Abstract
Background
The extent to which the composition and diversity of the oral microbiome varies with age is not clearly understood.
Methods
The 16S rRNA gene of subgingival plaque in 1219 women, aged 53–81 years, was sequenced and its taxonomy annotated against the Human Oral Microbiome Database (v.14.5). Composition of the subgingival microbiome was described in terms of centered log(2)-ratio (CLR) transformed OTU values, relative abundance, and prevalence. Correlations between microbiota abundance and age were evelauted using Pearson Product Moment correlations. P-values were corrected for multiple testing using the Bonferroni method.
Results
Of the 267 species identified overall, Veillonella dispar was the most abundant bacteria when described by CLR OTU (mean 8.3) or relative abundance (mean 8.9%); whereas Streptococcus oralis, Veillonella dispar and Veillonella parvula were most prevalent (100%, all) when described as being present at any amount. Linear correlations between age and several CLR OTUs (Pearson r = − 0.18 to 0.18), of which 82 (31%) achieved statistical significance (P < 0.05). The correlations lost significance following Bonferroni correction. Twelve species that differed across age groups (each corrected P < 0.05); 5 (42%) were higher in women ages 50–59 compared to ≥70 (corrected P < 0.05), and 7 (48%) were higher in women 70 years and older.
Conclusions
We identified associations between several bacterial species and age across the age range of postmenopausal women studied. Understanding the functions of these bacteria could identify intervention targets to enhance oral health in later life.
Keywords: Aging, Women, Oral Microbiome, Epidemiology
Background
The availability of high throughput metagenomics sequencing technology has allowed for deeper understanding of complex microbiota ecologies and their aggregate functional capacities within a defined microbiome [1, 2]. Marked differences in composition and function of microbiomes have been shown between various body sites among individuals [3, 4]. It has become increasingly clear that the microbiota and microbiome are correlated with both health and disease states in humans [5], and that the aging process could be an important determinant of these relationships [6, 7]. Aging is a complex, multifactorial process characterized by progressively lower resilience to stress, increased homeostatic imbalance, and greater susceptibility to pathologic insult and disease onset [8]. Changes in microbiome diversity and function have been observed with increasing age [9]. Alterations in the host environment that occur with physiologic aging processes could enable untoward shifts in relative abundance of commensal and pathogenic bacteria, and enhanced expression of pathogen genomes which, in turn, could heighten disease susceptibility. In support of this hypothesis are studies demonstrating links between human microbiomes and several diseases of aging including obesity, diabetes, heart disease, and certain cancers [5, 7, 10].
The oral microbiota comprise one of the most complex and diverse human microbiomes [3, 11, 12]. Oral bacteria have important functional roles that contribute to maintenance of oral health [13], to oral diseases such as caries and periodontitis in the setting of dysbiosis [14, 15], and potentially to systemic diseases of aging by way of bacterial translocation through ulcerated oral epithelium, aspiration, or ingestion [7, 16]. This could have important implications to public health given the rapid growth in numbers of older adults expected in coming decades.
Surprisingly, there exists a limited understanding of oral microbiota in aging populations. Feres et al. [17] conducted a comprehensive review of published literature and concluded that the majority of oral microbiome studies have included younger and middle-aged adults. Only a small number of studies have described the microbiome in older adults, among which sample sizes of adults 60 years and older tended to be, on average, modest (e.g., < 200), the majority of whom were men and were selected to have moderate to severe periodontitis [17–19]. A majority of previous studies have used low throughput microbial measurement techniques, such as microbial culture and targeted DNA probes, which result in an incomplete characterization of the oral microbiome composition and diversity in relationship to groups of men and women of differing ages. Recent investigations have extended these previous studies by using next generation sequencing methods, but again relatively small sample sizes (< 100) limited the contrasts that could be performed in relation to age in the majority of these studies [20–23].
Thus, at present, an incomplete understanding of the composition and characteristics of the oral microbiome exists in the context of aging, particularly in women. A critical step in advancing knowledge on how the oral microbiome relates with the frequency of oral (e.g., periodontitis) or systemic (e.g., breast cancer) diseases of aging, is to first understand the extent of the composition and how the microbiota vary with host characteristics, such as age. This information will be important in later understanding the interplay of the microbiome with pathogenic changes over time. Application of epidemiologic study methods to study populations not selected on disease status is a suggested approach to establish a foundational understanding of microbiome diversity expected in a population that then allow for hypotheses pertaining to disease-related variation that can then be accurately evaluated [24]. The objective of this current cross-sectional investigation was to describe the composition and diversity of the subgingival plaque microbiome and its relationship with age in a cohort of ambulatory postmenopausal women, aged 53–81 years, who were enrolled in an ongoing study from the community dwelling women without selection on periodontal health status at enrollment.
Methods
Participants
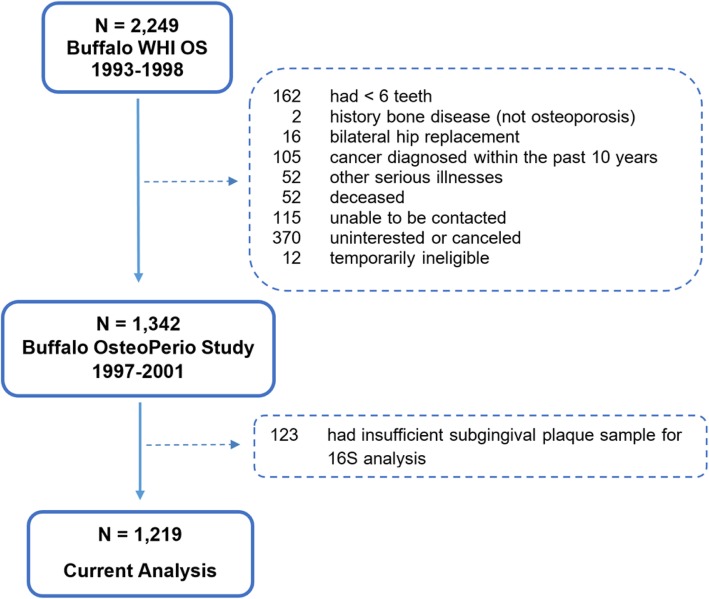
The present study included 1219 postmenopausal women enrolled in the Buffalo Osteoporosis and Periodontitis (OsteoPerio) Study, which is an ancillary study conducted at the Buffalo (NY) clinical center of the Women’s Health Initiative Observational Study (WHI OS). Participants provided written informed consent for all components of the studies, which were conducted in accord with the Helsinki Declaration on human subjects research. Experimental protocols for all aspects of the WHI study, the OsteoPerio Study, and the microbiome study detailed in this paper were approved by the Institutional Review Board at the University at Buffalo. This manuscript conforms to the STROBE guidelines for human observational studies. Details about recruitment, enrollment criteria, study implementation and measurements have been published for the WHI OS [25] and the OsteoPerio study [26, 27]. Briefly, 2249 postmenopausal women, ages 50–79, enrolled into the WHI OS at the Buffalo center between 1994 and 1998. Of these, 1362 enrolled into the OsteoPerio study 3 years later in 1997–2001 (mean age 66; range 53–81 years). Enrollment into the OsteoPerio study required at least 6 teeth present and no history of bone disease other than osteoporosis and no history of cancer in the previous 10 years. Women completed standardized questionnaires pertaining to demographic information, lifestyle habits, and personal health history, as well as undertaking a whole mouth oral examination conducted by trained and calibrated examiners. Neighborhood socioeconomic status was derived from questionnaire responses and census tract information [28]. Detailed descriptions of the oral examination measures and their reproducibility have been published [26]. Figure 1 shows a flow chart of participant enrollment into the OsteoPerio study.
Fig. 1.
Flow of participants into the Buffalo OsteoPerio Study
Subgingival plaque samples
A protocol for obtaining subgingival plaque samples was developed for this study and has been published [29]. Fine paper points – (#504; Henry Schein, Melville, NY) were placed in the gingival pockets of up to 12 pre-specified teeth (6 maxillary and 6 mandibular arch teeth) for 10 S. index teeth [3, 5, 7, 9, 12, 14, 19, 21, 23, 25, 28, 30, and] were usually sampled. Alternative teeth [2, 4, 8, 10, 13, 15, 18, 20, 24, 26, 29, 31, and] were used if the corresponding index tooth was missing. Paper points containing all subgingival plaque samples from each arch were placed directly into 4 mL lactated Ringer’s solution. The solution was taken to the lab where it was vortexed for dispersion of microorganisms, placed in cryogenic straws, frozen immediately at -80 °C and later placed in cryogenic tanks at -196 °C as previously described [29]. Before next generation sequencing, samples were placed in − 80 freezers and later thawed, with upper and lower arch samples combined into a single aliquot for the purpose of sequencing.
DNA isolation and purification
Genomic DNA was isolated using the QIAsymphony SP automated system (Qiagen, Valencia, CA) with the QIAsymphony DSP Virus/Pathogen Mini Kit (Qiagen, Valencia, CA) and the Complex200_V6_DSP protocol after enzymatic pretreatment. In detail, 500 μl of oral plaque solution contained in a barcoded 2 ml tube was equilibrated at room temperature (15-25 °C). Bacteria was pelleted by centrifugation at 5000×g for 10 min, resuspended in a 300 μl lysis solution (20 mg/ml lysozyme in 20 mM Tris-HCl, pH 8.0; 2 mM EDTA; 1.2% Triton X-100) and incubated at 37 °C for 30 min. Following incubation, tubes were briefly centrifuged to remove drops from inside the lid and then placed in the tube carrier of the QIAsymphony SP.
DNA extraction and purification was done according to the Qiasymphony DSP Virus /Pathogen Kit Instructions. Carrier RNA-AVE mixture was added to all samples for increased recovery of nucleic acids. After DNA purification, samples were eluted in a barcoded 96 well elution plate (Qiagen, Valencia, CA). All batches of samples were performed with DNA extraction negative controls and positive controls from a single large pool of mixed plaque samples.
16S rRNA amplification and sequencing
Metagenomic amplification of the extracted DNA for 16S amplification of the V3–V4 hypervariable region proceeded following the Illumina manufacturer protocol (Illumina Inc., San Diego, CA) with modifications developed for our study [30]. The Illumina protocol relies on limited cycle PCR for addition of Illumina sequencing adapters and dual-index barcodes to the 16S rRNA V3-V4 ampli. We also included as part of the 96-well plates, samples of the UltraClean DNA free PCR water (MO BIO Laboratories, Carlsbad, CA) and RNase/DNase free water (Ambion, Foster City, CA) as negative controls, and genomic DNA from microbial community HM-277D (microbial community B; BEI Resources; Manassas, VA) as a positive control during the amplification process. Metagenomic DNA was amplified using the 16S V3 (341F) forward and V4 (805R) reverse primer pairs with added Illumina adapter overhang nucleotide sequences. Amplicon PCR was completed with 42 μl of genomic DNA, 4 μl of amplicon PCR forward primer (5 μM), 4 μl of amplicon PCR reverse primer (5 μM), and 50 μl of 2x KAPA HiFi HotStart Ready Mix (KapaBiosystems) at 95 °C initial denaturation for 3 min, followed by 25 cycles of 95 °C for 30 s, 62.3 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min. Reactions were cleaned with Agencourt AMPure XP beads (Beckman Coulter Genomics, South Plainfield, NJ) according to the manufacturer’ s protocol.
Library generation was performed using 5 μl of amplicon PCR product DNA, 5 μl of Illumina Nextera XT Index Primer 1 (N7xx), 5 μl of Nextera XTIndex Primer 2 (S5xx), 25 μ l of 2x KAPA HiFi HotStart Ready Mix, and 10 μl of PCR-grade water (UltraClean MO BIO Laboratories, Inc.), with thermocycling at 95 °C for 3 min, followed by 8 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min. 16S metagenomic libraries were purified with Agencourt AMPure XP beads and quantified with Quant-iT PicoGreen. Nextera index primer sets (A, B, and C) were rotated for each batch to reduce sequence carryover between MiSeq runs.
Library quality control was performed with the Fragment Analyzer (Advanced Analytical Technologies, Inc., Ankeny, IA) to ascertain average size distribution. Generated 16S rRNA V3-V4 libraries were further quality-controlled using the following internal study criteria: 1. Library concentration of all negative(s) is < 5 ng/μl, 2. Participant samples have a fragment peak distribution with average size of ~ 600 bp, and 3. Negative controls yield a straight line when run in the Fragment Analyzer. If the above cutoffs were met, libraries were normalized and pooled to 4 nM based on PicoGreen concentrations. The pool of normalized libraries were then quantified with the NEBNext Library Quant Kit (New England Biolabs, Inc., Ipswich, M.), denatured with NaOH and diluted to a final concentration of 10 pM with a 20% PhiX (Illumina, Inc., San Diego, CA). 2 × 300 bp paired-end sequencing is performed in the Illumina MiSeq System (Illumina Inc., San Diego, CA) by multiplexing 96 samples per sequencing run with the MiSeq Reagent Kit.
Joining of Illumina paired-end reads were completed using Paired-End reAd mergeR (PEAR version 0.9.6). The percentage of successfully joined pair-end defined the “merge rate”; paired-end reads that could not be joined were removed from downstream analyses. Sequence quality filtering was done with the Fastx-Toolkit (V.0.013) to isolate reads with 90% of their bases having a score higher than Q30, which defined the “pass rate”; reads not meeting this criterion were removed. Primer sequences were trimmed based on the length of the forward and reverse sequencing primers. Following quality-filtering, reads were deduplicated by recording the number and type of identical sequences to reduce downstream processing time.
Taxonomy annotation was done with BLAST [31] at a 97% similarity, for species-level assignment approximation, against bacterial sequences from the HOMD version 14.5. Input query reads were given the same taxonomic label as the best hit in the reference sequence collection, defining the “hit count”; reads with no hits were excluded from downstream analyses. Sequences with the same labels were clustered into one OTU and the raw OTU table was constructed by combining absolute sequence abundances from the deduplication step, generated taxonomy annotations and manually generated metadata. We subsequently filtered the raw OTU table by discarding OTUs with a frequency < 0.02% of the total read count. At the preprocessing sequence analysis step we require a ‘Merge Rate’ ≥ 90%, ‘Pass Rate’ ≥ 60%, and ‘Hit Count’ per sample ≥ 3000.
Statistical analysis
For this analysis we used several approaches to characterize the composition and diversity of the subgingival microbiome and their relationships with age. Individual OTU counts were normalized using the centered log(2)-ratio (CLR) transformation. Gloor et al. [32] recommends the CLR transformation to account for the complex compositional data structure, to reduce the likelihood of spurious correlations, and to enhance the meaningfulness of subcomposition comparisons. A positive CLR OTU value for given taxon indicates a relatively higher amount than the overall composition mean, which is 0; a negative value indicates relatively lower amount. The fold-difference for a reported CLR OTU value relative to the compositional mean, can be determined by raising 2 to the power of the base 2 logarithm. For example, a mean CLR of 3, reflects an 8-fold (23) higher abundance compared to the compositional mean; a mean CLR of − 3 reflects an 8-fold lower abundance. The CLR distribution of each OTU was approximately normal and the variances in groups were similar by visual inspection. Alpha diversity was used to assess species richness and evenness across age categories. The rarefaction curve, bias-corrected Chao1 (richness), OTU count (richness), and Shannon entropy (evenness) values were calculated for each sample using scikit- bio v0.5.5. Beta diversity was evaluated using principal component analysis (PCA) [33]. T-tests were used to evaluate differences in alpha diversity, and PERMANOVA was used to evaluate differences in beta diversity, using SciPy v1.3.0. Comparisons of microbiota between age categories was performed using analysis of variance and evaluation of linear relationships between microbiota and age performed using Pearson product-moment correlations. We nominally defined correlations of |r| < 0.10 as weak, 0.10–0.49 as moderate, 0.50–0.70 as strong, and > 0.70 as very strong. We report uncorrected p-values and indicate which are statistically significant after Bonferroni correction for multiple testing.
To provide additional perspective and comparability with previous studies, we also describe microbiome composition and diversity according to conventional measures of relative abundance (the amount of a specific taxon relative to the total composition of the sample in which it is measured) and prevalence (presence of a taxon regardless of relative composition). To minimize the total number of hypothesis tests performed, formal comparisons using these measures were not conducted and these data are presented for descriptive purposes only.
Results
Characteristics of study group
Participant characteristics are shown for descriptive purposes in Table 1. Women in the present study were, on average, 66 years of age and the vast majority (97%) were Caucasian. Prevalence of current smoking (3%) and diabetes history (5.2%) was modest, and about half the group reported current use of hormone therapy. The group retained the majority of their natural teeth (mean, 23), the frequency of reported teeth brushing two or more times per day was high (77%) as was frequency of dental visits one or more times per year (91%). Mean pocket depth was 2.2 mm (range 1.2–3.8). As expected, prevalence of current smoking and current hormone therapy use declined with increasing age, and, prevalence of diabetes history was highest among the oldest women. The number of teeth present and frequency of dental visits declined with increasing age and, frequency of teeth brushing was higher in older than younger women. Both neighborhood socioeconomic status and mean pocket depth were similar across age groups.
Table 1.
Baseline characteristics of OsteoPerio Microbiome Study participants for the overall cohort and by age groups
| Characteristic | Overall (N = 1219) |
50–59 (N = 239) |
60–69 (N = 554) |
≥70 (N = 426) |
|---|---|---|---|---|
| Age (years), mean (SD) | 66.2 (7.0) | 56.7 (1.8) | 64.2 (2.9) | 74.1 (3.3) |
| Race-ethnicity: White, N (%) | 1187 (97.4) | 233 (97.5) | 537 (96.9) | 417 (97.9) |
| Neighborhood SES, mean (SD) | 76.2 (6.9) | 75.7 (7.5) | 76.6 (6.8) | 75.9 (6.7) |
| Smoking, N (%) | ||||
| Never | 642 (52.7) | 117 (48.9) | 280 (50.5) | 245 (57.6) |
| Former | 537 (44.1) | 107 (44.8) | 257 (46.4) | 173 (40.7) |
| Current | 39 (3.2) | 15 (6.3) | 17 (3.1) | 7 (1.7) |
| History of treated diabetes, N (%) | 63 (5.2) | 9 (3.8) | 26 (4.7) | 28 (6.6) |
| History of treated hypertension, N (%) | 392 (32.2) | 63 (26.4) | 148 (26.7) | 181 (42.5) |
| History of treated high cholesterol, N (%) | 201 (16.5) | 26 (10.9) | 76 (13.7) | 99 (23.4) |
| Hormone therapy use, N (%) | ||||
| Never | 390 (32.0) | 53 (22.2) | 158 (28.5) | 179 (42.0) |
| Former E-Alone | 132 (10.9) | 12 (5.0) | 50 (9.0) | 70 (16.5) |
| Former E + P | 111 (9.1) | 24 (10.1) | 58 (10.5) | 29 (6.8) |
| Current E-Alone | 307 (25.2) | 66 (27.7) | 136 (24.6) | 105 (24.7) |
| Current E + P | 277 (22.8) | 83 (34.9) | 152 (27.4) | 42 (9.9) |
| Years taking hormone therapya | 5.6 (7.3) | 4.6 (4.5) | 6.2 (7.0) | 5.5 (8.8) |
| Number of teeth present, mean (SD) | 23.2 (5.3) | 24.8 (4.0) | 23.6 (5.2) | 21.9 (5.8) |
| Brush teeth ≥2 times/day, N (%) | 942 (77.3) | 178 (74.5) | 422 (76.2) | 342 (80.3) |
| Floss teeth daily, N (%) | 529 (43.6) | 90 (37.7) | 247 (44.8) | 192 (45.4) |
| Dental visit ≥1 time/year, N (%) | 1114 (91.4) | 225 (94.1) | 504 (91.0) | 385 (90.4) |
| Mean Pocket Depth (mm), mean (SD) | 2.2 (0.4) | 2.2 (0.4) | 2.2 (0.4) | 2.1 (0.4) |
| Gingival Bleeding (%), mean (SD) | 34.4 (23.2) | 33.0 (23.5) | 34.5 (22.9) | 35.1 (23.4) |
SES socioeconomic statusn. See methods section for its definition and derivation, E estrogen, P progesti
aNever users coded as 0 years
Microbial community structure and composition
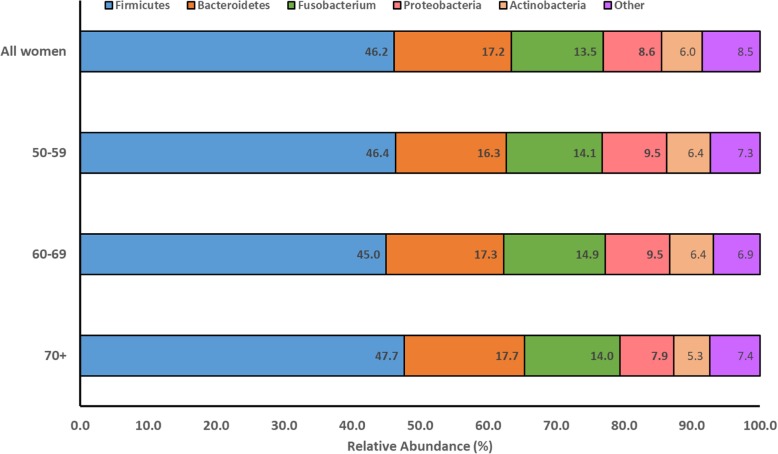
After filtering out OTUs < 0.02%, the total number of sequence reads for the overall cohort of 1219 women was 120,388,085 (mean reads per sample, 98,760; range 3034 to 1,080,317). Sequence reads per sample was somewhat higher with increasing age, with means (SDs) of 89,442 (71,698), 97,794 (86,908), and 105,243 (80,183) reads in women ages 50–59, 60–69, and ≥ 70 years, respectively. There were 267 microbial taxa identified in the subgingival plaque samples after filtering at 0.02%. The taxonomic classification and mean reads for each taxon overall and by age groups, are presented in Table 2. Of the 120,388,085 read, 46.2% were of the phylum Firmicutes, 17.2% Bacteroidetes, 13.5% Fusobacterium, 8.6% Proteobacteria, 6.0% Actinobacteria, and the remaining were among other phyla of < 4%, each (Fig. 2). The distribution of phyla was consistent across age groups. At the genus level, the highest mean relative abundance was for Veillonella (16.7%), followed by Streptococcus (14.2%), Fusobacterium (10.7%), Prevotella (8.6%), and Selenomonas (7.7%); relative abundance of the remaining genera was < 4%, each. This pattern was consistent across age groups. At the species level, among all women, the highest number of mean reads was for Veillonella dispar (Firmicutes phylum; mean, 8136) and Veillonella parvula (Firmicutes phylum; mean, 6262) (Table 2). Mean reads for each taxon increased across incremental age groups.
Table 2.
