Abstract
Background/Aim
During cancer progression metabolic reprogramming is observed in parallel to the alternation in transcriptional profiles of malignant cells. Recent studies suggest that metabolic isoenzymes of phosphofructokinase II (PFK-II) – PFKFB3 and PFKFB4, often induced in hypoxic environment, significantly contribute to enhancement of glucose metabolism and in consequence cancer progression.
Materials and methods
Using the publicly available data deposited in the R2 data base we performed a Kaplan–Meyer analysis for cancer patients divided into groups with high and low expression levels of PFKFB3/4, determined based on the median.
Results
Our data showed that high PFKFB3/4 expression significantly correlates with shorter overall survival in several cancers. Moreover, we found that neuroblastoma patients with poor overall survival and evidence free survival are characterized by high PFKFB3 and at the same time low PFKFB4 expression, whereas patients with high PFKFB4 expressions are characterized by significantly better overall survival/evidence free survival rates.
Conclusion
Our analysis clearly indicates that expression of PFKFB3/4 isoenzymes may have a key prognostic value for several cancers. What’s more, it seems that in neuroblastoma the prognostic value of PFK-II may be dependent on the relation between PFKFB3 and PFKFB4 isoenzyme expression, indicating that further studies analyzing the role of both cancer specific PFK-II isoenzymes are highly desired.
Keywords: Phosphofructokinase II, PFKFB3, PFKFB4, Cancer progression, Overall survival, Neuroblastoma
Background
Nowadays, the metabolic reprogramming of cancer cells is considered a very promising field for development of novel therapeutic strategies. Apparently, the most evident alterations in cancer metabolism are related to glucose breakdown. As initially observed by Otto Warburg, in comparison to normal cells, the cancer cells ATP is generated mainly by glycolysis, even in normoxic conditions [1, 2]. Although glycolysis is less efficient than oxidative phosphorylation, it allows the generation of ATP at a much faster pace, which is highly beneficial for proliferating cancer cells. Additionally, elevation of glycolysis enables sufficient production levels of glycolytic intermediates required for de novo nucleotide, lipid, amino acid, and NADPH synthesis. The role of enhanced glucose breakdown in carcinogenesis is well confirmed by studies, showing that attenuation of glycolysis inhibits growth and decreases survival of cancer cells [3, 4]. Without doubt, an attractive anti-cancer approach would be to target glycolysis by inhibiting the aforementioned enzymes crucial for this process. Nevertheless, to do so, there must be a significant difference in the particular enzyme expression and/or activity between cancer cells and normal proliferating cells. In fact, studies in recent years revealed that PFKFB3/PFKFB4 isoenzymes are excellent candidates for glycolysis targeting, especially in cancer cells [5, 6]. PFKFB3 and PFKFB4 are cancer specific isoenzymes of the bifunctional 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB) enzyme group, responsible for controlling the steady-state cytoplasmic levels of fructose-2,6-bisphosphate, which allosterically activates phosphofructokinase-I (PFK-I), the key enzyme that catalyzes the rate-limiting step of glycolysis [5, 7]. Several in vitro studies revealed that targeting PFKFB3 and PFKFB4 in cancer cells results in glycolysis inhibition and in consequence, attenuation of tumor growth [8–12]. Without doubt, it is worth noting that PFKFB4 and PFKFB3 isoenzymes are not only involved in glucose metabolism but may also regulate other processes crucial for carcinogenesis, having a multi-level anti-cancer effect [13–16]. Taking into consideration all of the aforementioned, and the fact that there is limited clinical data related to this topic, in our current paper, using publicly available data sets deposited in the R2 data base, we analyzed the influence of PFKFB3 and PFKFB4 expression on overall survival of 49 independent, available cancer data sets. The results of the analysis indicates the importance of PFKFB3/4 co-expression for the clinical outcome of cancer patients.
