Abstract
Dermaseptin B1 (DrsB1), an antimicrobial cationic 31 amino acid peptide, is produced by Phyllomedusa bicolor. In an attempt to enhance the antimicrobial efficacy of DrsB1, the DrsB1 encoding 93 bp sequence was either fused to the N or C terminus of sequence encoding chitin‐binding domain (CBD) of Avr4 gene from Cladosporium fulvum. Tobacco leaf disk explants were inoculated with Agrobacterium rhizogenes harboring pGSA/CBD‐DrsB1 and pGSA/DrsB1‐CBD expression vectors to produce hairy roots (HRs). Polymerase chain reaction (PCR) was employed to screen putative transgenic tobacco lines. Semi‐quantitative RT‐PCR and western blotting analysis indicated that the expression of recombinant genes were significantly higher, and recombinant proteins were produced in transgenic HRs. The recombinant proteins were extracted from the tobacco HRs and used against Pectobacterium carotovorum, Agrobacterium tumefaciens, Ralstonia solanacearum, and Xanthomonas campestris pathogenic bacteria and Alternaria alternata and Pythium sp. fungi. Two recombinant proteins had a statistically significant (p < 0.01) inhibitory effect on the growth and development of plant pathogens. The CBD‐DrsB1 recombinant protein demonstrated a higher antibacterial effect, whereas the DrsB1‐CBD recombinant protein demonstrated greater antifungal activity. Scanning electron microscopy images revealed that the structure of the fungal mycelia appeared segmented, adhered to each other, and crushed following the antimicrobial activity of the recombinant proteins. Due to the high antimicrobial activity of the recombinant proteins against plant pathogens, this strategy can be used to generate stable transgenic crop plants resistant to devastating plant pathogens.
Keywords: Agrobacterium rhizogenes, antimicrobial peptide, chitin‐binding domain, expression, hairy roots
Harnessing a chitin‐binding domain from Cladosporium fulvum Avr4 effector protein as fusion to the Dermaseptin B1 peptide enhances the antimicrobial activity of the peptide toward microbial pathogens.
1. INTRODUCTION
Plant pests and diseases are among the main factors reducing the production of agricultural products and diminishing their quality and yield as well as threatening food safety (Oerke, 2006). In developing countries, it is estimated that pests and diseases decrease the yield of crop plants by 30%–40% (Flood, 2010). Chemical control is often used to combat devastating plant pathogens. However, given the negative effects of chemical control on human health and the environment, and emergence of resistance by pathogens, it is necessary to employ safer and more sophisticated methods to cope with plant pathogens (Vidaver, 2002).
Plants activate their immune‐defense system when pathogens attack (Nguyen, Haney, & Vogel, 2011; Zasloff, 2002, 2006). For instance, following infection, plants express chitinases, glucanases and produce defensins and other antimicrobial peptides (AMPs) to fight pathogens (Bruce & Pickett, 2007). A broad spectrum of living organisms produces AMPs (Nguyen et al., 2011; Zasloff, 2002, 2006). AMPs, as an important part of host immune system, play a pivotal role in plant resistance toward pests and pathogenic microbes (Nguyen et al., 2011; Zasloff, 2002, 2006). Despite the considerable diversity among plant defensins, plant pathogens can still inflict considerable damage to plant yield and quality (Portieles et al., 2010).
Among more than 3,053 peptides listed in the antimicrobial peptide database so far (23‐01‐2019), peptides belonging to the Dermaseptin family show high antimicrobial activity and are mostly lytic to microbial pathogen (Reddy, Yedery, & Aranha, 2004). Dermaseptins are 23–35 amino acid long peptides produced and secreted by the skin glands of a number of frog families (Tossi, Sandri, & Giangaspero, 2000). Dermaseptins show a broad‐spectrum inhibitory effect against both gram‐negative and gram‐positive bacteria (Navon‐Venezia, Feder, Gaidukov, Carmeli, & Mor, 2002; Osusky, Osuska, Kay, & Misra, 2005; Yaron, Rydlo, Shachar, & Mor, 2003), yeasts (Coote, Holyoak, Bracey, Ferdinando, & Pearce, 1998), protozoa (Hernandez et al., 1992), and fungi (De Lucca, Bland, Jacks, Grimm, & Walsh, 1998).
Fungal cell wall plays a crucial role in fungal pathogenesis since it acts as the major interface with the host immune system (Hopke, Brown, Hall, & Wheeler, 2018), and fungal mutants affected in the biosynthesis of cell wall oligosaccharides are severely affected in their growth and pathogenicity (Bowman & Free, 2006). Chitin is one of the main structural oligosaccharides of the cell wall in various pathogenic fungi, playing a role of an important elicitor of innate defense responses in plants (Sánchez‐Vallet, Mesters, & Thomma, 2015). Through their hydrolytic activities, plant chitinases hydrolyze cell wall chitin eventually leading to cell death (Latgé, 2010; Latgé & Beauvais, 2014; Thomma, Nürnberger, & Joosten, 2011). To fight back, fungal pathogens employ various strategies, including changes in cell wall oligosaccharides, secreting effectors, and masking cell wall components (Fujikawa et al., 2012; Latgé & Beauvais, 2014). For example, Cladosporium fulvum produces and secretes Avr4 effectors at the infection site covering the cell wall chitin inhibiting plant hydrolytic enzymes (van den Burg, Harrison, Joosten, Vervoort, & de Wit, 2006). The Avr4 effector has a CBD binding to the chitin in fungal cell walls, decreasing host chitinases access to fungal cell walls and thus preventing fungal cell wall degradation (van den Burg et al., 2006).
Since correct packaging, disulfide bonds formation, accumulation of multi‐subunit proteins, and posttranslational modifications are performed by plant cells accurately, plant systems are used to produce eukaryotic recombinant proteins (Daniell, Streatfield, & Wycoff, 2001; Giddings, Allison, Brooks, & Carter, 2000). However, low production levels, high extraction costs, and limitations in the degree of recombinant protein purification are among major challenges facing recombinant protein production in plants (Borisjuk et al., 1999). The use of Agrobacterium rhizogenes‐mediated transformation to produce hairy roots (HRs) is one of the efficient strategies to produce recombinant proteins and secondary metabolites (Borisjuk et al., 1999).
The use of genetic engineering strategies to introduce genes encoding natural and synthetic antimicrobial peptides is a new approach in engineering resistance to a broad spectrum of pathogens (Zasloff, 2002). Various recombinant proteins and peptides have so far been introduced in HRs of different plant species, and their antimicrobial properties have been tested in vitro (Aleinein, Schäfer, & Wink, 2015; Moghadam, Niazi, Afsharifar, & Taghavi, 2016; Pham, Schäfer, & Wink, 2012). For instance, an anti‐HIV protein and an anti‐tumor protein MAP30, including ribosome‐inhibiting proteins were produced in tobacco HRs (Moghadam et al., 2016). Analysis of total proteins extracted from transgenic HRs indicated a strong antimicrobial activity against both gram‐positive and gram‐negative bacteria as well as pathogenic fungi (Moghadam et al., 2016). Chahardoli, Fazeli, and Ghabooli (2018) expressed a lactoferricin encoding protein in tobacco HR culture with potent antibacterial activity against Escherichia coli (Chahardoli et al., 2018). Introduction of ranalexin peptide in tobacco HRs resulted in efficient production of ranalaxin in HRs with strong inhibitory effect on multidrug resistant gram‐positive and gram‐negative pathogens such as Staphylococcus aureus, Streptococcus pyogenes, and E. coli and on Enterococcus strains resistant to vancomycin, suggesting that tobacco HR culture was a suitable system to produce ranalexin and other recombinant peptides (Aleinein et al., 2015).