Taxonomic classification and mean reads for the 267 bacteria identified, overall and by age groups
| Phyla | Class | Genus | Species | Age Categories (years) | |||
|---|---|---|---|---|---|---|---|
| Overall | 50–59 | 60–69 | ≥70 | ||||
| p__Actinobacteria | c__Actinobacteria | g__Actinobaculum | s__sp._oral_taxon_183 | 89.8 | 95.0 | 88.9 | 88.0 |
| p__Actinobacteria | c__Actinobacteria | g__Actinobaculum | s__sp._oral_taxon_848 | 48.2 | 37.3 | 52.9 | 48.2 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__gerencseriae | 68.6 | 60.5 | 66.7 | 75.6 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__israelii | 25.1 | 19.5 | 24.1 | 29.5 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__johnsonii | 106.0 | 104.9 | 105.2 | 107.6 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__massiliensis | 112.1 | 134.2 | 121.5 | 87.4 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__meyeri | 59.3 | 64.8 | 61.6 | 53.3 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__naeslundii | 453.1 | 482.6 | 447.6 | 443.6 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__oris | 225.7 | 236.6 | 218.8 | 228.5 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__sp._oral_taxon_169 | 195.8 | 251.2 | 183.7 | 180.4 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__sp._oral_taxon_170 | 60.4 | 44.8 | 59.8 | 70.0 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__sp._oral_taxon_171 | 88.2 | 86.5 | 89.2 | 87.9 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__sp._oral_taxon_178 | 27.0 | 23.9 | 26.8 | 29.0 |
| p__Actinobacteria | c__Actinobacteria | g__Actinomyces | s__sp._oral_taxon_180 | 119.0 | 121.7 | 123.6 | 111.5 |
| p__Actinobacteria | c__Actinobacteria | g__Bifidobacterium | s__dentium | 88.5 | 83.0 | 75.8 | 108.2 |
| p__Actinobacteria | c__Actinobacteria | g__Corynebacterium | s__durum | 121.3 | 153.3 | 120.2 | 104.7 |
| p__Actinobacteria | c__Actinobacteria | g__Corynebacterium | s__matruchotii | 1107 | 1095 | 1178 | 1020 |
| p__Actinobacteria | c__Actinobacteria | g__Microbacterium | s__flavescens | 1.5 | 1.8 | 1.6 | 1.1 |
| p__Actinobacteria | c__Actinobacteria | g__Rothia | s__aeria | 323.3 | 311.6 | 378.9 | 257.6 |
| p__Actinobacteria | c__Actinobacteria | g__Rothia | s__dentocariosa | 975.6 | 915.0 | 1039 | 927.8 |
| p__Actinobacteria | c__Actinobacteria | g__Rothia | s__mucilaginosa | 185.3 | 125.4 | 179.1 | 226.9 |
| p__Actinobacteria | c__Actinobacteria | g__Scardovia | s__wiggsiae | 68.7 | 74.1 | 69.7 | 64.4 |
| p__Actinobacteria | c__Coriobacteriia | g__Atopobium | s__parvulum | 77.9 | 57.2 | 78.3 | 88.8 |
| p__Actinobacteria | c__Coriobacteriia | g__Atopobium | s__rimae | 200.0 | 107.8 | 200.0 | 251.7 |
| p__Actinobacteria | c__Coriobacteriia | g__Atopobium | s__sp._oral_taxon_199 | 47.4 | 39.1 | 61.2 | 34.0 |
| p__Actinobacteria | c__Coriobacteriia | g__Atopobium | s__sp._oral_taxon_416 | 25.4 | 1.1 | 14.6 | 53.1 |
| p__Actinobacteria | c__Coriobacteriia | g__Olsenella | s__sp._oral_taxon_807 | 46.9 | 38.6 | 44.5 | 54.7 |
| p__Bacteroidetes | c__Bacteroidetes_[C-1] | g__Bacteroidetes_[G-5] | s__sp._oral_taxon_511 | 137.4 | 87.8 | 155.7 | 141.4 |
| p__Bacteroidetes | c__Bacteroidia | g__Alloprevotella | s__rava | 82.4 | 52.8 | 87.6 | 92.3 |
| p__Bacteroidetes | c__Bacteroidia | g__Alloprevotella | s__sp._oral_taxon_308 | 28.6 | 21.3 | 24.9 | 37.4 |
| p__Bacteroidetes | c__Bacteroidia | g__Alloprevotella | s__sp._oral_taxon_473 | 77.5 | 63.0 | 82.6 | 79.1 |
| p__Bacteroidetes | c__Bacteroidia | g__Alloprevotella | s__tannerae | 1562 | 1427 | 1580 | 1615 |
| p__Bacteroidetes | c__Bacteroidia | g__Bacteroidaceae_[G-1] | s__sp._oral_taxon_272 | 39.5 | 24.6 | 28.6 | 62.2 |
| p__Bacteroidetes | c__Bacteroidia | g__Bacteroidales_[G-2] | s__sp._oral_taxon_274 | 885.8 | 548.7 | 935.5 | 1010 |
| p__Bacteroidetes | c__Bacteroidia | g__Porphyromonas | s__catoniae | 104.1 | 111.4 | 115.8 | 84.7 |
| p__Bacteroidetes | c__Bacteroidia | g__Porphyromonas | s__endodontalis | 602.5 | 560.5 | 657.1 | 555.0 |
| p__Bacteroidetes | c__Bacteroidia | g__Porphyromonas | s__gingivalis | 1055 | 781.4 | 752.8 | 1603 |
| p__Bacteroidetes | c__Bacteroidia | g__Porphyromonas | s__sp._oral_taxon_275 | 46.0 | 29.0 | 62.6 | 33.9 |
| p__Bacteroidetes | c__Bacteroidia | g__Porphyromonas | s__sp._oral_taxon_278 | 39.2 | 22.1 | 43.1 | 43.6 |
| p__Bacteroidetes | c__Bacteroidia | g__Porphyromonas | s__sp._oral_taxon_279 | 310.5 | 255.9 | 315.5 | 334.7 |
| p__Bacteroidetes | c__Bacteroidia | g__Porphyromonas | s__sp._oral_taxon_284 | 183.2 | 187.5 | 187.8 | 174.9 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__baroniae | 50.3 | 29.2 | 60.5 | 48.8 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__buccae | 53.1 | 17.3 | 63.0 | 60.4 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__dentalis | 102.3 | 63.3 | 120.3 | 100.8 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__denticola | 773.5 | 501.4 | 724.4 | 989.9 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__histicola | 100.3 | 48.8 | 72.7 | 165.3 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__intermedia | 671.5 | 613.0 | 766.5 | 580.8 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__loescheii | 119.6 | 131.7 | 138.7 | 87.8 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__maculosa | 185.9 | 139.8 | 180.0 | 219.4 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__melaninogenica | 339.1 | 236.0 | 321.8 | 419.4 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__micans | 42.7 | 37.6 | 37.1 | 52.9 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__multiformis | 46.7 | 9.2 | 36.1 | 81.6 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__nigrescens | 1997 | 1719 | 1960 | 2200 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__oralis | 174.4 | 78.8 | 158.9 | 248.2 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__oris | 1968 | 1700 | 2093 | 1956 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__oulorum | 211.7 | 216.2 | 182.8 | 246.8 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__pallens | 94.4 | 91.5 | 71.4 | 125.9 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__pleuritidis | 579.6 | 478.7 | 606.4 | 601.4 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__saccharolytica | 73.7 | 52.8 | 80.9 | 76.1 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__salivae | 120.5 | 94.7 | 117.3 | 139.1 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_292 | 69.4 | 50.1 | 57.5 | 95.7 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_300 | 269.5 | 256.3 | 243.5 | 310.7 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_306 | 41.1 | 17.2 | 38.9 | 57.5 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_313 | 70.5 | 86.9 | 44.0 | 95.8 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_314 | 65.2 | 59.2 | 52.7 | 85.0 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_317 | 832.0 | 631.4 | 832.4 | 944.0 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_376 | 44.8 | 45.1 | 43.6 | 46.1 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_472 | 250.3 | 270.4 | 279.0 | 201.9 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_475 | 21.8 | 19.1 | 17.4 | 28.9 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__sp._oral_taxon_526 | 55.1 | 21.5 | 70.7 | 53.7 |
| p__Bacteroidetes | c__Bacteroidia | g__Prevotella | s__veroralis | 113.8 | 84.1 | 160.3 | 70.0 |
| p__Bacteroidetes | c__Bacteroidia | g__Tannerella | s__forsythia | 577.6 | 374.7 | 542.0 | 737.8 |
| p__Bacteroidetes | c__Bacteroidia | g__Tannerella | s__sp._oral_taxon_286 | 93.0 | 68.6 | 100.9 | 96.6 |
| p__Bacteroidetes | c__Bacteroidia | g__Tannerella | s__sp._oral_taxon_808 | 35.6 | 25.4 | 32.6 | 45.3 |
| p__Bacteroidetes | c__Flavobacteriia | g__Bergeyella | s__sp._oral_taxon_322 | 164.1 | 194.8 | 173.9 | 134.1 |
| p__Bacteroidetes | c__Flavobacteriia | g__Bergeyella | s__sp._oral_taxon_907 | 34.8 | 34.7 | 36.7 | 32.4 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__gingivalis | 502.9 | 619.2 | 471.7 | 478.3 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__granulosa | 597.4 | 513.3 | 629.6 | 602.6 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__leadbetteri | 614.5 | 534.6 | 619.4 | 653.0 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_323 | 37.7 | 26.7 | 40.2 | 40.5 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_324 | 33.8 | 19.5 | 32.3 | 43.8 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_326 | 258.0 | 222.7 | 281.8 | 246.9 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_332 | 64.5 | 104.3 | 60.6 | 47.3 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_336 | 159.5 | 127.8 | 157.8 | 179.6 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_338 | 62.3 | 62.5 | 56.7 | 69.5 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_380 | 29.6 | 23.3 | 40.3 | 19.1 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_412 | 43.7 | 42.0 | 46.4 | 41.2 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_864 | 76.3 | 76.5 | 80.4 | 70.9 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_902 | 45.4 | 51.0 | 43.5 | 44.7 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sp._oral_taxon_903 | 37.1 | 26.6 | 37.4 | 42.5 |
| p__Bacteroidetes | c__Flavobacteriia | g__Capnocytophaga | s__sputigena | 416.4 | 448.8 | 405.0 | 413.0 |
| p__Chloroflexi | c__Anaerolineae | g__Anaerolineae_[G-1] | s__sp._oral_taxon_439 | 59.1 | 30.1 | 54.2 | 81.8 |
| p__Firmicutes | c__Bacilli | g__Abiotrophia | s__defectiva | 104.2 | 135.7 | 109.9 | 79.0 |
| p__Firmicutes | c__Bacilli | g__Gemella | s__haemolysans | 338.8 | 412.1 | 338.9 | 297.4 |
| p__Firmicutes | c__Bacilli | g__Gemella | s__morbillorum | 603.8 | 670.7 | 622.2 | 542.2 |
| p__Firmicutes | c__Bacilli | g__Gemella | s__sanguinis | 44.3 | 33.8 | 36.2 | 60.7 |
| p__Firmicutes | c__Bacilli | g__Granulicatella | s__adiacens | 532.6 | 564.2 | 523.2 | 527.1 |
| p__Firmicutes | c__Bacilli | g__Granulicatella | s__elegans | 39.7 | 49.7 | 37.7 | 36.7 |
| p__Firmicutes | c__Bacilli | g__Lactobacillus | s__gasseri | 31.4 | 9.3 | 37.7 | 35.5 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__anginosus | 479.8 | 409.3 | 468.1 | 534.6 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__australis | 28.9 | 21.0 | 24.1 | 39.5 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__constellatus | 283.7 | 208.0 | 279.1 | 332.2 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__cristatus | 516.2 | 457.7 | 546.4 | 509.7 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__gordonii | 998.1 | 853.8 | 991.3 | 1088 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__intermedius | 897.1 | 1038 | 949.4 | 749.9 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__lactarius | 56.8 | 97.9 | 40.2 | 55.3 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__mutans | 530.8 | 392.2 | 500.4 | 648.0 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__oralis | 6725 | 8031 | 6651 | 6089 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__parasanguinis_I | 59.7 | 48.6 | 51.2 | 77.0 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__parasanguinis_II | 141.3 | 124.2 | 126.4 | 170.2 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__salivarius | 460.0 | 399.7 | 455.1 | 500.2 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__sanguinis | 1128 | 1441 | 1133 | 945.5 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__sinensis | 29.3 | 32.1 | 14.3 | 47.4 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__sobrinus | 52.0 | 2.5 | 19.9 | 121.7 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__sp._oral_taxon_056 | 90.0 | 102.7 | 82.4 | 92.7 |
| p__Firmicutes | c__Bacilli | g__Streptococcus | s__sp._oral_taxon_074 | 68.3 | 54.3 | 71.9 | 71.6 |
| p__Firmicutes | c__Clostridia | g__Butyrivibrio | s__sp._oral_taxon_080 | 24.6 | 14.1 | 35.9 | 15.9 |
| p__Firmicutes | c__Clostridia | g__Catonella | s__morbi | 230.1 | 228.9 | 222.5 | 240.8 |
| p__Firmicutes | c__Clostridia | g__Filifactor | s__alocis | 368.2 | 274.3 | 418.9 | 355.0 |
| p__Firmicutes | c__Clostridia | g__Johnsonella | s__ignava | 114.9 | 104.7 | 111.8 | 124.5 |
| p__Firmicutes | c__Clostridia | g__Johnsonella | s__sp._oral_taxon_166 | 32.7 | 17.7 | 43.8 | 26.7 |
| p__Firmicutes | c__Clostridia | g__Lachnoanaerobaculum | s__orale | 31.4 | 35.3 | 24.9 | 37.6 |
| p__Firmicutes | c__Clostridia | g__Lachnoanaerobaculum | s__saburreum | 152.3 | 124.4 | 153.0 | 167.0 |
| p__Firmicutes | c__Clostridia | g__Lachnoanaerobaculum | s__umeaense | 52.9 | 50.6 | 55.5 | 50.7 |
| p__Firmicutes | c__Clostridia | g__Lachnospiraceae_[G-3] | s__sp._oral_taxon_100 | 132.2 | 118.4 | 143.0 | 125.9 |
| p__Firmicutes | c__Clostridia | g__Lachnospiraceae_[G-8] | s__sp._oral_taxon_500 | 45.1 | 40.6 | 48.1 | 43.7 |
| p__Firmicutes | c__Clostridia | g__Oribacterium | s__sp._oral_taxon_078 | 112.2 | 68.2 | 102.9 | 149.0 |
| p__Firmicutes | c__Clostridia | g__Parvimonas | s__micra | 848.4 | 791.4 | 915.0 | 793.8 |
| p__Firmicutes | c__Clostridia | g__Parvimonas | s__sp._oral_taxon_393 | 265.1 | 361.3 | 250.6 | 230.0 |
| p__Firmicutes | c__Clostridia | g__Peptostreptococcaceae_[XI][G-1] | s__[Eubacterium]_infirmum | 38.9 | 32.2 | 32.2 | 51.2 |
| p__Firmicutes | c__Clostridia | g__Peptostreptococcaceae_[XI][G-5] | s__[Eubacterium]_saphenum | 87.3 | 55.6 | 107.1 | 79.2 |
| p__Firmicutes | c__Clostridia | g__Peptostreptococcaceae_[XI][G-6] | s__[Eubacterium]_nodatum | 64.3 | 60.2 | 55.8 | 77.7 |
| p__Firmicutes | c__Clostridia | g__Peptostreptococcaceae_[XI][G-7] | s__[Eubacterium]_yurii_subsps._yur | 146.2 | 167.1 | 151.3 | 127.9 |
| p__Firmicutes | c__Clostridia | g__Peptostreptococcaceae_[XI][G-9] | s__[Eubacterium]_brachy | 200.8 | 208.5 | 203.6 | 192.7 |
| p__Firmicutes | c__Clostridia | g__Peptostreptococcus | s__stomatis | 107.9 | 130.7 | 103.5 | 100.7 |
| p__Firmicutes | c__Clostridia | g__Pseudoramibacter | s__alactolyticus | 70.8 | 73.3 | 52.7 | 93.0 |
| p__Firmicutes | c__Clostridia | g__Ruminococcaceae_[G-1] | s__sp._oral_taxon_075 | 83.8 | 109.4 | 79.6 | 74.9 |
| p__Firmicutes | c__Clostridia | g__Shuttleworthia | s__satelles | 35.3 | 21.3 | 40.5 | 36.3 |
| p__Firmicutes | c__Clostridia | g__Stomatobaculum | s__longum | 55.2 | 47.4 | 52.5 | 63.0 |
| p__Firmicutes | c__Erysipelotrichia | g__Solobacterium | s__moorei | 42.1 | 40.4 | 41.0 | 44.6 |
| p__Firmicutes | c__Mollicutes | g__Mycoplasma | s__salivarium | 33.0 | 24.9 | 30.9 | 40.4 |
| p__Firmicutes | c__Negativicutes | g__Anaeroglobus | s__geminatus | 767.6 | 402.1 | 582.4 | 1214 |
| p__Firmicutes | c__Negativicutes | g__Centipeda | s__periodontii | 93.0 | 45.3 | 106.0 | 102.9 |
| p__Firmicutes | c__Negativicutes | g__Dialister | s__invisus | 612.2 | 466.3 | 625.5 | 676.8 |
| p__Firmicutes | c__Negativicutes | g__Dialister | s__pneumosintes | 225.5 | 179.8 | 224.8 | 252.1 |
| p__Firmicutes | c__Negativicutes | g__Megasphaera | s__micronuciformis | 208.0 | 136.7 | 212.5 | 242.2 |
| p__Firmicutes | c__Negativicutes | g__Megasphaera | s__sp._oral_taxon_123 | 158.7 | 166.4 | 138.4 | 180.8 |
| p__Firmicutes | c__Negativicutes | g__Mitsuokella | s__sp._oral_taxon_131 | 117.5 | 40.4 | 143.3 | 127.2 |
| p__Firmicutes | c__Negativicutes | g__Mitsuokella | s__sp._oral_taxon_521 | 31.2 | 11.1 | 13.3 | 65.7 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__artemidis | 738.5 | 720.4 | 712.4 | 782.7 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__dianae | 47.4 | 36.6 | 36.3 | 67.9 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__flueggei | 166.3 | 137.2 | 160.7 | 190.0 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__infelix | 272.6 | 194.0 | 268.0 | 322.7 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__noxia | 1502 | 1474 | 1514 | 1500 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_126 | 102.4 | 92.1 | 105.4 | 104.2 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_133 | 44.7 | 44.7 | 43.4 | 46.3 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_134 | 357.6 | 200.4 | 361.3 | 440.9 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_136 | 318.2 | 182.9 | 317.6 | 394.8 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_137 | 494.2 | 423.6 | 520.8 | 499.2 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_146 | 119.4 | 98.3 | 117.8 | 133.2 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_149 | 19.7 | 9.7 | 25.0 | 18.4 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_442 | 27.3 | 14.0 | 37.6 | 21.3 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_478 | 15.5 | 12.0 | 12.6 | 21.2 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_892 | 223.3 | 222.2 | 211.7 | 239.0 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_919 | 187.4 | 170.1 | 178.5 | 208.7 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_936 | 83.7 | 62.0 | 73.5 | 109.1 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sp._oral_taxon_937 | 22.1 | 14.6 | 22.5 | 25.7 |
| p__Firmicutes | c__Negativicutes | g__Selenomonas | s__sputigena | 3283 | 2195 | 2957 | 4319 |
| p__Firmicutes | c__Negativicutes | g__Veillonella | s__atypica | 586.0 | 380.8 | 569.5 | 722.5 |
| p__Firmicutes | c__Negativicutes | g__Veillonella | s__denticariosi | 201.2 | 265.6 | 159.9 | 218.6 |
| p__Firmicutes | c__Negativicutes | g__Veillonella | s__dispar | 8720 | 7556 | 8534 | 9615 |
| p__Firmicutes | c__Negativicutes | g__Veillonella | s__parvula | 6529 | 5964 | 6121 | 7376 |
| p__Firmicutes | c__Negativicutes | g__Veillonella | s__rogosae | 202.3 | 138.4 | 208.0 | 230.9 |
| p__Firmicutes | c__Negativicutes | g__Veillonella | s__sp._oral_taxon_780 | 109.3 | 147.1 | 100.8 | 99.2 |
| p__Firmicutes | c__Negativicutes | g__Veillonellaceae_[G-1] | s__sp._oral_taxon_129 | 54.4 | 27.0 | 45.9 | 81.0 |
| p__Firmicutes | c__Negativicutes | g__Veillonellaceae_[G-1] | s__sp._oral_taxon_145 | 59.1 | 39.5 | 58.4 | 70.9 |
| p__Firmicutes | c__Negativicutes | g__Veillonellaceae_[G-1] | s__sp._oral_taxon_150 | 295.0 | 204.8 | 251.6 | 402.1 |
| p__Firmicutes | c__Negativicutes | g__Veillonellaceae_[G-1] | s__sp._oral_taxon_155 | 265.6 | 194.6 | 212.4 | 374.6 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__naviforme | 826.5 | 725.4 | 813.9 | 899.6 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__nucleatum_subsp._animalis | 1650 | 1387 | 1575 | 1896 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__nucleatum_subsp._nucleatum | 223.2 | 165.8 | 149.4 | 351.4 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__nucleatum_subsp._polymorphum | 1439 | 1467 | 1499 | 1345 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__nucleatum_subsp._vincentii | 3930 | 3647 | 3798 | 4262 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__periodonticum | 87.3 | 57.2 | 88.7 | 102.2 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__sp._oral_taxon_203 | 1968 | 1576 | 2223 | 1857 |
| p__Fusobacteria | c__Fusobacteriia | g__Fusobacterium | s__sp._oral_taxon_370 | 34.1 | 36.1 | 37.0 | 29.2 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__buccalis | 399.0 | 365.5 | 436.8 | 368.7 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__goodfellowii | 22.9 | 26.0 | 24.7 | 18.7 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__hofstadii | 346.7 | 418.6 | 364.3 | 283.4 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__hongkongensis | 388.3 | 297.8 | 360.4 | 475.3 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__shahii | 387.0 | 312.2 | 250.1 | 607.1 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_212 | 253.4 | 237.4 | 264.5 | 248.1 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_215 | 107.2 | 117.0 | 107.5 | 101.2 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_219 | 36.9 | 37.3 | 35.4 | 38.6 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_223 | 78.4 | 39.0 | 82.6 | 95.1 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_225 | 244.7 | 299.5 | 328.2 | 105.5 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_392 | 195.9 | 187.9 | 212.8 | 178.3 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_417 | 363.2 | 241.0 | 408.8 | 372.5 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_498 | 210.4 | 135.1 | 176.6 | 296.4 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__sp._oral_taxon_879 | 52.3 | 57.8 | 31.4 | 76.5 |
| p__Fusobacteria | c__Fusobacteriia | g__Leptotrichia | s__wadei | 767.1 | 783.8 | 675.4 | 877.0 |
| p__Gracilibacteria_(GN02) | c__GN02_[C-2] | g__GN02_[G-2] | s__sp._oral_taxon_873 | 24.8 | 31.7 | 18.7 | 29.0 |
| p__Proteobacteria | c__Alphaproteobacteria | g__Bradyrhizobium | s__elkanii | 31.9 | 36.6 | 31.5 | 29.8 |
| p__Proteobacteria | c__Alphaproteobacteria | g__Brevundimonas | s__diminuta | 0.6 | 0.9 | 0.5 | 0.6 |
| p__Proteobacteria | c__Alphaproteobacteria | g__Porphyrobacter | s__tepidarius | 0.2 | 0.1 | 0.1 | 0.3 |
| p__Proteobacteria | c__Alphaproteobacteria | g__Sphingomonas | s__echinoides | 9.2 | 8.0 | 9.3 | 9.7 |
| p__Proteobacteria | c__Alphaproteobacteria | g__Sphingomonas | s__sp._oral_taxon_006 | 0.5 | 0.3 | 0.2 | 1.1 |
| p__Proteobacteria | c__Betaproteobacteria | g__Eikenella | s__corrodens | 264.4 | 284.9 | 279.0 | 233.9 |
| p__Proteobacteria | c__Betaproteobacteria | g__Kingella | s__denitrificans | 159.5 | 149.8 | 146.3 | 182.3 |
| p__Proteobacteria | c__Betaproteobacteria | g__Kingella | s__oralis | 311.9 | 324.2 | 285.2 | 339.8 |
| p__Proteobacteria | c__Betaproteobacteria | g__Lautropia | s__mirabilis | 145.1 | 173.9 | 144.3 | 129.9 |
| p__Proteobacteria | c__Betaproteobacteria | g__Leptothrix | s__sp._oral_taxon_025 | 0.3 | 0.6 | 0.4 | 0.2 |
| p__Proteobacteria | c__Betaproteobacteria | g__Neisseria | s__bacilliformis | 84.2 | 70.1 | 80.2 | 97.4 |
| p__Proteobacteria | c__Betaproteobacteria | g__Neisseria | s__elongata | 499.3 | 682.7 | 506.5 | 387.0 |
| p__Proteobacteria | c__Betaproteobacteria | g__Neisseria | s__flavescens | 440.2 | 302.3 | 352.9 | 631.0 |
| p__Proteobacteria | c__Betaproteobacteria | g__Neisseria | s__oralis | 376.2 | 558.0 | 272.4 | 409.2 |
| p__Proteobacteria | c__Betaproteobacteria | g__Neisseria | s__pharyngis | 70.1 | 86.1 | 41.5 | 98.3 |
| p__Proteobacteria | c__Betaproteobacteria | g__Neisseria | s__sicca | 730.6 | 656.1 | 784.9 | 701.7 |
| p__Proteobacteria | c__Betaproteobacteria | g__Neisseria | s__subflava | 174.1 | 125.9 | 194.7 | 174.4 |
| p__Proteobacteria | c__Betaproteobacteria | g__Ottowia | s__sp._oral_taxon_894 | 70.0 | 88.8 | 70.2 | 59.3 |
| p__Proteobacteria | c__Deltaproteobacteria | g__Desulfobulbus | s__sp._oral_taxon_041 | 139.1 | 91.1 | 132.8 | 174.4 |
| p__Proteobacteria | c__Epsilonproteobacteria | g__Campylobacter | s__concisus | 242.1 | 186.5 | 241.4 | 274.2 |
| p__Proteobacteria | c__Epsilonproteobacteria | g__Campylobacter | s__curvus | 39.6 | 46.6 | 31.6 | 46.1 |
| p__Proteobacteria | c__Epsilonproteobacteria | g__Campylobacter | s__gracilis | 858.3 | 734.1 | 849.7 | 939.1 |
| p__Proteobacteria | c__Epsilonproteobacteria | g__Campylobacter | s__showae | 496.9 | 422.2 | 521.4 | 507.0 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Aggregatibacter | s__actinomycetemcomitans | 49.2 | 68.1 | 46.4 | 42.2 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Aggregatibacter | s__aphrophilus | 312.9 | 232.7 | 392.3 | 254.5 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Aggregatibacter | s__paraphrophilus | 111.5 | 126.7 | 141.8 | 63.6 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Aggregatibacter | s__segnis | 233.8 | 295.3 | 259.7 | 165.6 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Aggregatibacter | s__sp._oral_taxon_458 | 158.1 | 130.8 | 151.6 | 181.8 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Aggregatibacter | s__sp._oral_taxon_513 | 61.5 | 35.1 | 85.1 | 45.7 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Cardiobacterium | s__hominis | 277.9 | 342.4 | 295.3 | 219.1 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Cardiobacterium | s__valvarum | 234.5 | 257.9 | 245.6 | 207.0 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Haemophilus | s__haemolyticus | 50.2 | 95.7 | 34.4 | 45.4 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Haemophilus | s__parahaemolyticus | 78.5 | 71.7 | 121.9 | 26.0 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Haemophilus | s__parainfluenzae | 1042 | 1480 | 940.9 | 927.4 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Haemophilus | s__sp._oral_taxon_036 | 75.0 | 82.2 | 71.0 | 76.2 |
| p__Proteobacteria | c__Gammaproteobacteria | g__Pseudomonas | s__fluorescens | 52.5 | 52.2 | 53.4 | 51.6 |
| p__SR1 | c__SR1_[C-1] | g__SR1_[G-1] | s__sp._oral_taxon_874 | 26.9 | 26.4 | 29.2 | 24.3 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_346 | 1103 | 901.9 | 1098 | 1223 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_347 | 100.0 | 143.7 | 105.6 | 68.1 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_348 | 106.9 | 75.8 | 123.5 | 102.9 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_349 | 1107 | 900.9 | 1062 | 1279 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_352 | 49.4 | 42.8 | 43.6 | 60.5 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_488 | 136.2 | 146.7 | 156.0 | 104.4 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_869 | 125.1 | 120.6 | 128.5 | 123.1 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-1] | s__sp._oral_taxon_952 | 665.5 | 706.5 | 669.6 | 637.1 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-2] | s__sp._oral_taxon_350 | 156.1 | 137.7 | 139.2 | 188.3 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-3] | s__sp._oral_taxon_351 | 33.2 | 21.1 | 35.6 | 36.9 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-5] | s__sp._oral_taxon_356 | 616.8 | 371.0 | 677.8 | 675.5 |
| p__Saccharibacteria_(TM7) | c__TM7_[C-1] | g__TM7_[G-6] | s__sp._oral_taxon_870 | 82.6 | 93.6 | 83.9 | 74.6 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__denticola | 372.7 | 246.3 | 374.0 | 442.1 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__lecithinolyticum | 65.5 | 62.1 | 65.8 | 67.0 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__maltophilum | 83.6 | 56.0 | 73.1 | 112.7 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__medium | 45.7 | 27.4 | 60.7 | 36.3 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__socranskii | 297.0 | 211.8 | 294.6 | 348.0 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__sp._oral_taxon_231 | 102.7 | 75.9 | 117.3 | 98.6 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__sp._oral_taxon_237 | 160.6 | 104.6 | 160.5 | 192.0 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__sp._oral_taxon_247 | 44.0 | 8.2 | 68.9 | 31.7 |
| p__Spirochaetes | c__Spirochaetia | g__Treponema | s__vincentii | 27.6 | 36.1 | 33.3 | 15.4 |
| p__Synergistetes | c__Synergistia | g__Fretibacterium | s__fastidiosum | 551.9 | 462.9 | 595.3 | 545.5 |
| p__Synergistetes | c__Synergistia | g__Fretibacterium | s__sp._oral_taxon_358 | 81.3 | 46.0 | 93.4 | 85.3 |
| p__Synergistetes | c__Synergistia | g__Fretibacterium | s__sp._oral_taxon_359 | 722.3 | 648.0 | 814.1 | 644.6 |
| p__Synergistetes | c__Synergistia | g__Fretibacterium | s__sp._oral_taxon_360 | 1247 | 818.0 | 1325 | 1385 |
| p__Synergistetes | c__Synergistia | g__Fretibacterium | s__sp._oral_taxon_361 | 54.1 | 4.5 | 61.8 | 71.9 |
| p__Synergistetes | c__Synergistia | g__Fretibacterium | s__sp._oral_taxon_362 | 212.8 | 161.8 | 233.9 | 213.9 |
| p__Synergistetes | c__Synergistia | g__Pyramidobacter | s__piscolens | 5.7 | 7.9 | 4.6 | 5.9 |
Fig. 2.
Distribution of phyla among the total reads identified, overall and according to categories of age. Numbers on chart are mean relative abundance. The “other” phylum category comprises 5 phyla ranging in frequency from 0.02 to 3.9%
For three known highly virulent periodontal pathogens, Porphyromonas ginigivalis (Bacteroidetes phylum), Tannerella forsythia (Bacteroidetes phylum), and Treponema denticola (Spirochaetes phylum), overall mean reads were 1055, 577.6, and 372.7, respectively; mean reads for each increased with age. Mean reads for bacteria typically associated with periodontal health (Streptococcus oralis, sanguinis and intermedius; Firmicutes phylum) were 6725, 1128, and 897; each decreasing across incremental age groups. To further evaluate the distribution of the two predominant phyla, we computed the Firmicutes-to-Bacteroidetes ratio by summing the mean reads separately within each of these phyla (Table 2) and then creating a ratio of these sums. The ratio was 1.56 among all women, and increased with age: 1.45 (50–69 years); 1.55 (60–69 years); and 1.61 (≥70 years).
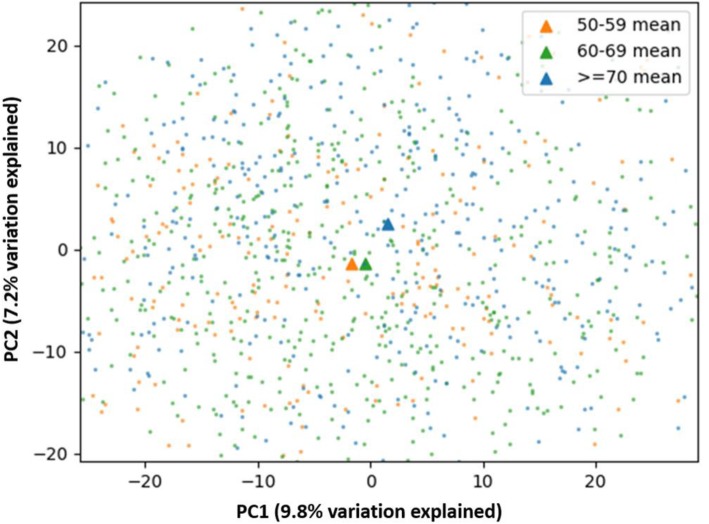
We next evaluated alpha (within-group) and beta (between-group) diversity of the bacterial species in the overall cohort and according to age categories. For alpha (within-group) diversity, mean (SD), OTU count richness, Chao1 richness, and Shannon entropy evenness were 165 (45.1), 185.0 (31.2), and 5.0 (0.7), respectively, among all women, and remained consistent across age categories (Fig. 3). Beta (between-group) diversity is shown in the PCA plot in Fig. 4. A Permutation MANOVA test yielded P = 0.001, suggesting that differences were present in mean vectors across age categories, despite unclear clustering in the PCA plot itself.
Fig. 3.