Materials and methods
R2 database
The R2 database (http://r2.amc.nl) is a simple to use web-based genomics analysis and visualization application developed at the Department of Oncogenomics in the Academic Medical Center (AMC) in Amsterdam, Netherlands [17]. It allows researchers to perform a range of analyses based on well annotated datasets. For the purpose of this study we have chosen the Kaplan–Meier analysis of PFKFB3/PFKFB4 gene expression in the panel of cancer data sets. The gene expression cutoff value was chosen as a median over the entire dataset to ensure all analyses were based on the same cutoff.
Data validation and statistical analysis
For validation purposes the gene expression and survival data of 498 neuroblastoma patients (GSE62564) were downloaded from the NCBI Gene Expression Omnibus (GEO), a public repository of microarray data [18]. Based on the median expression level of PFKFB4 and PFKFB3 genes, the patients were divided into four groups, namely: low PFKFB3/low PFKFB4 (G1), low PFKFB3/high PFKFB4 (G2), high PFKFB3/low PFKFB4 (G3) and high PFKFB3/high PFKFB4 (G4) respectively, the survival curves were plotted using the Kaplan–Meier method and compared between each other using the log-rank test with Bonferroni correction (p value below 0.05/6 was consider significant). Kaplan–Meyer analysis and univariate Cox proportional hazards regression were performed using GraphPad Prism 5.01 (GraphPad Software, La Jolla, CA, USA).
Results
PFKFB3 and PFKFB4 expression-based survival analysis in a panel of tumors using the R2 data base
Initially, using the R2 data base we analyzed the relation between PFKFB3 and PFKFB4 expression and the overall survival rate in the panel of cancers. The only criterium for this analysis was availability of overall survival and expression data. The analyzed panel composed of 49 data sets (Table 1), where some tumors were represented more than once. In every single data set, the patients were divided into two groups based on the median expression of PFKFB3 and/or PFKFB4. The difference in survival between these two groups was analyzed by a log rank test. Probability values below 0.05 were considered statistically significant and those between 0.05 and 0.1 were marked as an indication of a trend. Our analysis revealed that in the case of Glioma and Liver hepatocellular carcinoma patients with high PFKFB4 expression have significantly lower overall survival rates in comparison to patients with low expression of PFKFB4 (Table 1). As for PFKFB3, we have found that in kidney renal papillary cell carcinoma patients high expression of this isoenzyme was significantly correlated with lower overall survivability. Interestingly, in cervical squamous cell carcinoma we revealed that patients with high expression of both, PFKFB3 and PFKFB4 have significantly worse overall survival in comparison to the ones with low expression. Although the overexpressed PFKFB3/4 isoenzymes are considered oncogenic, mantle cell lymphoma, ovarian and pancreatic adenocarcinoma data sets have revealed that patients with low expression of PFKFB3 enzyme have significantly worse survival rates in comparison to the ones with high expression (Table 1).
