H domain, as suggested by experimental data compared to allosteric inhibitors. While interference with RAG1 and 2 recombination is associated with a negative impact on immune function, the inhibition of metnase or HTLV-1 integrase opens the way for the development of novel therapies for refractory cancers.
Keywords: HIV-1 integrase inhibitors, RAG1 and 2 recombination, transposases, computational analysis, metnase, raltegravir, elvitegravir, dolutegravir, styrylquinolines
1. Introduction
A mobile genetic element (MGE) is a fragment of genetic material that encodes an enzyme capable of moving or inserting it in another location in the same or another host genome. DNA transposases and retroviral integrases are among the most extensively studied MGEs. MGEs are present in all genomes sequenced thus far and comprise 3% of the human genome (1). MGEs play an essential role in genome evolution, actively driving rearrangements, insertions, deletions and translocations, while some transposases have evolved to be ‘domesticated’ by the host and perform functional specific tasks within the cell. As a result of selection pressure across millions of years, some parts of MGEs have evolved as non-coding gene modulators at the DNA or RNA level or code for proteins employed as defensive mechanisms against genetic instabilities or with an entirely new and sophisticated function, such as DNA repair (2,3).
A transposase is typically a multidomain enzyme, usually organized as a multimer in its active form. It possesses domains capable of specific interaction with DNA sequences flanking the mobilized fragment [terminal inverted repeats (TIRs)] and a catalytic core domain with nuclease activity, which performs a two-step process of DNA cleavage and transfer into the target DNA (4).
DNA transposons were first identified by Barbara McClintock almost 70 years ago while studying mutable loci in maize (5). Since then a wide variety of elements have been identified and classified. There are two major strategies for DNA transposition: ‘Copy and paste’, in which the transposon remains in place, but inserts a copy of itself into another location and ‘cut and paste’, in which the DNA fragment is excised from the original location and moves into a new one. DNA transposons can be classified according to the folding of their catalytic region and key mechanistic similarities: DDE(D) transposons, HUH transposons (6), serine transposons and tyrosine transposons (7). There are 19 superfamilies of ‘cut and paste’ elements and 11 of these share a DDE(D) triad (Asp, Asp and Glu) (8). DDE(D) elements include prokaryotic transposons, such as Tn5, Tn10, bacteriophage MuA and eukaryotic transposon families, such as hAT (Ac/Ds, Hermes), Transib, Tc1/Mariner (Mos1, Sleeping Beauty, Himar, Hsmar1 and 2, transposase domain of SETMAR) (7). Several transposase systems [such as Flp/FRT (9) or piggyBac (10)] are now part of the molecular toolkit for manipulating genes in vitro and in vivo.
DDE(D) transposons belong to the polynucleotide transferase superfamily, along with other evolutionarily-related metal-ion dependent enzymes, such as retroviral INs, RuvC resolvase, RNaseH, the Argonaut component of the RNA-induced silencing complex, reverse transcriptase (RT) and recombination-activating gene protein 1 (RAG1). Their reaction mechanism is driven by three negatively charged amino acids (Asp, Asp and Glu/Asp), which coordinate divalent metal ions and are located within a RNase H-like fold region of the catalytic core domain (4,11).
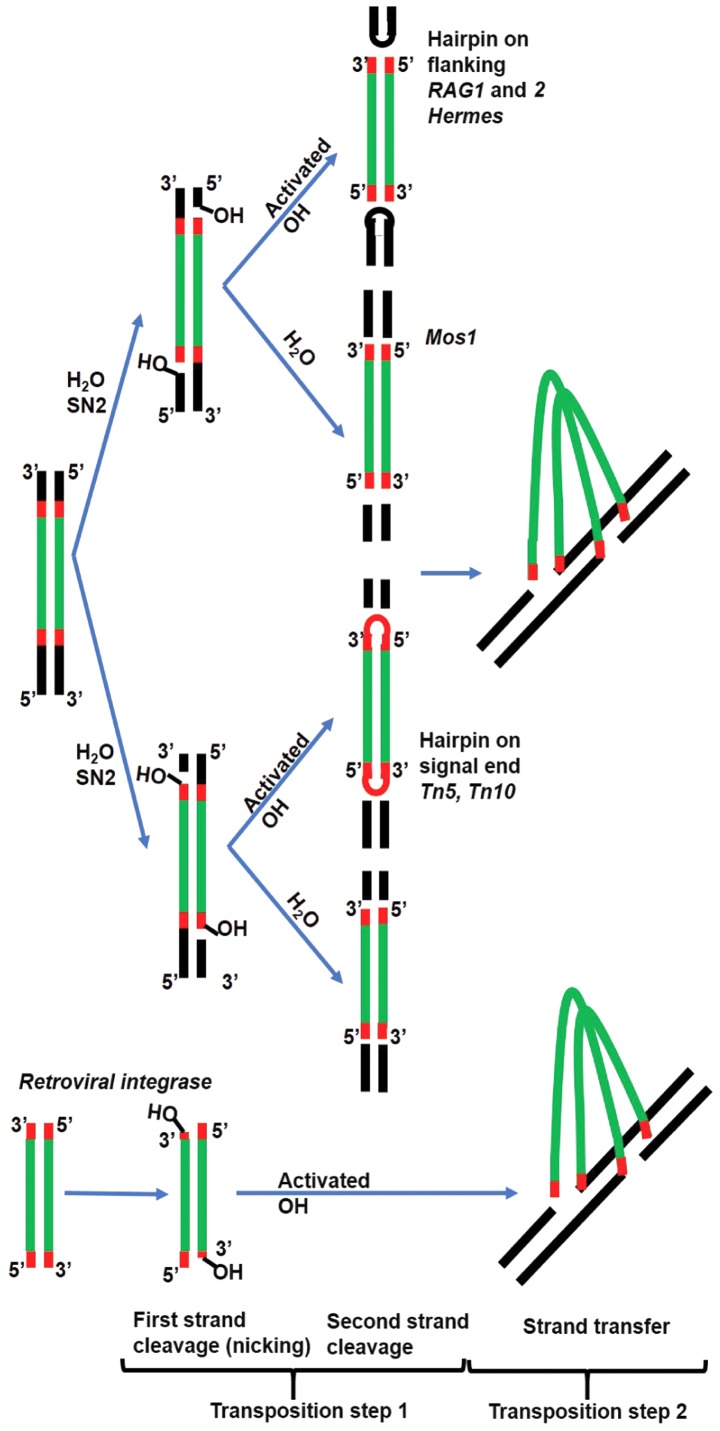
The precise positioning of the two metal ions in the DDE/(D) coordination system enables the nucleophilic substitution through a two-step mechanism. The first step (nicking), results in a reactive 3′OH group at both ends of the MGE. Each active hydroxyl will attack in the second step one strand of the target DNA [strand transfer (ST)] (Fig. 1). Reaction intermediates vary between DDE(D) polynucleotidyl transferases. Some cut only one DNA strand followed by joining the 3′donor end at the target site, while the 5′donor end remains in place, resulting in a branched DNA product that is resolved by DNA replication machinery (e.g., MuA and Tn3). In the case of retroviral INs, the 5′ flanking DNA is constituted only of a few bases and can be removed by repair enzymes. Others cut both strands with a hairpin intermediate on the excised fragment (e.g., Tn5 and Tn10) or on the flanking DNA (RAG1 and 2, and Hermes). The hairpin is opened through another hydrolytic reaction to free the 3′OH for ST, but in the case of RAG1 and 2, transposition does not normally occur and the hairpin is resolved by the non-homologous end joining (NHEJ) system by joining the flanking ends (Fig. 1) (12).
Figure 1.
Scheme illustrating the transposition reaction. In the first step the transposon is cut from its original place followed by pasting into another location in the genome (strand transfer). The initial cleavage occurs at each strand sequentially. The first strand is cleaved using a water molecule as a nucleophile is common for all DDE motif enzymes and releases an activated 3′ hydroxyl group on either the flanking DNA or on the signal end of the transposon (TIR). The second strand is cleaved by the activated OH resulting in a hairpin structure or by another water molecule. In the case of RAG recombinase, the hairpin is opened by NHEJ repair enzymes and the flanking ends are united, while the signal ends very rarely perform transposition and are instead united. In the case of retroviral IN, the vDNA is already independent and the activated OH proceeds directly to strand transfer. NHEJ, non-homologous end joining; RAG, recombination-activating gene protein.
The ST occurs in trans, only in the context of a protein-DNA paired complex, known as a synaptic complex (transpososome or intasome), featuring an oligomeric state of the protein bound to both ends of the mobile element. Accessory proteins are usually needed to stabilize the complex, and introduce bending and conformational changes in the DNA, thus making it accessible for the nuclease or guide the complex to the target site, by interacting with open chromatin (13).
The similarities between HIV-1 IN and domesticated transposases in terms of catalytic domain organization and the mechanism of DNA cleavage function as a double-edged sword. The non-specific inhibition of RAG1 by HIV-1 IN inhibitors can interfere with the assembly of immune system receptors (14). On the other hand, HIV-1 IN inhibitor interaction with the catalytic domain of metnase or other enzymes critical for viral replication [HTLV-1 IN (15), HMCV terminase (16)] offers new alternatives for medicinal chemistry.
This review summarizes the general common characteristics of DDE(D) polynucleotidyl transferases with emphasis on the particular case of RAG1 recombination and HIV-1 integration systems. In this context, we outline the off-target effects of HIV-1 IN inhibitors either with potential deleterious effects or on the contrary which can be exploited for the identification and optimization of novel treatment strategies. Moreover, we argue that computational analysis is a useful tool to predict and analyze possible off target interactions. This is illustrated herein by docking simulations revealing different affinities of HIV-1 IN inhibitors towards RAG1 catalytic domain. The calculated docking conformations explain the possible interaction between IN inhibitors already suggested by reported experimental data and illustrates the behavior of compounds with different IN inhibition mechanisms on RAG1 RNaseH. Such computational analysis is helpful for predicting the possible extent of compound interaction, not only with RAG1, but also with other DDE systems.