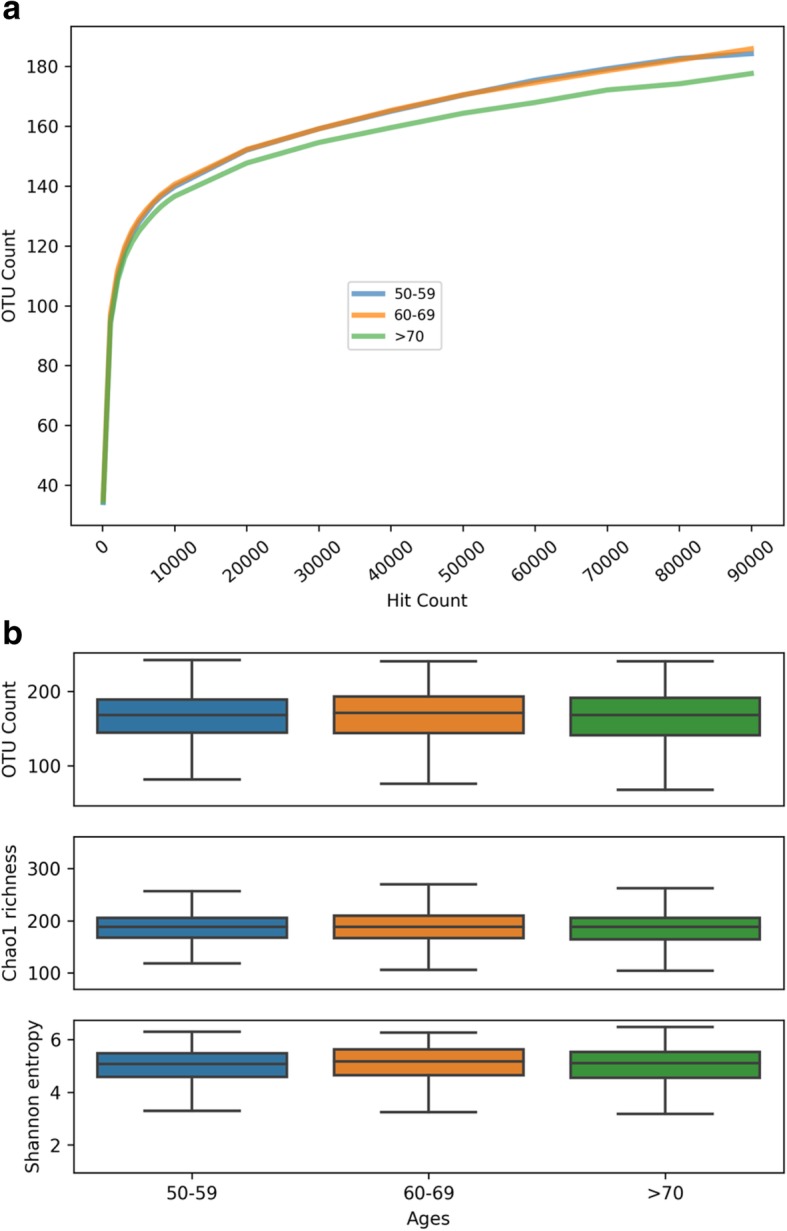
Alpha (within-group) diversity of identified taxa according to age groups. Panel a gives the rarefaction curve and Panel b gives measures of richness (Chao-1, P = 0.55; OTU counts, P = 0.35) and evenness (Shannon entropy, P = 0.42)
Fig. 4.
Beta (between-group) diversity of identified taxa according to age groups. Permutation MANOVA test yielded P = 0.001, suggesting differences are present in mean vectors (triangles) across age categories, despite unclear clustering in the PCA plot
Table 3 presents the top 20 highest and top 20 lowest OTUs based on their CLR mean for the overall cohort and according to age categories. Also shown in Table 3 are linear correlations between these OTUs and age, as well as an indication of whether or not the OTU has previously been cultured and named in the HOMD, and a notation of membership within the Socransky color complex of bacterial species previously identified using targeted methods [34]. Veillonella dispar (CLR mean, 8.3), S. Oralis (CLR mean 8.1), and Veillonella parvula (CLR mean, 7.6) had the greatest abundance, about a 256-fold (28) higher than the overall composition mean. There were 18 (90%) taxa with a 16-fold or greater (CLR mean ≥ 4) elevation in abundance based on CLR mean OTUs. Among the top 20 most abundant bacteria, 19 (95%) were previously named, whereas one (5%) was previously unnamed in HOMD. Among the top 20 taxa were bacteria previously associated with both periodontal health (S. oralis, sanguinis, gordonii, and intermedius) and periodontal disease (V. parvula; Fusobacterium nucleatum; Parvomonas micra; Prevotella nigrescens; Rothia dentocariosa; Actinomyces naeslundii). Ten of the top 20 bacteria were included in Socransky’s complex organization, with four (20%) from the yellow complex typically associated with healthy periodontium and four (20%) from the orange complex which is associated with periodontitis. Among taxa with reduced abundance, Porphyrobacter tepidarius (CLR mean, − 3.6), Sphingomonas sp._oral_taxon 006 (CLR mean, − 3.6), Pyramidobacter piscolens (CLR mean, − 3.5), Leptothirix sp._oral_taxon 025 (CLR mean, − 3.5), and Treponema sp._oral_taxon 247 (CLR mean, − 3.5) each had a 11-fold or lower abundance relative to the overall composition mean. Seven (35%) of the 20 least abundant bacteria have been previously named in HOMD; two (10%) are unnamed; and, 11 (55%) have been phylotyped, but as yet not named.
Table 3.
Top 20 highest* and lowest* mean CLR OTU for the overall cohort and by age categories, and their linear correlation with age
| Rank Order* | OTU Label | Culture Status | Socransky Complex | Overall Cohort (N = 1219) | Age Categories (years) | Linear Correlation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 50–59 (N = 239) | 60–69 (N = 554) | ≥70 (N = 426) | p-value | Pearson r | p-value | |||||
|
CLR OTU Mean (SE) |
CLR OTU Mean (SE) |
CLR OTU Mean (SE) |
CLR OTU Mean (SE) |
|||||||
| 20 Most Abundant Species | ||||||||||
| 1 | Veillonella dispar | N | – | 8.25 (0.06) | 8.23 (0.13) | 8.11 (0.09) | 8.45 (0.10) | 0.045 | 0.08 | 0.008 |
| 2 | Streptococcus oralis | N | Y | 8.06 (0.05) | 8.40 (0.11) | 8.06 (0.08) | 7.87 (0.09) | 0.002 | −0.10 | <.001 |
| 3 | Veillonella parvula | N | P | 7.60 (0.07) | 7.48 (0.15) | 7.45 (0.09) | 7.86 (0.11) | 0.014 | 0.10 | 0.001 |
| 4 | Fusobacterium nucleatum_subsp._vincentii | N | O | 6.43 (0.08) | 6.21 (0.17) | 6.48 (0.11) | 6.50 (0.13) | 0.356 | 0.02 | 0.587 |
| 5 | Selenomonas sputigena | N | – | 5.63 (0.08) | 5.34 (0.17) | 5.44 (0.12) | 6.03 (0.14) | 0.001 | 0.10 | <.001 |
| 6 | Fusobacterium nucleatum_subsp._animalis | N | – | 5.39 (0.07) | 5.29 (0.14) | 5.29 (0.10) | 5.57 (0.12) | 0.136 | 0.10 | 0.106 |
| 7 | Campylobacter gracilis | N | O | 5.19 (0.05) | 5.02 (0.11) | 5.15 (0.07) | 5.33 (0.09) | 0.052 | 0.10 | 0.016 |
| 8 | Fusobacterium nucleatum_subsp._polymorphum | N | O | 5.19 (0.07) | 5.36 (0.15) | 5.28 (0.09) | 4.97 (0.12) | 0.052 | −0.10 | 0.019 |
| 9 | Prevotella oris | N | – | 5.08 (0.09) | 5.25 (0.18) | 5.05 (0.13) | 5.03 (0.16) | 0.646 | −0.10 | 0.118 |
| 10 | Streptococcus sanguinis | N | Y | 4.91 (0.07) | 5.57 (0.15) | 5.04 (0.10) | 4.38 (0.13) | <.001* | −0.18 | <.001* |
| 11 | Corynebacterium matruchotii | N | – | 4.81 (0.07) | 4.87 (0.17) | 4.86 (0.10) | 4.70 (0.12) | 0.550 | −0.04 | 0.171 |
| 12 | Selenomonas noxia | N | – | 4.81 (0.08) | 4.63 (0.18) | 4.69 (0.11) | 5.06 (0.13) | 0.049 | 0.10 | <.001 |
| 13 | Prevotella nigrescens | N | O | 4.31 (0.10) | 4.48 (0.21) | 4.28 (0.15) | 4.26 (0.18) | 0.714 | − 0.03 | 0.370 |
| 14 | Parvimonas micra | N | – | 4.29 (0.08) | 4.08 (0.17) | 4.35 (0.11) | 4.33 (0.12) | 0.401 | 0.00 | 0.967 |
| 15 | Rothia dentocariosa | U | – | 4.28 (0.09) | 4.46 (0.19) | 4.38 (0.13) | 4.04 (0.15) | 0.113 | −0.10 | 0.008 |
| 16 | Fusobacterium sp._oral_taxon_203 | N | – | 4.23 (0.10) | 4.08 (0.22) | 4.36 (0.15) | 4.16 (0.17) | 0.497 | −0.01 | 0.713 |
| 17 | Streptococcus gordonii | N | Y | 4.19 (0.08) | 3.93 (0.18) | 4.15 (0.12) | 4.40 (0.13) | 0.090 | 0.04 | 0.039 |
| 18 | Granulicatella adiacens | N | – | 4.15 (0.06) | 4.23 (0.14) | 4.11 (0.09) | 4.15 (0.10) | 0.749 | 0.00 | 0.991 |
| 19 | Streptococcus intermedius | N | Y | 3.94 (0.10) | 4.33 (0.21) | 3.86 (0.15) | 3.81 (0.16) | 0.119 | −0.10 | 0.016 |
| 20 | Actinomyces naeslundii | N | B | 3.85 (0.06) | 4.00 (0.14) | 3.88 (0.09) | 3.72 (0.11) | 0.230 | −0.10 | 0.008 |
| 20 Least Abundant Species | ||||||||||
| 1 | Porphyrobacter tepidarius | N | – | − 3.58 (0.03) | −3.51 (0.08) | − 3.65 (0.05) | − 3.54 (0.06) | 0.216 | 0.01 | 0.869 |
| 2 | Sphingomonas sp._oral_taxon_006 | P | – | − 3.55 (0.04) | − 3.49 (0.08) | − 3.60 (0.05) | − 3.52 (0.06) | 0.407 | −0.00 | 0.989 |
| 3 | Pyramidobacter piscolens | P | – | − 3.53 (0.04) | − 3.44 (0.10) | − 3.60 (0.06) | − 3.47 (0.07) | 0.251 | − 0.01 | 0.919 |
| 4 | Leptothrix sp._oral_taxon_025 | P | – | −3.50 (0.04) | − 3.37 (0.08) | − 3.53 (0.06) | −3.53 (0.06) | 0.216 | −0.04 | 0.221 |
| 5 | Treponema sp._oral_taxon_247 | P | – | −3.45 (0.05) | −3.35 (0.09) | − 3.50 (0.07) | −3.44 (0.08) | 0.436 | − 0.02 | 0.566 |
| 6 | Atopobium sp._oral_taxon_416 | P | – | −3.38 (0.05) | −3.45 (0.09) | − 3.44 (0.07) | −3.27 (0.09) | 0.202 | 0.10 | 0.042 |
| 7 | Brevundimonas diminuta | N | – | −3.30 (0.04) | −3.17 (0.09) | − 3.35 (0.06) | −3.30 (0.07) | 0.232 | −0.03 | 0.306 |
| 8 | Prevotella multiformis | N | – | −3.08 (0.06) | −3.12 (0.11) | − 3.10 (0.09) | −3.05 (0.10) | 0.901 | 0.03 | 0.359 |
| 9 | GN02_[G-2] sp._oral_taxon_873 | P | – | −3.06 (0.05) | −3.08 (0.12) | − 3.03 (0.08) | −3.10 (0.09) | 0.851 | −0.00 | 0.927 |
| 10 | Streptococcus sobrinus | U | – | −3.04 (0.06) | −3.21 (0.10) | − 3.19 (0.09) | −2.75 (0.14) | 0.005 | 0.11 | <.001* |
| 11 | Aggregatibacter actinomycetemcomitans | P | – | − 3.01 (0.07) | − 2.97 (0.15) | − 3.09 (0.10) | − 2.94 (0.11) | 0.575 | 0.03 | 0.349 |
| 12 | Fretibacterium sp._oral_taxon_361 | P | – | −2.98 (0.06) | −3.18 (0.10) | −2.97 (0.09) | − 2.88 (0.11) | 0.207 | 0.02 | 0.469 |
| 13 | Butyrivibrio sp._oral_taxon_080 | P | – | −2.96 (0.06) | −2.85 (0.13) | − 2.90 (0.09) | −3.10 (0.09) | 0.193 | −0.10 | 0.038 |
| 14 | Lactobacillus gasseri | N | – | −2.95 (0.06) | −3.08 (0.12) | −3.09 (0.09) | −2.70 (0.12) | 0.012 | 0.10 | 0.002 |
| 15 | Mitsuokella sp._oral_taxon_521 | N | – | −2.90 (0.06) | −2.91 (0.12) | − 2.92 (0.08) | −2.87 (0.11) | 0.929 | 0.02 | 0.603 |
| 16 | Microbacterium flavescens | P | – | −2.89 (0.04) | −2.73 (0.09) | − 2.89 (0.06) | −2.97 (0.07) | 0.151 | −0.10 | 0.051 |
| 17 | Prevotella sp._oral_taxon_475 | N | – | −2.87 (0.06) | −2.81 (0.12) | − 2.84 (0.09) | −2.95 (0.09) | 0.575 | −0.04 | 0.157 |
| 18 | Neisseria pharyngis | N | – | −2.87 (0.06) | −2.81 (0.13) | − 2.95 (0.09) | −2.80 (0.11) | 0.479 | 0.01 | 0.760 |
| 19 | Fretibacterium sp._oral_taxon_358 | U | – | −2.84 (0.07) | −2.87 (0.14) | − 2.88 (0.10) | −2.77 (0.12) | 0.708 | 0.02 | 0.494 |
| 20 | Treponema medium | P | – | −2.79 (0.06) | −2.85 (0.14) | − 2.74 (0.10) | −2.81 (0.10) | 0.777 | 0.02 | 0.579 |
*OTUs are ranked according to mean CLR OTU. The 20 most abundant OTUs have positive mean CRL; the 20 least abundant OTUs have negative mean CLR
The CLR OTU can be interpreted as a log [2] fold-difference for the given species relative to the overall compositional geometric mean. A mean CLR of 3 indicates a 8-fold [23] higher abundance, and a mean CLR of −3 indicates a 8-fold lower abundance, relative to the overall compositional geometric mean
SE standard error, Pearson r the Pearson product-moment correlation coefficient
p-values: bolded are significant at alpha .05; asterisk are significant at alpha 0.05 after Bonferroni correction
Culture status annotation in HOMD: N = named; U = unnamed; P = phylotyped
Socransky complex32: R = red; O = orange; P = purple; G = green; Y = yellow; B = blue; −-- = not part of the Socransky classification
Linear correlations (Table 3) among the 20 most abundant bacteria ranged from r = − 0.18 to r = 0.10, with 11 (55%) of the correlations achieving statistical significance (uncorrected P < 0.05; bolded). After Bonferroni correction, only 1 (9%) of these remained statistically significant (S. sanguinis, r = − 0.18; corrected P < 0.001). Among the 20 least abundant bacteria, linear correlations ranged from − 0.10 to 0.11. Four (20%) correlations achieved statistical significance (uncorrected P < 0.05; bolded), of which 1 (25%) remained significant after Bonferonni correction (Streptococcus sobrinus, r = 0.11; corrected P < 0.001).
Differences in mean CLR across age categories achieved statistical significance (P < 0.05) for 8 (40%) of the 20 most abundant bacteria, of which only 1 (12.5%) remained significant following Bonferroni correction (S. sanguinis, corrected P < 0.001). Mean CLR differences across age categories among the least abundant bacteria were significant (P < 0.05) for two bacteria, neither of which remained significant after Bonferroni correction.
Table 4 presents the rank ordered mean CLR OTUs for all 267 taxa identified, as well as their linear correlations with age, culture status and Socransky classification. A total of 148 (55.4%) taxa had names previously annotated in the HOMD database, whereas 60 (22.5%) were unnamed and are OTUs potentially identifying new bacteria. In the overall cohort, 117 (43.8%) taxa demonstrated elevated abundance (CLR > 0), the remaining 150 (57.3%) demonstrating reduced abundance (CLR < 0), relative to the overall composition mean. Twenty eight (10.5%) taxa that demonstrated a 8-fold (i.e., 23) or greater elevation in abundance based on mean CLR OTUs. There were 15 (5.6%) taxa with a 8-fold lower abundance relative to the overall composition mean. Of the virulent periodontal pathogens included in Socransky’s classification, [34] only T. forsythia (mean CLR, 1.87) and F. nucleatum (mean CLR, 6.4) had an elevated abundance, whereas T. denticola (mean CLR, − 0.28), P. gingivalis (mean CLR, − 0.56), P. intermedia (mean CLR, − 1.36) were, on average, in lower abundance. Several bacteria associated with healthy periodontium were in higher abundance: S. oralis (mean CLR, 5.5), sanguinis (mean CLR, 3.4), gordonii (mean CLR, 2.8), and intermedius (mean CLR, 2.6); P. micra (mean CLR, 3.0).
Table 4.
Mean CLR OTUs for each of 267 bacterial species in the overall cohort and by age categories, and its linear correlation with age
| Rank Order* | OTU Label | Culture Status | Socransky Complex | Overall Cohort (N = 1219) |
Age Categories (years) | Linear Correlation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| CLR OTU Mean (SE) |
50–59 (N = 239) CLR OTU Mean (SE) |
60–69 (N = 554) CLR OTU Mean (SE) |
≥70 (N = 426) CLR OTU Mean (SE) |
p-value | Pearson r | p-value | ||||
| 1 | Veillonella dispar | N | – | 8.25 (0.06) | 8.23 (0.13) | 8.11 (0.09) | 8.45 (0.10) | 0.045 | 0.08 | 0.008 |
| 2 | Streptococcus oralis | N | Y | 8.06 (0.05) | 8.40 (0.11) | 8.06 (0.08) | 7.87 (0.09) | 0.002 | −0.10 | <.001 |
| 3 | Veillonella parvula | N | P | 7.60 (0.07) | 7.48 (0.15) | 7.45 (0.09) | 7.86 (0.11) | 0.014 | 0.10 | 0.001 |
| 4 | Fusobacterium nucleatum_subsp._vincentii | N | O | 6.43 (0.08) | 6.21 (0.17) | 6.48 (0.11) | 6.50 (0.13) | 0.356 | 0.02 | 0.587 |
| 5 | Selenomonas sputigena | N | – | 5.63 (0.08) | 5.34 (0.17) | 5.44 (0.12) | 6.03 (0.14) | 0.001 | 0.10 | <.001 |
| 6 | Fusobacterium nucleatum_subsp._animalis | N | – | 5.39 (0.07) | 5.29 (0.14) | 5.29 (0.10) | 5.57 (0.12) | 0.136 | 0.10 | 0.106 |
| 7 | Campylobacter gracilis | N | O | 5.19 (0.05) | 5.02 (0.11) | 5.15 (0.07) | 5.33 (0.09) | 0.052 | 0.10 | 0.016 |
| 8 | Fusobacterium nucleatum_subsp._polymorphum | N | O | 5.19 (0.07) | 5.36 (0.15) | 5.28 (0.09) | 4.97 (0.12) | 0.052 | −0.10 | 0.019 |
| 9 | Prevotella oris | N | – | 5.08 (0.09) | 5.25 (0.18) | 5.05 (0.13) | 5.03 (0.16) | 0.646 | −0.10 | 0.118 |
| 10 | Streptococcus sanguinis | N | Y | 4.91 (0.07) | 5.57 (0.15) | 5.04 (0.10) | 4.38 (0.13) | <.001* | −0.18 | <.001* |
| 11 | Corynebacterium matruchotii | N | – | 4.81 (0.07) | 4.87 (0.17) | 4.86 (0.10) | 4.70 (0.12) | 0.550 | −0.04 | 0.171 |
| 12 | Selenomonas noxia | N | – | 4.81 (0.08) | 4.63 (0.18) | 4.69 (0.11) | 5.06 (0.13) | 0.049 | 0.10 | <.001 |
| 13 | Prevotella nigrescens | N | O | 4.31 (0.10) | 4.48 (0.21) | 4.28 (0.15) | 4.26 (0.18) | 0.714 | −0.03 | 0.370 |
| 14 | Parvimonas micra | N | – | 4.29 (0.08) | 4.08 (0.17) | 4.35 (0.11) | 4.33 (0.12) | 0.401 | 0.00 | 0.967 |
| 15 | Rothia dentocariosa | U | – | 4.28 (0.09) | 4.46 (0.19) | 4.38 (0.13) | 4.04 (0.15) | 0.113 | −0.10 | 0.008 |
| 16 | Fusobacterium sp._oral_taxon_203 | N | – | 4.23 (0.10) | 4.08 (0.22) | 4.36 (0.15) | 4.16 (0.17) | 0.497 | −0.01 | 0.713 |
| 17 | Streptococcus gordonii | N | Y | 4.19 (0.08) | 3.93 (0.18) | 4.15 (0.12) | 4.40 (0.13) | 0.090 | 0.04 | 0.039 |
| 18 | Granulicatella adiacens | N | – | 4.15 (0.06) | 4.23 (0.14) | 4.11 (0.09) | 4.15 (0.10) | 0.749 | 0.00 | 0.991 |
| 19 | Streptococcus intermedius | N | Y | 3.94 (0.10) | 4.33 (0.21) | 3.86 (0.15) | 3.81 (0.16) | 0.119 | −0.10 | 0.016 |
| 20 | Actinomyces naeslundii | N | B | 3.85 (0.06) | 4.00 (0.14) | 3.88 (0.09) | 3.72 (0.11) | 0.230 | −0.10 | 0.008 |
| 21 | TM7_[G-1] sp._oral_taxon_346 | P | – | 3.78 (0.09) | 3.63 (0.19) | 3.90 (0.12) | 3.70 (0.16) | 0.421 | 0.00 | 0.917 |
| 22 | Haemophilus parainfluenzae | N | – | 3.74 (0.09) | 4.21 (0.19) | 3.73 (0.13) | 3.48 (0.15) | 0.014 | −0.10 | 0.004 |
| 23 | Fusobacterium naviforme | N | – | 3.58 (0.08) | 3.61 (0.18) | 3.62 (0.12) | 3.52 (0.14) | 0.852 | −0.03 | 0.294 |
| 24 | Dialister invisus | N | – | 3.51 (0.08) | 3.12 (0.16) | 3.50 (0.11) | 3.75 (0.13) | 0.013 | 0.10 | 0.003 |
| 25 | Capnocytophaga gingivalis | N | G | 3.36 (0.08) | 3.39 (0.18) | 3.45 (0.11) | 3.23 (0.13) | 0.423 | −0.04 | 0.172 |
| 26 | Streptococcus cristatus | N | – | 3.27 (0.07) | 3.17 (0.16) | 3.27 (0.11) | 3.32 (0.13) | 0.776 | 0.01 | 0.754 |
| 27 | Gemella morbillorum | N | – | 3.14 (0.09) | 3.37 (0.20) | 3.35 (0.13) | 2.73 (0.16) | 0.004 | −0.11 | <.001* |
| 28 | Streptococcus salivarius | U | – | 3.02 (0.08) | 3.05 (0.18) | 2.79 (0.13) | 3.29 (0.14) | 0.028 | 0.10 | 0.090 |
| 29 | TM7_[G-1] sp._oral_taxon_349 | N | – | 2.99 (0.10) | 2.71 (0.22) | 3.15 (0.14) | 2.95 (0.18) | 0.251 | 0.04 | 0.204 |
| 30 | Bacteroidales_[G-2] sp._oral_taxon_274 | N | – | 2.96 (0.10) | 2.47 (0.22) | 3.15 (0.14) | 3.00 (0.16) | 0.030 | 0.03 | 0.328 |
| 31 | Campylobacter showae | P | O | 2.89 (0.08) | 3.00 (0.17) | 2.81 (0.11) | 2.93 (0.14) | 0.609 | −0.01 | 0.685 |
| 32 | Veillonella atypica | N | – | 2.88 (0.09) | 2.47 (0.20) | 2.56 (0.13) | 3.53 (0.14) | <.001* | 0.16 | <.001* |
| 33 | Eikenella corrodens | N | – | 2.87 (0.07) | 3.00 (0.15) | 3.07 (0.10) | 2.53 (0.12) | 0.001 | −0.11 | <.001* |
| 34 | Capnocytophaga leadbetteri | N | – | 2.86 (0.09) | 2.87 (0.20) | 2.93 (0.13) | 2.78 (0.16) | 0.758 | −0.03 | 0.334 |
| 35 | Fretibacterium sp._oral_taxon_360 | N | – | 2.81 (0.11) | 2.29 (0.23) | 2.89 (0.16) | 3.00 (0.19) | 0.048 | 0.04 | 0.148 |
| 36 | Prevotella sp._oral_taxon_317 | N | – | 2.73 (0.10) | 2.79 (0.22) | 2.68 (0.16) | 2.76 (0.18) | 0.907 | −0.00 | 0.965 |
| 37 | Capnocytophaga granulosa | U | – | 2.72 (0.09) | 2.39 (0.21) | 2.84 (0.13) | 2.75 (0.16) | 0.189 | 0.04 | 0.152 |
| 38 | Alloprevotella tannerae | N | – | 2.71 (0.12) | 2.69 (0.24) | 2.78 (0.17) | 2.64 (0.20) | 0.865 | 0.01 | 0.788 |
| 39 | Kingella oralis | P | – | 2.66 (0.08) | 2.66 (0.19) | 2.65 (0.11) | 2.68 (0.14) | 0.983 | −0.01 | 0.847 |
| 40 | TM7_[G-1] sp._oral_taxon_952 | U | – | 2.58 (0.10) | 2.80 (0.22) | 2.83 (0.14) | 2.12 (0.16) | 0.002 | −0.10 | <.001 |
| 41 | Selenomonas artemidis | N | – | 2.51 (0.11) | 2.77 (0.24) | 2.39 (0.16) | 2.52 (0.19) | 0.419 | −0.00 | 0.957 |
| 42 | Campylobacter concisus | N | G | 2.41 (0.06) | 2.22 (0.14) | 2.37 (0.10) | 2.56 (0.11) | 0.152 | 0.10 | 0.036 |
| 43 | Actinomyces oris | N | B | 2.36 (0.08) | 2.63 (0.17) | 2.37 (0.11) | 2.18 (0.13) | 0.104 | −0.10 | 0.021 |
| 44 | Treponema socranskii | N | – | 2.22 (0.07) | 1.84 (0.17) | 2.16 (0.11) | 2.52 (0.13) | 0.004 | 0.10 | <.001 |
| 45 | Cardiobacterium hominis | N | – | 2.21 (0.08) | 2.50 (0.19) | 2.45 (0.12) | 1.75 (0.14) | <.001* | −0.11 | <.001 |
| 46 | Leptotrichia wadei | N | – | 2.15 (0.10) | 2.21 (0.23) | 1.94 (0.16) | 2.38 (0.18) | 0.159 | 0.03 | 0.305 |
| 47 | Anaeroglobus geminatus | P | – | 2.12 (0.10) | 1.32 (0.22) | 1.94 (0.15) | 2.79 (0.18) | <.001* | 0.17 | <.001* |
| 48 | Gemella haemolysans | N | – | 2.11 (0.09) | 2.42 (0.20) | 2.10 (0.13) | 1.96 (0.14) | 0.161 | −0.10 | 0.018 |
| 49 | Capnocytophaga sputigena | N | G | 2.11 (0.09) | 2.31 (0.21) | 2.23 (0.13) | 1.84 (0.17) | 0.113 | −0.10 | 0.094 |
| 50 | Rothia aeria | N | – | 2.11 (0.09) | 2.49 (0.20) | 2.42 (0.14) | 1.48 (0.15) | <.001* | −0.15 | <.001* |
| 51 | Bergeyella sp._oral_taxon_322 | N | – | 2.08 (0.07) | 2.38 (0.16) | 2.11 (0.10) | 1.87 (0.11) | 0.028 | −0.10 | 0.008 |
| 52 | Leptotrichia hongkongensis | N | – | 2.02 (0.09) | 2.02 (0.21) | 1.94 (0.14) | 2.13 (0.16) | 0.660 | 0.03 | 0.337 |
| 53 | Rothia mucilaginosa | U | – | 1.95 (0.08) | 1.79 (0.17) | 1.90 (0.11) | 2.09 (0.13) | 0.321 | 0.03 | 0.237 |
| 54 | Prevotella melaninogenica | N | – | 1.90 (0.08) | 1.75 (0.17) | 1.78 (0.12) | 2.14 (0.14) | 0.095 | 0.10 | 0.049 |
| 55 | Actinomyces sp._oral_taxon_169 | N | – | 1.89 (0.09) | 2.28 (0.20) | 1.96 (0.13) | 1.59 (0.15) | 0.016 | −0.10 | <.001 |
| 56 | Tannerella forsythia | N | R | 1.87 (0.10) | 1.50 (0.21) | 1.80 (0.14) | 2.18 (0.17) | 0.042 | 0.10 | 0.029 |
| 57 | Selenomonas sp._oral_taxon_136 | N | – | 1.86 (0.08) | 1.21 (0.18) | 1.76 (0.12) | 2.36 (0.15) | <.001* | 0.16 | <.001* |
| 58 | Catonella morbi | N | – | 1.86 (0.08) | 2.08 (0.17) | 1.89 (0.11) | 1.69 (0.13) | 0.184 | −0.10 | 0.020 |
| 59 | Prevotella denticola | N | – | 1.74 (0.10) | 1.46 (0.23) | 1.60 (0.15) | 2.10 (0.18) | 0.040 | 0.10 | 0.006 |