Table 1.
Analysis of the relation between PFKFB3 and PFKFB4 expression and the overall survival rate in the panel of cancers
| Cancer type | Patient number | Source | P value for PFKFB3 | P value for PFKFB4 |
|---|---|---|---|---|
| Acute myeloid leukemia | 422 | GEO ID:GSE37642 | 0.713 | 0.704 |
| Adrenocortical carcinoma | 79 | TCGA ID:ND | 0.083a | 0.076a |
| B-cell lymphoma | 414 | GEO ID:GSE10846 | 0.069b | 0.528 |
| Bladder urothelial carcinoma | 224 | GEO ID:GSE32894 | 0.508 | 0.331 |
| Bladder urothelial carcinoma | 408 | TCGA ID:BLCA | 0.555 | 0.352 |
| Breast | 159 | GEO ID:GSE1456 | 0.405 | 0.110 |
| Breast | 130 | GEO ID:GSE69031 | 0.527 | 0.913 |
| Breast invasive carcinoma | 1096 | TCGA ID:BRCA | 0.358 | 0.858 |
| Cervical squamous cell carcinoma | 292 | TCGA ID:CESC | 0.032a | 0.026a |
| Cholangiocarcinoma | 36 | TCGA ID:CHOL | 0.859 | 0.402 |
| Colon | 200 | GEO ID:GSE17538 | 0.041a | 0.775 |
| Esophageal adenocarcinomas | 70 | GEO ID:GSE19417 | 0.854 | 0.098b |
| Esophageal carcinoma | 183 | TCGA ID:ESCA | 0.814 | 0.384 |
| Ewing sarcoma | 85 | GEO ID:GSE63157 | 0.073a | 0.094a |
| Glioblastoma | 80 | GEO ID:GSE7696 | 1.000 | 1.000 |
| Glioblastoma | 377 | ND ID:ND | 0.525 | 0.286 |
| Glioblastoma | 504 | ND ID:ND | 0.964 | 0.387 |
| Glioma | 490 | GEO ID:GSE108474 | 0.378 | 1.2e−09a |
| Glioma | 273 | GEO ID:GSE16011 | 0.129 | 1.4e−10a |
| Glioma | 50 | GEO ID:GSE43378 | 0.730 | 0.028a |
| Glioma pediatric | 47 | GEO ID:GSE19578 | 0.092a | 0.625 |
| Intrinsic glioma subtypes | 95 | GEO ID:GSE43107 | 0.069b | 0.660 |
| Head neck squamous cell carcinoma | 520 | TCGA ID:HNSC | 0.640 | 0.875 |
| Kidney renal clear cell carcinoma | 533 | TCGA ID:KIRC | 0.464 | 0.542 |
| Kidney renal papillary cell carcinoma | 290 | TCGA ID:KIRP | 0.012a | 0.064a |
| Liver hepatocellular carcinoma | 371 | TCGA ID:LIHC | 0.058a | 6.6e−05a |
| Lung | 106 | GEO ID:GSE3141 | 0.819 | 0.100a |
| Lung adenocarcinoma | 515 | TCGA ID:LUAD | 0.814 | 0.961 |
| Tumor lymphoma | 162 | GEO ID:GSE58445 | 0.315 | 0.486 |
| Tumor lymphoma | 470 | GEO ID:GSE31312 | 0.431 | 0.555 |
| Mantle cell lymphoma | 122 | GEO ID:GSE93291 | 0.030b | 0.070a |
| Medulloblastoma | 612 | GEO ID:GSE85217 | 0.576 | 0.720 |
| Melanoma | 214 | GEO ID:GSE65904 | 0.633 | 0.048a |
| Metastatic melanoma | 44 | GEO ID:GSE19234 | 0.558 | 0.394 |
| Myeloma | 542 | GEO ID:GSE2658 | 0.078b | 0.514 |
| Neuroblastoma | 476 | GEO ID:GSE45547 | 1.8e−04a | 6.2e−04b |
| Neuroblastoma | 498 | GEO ID:GSE62564 | 6.8e−04a | 1.6e−06b |
| Neuroblastoma stage IV | 27 | GEO ID:GSE79910 | 0.939 | 0.325 |
| Neuroblastoma primary | 283 | GEO ID:GSE85047 | 0.090a | 3.3e−03b |
| Neuroblastoma | 247 | ND ID:ND | 0.272 | 6.1e−03b |
