Abstract
Abstract
Metaproteomics allows to decipher the structure and functionality of microbial communities. Despite its rapid development, crucial steps such as the creation of standardized protein search databases and reliable protein annotation remain challenging. To overcome those critical steps, we developed a new program named mPies (metaProteomics in environmental sciences). mPies allows the creation of protein databases derived from assembled or unassembled metagenomes, and/or public repositories based on taxon IDs, gene or protein names. For the first time, mPies facilitates the automatization of reliable taxonomic and functional consensus annotations at the protein group level, minimizing the well-known protein inference issue, which is commonly encountered in metaproteomics. mPies’ workflow is highly customizable with regards to input data, workflow steps, and parameter adjustment. mPies is implemented in Python 3/Snakemake and freely available on GitHub: https://github.com/johanneswerner/mPies/.
Reviewer
This article was reviewed by Dr. Wilson Wen Bin Goh.
Keywords: Bioinformatics, Metaproteomics, Microbial ecology, Protein annotation, Protein search database
Implementation
Background
Metaproteomics is a valuable method to link the taxonomic diversity and functions of microbial communities [1]. However, the use of metaproteomics still faces methodological challenges and lacks of standardisation [2]. The creation of relevant protein search databases and protein annotation remain hampered by the inherent complexity of microbial communities [3].
Protein search databases can be created based on reads or contigs derived from metagenomic and/or metatranscriptomic data [4, 5]. Public repositories such as Ensembl [6], NCBI [7] or UniProtKB [8] can also be used as search databases but it is necessary to apply relevant filters (e.g. based on the habitat or the taxonomic composition) in order to decrease computation time and false discovery rate [4]. Until now, no tool exists that either creates taxonomic or functional subsets of public repositories or combines different protein databases in order to optimize the total number of identified proteins.
The so-called protein inference issue occurs when the same peptide sequence is found in multiple proteins, thus leading to inaccurate taxonomic and functional interpretation [9]. To address this issue, protein identification software tools such as ProteinPilot (Pro Group algorithm) [10], Prophane [11] or MetaProteomeAnalyzer [12] perform automatic grouping of homologous protein sequences. Interpreting protein groups can be challenging especially in complex microbial community where redundant proteins can be found in a broad taxonomic range. A well-known strategy to deal with homologous protein sequences is to calculate the lowest common ancestor (LCA). For instance, MEGAN performs taxonomic binning by assigning sequences on the nodes of the NCBI taxonomy and calculates the LCA on the best alignment hit [13]. However, another crucial challenge related to protein annotation still remains: protein sequences annotation often relies on alignment programs automatically retrieving the first hit only [14]. The reliability of this approach is hampered by the existence of taxonomic and functional discrepancies among the top alignment results with very low e-values [5]. Here, we present mPies, a new highly customizable program that allows the creation of protein search databases and performs post-search protein consensus annotation, thus facilitating biological interpretation.
Workflow design
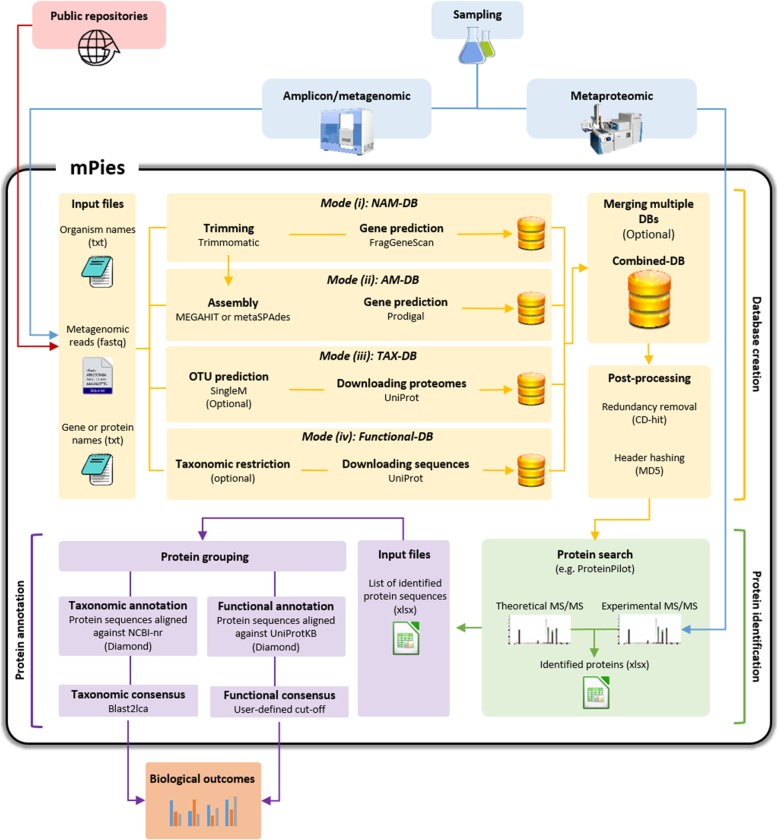