In this study, we showed that fusion of dermaseptin B1 (DrsB1) antimicrobial peptide to the chitin‐binding domain (CBD) of Avr4 protein from C. fulvum enhanced the antibacterial activity of DrsB1 peptide, suggesting that CBD might facilitate DrsB1 peptide access to the fungal plasma membrane, leading to cell membrane rupture and deformation.
2. MATERIALS AND METHODS
2.1. Expression cassettes
Ninety‐three nucleotide‐long DNA sequence encoding the DrsB1 antimicrobial peptide (UniProtKB accession number P80282) was codon optimized and fused either to the C or the N terminus of the 192 nucleotides‐long sequence encoding the CBD of Avr4 effector gene of C. fulvum (GenBank accession number CAA69643.1). Two recombinant constructs were chemically synthesized and cloned in two pUC cloning vectors (Biomatik, Canada). Sequence encoding 18 amino acids long signal peptide (SP) of Avr4 gene was also fused to the 5′ end of the constructs to ensure secretion of recombinant proteins in the apoplastic space. Moreover, the cleavage sites of NcoI and BamHI restriction enzymes were engineered at the 5′ and the 3′ end of the recombinant genes for cloning purposes (Figure 1). The rice chitinase helix‐forming linker (EAAAK)4 sequence was used to fuse DrsB1 to CBD. The pUC vectors were digested with NcoI and BamHI restriction enzymes, and recombinant fragments were subcloned in the pGSA1285 expression vector, resulting in pGSA1285/CBD‐DrsB1 and pGSA1285/DrsB1‐CBD vectors (Figure 1). The recombinant genes were driven by cauliflower mosaic virus 35S (3×) promoter. The Arg‐Gly‐Ser‐(His)6 sequence was engineered after SP to identify the recombinant proteins (Figure 1).
Figure 1.
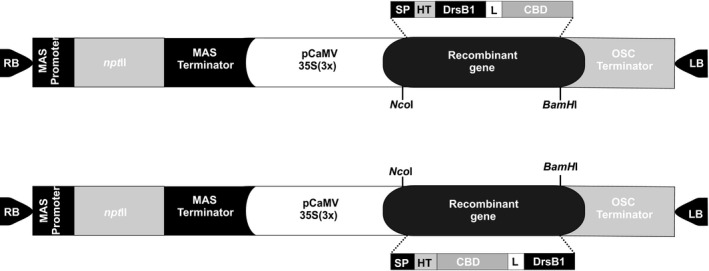
The schematic illustration of the expression vectors used for the recombinant protein production in tobacco hairy roots. MAS: Mannopine synthase, npt II: Neomycin phosphotransferase II, CaMV35S: Cauliflower mosaic virus 35S promoter, OSC: Octopine synthase, SP: Avr4 signal peptide, HT: Histidine tag (RGS‐(His)6), L: (EAAAK)4 Linker, CBD: chitin binding domain, RB: right border and LB: left border
2.2. Agrobacterium rhizogenes‐mediated transformation
Tobacco (Nicotiana tabacum) Xanthi cultivar seeds were disinfected in a detergent solution (5% sodium hypochlorite and Triton X‐100) for 10 min and then washed three times with distilled water to remove detergent residues. The seeds were then germinated on MS culture medium under 16 hr light/8 hr dark photoperiod at 24 ± 2°C. Two A. rhizogenes (ATCC15834) clones harboring the pGSA1285/CBD‐DrsB1 and pGSA1285/DrsB1‐CBD expression vectors were used to inoculate tobacco leaf disks. Briefly, sterilized 3‐week‐old tobacco leaf disks (1 cm2) were cut and put in an A. rhizogenes two‐day inoculation suspension for 10 min. The inoculated leaves were dried using a sterile filter paper and then placed on the hormone and antibiotic‐free MS culture medium. The inoculated leaf disks were incubated at 24 ± 2°C for 2–3 days in dark and were eventually transferred to the selective medium containing kanamycin (50 mg/L) and cefotaxime (200 mg/L) for HR induction. The explants were regularly subcultured once every 2 weeks until root formation (Tempe & Casse‐Delbart, 2012).
2.3. Genomic DNA extraction and screening of putative transgenic HRs
Genomic DNA was extracted from putative transgenic and control HRs, using the CTAB method (Gawel & Jarret, 1991). In order to demonstrate that the HRs are free of A. rhizogenes cells, the polymerase chain reaction (PCR) was performed using the VirG specific primers (VirGF:5′‐CCGCGGTCAGCCGCAATTCT‐3′; VirGR:5′‐CTGCACGTCCGCGTCAAAGAAATA‐3′). The presence of the T‐DNA in HRs was confirmed by PCR amplification of the rolC gene by using rolC specific primers (rolCF:5′‐CTCCTGACATCAAAACTCGTC‐3′, rolCR:5′‐TGCTTCGAGTTATGGGTACA‐3′). Finally, to screen the putative transgenic HRs lines, the DrsB1 specific primers (DrsF:5′‐GCTAAGGCTATGTGGAAGGATG‐3′, DrsR:5′‐ATTGAGAAATAGTATCAGCAACAGC‐3′) were used. In each PCR, the initial denaturation was carried out once at 94°C for 5 min followed by 35 cycles of denaturation (at 94°C for 1 min), annealing (at 59°C, 53°C and 59°C, respectively for 30 s), extension (at 72°C for 30 s to one min), and a final extension at 72°C for 5 min.
2.4. Expression analysis of the recombinant genes
Total RNA was extracted following lithium chloride method (Li, Wang, Sun, & Li, 2011) followed by DNase treatment to remove genomic DNA contamination. cDNA was synthesized using a cDNA synthesis kit (Thermo Fisher Scientific Inc., #k1622). Semi‐quantitative RT‐PCR analysis was performed according to Nazarian Firouzabadi et al. (2007) employing the DrsB1 specific primers (RtDrsF: GCTAAGGCTATGTGGAAGGATG; RtDrsR: ATTGAGAAATAGTATCAGCAACAGC). The elongation factor 1α (elf‐1α) gene was used as an internal control (Nazarian Firouzabadi et al., 2007).
2.5. Total protein extraction from HRs
One‐gram HR tissue from transgenic and control lines were ground in liquid nitrogen and homogenized in a potassium phosphate buffer (50 mM at pH7, 1 mM phenylmethylsulfonyl fluoride). The homogenized samples were vigorously vortexed for 2–5 min and centrifuged at 13,000 rpm for 30 min at 4°C, and the supernatants were filtered using 0.45 µm membranes (Stone & Gifford, 1997). Furthermore, total protein was also extracted from a DrsB1‐expressing (DrsB1‐04) transgenic HR with no CBD fused as a control (Alibakhshi, 2017). The concentration of extracted proteins was measured using the Bradford method (Bradford, 1976), and proteins were stored at −20°C.