| Neuroblastoma | 88 | GEO ID:GSE16476 | 0.398 | 0.078b |
| Ovarian | 75 | GEO ID:GSE63885 | 3.9e−03b | 0.941 |
| Ovarian adenocarcinoma | 107 | GEO ID:GSE26193 | 0.371 | 0.302 |
| Pancreatic subtypes | 96 | ND ID:ND | 0.070a | 0.058a |
| Pancreatic adenocarcinoma | 146 | TCGA ID:PAAD | 0.016b | 0.928 |
| Pancreatic ductal adenocarcinoma | 102 | GEO ID:GSE21501 | 0.615 | 0.598 |
| Skin cutaneous melanoma | 468 | TCGA ID:SKCM | 0.192 | 0.493 |
| Stomach adenocarcinoma | 415 | TCGA ID:STAD | 0.218 | 0.738 |
| Thymoma | 120 | TCGA ID:THYM | 0.952 | 0.113 |
Bold italic indicates the p value below 0.05
italic indicates the p value between 0.05 and 0.1
ND no data
aHigh is worse
bHigh is better
Analysis of the relation between PFKFB3 and PFKFB4 expression in neuroblastoma patients
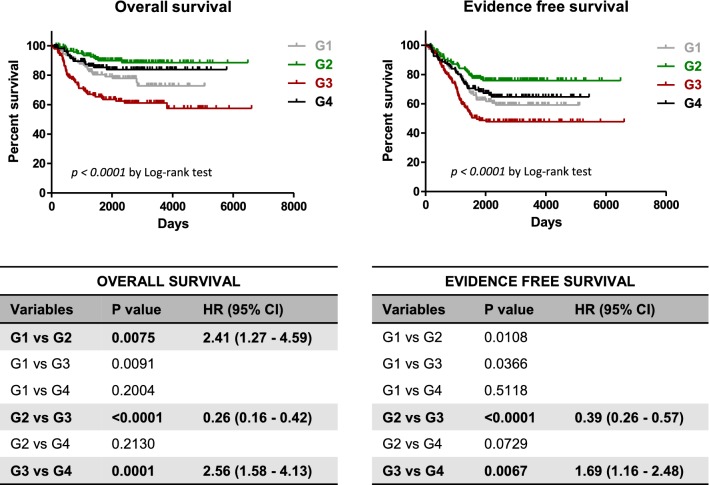
Without doubt, the most interesting data was obtained through neuroblastoma data sets. Surprisingly, in two independent cohorts we observed that high expression of PFKFB3 negatively influences the overall survival, whereas in five independent data sets patients with high expression of PFKFB4 had significantly better OS (Table 1). In order to determine the influence of PFKFB3/4 co-expression on survival rate, the gene expression data were downloaded (GSE62564) and neuroblastoma patients were divided on four groups based on the median PFKFB3/PFKFB4 expression (Fig. 1), as described in Materials and Methods. As determined in Fig. 2, our analysis clearly indicates that patients with high PFKFB4 expression (G2 and G4 groups) have a significantly better overall survival, which might suggest that regardless of the PFKFB3 level of expression (high in G4 group and low in G2), PFKFB4 has a positive impact on the patients survival rate. The worst survival prognosis could be observed for the patients with the dominant PFKFB3 expression (G3 group), which clearly indicates that low expression of PFKFB4, and high expression of PFKFB3 at the same time has a negative impact on the survival of neuroblastoma patients. Importantly, the same correlation was observed for evidence free survival. This suggests that if the high expression of the PFKFB3 isoenzyme is not balanced by the expression of PFKFB4, then the patients show much worse survival rate. Subgroup analysis was further extended, and univariate Cox proportional hazards regression was performed. As determined in Fig. 2 lower panel, there is a better overall survival and evidence free survival rate in the group of patients with dominant PFKFB4 (G2 group) in comparison to the group of patients with dominant PFKFB3 (G3 group), with hazard rates of 0.26 and 0.39 respectively. What is more, group G3 presented worse overall survival and evidence free survival in comparison to the G4 group (high PFKFB3/high PFKFB4), with hazard ratios of 2.56 and 1.69 respectively, which indicates that negative impact of PFKFB3 expression can be offset by the expression of PFKFB4.
Fig. 1.