mPies provides multiple options for optimizing metaproteomic analysis within a standardized and automatized workflow (Fig. 1). mPies is written in Python 3.6, uses the workflow management system Snakemake [15] and relies on Bioconda [16] to ensure reproducibility. mPies can run in up to four different modes to create databases (DBs) for protein search using amplicon/metagenomic and/or public repositories data: (i) non-assembled metagenome-derived DB, (ii) assembled metagenome-derived DB, (iii) taxonomy-derived DB, and (iv) functional-derived DB. After protein identification, mPies can automatically compute sequence alignment-based consensus annotation at protein group level. By taking into account multiple alignment hits for reliable taxonomic and functional inference, mPies limits the protein inference issue and allows more relevant biological interpretation of metaproteomes from diverse environments.
Fig. 1.
Workflow of mPies
Mode (i): Non-assembled metagenome-derived DB
In mode (i), mPies trims metagenomic raw reads (fastq files) with Trimmomatic [17], and predicts partial genes with FragGeneScan [18] which are built into the protein DB.
Mode (ii): Assembled metagenome-derived DB
In mode (ii), trimmed metagenomic reads are assembled either with MEGAHIT [19] or metaSPAdes [20]. The genes are subsequently called with Prodigal [21]. The utilization of Snakemake allows easy adjustment of the assembly and gene calling parameters.
Mode (iii): Taxonomy-derived DB
In mode (iii), mPies extracts the taxonomic information derived from the metagenomic raw data and downloads the corresponding proteomes from UniProt. To do so, mPies uses SingleM [22] to predict OTUs from the metagenomic reads. Subsequently, a non-redundant list of taxon IDs corresponding to the taxonomic diversity of the observed habitat is generated. Finally, mPies retrieves all available proteomes for each taxon ID from UniProt. It is noteworthy that the taxonomy-derived DB can be generated from 16S amplicon data or a user-defined list.
Mode (iv): Functional-derived DB
Mode (iv) is a variation of mode (iii) which allows to create DBs that target specific functional processes (e.g. carbon fixation or sulphur cycle) instead of downloading entire proteomes for taxonomic ranks. For that purpose, mPies requires a list of gene or protein names as input and downloads all the corresponding protein sequences from UniProt. Taxonomic restriction can be defined (e.g. Proteobacteria-related sequences only) for highly specific DB creation.
Post-processing
If more than one mode was selected for protein DB generation, all proteins are merged into one combined protein search DB. Duplicated protein sequences (default: sequence similarity 100%) are removed with CD-HIT [23]. All protein headers are hashed (default: MD5) to obtain uniform headers and to reduce the file size for the final protein search database in order to keep the memory requirements of downstream analysis low.
Protein annotation
mPies facilitates taxonomic and functional consensus annotation at protein level. After protein identification, each protein is aligned with Diamond [24] against NCBI-nr [7] for the taxonomic annotation. For the functional prediction, proteins are aligned against UniProt (Swiss-Prot or TrEMBL) [8] and COG [25]. The alignment hits (default: retained aligned sequences = 20, bitscore ≥80) are automatically retrieved for consensus taxonomic and functional annotation, for which the detailed strategies are provided below.
The taxonomic consensus annotation uses the alignment hits against NCBI-nr and applies the LCA algorithm to retrieve a taxonomic annotation for each protein group (protein grouping comprises the assignment of multiple peptides to the same protein and is facilitated by proteomics software) as described by Huson et al. [13]. For the functional consensus, the alignment hits against UniProt and/or COG are used to extract the most frequent functional annotation per protein group within their systematic recommended names. This is the first time that a metaproteomics tool includes this critical step, as previously only the first alignment hit was kept. In order to ensure the most accurate annotation, a minimum of 20 best alignment hits should be kept for consensus annotation. Nevertheless, this parameter is customizable and and this number could be modified.