2.6. Purification of the recombinant proteins and Western blot analysis
To purify the expressed recombinant proteins, total protein isolated from transgenic and controls was transferred to a chromatography column containing the PrepEase Ni‐IDA resin. The column was prewashed with 300 mM NaCl and then with 50 mM NaH2O4. The purified recombinant proteins were removed from the column by rinsing the column, using 250 mM imidazole. Purified proteins were then loaded on 14% acrylamide gel and electrophoresed at 150 V. The recombinant proteins were electroblotted by using a Mini‐Protean II Multiscreen Apparatus (Bio‐Rad). The nitrocellulose blot was blocked for 1 hr, using tris buffered saline (TBS) containing 5% powdered milk. The blots were washed three times with TBS buffer and then exposed to 1:2000 dilution of mouse monoclonal anti‐His antibody at 37°C for 1 hr followed by 3,3′‐diaminobenzidine tetrahydrochloride (DAB) detection according to the manufacturer's instructions (Bollag, Edelstein, & Rozycki, 1996).
2.7. Antimicrobial activity of the recombinant proteins
Antimicrobial activities of the recombinant proteins were determined by studying the activity of the total protein extracted from transgenic HRs against the gram‐negative plant bacteria, including Agrobacterium tumefaciens (PTCC 1654), Pectobacterium carotovorum subsp. carotovorum (PTCC 1675), Ralstonia solanacearum (ATCC 11696), and Xanthomonas campestris (PTCC 1473) using the disk diffusion method (Bauer, Kirby, Sherris, & Turck, 1966; Mangena & Muyima, 1999). Bacterial cultures were provided by the Department of Plant Protection, Faculty of Agriculture, Lorestan University, Iran. Briefly, 100 µl of 21 hr old culture of the bacterial suspensions of half‐MacFarland (1.5 × 108 cfu/ml) was poured onto the surface of the nutrient agar culture medium and spread by using a sterilized cotton swab. Next, 50 µl of the filter sterilized extracted fusion proteins (60 μg/ml) were added to each of the sterile 6‐mm disks. The disks containing the recombinant proteins were then placed on the bacterial culture medium. The Petri dishes were maintained at 4°C for 30 min to fix the proteins in the disks and then incubated at 28°C or 30°C (depending on the bacterium type) for 16 hr. In order to determine the antimicrobial activity of fusion proteins, the diameter of inhibition zones surrounding the disks was measured in triplicates in millimeter (r zone − r disk). It must be mentioned that the gentamicin disk (10 μg/disk) was used as the positive control and the proteins extracted from the non‐transgenic HRs as the negative control. Statistical analysis of the treatments was conducted using MSTATC and SAS 9.1 softwares in a factorial experiment using a completely randomized design with three replications. If there were significant differences between the treatments, Duncan's multiple range test at the relevant significant level was used to compare the means.
2.8. Minimum inhibitory concentrations of the recombinant proteins
The microdilution method was used to determine the minimum inhibitory concentrations (MICs) of the recombinant proteins according to Che et al. (2011). Briefly, five different protein concentrations (5.62, 11.25, 22.5, 45, and 90 μg/ml) of the purified recombinant proteins were tested against bacterial suspensions in 96‐well plates with serial dilutions in a final volume of 200 μl. Twenty microliters of the bacterial suspensions (1.5 × 108 cells) was added to each well and the plates were maintained at 28°C or 37°C for 24 hr. The MIC was measured by the lowest concentration with no visible growth. LB broth was used as the negative control and bacterial culture without addition of antimicrobial recombinant proteins was used as the positive control. All experiments were designed and performed in triplicates for each bacterial species (Che et al., 2011).
2.9. Antifungal activity of the recombinant proteins
Antifungal activity of the recombinant proteins was evaluated by mixing the recombinant proteins to the growth medium. Recombinant proteins were added in PDA medium to the final concentration of 50 μg/ml. Ten‐millimeter plugs of young fungal cultures were kept in the center of 9 cm PDA containing Petri plates. Fungal mycelia diameters were measured at 4–7 days after inoculation at 25°C. The following equation was used to calculate the percentage of mycelium growth inhibition:
where C is the percentage of mycelium growth inhibition, W is the radial growth zone diameter in the control, and T represents the diameter of the radial growth zone in the treatment.
For fungal MIC assessment, various concentrations (5, 10, 20, 30, 40, and 50 µg/ml) of the purified recombinant proteins were mixed with the fungal spore suspension (1 × 108 cfu/ml) in a final volume of 100 µl. After 24 hr, germinated conidia and spores were counted under a light microscope. The MIC values were defined as the lowest concentration of recombinant proteins required for complete suppression of fungal spore and conidia germination (Yevtushenko et al., 2005).
2.10. Scanning electron microscopy analysis
Alternaria alternata (PTCC 5,224) and Pythium sp. fungi were cultured on the PDA medium for 4 days. Fungal plugs (1 cm2) were injected with each of the recombinant proteins (50 μg/ml) and incubated at 25°C for 24 hr followed by freezing at −80°C for 24 hr. The frozen plugs were then put in a freeze‐drying machine for 4 hr. The fixed samples were coated with gold nanoparticles using a Desk Sputter Coater–DSRI, and finally scanned with an electron microscope (FE‐SEM, Tescan Mira3 LMU, at HV = 20 kV).
3. RESULTS
3.1. Molecular analysis of transgenic plants
Transgenic HRs were produced and designated as CBD‐DrsB1‐XX and DrsB1‐CBD‐XX based on the type of the construct employed to express the recombinant proteins, XX represents the transgenic line number.
Polymerase chain reaction analysis of putative transgenic HRs using rolC and DrsB1 peptide specific primers resulted in amplification of 600 bp and 100 bp PCR products, respectively, suggesting that HRs are transgenic, whereas no PCR product was amplified in non‐transgenic HRs. Moreover, PCR analysis using the VirG specific primers did not lead to any amplification, ruling out possible A. rhizogenes contamination (Appendix Figure A1).
Expression of the recombinant genes was determined using semi‐quantitative RT‐PCR analysis according to Nazarian Firouzabadi et al. (2007). A PCR product with the approximate 100 bp size was amplified from the transgenic HRs lines, suggesting that the recombinant genes are transcribed. No fragment was observed in the non‐transgenic HRs controls. Considering the intensity of mRNA transcript of both recombinant genes and the elf‐1α as the housekeeping gene, no obvious difference was noticed between different transgenic HR lines regarding the level of the expression of recombinant genes (Figure 2a). In the selected transformants, the presence of CBD‐DrsB1 and DrsB1‐CBD recombinant proteins was analyzed using Western blotting analysis. The recombinant proteins were produced in the HRs, whereas no traces of such proteins were found in the non‐transgenic HRs (Figure 2b).
Figure 2.