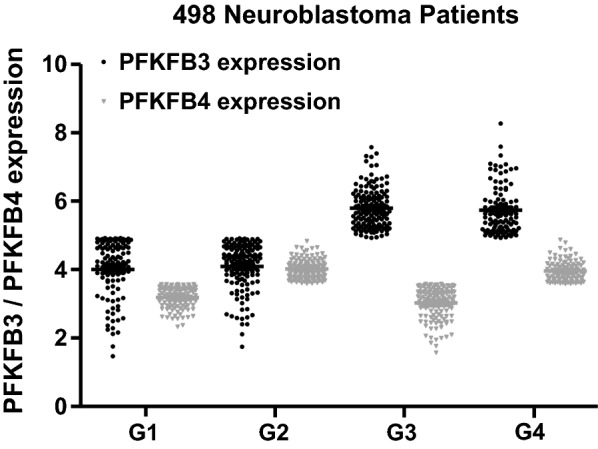
Analysis of PFKFB3 and PFKFB4 mRNA expression in neuroblastoma patients using a publicly available data set GSE62564. 498 neuroblastoma patients were classified in accordance to their PFKFB3 and PFKFB4 mRNA expression. The patients were divided into four expression groups (G1—low PFKFB3/low PFKFB4, G2—low-PFKFB3/highPFKFB4, G3—high PFKFB3/low PFKFB4, G4—high PFKFB3/high PFKFB4) based on the median
Fig. 2.
PFKFB3/PFKFB4 expression profile influences the prognosis of neuroblastoma patients. Upper panel: The publicly available data set GSE62564 was used to analyze the overall survival and evidence free survival in four expression groups G1, G2, G3, G4. Kaplan–Meier curves are presents for overall survival and evidence free survival. Lower panel: Survival curves comparison using the log-rank test with Bonferroni correction (p value below 0.0083 was considered significant). Univariate hazard ratios with confidence intervals were obtained by Cox regression analysis
Discussion
Currently, several literature data indicates PFK-II as a potential marker of cancer prognosis [19–25]. Nevertheless, up to this day, most research on PFK-II refers to one of the two cancer specific isoenzymes PFKFB3 or PFKFB4 [8–12, 19, 20, 22–25], undoubtedly neglecting the importance of co-expression of individual isoenzymes. Our analysis of publicly available data sets unquestionably indicates that in several tumors, high PFKFB3 and/or PFKFB4 expression correlates with poor survivability. Importantly, our analysis confirmed the previously published negative impact of PFKFB4 on the prognosis of glioma patients [23] and negative impact of PFKFB3 on the prognosis of hepatocellular cancer [24]. Interestingly, recently published data also revealed the negative impact of PFKFB4 on breast and bladder cancer patients but it was not noticable in the data sets we have analyzed. Nevertheless, Li et al. [22] and Ling et al. [20] analyzed the expression of PFKFB3 and PFKFB4 at the protein level in lung and breast cancer, which definitely may explain the discrepancy between the outcomes. Moreover, it may suggest that the same analysis, as we have done so far, could be performed at the protein level revealing more tumors for which expression of PFKFB3/PFKFB4 constitutes a potent prognostic marker. Undoubtedly, attention should be paid to the fact that in the case of neuroblastoma solely high expression of isoenzyme PFKFB3 correlates with poor prognosis whereas solely high expression of PFKFB4 is a positive prognostic factor for these patients. According to our current knowledge, there is no single study analyzing the influence of PFK-II on neuroblastoma patients’ prognosis, whereas our analysis clearly indicates that cancer specific isoenzymes may have opposite effects on prognosis in this group of specific cancer patients.