2. Docking simulations and conformations
Docking simulations were performed using Autodock Vina software (17) on the RAG1 dimer structure selected from the published crystal of RAG1&2 complex (PDB accession code 4WWX) (18). The 3D structure of the ligands was constructed with Avogadro (19). The protein structure was maintained rigid and the ligand was flexible having all the rotatable bonds set free. The protein and ligand were prepared using AutoDock Tools and Gasteiger partial charges were computed. The search space was defined by the coordinates x=21.488, y=63.789, z=58.445 and the dimensions 100 Å on × scale, 96 Å on y scale and 112 Å on z scale. To improve the sampling of the energy landscape and to increase the probability of finding deep energy minima, the exhaustiveness parameter was set to 50, given a default value of the program of only 8. Five rounds of simulation were performed for each compound, each resulting in 20 output conformations. The conformations were sorted and clustered based on the docking regions in the protein and their energies. Only conformations within 5 Å of the DDE motif were selected for analysis. The results were rendered in PyMol (20).
3. Recombination protein RAG: A domesticated transposase
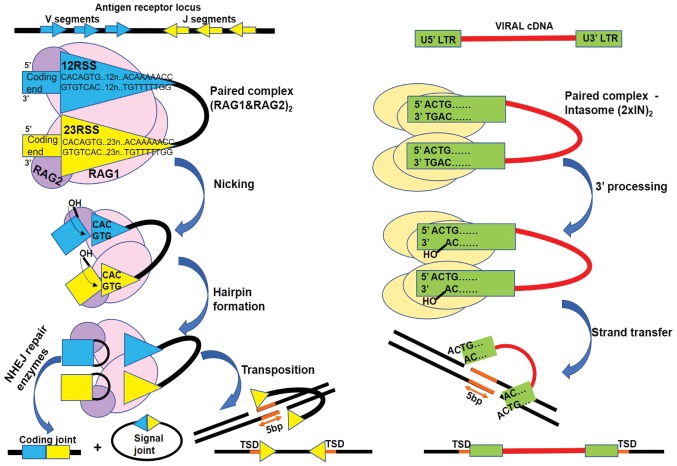
One illustrating example of the transposase domestication is the emergence of RAG1 and RAG2, key enzymes for generating the diverse repertoire of adaptive immune system effectors. The gene encoding the variable region of immunoglobulins and T cell receptors (TCR) is assembled in a combinatorial manner from 3 segments, namely V (variable), D (diversity) and J (joining), each originating from clusters or loci located at large distances in the genome. This process is termed V(D)J recombination and is catalyzed by RAG1 in complex with RAG2 and other accessory proteins, which recognize and cleave at specific sequences termed recombination signal sequence (RSS) bordering the V, D and J segments. An RSS sequence has a 9 bp region (nonamer) rich in A/T and a conserved 7 bp region (heptamer) separated by less conserved 12 or 23 bp (12 RSS and 23 RSS). The heptamer is followed by the coding V, D or J segment and recombination occurs in an orderly and tightly regulated manner, always between a segment flanked by a 12RSS and another flanked by a 23RSS (12/23 rule). The Ig heavy chain and TCR β chain loci are the first to be recombined by bringing together a D next to a J, followed by a V next to the pre-assembled DJ. Subsequently, the variable region of the Ig light chain and TCR α genes are assembled from the V and J segments. The functional unit for concerted recombination is a heterotetramer, containing two RAG1 subunits each in association with a RAG2. The heterotetramer binds a pair of 12/23 RSS and forms the synaptic or paired complex, facilitated by DNA bending performed by accessory protein high mobility group protein B (HMGB)1 or 2. Transesterification is performed by the RAG1 catalytic domain, while RAG2 is involved in heptamer binding, chromatin targeting and the stability of the reaction intermediate complexes. RAG1 introduces at both 12 and 23 RSS, a single strand cleavage between the heptamer and the V, D or J coding sequence with the release of a 3′OH on the coding flank and the free phosphate on the heptamer (signal end). In the following step, the active hydroxyl becomes a nucleophile and attacks the second strand of the coding strand, forming a cyclic phosphate ester intermediate structure termed a ‘hairpin’. In terms of the DDE(D) transposases classic mechanism, this is considered the ST step (Fig. 2). The RAG1 and 2 post-cleavage complex maintains the recessed DNA ends in close proximity, while repair enzymes belonging to NHEJ pathway open the hairpins and attach to each other the two coding ends with the deletion or addition of nucleotides and also the two signal ends, respectively, typically without modifications (signal end joint) (12). In the description below, we refer to mouse RAG residue numbers (14,15).
Figure 2.
RAG-mediated recombination (left) and HIV-1 IN mediated retroviral integration (right) share a two-step transesterification mechanism. The RAG complex recognizes specific sequences named recognition signal sequences (RSS). RSS flank the segments (coding segments) to be recombined in the antigen receptor loci. IN recognizes sequences flanking the long terminal repeats of the viral cDNA (U5′LTR and U3′LTR). Both enzymes are active in the context of a paired complex with DNA: synaptic complex heterotetrameric (RAG1 and 2)2 or intasome homotetrameric (2×IN)2. In the first step RAG1 and 2 cleaves one DNA strand between the heptamers and the coding segments ends, with the release of a reactive 3′ hydroxyl on the coding end. Similarly, IN cleaves one DNA strand at the end of LTR, near the conserved CA dinucleotide, with the removal of the last two 3′ nucleotides of LTR ends and the release of a reactive 3′ hydroxyl (3′ processing). In the second step the viral cDNA reactive ends attack the double stranded target DNA and the 3′ viral DNA ends are united with 3′ target strand. By contrast, RAG generated reactive hydroxyl, attacks the second coding end strand forming a structure called hairpin and detaching the RSS ends (signal ends). Cellular repair enzymes open the hairpins and fuse together the coding segments and in a similar fashion they repair the gaps near the inserted viral DNA. In vivo RAG mediated transposition events of signal ends are highly uncommon, but they result in a 5-bp target site duplication (TSD) similarly to HIV-1 IN strand transfer product. The TSD arises from the 5 nucleotides in the target DNA separating the insertion sites of LTRs and RSS, respectively. NHEJ, non-homologous end joining; RAG, recombination-activating gene protein.
RAG1 is a 1,040 amino acid protein divided into three main domains: The N-terminal domain (1–383), core domain (384–1008) and a short C-terminal domain (1009–1040). RAG2 is a 527 amino acids protein, essential for the proper function of RAG1, comprised of a core region (1–387) and a C-terminal domain (388–527). The most extensively studied regions of RAG proteins are the core domains, defined as the minimum portion of the proteins capable of performing V(D)J recombination. Their structure and conformational changes have been recently illustrated by X-ray and cryo-EM studies (16,17). The N-terminal (NTD) and C-terminal (CTD) domains have regulatory functions and stabilize the protein-DNA complex. RAG1 NTD contains a RING finger domain (264–389), which has E3 ubiquitin-ligase properties and ubiquitylates histone H3 (24). It also has three conserved cysteine pairs that form a Zn2+ binding site (ZnA). RAG1 possesses a complex core region further subdivided into functional subdomains. At the NTD, a series of three helices from each monomer intertwine to form the nonamer binding domain, essential for catalysis (NBD, 391–459) connected via a linker to the dimerization and DNA binding domain (DDBD, 460–515). This is followed by pre RNaseH (515–588) and the RNaseH domains (589–719). The highly helical region separating the last Glu962 from the rest of the catalytic triad contains a pair of cysteines (Cys727 and Cys730) and a pair of histidines (His937 and His942), forming the second Zn2+ binding site (ZnB). RAG2 folds into a 6-bladed β-propeller structure. RAG2 establishes contacts with the RAG1 preR, RNaseH and ZnB domains, through a well conserved interface. RAG2 CTD contains a plant homeodomain finger (PHD) thought to guide the complex to accessible DNA areas of open chromatin by binding to the lysine 4 of the trimethylated histone H3 (25).
RAG1 shares a number of similarities with DNA DDE(D) transposases and retroviral INs in terms of reaction mechanism, intermediates and functional motifs. Double strand cleavage via a hairpin intermediate on the flanking DNA ends is also performed by hAT transposases (Hermes). Following its recruitment, the rag1 gene evolves under positive selection away from transposase origins, losing the ability to perform transposition, but instead developing as part of a strictly regulated recombination machinery which minimizes random and deleterious cleavage within the genome. This argument is further supported by recent research which identified ProtoRAG in cephalochordate amphioxus, a transposon intermediate in the evolution and molecular taming of RAG (26). During chordate development, the RAG transposase ancestor undergoes critical changes that transform it in jawed vertebrates into a recombinase, which favors the joining of excised DNA rather than its insertion. It has been demonstrated that RAG1 residues Arg848, Glu649 and RAG2 acidic hinge (amino-acids 362–383) suppress transposition in vivo (27).