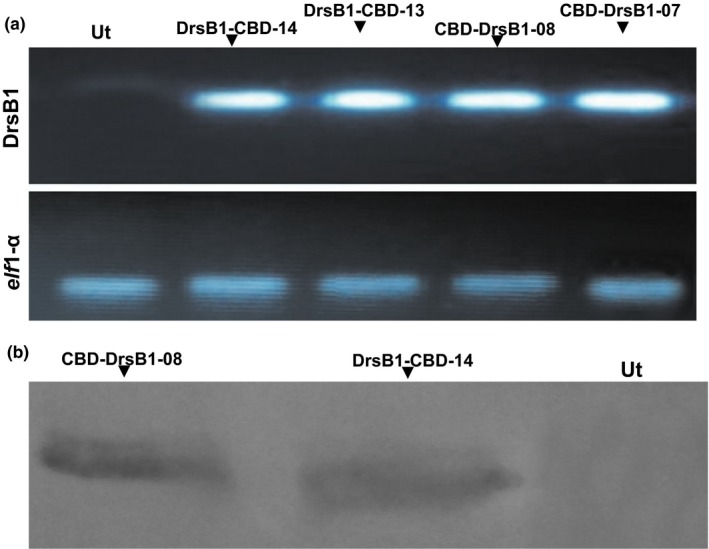
Semi‐quantitative RT‐PCR (a) and western blotting (b) analysis of the transgenic and control hairy roots (HRs). The polymerase chain reaction (PCR) products of the elf1‐α housekeeping gene were used to compare the level of mRNA transcripts of different transgenic HRs
3.2. Antimicrobial activity of the recombinant proteins
Sixty micrograms per milliliter (60 μg/ml) total protein from the CBD‐DrsB1‐8, CBD‐DB1‐14, and DrsB1‐04 lines (Alibakhshi, 2017) were used to compare the activity of recombinant proteins against four pathogenic bacteria. Total protein from transgenic HRs lines had a significant (p < 0.01) higher antibacterial activity than total protein extracted from non‐transgenic HRs. Interestingly, the total protein from transgenic HRs lines exhibited a higher activity than DrsB1‐04 transgenic line (Table 1).
Table 1.
The effect of recombinant fusion proteins on four bacteria species. The inhibition zone diameter is presented as the mean ± standard deviation of three replicates
| Diameter of the inhibition zone (mm) | ||||
|---|---|---|---|---|
| Transgenic line/antibiotics | Agrobacterium tumefaciens | Pectobacterium carotovorum | Ralstonia solanacearum | Xanthomonas campestris |
| CBD‐DrsB1‐08 | 13.33 ± 0.5d | 21. 60 ± 0.6ab | 7.60 ± 0.5ef | 8.30 ± 0.5e |
| DrsB1‐CBD‐14 | 12.00 ± 0.5d | 13.40 ± 0.6d | 3.50 ± 0.3gh | 2.10 ± 0.3hi |
| DrsB1‐04 | 3.60 ± 0.6gh | 5.00 ± 0.5fg | 1.20 ± 0.2hi | 0i |
| Ut | 0 | 0 | 0 | 0 |
| Gentamicin (10 μg) | 21.30 ± 0.5ab | 22.30 ± 0.5a | 18.80 ± 0.5bc | 16.60 ± 0.5c |
Means that do not share the same alphabetic superscript are significantly (p < 0.01) different according to Duncan's multiple range test.
Ut: non‐transgenic control line.
The antibacterial activity of the recombinant proteins was assayed for bactericidal activity against four gram‐negative bacteria. The recombinant proteins showed a stronger activity against P. carotovorum and A. tumefaciens bacteria than R. solanacearum and X. campestris bacteria (Figure 3). The recombinant protein of CBD‐DrsB1‐8 transgenic HR line demonstrated a stronger inhibition activity against P. carotovorum in comparison to that of DrsB1‐CBD‐14 transgenic line. Moreover, the inhibitory activity of the CBD‐DrsB1‐8 and DrsB1‐CBD‐14 lines was significantly (p < 0.01) higher than the DrsB1‐04 transgenic line (Table 1).
Figure 3.
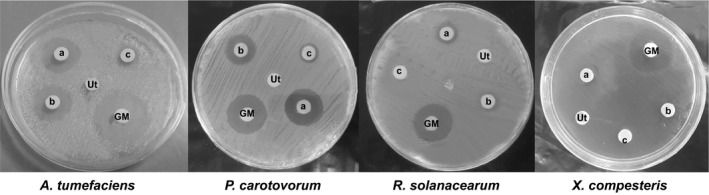
The antimicrobial activities of the total proteins derived from the transgenic and control hairy root lines. (a) CBD‐DrsB1‐08 line, (b) DrsB1‐CBD‐14 line, (c) DrsB1‐04 line, Ut: non‐transgenic control plant and GM: Gentamicin (10 μg/disk)
The antifungal activity of the recombinant proteins was assayed in triplicates against A. alternata and Pythium sp. fungi. Total protein isolated from transgenic HRs exhibited a significant (p < 0.01) antifungal activity against both fungi in comparison to non‐transgenic control HRs. Additionally, the inhibitory effect of the two recombinant proteins with the CBD was significantly (p < 0.01) stronger than that of the DrsB1‐04 peptide. Among the two recombinant proteins, DrsB1‐CBD exhibited a higher inhibitory effect against A. alternata than the CBD‐DrsB1 recombinant protein, whereas the two recombinant proteins had a similar inhibitory effect (p > 0.05) on Pythium sp. growth (Figure 4).
Figure 4.
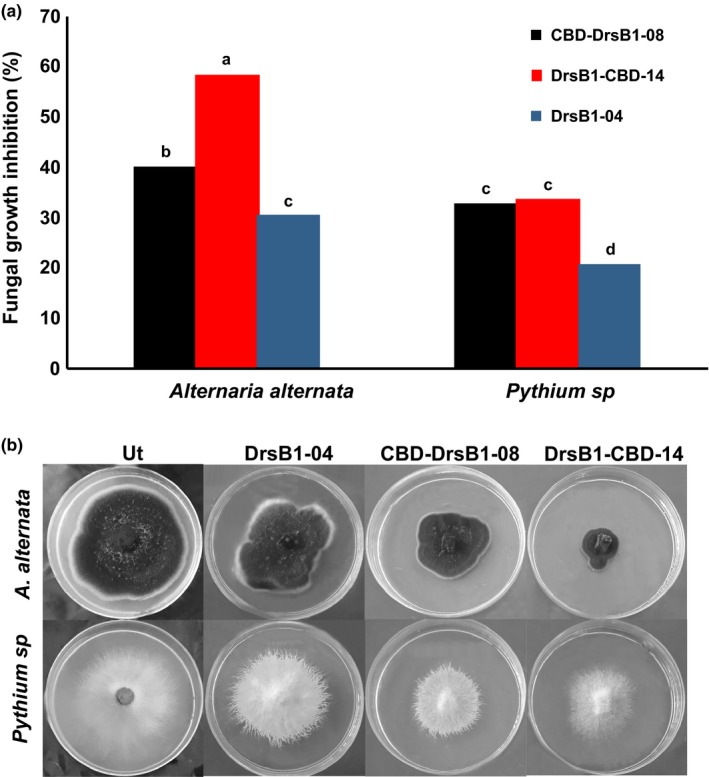
(a) The diagram showing the effect of recombinant proteins on fungal growth for Alternaria alternata and Pythium sp. (b) Antifungal activity of non‐transgenic control and recombinant proteins isolated from different transgenic HR lines. Ut: non‐transgenic control line
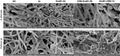
Alternaria alternata and Pythium sp. fungi were treated with the recombinant proteins to study effects of antifungal activity on the cellular structures of fungi. It was found that the growth and development of A. alternata hyphae slowed or stopped under the influence of the two types of recombinant proteins. SEM images demonstrated that A. alternata spores were deformed and shrunken, and apparently, their contents leaked out. As observed, the DrsB1‐CBD recombinant protein had greater destructive effects on this fungus spores than the CBD‐DrsB1 recombinant protein. Additionally, deformation and adhesion of Pythium sp. mycelia were observed. Both recombinant proteins had similar effects on the structure of Pythium sp. mycelia (Figure 5).