Although both genes code for phosphofructokinase II, according to the literature, kinase activity dominates in the PFKFB3 isoenzyme [26, 27], enhancing the glycolytic breakdown of glucose, whereas PFKFB4 seems to have higher FBPase-2 activity [5, 28], stimulating the flow of glucose toward the pentose phosphate pathway, providing the source of NADPH, crucial as a reducing factor for lipid biosynthesis and ROS-detoxifying enzymes. Altogether, we speculate, that the group with high PFKFB3 expression and low PFKFB4 may be characterized by higher glycolysis rates in relation to other groups whereas the PFKFB4 solely high group will have the lower glycolytic activity in comparison to other patients. If PFKFB3 dependent glycolytic cell activity in neuroblastoma indeed has significant impact on neuroblastoma cell proliferation [29], as determined by Almeida et al., one would expect to have opposite effects of PFKFB3 and PFKFB4 expression on patient survival as observed in our study. In addition, the analysis of the impact of isoenzymes on patients’ survival rates should also take into account that both, PFKFB3 and PFKFB4 are vastly involved in other important biological processes in a non-glycolysis-dependent manner [13–16, 30–32], which could also explain their opposite effect on neuroblastoma patients’ prognosing. For instance, it has been shown that PFKFB3 can be localized in the nucleus, which results in proliferation enhancement without increasing of the glycolytic rate. As reported by Yalcin et al. both, kinase activity of PFKFB3 and nuclear localization are needed for its effect on cancer cell proliferation [32]. As for PFKFB4, it has been reported that this enzyme is somewhat involved, be it directly or indirectly, in phosphorylation of the CBP-interacting domain of oncogenic steroid receptor coactivator-3 (SRC-3), enhancing its transcriptional activity, which results in higher expression of its target genes; transketolase, adenosine monophosphate deaminase-1 (AMPD1) and xanthine dehydrogenase (XDH), involved in the metabolism of nucleotides [13]. Regardless of the molecular mechanism of action of PFKFB3/4 isoenzymes our data clearly indicates that in analyzing the effect of PFK-II on the prognosis of cancer patients, the expression of both PFKFB3/4 isozymes should be considered. This is particularly important in the case of potential use of PFK-II inhibitors, because, as our analysis has undoubtedly shown, the effectiveness of inhibition may be dependent on the suitable inhibitor application for a specific isoenzyme overexpressed in specific cancer cells. What is more, our analysis indicates worryingly, that application of PFKFB4 inhibitors for neuroblastoma patients characterized by overexpression of both isoenzymes could even have a negative impact on the patient prognosis. Consequently, the results of our analysis provide important insight for future clinical oncology by presenting the importance of metabolic enzymes as a likely destination for modern targeted anti-tumor therapy.
Acknowledgements
Not applicable.
Abbreviations
- AMPD1
adenosine monophosphate deaminase 1
- ATP
adenosine triphosphate
- GEO
gene expression Omnibus
- FBPase-2
fructose-2,6-biphosphatase
- NADPH
dihydronicotinamide-adenine dinucleotide phosphate
- OS
overall survival
- PFK-I
phosphofructokinase I
- PFK-II
phosphofructokinase II
- PFKFB
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase
- PFKFB3
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
- PFKFB4
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
- ROS
reactive oxygen species
- SRC-3
steroid receptor coactivator-3
- XDH
xanthine dehydrogenase
Authors’ contributions
SET, KL and KAKP performed analysis and interpreted the data. SET, MJM and KAKP wrote the manuscript. All authors read and approved the final manuscript.
Funding
Work presented in this paper was supported by the National Science Centre (Poland) grant number 2018/31/N/NZ5/03561. Work presented in this paper was partially supported by a grant from the National Committee for Scientific Research with nr N41/DBS/000114.
Availability of data and materials