| 60 | Neisseria sicca | N | – | 1.72 (0.11) | 1.58 (0.25) | 1.88 (0.16) | 1.60 (0.18) | 0.416 | −0.01 | 0.678 |
| 61 | Peptostreptococcaceae_[XI][G-9] [Eubacterium]_brac | U | – | 1.70 (0.08) | 2.08 (0.16) | 1.72 (0.12) | 1.47 (0.14) | 0.030 | −0.10 | 0.003 |
| 62 | Neisseria elongata | N | – | 1.63 (0.10) | 1.95 (0.25) | 1.81 (0.15) | 1.22 (0.17) | 0.012 | −0.10 | 0.008 |
| 63 | Cardiobacterium valvarum | U | – | 1.49 (0.09) | 1.60 (0.21) | 1.63 (0.13) | 1.26 (0.14) | 0.137 | −0.10 | 0.044 |
| 64 | Porphyromonas sp._oral_taxon_279 | P | – | 1.49 (0.09) | 1.46 (0.20) | 1.62 (0.13) | 1.34 (0.16) | 0.394 | −0.02 | 0.494 |
| 65 | Selenomonas infelix | U | – | 1.48 (0.08) | 1.56 (0.17) | 1.44 (0.12) | 1.50 (0.15) | 0.871 | −0.01 | 0.709 |
| 66 | Leptotrichia sp._oral_taxon_212 | U | – | 1.44 (0.09) | 1.63 (0.21) | 1.64 (0.13) | 1.08 (0.16) | 0.013 | −0.10 | 0.011 |
| 67 | Actinomyces sp._oral_taxon_180 | N | – | 1.44 (0.07) | 1.54 (0.15) | 1.45 (0.10) | 1.38 (0.11) | 0.704 | −0.03 | 0.235 |
| 68 | Prevotella sp._oral_taxon_300 | N | – | 1.43 (0.09) | 1.31 (0.19) | 1.24 (0.13) | 1.75 (0.15) | 0.025 | 0.10 | 0.007 |
| 69 | Prevotella maculosa | N | – | 1.42 (0.08) | 1.07 (0.17) | 1.32 (0.11) | 1.75 (0.13) | 0.003 | 0.10 | 0.001 |
| 70 | Streptococcus mutans | N | – | 1.39 (0.11) | 0.98 (0.24) | 1.19 (0.16) | 1.87 (0.20) | 0.005 | 0.10 | <.001 |
| 71 | Actinomyces massiliensis | N | B | 1.38 (0.07) | 1.68 (0.17) | 1.50 (0.10) | 1.05 (0.11) | 0.002 | −0.12 | <.001* |
| 72 | Fretibacterium fastidiosum | N | – | 1.30 (0.10) | 1.12 (0.22) | 1.25 (0.15) | 1.47 (0.17) | 0.393 | 0.04 | 0.173 |
| 73 | Veillonellaceae_[G-1] sp._oral_taxon_150 | N | – | 1.23 (0.09) | 0.74 (0.20) | 1.00 (0.13) | 1.81 (0.16) | <.001* | 0.14 | <.001* |
| 74 | Neisseria flavescens | N | – | 1.23 (0.10) | 0.98 (0.21) | 1.28 (0.14) | 1.30 (0.17) | 0.425 | 0.02 | 0.533 |
| 75 | Prevotella oulorum | P | – | 1.22 (0.09) | 1.23 (0.20) | 1.00 (0.13) | 1.50 (0.15) | 0.041 | 0.10 | 0.059 |
| 76 | Streptococcus anginosus | N | Y | 1.17 (0.11) | 1.03 (0.23) | 0.92 (0.16) | 1.56 (0.19) | 0.024 | 0.10 | 0.013 |
| 77 | Lachnoanaerobaculum saburreum | N | – | 1.07 (0.08) | 0.85 (0.18) | 1.12 (0.12) | 1.13 (0.14) | 0.384 | 0.04 | 0.208 |
| 78 | Veillonellaceae_[G-1] sp._oral_taxon_155 | N | – | 1.00 (0.09) | 0.66 (0.20) | 0.84 (0.13) | 1.39 (0.15) | 0.004 | 0.10 | <.001 |
| 79 | Selenomonas sp._oral_taxon_892 | L | – | 0.98 (0.08) | 1.18 (0.19) | 1.05 (0.12) | 0.76 (0.15) | 0.148 | −0.10 | 0.030 |
| 80 | Actinomyces johnsonii | N | B | 0.97 (0.07) | 1.18 (0.16) | 0.92 (0.11) | 0.92 (0.12) | 0.365 | −0.04 | 0.229 |
| 81 | Selenomonas sp._oral_taxon_137 | U | – | 0.95 (0.11) | 0.95 (0.25) | 1.21 (0.17) | 0.61 (0.20) | 0.064 | −0.04 | 0.145 |
| 82 | Corynebacterium durum | U | – | 0.93 (0.08) | 1.53 (0.19) | 1.07 (0.12) | 0.42 (0.13) | <.001* | −0.15 | <.001* |
| 83 | Lautropia mirabilis | U | – | 0.88 (0.09) | 1.28 (0.21) | 0.99 (0.13) | 0.53 (0.15) | 0.006 | −0.10 | <.001 |
| 84 | Veillonella rogosae | N | – | 0.87 (0.09) | 1.06 (0.20) | 1.08 (0.13) | 0.49 (0.16) | 0.010 | −0.10 | <.001 |
| 85 | Leptotrichia sp._oral_taxon_417 | N | – | 0.86 (0.09) | 0.41 (0.20) | 0.95 (0.13) | 1.01 (0.16) | 0.048 | 0.10 | 0.006 |
| 86 | Selenomonas flueggei | N | – | 0.81 (0.08) | 0.58 (0.18) | 0.81 (0.12) | 0.93 (0.14) | 0.317 | 0.04 | 0.122 |
| 87 | Selenomonas sp._oral_taxon_134 | P | – | 0.77 (0.10) | 0.55 (0.21) | 0.75 (0.14) | 0.94 (0.18) | 0.367 | 0.03 | 0.251 |
| 88 | Streptococcus parasanguinis_II | U | – | 0.74 (0.09) | 0.58 (0.19) | 0.49 (0.13) | 1.15 (0.15) | 0.003 | 0.11 | <.001* |
| 89 | Megasphaera micronuciformis | N | – | 0.70 (0.08) | 0.47 (0.19) | 0.46 (0.13) | 1.14 (0.14) | <.001 | 0.12 | <.001* |
| 90 | Capnocytophaga sp._oral_taxon_336 | N | – | 0.69 (0.09) | 0.68 (0.19) | 0.69 (0.13) | 0.71 (0.16) | 0.991 | 0.02 | 0.480 |
| 91 | Oribacterium sp._oral_taxon_078 | U | – | 0.66 (0.08) | −0.03 (0.17) | 0.51 (0.11) | 1.26 (0.14) | <.001* | 0.18 | <.001* |
| 92 | Actinomyces sp._oral_taxon_171 | P | – | 0.66 (0.08) | 0.82 (0.18) | 0.66 (0.12) | 0.57 (0.13) | 0.527 | −0.10 | 0.090 |
| 93 | Lachnospiraceae_[G-3] sp._oral_taxon_100 | U | – | 0.59 (0.08) | 0.60 (0.18) | 0.73 (0.12) | 0.41 (0.14) | 0.200 | −0.03 | 0.239 |
| 94 | Parvimonas sp._oral_taxon_393 | U | – | 0.58 (0.11) | 1.11 (0.24) | 0.63 (0.16) | 0.22 (0.17) | 0.010 | −0.12 | <.001* |
| 95 | Actinomyces gerencseriae | N | B | 0.56 (0.07) | 0.49 (0.17) | 0.37 (0.11) | 0.86 (0.13) | 0.012 | 0.10 | 0.008 |
| 96 | Dialister pneumosintes | P | – | 0.55 (0.09) | 0.64 (0.20) | 0.42 (0.14) | 0.65 (0.16) | 0.477 | 0.03 | 0.357 |
| 97 | Kingella denitrificans | N | – | 0.54 (0.09) | 0.21 (0.20) | 0.45 (0.13) | 0.84 (0.15) | 0.025 | 0.10 | 0.024 |
| 98 | Porphyromonas endodontalis | N | – | 0.52 (0.12) | 1.09 (0.27) | 0.56 (0.17) | 0.13 (0.20) | 0.015 | −0.10 | <.001 |
| 99 | TM7_[G-5] sp._oral_taxon_356 | P | – | 0.49 (0.11) | 0.30 (0.23) | 0.61 (0.16) | 0.43 (0.19) | 0.517 | 0.02 | 0.580 |
| 100 | Selenomonas sp._oral_taxon_919 | U | – | 0.47 (0.08) | 0.72 (0.18) | 0.45 (0.12) | 0.35 (0.15) | 0.287 | −0.10 | 0.096 |
| 101 | Leptotrichia buccalis | N | – | 0.45 (0.10) | 0.23 (0.23) | 0.77 (0.14) | 0.16 (0.16) | 0.011 | −0.03 | 0.375 |
| 102 | Prevotella sp._oral_taxon_472 | N | – | 0.45 (0.10) | 0.65 (0.23) | 0.80 (0.15) | −0.13 (0.17) | <.001* | −0.10 | <.001 |
| 103 | Capnocytophaga sp._oral_taxon_326 | P | – | 0.41 (0.10) | 0.38 (0.23) | 0.62 (0.15) | 0.15 (0.17) | 0.121 | −0.10 | 0.080 |
| 104 | Streptococcus constellatus | N | O | 0.35 (0.10) | 0.25 (0.21) | 0.18 (0.15) | 0.63 (0.17) | 0.114 | 0.04 | 0.139 |
| 105 | Prevotella salivae | N | – | 0.34 (0.08) | −0.22 (0.18) | 0.32 (0.12) | 0.69 (0.14) | <.001 | 0.15 | <.001* |
| 106 | Leptotrichia sp._oral_taxon_392 | U | – | 0.33 (0.09) | 0.49 (0.20) | 0.58 (0.13) | − 0.08 (0.15) | 0.003 | −0.10 | 0.001 |
| 107 | Actinobaculum sp._oral_taxon_183 | N | – | 0.32 (0.08) | 0.29 (0.17) | 0.28 (0.12) | 0.40 (0.14) | 0.765 | 0.01 | 0.742 |
| 108 | Neisseria oralis | U | – | 0.31 (0.11) | 0.47 (0.25) | 0.35 (0.15) | 0.19 (0.18) | 0.618 | −0.04 | 0.133 |
| 109 | Atopobium rimae | N | – | 0.21 (0.09) | −0.08 (0.19) | 0.17 (0.13) | 0.43 (0.16) | 0.124 | 0.10 | 0.108 |
| 110 | Leptotrichia hofstadii | N | – | 0.20 (0.10) | 0.10 (0.22) | 0.27 (0.15) | 0.17 (0.17) | 0.804 | 0.01 | 0.743 |
| 111 | Streptococcus sp._oral_taxon_074 | N | – | 0.15 (0.07) | 0.15 (0.16) | 0.25 (0.11) | 0.02 (0.12) | 0.340 | −0.04 | 0.154 |
| 112 | Fretibacterium sp._oral_taxon_359 | L | – | 0.15 (0.11) | 0.14 (0.25) | 0.04 (0.16) | 0.30 (0.18) | 0.537 | 0.02 | 0.552 |
| 113 | Leptotrichia shahii | U | – | 0.11 (0.10) | 0.30 (0.22) | −0.09 (0.15) | 0.26 (0.19) | 0.209 | 0.03 | 0.298 |
| 114 | Tannerella sp._oral_taxon_286 | U | – | 0.11 (0.07) | − 0.15 (0.16) | 0.26 (0.10) | 0.06 (0.13) | 0.101 | 0.03 | 0.370 |
| 115 | Porphyromonas sp._oral_taxon_284 | N | – | 0.08 (0.10) | 0.14 (0.21) | 0.22 (0.14) | − 0.14 (0.16) | 0.223 | −0.10 | 0.080 |
| 116 | Peptostreptococcaceae_[XI][G-7] [Eubacterium]_yuri | P | – | 0.05 (0.10) | 0.50 (0.21) | 0.19 (0.14) | −0.40 (0.16) | 0.001 | −0.14 | <.001* |
| 117 | Selenomonas sp._oral_taxon_146 | L | – | 0.04 (0.08) | 0.04 (0.18) | − 0.03 (0.12) | 0.14 (0.14) | 0.659 | 0.014 | 0.633 |
| 118 | Fusobacterium periodonticum | N | O | −0.02 (0.07) | −0.12 (0.16) | 0.13 (0.11) | −0.17 (0.13) | 0.174 | −0.02 | 0.488 |
| 119 | Actinomyces meyeri | L | B | −0.04 (0.08) | 0.29 (0.18) | 0.05 (0.12) | − 0.35 (0.13) | 0.010 | −0.11 | <.001* |
| 120 | Leptotrichia sp._oral_taxon_215 | N | – | − 0.05 (0.08) | 0.16 (0.18) | 0.01 (0.12) | − 0.24 (0.14) | 0.157 | −0.04 | 0.170 |
| 121 | Prevotella oralis | N | – | − 0.06 (0.10) | −0.34 (0.20) | − 0.13 (0.14) | 0.20 (0.17) | 0.105 | 0.10 | 0.030 |
| 122 | Prevotella pleuritidis | N | – | − 0.10 (0.12) | −0.07 (0.27) | 0.09 (0.18) | −0.38 (0.20) | 0.211 | −0.04 | 0.190 |
| 123 | Abiotrophia defectiva | N | – | −0.15 (0.09) | −0.05 (0.19) | − 0.01 (0.13) | − 0.38 (0.14) | 0.138 | − 0.10 | 0.028 |
| 124 | Gemella sanguinis | N | – | −0.19 (0.07) | −0.16 (0.15) | − 0.35 (0.10) | − 0.00 (0.12) | 0.077 | 0.04 | 0.204 |
| 125 | Fusobacterium nucleatum_subsp._nucleatum | N | O | −0.25 (0.08) | −0.44 (0.17) | − 0.30 (0.11) | − 0.08 (0.14) | 0.209 | 0.03 | 0.259 |
| 126 | Atopobium parvulum | N | – | − 0.25 (0.08) | −0.28 (0.17) | − 0.47 (0.11) | 0.06 (0.14) | 0.009 | 0.10 | 0.005 |
| 127 | Aggregatibacter sp._oral_taxon_458 | N | – | − 0.25 (0.09) | − 0.11 (0.19) | − 0.17 (0.13) | − 0.44 (0.15) | 0.309 | −0.10 | 0.047 |
| 128 | Streptococcus parasanguinis_I | N | – | − 0.26 (0.08) | −0.29 (0.17) | − 0.48 (0.11) | 0.05 (0.13) | 0.008 | 0.10 | 0.008 |
| 129 | Aggregatibacter aphrophilus | N | – | − 0.27 (0.11) | −0.18 (0.25) | − 0.01 (0.17) | − 0.66 (0.18) | 0.027 | − 0.10 | 0.015 |
| 130 | Treponema denticola | N | R | −0.28 (0.10) | −0.16 (0.22) | − 0.25 (0.15) | − 0.39 (0.18) | 0.692 | − 0.04 | 0.211 |
| 131 | Prevotella saccharolytica | U | – | − 0.37 (0.07) | − 0.38 (0.16) | − 0.26 (0.11) | − 0.51 (0.13) | 0.336 | − 0.01 | 0.663 |
| 132 | TM7_[G-1] sp._oral_taxon_488 | P | – | − 0.39 (0.10) | − 0.39 (0.22) | − 0.11 (0.15) | − 0.76 (0.16) | 0.015 | −0.04 | 0.132 |
| 133 | Johnsonella ignava | U | – | − 0.41 (0.09) | −0.50 (0.21) | − 0.34 (0.14) | − 0.45 (0.16) | 0.783 | − 0.01 | 0.783 |
| 134 | Actinomyces israelii | N | B | −0.47 (0.06) | −0.65 (0.13) | − 0.52 (0.09) | − 0.30 (0.11) | 0.091 | 0.10 | 0.012 |
| 135 | Lachnoanaerobaculum umeaense | P | – | − 0.47 (0.07) | −0.35 (0.17) | − 0.39 (0.11) | − 0.65 (0.12) | 0.198 | −0.03 | 0.316 |
| 136 | Streptococcus sp._oral_taxon_056 | U | – | − 0.49 (0.08) | −0.38 (0.19) | − 0.37 (0.12) | − 0.73 (0.14) | 0.127 | −0.04 | 0.050 |
| 137 | Olsenella sp._oral_taxon_807 | N | – | − 0.49 (0.07) | −0.93 (0.15) | − 0.50 (0.10) | − 0.25 (0.12) | 0.002 | 0.11 | <.001* |
| 138 | Solobacterium moorei | U | – | − 0.51 (0.06) | −0.49 (0.15) | − 0.60 (0.09) | − 0.42 (0.11) | 0.437 | 0.04 | 0.221 |
| 139 | Treponema maltophilum | P | – | − 0.53 (0.08) | −0.78 (0.16) | − 0.59 (0.11) | − 0.32 (0.13) | 0.070 | 0.10 | 0.040 |
| 140 | TM7_[G-1] sp._oral_taxon_348 | N | – | − 0.53 (0.08) | − 0.66 (0.17) | − 0.29 (0.13) | − 0.77 (0.14) | 0.026 | − 0.03 | 0.373 |
| 141 | Porphyromonas gingivalis | N | R | −0.56 (0.12) | −0.70 (0.26) | − 0.60 (0.17) | − 0.43 (0.22) | 0.688 | 0.03 | 0.348 |
| 142 | Centipeda periodontii | N | – | − 0.60 (0.08) | − 0.94 (0.15) | − 0.55 (0.11) | − 0.47 (0.13) | 0.074 | 0.06 | 0.031 |
| 143 | Selenomonas sp._oral_taxon_126 | N | – | − 0.60 (0.08) | −0.77 (0.18) | − 0.56 (0.12) | − 0.56 (0.14) | 0.588 | 0.03 | 0.386 |
| 144 | Desulfobulbus sp._oral_taxon_041 | U | – | −0.66 (0.10) | −1.02 (0.20) | − 0.70 (0.14) | − 0.40 (0.17) | 0.064 | 0.10 | 0.027 |
| 145 | Alloprevotella rava | U | – | −0.68 (0.08) | −0.78 (0.18) | − 0.64 (0.12) | − 0.69 (0.14) | 0.809 | 0.02 | 0.464 |
| 146 | Actinobaculum sp._oral_taxon_848 | P | – | − 0.70 (0.08) | −0.83 (0.17) | − 0.82 (0.11) | −0.46 (0.13) | 0.080 | 0.10 | 0.016 |
| 147 | Stomatobaculum longum | N | – | − 0.70 (0.07) | − 0.95 (0.16) | − 0.76 (0.11) | −0.48 (0.13) | 0.054 | 0.11 | <.001 |
| 148 | Prevotella histicola | N | – | − 0.75 (0.09) | − 0.96 (0.18) | −1.04 (0.13) | − 0.26 (0.16) | <.001 | 0.11 | <.001* |
| 149 | Aggregatibacter segnis | U | – | − 0.79 (0.10) | − 0.42 (0.22) | − 0.69 (0.14) | −1.13 (0.15) | 0.020 | − 0.11 | <.001* |
| 150 | Leptotrichia sp._oral_taxon_225 | N | – | − 0.80 (0.10) | − 0.43 (0.22) | − 0.60 (0.15) | − 1.25 (0.15) | 0.002 | − 0.11 | <.001* |
| 151 | Ruminococcaceae_[G-1] sp._oral_taxon_075 | N | – | − 0.80 (0.08) | − 0.79 (0.17) | − 0.69 (0.12) | −0.94 (0.13) | 0.350 | −0.03 | 0.341 |
| 152 | Peptostreptococcus stomatis | U | – | − 0.80 (0.09) | −0.50 (0.19) | − 0.77 (0.13) | − 1.02 (0.14) | 0.085 | − 0.10 | 0.032 |
| 153 | Porphyromonas catoniae | P | – | − 0.92 (0.09) | − 0.50 (0.20) | − 0.79 (0.13) | −1.31 (0.14) | 0.002 | − 0.10 | <.001 |
| 154 | Actinomyces sp._oral_taxon_178 | P | – | −0.93 (0.06) | − 1.15 (0.14) | − 0.94 (0.09) | − 0.78 (0.10) | 0.099 | 0.05 | 0.080 |
| 155 | Prevotella pallens | N | – | − 0.93 (0.08) | −1.00 (0.17) | − 1.05 (0.11) | −0.73 (0.14) | 0.191 | 0.10 | 0.054 |
| 156 | Filifactor alocis | U | – | − 0.94 (0.10) | −0.66 (0.23) | − 0.98 (0.15) | − 1.05 (0.18) | 0.382 | − 0.10 | 0.077 |
| 157 | Selenomonas sp._oral_taxon_936 | P | – | −0.96 (0.08) | − 0.91 (0.17) | −1.05 (0.12) | − 0.87 (0.14) | 0.578 | 0.03 | 0.323 |
| 158 | TM7_[G-1] sp._oral_taxon_347 | N | – | − 0.97 (0.09) | −0.64 (0.20) | − 0.95 (0.14) | −1.17 (0.14) | 0.107 | − 0.10 | 0.071 |
| 159 | Haemophilus sp._oral_taxon_036 | N | – | −1.01 (0.09) | − 0.65 (0.19) | − 1.00 (0.13) | − 1.21 (0.15) | 0.075 | − 0.10 | 0.002 |
| 160 | Prevotella sp._oral_taxon_292 | P | – | − 1.06 (0.08) | − 1.34 (0.17) | − 1.15 (0.12) | −0.79 (0.15) | 0.040 | 0.10 | 0.001 |
| 161 | Leptotrichia sp._oral_taxon_498 | N | – | − 1.07 (0.09) | −1.28 (0.20) | − 1.20 (0.13) | −0.77 (0.16) | 0.059 | 0.10 | 0.013 |
| 162 | Capnocytophaga sp._oral_taxon_338 | U | – | − 1.08 (0.09) | −1.17 (0.20) | − 1.12 (0.13) | −0.98 (0.15) | 0.671 | 0.01 | 0.808 |
| 163 | Alloprevotella sp._oral_taxon_308 | N | – | − 1.14 (0.07) | −0.90 (0.14) | −1.28 (0.10) | − 1.10 (0.12) | 0.111 | −0.01 | 0.799 |
| 164 | Treponema sp._oral_taxon_231 | P | – | − 1.15 (0.09) | −1.04 (0.18) | − 1.03 (0.13) | − 1.36 (0.15) | 0.206 | − 0.10 | 0.074 |
| 165 | TM7_[G-1] sp._oral_taxon_352 | N | – | − 1.15 (0.07) | − 1.31 (0.15) | − 1.11 (0.10) | − 1.11 (0.12) | 0.545 | 0.04 | 0.173 |
| 166 | Prevotella buccae | U | – | − 1.23 (0.07) | −1.71 (0.14) | − 1.36 (0.11) | − 0.78 (0.12) | <.001* | 0.14 | <.001* |
| 167 | Prevotella loescheii | N | – | −1.24 (0.09) | −0.96 (0.21) | −0.97 (0.14) | − 1.74 (0.15) | <.001 | − 0.10 | 0.001 |
| 168 | Capnocytophaga sp._oral_taxon_864 | U | – | − 1.27 (0.08) | −0.98 (0.20) | −1.15 (0.13) | − 1.57 (0.13) | 0.020 | − 0.10 | 0.005 |
| 169 | Actinomyces sp._oral_taxon_170 | U | – | −1.29 (0.08) | − 0.99 (0.19) | − 1.16 (0.13) | − 1.62 (0.13) | 0.011 | − 0.10 | <.001 |
| 170 | Lachnoanaerobaculum orale | U | – | − 1.31 (0.07) | − 1.09 (0.16) | − 1.46 (0.09) | −1.23 (0.12) | 0.085 | 0.02 | 0.513 |
| 171 | Ottowia sp._oral_taxon_894 | N | – | − 1.31 (0.08) | −1.44 (0.18) | − 1.24 (0.12) | − 1.33 (0.14) | 0.662 | −0.00 | 0.938 |
| 172 | Streptococcus lactarius | N | – | − 1.32 (0.07) | − 1.06 (0.15) | − 1.46 (0.10) | − 1.30 (0.12) | 0.097 | −0.02 | 0.414 |
| 173 | Peptostreptococcaceae_[XI][G-1] [Eubacterium]_infi | P | – | − 1.33 (0.07) | − 1.59 (0.15) | − 1.41 (0.09) | − 1.08 (0.12) | 0.014 | 0.10 | 0.002 |
| 174 | Leptotrichia sp._oral_taxon_219 | N | – | − 1.34 (0.07) | −1.39 (0.17) | − 1.27 (0.11) | −1.39 (0.12) | 0.736 | −0.01 | 0.717 |
| 175 | Prevotella intermedia | P | O | −1.36 (0.11) | −1.31 (0.25) | − 1.35 (0.17) | − 1.41 (0.18) | 0.943 | −0.04 | 0.181 |
| 176 | Veillonella denticariosi | U | – | − 1.37 (0.09) | − 1.51 (0.19) | − 1.39 (0.13) | − 1.27 (0.15) | 0.621 | 0.04 | 0.156 |
| 177 | Fretibacterium sp._oral_taxon_362 | N | – | − 1.43 (0.08) | −1.48 (0.19) | − 1.43 (0.12) | − 1.42 (0.14) | 0.965 | 0.00 | 0.888 |
| 178 | Bacteroidetes_[G-5] sp._oral_taxon_511 | U | – | − 1.47 (0.09) | − 1.44 (0.19) | − 1.43 (0.14) | − 1.54 (0.15) | 0.846 | −0.03 | 0.378 |
| 179 | Prevotella sp._oral_taxon_313 | P | – | − 1.49 (0.08) | −1.14 (0.18) | − 1.75 (0.11) | − 1.35 (0.14) | 0.007 | 0.02 | 0.431 |
| 180 | TM7_[G-3] sp._oral_taxon_351 | P | – | − 1.50 (0.06) | −1.84 (0.13) | − 1.39 (0.10) | − 1.45 (0.11) | 0.029 | 0.10 | 0.018 |
| 181 | Streptococcus australis | N | – | − 1.51 (0.07) | −1.62 (0.15) | − 1.57 (0.11) | − 1.38 (0.13) | 0.373 | 0.03 | 0.295 |
| 182 | Granulicatella elegans | N | – | − 1.59 (0.07) | −1.18 (0.17) | − 1.72 (0.11) | −1.65 (0.12) | 0.023 | −0.10 | 0.067 |
| 183 | TM7_[G-1] sp._oral_taxon_869 | P | – | − 1.60 (0.09) | −1.64 (0.20) | − 1.55 (0.14) | − 1.64 (0.15) | 0.890 | 0.01 | 0.663 |
| 184 | Capnocytophaga sp._oral_taxon_902 | N | – | −1.65 (0.08) | − 1.56 (0.19) | − 1.68 (0.12) | −1.67 (0.14) | 0.860 | −0.04 | 0.220 |
| 185 | Prevotella dentalis | P | – | −1.66 (0.08) | −1.65 (0.19) | − 1.82 (0.12) | −1.47 (0.14) | 0.180 | 0.03 | 0.326 |
| 186 | Campylobacter curvus | N | – | − 1.67 (0.07) | −1.57 (0.17) | − 1.79 (0.10) | − 1.58 (0.13) | 0.365 | 0.03 | 0.335 |
| 187 | Pseudoramibacter alactolyticus | P | – | − 1.68 (0.08) | − 2.18 (0.17) | −1.73 (0.12) | − 1.35 (0.15) | 0.001 | 0.12 | <.001* |
| 188 | TM7_[G-6] sp._oral_taxon_870 | N | – | − 1.71 (0.08) | −1.47 (0.18) | − 1.69 (0.12) | − 1.85 (0.13) | 0.220 | −0.10 | 0.085 |
| 189 | Veillonellaceae_[G-1] sp._oral_taxon_129 | N | – | − 1.71 (0.08) | − 1.96 (0.16) | − 1.80 (0.11) | − 1.46 (0.14) | 0.042 | 0.10 | 0.010 |
| 190 | Tannerella sp._oral_taxon_808 | U | – | − 1.75 (0.07) | −2.03 (0.14) | −1.70 (0.10) | − 1.65 (0.12) | 0.104 | 0.10 | 0.011 |
| 191 | Peptostreptococcaceae_[XI][G-6] [Eubacterium]_noda | L | – | − 1.75 (0.08) | − 1.67 (0.18) | − 1.90 (0.11) | −1.61 (0.14) | 0.238 | 0.02 | 0.425 |
| 192 | Mitsuokella sp._oral_taxon_131 | N | – | − 1.79 (0.09) | −2.18 (0.17) | − 1.96 (0.13) | − 1.36 (0.16) | <.001 | 0.14 | <.001* |
| 193 | Mycoplasma salivarium | P | – | − 1.80 (0.07) | −1.77 (0.15) | − 1.87 (0.11) | − 1.71 (0.12) | 0.579 | 0.03 | 0.302 |
| 194 | Haemophilus haemolyticus | P | – | − 1.81 (0.08) | −1.30 (0.19) | − 1.92 (0.12) | − 1.94 (0.12) | 0.006 | −0.10 | 0.043 |
| 195 | Bergeyella sp._oral_taxon_907 | P | – | −1.81 (0.07) | −1.86 (0.15) | − 1.72 (0.10) | − 1.92 (0.12) | 0.405 | − 0.01 | 0.765 |
| 196 | TM7_[G-2] sp._oral_taxon_350 | N | – | − 1.82 (0.09) | − 1.86 (0.18) | − 1.82 (0.13) | − 1.80 (0.15) | 0.968 | 0.02 | 0.412 |
| 197 | Prevotella veroralis | U | – | − 1.82 (0.08) | −1.73 (0.18) | − 1.78 (0.13) | − 1.93 (0.13) | 0.582 | −0.02 | 0.413 |
| 198 | Aggregatibacter paraphrophilus | N | – | − 1.82 (0.09) | −1.48 (0.21) | − 1.69 (0.14) | −2.19 (0.13) | 0.007 | −0.10 | <.001 |
| 199 | Leptotrichia sp._oral_taxon_223 | P | – | − 1.82 (0.08) | −1.71 (0.17) | − 1.98 (0.11) | − 1.69 (0.14) | 0.206 | 0.02 | 0.499 |
| 200 | Bifidobacterium dentium | N | – | − 1.85 (0.08) | −2.10 (0.18) | −2.12 (0.12) | −1.37 (0.15) | <.001* | 0.15 | <.001* |
| 201 | Alloprevotella sp._oral_taxon_473 | U | – | − 1.87 (0.08) | −1.63 (0.17) | − 1.88 (0.12) | − 1.98 (0.13) | 0.300 | −0.10 | 0.030 |
| 202 | Veillonellaceae_[G-1] sp._oral_taxon_145 | U | – | − 1.88 (0.08) | −1.98 (0.16) | − 1.83 (0.12) | − 1.88 (0.14) | 0.768 | 0.02 | 0.551 |
| 203 | Neisseria bacilliformis | N | – | −1.89 (0.09) | −1.82 (0.20) | − 1.90 (0.13) | − 1.93 (0.14) | 0.906 | −0.04 | 0.194 |
| 204 | Capnocytophaga sp._oral_taxon_412 | N | – | − 1.89 (0.08) | −1.74 (0.18) | − 1.81 (0.12) | −2.09 (0.13) | 0.173 | − 0.04 | 0.162 |
| 205 | Neisseria subflava | N | – | − 1.89 (0.09) | − 1.84 (0.18) | −2.04 (0.13) | − 1.73 (0.16) | 0.267 | 0.10 | 0.118 |