Figure 5.
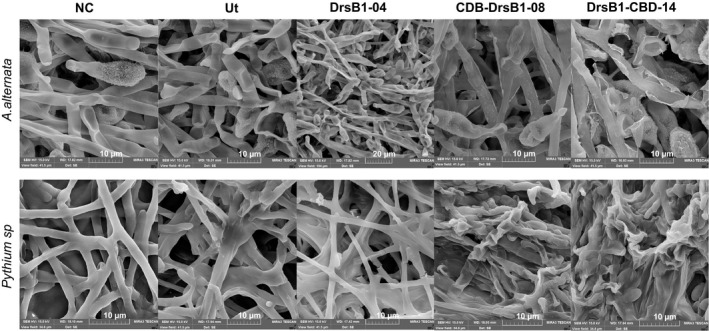
Electron microscopy images of Alternaria alternata (upper panel) and Pythium sp. (lower panel) hyphae treated with the recombinant proteins (50 μg/ml). Scale bars are indicated in µm for each image. Ut: non‐transgenic control line and NC: nontreated control
The MIC, the lowest concentration of recombinant fusion proteins inhibiting the pathogen growth, was determined in a microdilution assay. The MIC of the recombinant fusion proteins was in the range of 45–90 μg/ml, whereas a higher concentration of DrsB1 peptide (90 μg/ml) was needed to inhibit bacterial growth in vitro (Alibakhshi, 2017). The MIC value of recombinant fusion proteins for P. carotovorum and A. tumefaciens was lower (45 μg/ml) in comparison to the MIC value for R. solanacearum and X. campestris (90 μg/ml) bacteria. Interestingly, the MIC of the recombinant fusion proteins was significantly different for A. alternata, suggesting the position of DrsB1 peptide may have an effect in the overall activity of the recombinant proteins. A lower (10 µg/ml) concentration of the DrsB1‐CBD recombinant fusion protein was needed to completely inhibit the A. alternata conidia germination in comparison with that of CBD‐DrsB1 (20 µg/ml) recombinant fusion protein. Furthermore, a MIC of 30 µg/ml of the DrsB1 peptide was sufficient to inhibit A. alternata conidia germinations, suggesting that fusion of DrsB1 to the CBD enhanced the antifungal activity of DrsB1 peptide. Interestingly, DrsB1‐CBD (20 µg/ml) recombinant protein had a better antifungal activity against Pythium sp. than CBD‐DrsB1 (30 µg/ml) and DrsB1 (40 µg/ml).
4. DISCUSSION
Over the past few decades, various approaches have been employed in molecular biology to improve and increase plant resistance to a broad spectrum of plant pathogens (Cao, Li, & Dong, 1998). For instance, expression of fungal and bacterial cell wall‐degrading enzymes (chitinases), expression of pathogenesis‐related proteins, increase in production of host proteins and metabolites involved in plant defense pathways, and expression of plant antimicrobial proteins and peptides have been reported (Punja, 2001).
All living organisms produce different classes of antimicrobial peptides as a part of their innate immune system to combat pathogens (Hancock & Scott, 2000; Holaskova, Galuszka, Frebort, & Oz, 2015). To enhance the antibacterial activity of natural peptides, researchers design and synthesize new variants or recombinant peptides for pharmaceutical and agricultural industries (Melo, Ferre, & Castanho, 2009; Yevtushenko & Misra, 2012; Yevtushenko et al., 2005). In this study, the DrsB1 peptide was fused to the CBD of the Avr4 gene from C. fulvum so that the recombinant protein could bind to the fungal cell wall as well as the peptidoglycans of bacterial cell walls perturbing the integrity of cell wall components. Although the DrsB1 peptide exhibited strong in vitro antimicrobial activity against bacteria, fungi, protozoans, and yeasts, the antibacterial activity was significantly increased when the N‐terminal region of the DrsB1 peptide and MsrA2 analog was manipulated to bind to the negatively charged lipids (Osusky et al., 2005). The expression of modified peptides in potato and tobacco plants led to the production of transgenic lines with enhanced resistance to a number of devastating plant pathogens. Interestingly, the recombinant proteins extracted from transgenic HRs in this study, exhibited a significant (p < 0.01) antibacterial activity against plant pathogenic gram‐negative bacteria. Similarly, Badrhadad, Nazarian‐Firouzabadi, and Ismaili (2018) provided strong evidence that fusion of an alfalfa antibacterial peptide to rice chitinase CBD inhibited the growth and development of plant pathogens (Badrhadad et al., 2018).
The inhibitory concentrations of the recombination proteins varied significantly for different bacteria and fungi. It was found that the fungi are more sensitive to recombinant proteins than bacteria. The susceptibility of bacteria and fungi to recombinant proteins may be attributed to the variation of different cell components present in bacteria and fungi (Marcos, Muñoz, Pérez‐Payá, Misra, & López‐García, 2008).
Although the mechanism by which antimicrobial peptides attack pathogens is not fully understood, the mode of action may involve interaction of charged components (Nguyen et al., 2011; Toke, 2005). A relatively higher positive charge of the recombinant peptides and the binding affinity of the CBD toward cell wall building blocks may accumulate more DrsB1 peptide on the surface of the pathogen leading to effective interaction between positively charged recombinant peptides and the negatively charged membrane surface of the pathogens. It is documented that the positive charge in the hydrophobic part of cationic peptides is essential for their antimicrobial activity (Yin, Edwards, Li, Yip, & Deber, 2012). In other words, fusion of the CBD from Avr4 along with addition of histidine residues increases the antimicrobial activity of the recombinant proteins against pathogenic microbes. There seems to be a relationship between the chitin‐binding capability and antimicrobial activity of the CY‐AMP peptide against the gram‐positive bacteria Lactococcus lactis, Streptococcus mutans, and Clavibacter michiganensis, the gram‐negative Erwinia carotovora and Enterobacter cloacae, and the fungi Fusarium oxysporum and Geotrichum candidum (Yokoyama et al., 2009). Mutations in the CBD of the CY‐AMP peptide led to a decrease in chitin‐binding ability and hence antifungal activity, whereas the antibacterial activity of CY‐AMP against gram‐positive and gram‐negative bacteria did not change in comparison with that of the wild‐type peptide (Yokoyama et al., 2009). It is noteworthy that the antifungal activity of a barley chitinase mutated at the important catalytic domain of acidic residues declined by 75% in comparison with that of wild‐type chitinase (Andersen, Jensen, Robertus, Robert, & Skriver, 1997). Overall, it can be concluded that the antifungal activity of the recombinant proteins increases as CBD shows intrinsic affinity for chitin. Therefore, the recombinant proteins in this study had a relatively higher inhibitory effect against A. alternata in comparison to Pythium sp. Furthermore, the presence of chitin in the cell wall seems to be crucial for antifungal activity of the recombinant proteins (Yan et al., 2008). The results of a similar study indicated that a protein bound to a chitin‐binding protein with antifungal activity from Moringa oleifera seeds (MO‐CBP3) had a strong antifungal activity against Fusarium solani, F. oxysporum, but did not inhibit the growth and germination of Pythium oligandrum, suggesting that adhesion to the chitin is vital for antibacterial activity (Gifoni et al., 2012).