The datasets used and/or analysed during the current study are available in the R2 database repository, https://hgserver1.amc.nl/cgi-bin/r2/main.cgi, date of access 12.02.2019. Gene expression and survival data of 498 neuroblastoma patients were downloaded from the NCBI Gene Expression Omnibus (GEO), a public repository of microarray data, dataset ID: GSE62564.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author(s) declare(s) that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Warburg O. Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften. 1924;12(50):1131–1137. doi: 10.1007/BF01504608. [DOI] [Google Scholar]
- 2.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu L, Chen X, Sun X, Wang L, Chen S. The glycolytic switch in tumors: how many players are involved? J Cancer. 2017;8(17):3430–3440. doi: 10.7150/jca.21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5):e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi M, Ban Y, Tan Y, Xiong W, Li G, Xiang B. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: a pair of valves for fine-tuning of glucose metabolism in human cancer. Mol Metab. 2019;20:1–13. doi: 10.1016/j.molmet.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi L, Pan H, Liu Z, Xie J, Han W. Roles of PFKFB3 in cancer. Signal Transduct Target Ther. 2017;2:17044. doi: 10.1038/sigtrans.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kocemba KA, Dulinska-Litewka J, Wojdyla KL, Pekala PA. The role of 6-phosphofructo-2-kinase (PFK-2)/fructose 2,6-bisphosphatase (FBPase-2) in metabolic reprogramming of cancer cells. Postepy Hig Med Dosw (Online) 2016;70:938–950. doi: 10.5604/17322693.1218187. [DOI] [PubMed] [Google Scholar]
- 8.Li HM, Yang JG, Liu ZJ, Wang WM, Yu ZL, Ren JG, Chen G, Zhang W, Jia J. Blockage of glycolysis by targeting PFKFB3 suppresses tumor growth and metastasis in head and neck squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36(1):7. doi: 10.1186/s13046-016-0481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neal J, Clem A, Reynolds L, Dougherty S, Imbert-Fernandez Y, Telang S, Chesney J, Clem BF. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) suppresses glucose metabolism and the growth of HER2 + breast cancer. Breast Cancer Res Treat. 2016;160(1):29–40. doi: 10.1007/s10549-016-3968-8. [DOI] [PubMed] [Google Scholar]
- 10.Cantelmo AR, Conradi LC, Brajic A, Goveia J, Kalucka J, Pircher A, Chaturvedi P, Hol J, Thienpont B, Teuwen LA, et al. Inhibition of the glycolytic activator PFKFB3 in endothelium induces tumor vessel normalization, impairs metastasis, and improves chemotherapy. Cancer Cell. 2016;30(6):968–985. doi: 10.1016/j.ccell.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesney J, Clark J, Lanceta L, Trent JO, Telang S. Targeting the sugar metabolism of tumors with a first-in-class 6-phosphofructo-2-kinase (PFKFB4) inhibitor. Oncotarget. 2015;6(20):18001–18011. doi: 10.18632/oncotarget.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor C, Mannion D, Miranda F, Karaminejadranjbar M, Herrero-Gonzalez S, Hellner K, Zheng Y, Bartholomeusz G, Bast RC, Jr, Ahmed AA. Loss of PFKFB4 induces cell death in mitotically arrested ovarian cancer cells. Oncotarget. 2017;8(11):17960–17980. doi: 10.18632/oncotarget.14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta S, Rajapakshe K, Zhu B, Nikolai BC, Yi P, Putluri N, Choi JM, Jung SY, Coarfa C, Westbrook TF, et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature. 2018;556(7700):249–254. doi: 10.1038/s41586-018-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia W, Zhao X, Zhao L, Yan H, Li J, Yang H, Huang G, Liu J. Non-canonical roles of PFKFB3 in regulation of cell cycle through binding to CDK4. Oncogene. 2018;37(13):1685–1698. doi: 10.1038/s41388-017-0072-4. [DOI] [PubMed] [Google Scholar]
- 15.Yan S, Wei X, Xu S, Sun H, Wang W, Liu L, Jiang X, Zhang Y, Che Y. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform 3 spatially mediates autophagy through the AMPK signaling pathway. Oncotarget. 2017;8(46):80909–80922. doi: 10.18632/oncotarget.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yalcin A, Clem BF, Imbert-Fernandez Y, Ozcan SC, Peker S, O’Neal J, Klarer AC, Clem AL, Telang S, Chesney J. 6-Phosphofructo-2-kinase (PFKFB3) promotes cell cycle progression and suppresses apoptosis via Cdk1-mediated phosphorylation of p27. Cell Death Dis. 2014;5:e1337. doi: 10.1038/cddis.2014.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R2: Genomics analysis and visualization platform (http://r2.amc.nl).