Conclusions
The field of metaproteomics has rapidly expanded in recent years and has led to valuable insights in the understanding of microbial community structure and functioning. In order to cope with metaproteomic limitations, new tools development and workflow standardization are of urgent needs. With regards to the diversity of the technical approaches found in the literature which are responsible for methodological inconsistencies and interpretation biases across metaproteomic studies, we developed the open-source program mPies. It proposes a standardized and reproducible workflow that allows customized protein search DB creation and reliable taxonomic and functional protein annotations. mPies facilitates biological interpretation of metaproteomics data and allows unravelling microbial community complexity.
Reviewer’s comments
Wilson Wen Bin Goh PhD, School of Biological Sciences, Nanyang Technological University
Reviewer summary
Metaproteomics is a growing area. Although its sister discipline, metagenomics is relatively more mature, metaproteomics is expected to be harder due to the indirect means of assaying peptide information based on the MS. There is a lack of tools for performing metaproteomics analysis. And so, I think the author’s pipelines adds a useful resource. The manuscript is well-written, and to the point, I have no points to add regarding grammar and spell proofing.
Authors response: We thank Dr. Wilson Wen Bin Goh for his overall very positive review.
Reviewer recommendations to authors
The manuscript runs a bit on the short. While I appreciate the conciseness, I think to get more people interested, inclusion of a case study on application, or possible generic user-routes to get people jumping in and tinkering would be great. I particularly like the idea of integrating functional consensus information automatically with a protein group. I think this helps to establish the coherence of a protein group. For example, in the case of OpenMS, some examples of workflows https://www.openms.de/workflows/, help readers understand the usefulness of the pipelines, and how to integrate it with their needs. As Biology Direct is not a bioinformatics journal per se, this addition would help the readership.
Authors response: We would like to thank the Reviewer for this comment. We agree with the Reviewer’s suggestion and improved the visualization of the overall metaproteomics worfkow using mPies from data generation to biological interpretation (Fig. 1). We also provided copy-paste usage examples, with test-data, on the GitHub repository to get people started quickly, thus maximizing the use of mPies by the widest community.
Minor issues
Looking at the protein annotation figure, is the max of 20 a fixed number? Can this be changed? As for most frequent protein name, is it based on SwissProt ID or the gene symbol?
Authors response: The value for maximum target sequences is adaptable, as are most parameters in the Snakemake workflow. Based on our experience on several (not-yet-published) in-house datasets, 20 is significantly more robust than lower values (tested: 10, 20, 50, 100); higher values do not capture significantly more functions. Depending on the studied environment and available reference data, a higher value for consensus annotations might be useful, although we recommend to never use a value lower than 20 to limit the influence of outliers and false positives.
The most frequent protein name is not a gene ID but the “recommended” UniProt protein name, which we use for consensus calculation.
We adapted the respective sentences in the revised manuscript.
Availability and requirements
Project name: mPies
Project homepage: https://github.com/johanneswerner/mPies/
Operating system: Linux
Programming language: Python 3.6
Other requirements: Snakemake, bioconda
License: GNU GPL v3.0
Any restrictions to use by non-academics: none.
Acknowledgements
The authors acknowledge the use of de.NBI cloud and the support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen and the Federal Ministry of Education and Research (BMBF) through grant no 031 A535A. JW and JK want to personally acknowledge Manuel Prinz and Katrin Leinweber (Technische Informationsbibliothek, TIB.eu) for code review, critical thoughts, and software publication advice.
Authors’ contributions
JW, SMS, and AG designed mPies. JW developed and implemented mPies, JK contributed valuable discussions with regards to the software design of mPies and performed code review. JW, AG and SMS wrote substantial parts of the manuscript, all authors performed proofreading and approved the final version of the manuscript.
Funding
This work was supported by the Royal Society funded Research Grant [RG160594]. The publication of this article was funded by the Open Access Fund of the Leibniz Association.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Johannes Werner, Email: johannes.werner@io-warnemuende.de.
Augustin Géron, Email: a.f.geron@stir.ac.uk.