| 206 | Veillonella sp._oral_taxon_780 | N | – | − 1.94 (0.08) | − 1.46 (0.20) | − 1.93 (0.12) | − 2.20 (0.12) | 0.004 | −0.10 | <.001 |
| 207 | Lachnospiraceae_[G-8] sp._oral_taxon_500 | N | – | − 1.94 (0.07) | −1.91 (0.16) | −2.03 (0.10) | − 1.83 (0.12) | 0.443 | 0.01 | 0.695 |
| 208 | Capnocytophaga sp._oral_taxon_323 | P | – | −1.94 (0.08) | −1.99 (0.17) | − 1.93 (0.12) | − 1.94 (0.14) | 0.966 | − 0.01 | 0.731 |
| 209 | Scardovia wiggsiae | P | – | −1.94 (0.09) | − 2.07 (0.19) | − 2.11 (0.12) | − 1.66 (0.16) | 0.061 | 0.10 | 0.009 |
| 210 | Sphingomonas echinoides | N | – | − 1.95 (0.06) | −1.75 (0.14) | − 1.89 (0.10) | − 2.15 (0.11) | 0.056 | −0.10 | <.001 |
| 211 | SR1_[G-1] sp._oral_taxon_874 | U | – | − 1.99 (0.07) | −1.76 (0.15) | − 1.88 (0.10) | −2.26 (0.11) | 0.008 | −0.10 | 0.006 |
| 212 | Treponema lecithinolyticum | U | – | −2.01 (0.08) | −1.98 (0.17) | − 1.90 (0.12) | − 2.18 (0.13) | 0.302 | −0.10 | 0.068 |
| 213 | Prevotella baroniae | P | – | −2.04 (0.07) | −2.20 (0.15) | − 2.03 (0.11) | − 1.96 (0.13) | 0.490 | 0.04 | 0.209 |
| 214 | Pseudomonas fluorescens | P | – | − 2.05 (0.07) | − 2.12 (0.15) | − 2.07 (0.11) | −1.98 (0.13) | 0.767 | 0.03 | 0.334 |
| 215 | Shuttleworthia satelles | P | – | − 2.08 (0.07) | − 2.27 (0.14) | − 2.21 (0.10) | −1.82 (0.12) | 0.016 | 0.10 | <.001 |
| 216 | Prevotella sp._oral_taxon_306 | U | −2.08 (0.07) | −2.32 (0.14) | − 2.21 (0.10) | −1.79 (0.13) | 0.010 | 0.10 | <.001 | |
| 217 | Porphyromonas sp._oral_taxon_275 | N | – | − 2.11 (0.07) | −2.09 (0.16) | − 2.01 (0.12) | − 2.26 (0.12) | 0.321 | −0.04 | 0.184 |
| 218 | Streptococcus sinensis | P | – | − 2.14 (0.06) | − 1.95 (0.13) | − 2.30 (0.08) | − 2.05 (0.11) | 0.050 | 0.02 | 0.517 |
| 219 | Anaerolineae_[G-1] sp._oral_taxon_439 | N | – | − 2.14 (0.08) | − 2.47 (0.15) | − 2.22 (0.11) | −1.85 (0.14) | 0.008 | 0.12 | <.001* |
| 220 | Bacteroidaceae_[G-1] sp._oral_taxon_272 | N | – | − 2.15 (0.07) | − 2.48 (0.15) | − 2.21 (0.10) | −1.87 (0.13) | 0.007 | 0.10 | <.001 |
| 221 | Prevotella micans | N | – | − 2.15 (0.07) | −2.24 (0.14) | − 2.18 (0.10) | − 2.08 (0.12) | 0.658 | 0.02 | 0.452 |
| 222 | Leptotrichia goodfellowii | U | – | − 2.17 (0.07) | −1.90 (0.17) | −2.14 (0.11) | − 2.36 (0.12) | 0.080 | −0.10 | 0.008 |
| 223 | Capnocytophaga sp._oral_taxon_903 | N | – | − 2.17 (0.08) | −2.15 (0.17) | − 2.12 (0.11) | − 2.25 (0.13) | 0.751 | − 0.01 | 0.731 |
| 224 | Peptostreptococcaceae_[XI][G-5] [Eubacterium]_saph | U | – | − 2.18 (0.08) | −2.46 (0.15) | − 2.13 (0.12) | − 2.08 (0.13) | 0.195 | 0.04 | 0.229 |
| 225 | Treponema sp._oral_taxon_237 | P | – | − 2.20 (0.08) | −1.68 (0.19) | −2.21 (0.12) | − 2.46 (0.13) | 0.003 | −0.11 | <.001* |
| 226 | Fusobacterium sp._oral_taxon_370 | U | – | − 2.20 (0.06) | −2.25 (0.12) | − 2.08 (0.09) | − 2.32 (0.10) | 0.176 | −0.04 | 0.176 |
| 227 | Atopobium sp._oral_taxon_199 | U | – | − 2.26 (0.07) | −2.19 (0.16) | − 2.18 (0.11) | − 2.40 (0.12) | 0.334 | −0.02 | 0.433 |
| 228 | Prevotella sp._oral_taxon_314 | N | – | − 2.28 (0.07) | − 2.29 (0.16) | − 2.35 (0.11) | − 2.18 (0.13) | 0.605 | 0.02 | 0.519 |
| 229 | Capnocytophaga sp._oral_taxon_324 | U | – | − 2.28 (0.07) | −2.43 (0.15) | − 2.24 (0.11) | − 2.25 (0.12) | 0.588 | 0.03 | 0.256 |
| 230 | Haemophilus parahaemolyticus | N | – | − 2.29 (0.08) | −1.86 (0.18) | −2.23 (0.12) | − 2.61 (0.11) | 0.002 | −0.11 | <.001* |
| 231 | Porphyromonas sp._oral_taxon_278 | U | – | − 2.29 (0.07) | −2.40 (0.14) | − 2.24 (0.11) | − 2.30 (0.12) | 0.716 | 0.00 | 0.977 |
| 232 | Selenomonas sp._oral_taxon_937 | U | – | − 2.35 (0.06) | −2.34 (0.12) | − 2.46 (0.09) | − 2.22 (0.11) | 0.203 | 0.03 | 0.386 |
| 233 | Selenomonas dianae | N | – | − 2.36 (0.07) | − 2.52 (0.14) | − 2.27 (0.10) | − 2.38 (0.11) | 0.367 | 0.02 | 0.569 |
| 234 | Selenomonas sp._oral_taxon_133 | U | – | − 2.37 (0.08) | − 2.25 (0.18) | − 2.40 (0.11) | − 2.41 (0.13) | 0.719 | −0.02 | 0.396 |
| 235 | Selenomonas sp._oral_taxon_478 | P | – | − 2.49 (0.06) | −2.54 (0.13) | − 2.46 (0.09) | − 2.49 (0.10) | 0.882 | 0.03 | 0.306 |
| 236 | Capnocytophaga sp._oral_taxon_332 | U | – | − 2.50 (0.07) | −2.26 (0.16) | − 2.42 (0.11) | − 2.72 (0.11) | 0.043 | −0.10 | 0.043 |
| 237 | Megasphaera sp._oral_taxon_123 | N | – | − 2.51 (0.08) | − 2.06 (0.19) | − 2.51 (0.12) | − 2.77 (0.13) | 0.009 | − 0.10 | 0.002 |
| 238 | Bradyrhizobium elkanii | U | – | − 2.54 (0.08) | −2.37 (0.17) | − 2.68 (0.11) | − 2.44 (0.13) | 0.209 | −0.00 | 0.900 |
| 239 | Prevotella sp._oral_taxon_376 | P | – | − 2.54 (0.07) | −2.22 (0.17) | − 2.49 (0.11) | − 2.79 (0.12) | 0.015 | −0.10 | <.001 |
| 240 | Prevotella sp._oral_taxon_526 | P | – | − 2.56 (0.07) | −2.59 (0.14) | − 2.57 (0.10) | − 2.52 (0.12) | 0.923 | 0.01 | 0.736 |
| 241 | Selenomonas sp._oral_taxon_442 | U | – | − 2.58 (0.05) | − 2.78 (0.11) | − 2.52 (0.09) | − 2.53 (0.09) | 0.177 | 0.10 | 0.065 |
| 242 | Leptotrichia sp._oral_taxon_879 | U | – | − 2.60 (0.07) | − 2.56 (0.16) | − 2.62 (0.10) | −2.59 (0.11) | 0.951 | − 0.02 | 0.597 |
| 243 | Selenomonas sp._oral_taxon_149 | P | – | − 2.61 (0.06) | − 2.75 (0.12) | − 2.70 (0.09) | − 2.42 (0.10) | 0.048 | 0.10 | 0.004 |
| 244 | Aggregatibacter sp._oral_taxon_513 | U | – | − 2.66 (0.07) | − 2.64 (0.14) | − 2.52 (0.11) | − 2.85 (0.11) | 0.107 | − 0.03 | 0.368 |
| 245 | Treponema vincentii | N | – | − 2.67 (0.06) | −2.55 (0.13) | − 2.60 (0.09) | − 2.83 (0.09) | 0.140 | −0.04 | 0.125 |
| 246 | Capnocytophaga sp._oral_taxon_380 | N | – | − 2.70 (0.07) | −2.65 (0.14) | − 2.59 (0.11) | − 2.88 (0.11) | 0.132 | −0.10 | 0.107 |
| 247 | Johnsonella sp._oral_taxon_166 | U | – | − 2.78 (0.06) | −2.96 (0.12) | − 2.69 (0.10) | − 2.80 (0.11) | 0.292 | −0.01 | 0.855 |
| 248 | Treponema medium | P | – | −2.79 (0.06) | − 2.85 (0.14) | − 2.74 (0.10) | −2.81 (0.10) | 0.777 | 0.02 | 0.579 |
| 249 | Fretibacterium sp._oral_taxon_358 | U | – | − 2.84 (0.07) | −2.87 (0.14) | − 2.88 (0.10) | − 2.77 (0.12) | 0.708 | 0.02 | 0.494 |
| 250 | Neisseria pharyngis | N | – | − 2.87 (0.06) | −2.81 (0.13) | − 2.95 (0.09) | − 2.80 (0.11) | 0.479 | 0.01 | 0.760 |
| 251 | Prevotella sp._oral_taxon_475 | N | – | − 2.87 (0.06) | −2.81 (0.12) | − 2.84 (0.09) | − 2.95 (0.09) | 0.575 | −0.04 | 0.157 |
| 252 | Microbacterium flavescens | P | – | − 2.89 (0.04) | − 2.73 (0.09) | − 2.89 (0.06) | −2.97 (0.07) | 0.151 | −0.10 | 0.051 |
| 253 | Mitsuokella sp._oral_taxon_521 | N | – | − 2.90 (0.06) | −2.91 (0.12) | − 2.92 (0.08) | − 2.87 (0.11) | 0.929 | 0.02 | 0.603 |
| 254 | Lactobacillus gasseri | N | – | − 2.95 (0.06) | −3.08 (0.12) | − 3.09 (0.09) | −2.70 (0.12) | 0.012 | 0.10 | 0.002 |
| 255 | Butyrivibrio sp._oral_taxon_080 | P | – | − 2.96 (0.06) | −2.85 (0.13) | − 2.90 (0.09) | −3.10 (0.09) | 0.193 | −0.10 | 0.038 |
| 256 | Fretibacterium sp._oral_taxon_361 | P | – | − 2.98 (0.06) | − 3.18 (0.10) | − 2.97 (0.09) | − 2.88 (0.11) | 0.207 | 0.02 | 0.469 |
| 257 | Aggregatibacter actinomycetemcomitans | P | – | − 3.01 (0.07) | −2.97 (0.15) | −3.09 (0.10) | −2.94 (0.11) | 0.575 | 0.03 | 0.349 |
| 258 | Streptococcus sobrinus | U | – | − 3.04 (0.06) | −3.21 (0.10) | − 3.19 (0.09) | −2.75 (0.14) | 0.005 | 0.11 | <.001* |
| 259 | GN02_[G-2] sp._oral_taxon_873 | P | – | − 3.06 (0.05) | −3.08 (0.12) | − 3.03 (0.08) | − 3.10 (0.09) | 0.851 | −0.00 | 0.927 |
| 260 | Prevotella multiformis | N | – | − 3.08 (0.06) | − 3.12 (0.11) | − 3.10 (0.09) | − 3.05 (0.10) | 0.901 | 0.03 | 0.359 |
| 261 | Brevundimonas diminuta | N | – | − 3.30 (0.04) | − 3.17 (0.09) | − 3.35 (0.06) | − 3.30 (0.07) | 0.232 | −0.03 | 0.306 |
| 262 | Atopobium sp._oral_taxon_416 | P | – | − 3.38 (0.05) | − 3.45 (0.09) | − 3.44 (0.07) | − 3.27 (0.09) | 0.202 | 0.10 | 0.042 |
| 263 | Treponema sp._oral_taxon_247 | P | – | − 3.45 (0.05) | − 3.35 (0.09) | − 3.50 (0.07) | − 3.44 (0.08) | 0.436 | − 0.02 | 0.566 |
| 264 | Leptothrix sp._oral_taxon_025 | P | – | − 3.50 (0.04) | −3.37 (0.08) | − 3.53 (0.06) | − 3.53 (0.06) | 0.216 | − 0.04 | 0.221 |
| 265 | Pyramidobacter piscolens | P | – | − 3.53 (0.04) | − 3.44 (0.10) | − 3.60 (0.06) | −3.47 (0.07) | 0.251 | − 0.01 | 0.919 |
| 266 | Sphingomonas sp._oral_taxon_006 | P | – | − 3.55 (0.04) | −3.49 (0.08) | − 3.60 (0.05) | − 3.52 (0.06) | 0.407 | − 0.00 | 0.989 |
| 267 | Porphyrobacter tepidarius | N | – | −3.58 (0.03) | −3.51 (0.08) | − 3.65 (0.05) | −3.54 (0.06) | 0.216 | 0.01 | 0.869 |
Rank order, OTUs are sorted high to low, with most abundant OTU at the top of the table
The CLR OTU is interpreted as a log [2] fold-difference for the given species relative to the overall compositional geometric mean. A mean CLR of 3 indicates a 8-fold [23] higher abundance, and a mean CLR of − 3 indicates a 8-fold lower abundance, relative to the overall compositional geometric mean
SE standard error, Pearson r the Pearson product-moment correlation coefficient
p-values: bolded are significant at alpha 0.05; asterisk are significant at alpha 0.05 after Bonferroni correction
Culture status: N = named; U = unnamed; P = phylotyped
Socransky complex32: R = red; O = orange; P = purple; G = green; Y = yellow; B = blue; −-- = not part of the Socransky classification
Among all 1219 women (Table 4), Pearson correlations ranged from r = − 0.18 to r = 0.18. Eighty two (31%) taxa were significantly correlated with age (uncorrected P < 0.05; bolded), of which 28 (34.2%) remained significant after Bonferroni correction. The largest positive correlation was with Oribacterium sp._oral_taxon 078 (r = 0.18; corrected P < 0.001); the most negative correlation was with Strep. sanguinis (r = − 0.18; corrected P < .001). Correlations between established pathogenic bacteria from Socransky’s complex [34] and age were of weak (T. denticola, r = − 0.04; P. gingivalis, r = 0.03; F. nucleatum, r = 0.03) to moderate (Fusobacterium nucleatum polymorphum, r = − 0.10; T. forsythia, r = 0.10) magnitude. Bacteria associated with healthy periodontium were correlated with age on a similar (S. oralis, r = − 0.10; intermedius r = − 0.10; mutans, r = 0.10) or somewhat stronger (S. sanguinis, r = − 0.16) magnitude.
Ninety (33.7%) bacteria were observed to be significantly different across age categories (uncorrected P < 0.05; bolded in Table 4), of which 12 (13.3%) remained significant after Bonferroni correction (corrected P < 0.05). Fig. 5 presents box-and-whisker plots depicting the variability of CLR OTUs for the 12 bacteria that were significantly different across age groups (corrected P < 0.001). Of these 12 bacteria, 7 were significantly higher in older than younger women; whilst the remaining 5 were higher in the younger women. Bifidobacterium dentium showed the greatest difference (0.73 CLR OTU units) between age groups among the bacteria observed to be higher in older women, whereas S. sanguinis showed the largest difference (1.19 CLR OTU units) between age groups for bacteria higher in younger women.
Fig. 5.
Box plots of mean CLR OTUs that differed between age categories (corrected P < 0.05). Box reflects the 25th, 50th, and 75th percentile CLR OTU; whiskers reflect the range of CLR OTU
Tables 5 and 6 present additional measures used in previous studies to characterize the oral microbiome. Relative abundance for the overall cohort and according to age categories is shown in Table 5, ranked high to low, with dashed lines denoting the top 20 taxa. V. dispar demonstrated the highest relative abundance (mean, 8.9%), and the remaining bacterial order is quite similar to the top 20 when ordered according to CLR mean OTU (Table 2). Patterns of relative abundance across age categories also were generally comparable to those observed for CLR mean OTUs. Bacterial prevalence (present at any abundance) is shown in Table 6, for which a slightly different ordering is noted for the top 20 most prevalent bacteria compared to those ordered on CLR OTU or relative abundance. There were 12 (4.5%) bacteria prevalent at 99% or higher and 3 (1.1%) present in all samples (S. oralis, V. dispar and parvula). Differences in prevalence across age categories were modest. Among all women, prevalence of established pathogenic bacteria in Socransky’s complex was 86.8, 82.9 56.6, and 55.1% for F. nucleatum, T. forsythia, T. denticola, and P. gingivalis, respectively. Prevalence of T. forsythia did not vary consistently with age, whereas prevalence of F. nucleatum and T. denticola tended to decline with age and P. gingivalis tended to increase with age.
Table 5.
Mean Relative Abundance for each of 267 bacterial species, overall and by age categories
| Rank Ordera | OTU label | Overall (N = 1219) |
50–59 (N = 239) |
60–69 (N = 554) |
≥70 (N = 426) |
|---|---|---|---|---|---|
| Relative Abundance (%) Mean (SE) |
Relative Abundance (%) Mean (SE) |
Relative Abundance (%) Mean (SE) |
Relative Abundance (%) Mean (SE) |
||
| 1 | Veillonella dispar | 8.94 (0.26) | 8.66 (0.52) | 8.52 (0.37) | 9.64 (0.48) |
| 2 | Streptococcus oralis | 7.68 (0.24) | 9.52 (0.60) | 7.95 (0.39) | 6.29 (0.31) |
| 3 | Veillonella parvula | 6.57 (0.22) | 6.14 (0.44) | 6.07 (0.30) | 7.46 (0.42) |
| 4 | Fusobacterium nucleatum_subsp._vincentii | 4.00 (0.14) | 3.82 (0.32) | 4.11 (0.21) | 3.97 (0.23) |
| 5 | Selenomonas sputigena | 2.94 (0.12) | 2.23 (0.20) | 2.66 (0.16) | 3.70 (0.25) |
| 6 | Fusobacterium sp._oral_taxon_203 | 2.35 (0.13) | 2.15 (0.28) | 2.63 (0.21) | 2.09 (0.20) |
| 7 | Prevotella oris | 1.85 (0.07) | 1.81 (0.14) | 1.89 (0.11) | 1.81 (0.10) |
| 8 | Prevotella nigrescens | 1.78 (0.07) | 1.64 (0.13) | 1.81 (0.11) | 1.81 (0.12) |
| 9 | Fusobacterium nucleatum_subsp._animalis | 1.65 (0.06) | 1.54 (0.13) | 1.63 (0.09) | 1.74 (0.09) |
| 10 | Selenomonas noxia | 1.48 (0.07) | 1.52 (0.18) | 1.45 (0.11) | 1.50 (0.11) |
| 11 | Fusobacterium nucleatum_subsp._polymorphum | 1.42 (0.04) | 1.57 (0.11) | 1.50 (0.07) | 1.24 (0.07) |
| 12 | Rothia dentocariosa | 1.41 (0.10) | 1.43 (0.17) | 1.60 (0.18) | 1.16 (0.13) |
| 13 | Streptococcus sanguinis | 1.40 (0.07) | 1.83 (0.14) | 1.40 (0.09) | 1.16 (0.13) |
| 14 | Alloprevotella tannerae | 1.39 (0.07) | 1.22 (0.16) | 1.46 (0.12) | 1.41 (0.11) |
| 15 | Haemophilus parainfluenzae | 1.21 (0.09) | 1.62 (0.24) | 1.15 (0.13) | 1.07 (0.14) |
| 16 | Corynebacterium matruchotii | 1.17 (0.05) | 1.34 (0.13) | 1.24 (0.07) | 0.99 (0.06) |
| 17 | Fretibacterium sp._oral_taxon_360 | 1.15 (0.07) | 0.89 (0.13) | 1.24 (0.11) | 1.19 (0.10) |
| 18 | Streptococcus intermedius | 1.05 (0.05) | 1.27 (0.12) | 1.13 (0.08) | 0.84 (0.05) |
| 19 | TM7_[G-1] sp._oral_taxon_349 | 1.04 (0.06) | 0.97 (0.14) | 0.97 (0.07) | 1.18 (0.14) |
| 20 | Streptococcus gordonii | 1.03 (0.05) | 0.98 (0.15) | 1.01 (0.07) | 1.09 (0.10) |
| 21 | TM7_[G-1] sp._oral_taxon_346 | 1.02 (0.05) | 1.01 (0.11) | 1.05 (0.07) | 1.00 (0.08) |
| 22 | Porphyromonas gingivalis | 0.94 (0.10) | 0.95 (0.27) | 0.74 (0.10) | 1.21 (0.19) |
| 23 | Campylobacter gracilis | 0.94 (0.02) | 0.90 (0.06) | 0.96 (0.04) | 0.95 (0.04) |
| 24 | Fusobacterium naviforme | 0.89 (0.06) | 0.91 (0.13) | 0.92 (0.10) | 0.83 (0.07) |
| 25 | Parvimonas micra | 0.88 (0.03) | 0.87 (0.07) | 0.96 (0.05) | 0.78 (0.05) |
| 26 | Neisseria sicca | 0.80 (0.07) | 0.83 (0.21) | 0.87 (0.11) | 0.69 (0.09) |
| 27 | Leptotrichia wadei | 0.79 (0.06) | 0.81 (0.15) | 0.74 (0.08) | 0.86 (0.11) |
| 28 | Selenomonas artemidis | 0.79 (0.05) | 0.87 (0.12) | 0.75 (0.07) | 0.79 (0.08) |
| 29 | Bacteroidales_[G-2] sp._oral_taxon_274 | 0.79 (0.04) | 0.64 (0.08) | 0.85 (0.07) | 0.79 (0.06) |
| 30 | Prevotella sp._oral_taxon_317 | 0.77 (0.04) | 0.67 (0.07) | 0.80 (0.05) | 0.78 (0.06) |
| 31 | Anaeroglobus geminatus | 0.67 (0.04) | 0.43 (0.08) | 0.58 (0.06) | 0.92 (0.09) |
| 32 | Capnocytophaga granulosa | 0.65 (0.03) | 0.63 (0.09) | 0.66 (0.05) | 0.64 (0.06) |
| 33 | Streptococcus salivarius | 0.64 (0.05) | 0.61 (0.09) | 0.67 (0.09) | 0.64 (0.07) |
| 34 | TM7_[G-1] sp._oral_taxon_952 | 0.64 (0.03) | 0.72 (0.08) | 0.71 (0.05) | 0.50 (0.05) |
| 35 | Prevotella denticola | 0.62 (0.05) | 0.49 (0.07) | 0.57 (0.08) | 0.76 (0.09) |
| 36 | Capnocytophaga leadbetteri | 0.61 (0.03) | 0.59 (0.07) | 0.63 (0.05) | 0.61 (0.07) |
| 37 | Gemella morbillorum | 0.61 (0.02) | 0.69 (0.07) | 0.65 (0.04) | 0.52 (0.04) |
| 38 | Veillonella atypica | 0.61 (0.05) | 0.50 (0.09) | 0.55 (0.07) | 0.75 (0.08) |
| 39 | Streptococcus mutans | 0.60 (0.05) | 0.47 (0.09) | 0.51 (0.07) | 0.80 (0.10) |
| 40 | Fretibacterium sp._oral_taxon_359 | 0.60 (0.06) | 0.77 (0.17) | 0.60 (0.09) | 0.49 (0.07) |
| 41 | Prevotella intermedia | 0.59 (0.06) | 0.62 (0.15) | 0.63 (0.09) | 0.52 (0.09) |
| 42 | Dialister invisus | 0.57 (0.02) | 0.47 (0.04) | 0.60 (0.04) | 0.60 (0.04) |
| 43 | Granulicatella adiacens | 0.56 (0.02) | 0.65 (0.05) | 0.55 (0.03) | 0.53 (0.03) |
| 44 | Porphyromonas endodontalis | 0.55 (0.04) | 0.63 (0.08) | 0.57 (0.05) | 0.49 (0.06) |
| 45 | Streptococcus cristatus | 0.55 (0.03) | 0.51 (0.07) | 0.59 (0.05) | 0.53 (0.04) |
| 46 | Capnocytophaga gingivalis | 0.55 (0.03) | 0.60 (0.07) | 0.55 (0.04) | 0.51 (0.04) |
| 47 | Prevotella pleuritidis | 0.53 (0.04) | 0.51 (0.08) | 0.56 (0.06) | 0.51 (0.06) |
| 48 | Neisseria elongata | 0.53 (0.04) | 0.66 (0.10) | 0.55 (0.05) | 0.43 (0.07) |
| 49 | Actinomyces naeslundii | 0.52 (0.02) | 0.62 (0.05) | 0.53 (0.03) | 0.46 (0.03) |
| 50 | Selenomonas sp._oral_taxon_137 | 0.51 (0.04) | 0.51 (0.09) | 0.52 (0.05) | 0.50 (0.07) |
| 51 | Tannerella forsythia | 0.49 (0.03) | 0.42 (0.06) | 0.46 (0.04) | 0.58 (0.05) |
| 52 | TM7_[G-5] sp._oral_taxon_356 | 0.47 (0.03) | 0.36 (0.07) | 0.51 (0.06) | 0.48 (0.06) |
| 53 | Campylobacter showae | 0.46 (0.02) | 0.44 (0.04) | 0.47 (0.04) | 0.46 (0.03) |
| 54 | Fretibacterium fastidiosum | 0.46 (0.03) | 0.54 (0.10) | 0.45 (0.04) | 0.42 (0.04) |
| 55 | Rothia aeria | 0.45 (0.03) | 0.45 (0.05) | 0.55 (0.06) | 0.33 (0.04) |
| 56 | Capnocytophaga sputigena | 0.45 (0.03) | 0.47 (0.05) | 0.44 (0.04) | 0.46 (0.05) |
| 57 | Gemella haemolysans | 0.44 (0.04) | 0.54 (0.08) | 0.45 (0.07) | 0.38 (0.06) |
| 58 | Leptotrichia hongkongensis | 0.43 (0.03) | 0.36 (0.04) | 0.44 (0.05) | 0.44 (0.05) |
| 59 | Neisseria flavescens | 0.42 (0.04) | 0.29 (0.06) | 0.38 (0.06) | 0.55 (0.09) |
| 60 | Neisseria oralis | 0.42 (0.05) | 0.59 (0.15) | 0.32 (0.04) | 0.45 (0.11) |
| 61 | Streptococcus anginosus | 0.42 (0.03) | 0.38 (0.05) | 0.38 (0.04) | 0.49 (0.06) |
| 62 | Aggregatibacter aphrophilus | 0.37 (0.04) | 0.33 (0.05) | 0.45 (0.07) | 0.28 (0.05) |
| 63 | Kingella oralis | 0.36 (0.02) | 0.41 (0.05) | 0.31 (0.02) | 0.39 (0.04) |
| 64 | Leptotrichia shahii | 0.36 (0.04) | 0.32 (0.07) | 0.26 (0.03) | 0.50 (0.08) |
| 65 | Leptotrichia buccalis | 0.35 (0.03) | 0.36 (0.07) | 0.40 (0.05) | 0.29 (0.05) |
| 66 | Selenomonas sp._oral_taxon_134 | 0.34 (0.02) | 0.23 (0.04) | 0.32 (0.03) | 0.43 (0.05) |
| 67 | Leptotrichia hofstadii | 0.33 (0.04) | 0.42 (0.11) | 0.34 (0.05) | 0.27 (0.05) |
| 68 | Leptotrichia sp._oral_taxon_417 | 0.32 (0.03) | 0.23 (0.04) | 0.36 (0.05) | 0.32 (0.05) |