RAG-mediated double strand breaks have been found to be involved in generating translocations responsible for T cell and B cell lymphomas, along with activation-induced deaminase (AID) (28). In human T cells, deletions can arise between two RAG-mediated DSB or translocations between one RAG-mediated DSB and a DSB from another source. Despite its high substrate specificity, RAG is known to also rarely bind to RSS-mimicking sequences (cryptic RSS), all of which have in common the first three 5′-CAC-3′ nucleotides of the heptamer. It has been estimated that there are around 10 million such RAG cleavages in the human genome, which may play an important role in lymphoid tumor development (29). Off-target cleavage at the CAC motif leads to genomic instability (deletions, insertions and translocations) in acute lymphoblastic leukemia pre-B cells, due to the continuous expression of RAG (30). However, it is very uncommon for RAG to perform transposition in vivo in normal cells (31). RAG transposition, identified as the reinsertion of the DNA piece flanked by signal ends (Fig. 2), has been demonstrated to occur in vitro. Reddy et al reported a transposition frequency of one to every 50,000 V(D)J recombination events, in a murine preB cell line (32).
4. RAG recombination versus retroviral integration
The survival and proliferation of retroviruses depends on the integration of their genetic material into the host genome and the exploitation of cellular enzymatic equipment to synthesize viral proteins. The genetic material of retroviruses is represented by two molecules of plus sense RNA, which is released into the cytoplasm following capsid fusion with the cellular membrane, as part of a nucleoprotein complex along with reverse-transcriptase and IN enzymes and other viral proteins. Viral cDNA (vDNA) synthesis occurs through reverse transcription in the cytoplasm, resulting in a pre-integration complex (PIC), which will be further be transported into the nucleus for subsequent insertion into the chromosomal DNA (33). IN is one of the main components of PIC responsible for coordinated DNA integration, which make it an attractive pharmacological target for antiretroviral therapy (ART). Recent studies have demonstrated that IN also plays critical roles in the viral life cycle, apart from integration (34,35). It has been shown that certain mutations in the IN gene can disrupt viral particle assembly and nuclear import. IN follows the classical transesterification process of DDE(D) transposases with the difference that it can only use linear DNA as a substrate flanked by sequences called long terminal repeats (LTRs) and cannot engage DNA already inserted in the genome. Concerted integration of the two vDNA ends takes place in the context of a homotetrameric complex (intasome), in which one IN dimer binds one LTR end (U5′LTR and U3′LTR respectively). 3′Processing (3′P) occurs invariably at a conserved 5′-CAGT-3′, using a water molecule as a nucleophile, with the removal of GT end and the release of adenosine's reactive 3′OH. For the ST PIC is transported to the nucleus, where 3′OH becomes the nucleophile for the single strand hydrolysis of the target DNA (tDNA). The two vDNA ends are inserted simultaneously in a staggered fashion. Similar to RAG1-mediated transposition, a 5 bp TSD emerges after the repair of the resulting branched intermediate. Interestingly, in the case of RAG1, only the first three nucleotides (CAC) of the heptamer sequence establish specific protein-nucleotide interactions via hydrogen bonds. The CA- and TG-rich sequences observed in the RSS consensus heptamer, U5′LTR and U3′LTR promote backbone deformation, DNA unwinding and facilitate cleavage which can explain their high degree of conservation among polynucleotidyl transferases recognition sequences (Fig. 2).
HIV-1 IN is a 288 amino acid protein, divided into three main functional domains: Amino acids 1–49 (NTD), amino acids 50–212 catalytic core domain (CCD) and amino acids 213–288 (CTD). For concerted integration, IN is organized as a dimer of dimers, each dimer binding to one LTR end and tDNA. Only inner protomers are involved in catalysis, while outer protomers are mostly contacting vDNA. NTD contains three consecutive, antiparallel α-helices (α1, α2 and α3), slightly tilted with respect to each other, which harbor a Zn binding motif His12, His16, Cys40 and C43 at their tip. The inner protomer NTD contacts the CCD of the other inner protomer, creating the tetramerization interface in a configuration which wraps the two IN dimers around the DNA. CCD adopts a SH3 like fold (Src homology 3 domain), which was initially described in the tyrosine kinase of Rous sarcoma virus (36). This fold brings together 5 antiparallel β-sheets packed around each other in a barrel configuration. CTD establishes contacts mainly with vDNA through Glu246, Ala248, Lys266 and a series of arginines (Arg228, Arg231 and Arg263).
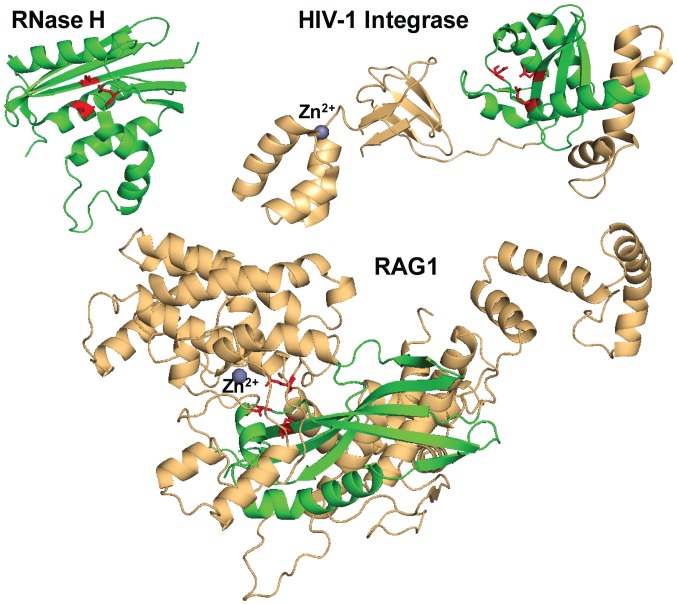
The hallmark RNase H-like fold of DDE(D) polynucleotidyl transferases catalytic domain contains a series of β-sheets, the first 3 antiparallel and consecutive, the others parallel, separated and flanked by 4 α-helices positioned around the β-sheets. This fold was first identified in 1990 by Yang et al, in ribonuclease H from E. coli, β1-β2-β3-α1-β4-α2-α3-β5-α4 (37). IN closely follows this fold in both distribution and orientation of the α and β structures (β1-β2-β3-α4-β4-α5-α6-β5-α7), while in the case of RAG1, there are some differences: There are only two α-helices and four initial consecutive β-sheets, 3 of which are antiparallel, while the following two β-sheets are not separated by α-helices and are instead parallel (β3-β4-β5-β6-α8-β7-β8-α9) (Fig. 3). The DDE(D) motif is the common denominator for this superfamily, imposing the catalytic mechanism of transesterification; however, the spacing of the acidic residues within the RNase H-like fold and overall catalytic core varies according to the protein domain organization and intasome functional assembly. Various intasome structures have evolved different ways to accommodate the substrate DNA with the same purpose of precisely aligning the scissile phosphate in the Mg2+ coordination sphere. Thus, HIV-1 IN Asp64, Asp116 and Glu152 are spaced in a similar manner to other transposases. However, the last catalytic residue of the triad Asp600, Asp708 and Glu962 of mouse RAG1 is separated by a sequence of α-helices from the main RNase H-like fold. The same particularity can be observed in Hermes transposase, where the last Glu is separated by an α-helical insertion domain of almost 300 amino acids from the rest of the catalytic residues.
Figure 3.
RNase H-like fold-crystal structure of E. coli ribonuclease H [PDB accession code 1RNH (37)], structure of HIV-1 IN [PDB accession code 5U1C (90)] and RAG1 structure from the crystal RAG1 and 2 complex [PDB accession code 4WWX (18)]. HIV-1 IN and RAG1 share the RNase H-like fold (green) harboring the DDE catalytic motif (red). Note: The HIV-1 IN structure reported (90) has an E152Q mutation, within the DDE motif.
5. IN inhibitors
The search for more efficient antiretroviral drugs is focusing lately on molecules able to prevent viral infection and spread such as fusion inhibitors (38) and INs. IN has emerged as an important antiretroviral therapeutic target due to its key role in the early steps of infection, immediately after the virus enters the cell. IN inhibitors prevent the insertion of the genetic material into the infected cell genome and thus limit the number of latent reservoirs and the viral spread. α,γ-diketo acid derivatives were discovered independently by Shinoghi and Merck in 1999 (39). These compounds provide a successful strategy for inhibition via chelation, metal-dependent viral endonucleases, such as HIV IN, RT RNase H and hepatitis C virus polymerase. The first selective IN ST inhibitors (INSTI) developed were two α,γ-diketo acid derivatives, L-731,988 and L-708,906, that proved to be active against virus spread in cell culture assays (40). A large series of derivatives based on the acetylpyruvic acid scaffold were developed as less toxic, highly active IN inhibitors, and with improved biopharmaceutical properties. These compounds are generally known as diketo acids, even if chemically most of them have a keto-enol structure or even lack this chemical moiety (Fig. 4A).
Figure 4.
Structure of IN inhibitors discussed in this review. (A) Diketo acid INSTIs, (B) styrylquinoline derivative FZ41, (C) allosteric inhibitors tert-butoxy-(4-phenyl-quinolin-3-yl)-acetic acid derivatives and chemically related compounds.
The majority of INSTIs are characterized by a chelating moiety, usually containing three coplanar O or N atoms. They also have an aromatic hydrophobic group thought to displace the activated adenine after 3′P, rendering it less accessible for ST (Fig. 4A). The binding of diketo acid INSTIs occurs more significantly in the assembled intasome, after the 3′P step, due to their π stacking contacts with the bases adjacent to the activated adenine. Thus, they are more potent inhibitors of ST than of 3′P step.