- 18.Edgar R, Domrachev M, Lash AE. Gene Expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trojan SE, Piwowar M, Ostrowska B, Laidler P, Kocemba-Pilarczyk KA. Analysis of malignant melanoma cell lines exposed to hypoxia reveals the importance of PFKFB4 overexpression for disease progression. Anticancer Res. 2018;38(12):6745–6752. doi: 10.21873/anticanres.13044. [DOI] [PubMed] [Google Scholar]
- 20.Yao L, Wang L, Cao ZG, Hu X, Shao ZM. High expression of metabolic enzyme PFKFB4 is associated with poor prognosis of operable breast cancer. Cancer Cell Int. 2019;19:165. doi: 10.1186/s12935-019-0882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng F, Li Q, Sun JY, Luo Y, Chen M, Bao Y. PFKFB3 is involved in breast cancer proliferation, migration, invasion and angiogenesis. Int J Oncol. 2018;52(3):945–954. doi: 10.3892/ijo.2018.4257. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Liu J, Qian L, Ke H, Yao C, Tian W, Liu Y, Zhang J. Expression of PFKFB3 and Ki67 in lung adenocarcinomas and targeting PFKFB3 as a therapeutic strategy. Mol Cell Biochem. 2018;445(1–2):123–134. doi: 10.1007/s11010-017-3258-8. [DOI] [PubMed] [Google Scholar]
- 23.Goidts V, Bageritz J, Puccio L, Nakata S, Zapatka M, Barbus S, Toedt G, Campos B, Korshunov A, Momma S, et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene. 2012;31(27):3235–3243. doi: 10.1038/onc.2011.490. [DOI] [PubMed] [Google Scholar]
- 24.Shi WK, Zhu XD, Wang CH, Zhang YY, Cai H, Li XL, Cao MQ, Zhang SZ, Li KS, Sun HC. PFKFB3 blockade inhibits hepatocellular carcinoma growth by impairing DNA repair through AKT. Cell Death Dis. 2018;9(4):428. doi: 10.1038/s41419-018-0435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yun SJ, Jo SW, Ha YS, Lee OJ, Kim WT, Kim YJ, Lee SC, Kim WJ. PFKFB4 as a prognostic marker in non-muscle-invasive bladder cancer. Urol Oncol. 2012;30(6):893–899. doi: 10.1016/j.urolonc.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Kim SG, Manes NP, El-Maghrabi MR, Lee YH. Crystal structure of the hypoxia-inducible form of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3): a possible new target for cancer therapy. J Biol Chem. 2006;281(5):2939–2944. doi: 10.1074/jbc.M511019200. [DOI] [PubMed] [Google Scholar]
- 27.Sakakibara R, Kato M, Okamura N, Nakagawa T, Komada Y, Tominaga N, Shimojo M, Fukasawa M. Characterization of a human placental fructose-6-phosphate, 2-kinase/fructose-2,6-bisphosphatase. J Biochem. 1997;122(1):122–128. doi: 10.1093/oxfordjournals.jbchem.a021719. [DOI] [PubMed] [Google Scholar]
- 28.Rider MH, Bertrand L, Vertommen D, Michels PA, Rousseau GG, Hue L. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: head-to-head with a bifunctional enzyme that controls glycolysis. Biochem J. 2004;381(Pt 3):561–579. doi: 10.1042/BJ20040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almeida A, Bolanos JP, Moncada S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci U S A. 2010;107(2):738–741. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z, Hunter T. Metabolic kinases moonlighting as protein kinases. Trends Biochem Sci. 2018;43(4):301–310. doi: 10.1016/j.tibs.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klarer AC, O’Neal J, Imbert-Fernandez Y, Clem A, Ellis SR, Clark J, Clem B, Chesney J, Telang S. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) induces autophagy as a survival mechanism. Cancer Metab. 2014;2(1):2. doi: 10.1186/2049-3002-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yalcin A, Clem BF, Simmons A, Lane A, Nelson K, Clem AL, Brock E, Siow D, Wattenberg B, Telang S, et al. Nuclear targeting of 6-phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinases. J Biol Chem. 2009;284(36):24223–24232. doi: 10.1074/jbc.M109.016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available in the R2 database repository, https://hgserver1.amc.nl/cgi-bin/r2/main.cgi, date of access 12.02.2019. Gene expression and survival data of 498 neuroblastoma patients were downloaded from the NCBI Gene Expression Omnibus (GEO), a public repository of microarray data, dataset ID: GSE62564.