| 69 | Cardiobacterium hominis | 0.32 (0.02) | 0.47 (0.08) | 0.33 (0.02) | 0.23 (0.02) |
| 70 | Selenomonas sp._oral_taxon_136 | 0.31 (0.02) | 0.20 (0.03) | 0.29 (0.03) | 0.40 (0.04) |
| 71 | Eikenella corrodens | 0.31 (0.01) | 0.34 (0.03) | 0.33 (0.02) | 0.25 (0.02) |
| 72 | Prevotella melaninogenica | 0.30 (0.02) | 0.25 (0.03) | 0.28 (0.03) | 0.36 (0.04) |
| 73 | Porphyromonas sp._oral_taxon_279 | 0.30 (0.02) | 0.28 (0.04) | 0.32 (0.04) | 0.30 (0.03) |
| 74 | Treponema denticola | 0.28 (0.02) | 0.26 (0.05) | 0.28 (0.03) | 0.31 (0.04) |
| 75 | Filifactor alocis | 0.28 (0.02) | 0.29 (0.05) | 0.29 (0.04) | 0.27 (0.04) |
| 76 | Actinomyces oris | 0.28 (0.02) | 0.31 (0.04) | 0.27 (0.02) | 0.27 (0.03) |
| 77 | Parvimonas sp._oral_taxon_393 | 0.28 (0.02) | 0.36 (0.05) | 0.28 (0.02) | 0.23 (0.02) |
| 78 | Veillonellaceae_[G-1] sp._oral_taxon_150 | 0.27 (0.02) | 0.22 (0.04) | 0.23 (0.02) | 0.35 (0.03) |
| 79 | Actinomyces sp._oral_taxon_169 | 0.27 (0.02) | 0.39 (0.06) | 0.26 (0.02) | 0.22 (0.03) |
| 80 | Leptotrichia sp._oral_taxon_212 | 0.27 (0.02) | 0.28 (0.03) | 0.28 (0.02) | 0.24 (0.04) |
| 81 | Rothia mucilaginosa | 0.27 (0.02) | 0.21 (0.03) | 0.28 (0.04) | 0.29 (0.03) |
| 82 | Selenomonas infelix | 0.27 (0.02) | 0.21 (0.02) | 0.27 (0.03) | 0.29 (0.03) |
| 83 | Cardiobacterium valvarum | 0.25 (0.01) | 0.31 (0.04) | 0.26 (0.02) | 0.21 (0.02) |
| 84 | Capnocytophaga sp._oral_taxon_326 | 0.25 (0.02) | 0.26 (0.04) | 0.28 (0.03) | 0.22 (0.03) |
| 85 | Prevotella sp._oral_taxon_472 | 0.25 (0.02) | 0.30 (0.05) | 0.27 (0.02) | 0.19 (0.02) |
| 86 | Treponema socranskii | 0.25 (0.01) | 0.22 (0.02) | 0.24 (0.01) | 0.27 (0.02) |
| 87 | Streptococcus constellatus | 0.25 (0.02) | 0.19 (0.03) | 0.25 (0.03) | 0.28 (0.03) |
| 88 | Veillonella rogosae | 0.24 (0.02) | 0.21 (0.04) | 0.27 (0.04) | 0.23 (0.03) |
| 89 | Fusobacterium nucleatum_subsp._nucleatum | 0.24 (0.03) | 0.21 (0.06) | 0.19 (0.04) | 0.34 (0.07) |
| 90 | Prevotella sp._oral_taxon_300 | 0.23 (0.01) | 0.24 (0.03) | 0.20 (0.01) | 0.27 (0.03) |
| 91 | Veillonellaceae_[G-1] sp._oral_taxon_155 | 0.23 (0.02) | 0.20 (0.03) | 0.20 (0.02) | 0.28 (0.03) |
| 92 | Campylobacter concisus | 0.22 (0.01) | 0.20 (0.02) | 0.23 (0.01) | 0.23 (0.02) |
| 93 | Leptotrichia sp._oral_taxon_225 | 0.22 (0.02) | 0.26 (0.06) | 0.28 (0.04) | 0.12 (0.03) |
| 94 | Selenomonas sp._oral_taxon_892 | 0.22 (0.01) | 0.24 (0.04) | 0.21 (0.02) | 0.21 (0.02) |
| 95 | Catonella morbi | 0.21 (0.01) | 0.23 (0.02) | 0.21 (0.01) | 0.19 (0.01) |
| 96 | Aggregatibacter segnis | 0.20 (0.02) | 0.21 (0.05) | 0.24 (0.04) | 0.15 (0.02) |
| 97 | Prevotella oulorum | 0.20 (0.01) | 0.21 (0.03) | 0.18 (0.01) | 0.22 (0.02) |
| 98 | Leptotrichia sp._oral_taxon_392 | 0.19 (0.01) | 0.20 (0.04) | 0.22 (0.02) | 0.16 (0.02) |
| 99 | Dialister pneumosintes | 0.19 (0.01) | 0.18 (0.03) | 0.19 (0.02) | 0.20 (0.02) |
| 100 | Veillonella denticariosi | 0.19 (0.03) | 0.17 (0.06) | 0.17 (0.04) | 0.22 (0.07) |
| 101 | Peptostreptococcaceae_[XI][G-9] [Eubacterium]_brac | 0.19 (0.01) | 0.21 (0.02) | 0.19 (0.01) | 0.17 (0.01) |
| 102 | Megasphaera micronuciformis | 0.19 (0.02) | 0.15 (0.03) | 0.17 (0.02) | 0.23 (0.04) |
| 103 | Lautropia mirabilis | 0.18 (0.01) | 0.23 (0.03) | 0.20 (0.02) | 0.14 (0.01) |
| 104 | Streptococcus parasanguinis_II | 0.18 (0.02) | 0.15 (0.03) | 0.17 (0.02) | 0.21 (0.03) |
| 105 | Porphyromonas sp._oral_taxon_284 | 0.18 (0.01) | 0.18 (0.02) | 0.19 (0.02) | 0.17 (0.02) |
| 106 | Selenomonas sp._oral_taxon_919 | 0.18 (0.01) | 0.18 (0.02) | 0.17 (0.02) | 0.20 (0.03) |
| 107 | Bergeyella sp._oral_taxon_322 | 0.18 (0.01) | 0.22 (0.02) | 0.19 (0.01) | 0.14 (0.01) |
| 108 | Leptotrichia sp._oral_taxon_498 | 0.18 (0.02) | 0.14 (0.03) | 0.16 (0.03) | 0.22 (0.04) |
| 109 | Kingella denitrificans | 0.17 (0.01) | 0.17 (0.04) | 0.17 (0.02) | 0.18 (0.02) |
| 110 | Prevotella maculosa | 0.17 (0.01) | 0.16 (0.02) | 0.16 (0.01) | 0.18 (0.01) |
| 111 | Atopobium rimae | 0.17 (0.01) | 0.12 (0.02) | 0.15 (0.02) | 0.21 (0.03) |
| 112 | Corynebacterium durum | 0.17 (0.01) | 0.24 (0.03) | 0.16 (0.01) | 0.13 (0.03) |
| 113 | TM7_[G-1] sp._oral_taxon_488 | 0.17 (0.02) | 0.23 (0.07) | 0.18 (0.02) | 0.11 (0.01) |
| 114 | Capnocytophaga sp._oral_taxon_336 | 0.17 (0.01) | 0.15 (0.02) | 0.17 (0.01) | 0.17 (0.02) |
| 115 | Selenomonas flueggei | 0.16 (0.01) | 0.15 (0.03) | 0.16 (0.02) | 0.18 (0.03) |
| 116 | Aggregatibacter sp._oral_taxon_458 | 0.16 (0.02) | 0.14 (0.02) | 0.16 (0.02) | 0.17 (0.03) |
| 117 | Fretibacterium sp._oral_taxon_362 | 0.16 (0.02) | 0.15 (0.04) | 0.18 (0.04) | 0.14 (0.03) |
| 118 | Neisseria subflava | 0.16 (0.02) | 0.14 (0.04) | 0.16 (0.04) | 0.17 (0.03) |
| 119 | Peptostreptococcaceae_[XI][G-7] [Eubacterium]_yuri | 0.16 (0.01) | 0.18 (0.02) | 0.17 (0.01) | 0.12 (0.01) |
| 120 | TM7_[G-1] sp._oral_taxon_869 | 0.16 (0.03) | 0.15 (0.04) | 0.20 (0.06) | 0.11 (0.02) |
| 121 | Lachnoanaerobaculum saburreum | 0.15 (0.01) | 0.14 (0.02) | 0.15 (0.01) | 0.15 (0.01) |
| 122 | Prevotella oralis | 0.15 (0.01) | 0.09 (0.01) | 0.13 (0.01) | 0.20 (0.02) |
| 123 | TM7_[G-2] sp._oral_taxon_350 | 0.15 (0.02) | 0.12 (0.03) | 0.14 (0.02) | 0.17 (0.04) |
| 124 | Megasphaera sp._oral_taxon_123 | 0.14 (0.03) | 0.17 (0.05) | 0.12 (0.03) | 0.15 (0.06) |
| 125 | Aggregatibacter paraphrophilus | 0.14 (0.02) | 0.18 (0.06) | 0.18 (0.03) | 0.07 (0.02) |
| 126 | Desulfobulbus sp._oral_taxon_041 | 0.14 (0.01) | 0.11 (0.02) | 0.13 (0.02) | 0.17 (0.02) |
| 127 | Actinomyces massiliensis | 0.14 (0.01) | 0.18 (0.02) | 0.15 (0.02) | 0.10 (0.01) |
| 128 | Johnsonella ignava | 0.13 (0.01) | 0.13 (0.02) | 0.14 (0.01) | 0.13 (0.01) |
| 129 | Prevotella loescheii | 0.13 (0.01) | 0.15 (0.03) | 0.15 (0.02) | 0.09 (0.02) |
| 130 | Lachnospiraceae_[G-3] sp._oral_taxon_100 | 0.13 (0.01) | 0.12 (0.01) | 0.14 (0.01) | 0.12 (0.01) |
| 131 | Actinomyces johnsonii | 0.13 (0.01) | 0.12 (0.01) | 0.13 (0.02) | 0.12 (0.02) |
| 132 | TM7_[G-1] sp._oral_taxon_347 | 0.12 (0.02) | 0.16 (0.05) | 0.14 (0.02) | 0.08 (0.02) |
| 133 | Bacteroidetes_[G-5] sp._oral_taxon_511 | 0.12 (0.01) | 0.10 (0.02) | 0.13 (0.02) | 0.12 (0.02) |
| 134 | Actinomyces sp._oral_taxon_180 | 0.12 (0.01) | 0.13 (0.01) | 0.12 (0.01) | 0.10 (0.01) |
| 135 | Prevotella salivae | 0.12 (0.01) | 0.09 (0.02) | 0.12 (0.01) | 0.13 (0.02) |
| 136 | Actinomyces sp._oral_taxon_171 | 0.12 (0.01) | 0.12 (0.02) | 0.12 (0.01) | 0.11 (0.01) |
| 137 | Selenomonas sp._oral_taxon_146 | 0.12 (0.01) | 0.12 (0.02) | 0.11 (0.01) | 0.12 (0.01) |
| 138 | Abiotrophia defectiva | 0.12 (0.01) | 0.14 (0.03) | 0.13 (0.01) | 0.09 (0.01) |
| 139 | Pseudomonas fluorescens | 0.11 (0.05) | 0.07 (0.04) | 0.15 (0.10) | 0.10 (0.04) |
| 140 | Veillonella sp._oral_taxon_780 | 0.11 (0.02) | 0.16 (0.06) | 0.12 (0.04) | 0.08 (0.03) |
| 141 | Porphyromonas catoniae | 0.11 (0.01) | 0.13 (0.03) | 0.13 (0.02) | 0.08 (0.01) |
| 142 | Mitsuokella sp._oral_taxon_131 | 0.11 (0.02) | 0.05 (0.01) | 0.11 (0.03) | 0.14 (0.04) |
| 143 | Actinobaculum sp._oral_taxon_183 | 0.11 (0.01) | 0.12 (0.02) | 0.11 (0.01) | 0.09 (0.01) |
| 144 | Oribacterium sp._oral_taxon_078 | 0.11 (0.01) | 0.07 (0.01) | 0.10 (0.01) | 0.14 (0.01) |
| 145 | Treponema sp._oral_taxon_237 | 0.11 (0.01) | 0.11 (0.03) | 0.10 (0.02) | 0.11 (0.02) |
| 146 | Haemophilus sp._oral_taxon_036 | 0.10 (0.01) | 0.11 (0.03) | 0.10 (0.02) | 0.09 (0.02) |
| 147 | Leptotrichia sp._oral_taxon_215 | 0.10 (0.01) | 0.12 (0.02) | 0.10 (0.01) | 0.08 (0.01) |
| 148 | Prevotella histicola | 0.10 (0.01) | 0.06 (0.01) | 0.08 (0.02) | 0.15 (0.03) |
| 149 | TM7_[G-1] sp._oral_taxon_348 | 0.10 (0.01) | 0.08 (0.01) | 0.12 (0.01) | 0.09 (0.01) |
| 150 | Peptostreptococcus stomatis | 0.10 (0.01) | 0.11 (0.02) | 0.10 (0.01) | 0.08 (0.01) |
| 151 | Prevotella veroralis | 0.10 (0.01) | 0.08 (0.02) | 0.12 (0.02) | 0.08 (0.02) |
| 152 | Streptococcus sp._oral_taxon_056 | 0.10 (0.01) | 0.11 (0.02) | 0.09 (0.01) | 0.09 (0.01) |
| 153 | Selenomonas sp._oral_taxon_126 | 0.10 (0.01) | 0.09 (0.02) | 0.10 (0.01) | 0.09 (0.01) |
| 154 | Treponema sp._oral_taxon_231 | 0.09 (0.01) | 0.07 (0.01) | 0.10 (0.01) | 0.09 (0.01) |
| 155 | Neisseria bacilliformis | 0.09 (0.01) | 0.11 (0.04) | 0.08 (0.01) | 0.08 (0.02) |
| 156 | Actinomyces gerencseriae | 0.09 (0.01) | 0.09 (0.01) | 0.08 (0.01) | 0.09 (0.01) |
| 157 | Prevotella dentalis | 0.08 (0.01) | 0.08 (0.02) | 0.08 (0.02) | 0.08 (0.01) |
| 158 | Fusobacterium periodonticum | 0.08 (0.01) | 0.06 (0.01) | 0.09 (0.01) | 0.09 (0.01) |
| 159 | Prevotella pallens | 0.08 (0.01) | 0.08 (0.02) | 0.06 (0.01) | 0.11 (0.02) |
| 160 | Centipeda periodontii | 0.08 (0.01) | 0.05 (0.01) | 0.10 (0.01) | 0.08 (0.01) |
| 161 | Scardovia wiggsiae | 0.08 (0.01) | 0.09 (0.03) | 0.07 (0.02) | 0.09 (0.02) |
| 162 | Streptococcus sp._oral_taxon_074 | 0.08 (0.01) | 0.07 (0.01) | 0.09 (0.01) | 0.07 (0.01) |
| 163 | Haemophilus parahaemolyticus | 0.08 (0.02) | 0.09 (0.03) | 0.11 (0.03) | 0.04 (0.02) |
| 164 | Tannerella sp._oral_taxon_286 | 0.08 (0.00) | 0.06 (0.01) | 0.08 (0.01) | 0.08 (0.01) |
| 165 | Selenomonas sp._oral_taxon_936 | 0.08 (0.01) | 0.06 (0.01) | 0.07 (0.01) | 0.10 (0.02) |
| 166 | Actinomyces sp._oral_taxon_170 | 0.08 (0.01) | 0.06 (0.01) | 0.10 (0.02) | 0.06 (0.02) |
| 167 | Ruminococcaceae_[G-1] sp._oral_taxon_075 | 0.08 (0.01) | 0.09 (0.02) | 0.08 (0.01) | 0.07 (0.01) |
| 168 | Bifidobacterium dentium | 0.08 (0.01) | 0.07 (0.02) | 0.06 (0.01) | 0.10 (0.02) |
| 169 | Alloprevotella rava | 0.08 (0.01) | 0.07 (0.01) | 0.07 (0.01) | 0.09 (0.01) |
| 170 | Atopobium parvulum | 0.08 (0.01) | 0.06 (0.01) | 0.06 (0.01) | 0.11 (0.02) |
| 171 | Ottowia sp._oral_taxon_894 | 0.08 (0.01) | 0.08 (0.02) | 0.09 (0.01) | 0.06 (0.01) |
| 172 | Prevotella sp._oral_taxon_313 | 0.07 (0.01) | 0.09 (0.02) | 0.05 (0.01) | 0.10 (0.03) |
| 173 | TM7_[G-6] sp._oral_taxon_870 | 0.07 (0.01) | 0.08 (0.02) | 0.08 (0.01) | 0.06 (0.01) |
| 174 | Capnocytophaga sp._oral_taxon_864 | 0.07 (0.01) | 0.08 (0.01) | 0.08 (0.01) | 0.05 (0.01) |
| 175 | Actinomyces meyeri | 0.07 (0.00) | 0.08 (0.01) | 0.08 (0.01) | 0.06 (0.00) |
| 176 | Leptotrichia sp._oral_taxon_223 | 0.07 (0.01) | 0.05 (0.01) | 0.07 (0.02) | 0.09 (0.02) |
| 177 | Capnocytophaga sp._oral_taxon_338 | 0.07 (0.00) | 0.07 (0.01) | 0.07 (0.01) | 0.07 (0.01) |
| 178 | Streptococcus parasanguinis_I | 0.07 (0.01) | 0.06 (0.01) | 0.06 (0.01) | 0.08 (0.01) |
| 179 | Pseudoramibacter alactolyticus | 0.07 (0.01) | 0.10 (0.05) | 0.05 (0.01) | 0.07 (0.01) |
| 180 | Haemophilus haemolyticus | 0.07 (0.01) | 0.15 (0.05) | 0.05 (0.01) | 0.05 (0.01) |
| 181 | Neisseria pharyngis | 0.07 (0.02) | 0.06 (0.03) | 0.04 (0.02) | 0.10 (0.05) |
| 182 | Treponema maltophilum | 0.07 (0.00) | 0.06 (0.01) | 0.06 (0.00) | 0.08 (0.01) |
| 183 | Bradyrhizobium elkanii | 0.07 (0.02) | 0.12 (0.06) | 0.05 (0.01) | 0.05 (0.02) |
| 184 | Alloprevotella sp._oral_taxon_473 | 0.07 (0.01) | 0.05 (0.01) | 0.08 (0.02) | 0.06 (0.01) |
| 185 | Streptococcus lactarius | 0.06 (0.01) | 0.11 (0.04) | 0.05 (0.01) | 0.06 (0.01) |
| 186 | Prevotella sp._oral_taxon_292 | 0.06 (0.00) | 0.05 (0.01) | 0.06 (0.01) | 0.08 (0.01) |
| 187 | Capnocytophaga sp._oral_taxon_332 | 0.06 (0.01) | 0.08 (0.04) | 0.07 (0.01) | 0.05 (0.01) |
| 188 | Prevotella saccharolytica | 0.06 (0.00) | 0.06 (0.01) | 0.07 (0.01) | 0.06 (0.00) |
| 189 | Leptotrichia sp._oral_taxon_879 | 0.06 (0.02) | 0.14 (0.11) | 0.03 (0.01) | 0.05 (0.01) |
| 190 | Prevotella sp._oral_taxon_314 | 0.06 (0.01) | 0.05 (0.02) | 0.05 (0.01) | 0.07 (0.01) |
| 191 | Actinobaculum sp._oral_taxon_848 | 0.06 (0.00) | 0.04 (0.01) | 0.06 (0.01) | 0.06 (0.01) |
| 192 | Fretibacterium sp._oral_taxon_358 | 0.06 (0.01) | 0.04 (0.02) | 0.06 (0.02) | 0.06 (0.01) |
| 193 | Streptococcus sobrinus | 0.06 (0.01) | 0.00 (0.00) | 0.03 (0.01) | 0.12 (0.04) |
| 194 | Aggregatibacter actinomycetemcomitans | 0.06 (0.01) | 0.06 (0.02) | 0.06 (0.02) | 0.05 (0.02) |
| 195 | Treponema lecithinolyticum | 0.06 (0.01) | 0.06 (0.01) | 0.05 (0.01) | 0.06 (0.01) |
| 196 | Capnocytophaga sp._oral_taxon_902 | 0.06 (0.01) | 0.08 (0.02) | 0.05 (0.01) | 0.05 (0.01) |
| 197 | Peptostreptococcaceae_[XI][G-5] [Eubacterium]_saph | 0.06 (0.01) | 0.04 (0.01) | 0.07 (0.01) | 0.05 (0.01) |
| 198 | Lachnoanaerobaculum umeaense | 0.06 (0.00) | 0.06 (0.01) | 0.06 (0.01) | 0.05 (0.01) |
| 199 | Peptostreptococcaceae_[XI][G-6] [Eubacterium]_noda | 0.05 (0.00) | 0.06 (0.01) | 0.05 (0.01) | 0.06 (0.01) |
| 200 | Veillonellaceae_[G-1] sp._oral_taxon_145 | 0.05 (0.01) | 0.04 (0.01) | 0.05 (0.01) | 0.06 (0.01) |
| 201 | TM7_[G-1] sp._oral_taxon_352 | 0.05 (0.01) | 0.05 (0.01) | 0.05 (0.01) | 0.06 (0.01) |
| 202 | Capnocytophaga sp._oral_taxon_412 | 0.05 (0.01) | 0.05 (0.01) | 0.05 (0.01) | 0.06 (0.01) |
| 203 | Gemella sanguinis | 0.05 (0.00) | 0.04 (0.01) | 0.05 (0.01) | 0.06 (0.01) |
| 204 | Aggregatibacter sp._oral_taxon_513 | 0.05 (0.01) | 0.03 (0.02) | 0.07 (0.01) | 0.04 (0.01) |
| 205 | Selenomonas sp._oral_taxon_133 | 0.05 (0.01) | 0.06 (0.02) | 0.05 (0.01) | 0.05 (0.01) |
| 206 | Granulicatella elegans | 0.05 (0.01) | 0.06 (0.01) | 0.05 (0.01) | 0.05 (0.01) |
| 207 | Veillonellaceae_[G-1] sp._oral_taxon_129 | 0.05 (0.01) | 0.03 (0.01) | 0.04 (0.01) | 0.07 (0.02) |
| 208 | Selenomonas dianae | 0.05 (0.01) | 0.04 (0.02) | 0.04 (0.01) | 0.07 (0.03) |
| 209 | Prevotella sp._oral_taxon_526 | 0.05 (0.01) | 0.02 (0.01) | 0.06 (0.02) | 0.05 (0.01) |
| 210 | Porphyromonas sp._oral_taxon_275 | 0.05 (0.01) | 0.04 (0.01) | 0.06 (0.01) | 0.03 (0.01) |
| 211 | Anaerolineae_[G-1] sp._oral_taxon_439 | 0.05 (0.01) | 0.04 (0.01) | 0.04 (0.01) | 0.06 (0.01) |
| 212 | Stomatobaculum longum | 0.05 (0.00) | 0.04 (0.01) | 0.04 (0.00) | 0.05 (0.01) |
| 213 | Olsenella sp._oral_taxon_807 | 0.04 (0.00) | 0.04 (0.01) | 0.04 (0.00) | 0.05 (0.00) |
| 214 | Capnocytophaga sp._oral_taxon_323 | 0.04 (0.00) | 0.03 (0.01) | 0.05 (0.01) | 0.05 (0.01) |
| 215 | Capnocytophaga sp._oral_taxon_903 | 0.04 (0.00) | 0.03 (0.01) | 0.04 (0.01) | 0.05 (0.01) |
| 216 | Fretibacterium sp._oral_taxon_361 | 0.04 (0.01) | 0.01 (0.00) | 0.05 (0.02) | 0.05 (0.02) |
| 217 | Leptotrichia sp._oral_taxon_219 | 0.04 (0.00) | 0.05 (0.02) | 0.04 (0.00) | 0.04 (0.01) |
| 218 | Prevotella sp._oral_taxon_306 | 0.04 (0.01) | 0.04 (0.02) | 0.03 (0.01) | 0.06 (0.01) |
| 219 | Atopobium sp._oral_taxon_199 | 0.04 (0.01) | 0.04 (0.01) | 0.04 (0.01) | 0.03 (0.01) |
| 220 | Capnocytophaga sp._oral_taxon_324 | 0.04 (0.01) | 0.03 (0.01) | 0.04 (0.01) | 0.05 (0.01) |
| 221 | Prevotella buccae | 0.04 (0.00) | 0.02 (0.00) | 0.04 (0.01) | 0.05 (0.01) |
| 222 | Prevotella baroniae | 0.04 (0.00) | 0.02 (0.01) | 0.04 (0.01) | 0.04 (0.01) |
| 223 | Lachnospiraceae_[G-8] sp._oral_taxon_500 | 0.04 (0.00) | 0.04 (0.01) | 0.04 (0.01) | 0.04 (0.00) |
| 224 | Prevotella sp._oral_taxon_376 | 0.04 (0.00) | 0.05 (0.01) | 0.04 (0.01) | 0.03 (0.01) |
| 225 | Porphyromonas sp._oral_taxon_278 | 0.04 (0.01) | 0.03 (0.01) | 0.04 (0.01) | 0.04 (0.01) |
| 226 | Solobacterium moorei | 0.04 (0.00) | 0.04 (0.00) | 0.04 (0.00) | 0.04 (0.00) |
| 227 | Campylobacter curvus | 0.03 (0.00) | 0.05 (0.01) | 0.03 (0.01) | 0.03 (0.01) |
| 228 | Prevotella micans | 0.03 (0.00) | 0.03 (0.01) | 0.03 (0.01) | 0.04 (0.01) |
| 229 | Bacteroidaceae_[G-1] sp._oral_taxon_272 | 0.03 (0.00) | 0.03 (0.01) | 0.03 (0.00) | 0.04 (0.01) |
| 230 | Streptococcus australis | 0.03 (0.00) | 0.03 (0.01) | 0.03 (0.00) | 0.04 (0.01) |
| 231 | Tannerella sp._oral_taxon_808 | 0.03 (0.00) | 0.02 (0.00) | 0.03 (0.00) | 0.04 (0.01) |
| 232 | Bergeyella sp._oral_taxon_907 | 0.03 (0.00) | 0.03 (0.01) | 0.04 (0.01) | 0.03 (0.00) |
| 233 | Prevotella multiformis | 0.03 (0.01) | 0.01 (0.00) | 0.03 (0.01) | 0.05 (0.02) |
| 234 | Fusobacterium sp._oral_taxon_370 | 0.03 (0.01) | 0.02 (0.01) | 0.04 (0.01) | 0.03 (0.01) |
| 235 | Capnocytophaga sp._oral_taxon_380 | 0.03 (0.01) | 0.03 (0.01) | 0.04 (0.01) | 0.03 (0.01) |
| 236 | Peptostreptococcaceae_[XI][G-1] [Eubacterium]_infi | 0.03 (0.00) | 0.03 (0.01) | 0.03 (0.00) | 0.04 (0.00) |
| 237 | Treponema medium | 0.03 (0.00) | 0.03 (0.01) | 0.04 (0.01) | 0.02 (0.01) |
| 238 | Lachnoanaerobaculum orale | 0.03 (0.00) | 0.04 (0.01) | 0.02 (0.00) | 0.03 (0.01) |
| 239 | Alloprevotella sp._oral_taxon_308 | 0.03 (0.00) | 0.03 (0.00) | 0.03 (0.00) | 0.03 (0.01) |
| 240 | SR1_[G-1] sp._oral_taxon_874 | 0.03 (0.00) | 0.03 (0.00) | 0.03 (0.01) | 0.02 (0.00) |
| 241 | Actinomyces israelii | 0.03 (0.00) | 0.03 (0.00) | 0.03 (0.00) | 0.03 (0.00) |
| 242 | Shuttleworthia satelles | 0.03 (0.00) | 0.02 (0.01) | 0.03 (0.01) | 0.04 (0.01) |
| 243 | Mycoplasma salivarium | 0.03 (0.00) | 0.02 (0.00) | 0.03 (0.00) | 0.03 (0.00) |
| 244 | Leptotrichia goodfellowii | 0.03 (0.00) | 0.04 (0.01) | 0.03 (0.01) | 0.02 (0.00) |
| 245 | Streptococcus sinensis | 0.03 (0.00) | 0.03 (0.01) | 0.02 (0.00) | 0.04 (0.01) |
| 246 | TM7_[G-3] sp._oral_taxon_351 | 0.03 (0.00) | 0.02 (0.00) | 0.03 (0.00) | 0.03 (0.00) |
| 247 | Mitsuokella sp._oral_taxon_521 | 0.03 (0.01) | 0.01 (0.01) | 0.02 (0.01) | 0.04 (0.02) |
| 248 | Selenomonas sp._oral_taxon_442 | 0.03 (0.01) | 0.01 (0.00) | 0.04 (0.01) | 0.02 (0.01) |
| 249 | Actinomyces sp._oral_taxon_178 | 0.03 (0.00) | 0.02 (0.00) | 0.03 (0.00) | 0.03 (0.00) |
| 250 | Johnsonella sp._oral_taxon_166 | 0.02 (0.00) | 0.02 (0.01) | 0.03 (0.00) | 0.02 (0.00) |
| 251 | Lactobacillus gasseri | 0.02 (0.01) | 0.01 (0.00) | 0.02 (0.01) | 0.03 (0.01) |
| 252 | Treponema sp._oral_taxon_247 | 0.02 (0.01) | 0.01 (0.01) | 0.03 (0.01) | 0.02 (0.01) |
| 253 | GN02_[G-2] sp._oral_taxon_873 | 0.02 (0.00) | 0.03 (0.02) | 0.02 (0.01) | 0.02 (0.01) |
| 254 | Selenomonas sp._oral_taxon_937 | 0.02 (0.00) | 0.01 (0.00) | 0.02 (0.01) | 0.02 (0.00) |
| 255 | Atopobium sp._oral_taxon_416 | 0.02 (0.01) | 0.00 (0.00) | 0.01 (0.01) | 0.04 (0.02) |
| 256 | Sphingomonas echinoides | 0.02 (0.00) | 0.02 (0.01) | 0.02 (0.00) | 0.02 (0.01) |
| 257 | Butyrivibrio sp._oral_taxon_080 | 0.02 (0.00) | 0.02 (0.01) | 0.02 (0.01) | 0.02 (0.00) |
| 258 | Treponema vincentii | 0.02 (0.00) | 0.02 (0.00) | 0.03 (0.00) | 0.01 (0.00) |
| 259 | Selenomonas sp._oral_taxon_149 | 0.02 (0.00) | 0.01 (0.00) | 0.02 (0.01) | 0.02 (0.00) |
| 260 | Selenomonas sp._oral_taxon_478 | 0.02 (0.00) | 0.01 (0.00) | 0.01 (0.00) | 0.02 (0.01) |
| 261 | Prevotella sp._oral_taxon_475 | 0.02 (0.00) | 0.02 (0.01) | 0.02 (0.00) | 0.02 (0.01) |
| 262 | Pyramidobacter piscolens | 0.01 (0.00) | 0.01 (0.01) | 0.00 (0.00) | 0.01 (0.00) |
| 263 | Microbacterium flavescens | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) |
| 264 | Brevundimonas diminuta | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) |
| 265 | Sphingomonas sp._oral_taxon_006 | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) |
| 266 | Leptothrix sp._oral_taxon_025 | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) |
| 267 | Porphyrobacter tepidarius | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) |
aBacteria in the table are rank ordered according to their mean relative abundance (%) in the overall cohort
Dashed line inserted below the top 20 taxa
Table 6.