Jules Kerssemakers, Email: j.kerssemakers@dkfz-heidelberg.de.
Sabine Matallana-Surget, Email: sabine.matallanasurget@stir.ac.uk.
References
- 1.Wilmes P, Bond PL. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ Microbiol. 2004;6(9):911–920. doi: 10.1111/j.1462-2920.2004.00687.x. [DOI] [PubMed] [Google Scholar]
- 2.Matallana-Surget S, Jagtap PD, Griffin TJ, Beraud M, Wattiez R. Comparative Metaproteomics to study environmental changes. In: Nagarajan M, editor. Metagenomics – perspectives, methods, and applications. 2018. pp. 327–363. [Google Scholar]
- 3.Heyer R, Schallert K, Zoun R, Becher B, Saake G, Beendorf D. Challenges and perspectives of metaproteomic data analysis. J Biotechnol. 2017;261:24–36. doi: 10.1016/j.jbiotec.2017.06.1201. [DOI] [PubMed] [Google Scholar]
- 4.Tanca A, Palomba A, Fraumene C, Pagnozzi D, Manghina V, Deligios M, et al. The impact of sequence database choice on metaproteomic results in gut microbiota studies. Microbiome. 2016;4:51. doi: 10.1186/s40168-016-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timmins-Schiffman E, May DH, Mikan M, Riffle M, Frazar C, Harvey HR, et al. Critical decisions in metaproteomics: achieving high confidence protein annotations in a sea of unknowns. ISME J. 2017;11(2):309–314. doi: 10.1038/ismej.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zerbino DR, Achuthan P, Akanni W, Ridwan Amode M, Berrell D, Bhqi J, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resource Coordinators NCBI. Database resources of the National Center for biotechnology information. Nucleic Acids Res. 2018;46(Database issue):D8–13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The UniProt Consortium UniProt: the universal protein knowledge base. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst F-A, Lünsmann V, Kjeldal H, Jehmlich N, Tholey A, von Bergen M, et al. Enhancing metaproteomics—the value of models and defined environmental microbial systems. Proteomics. 2016;16(5):783–798. doi: 10.1002/pmic.201500305. [DOI] [PubMed] [Google Scholar]
- 10.AbSciex. Understanding the Pro Group™ Algorithm. https://sciex.com/Documents/manuals/proteinPilot-ProGroup-Algorithm.pdf, Accessed on 2019-06-12.
- 11.Schneider T, Schmid E, de Castro JV, Jr, Cardinale M, Eberl L, Grube M, et al. Structure and function of the symbiosis partners of the lung lichen (Lobaria pulmonaria L. Hoffm.) analyzed by metaproteomics. Proteomics. 2011;11:2752–2756. doi: 10.1002/pmic.201000679. [DOI] [PubMed] [Google Scholar]
- 12.Muth T, Behne A, Heyer R, Kohrs F, Benndorf D, Hoffmann M, et al. The MetaProteomeAnalyzer: a powerful open-source software suite for metaproteomics data analysis and interpretation. J Proteome Res. 2015;14(3):1557–1565. doi: 10.1021/pr501246w. [DOI] [PubMed] [Google Scholar]
- 13.Huson DH, Beier S, Flade I, Gorska A, El-Hadidi M, Mitra S, et al. Megan community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. 2016;12(6):e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pible O, Armengaud J. Improving the quality of genome, protein sequence, and taxonomy databases: a prerequisite for microbiome meta-omics 2.0. Proteomics. 2015;15:3418–3423. doi: 10.1002/pmic.201500104. [DOI] [PubMed] [Google Scholar]
- 15.Köster J, Rahmann S. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics. 2012;28(19):2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 16.Grüning B, Dale R, Sjödin A, Chapman BA, Rowe J, Tomkins-Tinch CH, et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods. 2018;15(7):475–476. doi: 10.1038/s41592-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumine sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38(20):e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D, Liu CM, Luo R, Sadakane K, Lam TW. Megahit: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics. 2015;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 20.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. Metaspades: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyatt D, Chen KL, Locascio PF, Lanl ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodcroft B. Singlem. 2018. [Google Scholar]
- 23.Fu L, Niu B, Zhu Z, Wu S, Li W. Cd-hit: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using diamond. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 25.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the cog database. Nucleic Acids Res. 2015;43(Database issue):D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.