Raltegravir (RAL) (41) was developed based on the diketo acids, although chemically it has a distinct structure being a N-methyl-4-hydroxypyrimidinone-carboxamide derivative. It was the first FDA-approved INSTI for treatment-experienced adult patients and treatment-naive patients (42). Despite its effectiveness, RAL has a low genetic barrier as a large number of resistance mutations, such as E92QV/N155H, T97A/Y143CHR, and G140CS/Q148HKR have been described (43). The structural simplification of the diketo acid scaffold led to the development of 4-quinolone-3-carboxylic acids which selected elvitegravir (EVG), a potent, once-daily dosing INSTI (44). EVG was first approved in 2012 by the FDI as part of a fixed dose combination and in 2014 as independent formulation for the treatment of HIV-1 infection in treatment-experienced adults in combination with other antiretrovirals (42). EVG is metabolized by CYP3A and thus it is usually associated with CYP3A inhibitors, such as ritonavir or cobicistat (45). EVG shares a similar spectrum of resistance mutation as RAL with some exceptions: Y143/CR, T97A/Y143CR respond to EVG, but are resistant to RAL, while E138K/Q148H and T66I/R263K are susceptible to RAL, but moderately to highly resistant to EVG (43). Second generation INSTIs were developed in order to have a high barrier genetic resistance and reduced cross-resistance (46). The first representative of this category was dolutegravir (DTG), approved by the FDA in 2013, in ARV regimens of both treatment-experienced and treatment-naïve patients (42). DTG acts in a similar manner to first generation INSTIs via three coplanar oxygen atoms which coordinate the active Mg2+ (47) and a difluorobenzyl group which dislocates the 3′activated nucleotide. DTG displays higher potency and is active on most previously reported resistance mutants due to intimate interactions within the catalytic core binding pocket involving a cytidine base next to the activated nucleotide and E152, Q146 residues (40,41). This is evidenced by a 8-fold longer dissociation rate from the IN-DNA complex compared to RAL (50). The most relevant resistance pathways are E138 and Q148 substitutions, where E138K/Q148K in particular is associated with low DTG susceptibility (43). Several other dolutegravir analogues are in current development: S/GSK-1265744 (cabotegravir), formulated as long-acting injectable drug (51) is now under clinical trials (ClinicalTrials.gov Identifier: NCT00920426) and GS-9883 (bictegravir) (52) with similar antiviral potency and improved activity on INSTI-associated IN variants is undergoing phase III clinical trials (ClinicalTrials.gov Identifier: NCT02607930).
Initially designed as polyhydroxylated metal chelators, styrylquinoline derivatives (SQLs) were identified as potent in vitro IN inhibitors of both 3′P and ST steps and reduced HIV-1 replication in cell culture (53). Unlike INSTIs which have a much more potent and specific inhibition on ST compared to 3′P and only bind to IN in complex with vDNA, the SQL in vitro IC50 values for 3′P and ST are usually in the same range or lower for 3′P and they are inactive if added before the assembly of IN-LTR complex (54). SQLs, such as FZ41, KHD161 or FZ55 are competitors of the 3′P reaction and for FZ41 it has been demonstrated that it interferes with DNA binding (Fig. 4B) (55). The SQL binding pocket has not been confirmed by crystallographic data; however, resistance mutations and docking studies have indicated they interact within the CCD with residues involved in vDNA binding. V164I (CCD), V249I and C280Y (CTD) have been reported as resistance mutations to FZ41 (56). Docking studies of KHD161 and FZ55 (56) pointed to 3 possible locations for the compounds, two of which were situated around V165 where the compounds establish contacts with critical residues contacting vDNA (57) (K156, K160, S119, N120, N117) and also located at dimer interface (K186, K188) (57). A later docking study on FZ41 also confirmed interaction with CCD residues situated near protein-protein interface (58).
In parallel with developing inhibitors of the catalytic site, the dimer interface between the two CCDs has been acknowledged as a valid target for small molecule inhibitors. Tetra-acetylated chicoric acid (59) and 1-pyrrolidineacetamide (60) are some of the first compounds discovered to engage residues involved in protein-protein interaction (K103, K173 and T174) and prevent vDNA binding. Moreover, at the IN dimer interface lays the binding site for lens epithelium-derived growth factor (LEDGF), one of the most relevant cellular binding partners of IN. LEDGF, also known as human transcriptional activator p75 is a 530-amino acid nuclear protein associated with transcriptional activation (61). Interaction with LEDGF is a specific property of lentiviruses and it has been demonstrated that it plays a role in the chromatin tropism of PIC (62), viral replication and protects IN from proteasome degradation (63). LEDGF also plays an important role in IN oligomerization. IN binding to LEDGF before vDNA results in inactive tetramers via allosteric alterations, which is supported by studies in which the overexpression of LEDGF has been shown to inhibit viral infectivity (64). LEDGF contains a small domain of interaction with IN (amino acids 347–429), made out of a bundle of 5 helices which contacts a pocket at the IN CCD dimerization interface, in the vicinity of RNaseH domain, establishing contacts with Q168, T174, R166, E170, H171 of inner monomer and W131, W132, A128, T125, L102, Q95 of the outer monomer (65).
This inspired researchers into finding an alternative inhibitory mechanism to INSTIs, based on the LEDGF binding pocket, leading to the development of allosteric IN inhibitors (ALLINIs). Phenoxymethyl-benzoic acid derivative D77 was among the first for which this mechanism of inhibition was demonstrated (66). The most successful class to date is quinoline-based acetic acid derivatives. Structure-based virtual screening approach targeted at LEDGF-IN binding pocket has yielded 2-(quinolin-3-yl) acetic acid derivatives that block the IN-LEDGF interaction (LEDGINs) (67). Tert-butoxy-(4-aryl-quinolin-3-yl)-acetic acid derivative (tBQA) and chemically related compounds, such as BI224436 (68), GSK1264, GSK002 (34) and GS-A (69) derivatives, are among the most successful ALLINIs (Fig. 4C). Although thought in the beginning to act differently, LEDGINs and tBQAs share the same complex mechanism beyond disrupting IN-LEDGF interaction. Both inhibit the integration of competent IN-DNA complex assembly, promote IN aggregation by the formation of high order oligomers and at a later stage, inhibit viral particle assembly and maturation (34). Crystal structures of the compounds at their binding site, as well as resistant IN mutants have revealed critical residues for their activity, such as A128, E170, H171, W131, T174 and T125 which facilitate stronger interactions between the two dimers and also between dimer units, leading to IN aggregation (61,62). Notably, ALLINI resistance mutations at residues 226, 235, 264 and 266 have highlighted the role CTD plays in protein assembly in the absence of vDNA (34).
6. HIV-1 IN inhibitors potentially interfere with RAG-mediated recombination
Before RAL was introduced as part of highly active ART, the prospective that IN inhibitors could also interfere with RAG activity, due to its similarities to IN, was considered. In 2002 Melek et al published an in vitro study on the effects of 5-CITEP and one of the most active β diketo acids to date, L-708,906 (70). 5-CITEP was demonstrated to be a less efficient RAG inhibitor than L-708,906 by 10-fold in all assays. The former has exhibited an IC50 value of 2.1 µM in a ST assay, while the later has exhibited an IC50 value of 0.1 µM (71) and a 2.5 µM IC50 value in a HIV-1 infectivity assay (40). L-708,906 inhibits several steps of RAG catalysis in vitro: Nicking, subsequent hairpin formation and the disintegration reaction, with an IC50 value of 20 µM, when the compound is incubated with IN prior to the addition of the divalent cation. The authors concluded that L-708,906 interferes with RAG binding to its RSS substrate when added prior to the metal ion cofactor and alters the complex if added after, although in this case, the enzyme remains partially competent for cleavage in the presence of Mg2+. Hairpin formation has been found to be more sensitive to the compound presence than nicking. However, in contrast with the IN inhibitory mechanism, the compound does not interfere with the transposition, the equivalent of ST step, nor with non-specific target DNA binding (70).
A later study indicated a similar inhibition pattern on enzymatic 12RSS binding, nicking and hairpin formation for EVG, which displayed a Kd=32.53+2.9 µM corresponding to 1:1 binding to RAG1 central domain (amino acids 528–760) as determined by biolayer interferometry assay. EVG induces the impairment of RAG function as also confirmed in the context of an episomal assay on Nalm6 cell culture (B cell precursor leukemia cell line) transfected with plasmids bearing an artificial 12/23RSS substrate and an antibiotic resistance reporter gene. In cells exposed to 1 µM EVG signal joint formation was reduced by 5.9-fold and coding joint formation by 8.2-fold, respectively. Moreover, the percentage of the CD45+CD25+ B cell population displayed a significant decrease in Balb/c mice treated with EVG and cobicistat (30 mg/kg body weight), but only in 11 out of 16 mice, while the other 5 remained unaffected. This decrease could be linked to RAG activity, although the results warrant further investigation as other mechanisms may also be involved. The same authors reported only limited enzymatic inhibitory activity for RAL, which was not pursued further in cell culture and in vivo assays. However, they reported RAL binding to IN central domain (amino acids 528–760) using the same circular dichroism assay as for EVG (14).
The safety and efficacy of INSTIs currently available for ART is monitored by clinical studies and case reports. This class of antiretrovirals has been considered well tolerated and highly effective in reducing viral loads in adults (72). However, a study conducted between 2006–2008 involving 78 patients following new ART revealed an increase in the incidence of non-Hodgkin lymphoma (NHL) by 20-fold and a rapid onset of NHL following treatment initiation; he virologic response was greater than in patients without NHL which could facilitate abnormal proliferation pathways in lymphocytes (73). The development of NHL could be associated with severe immunosuppression or complications from immune reconstitution inflammatory syndrome in treatment-experienced patients receiving new ART. Nucleos(t)ide reverse transcriptase inhibitors and RAL were the only common antiretrovirals taken by all patients with NHL included in the study. Previous studies have also indicated a high risk of malignancy associated with RAL, NHL being the most common (74). However, NHL could also be associated in these cases with the presence of EBV, which is responsible for dysfunctional T cell activity, or due to other HIV-1 induced oncogenic mechanisms (75). Huhn et al suggested that an enhanced cytokine response and/or impaired T cell function in conjunction with alterations in the RAG1 and 2 recombination induced by INSTIs could account for the proliferation of abnormal lymphocytes (73).