In conclusion, results of the present study indicated that the cell wall chitin can be targeted to control plant pathogenic fungi containing chitin (Yan et al., 2008). Accumulation of DrsB1 peptide on the surface of fungal pathogens in a carpet‐like manner (Pouny, Rapaport, Mor, Nicolas, & Shai, 1992; Shai, 1999) may result in instability of the fungal cell, eventually leading to the fungal cell death (Figure 5). The DrsB1‐CBD recombinant protein had a higher inhibitory effect than CBD‐DrsB1 recombinant protein, suggesting that DrsB1 peptide may interfere with CBD affinity for cell wall chitin, leading to a lower concentration of the CBD‐DrsB1 recombinant protein at the cell wall surface. It is noteworthy that no plant chitinase has so far been identified with the CBD at the C‐terminal part of the main catalytic domains (Beintema, 1994; Iseli, Boller, & Neuhaus, 1993). Due to the high antimicrobial activity of the recombinant proteins of this study, it would be interesting to introduce the recombinant genes to crop plants and generate resistant lines to devastating plant pathogens.
CONFLICT OF INTERESTS
None declared.
AUTHORS CONTRIBUTION
MK performed the experiments. F.N‐F designed the experiments, wrote the manuscript. A.A and RSK helped in designing some of the experiments.
ETHICS STATEMENT
None required.
ACKNOWLEDGEMENTS
This study was supported by the Lorestan University research grant. We appreciate Laboratory of plant pathology at Lorestan and Gillan universities for their excellent assistance. We are also grateful to Dr. Dhananjay Dhokane from Reliance Industries for reading our manuscript.
APPENDIX 1.
1.1.
Figure A1.
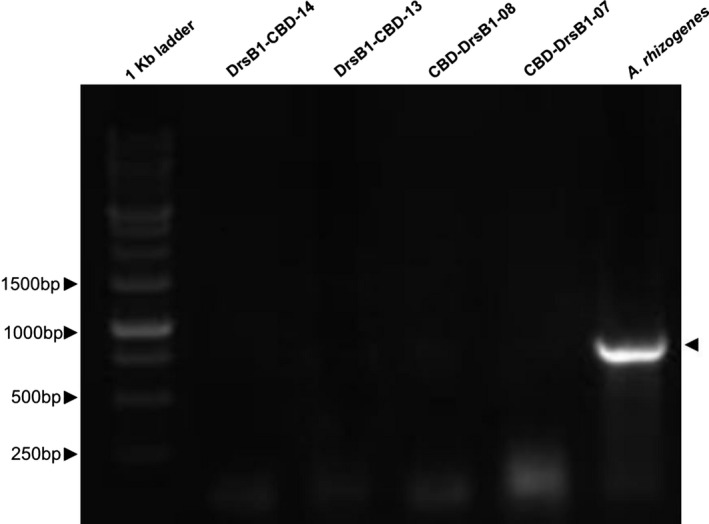
PCR analysis of tobacco hairy roots obtained after transformation with A. rhizogenes. DNA isolated from A. rhizogenes as positive control. Arrow shows amplified fragments of virG
Khademi M, Nazarian‐Firouzabadi F, Ismaili A, Shirzadian Khorramabad R. Targeting microbial pathogens by expression of new recombinant dermaseptin peptides in tobacco. MicrobiologyOpen. 2019;8:e837 10.1002/mbo3.837
DATA ACCESSIBILITY
All data used in this study are presented in the manuscript.
REFERENCES
- Aleinein, R. , Schäfer, H. , & Wink, M. (2015). Rhizosecretion of the recombinant antimicrobial peptide ranalexin from transgenic tobacco hairy roots. Journal of Botanical Sciences, 1, 45–55. [Google Scholar]
- Alibakhshi, A. (2017). Expression of a Dermaseptin B1 peptide in tobacco to produce transgenic plants resistance to pathogens. (MS.c Degree), Iran: Lorestan University. [Google Scholar]
- Andersen, M. D. , Jensen, A. , Robertus, J. D. , Robert, L. , & Skriver, K. (1997). Heterologous expression and characterization of wild‐type and mutant forms of a 26 kDa endochitinase from barley (Hordeum vulgare L.). Biochemical Journal, 322(3), 815–822. 10.1042/bj3220815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrhadad, A. , Nazarian‐Firouzabadi, F. , & Ismaili, A. (2018). Fusion of a chitin‐binding domain to an antibacterial peptide to enhance resistance to Fusarium solani in tobacco (Nicotiana tabacum). 3 Biotech, 8(9), 391 10.1007/s13205-018-1416-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, A. , Kirby, W. , Sherris, J. C. , & Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology, 45(4), 493–496. 10.1093/ajcp/45.4_ts.493 [DOI] [PubMed] [Google Scholar]
- Beintema, J. J. (1994). Structural features of plant chitinases and chitin‐binding proteins. FEBS Letters, 350(2–3), 159–163. 10.1016/0014-5793(94)00753-5 [DOI] [PubMed] [Google Scholar]
- Bollag, D. M. , Edelstein, S. J. , & Rozycki, M. D. (1996). Proteins methods, 2nd ed. New York, NY: John Wiley & Sons. [Google Scholar]
- Borisjuk, N. V. , Borisjuk, L. G. , Logendra, S. , Petersen, F. , Gleba, Y. , & Raskin, I. (1999). Production of recombinant proteins in plant root exudates. NatureBiotechnology, 17(5), 466–469. 10.1016/10.1038/8643 [DOI] [PubMed] [Google Scholar]
- Bowman, S. M. , & Free, S. J. (2006). The structure and synthesis of the fungal cell wall. BioEssays, 28(8), 799–808. 10.1016/10.1002/bies.20441 [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry, 72(1–2), 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Bruce, T. J. , & Pickett, J. A. (2007). Plant defence signalling induced by biotic attacks. Current Opinion in Plant Biology, 10(4), 387–392. 10.1016/j.pbi.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Cao, H. , Li, X. , & Dong, X. (1998). Generation of broad‐spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proceedings of the National Academy of Sciences, 95(11), 6531–6536. 10.1073/pnas.95.11.6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahardoli, M. , Fazeli, A. , & Ghabooli, M. (2018). Recombinant production of bovine Lactoferrin‐derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiology and Biochemistry, 123, 414–421. 10.1016/j.plaphy.2017.12.037 [DOI] [PubMed] [Google Scholar]
- Che, Y. Z. , Li, Y. R. , Zou, H. S. , Zou, L. F. , Zhang, B. , & Chen, G. Y. (2011). A novel antimicrobial protein for plant protection consisting of a Xanthomonas oryzae harpin and active domains of cecropin A and melittin. Microbial Biotechnology, 4(6), 777–793. 10.1111/j.1751-7915.2011.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote, P. J. , Holyoak, C. D. , Bracey, D. , Ferdinando, D. P. , & Pearce, J. A. (1998). Inhibitory action of a truncated derivative of the amphibian skin peptide dermaseptin s3 on Saccharomyces cerevisiae. Antimicrobial Agents and Chemotherapy, 42(9), 2160–2170. 10.1128/AAC.42.9.2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Streatfield, S. J. , & Wycoff, K. (2001). Medical molecular farming: Production of antibodies, biopharmaceuticals and edible vaccines in plants. Trends in Plant Science, 6(5), 219–226. 10.1016/S1360-1385(01)01922-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucca, A. , Bland, J. , Jacks, T. , Grimm, C. , & Walsh, T. (1998). Fungicidal and binding properties of the natural peptides cecropin B and dermaseptin. Medical Mycology, 36(5), 291–298. 10.1080/02681219880000461 [DOI] [PubMed] [Google Scholar]