Prevalence (present, absent) for each of 267 bacterial species identified, overall and by age categories
| Rank Ordera | OTU Label | Age categories (years) | |||
|---|---|---|---|---|---|
| Overall (N = 1219) |
50–59 (N = 239) |
60–69 (N = 554) |
≥70 (N = 426) |
||
| % | % | % | % | ||
| 1 | Streptococcus oralis | 100 | 100 | 100 | 100 |
| 1 | Veillonella dispar | 100 | 100 | 100 | 100 |
| 1 | Veillonella parvula | 100 | 100 | 100 | 100 |
| 4 | Selenomonas sputigena | 99.8 | 99.6 | 99.8 | 99.8 |
| 5 | Fusobacterium nucleatum_subsp._vincentii | 99.6 | 99.6 | 99.6 | 99.5 |
| 6 | Fusobacterium nucleatum_subsp._animalis | 99.3 | 99.6 | 99.3 | 99.3 |
| 6 | Granulicatella adiacens | 99.3 | 99.2 | 99.5 | 99.3 |
| 8 | Streptococcus sanguinis | 99.3 | 100 | 99.3 | 99.1 |
| 9 | Rothia dentocariosa | 99.2 | 99.2 | 99.3 | 99.1 |
| 10 | Campylobacter gracilis | 99.1 | 99.6 | 98.9 | 99.1 |
| 11 | Selenomonas noxia | 99.0 | 98.7 | 99.1 | 99.1 |
| 11 | Streptococcus gordonii | 99.0 | 98.7 | 98.9 | 99.3 |
| 13 | Fusobacterium nucleatum_subsp._polymorphum | 98.6 | 99.2 | 98.9 | 97.9 |
| 14 | Fusobacterium sp._oral_taxon_203 | 98.4 | 98.7 | 98.0 | 98.8 |
| 14 | Streptococcus cristatus | 98.4 | 98.7 | 98.2 | 98.6 |
| 14 | Fusobacterium naviforme | 98.4 | 99.2 | 98.6 | 97.7 |
| 17 | Actinomyces naeslundii | 98.0 | 97.9 | 98.2 | 97.9 |
| 18 | Corynebacterium matruchotii | 97.5 | 96.7 | 97.3 | 98.1 |
| 19 | Prevotella oris | 97.2 | 97.5 | 97.3 | 96.9 |
| 20 | Actinomyces oris | 97.1 | 97.5 | 96.9 | 97.2 |
| 21 | Streptococcus salivarius | 97.0 | 96.7 | 96.8 | 97.7 |
| 21 | Parvimonas micra | 97.0 | 95.8 | 96.9 | 97.7 |
| 23 | Haemophilus parainfluenzae | 96.9 | 98.7 | 97.3 | 95.3 |
| 24 | Streptococcus intermedius | 96.8 | 97.9 | 95.5 | 97.9 |
| 25 | Veillonella atypica | 96.4 | 95.0 | 95.7 | 98.1 |
| 26 | Rothia mucilaginosa | 96.1 | 96.2 | 96.2 | 96.0 |
| 27 | Dialister invisus | 95.4 | 95.8 | 94.6 | 96.2 |
| 28 | Campylobacter concisus | 95.2 | 95.8 | 94.8 | 95.3 |
| 29 | Capnocytophaga gingivalis | 94.9 | 95.8 | 94.9 | 94.4 |
| 29 | TM7_[G-1] sp._oral_taxon_346 | 94.9 | 95.0 | 96.0 | 93.4 |
| 31 | Prevotella nigrescens | 94.2 | 95.0 | 94.2 | 93.7 |
| 32 | Kingella oralis | 93.0 | 92.9 | 94.2 | 91.5 |
| 33 | Bergeyella sp._oral_taxon_322 | 92.7 | 92.9 | 91.9 | 93.7 |
| 34 | Gemella haemolysans | 92.5 | 91.6 | 94.2 | 90.8 |
| 35 | Actinomyces sp._oral_taxon_169 | 92.3 | 93.7 | 93.0 | 90.6 |
| 36 | Actinomyces sp._oral_taxon_180 | 92.1 | 94.1 | 92.2 | 90.8 |
| 36 | Eikenella corrodens | 92.1 | 95.4 | 92.6 | 89.7 |
| 38 | Prevotella melaninogenica | 91.6 | 92.1 | 90.6 | 92.7 |
| 38 | TM7_[G-1] sp._oral_taxon_349 | 91.6 | 92.9 | 92.4 | 89.7 |
| 40 | Campylobacter showae | 91.3 | 91.6 | 91.0 | 91.5 |
| 41 | Actinomyces johnsonii | 91.1 | 91.6 | 91.7 | 90.1 |
| 42 | Capnocytophaga granulosa | 90.8 | 89.5 | 92.4 | 89.4 |
| 43 | Selenomonas sp._oral_taxon_136 | 90.7 | 89.1 | 90.4 | 92.0 |
| 44 | Bacteroidales_[G-2] sp._oral_taxon_274 | 90.4 | 88.3 | 91.5 | 90.1 |
| 45 | Selenomonas artemidis | 90.3 | 90.4 | 90.4 | 90.1 |
| 46 | Capnocytophaga leadbetteri | 90.2 | 90.0 | 91.7 | 88.5 |
| 46 | Actinomyces massiliensis | 90.2 | 92.1 | 91.3 | 87.6 |
| 48 | Fretibacterium sp._oral_taxon_360 | 90.0 | 91.2 | 90.4 | 88.7 |
| 49 | Gemella morbillorum | 89.8 | 89.5 | 90.3 | 89.4 |
| 50 | Streptococcus sp._oral_taxon_074 | 89.3 | 87.0 | 91.0 | 88.3 |
| 51 | Rothia aeria | 89.2 | 90.0 | 91.9 | 85.2 |
| 52 | Treponema socranskii | 88.8 | 88.7 | 88.4 | 89.4 |
| 53 | Leptotrichia hongkongensis | 88.7 | 85.4 | 89.2 | 89.9 |
| 54 | Streptococcus mutans | 88.4 | 84.9 | 88.1 | 90.8 |
| 54 | Cardiobacterium hominis | 88.4 | 91.6 | 89.2 | 85.4 |
| 54 | TM7_[G-1] sp._oral_taxon_952 | 88.4 | 90.0 | 90.3 | 85.0 |
| 57 | Actinomyces gerencseriae | 87.9 | 84.9 | 87.0 | 90.6 |
| 58 | Leptotrichia wadei | 87.7 | 86.6 | 87.0 | 89.2 |
| 59 | Anaeroglobus geminatus | 87.6 | 89.1 | 86.5 | 88.3 |
| 60 | Streptococcus parasanguinis_II | 87.4 | 87.4 | 84.3 | 91.5 |
| 61 | Alloprevotella tannerae | 87.3 | 90.0 | 85.6 | 88.0 |
| 62 | Actinomyces sp._oral_taxon_171 | 87.2 | 88.7 | 87.4 | 86.2 |
| 62 | Capnocytophaga sputigena | 87.2 | 88.7 | 89.0 | 84.0 |
| 64 | Fusobacterium nucleatum_subsp._nucleatum | 86.8 | 90.8 | 86.5 | 85.0 |
| 65 | Catonella morbi | 86.6 | 87.0 | 87.2 | 85.7 |
| 66 | Corynebacterium durum | 85.6 | 88.7 | 86.8 | 82.2 |
| 66 | Selenomonas infelix | 85.6 | 88.7 | 85.6 | 83.8 |
| 68 | Prevotella denticola | 85.5 | 82.8 | 84.7 | 88.0 |
| 69 | Prevotella sp._oral_taxon_317 | 85.2 | 88.3 | 83.6 | 85.4 |
| 70 | Leptotrichia sp._oral_taxon_212 | 84.8 | 87.4 | 87.0 | 80.5 |
| 71 | Porphyromonas sp._oral_taxon_279 | 84.5 | 83.7 | 85.7 | 83.3 |
| 72 | Neisseria sicca | 83.5 | 80.8 | 86.3 | 81.5 |
| 73 | Gemella sanguinis | 83.1 | 85.4 | 80.9 | 84.7 |
| 74 | Peptostreptococcaceae_[XI][G-9] [Eubacterium]_brac | 83.0 | 87.4 | 82.5 | 81.2 |
| 75 | Tannerella forsythia | 82.9 | 82.0 | 83.8 | 82.4 |
| 75 | Neisseria flavescens | 82.9 | 77.8 | 85.4 | 82.4 |
| 77 | Prevotella sp._oral_taxon_300 | 82.5 | 80.8 | 81.4 | 85.0 |
| 78 | Prevotella maculosa | 82.3 | 80.8 | 81.4 | 84.3 |
| 79 | Neisseria elongata | 82.2 | 80.3 | 84.1 | 80.8 |
| 80 | Selenomonas flueggei | 82.0 | 79.9 | 82.9 | 81.9 |
| 81 | Selenomonas sp._oral_taxon_892 | 81.8 | 85.4 | 83.6 | 77.5 |
| 82 | Actinomyces israelii | 81.7 | 79.9 | 80.9 | 83.8 |
| 83 | Oribacterium sp._oral_taxon_078 | 81.4 | 78.2 | 80.1 | 84.7 |
| 84 | Lachnoanaerobaculum saburreum | 81.3 | 78.7 | 82.5 | 81.2 |
| 85 | Megasphaera micronuciformis | 81.1 | 79.1 | 78.5 | 85.4 |
| 85 | Prevotella salivae | 81.1 | 73.2 | 80.9 | 85.7 |
| 87 | Streptococcus parasanguinis_I | 80.6 | 80.3 | 78.2 | 84.0 |
| 88 | Cardiobacterium valvarum | 80.5 | 81.2 | 82.1 | 77.9 |
| 89 | Prevotella oulorum | 80.1 | 82.8 | 77.6 | 81.9 |
| 89 | Veillonellaceae_[G-1] sp._oral_taxon_150 | 80.1 | 78.2 | 79.1 | 82.4 |
| 91 | Leptotrichia sp._oral_taxon_417 | 79.7 | 74.5 | 80.3 | 81.7 |
| 92 | Veillonella rogosae | 79.6 | 82.4 | 82.7 | 73.9 |
| 93 | Streptococcus anginosus | 79.5 | 81.2 | 78.0 | 80.5 |
| 94 | Veillonellaceae_[G-1] sp._oral_taxon_155 | 79.4 | 77.4 | 77.8 | 82.6 |
| 95 | Fretibacterium fastidiosum | 79.2 | 79.1 | 80.1 | 78.2 |
| 96 | Lautropia mirabilis | 78.2 | 82.8 | 78.9 | 74.6 |
| 97 | Actinobaculum sp._oral_taxon_183 | 78.0 | 78.7 | 78.3 | 77.2 |
| 98 | Fusobacterium periodonticum | 77.7 | 74.9 | 82.1 | 73.5 |
| 99 | Selenomonas sp._oral_taxon_919 | 77.2 | 84.1 | 75.6 | 75.4 |
| 100 | Selenomonas sp._oral_taxon_137 | 76.8 | 80.3 | 76.9 | 74.6 |
| 101 | Atopobium parvulum | 76.5 | 76.2 | 73.8 | 80.3 |
| 101 | Streptococcus lactarius | 76.5 | 80.3 | 75.1 | 76.1 |
| 103 | Kingella denitrificans | 76.3 | 72.0 | 76.0 | 79.1 |
| 104 | Capnocytophaga sp._oral_taxon_336 | 75.1 | 77.4 | 75.5 | 73.5 |
| 105 | Lachnospiraceae_[G-3] sp._oral_taxon_100 | 73.7 | 77.0 | 73.6 | 72.1 |
| 106 | Atopobium rimae | 73.3 | 72.4 | 73.1 | 74.2 |
| 107 | Streptococcus constellatus | 73.0 | 72.8 | 71.7 | 74.9 |
| 108 | Tannerella sp._oral_taxon_286 | 72.8 | 69.9 | 74.9 | 71.8 |
| 108 | Selenomonas sp._oral_taxon_134 | 72.8 | 75.7 | 72.9 | 70.9 |
| 110 | Leptotrichia buccalis | 72.0 | 64.9 | 76.0 | 70.9 |
| 111 | Selenomonas sp._oral_taxon_146 | 71.9 | 73.2 | 72.9 | 69.7 |
| 112 | Leptotrichia sp._oral_taxon_215 | 71.8 | 75.3 | 72.4 | 69.0 |
| 113 | Leptotrichia sp._oral_taxon_392 | 71.4 | 74.5 | 74.9 | 65.0 |
| 114 | TM7_[G-5] sp._oral_taxon_356 | 71.2 | 74.5 | 72.2 | 68.1 |
| 115 | Dialister pneumosintes | 70.6 | 74.1 | 70.2 | 69.2 |
| 115 | Prevotella sp._oral_taxon_472 | 70.6 | 71.5 | 75.3 | 64.1 |
| 117 | Streptococcus sp._oral_taxon_056 | 70.6 | 69.0 | 73.3 | 68.1 |
| 118 | Fretibacterium sp._oral_taxon_359 | 70.5 | 66.9 | 71.1 | 71.6 |
| 119 | Solobacterium moorei | 70.4 | 69.0 | 70.0 | 71.6 |
| 120 | Abiotrophia defectiva | 70.1 | 69.9 | 72.9 | 66.7 |
| 121 | Lachnoanaerobaculum umeaense | 69.3 | 69.9 | 70.2 | 67.8 |
| 122 | Parvimonas sp._oral_taxon_393 | 69.2 | 69.9 | 70.2 | 67.4 |
| 123 | Actinomyces meyeri | 67.8 | 71.1 | 69.7 | 63.4 |
| 124 | Centipeda periodontii | 67.7 | 64.0 | 69.1 | 67.8 |
| 125 | Leptotrichia shahii | 67.6 | 72.4 | 66.6 | 66.2 |
| 126 | Olsenella sp._oral_taxon_807 | 66.9 | 61.9 | 67.1 | 69.2 |
| 127 | Actinobaculum sp._oral_taxon_848 | 66.3 | 64.9 | 66.1 | 67.4 |
| 128 | Leptotrichia hofstadii | 66.0 | 63.6 | 68.2 | 64.6 |
| 128 | Neisseria oralis | 66.0 | 66.1 | 69.0 | 62.2 |
| 128 | Prevotella histicola | 66.0 | 62.3 | 62.8 | 72.1 |
| 131 | Actinomyces sp._oral_taxon_178 | 65.8 | 61.1 | 65.3 | 69.0 |
| 132 | Stomatobaculum longum | 65.7 | 62.3 | 63.7 | 70.2 |
| 132 | TM7_[G-1] sp._oral_taxon_352 | 65.7 | 60.3 | 70.6 | 62.4 |
| 134 | Capnocytophaga sp._oral_taxon_326 | 65.4 | 64.4 | 68.1 | 62.4 |
| 135 | Prevotella oralis | 64.5 | 63.6 | 65.2 | 64.1 |
| 136 | Streptococcus sinensis | 63.9 | 65.7 | 62.1 | 65.3 |
| 137 | Alloprevotella sp._oral_taxon_308 | 63.5 | 68.6 | 61.9 | 62.7 |
| 138 | Porphyromonas sp._oral_taxon_284 | 63.3 | 64.4 | 66.8 | 58.2 |
| 138 | Prevotella pallens | 63.3 | 63.2 | 61.7 | 65.5 |
| 140 | Aggregatibacter sp._oral_taxon_458 | 63.2 | 65.7 | 65.3 | 58.9 |
| 141 | Porphyromonas endodontalis | 63.1 | 69.5 | 63.7 | 58.7 |
| 142 | Prevotella saccharolytica | 63.0 | 65.3 | 65.0 | 59.2 |
| 143 | Selenomonas sp._oral_taxon_126 | 62.0 | 62.3 | 63.4 | 60.1 |
| 144 | Lachnoanaerobaculum orale | 61.1 | 63.2 | 58.5 | 63.4 |
| 145 | Treponema maltophilum | 61.0 | 59.0 | 62.3 | 60.3 |
| 146 | Peptostreptococcaceae_[XI][G-7] [Eubacterium]_yuri | 60.2 | 64.0 | 62.3 | 55.4 |
| 147 | Veillonella denticariosi | 60.1 | 56.5 | 60.8 | 61.3 |
| 148 | Sphingomonas echinoides | 59.8 | 64.9 | 61.7 | 54.5 |
| 149 | Streptococcus australis | 59.7 | 57.3 | 59.0 | 62.0 |
| 150 | TM7_[G-1] sp._oral_taxon_348 | 59.6 | 57.7 | 63.4 | 55.9 |
| 151 | TM7_[G-1] sp._oral_taxon_488 | 57.8 | 56.9 | 61.2 | 54.0 |
| 152 | Ruminococcaceae_[G-1] sp._oral_taxon_075 | 56.9 | 58.2 | 59.7 | 52.6 |
| 153 | Selenomonas sp._oral_taxon_936 | 56.8 | 58.2 | 56.3 | 56.8 |
| 154 | Prevotella buccae | 56.7 | 49.4 | 54.7 | 63.4 |
| 155 | Leptotrichia sp._oral_taxon_225 | 56.4 | 60.3 | 58.7 | 51.4 |
| 155 | Prevotella pleuritidis | 56.4 | 55.2 | 59.4 | 53.3 |
| 157 | Alloprevotella rava | 56.2 | 57.7 | 56.9 | 54.5 |
| 158 | Aggregatibacter aphrophilus | 56.1 | 57.7 | 59.6 | 50.7 |
| 159 | Treponema denticola | 56.0 | 59.4 | 58.1 | 51.4 |
| 160 | Peptostreptococcus stomatis | 55.9 | 64.0 | 55.1 | 52.3 |
| 161 | Pseudomonas fluorescens | 55.5 | 53.6 | 57.0 | 54.5 |
| 162 | Actinomyces sp._oral_taxon_170 | 55.2 | 60.3 | 57.0 | 50.0 |
| 163 | Porphyromonas gingivalis | 55.1 | 51.0 | 56.3 | 55.9 |
| 164 | Haemophilus sp._oral_taxon_036 | 55.0 | 59.4 | 54.7 | 52.8 |
| 165 | Johnsonella ignava | 54.6 | 54.0 | 56.3 | 52.8 |
| 166 | Porphyromonas catoniae | 54.4 | 57.7 | 56.5 | 49.8 |
| 167 | Peptostreptococcaceae_[XI][G-1] [Eubacterium]_infi | 54.2 | 47.3 | 54.7 | 57.5 |
| 168 | Prevotella sp._oral_taxon_292 | 53.8 | 49.4 | 54.3 | 55.6 |
| 169 | Fretibacterium sp._oral_taxon_362 | 53.6 | 53.6 | 53.8 | 53.3 |
| 170 | TM7_[G-1] sp._oral_taxon_347 | 53.2 | 58.2 | 54.3 | 49.1 |
| 171 | Leptotrichia sp._oral_taxon_219 | 52.7 | 49.4 | 54.2 | 52.6 |
| 172 | Aggregatibacter segnis | 52.1 | 56.1 | 54.9 | 46.2 |
| 173 | TM7_[G-3] sp._oral_taxon_351 | 51.9 | 45.6 | 55.4 | 50.9 |
| 174 | Fusobacterium sp._oral_taxon_370 | 51.1 | 45.6 | 55.6 | 48.4 |
| 175 | Prevotella sp._oral_taxon_313 | 50.5 | 53.1 | 46.4 | 54.2 |
| 176 | Prevotella loescheii | 50.4 | 52.3 | 54.5 | 43.9 |
| 177 | Desulfobulbus sp._oral_taxon_041 | 50.3 | 48.1 | 50.9 | 50.7 |
| 178 | Capnocytophaga sp._oral_taxon_864 | 49.1 | 51.9 | 52.0 | 43.9 |
| 179 | Ottowia sp._oral_taxon_894 | 48.4 | 49.4 | 50.2 | 45.5 |
| 180 | Microbacterium flavescens | 48.0 | 50.2 | 49.5 | 44.8 |
| 181 | Leptotrichia sp._oral_taxon_498 | 47.8 | 42.3 | 48.2 | 50.5 |
| 182 | Granulicatella elegans | 47.3 | 52.7 | 45.5 | 46.5 |
| 183 | Filifactor alocis | 46.8 | 50.2 | 46.8 | 44.8 |
| 184 | Campylobacter curvus | 46.2 | 45.6 | 46.4 | 46.2 |
| 185 | Mycoplasma salivarium | 45.0 | 47.7 | 45.3 | 43.2 |
| 186 | Capnocytophaga sp._oral_taxon_338 | 44.8 | 42.7 | 44.4 | 46.5 |
| 187 | Capnocytophaga sp._oral_taxon_902 | 44.3 | 43.1 | 46.6 | 42.0 |
| 188 | Treponema sp._oral_taxon_231 | 43.6 | 51.5 | 45.3 | 37.1 |
| 189 | Tannerella sp._oral_taxon_808 | 43.1 | 39.3 | 45.7 | 41.8 |
| 190 | Prevotella intermedia | 42.8 | 40.2 | 43.3 | 43.7 |
| 191 | TM7_[G-6] sp._oral_taxon_870 | 42.7 | 43.9 | 44.2 | 39.9 |
| 192 | Mitsuokella sp._oral_taxon_131 | 42.2 | 35.6 | 41.0 | 47.7 |
| 193 | Haemophilus haemolyticus | 42.0 | 47.3 | 41.3 | 39.9 |
| 194 | Prevotella veroralis | 41.7 | 44.4 | 40.8 | 41.3 |
| 195 | Veillonellaceae_[G-1] sp._oral_taxon_129 | 40.9 | 37.7 | 40.8 | 43.0 |
| 195 | Aggregatibacter paraphrophilus | 40.9 | 45.2 | 42.2 | 36.6 |
| 197 | Pseudoramibacter alactolyticus | 40.9 | 33.5 | 41.5 | 44.1 |
| 198 | Bergeyella sp._oral_taxon_907 | 40.6 | 39.7 | 42.8 | 38.3 |
| 199 | Bifidobacterium dentium | 40.5 | 33.9 | 37.9 | 47.7 |
| 199 | TM7_[G-1] sp._oral_taxon_869 | 40.5 | 41.8 | 41.5 | 38.5 |
| 201 | Prevotella sp._oral_taxon_306 | 40.3 | 36.0 | 39.5 | 43.7 |
| 202 | Prevotella dentalis | 39.3 | 40.2 | 36.5 | 42.5 |
| 203 | Shuttleworthia satelles | 39.2 | 34.7 | 37.5 | 43.9 |
| 204 | Veillonella sp._oral_taxon_780 | 39.1 | 40.6 | 42.4 | 34.0 |
| 205 | Capnocytophaga sp._oral_taxon_412 | 38.7 | 38.9 | 41.5 | 35.0 |
| 206 | Neisseria subflava | 38.6 | 39.3 | 37.4 | 39.7 |
| 206 | Scardovia wiggsiae | 38.6 | 37.2 | 37.7 | 40.4 |
| 208 | Leptotrichia sp._oral_taxon_223 | 37.8 | 42.3 | 36.5 | 37.1 |
| 209 | Peptostreptococcaceae_[XI][G-6] [Eubacterium]_noda | 37.2 | 35.6 | 37.0 | 38.5 |
| 209 | Selenomonas sp._oral_taxon_937 | 37.2 | 36.4 | 35.2 | 40.1 |
| 211 | Bacteroidetes_[G-5] sp._oral_taxon_511 | 36.8 | 41.0 | 37.2 | 33.8 |
| 212 | Capnocytophaga sp._oral_taxon_323 | 36.7 | 36.0 | 37.4 | 36.2 |
| 213 | Selenomonas sp._oral_taxon_149 | 36.4 | 30.5 | 33.8 | 43.2 |
| 213 | Selenomonas sp._oral_taxon_442 | 36.4 | 30.1 | 37.5 | 38.5 |
| 215 | SR1_[G-1] sp._oral_taxon_874 | 36.3 | 39.3 | 40.3 | 29.6 |
| 216 | Porphyromonas sp._oral_taxon_275 | 36.0 | 36.4 | 37.7 | 33.6 |
| 217 | Selenomonas sp._oral_taxon_478 | 35.9 | 31.4 | 38.4 | 35.2 |
| 218 | Alloprevotella sp._oral_taxon_473 | 35.8 | 39.7 | 36.8 | 32.4 |
| 219 | Neisseria bacilliformis | 35.5 | 35.1 | 35.4 | 35.9 |
| 220 | Capnocytophaga sp._oral_taxon_324 | 35.4 | 32.6 | 38.1 | 33.3 |
| 221 | Veillonellaceae_[G-1] sp._oral_taxon_145 | 35.2 | 35.1 | 36.1 | 34.0 |
| 222 | Atopobium sp._oral_taxon_199 | 34.9 | 36.0 | 36.1 | 32.6 |
| 223 | Lachnospiraceae_[G-8] sp._oral_taxon_500 | 34.8 | 33.1 | 35.7 | 34.5 |
| 224 | TM7_[G-2] sp._oral_taxon_350 | 34.6 | 34.7 | 34.7 | 34.5 |
| 225 | Bacteroidaceae_[G-1] sp._oral_taxon_272 | 33.1 | 26.8 | 33.9 | 35.7 |
| 225 | Capnocytophaga sp._oral_taxon_903 | 33.1 | 31.8 | 36.1 | 30.0 |
| 225 | Selenomonas dianae | 33.1 | 28.9 | 35.0 | 32.9 |
| 228 | Prevotella baroniae | 33.0 | 32.2 | 33.4 | 32.9 |
| 229 | Leptotrichia goodfellowii | 32.2 | 37.2 | 34.1 | 27.0 |
| 230 | Prevotella micans | 31.4 | 29.3 | 32.7 | 31.0 |
| 231 | Porphyromonas sp._oral_taxon_278 | 31.0 | 31.8 | 31.9 | 29.3 |
| 232 | Prevotella sp._oral_taxon_314 | 30.8 | 30.1 | 31.4 | 30.5 |
| 233 | Haemophilus parahaemolyticus | 30.3 | 37.2 | 31.8 | 24.4 |
| 234 | Anaerolineae_[G-1] sp._oral_taxon_439 | 30.2 | 25.5 | 30.0 | 33.1 |
| 235 | Selenomonas sp._oral_taxon_133 | 30.1 | 28.9 | 30.5 | 30.3 |
| 236 | Treponema lecithinolyticum | 29.5 | 30.1 | 31.2 | 26.8 |
| 237 | Peptostreptococcaceae_[XI][G-5] [Eubacterium]_saph | 28.3 | 22.2 | 31.4 | 27.7 |
| 238 | Treponema sp._oral_taxon_237 | 27.0 | 32.2 | 28.9 | 21.6 |
| 239 | Neisseria pharyngis | 25.8 | 25.1 | 25.5 | 26.8 |
| 240 | Leptotrichia sp._oral_taxon_879 | 25.4 | 25.9 | 25.6 | 24.9 |
| 241 | Brevundimonas diminuta | 25.3 | 27.6 | 25.8 | 23.2 |
| 242 | Bradyrhizobium elkanii | 24.9 | 26.4 | 22.7 | 26.8 |
| 242 | Capnocytophaga sp._oral_taxon_332 | 24.9 | 29.7 | 26.2 | 20.4 |
| 244 | Megasphaera sp._oral_taxon_123 | 24.5 | 32.2 | 24.2 | 20.7 |
| 245 | Capnocytophaga sp._oral_taxon_380 | 22.2 | 20.5 | 24.9 | 19.7 |
| 246 | Treponema vincentii | 22.0 | 23.0 | 23.5 | 19.5 |
| 247 | Prevotella sp._oral_taxon_376 | 21.4 | 26.8 | 23.5 | 15.7 |
| 248 | Prevotella sp._oral_taxon_526 | 21.1 | 21.8 | 21.7 | 20.0 |
| 249 | Aggregatibacter sp._oral_taxon_513 | 20.2 | 21.8 | 23.1 | 15.5 |
| 250 | Lactobacillus gasseri | 19.9 | 18.0 | 17.9 | 23.7 |
| 251 | Fretibacterium sp._oral_taxon_358 | 19.3 | 16.7 | 19.3 | 20.7 |
| 252 | Mitsuokella sp._oral_taxon_521 | 18.7 | 18.4 | 19.9 | 17.4 |
| 253 | Fretibacterium sp._oral_taxon_361 | 18.5 | 15.5 | 19.1 | 19.2 |
| 254 | Prevotella sp._oral_taxon_475 | 18.2 | 19.2 | 20.0 | 15.3 |
| 255 | Treponema medium | 18.1 | 16.3 | 19.5 | 17.4 |
| 256 | Johnsonella sp._oral_taxon_166 | 16.7 | 12.6 | 18.6 | 16.7 |
| 257 | Streptococcus sobrinus | 15.9 | 13.8 | 14.4 | 19.0 |
| 258 | Prevotella multiformis | 15.5 | 16.3 | 15.5 | 15.0 |
| 259 | Butyrivibrio sp._oral_taxon_080 | 14.5 | 15.1 | 15.9 | 12.4 |
| 260 | GN02_[G-2] sp._oral_taxon_873 | 13.7 | 10.9 | 15.0 | 13.6 |
| 261 | Aggregatibacter actinomycetemcomitans | 12.3 | 12.1 | 11.4 | 13.6 |
| 262 | Leptothrix sp._oral_taxon_025 | 10.9 | 13.4 | 11.4 | 8.9 |
| 263 | Atopobium sp._oral_taxon_416 | 8.7 | 5.4 | 8.5 | 10.8 |
| 264 | Sphingomonas sp._oral_taxon_006 | 7.2 | 5.9 | 7.9 | 7.0 |
| 265 | Porphyrobacter tepidarius | 6.9 | 7.1 | 6.1 | 7.7 |
| 266 | Treponema sp._oral_taxon_247 | 4.8 | 6.3 | 4.5 | 4.2 |
| 267 | Pyramidobacter piscolens | 4.0 | 2.9 | 3.8 | 4.9 |
aBacteria in the table are rank ordered according to their prevalence in the overall cohort
Dashed line inserted below top 20 taxa
Discussion
The objective of the present study was to characterize, using high throughput sequencing of the 16S rRNA bacterial gene, the subgingival microbiome in relation to age among community-dwelling postmenopausal women, aged 53–81 years, whose selection into the study was not conditioned on presence or severity of periodontitis. We identified 267 taxa, of which 55% had previously been named within the HOMD database. The remaining previously unnamed OTUs could potentially identify novel microbiota residing in human subgingival biofilm, new discovery that could have important implications to periodontal microbiology [17, 34, 35]. The majority of taxa identified in our study fell within the four major human bacterial phyla (Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria) determined in the HMP [36] and by others [3, 23] including the oral microbiome [21, 22, 37–39]. As in other studies on the oral microbiome [21, 22, 38–41], the most abundant phyla in our study were Firmicutes, Bacteroidetes, Fusobacteria and Proteobacteria, accounting for 46, 17, 14, and 9%, respectively, of the 265 taxa identified. The Firmicutes-to-Bacteroidetes ratio has been suggested as a possible indicator of the overall status of a microbial habitat in aging humans [6]. Previous studies on the gut microbiome have shown a lower ratio in older compared with younger individuals [42, 43]. In contrast, we observed a tendency toward higher Firmicutes-to-Bacteroidetes ratios across incremental age groups. In so much as some of the most virulent and well established periodontal pathogens (e.g., P. gingivalis, T. forsythia, T. denticola) reside in the phylum Bacteroides, whereas bacteria associated with a healthy periodontium (S. sanguis, oralis) reside in the phylum Firmicutes, a higher F-B ratio in the present cohort of aging women might be expected given the relatively small mean probing pocket depth (2.2 mm) overall, and lack of difference in this clinical measure of periodontitis across age groups. Whether the F-B ratio has similar relevance in the oral microbiome as has been reported previously for the gut microbiome requires further investigation.