Possible immunological adverse reactions associated with V(D)J recombination interference and associated with INSTI therapy should also be investigated in the context of developing immune response in newborns and children. IN inhibitors are approved for children beginning from 4 weeks old (RAL) or 6 years old (EVG/cobicistat/emtricitabine/tenofovir and DTG). Through their mechanism of preventing viral DNA integration, INSTIs could potentially prevent infections of newborns from mothers infected with HIV-1. The significant benefit, safety and pharmacokinetics of RAL in this case are being evaluated during a phase I clinical trial initiated in 2013 and expected to be completed in 2020 (ClinicalTrials.gov Identifier: NCT01780831). The GS-US-292-0106 ongoing study (ClinicalTrials.gov Identifier: NCT01854775) is currently evaluating the pharmacokinetics, antiviral activity and safety of the EVG/Cobicistat/Emtricitabine/Tenofovir alafenamide fumarate (GENVOYA®) in pediatric patients <18 years of age. Based on preliminary results, the FDA reported a decline in CD4+ counts in 23 children between 6 and 12 years of age, beginning with the second week of treatment and persisting up to 24 weeks. After 48 weeks the CD+ decline is less than the one observed initially. The etiology remains unclear and further data are required for a conclusive evaluation (76).
7. Docking simulations of IN inhibitors on RAG1 dimer
Computational approaches are useful to estimate and compare the affinities of different compounds towards an enzyme and also to predict a potential binding pocket and an associated mechanism. Docking analysis can be applied to verify the IN inhibitor predilection for binding within RNase H-like fold catalytic domains and associate it with potential off-target effects observed in vivo.
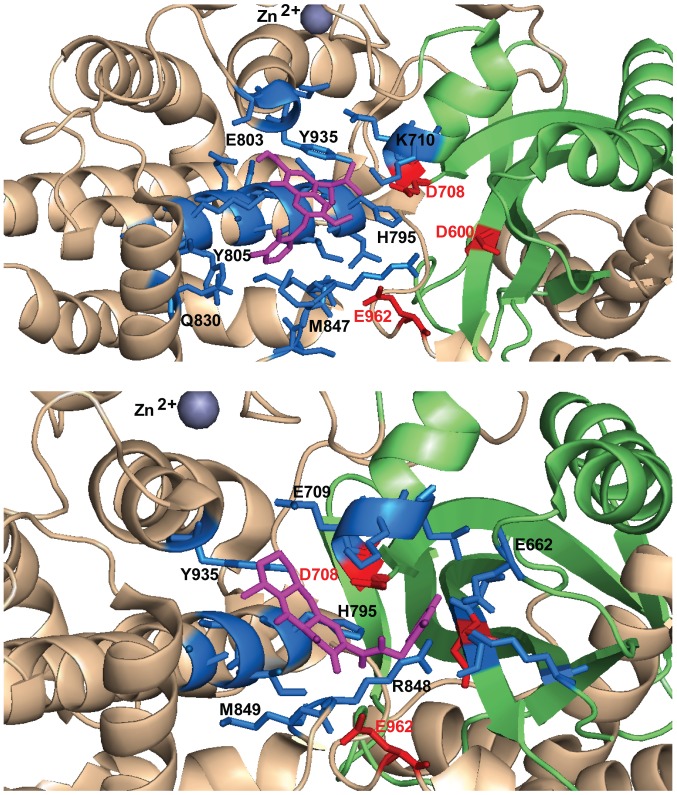
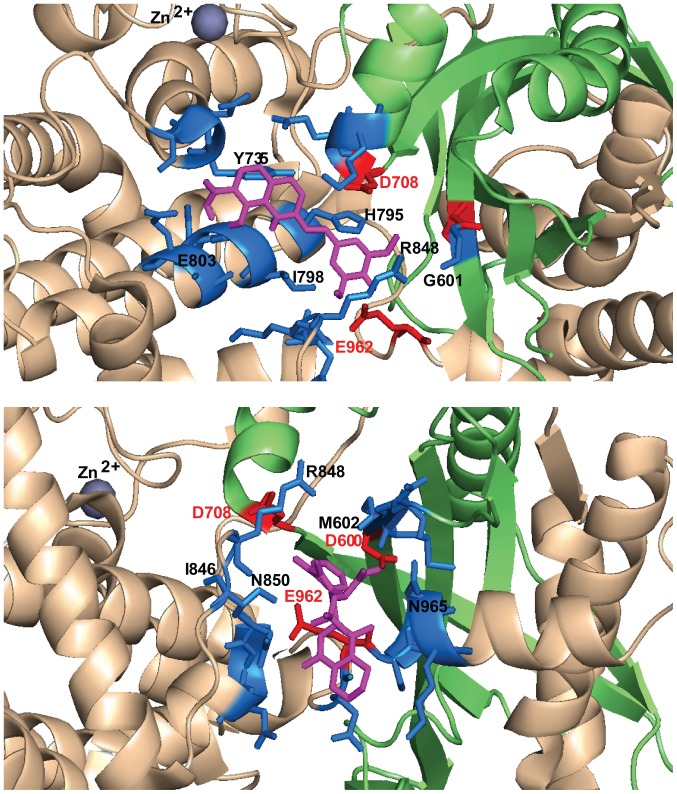
To exemplify this argument, we selected four diketo acid INSTIs (RAL, EVG, DTG and L-708,906), one styrylquinoline (FZ41) and three allosteric inhibitors (GSK1264, GS-A and BI224436) and performed docking simulations on RAG1 dimer (amino acids 391–1006) in apo form, from the previously reported crystal structure of RAG1 and 2 tetrameric complex at resolution 3.2 Å (PDB accession code 4WWX) (18). For each compound 5 rounds of simulation were performed, corresponding to 100 possible binding configurations. We focused on the configurations located in RNase H-like fold, in the vicinity of the DDE catalytic motif, within maximum 5 Å of any of the catalytic residues and the following analysis will refer exclusively to this binding pocket. Docking configurations within this pocket also displayed the highest calculated affinity corresponding to the lowest free energy (kcal/mol) of all 100 configurations, in the case of INSTIs and FZ41, but not in the case of ALLINIs. The pocket involves residues located on the first β-sheet of the RNase H fold, near D600 (between 599 and 604), on the last helix of the the RNase H fold, near D708 (708–711), on the helical region between RNase H domain and the last catalytic residue E962 (795–806, 848–852, 933–935) and on the loop around E962 (961–969) (Table I).
Table I.
Residues which define the binding pocket from docking simulations within RAG1 catalytic core domain.
| Compound | Catalytic core pocket residues within 5 Å of compound | Contacting residues (H-bonding, stacking, electrostatic or hydrophobic interactions) |
|---|---|---|
| Raltegravir | i) D600, G601, M602, G603, D604, K618, R621, E662, D708, K710, L711, L794, H795, I798, R848, M849, N858, S958, E959, E962, S963 | G601, K618, E662, D708, H795, I798, R848, S958, E962 |
| ii) D600, G601, E662, D708, E709, K710, H795, I798, G799, M800, A802, E803, R848, M849, T933, N934, Y935, E962 | G601, D708, H795, G799, M849, T933, E962 | |
| Elvitegravir | D708, E709, K710, L794, H795, L796, I798, G799, N800, A802, E803, Y805, K806, Q809, Q830, I846, M847, R848, M849, T933, N934, Y935 | D708, K710, H795, E803, Y805, K806, N809, Q830, M847, Y935 |
| Dolutegravir | C599, D600, G601, K618, E662, D708, E709, K710, L711, H795, I798, G799, A802, R848, M849, Y935, E962 | G601, E662, D708, E709, H795, R848, Y935 |
| L-708,906 | D708, E709, H795, I798, G799, N800, A802, E803, Y805, K806, I846, M847, R848, M849, T933, N934, Y935, E962 | D708, H795, Y805, M847, R848, Y935 |
| FZ41 | G601, D708, E709, K710, H795, I798, G799, N800, A802, E803, R848, M849, N850, T933, N934, Y935, E962 | G601, D708, H795, I798, G799, E803, R848, Y935, E962 |
| GSK1264 | D600, G601, M602, G603, D604, I846, R848, N850, G851, N852, N961, E962, S963, G964, N965, K966 | M602, D604, R848, N850, N861, E962, N965, K966 |
| GS-A | D600, G601, M602, G603, D604, N850, N852, N961, E962, S963, G964, N965, K966, R969 | N961, E962, S963, N965 |
| BI224436 | D600, G601, M602, G603, D604, I846, M847, R848, M849, N850, G851, N852, N961, E962, N965, R969 | D600, G601, M602, M847, R848, N850, E962, N965 |
Catalytic residues are depicted in bold font.