- Flood, J. (2010). The importance of plant health to food security. Food Security, 2(3), 215–231. 10.1007/s12571-010-0072-5 [DOI] [Google Scholar]
- Fujikawa, T. , Sakaguchi, A. , Nishizawa, Y. , Kouzai, Y. , Minami, E. , Yano, S. , … Nishimura, M. (2012). Surface α‐1,3‐glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Path, 8(8), e1002882 10.1371/journal.ppat.1002882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawel, N. , & Jarret, R. (1991). A modified CTAB DNA extraction procedure for Musa and Ipomoea . Plant Molecular Biology Reporter, 9(3), 262–266. 10.1007/BF02672076 [DOI] [Google Scholar]
- Giddings, G. , Allison, G. , Brooks, D. , & Carter, A. (2000). Transgenic plants as factories for biopharmaceuticals. NatureBiotechnology, 18(11), 1151–1155. 10.1038/81132 [DOI] [PubMed] [Google Scholar]
- Gifoni, J. M. , Oliveira, J. T. , Oliveira, H. D. , Batista, A. B. , Pereira, M. L. , Gomes, A. S. , & Vasconcelos, I. M. (2012). A novel chitin‐binding protein from Moringa oleifera seed with potential for plant disease control. Peptide Science, 98(4), 406–415. 10.1002/bip.22068 [DOI] [PubMed] [Google Scholar]
- Hancock, R. E. , & Scott, M. G. (2000). The role of antimicrobial peptides in animal defenses. Proceedings of the National Academy of Sciences, 97(16), 8856–8861. 10.1073/pnas.97.16.8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, C. , Mor, A. , Dagger, F. , Nicolas, P. , Hernandez, A. , Benedetti, E. , & Dunia, I. (1992). Functional and structural damage in Leishmania mexicana exposed to the cationic peptide dermaseptin. European Journal of Cell Biology, 59(2), 414–424. [PubMed] [Google Scholar]
- Holaskova, E. , Galuszka, P. , Frebort, I. , & Oz, M. T. (2015). Antimicrobial peptide production and plant‐based expression systems for medical and agricultural biotechnology. BiotechnologyAdvances, 33(6), 1005–1023. 10.1016/j.biotechadv [DOI] [PubMed] [Google Scholar]
- Hopke, A. , Brown, A. J. , Hall, R. A. , & Wheeler, R. T. (2018). Dynamic Fungal cell wall architecture in stress adaptation and immune evasion. Trends in Microbiology, 26(4), 284–295. 10.1016/j.tim.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli, B. , Boller, T. , & Neuhaus, J.‐M. (1993). The N‐terminal cysteine‐rich domain of tobacco class I chitinase is essential for chitin binding but not for catalytic or antifungal activity. Plant Physiology, 103(1), 221–226. 10.1104/pp.103.1.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé, J. P. (2010). Tasting the fungal cell wall. Cellular Microbiology, 12(7), 863–872. 10.1111/j.1462-5822.2010.01474.x [DOI] [PubMed] [Google Scholar]
- Latgé, J.‐P. , & Beauvais, A. (2014). Functional duality of the cell wall. Current Opinion in Microbiology, 20, 111–117. 10.1016/j.mib.2014.05.009 [DOI] [PubMed] [Google Scholar]
- Li, X. , Wang, C. , Sun, H. , & Li, T. (2011). Establishment of the total RNA extraction system for lily bulbs with abundant polysaccharides. African Journal of Biotechnology, 10(78), 17907–17915. 10.5897/AJB10.2523 [DOI] [Google Scholar]
- Mangena, T. , & Muyima, N. (1999). Comparative evaluation of the antimicrobial activities of essential oils of Artemisia afra, Pteronia incana and Rosmarinus officinalis on selected bacteria and yeast strains. Letters in Applied Microbiology, 28(4), 291–296. 10.1046/j.1365-2672.1999.00525.x [DOI] [PubMed] [Google Scholar]
- Marcos, J. F. , Muñoz, A. , Pérez‐Payá, E. , Misra, S. , & López‐García, B. (2008). Identification and rational design of novel antimicrobial peptides for plant protection. Annual Review of Phytopathology, 46, 273–301. 10.1146/annurev.phyto.121307.094843 [DOI] [PubMed] [Google Scholar]
- Melo, M. N. , Ferre, R. , & Castanho, M. A. (2009). Antimicrobial peptides: Linking partition, activity and high membrane‐bound concentrations. Nature Reviews Microbiology, 7(3), 245 10.1038/nrmicro2095 [DOI] [PubMed] [Google Scholar]
- Moghadam, A. , Niazi, A. , Afsharifar, A. , & Taghavi, S. M. (2016). Expression of a recombinant anti‐HIV and anti‐tumor protein, MAP30, in nicotiana tobacum hairy roots: A pH‐stable and thermophilic antimicrobial protein. PLoS ONE, 11(7), e0159653 10.1371/journal.pone.0159653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon‐Venezia, S. , Feder, R. , Gaidukov, L. , Carmeli, Y. , & Mor, A. (2002). Antibacterial properties of dermaseptin S4 derivatives with in vivo activity. Antimicrobial Agents and Chemotherapy, 46(3), 689–694. 10.1128/AAC.46.3.689-694.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian Firouzabadi, F. , Kok‐Jacon, G. A. , Vincken, J.‐P. , Ji, Q. , Suurs, L. C. , & Visser, R. G. (2007). Fusion proteins comprising the catalytic domain of mutansucrase and a starch‐binding domain can alter the morphology of amylose‐free potato starch granules during biosynthesis. Transgenic Research, 16(5), 645–656. 10.1007/s11248-006-9053-z [DOI] [PubMed] [Google Scholar]
- Nguyen, L. T. , Haney, E. F. , & Vogel, H. J. (2011). The expanding scope of antimicrobial peptide structures and their modes of action. Trends in Biotechnology, 29(9), 464–472. 10.1016/j.tibtech.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Oerke, E.‐C. (2006). Crop losses to pests. The Journal of Agricultural Science, 144(1), 31–43. 10.1017/S0021859605005708 [DOI] [Google Scholar]