The most abundant genus in our cohort was Veillonella, followed by Streptococcus, Fusobacterium, and Prevotella, with little variation in the distribution across age categories. Previous studies using targeted methods for measuring oral microbiota found substantially elevated abundance of Actinomyces and Fusobacterium genera in older adults [17, 38], which was not the case in our study (Actinomyces, overall: 1%, 70–79 years: 2%; Fusobacterium, overall: 11%, 70–79 years 10%) when using untargeted high-throughput sequencing. Other studies that measured the oral microbiome using 16S rRNA sequencing have reported the distribution of genera. Among community-dwelling adults (mean age 83; 61% women), analysis of salivary microbiome revealed Prevotella (22%) was most abundant, followed by Neisseria (12%), Veillonella (10%), and Streptococcus (8%) [22]. In another study on the salivary microbiome in Mexican American women, aged 50 and older, Hoffman et al. [21] reported that Streptococcus was most abundant (37%), followed by Prevotella (11%), Haemophilus (10%), and Veillonella (6%). Among Alaskan adults, aged 20–40 years, Streptococcus (28%) and Prevotella (27%) were by far most abundant, followed by Rothia (11%) and Veillonella (8%) [41]. Variation of microbial genera with age was not reported in these previous studies. Notwithstanding, there does appear to be some consistency across studies using culture-independent sequencing methods, including ours, in that Streptococcus, Prevotella, and Veillonella are abundant microbial genera commonly observed in the adult human oral microbiome.
Our primary analysis on microbial species composition and variation with age was based on CLR transformed OTUs taking into account the complex compositional structure of microbiome data [32]. The top 20 most abundant bacterial species had CLR means from 3.85 to 8.25, indicating these species were 14- to 304-fold (i.e., 23.85 to 28.25) more abundant than the overall composition mean (Table 3). V. dispar, S. oralis, and V. parvula were the top three most abundant species, each with CLR means > 7. V. dispar and parvula are gram-negative anaerobic bacteria commonly found in the human oral cavity [44], and have been associated with caries and periodontitis [34]. Evidence suggests V. parvula synergizes with Lachnoanaerobaculum (Eubacterium) saburreum, and the energy it produces, as a critical part of human subgingival biofilm formation [45]. L. saburreum was found at a relatively small, but elevated, abundance in our cohort (mean CLR, 1.07). Both bacterial species were positively correlated with age in our study, with a stronger correlation for V. parvula (r = 0.10) compared with L. saburreum (r = 0.04). S. oralis, in contrast, tends to be abundant in soft tissues of healthy periodontium [34], and as such was an original component in Socransky’s “yellow complex” defined using the checkerboard DNA-DNA hybridization method. S. oralis abundance has been shown to decline in the setting of experimental subgingival biofilm growth [34], which suggests it might be a key bacterium involved with the shift from a healthy to disease subgingival microbial ecology leading to periodontitis. The correlations with age for V. parvula (r = 0.10) and S. oralis (r = − 0.10) observed in the present study suggest that age could be a potential host factor contributing to susceptibility for untoward shifts in the human subgingival microbial ecology. Chronological age, per se, however, may not be the biologically relevant effector of shifts in microbial ecology. Rather, the tendency of aging to be associated with chronic immune function decline and upregulated proinflammatory signaling [8], referred to as “inflamm-aging” by Franceshi and coworkers [46] is likely a culpable perturbation of colonizing microbiota. Consistent with this hypothesis are results from studies of experimentally induced gingivitis, which demonstrated markedly greater amounts and severity of biofilm development in older than younger adults, despite no obvious differences in microbial compositional characteristics of the biofilm between age groups [17].
In the present study, 12 bacterial species differed significantly across age groups (Fig. 5). The largest difference in bacteria elevated in older adults was for B. dentium (phylum Actinobacteria), an anaerobe that has strong adhesion capacity, tolerates highly acidic conditions, and has been associated with human dental caries [47], but also with suppression of P. gingivalis, a virulent periodontitis pathogen [48]. This might partially explain why P. gingivalis was in relatively low abundance in our cohort of older women. Anaeroglobus geminatus (phylum Firmicutes) also demonstrated a rather large elevation in older compared with younger adults in our cohort. This bacterium has an identified role in perturbing a shift in the subgingival microbial ecology that favors development of periodontitis [49]. There was no difference in mean pocket depth measures among age groups in our cohort of older women, among whom prevalence of major risk factors for periodontitis, smoking and diabetes, also were low. However, it is conceivable that higher abundance of B. dentium and A. geminatus in the older age group could be reflective of an ongoing subgingival microbial community shift that leads to increased susceptibility to periodontitis progression in these women over time. Longitudinal analyses are required to confirm this hypothesis.
S. sanguinis and Corynebacterium durum showed the largest differences in bacteria between age groups among those elevated in younger women (Fig. 5). S. sanguinis (phylum Firmicutes) is a gram-positive anaerobe that is abundant in healthy periodontium [34] and plays a role in modifying the environment on oral surfaces such as to suppress growth of other Streptococci bacteria involved with oral diseases, such as S. mutans which is a causal agent in human carries [50]. S. sanguinis also might play a role in the shift of subgingival microbiota from a healthy to a disease ecology, serving as an adhesion site for virulent periodontal pathogens, such as P. gingivalis and F. nucleatum [50], each of which were in relatively low abundance in the present study. The role that C. durum (phylum Actinobacteria), also a gram-positive bacterium, might have in the subginigival microbial ecology is not entirely clear. Elevations of this bacterium originally was identified in bronchial wash solution and implicated in maintaining a healthy respiratory tract [51] and later, it’s reduction in saliva was associated with halitosis [52] and celiac disease [53]. Given it’s propensity to produce acid from available sugar compounds in saliva [54], and perhaps in other oral fluids including the gingival crevice, it is possible that this bacterium has a role in establishing or maintaining pH of the gingival pocket at a level commensurate with survival of other bacteria associated with periodontal health, such as S. sanguinis.
The vast majority of studies using untargeted high-throughput sequencing methods of the oral microbiome have reported measures of relative abundance or prevalence when describing microbial composition. Our primary measure for analysis of microbiota abundance in was the centered log-transformed ratio (CLR) OTU, as recommended by Gloor and coworkers [32]. While the basic cross-sectional findings of the present study were generally consistent when based on mean CLR OTUs, relative abundance, and prevalence, we believe that the CLR approach is the method of choice. Compositional data are vectors of non-negative numbers that sum to a fixed value, a constraint that can lead to spurious correlations. Subsequent work by Aitchison and colleagues yielded a set of log-ratio transformations that alleviate the sum-constraint burden, provide a consistent variance-covariance structure, and ensure that statistical results show consistency over subcompositions and OTU permutations [32, 55]. Subcompositional consistency, in particular, is necessary for the fundamental scientific concept of reproducibility across studies. The application of methods which ignore the compositional structure of microbiome data, like simple proportions (e.g., relative abundance, prevalent, or rarefaction) can lead to false positive associations and inferences [32]. In addition, the CLR transformation does not reduce the dimensionality of the dataset, maintaining the correspondence between transformed variables and OTUs, and easing the interpretation of conventional statistical tests, such as bivariate correlations and analysis of variance. Given the recent growth in microbiome research, the plethora of published studies that used different analytic methods, and the potential impact that continued investigation of the human microbiome could have on future understanding of disease etiology and therapeutics [24], the need for standardization of methods for analyzing and reporting microbiome data is paramount.
The present study has both strengths and limitations that need be considered when interpreting and generalizing its findings. Strengths include the large sample size of community-dwelling older postmenopausal women whose selection into the study was not conditioned on periodontitis presence or severity. Of the published studies reporting on the oral microbiome in older adults, the vast majority included relatively small sample sizes (e.g., < 100) and individuals that were selected to have either periodontal health or disease, often recruited from dental or other healthcare settings [17, 18, 22, 37, 40, 56]. Understanding the epidemiology of oral microbiota composition and its association with host characteristics in a more general community setting is a critical foundational step for evaluating associations between oral microbiome and both oral and systemic disease, as well as response to therapeutic intervention [24]. Previous oral microbiome studies on older adults relied largely on targeted low-throughput methods for characterizing oral microbiota [17, 18, 38, 56]. The limitations of these methods have been discussed elsewhere [17, 57]. Only recently have studies, including ours, utilized state-of-the science untargeted high-throughput next generation sequencing methods to investigate the oral microbiome in adults in middle- and older ages [21–23, 39–41]. This not only allows for greater sensitivity in characterizing the complexity of oral microbial communities, but also for potential discovery of new previously unidentified microbiota, which is essential to deeper understanding of the oral microbiome [17, 35]. Weaknesses of the present study include its cross-sectional design, which precludes temporal understanding of the relationship between aging and formation of the observed oral microbiome. The cross-sectional nature of our results precludes causal inferences regarding the relationship between age and the subgingival microbial composition and diversity. Using means to describe complex data, such as the subgingival microbiome, is helpful for descriptive purposes and ease of understanding, however they do not provide insight on between-subject variability nor do they allow for understanding of shifts between healthy and disease ecologies [34]. Prospective studies are needed using statistical methods appropriate for quantifying changes in microbiota between groups differing on host characteristics, such as aging or periodontal disease onset and progression, or in response to therapeutic intervention. The present cross-sectional observations, such as the significant differences in CLR mean OTUs between older and younger women (Fig. 5), could inform development of hypotheses for testing in a prospective study design. Lastly, we were not able to determine the functional attributes of the particularly abundant or sparse microbiota identified in our older cohort of women, nor of the bacterium that differed in abundance between older and younger women. It is becoming clearer that the functions determined by the genes expressed by microbiota are likely more influential on health or disease states than is the microbial composition [1, 15, 17]. Because aging is a non-modifiable host characteristic intimately involved with both structural and functional changes in the human body over the adult lifespan, the relationship between age and microbial function is of high interest [7].
Conclusion
We conclude that in a large cross-sectional analysis on the subgingival microbiome in postmenopausal women, aged 53–81 years, who were not selected on the basis of periodontitis status, a diverse subgingival microbiome was present and several bacterial species were correlated with age across the age range studied. Twelve microbiota were identified that differed significantly in abundance between women aged 50–59 versus 70 and older. Prospective data are needed to characterize the temporal relation between aging and shifts or stability in the abundance and pattern of subgingival microbiota observed herein to better elucidate the role, if any, that aging has on the oral microbiome. Age alone, however, does not determine the human subgingival microbiome. Other factors, including senescence of tissues and functions, side effects of medication use, status of the gingiva and dentition, systemic diseases, oral hygiene and behavioral habits, are thought to influence the microbiome. The extensive cross-sectional observations reported here provide a starting point and direction to define a targeted subset of bacteria that appear be related with age for further analysis in which issues such as confounding or interaction with the above and other factors can be evaluated with greater statistical efficiency involving fewer tests to correct for false discovery. This will be the focus of a forthcoming manuscript from our longitudinal cohort. Additional understanding about the functions of bacteria that differ with age in later life could identify intervention targets for enhanced oral health and, possibly control of other diseases.
Acknowledgements
Not applicable.
Abbreviations
- CLR
Centered-log(2)-ratio
- OTU
Operational Taxonomic Unit
Authors’ contributions
Conception, design, and acquisition of data and biologic samples in the original studies (JWW, RJG). Microbiome laboratory (MJB, YS, MT, DIM). Data analysis and interpretation (LL, DIM, KMH, CAA, MJL, MJB, WZ, YS, JWW, RJG). Manuscript drafting and editing (MJL, KMH, CAA, DIM, JWW, RJG, YS, MJB, WZ, LL, HB, AEM). Final Approval of manuscript (MJL, RJG, MJB, DIM, LL, KMH, CAA, WZ, YS, MT, HB, AEM, JWW). Agreed to be accountable for all aspects of the work ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved (MJL, JWW).
Funding
National Heart, Lung, and Blood Institute contract N01WH32122; National Institute for Dental and Craniofacial Research Grants: DE13505, DE4898, DE022654, and DE024523; National Institute of Allergy and Infectious Diseases R01Al125982, U.S. Army Reserve Medical Corps Grant: DAMD17–96-1-6319; Feasibility Study Award (AS382) from the Women’s Health Initiative Program. Decisions concerning design and conduct of the study, collection, management, analysis, and interpretation of the data, preparation, review, and approval of the manuscript, and the decision to submit the manuscript for publication resided with committees comprising WHI investigators that included NHLBI and NIDCR representatives. The contents of the manuscript are solely the responsibility of the authors.
Availability of data and materials
Data that support the findings of this study are available from the authors upon reasonable request and with permission of the U.S. Women’s Health Initiative program.
Ethics approval and consent to participate
Participants provided written informed consent for all components of the studies, which were conducted in accord with the Helsinki Declaration on human subjects research. Experimental protocols for the WHI study, the OsteoPerio study, and microbiome study detailed in this paper were approved by the Institutional Review Board at the University at Buffalo.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Prior to publication of this work, Dr. Robert Genco passed away unexpectedly. The manuscript is dedicated to his lasting memory as a pioneering scientist, teacher, mentor, colleague, and sorely missed friend. Rest in peace, Bob.
Contributor Information
Michael J. LaMonte, Phone: 716-829-5379, Email: mlamonte@buffalo.edu
Michael J. Buck, Email: mjbuck@buffalo.edu
Daniel I. McSkimming, Email: dim@buffalo.edu
Lu Li, Email: lli59@buffalo.edu.
Kathleen M. Hovey, Email: koreilly@buffalo.edu
Christopher A. Andrews, Email: candrews@buffalo.edu
Wei Zheng, Email: zhengw@buffalo.edu.
Yijun Sun, Email: yijunsun@buffalo.edu.
Amy E. Millen, Email: aemillen@buffalo.edu
Maria Tsompana, Email: mtsompana@buffalo.edu.
Hailey R. Banack, Email: hrbanack@buffalo.edu
Jean Wactawski-Wende, Email: jww@bufalo.ed.
References
- 1.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550(7674):61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin Chem. 2009;55(5):856–866. doi: 10.1373/clinchem.2008.107565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Human MJR, Strains C, Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328(5981):994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P. Aging of the human metaorganism: the microbial counterpart. Age (Dordr) 2012;34(1):247–267. doi: 10.1007/s11357-011-9217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zapata HJ, Quagliarello VJ. The microbiota and microbiome in aging: potential implications in health and age-related diseases. J Am Geriatr Soc. 2015;63(4):776–781. doi: 10.1111/jgs.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinert BT, Timiras PS. Invited review: theories of aging. J Appl Physiol (1985) 2003;95(4):1706–1716. doi: 10.1152/japplphysiol.00288.2003. [DOI] [PubMed] [Google Scholar]
- 9.Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY) 2013;5(12):902–12. doi: 10.18632/aging.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, et al. Oral Microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77(23):6777–6787. doi: 10.1158/0008-5472.CAN-17-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4(8):962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi N. Oral Microbiome metabolism: from "who are they?" to "what are they doing?". J Dent Res. 2015;94(12):1628–1637. doi: 10.1177/0022034515606045. [DOI] [PubMed] [Google Scholar]
- 14.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13(12):589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang GP. Defining functional signatures of dysbiosis in periodontitis progression. Genome Med. 2015;7(1):40. doi: 10.1186/s13073-015-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han YW, Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013;92(6):485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feres M, Teles F, Teles R, Figueiredo LC, Faveri M. The subgingival periodontal microbiota of the aging mouth. Periodontol 2000. 2016;72(1):30–53. doi: 10.1111/prd.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL, Jr, et al. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25(5):346–353. doi: 10.1111/j.1600-051X.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 19.Zawadzki PJ, Perkowski K, Padzik M, Mierzwinska-Nastalska E, Szaflik JP, Conn DB, et al. Examination of Oral microbiota diversity in adults and older adults as an approach to prevent spread of risk factors for Human infections. Biomed Res Int. 2017;2017:8106491. doi: 10.1155/2017/8106491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asakawa M, Takeshita T, Furuta M, Kageyama S, Takeuchi K, Hata J, et al. Tongue microbiota and Oral health status in community-dwelling elderly adults. mSphere. 2018;3(4):e00332-18. [DOI] [PMC free article] [PubMed]
- 21.Hoffman KL, Hutchinson DS, Fowler J, Smith DP, Ajami NJ, Zhao H, et al. Oral microbiota reveals signs of acculturation in Mexican American women. PLoS One. 2018;13(4):e0194100. doi: 10.1371/journal.pone.0194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa T, Hirose Y, Honda-Ogawa M, Sugimoto M, Sasaki S, Kibi M, et al. Composition of salivary microbiota in elderly subjects. Sci Rep. 2018;8(1):414. doi: 10.1038/s41598-017-18677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibagaki N, Suda W, Clavaud C, Bastien P, Takayasu L, Iioka E, et al. Aging-related changes in the diversity of women's skin microbiomes associated with oral bacteria. Sci Rep. 2017;7(1):10567. doi: 10.1038/s41598-017-10834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foxman B, Seitz SM, Rothenberg R. Epidemiology and the microbiome. Ann Epidemiol. 2016;26(5):386–387. doi: 10.1016/j.annepidem.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langer RD, White E, Lewis CE, Kotchen JM, Hendrix SL, Trevisan M. The Women's Health Initiative observational study: baseline characteristics of participants and reliability of baseline measures. Ann Epidemiol. 2003;13(9 Suppl):S107–S121. doi: 10.1016/S1047-2797(03)00047-4. [DOI] [PubMed] [Google Scholar]
- 26.LaMonte MJ, Hovey KM, Genco RJ, Millen AE, Trevisan M, Wactawski-Wende J. Five-year changes in periodontal disease measures among postmenopausal females: the Buffalo OsteoPerio study. J Periodontol. 2013;84(5):572–584. doi: 10.1902/jop.2012.120137. [DOI] [PubMed] [Google Scholar]
- 27.Wactawski-Wende J, Hausmann E, Hovey K, Trevisan M, Grossi S, Genco RJ. The association between osteoporosis and alveolar crestal height in postmenopausal women. J Periodontol. 2005;76(11 Suppl):2116–2124. doi: 10.1902/jop.2005.76.11-S.2116. [DOI] [PubMed] [Google Scholar]
- 28.Gordon JH, LaMonte MJ, Genco RJ, Zhao J, Cimato TR, Hovey KM, et al. Association of clinical measures of periodontal disease with blood pressure and hypertension among postmenopausal women. J Periodontol. 2018;89(10):1193–1202. doi: 10.1002/JPER.17-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan RM, Genco RJ, Wilding GE, Hovey KM, Trevisan M, Wactawski-Wende J. Bacterial species in subgingival plaque and oral bone loss in postmenopausal women. J Periodontol. 2007;78(6):1051–1061. doi: 10.1902/jop.2007.060436. [DOI] [PubMed] [Google Scholar]
- 30.Zheng W, Tsompana M, Ruscitto A, Sharma A, Genco R, Sun Y, et al. An accurate and efficient experimental approach for characterization of the complex oral microbiota. Microbiome. 2015;3:48. doi: 10.1186/s40168-015-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Gloor GB, Wu JR, Pawlowsky-Glahn V, Egozcue JJ. It's all relative: analyzing microbiome data as compositions. Ann Epidemiol. 2016;26(5):322–329. doi: 10.1016/j.annepidem.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontology 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Chaparro PJ, Goncalves C, Figueiredo LC, Faveri M, Lobao E, Tamashiro N, et al. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 2014;93(9):846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6(6):1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preza D, Olsen I, Willumsen T, Grinde B, Paster BJ. Diversity and site-specificity of the oral microflora in the elderly. Eur J Clin Microbiol Infect Dis. 2009;28(9):1033–1040. doi: 10.1007/s10096-009-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10(10):2435–2446. doi: 10.1038/ismej.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang S, Yang F, Zeng X, Chen J, Li R, Wen T, et al. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health. 2011;11:33. doi: 10.1186/1472-6831-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Quinque D, Horz HP, Li M, Rzhetskaya M, Raff JA, et al. Comparative analysis of the human saliva microbiome from different climate zones: Alaska, Germany, and Africa. BMC Microbiol. 2014;14:316. doi: 10.1186/s12866-014-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato T, Matsuyama J, Sato M, Hoshino E. Differentiation of Veillonella atypica, Veillonella dispar and Veillonella parvula using restricted fragment-length polymorphism analysis of 16S rDNA amplified by polymerase chain reaction. Oral Microbiol Immunol. 1997;12(6):350–353. doi: 10.1111/j.1399-302X.1997.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 45.Mashimo PA, Murayama Y, Reynolds H, Mouton C, Ellison SA, Genco RJ. Eubacterium saburreum and Veillonella parvula: a symbiotic association or oral strains. J Periodontol. 1981;52(7):374–379. doi: 10.1902/jop.1981.52.7.374. [DOI] [PubMed] [Google Scholar]
- 46.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 47.Toh H, Hayashi J, Oshima K, Nakano A, Takayama Y, Takanashi K, et al. Complete genome sequence of Bifidobacterium dentium strain JCM 1195T, isolated from Human dental caries. Genome Announc. 2015;3(2):e00284-15. [DOI] [PMC free article] [PubMed]
- 48.Jasberg H, Soderling E, Endo A, Beighton D, Haukioja A. Bifidobacteria inhibit the growth of Porphyromonas gingivalis but not of Streptococcus mutans in an in vitro biofilm model. Eur J Oral Sci. 2016;124(3):251–258. doi: 10.1111/eos.12266. [DOI] [PubMed] [Google Scholar]
- 49.Bao K, Bostanci N, Thurnheer T, Belibasakis GN. Proteomic shifts in multi-species oral biofilms caused by Anaeroglobus geminatus. Sci Rep. 2017;7(1):4409. doi: 10.1038/s41598-017-04594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu B, Macleod LC, Kitten T, Xu P. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol. 2018;13:915–932. doi: 10.2217/fmb-2018-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riegel P, Heller R, Prevost G, Jehl F, Monteil H. Corynebacterium durum sp. nov., from human clinical specimens. Int J Syst Bacteriol. 1997;47(4):1107–1111. doi: 10.1099/00207713-47-4-1107. [DOI] [PubMed] [Google Scholar]
- 52.Haraszthy VI, Zambon JJ, Sreenivasan PK, Zambon MM, Gerber D, Rego R, et al. Identification of oral bacterial species associated with halitosis. J Am Dent Assoc(1939) 2007;138(8):1113–1120. doi: 10.14219/jada.archive.2007.0325. [DOI] [PubMed] [Google Scholar]
- 53.Francavilla R, Ercolini D, Piccolo M, Vannini L, Siragusa S, De Filippis F, et al. Salivary microbiota and metabolome associated with celiac disease. Appl Environ Microbiol. 2014;80(11):3416–3425. doi: 10.1128/AEM.00362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuzukibashi O, Uchibori S, Shinozaki-Kuwahara N, Kobayashi T, Takada K, Hirasawa M. A selective medium for the isolation of Corynebacterium species in oral cavities. J Microbiol Methods. 2014;104:67–71. doi: 10.1016/j.mimet.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Aitchison J. The statistical analysis of compositional data. London: Chapman and Hall; 1986. [Google Scholar]
- 56.Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000;27(9):648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- 57.Dahlen G, Preus HR, Baelum V. Methodological issues in the quantification of subgingival microorganisms using the checkerboard technique. J Microbiol Methods. 2015;110:68–77. doi: 10.1016/j.mimet.2015.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data that support the findings of this study are available from the authors upon reasonable request and with permission of the U.S. Women’s Health Initiative program.