RAL emerged as the compound with the highest frequency of docking in the described area (83%), followed by DTG (57%), L-708,906 (43%), EVG and FZ41 (41%) (Table II). The highest calculated affinity belonged to docking configurations in this pocket for DTG and RAL (−8.2 kcal/mol and −8.1 kcal/mol, respectively), followed by FZ41 (−7.8 kcal/mol), EVG (−7.5 kcal/mol) and L-708,906 (−7.2 kcal/mol). ALLINIs had the lowest frequency of docking in the selected pocket, between 26 and 31% and with a corresponding low affinity, between −7.1 kcal/mol and −6.6 kcal/mol (Table II). This is not a surprising result taking into consideration that diketo acid derivatives and styrylquinolines were designed to bind in the vicinity of the DDE catalytic domain of IN, while ALLINIs have an affinity for the protein-protein interface. RAL docking configurations were categorized into two clusters (Tables I and II). Cluster no. 1 contains configurations which protrude more deeply in the space between the DDE residues (Fig. 5). Representative conformations for ALLINIs are oriented differently compared to diketo acids (Figs. 5 and 6) and FZ41, closer to the D600 and E962, particularly oriented towards the loop containing E962 and the helix that follows it (Fig. 7). Diketo acid derivatives and FZ41 all establish possible contacts with D708, while ALLINIs configurations allow contacts preferentially with E962 (Table I). In the selected docking conformations, D708 forms hydrogen bonds with RAL (3.2 Å), EVG (5 Å), DTG (4.3 Å), L-708,906 (3.1 Å), FZ41 (4.5 Å). E962 forms hydrogen bonds or electrostatic interactions with RAL (3.6 Å), FZ41 (3.9 Å), GSK1264 (3.3 Å), GS-A (2.7 Å) and BI224436 (4.1 Å). A possible hydrogen bond with D600 was observed only for BI224436 conformation (4.9 Å).
Table II.
Maximum apparent affinities and frequency of docking conformations within 5 Å of the RAG1 DDE motif.
Figure 5.
Selected docking conformations for raltegravir (magenta). The binding pocket is defined by all the residues within 5 Å (blue). The top image illustrates docking conformations corresponding to cluster number 1 (top image) and cluster number 2 (bottom image). DDE motif residues are depicted in red and the RNase H fold is depicted in green.
Figure 6.
Selected docking conformations for elvitegravir (magenta, upper image) and DTG (magenta, bottom image). The binding pocket is defined by all the residues within 5 Å (blue). DDE motif residues are depicted in red and the RNase H fold is depicted in green.
Figure 7.
Selected docking conformations for FZ41 (magenta, upper image) and GSK1264 (magenta, bottom image). The binding pocket is defined by all the residues within 5 Å (blue). DDE motif residues are depicted in red and the RNase H fold is depicted in green.
The compounds dock in an area critical for RSS binding. K608, H609, G610, S611, G851, N852, R855, L794, S963 and E959 are important residues for positioning the heptamer (21). N934 and T933 are part of the loop that binds the first 5 nucleotides of the coding DNA (21). Notably, R848 is a common contact for most of the compounds interacting through possible hydrogen bonds with RAL (4.3 Å), DTG (4.2 Å), FZ41 (3.4 Å), L-708,906 (4.6 Å), GSK1264 (4 Å), BI224436 (3.6 Å). R848 is a key residue on the 848–855 loop of Zn finger domain and it is important for heptamer CAC recognition. Upon hairpin formation, R848 modifies its position and helps in substrate orientation by forming π-cation interactions with the activated nucleotide and electrostatic interactions with the scissile phosphate (21). This residue is also involved in suppressing RAG-mediated transposition (27). H795 establishes possible hydrogen bonds or stacking contacts with RAL (3.6 Å), EVG (2.5 Å), DTG (3.9 Å), L-708,906 (3.1 Å) and FZ41 (3.6 Å), but not with ALLINIs. Mutations of this residue have been previously shown to alter RAG catalytic activity in both 3′nicking and hairpin formation steps, but not DNA binding (77).
8. HIV-1 IN inhibitors interfere with various polynucleotidyl transferases
During the development of IN inhibitors, transposases were considered as a surrogate model for HIV-1 based on the structural similarity of the catalytic RNase H-like fold domain. Among the first well characterized transposase-DNA complexes was Tn5 (78), which made it a suitable model enzyme for the identification of potential anti-HIV-1 IN candidates. This strategy was employed for the screening of 16,000 compounds and the subsequent identification of 20 Tn5 inhibitors, of which 6 also inhibited HIV-1 IN 3′P, namely coumarin dimers, cinnamoyl derivatives and a chlorinated bithionol sulfoxide, with IC50 values between 9 and 32 µM. Most compounds inhibited Tn5 more potently than HIV-1 IN (79). Furthermore, six diketo acid derivatives are able to interfere with paired complex formation and with both donor DNA cleavage and ST steps of the reaction. It has been suggested that the compounds bind at or near the active site, independently of Mg2+ in the case of three of them (80). Of note, L-708,906 does not interfere with Tn10 transposase activity, which indicates that despite of the similar RNaseH fold like catalytic core, not all transposases are susceptible to inhibition by diketo acid compounds (70).
RAL is able to inhibit other retroviral INs and transposases (81). It has been demonstrated that RAL is able to play a part in triggering and exacerbating autoimmune disease in mice, by interacting with endogenous retroelement INs, thus leading to accumulation of pre-integration cDNA (82). Certain autoimmune diseases, such as systemic lupus erythematosus have been associated with the accumulation of cDNA in the cytoplasm and the activation of the type I interferon response (83). The co-crystal structure of RAL in complex with mariner transposase Mos1 (PDB accession code 4MDB) revealed the compound's versatility in interacting with the catalytic core pocket of the apo enzyme, by adopting a very distinct, more compact conformation, than the extended one seen in the PFV intasome (PDB accession code 3OYA). Diffraction data have revealed how RAL's three coplanar oxygen atoms compete with the enzyme's DDD motif (D156, D249 and D284) for the binding of the two divalent metal cations. RAL also inhibited Mos1 enzymatic activity in vitro, with an estimated IC50 value between 60 and 70 µM on the first cleavage step and a significantly higher potency on ST (IC50 ~2 µM). This suggests that RAL binds more efficiently to the transpososome and can adopt yet a different and more efficient spatial configuration than in the absence of DNA. By contrast, EVG does not bind to Mos1 and it does not interfere with its activity (84).
Mos1 shares 48.4% sequence similarity to the SETMAR transposase catalytic domain, also a member of the Tc1/mariner family. SETMAR, also known as metnase, is another example of transposase domestication, a protein present only in anthropoid lineage, resulting from the fusion of a SET histone methylase domain and a Hsmar1 mobile element domain. Metnase performs a variety of functions linked to DNA repair mechanisms through the NHEJ complex and exogenous DNA integration (3). The SET domain methylates histones in the vicinity of DSBs, thus stabilizing components of NHEJ machinery, enhances DNA repair and suppresses chromosomal translocations. The transposase domain is able to bind TIRs, performs 5′nicking and has a modified DDN catalytic motif (D483, D575 and N610), which does not allow double-strand cleavage and mediates transposition with very low frequency. Metnase enhances topoisomerase IIα activity and promotes the restart of stalled replication forks. Metnase overexpression has been associated with an increase in HIV-1 cDNA integration [reviewed in (85)]. Due to its genomic stabilizing and repair properties, metnase overexpression is linked to resistance to chemotherapy in cancer cells (metnase mediates resistance to topoisomerase II inhibitors in breast cancer cells; metnase mediates chromosome decatenation in acute leukemia cells). Both RAL and EVG have been identified as active enzymatic inhibitors of metnase in vitro 5′ cleavage at 2 µM (86).
Human cytomegalovirus [(HMCV), belonging to the herpesvirus family] terminase is a 2-subunit protein with a N-terminal U56 ATPase domain and C-terminal UL89 nuclease domain and is responsible for cutting long genomic DNA head-to-tail concatemers into individual units of genomic DNA to be singly packed into viral capsids. The structure of the herpesvirus packaging terminase UL89 nuclease C-terminal subunit (UL89C) revealed a RNase H-like fold responsible for DNA cleavage, with a DED motif (D463, E534 and D651), which coordinates two metal divalent cations. In the presence of Mn2+ UL89C leads to full DNA degradation, while in the presence of Mg2+ the single strand nicking step is predominant. Of note, RAL inhibits UL89C double-strand cleavage activity in vitro more efficiently than nicking activity, beginning from 1 µM. By contrast, EVG does not show any interference with UL89C under similar conditions (16). HMCV infection can be life-threatening for immunocompromised patients and can cause serious birth defects. The versatility of INSTI metal ion chelation mechanism is a source of inspiration and opens the way for the design of novel DNA packaging inhibitors.
Human T-lymphotropic virus 1 (HTLV-1) belongs to the same Orthoretrovirinae subfamily as HIV-1. The HTLV-1 infection of CD4+ lymphocytes (to a lesser degree CD8+) is associated with clonal expansion and adult T-cell leukemia/lymphoma, a very aggressive and often treatment refractory form of cancer or tropical spastic paraparesis/HTLV-1-associated myelopathy (TSP/HAM). Both have a very poor disease prognosis and efficient targeted therapy still remains a challenge (87). Given the similarities between the two viruses, it was hypothesized that HIV-1 IN inhibitors can also function as HTLV-1 IN inhibitors. Indeed, representatives from both diketo acid INSTI and SQLs class have been demonstrated to interfere in vitro with ST reaction catalyzed by HTLV-1 IN. While L-731,988 and L-839,616 diketo acid derivatives maintain a similar potency as observed on HIV-1 IN (0.051 and 0.069 µM), four SQLs (KH161, KH211, FZ41, FZ149) displayed IC50s in micromolar range (minimum 4.9 µM and maximum 7.4 µM), almost three times less potent than on HIV-1 IN. The results were confirmed ex vivo only for L-731,988 by assessing the number of integration events which occur in infected cells in culture (88). RAL and the diketo acid derivative, MK-2048, were later investigated in ex vivo in cell-free and cell-to-cell HTLV-1 infectivity models. Under these conditions, both compounds significantly reduced virus transmission, with IC50 values of 35 and 1 nM, respectively (15). In 2013, a phase I clinical trial investigating the effects of RAL on HTLV-1 proviral load in patients with HTLV-1 TSP/HAM was initiated (ClinicalTrials.gov Identifier: NCT01867320). EVG is also active on a range of retroviral infections. EVG and L-870,810 potently inhibited murine leukemia virus and simian virus replication in cell culture with EC50 values in nanomolar range (89).