- Osusky, M. , Osuska, L. , Kay, W. , & Misra, S. (2005). Genetic modification of potato against microbial diseases: In vitro and in planta activity of a dermaseptin B1 derivative, MsrA2. Theoretical and Applied Genetics, 111(4), 711–722. 10.1007/s00122-005-2056-y [DOI] [PubMed] [Google Scholar]
- Pham, N. B. , Schäfer, H. , & Wink, M. (2012). Production and secretion of recombinant thaumatin in tobacco hairy root cultures. BiotechnologyJournal, 7(4), 537–545. 10.1007/s00122-005-2056-y [DOI] [PubMed] [Google Scholar]
- Portieles, R. , Ayra, C. , Gonzalez, E. , Gallo, A. , Rodriguez, R. , Chacón, O. , … Borrás‐Hidalgo, O. (2010). NmDef02, a novel antimicrobial gene isolated from Nicotiana megalosiphon confers high‐level pathogen resistance under greenhouse and field conditions. Plant Biotechnology Journal, 8(6), 678–690. 10.1111/j.1467-7652.2010.00501.x [DOI] [PubMed] [Google Scholar]
- Pouny, Y. , Rapaport, D. , Mor, A. , Nicolas, P. , & Shai, Y. (1992). Interaction of antimicrobial dermaseptin and its fluorescently labeled analogs with phospholipid membranes. Biochemistry, 31(49), 12416–12423. 10.1021/bi00164a017 [DOI] [PubMed] [Google Scholar]
- Punja, Z. K. (2001). Genetic engineering of plants to enhance resistance to fungal pathogens—A review of progress and future prospects. Canadian Journal of Plant Pathology, 23(3), 216–235. 10.1080/07060660109506935 [DOI] [Google Scholar]
- Reddy, K. , Yedery, R. , & Aranha, C. (2004). Antimicrobial peptides: Premises and promises. International Journal of Antimicrobial Agents, 24(6), 536–547. 10.1016/j.ijantimicag.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , Mesters, J. R. , & Thomma, B. P. (2015). The battle for chitin recognition in plant‐microbe interactions. FEMS Microbiology Reviews, 39(2), 171–183. 10.1093/femsre/fuu003 [DOI] [PubMed] [Google Scholar]
- Shai, Y. (1999). Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α‐helical antimicrobial and cell non‐selective membrane‐lytic peptides. Biochimica Et Biophysica Acta (BBA)—Biomembranes, 1462(1), 55–70. 10.1016/S0005-2736(99)00200-X [DOI] [PubMed] [Google Scholar]
- Stone, S. L. , & Gifford, D. J. (1997). Structural and biochemical changes in loblolly pine (Pinus taeda L.) seeds during germination and early‐seedling growth. I. Storage protein reserves. International Journal of Plant Sciences, 158(6), 727–737. 10.1086/297484 [DOI] [Google Scholar]
- Tempe, J. , & Casse‐Delbart, F. (2012). Plant gene vectors and genetic transformation: Agrobacterium Ri plasmids. CellCulture and Somatic Cell Genetics of Plants, 6, 25–49. [Google Scholar]
- Thomma, B. P. , Nürnberger, T. , & Joosten, M. H. (2011). Of PAMPs and effectors: The blurred PTI‐ETI dichotomy. The Plant Cell, 23(1), 4–15. 10.1105/tpc.110.082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toke, O. (2005). Antimicrobial peptides: New candidates in the fight against bacterial infections. Peptide Science: Original Research on Biomolecules, 80(6), 717–735. 10.1002/bip.20286 [DOI] [PubMed] [Google Scholar]
- Tossi, A. , Sandri, L. , & Giangaspero, A. (2000). Amphipathic, α‐helical antimicrobial peptides. Peptide Science, 55(1), 4–30. 10.1002/1097-0282 [DOI] [PubMed] [Google Scholar]
- van den Burg, H. A. , Harrison, S. J. , Joosten, M. H. , Vervoort, J. , & de Wit, P. J. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Molecular Plant‐Microbe Interactions, 19(12), 1420–1430. 10.1094/MPMI-19-1420 [DOI] [PubMed] [Google Scholar]
- Vidaver, A. K. (2002). Uses of antimicrobials in plant agriculture. Clinical Infectious Diseases, 34(s3), S107–S110. 10.1086/340247 [DOI] [PubMed] [Google Scholar]
- Yan, R. , Hou, J. , Ding, D. , Guan, W. , Wang, C. , Wu, Z. , & Li, M. (2008). In vitro antifungal activity and mechanism of action of chitinase against four plant pathogenic fungi. Journal of Basic Microbiology, 48(4), 293–301. 10.1002/jobm [DOI] [PubMed] [Google Scholar]
- Yaron, S. , Rydlo, T. , Shachar, D. , & Mor, A. (2003). Activity of dermaseptin K4–S4 against foodborne pathogens. Peptides, 24(11), 1815–1821. 10.1016/j.peptides [DOI] [PubMed] [Google Scholar]
- Yevtushenko, D. P. , & Misra, S. (2012). Transgenic expression of antimicrobial peptides in plants: Strategies for enhanced disease resistance, improved productivity, and production of therapeutics In Rajasekaran K., Cary J. W., Jaynes J. M. & Montesinos E. (Eds.), Small wonders: Peptides for disease control (pp. 445–458). Washington, DC: ACS Publications; 10.1021/bk-2012-1095.ch021 [DOI] [Google Scholar]
- Yevtushenko, D. P. , Romero, R. , Forward, B. S. , Hancock, R. E. , Kay, W. W. , & Misra, S. (2005). Pathogen‐induced expression of a cecropin A‐melittin antimicrobial peptide gene confers antifungal resistance in transgenic tobacco. Journal of Experimental Botany, 56(416), 1685–1695. 10.1093/jxb/eri165 [DOI] [PubMed] [Google Scholar]
- Yin, L. M. , Edwards, M. A. , Li, J. , Yip, C. M. , & Deber, C. M. (2012). Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide‐membrane interactions. Journal of Biological Chemistry, 287(10), 7738–7745. 10.1074/jbc.M111.303602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, S. , Iida, Y. , Kawasaki, Y. , Minami, Y. , Watanabe, K. , & Yagi, F. (2009). The chitin‐binding capability of Cy‐AMP1 from cycad is essential to antifungal activity. Journal of Peptide Science, 15(7), 492–497. 10.1002/psc.1147 [DOI] [PubMed] [Google Scholar]
- Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. Nature, 415(6870), 389–395. 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- Zasloff, M. (2006). Defending the epithelium. NatureMedicine, 12(6), 607–608. 10.1038/nm0606-607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this study are presented in the manuscript.