9. Conclusions
Metal-dependent catalytic amino acid triad assembled by a RNAse H-like fold is a highly efficient and versatile feature of polynucleotidyl transferases, relating enzymes with very different cellular functions, present in all organisms, from viruses to humans. On one hand, these structural and mechanistic similarities between retroviral INs, ancient transposons and RAG1 recombinase have revealed a complex process of domestication during species evolution. On the other hand, it has prompted more detailed investigations into compounds acting as inhibitors within the DDE(D) center.
Docking simulations have shown that IN inhibitors designed to interfere with HIV-1 IN DDE motif also display affinity towards the RAG1 catalytic pocket. The selected docking conformations revealed interactions with key residues, such as D708, E962, R848 and H795 which can explain compound interference with RAG-mediated DNA cleavage demonstrated by previously reported in vitro and ex vivo experiments (14,70) However, given that these simulations were performed on RAG1 dimer complex without DNA, it is possible that the identified binding pockets for the compounds are not accessible upon synaptic complex formation. This argument is supported by the fact that L-708,906 inhibits RAG cleavage more potently when added to the protein before substrate (70). As indicated herein, the styrylquinoline derivative FZ41 displays a similar affinity to diketo acid INSTIs, and tBQA allosteric inhibitors have a significantly lower affinity towards the catalytic center.
Diketo acid INSTIs are able to bind and inhibit the activity of various polynucleotide transferases, despite the different architecture of their enzyme-DNA complex. While their potency and selectivity for certain enzymes differ, it is suggested that they all interact within the common RNase H-like fold domain, harboring the catalytic DDE(D) motif. IN inhibitors interaction with domesticated transposases, such as RAG1 and metnase prompts researchers to be watchful on possible immunological adverse events in patients, but also opens possibilities for the identification of novel adjuvant therapies for treatment refractory cancers. Moreover, the inhibition of related retroviral INs, such as HLTV-1 and other nucleases depending on a RNase H-like fold, such as HMCV terminase, can expand INSTIs disease spectrum and offer lead compounds for the development of more efficient novel targeted drugs.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
MGM performed the literature research, participated in writing the manuscript and performed the computational analysis. MS assisted in the computational analysis and critical review of the literature research. GMN and DM were involved in the design of the review, participated in the writing of the manuscript and the critical selection of the literature. DAS and AT were involved in the design of the review. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that they have no competing interests.
References
- 1.Pace JK, II, Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jangam D, Feschotte C, Betrán E. Transposable Element Domestication As an Adaptation to Evolutionary Conflicts. Trends Genet. 2017;33:817–831. doi: 10.1016/j.tig.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci USA. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hickman AB, Dyda F. DNA Transposition at Work. Chem Rev. 2016;116:12758–12784. doi: 10.1021/acs.chemrev.6b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCLINTOCK B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B. Breaking and joining single-stranded DNA: The HUH endonuclease superfamily. Nat Rev Microbiol. 2013;11:525–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 8.Yuan YW, Wessler SR. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci USA. 2011;108:7884–7889. doi: 10.1073/pnas.1104208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacroix C, Giovannini D, Combe A, Bargieri DY, Späth S, Panchal D, Tawk L, Thiberge S, Carvalho TG, Barale JC, et al. FLP/FRT-mediated conditional mutagenesis in pre-erythrocytic stages of Plasmodium berghei. Nat Protoc. 2011;6:1412–1428. doi: 10.1038/nprot.2011.363. [DOI] [PubMed] [Google Scholar]
- 10.Ye J, Hong J, Ye F. Reprogramming rat embryonic fibroblasts into induced pluripotent stem cells using transposon vectors and their chondrogenic differentiation in vitro. Mol Med Rep. 2015;11:989–994. doi: 10.3892/mmr.2014.2793. [DOI] [PubMed] [Google Scholar]
- 11.Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 12.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 13.Dai Y, Wong B, Yen Y-M, Oettinger MA, Kwon J, Johnson RC. Determinants of HMGB proteins required to promote RAG1/2-recombination signal sequence complex assembly and catalysis during V(D)J recombination. Mol Cell Biol. 2005;25:4413–4425. doi: 10.1128/MCB.25.11.4413-4425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishana M, Nilavar NM, Kumari R, Pandey M, Raghavan SC. HIV integrase inhibitor, Elvitegravir, impairs RAG functions and inhibits V(D)J recombination. Cell Death Dis. 2017;8:e2852. doi: 10.1038/cddis.2017.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seegulam ME, Ratner L. Integrase inhibitors effective against human T-cell leukemia virus type 1. Antimicrob Agents Chemother. 2011;55:2011–2017. doi: 10.1128/AAC.01413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadal M, Mas PJ, Blanco AG, Arnan C, Solà M, Hart DJ, Coll M. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc Natl Acad Sci USA. 2010;107:16078–16083. doi: 10.1073/pnas.1007144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avogadro, corp-author. Avogadro: an open-source molecular builder and visualization tool. Version 1.0.3. http://AvogadroOpenmoleculesNet/ 2012
- 20.DeLano WL. Schrödinger LLC; New York, NY: 2002. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- 21.Kim MS, Chuenchor W, Chen X, Cui Y, Zhang X, Zhou ZH, Gellert M, Yang W. Cracking the DNA Code for V(D)J Recombination. Mol Cell. 2018;70:358–370.e4. doi: 10.1016/j.molcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 23.Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grazini U, Zanardi F, Citterio E, Casola S, Goding CR, McBlane F. The RING domain of RAG1 ubiquitylates histone H3: A novel activity in chromatin-mediated regulation of V(D)J joining. Mol Cell. 2010;37:282–293. doi: 10.1016/j.molcel.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Matthews AGW, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, et al. Transposon molecular domestication and the evolution of the RAG recombinase. Nature. 2019;569:79–84. doi: 10.1038/s41586-019-1093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang YH, Son CY, Lee CH, Ryu CJ. Aberrant V(D)J cleavages in T cell receptor β enhancer- and p53-deficient lymphoma cells. Oncol Rep. 2010;23:1463–1468. doi: 10.3892/or_00000785. [DOI] [PubMed] [Google Scholar]
- 29.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997;17:3125–3136. doi: 10.1128/MCB.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 2003;22:1381–1388. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy YVR, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Wu S, Wang J, Li W, Lin Y, Ji C, Xue J, Chen J. Evaluation of the interactions of HIV-1 integrase with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. Int J Mol Med. 2012;30:1053–1060. doi: 10.3892/ijmm.2012.1088. [DOI] [PubMed] [Google Scholar]
- 34.Gupta K, Turkki V, Sherrill-Mix S, Hwang Y, Eilers G, Taylor L, McDanal C, Wang P, Temelkoff D, Nolte RT, et al. Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase. PLoS Biol. 2016;14:e1002584. doi: 10.1371/journal.pbio.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lusic M, Siliciano RF. Nuclear landscape of HIV-1 infection and integration. Nat Rev Microbiol. 2017;15:69–82. doi: 10.1038/nrmicro.2017.22. [DOI] [PubMed] [Google Scholar]
- 36.Chen JC-H, Krucinski J, Miercke LJW, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: A model for viral DNA binding. Proc Natl Acad Sci USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 38.Venanzi Rullo E, Ceccarelli M, Condorelli F, Facciolà A, Visalli G, D'Aleo F, Paolucci I, Cacopardo B, Pinzone MR, Di Rosa M, et al. Investigational drugs in HIV: Pros and cons of entry and fusion inhibitors (Review) Mol Med Rep. 2019;19:1987–1995. doi: 10.3892/mmr.2019.9840. [DOI] [PubMed] [Google Scholar]
- 39.Wai JS, Egbertson MS, Payne LS, Fisher TE, Embrey MW, Tran LO, Melamed JY, Langford HM, Guare JP, Jr, Zhuang L, et al. 4-Aryl-2,4-dioxobutanoic acid inhibitors of HIV-1 integrase and viral replication in cells. J Med Chem. 2000;43:4923–4926. doi: 10.1021/jm000176b. [DOI] [PubMed] [Google Scholar]
- 40.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 41.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 42.U.S. Food & Drug Administration, corp-author. HIV Timeline and History of Approvals. https://www.fda.gov/patients/hivaids/hiv-timeline-and-history-approvals. [Aug 1;2018 ];
- 43.Di Santo R. Inhibiting the HIV integration process: Past, present, and the future. J Med Chem. 2014;57:539–566. doi: 10.1021/jm400674a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, et al. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem. 2006;49:1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 45.Lee JSF, Calmy A, Andrieux-Meyer I, Ford N. Review of the safety, efficacy, and pharmacokinetics of elvitegravir with an emphasis on resource-limited settings. HIV AIDS (Auckl) 2012;4:5–15. doi: 10.2147/HIV.S20993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnhart M, Shelton JD. ARVs: The next generation. Going boldly together to new frontiers of HIV treatment. Glob Health Sci Pract. 2015;3:1–11. doi: 10.9745/GHSP-D-14-00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johns BA, Kawasuji T, Weatherhead JG, Taishi T, Temelkoff DP, Yoshida H, Akiyama T, Taoda Y, Murai H, Kiyama R, et al. Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744) J Med Chem. 2013;56:5901–5916. doi: 10.1021/jm400645w. [DOI] [PubMed] [Google Scholar]
- 48.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hare S, Smith SJ, Métifiot M, Jaxa-Chamiec A, Pommier Y, Hughes SH, Cherepanov P. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572) Mol Pharmacol. 2011;80:565–572. doi: 10.1124/mol.111.073189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hightower KE, Wang R, Deanda F, Johns BA, Weaver K, Shen Y, Tomberlin GH, Carter HL, III, Broderick T, Sigethy S, et al. Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes. Antimicrob Agents Chemother. 2011;55:4552–4559. doi: 10.1128/AAC.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshinaga T, Kobayashi M, Seki T, Miki S, Wakasa-Morimoto C, Suyama-Kagitani A, Kawauchi-Miki S, Taishi T, Kawasuji T, Johns BA, et al. Antiviral characteristics of GSK1265744, an HIV integrase inhibitor dosed orally or by long-acting injection. Antimicrob Agents Chemother. 2015;59:397–406. doi: 10.1128/AAC.03909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsiang M, Jones GS, Goldsmith J, Mulato A, Hansen D, Kan E, Tsai L, Bam RA, Stepan G, Stray KM, et al. Antiviral activity of bictegravir (GS-9883), a novel potent HIV-1 integrase strand transfer inhibitor with an improved resistance profile. Antimicrob Agents Chemother. 2016;60:7086–7097. doi: 10.1128/AAC.01474-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mekouar K, Mouscadet JF, Desmaële D, Subra F, Leh H, Savouré D, Auclair C, d'Angelo J. Styrylquinoline derivatives: A new class of potent HIV-1 integrase inhibitors that block HIV-1 replication in CEM cells. J Med Chem. 1998;41:2846–2857. doi: 10.1021/jm980043e. [DOI] [PubMed] [Google Scholar]
- 54.Deprez E, Barbe S, Kolaski M, Leh H, Zouhiri F, Auclair C, Brochon JC, Le Bret M, Mouscadet JF. Mechanism of HIV-1 integrase inhibition by styrylquinoline derivatives in vitro. Mol Pharmacol. 2004;65:85–98. doi: 10.1124/mol.65.1.85. [DOI] [PubMed] [Google Scholar]
- 55.Han Y-S, Xiao W-L, Quashie PK, Mesplède T, Xu H, Deprez E, Delelis O, Pu JX, Sun HD, Wainberg MA. Development of a fluorescence-based HIV-1 integrase DNA binding assay for identification of novel HIV-1 integrase inhibitors. Antiviral Res. 2013;98:441–448. doi: 10.1016/j.antiviral.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Bonnenfant S, Thomas CM, Vita C, Subra F, Deprez E, Zouhiri F, Desmaële D, D'Angelo J, Mouscadet JF, Leh H. Styrylquinolines, integrase inhibitors acting prior to integration: A new mechanism of action for anti-integrase agents. J Virol. 2004;78:5728–5736. doi: 10.1128/JVI.78.11.5728-5736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Passos DO, Li M, Yang R, Rebensburg SV, Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R, Lyumkis D. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science. 2017;355:89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quashie PK, Han YS, Hassounah S, Mesplède T, Wainberg MA. Structural studies of the HIV-1 integrase protein: Compound screening and characterization of a DNA-binding inhibitor. PLoS One. 2015;10:e0128310. doi: 10.1371/journal.pone.0128310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shkriabai N, Patil SS, Hess S, Budihas SR, Craigie R, Burke TR, Jr, Le Grice SF, Kvaratskhelia M. Identification of an inhibitor-binding site to HIV-1 integrase with affinity acetylation and mass spectrometry. Proc Natl Acad Sci USA. 2004;101:6894–6899. doi: 10.1073/pnas.0400873101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du L, Zhao YX, Yang LM, Zheng YT, Tang Y, Shen X, Jiang HL. Symmetrical 1-pyrrolidineacetamide showing anti-HIV activity through a new binding site on HIV-1 integrase. Acta Pharmacol Sin. 2008;29:1261–1267. doi: 10.1111/j.1745-7254.2008.00863.x. [DOI] [PubMed] [Google Scholar]
- 61.Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 63.Llano M, Delgado S, Vanegas M, Poeschla EM. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J Biol Chem. 2004;279:55570–55577. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- 64.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J Virol. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cherepanov P, Ambrosio ALB, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du L, Zhao Y, Chen J, Yang L, Zheng Y, Tang Y, Shen X, Jiang H. D77, one benzoic acid derivative, functions as a novel anti-HIV-1 inhibitor targeting the interaction between integrase and cellular LEDGF/p75. Biochem Biophys Res Commun. 2008;375:139–144. doi: 10.1016/j.bbrc.2008.07.139. [DOI] [PubMed] [Google Scholar]
- 67.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 68.Fader LD, Malenfant E, Parisien M, Carson R, Bilodeau F, Landry S, Pesant M, Brochu C, Morin S, Chabot C, et al. Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1. ACS Med Chem Lett. 2014;5:422–427. doi: 10.1021/ml500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, et al. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J Biol Chem. 2012;287:21189–21203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melek M, Jones JM, O'Dea MH, Pais G, Burke TR, Jr, Pommier Y, Neamati N, Gellert M. Effect of HIV integrase inhibitors on the RAG1/2 recombinase. Proc Natl Acad Sci USA. 2002;99:134–137. doi: 10.1073/pnas.012610699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H, Davies DR. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: A platform for antiviral drug design. Proc Natl Acad Sci USA. 1999;96:13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Miguel R, Montejano R, Stella-Ascariz N, Arribas JR. A safety evaluation of raltegravir for the treatment of HIV. Expert Opin Drug Saf. 2018;17:217–223. doi: 10.1080/14740338.2018.1411903. [DOI] [PubMed] [Google Scholar]
- 73.Huhn GD, Badri S, Vibhakar S, Tverdek F, Crank C, Lubelchek R, Max B, Simon D, Sha B, Adeyemi O, et al. Early development of non-hodgkin lymphoma following initiation of newer class antiretroviral therapy among HIV-infected patients - implications for immune reconstitution. AIDS Res Ther. 2010;7:44. doi: 10.1186/1742-6405-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steigbigel RT, Cooper DA, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Gatell JM, Rockstroh JK, et al. BENCHMRK Study Teams Raltegravir with optimized background therapy for resistant HIV-1 infection. N Engl J Med. 2008;359:339–354. doi: 10.1056/NEJMoa0708975. [DOI] [PubMed] [Google Scholar]
- 75.Barbaro G, Barbarini G. HIV infection and cancer in the era of highly active antiretroviral therapy (Review) Oncol Rep. 2007;17:1121–1126. [PubMed] [Google Scholar]
- 76.Gillead Sciences. sNDA 207561/S-014. Genvoya (elvitegravir/cobicistat/emtricitabine/tenofovir alafenamide) Clinical and Cross-Discipline Team Leader Review [Google Scholar]
- 77.Huye LE, Purugganan MM, Jiang M-M, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol Cell Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 79.Ason B, Knauss DJ, Balke AM, Merkel G, Skalka AM, Reznikoff WS. Targeting Tn5 transposase identifies human immunodeficiency virus type 1 inhibitors. Antimicrob Agents Chemother. 2005;49:2035–2043. doi: 10.1128/AAC.49.5.2035-2043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Czyz A, Stillmock KA, Hazuda DJ, Reznikoff WS. Dissecting Tn5 transposition using HIV-1 integrase diketoacid inhibitors. Biochemistry. 2007;46:10776–10789. doi: 10.1021/bi7006542. [DOI] [PubMed] [Google Scholar]
- 81.Koh Y, Matreyek KA, Engelman A. Differential sensitivities of retroviruses to integrase strand transfer inhibitors. J Virol. 2011;85:3677–3682. doi: 10.1128/JVI.02541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beck-Engeser GB, Eilat D, Harrer T, Jäck HM, Wabl M. Early onset of autoimmune disease by the retroviral integrase inhibitor raltegravir. Proc Natl Acad Sci USA. 2009;106:20865–20870. doi: 10.1073/pnas.0908074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wolkowicz UM, Morris ER, Robson M, Trubitsyna M, Richardson JM. Structural basis of Mos1 transposase inhibition by the anti-retroviral drug Raltegravir. ACS Chem Biol. 2014;9:743–751. doi: 10.1021/cb400791u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shaheen M, Williamson E, Nickoloff J, Lee SH, Hromas R. Metnase/SETMAR: A domesticated primate transposase that enhances DNA repair, replication, and decatenation. Genetica. 2010;138:559–566. doi: 10.1007/s10709-010-9452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williamson EA, Damiani L, Leitao A, Hu C, Hathaway H, Oprea T, Sklar L, Shaheen M, Bauman J, Wang W, et al. Targeting the transposase domain of the DNA repair component Metnase to enhance chemotherapy. Cancer Res. 2012;72:6200–6208. doi: 10.1158/0008-5472.CAN-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marino-Merlo F, Mastino A, Grelli S, Hermine O, Bazarbachi A, Macchi B. Future Perspectives on Drug Targeting in Adult T Cell Leukemia-Lymphoma. Front Microbiol. 2018;9:925. doi: 10.3389/fmicb.2018.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rabaaoui S, Zouhiri F, Lançon A, Leh H, d'Angelo J, Wattel E. Inhibitors of strand transfer that prevent integration and inhibit human T-cell leukemia virus type 1 early replication. Antimicrob Agents Chemother. 2008;52:3532–3541. doi: 10.1128/AAC.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, et al. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137) J Virol. 2008;82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Passos DO, Li M, Yang R, Rebensburg SV, Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R, Lyumkis D. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science. 2017;355:89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.