Abstract
Aeromonas is recognized as a human pathogen following ingestion of contaminated food and water. One major problem in Aeromonas identification is that certain species are phenotypically very similar. The antimicrobial resistance is another significant challenge worldwide. We therefore aimed to use mass spectrometry technology for identification and discrimination of Aeromonas species and to screen the antimicrobial resistance of Aeromonas hydrophila (A. hydrophila). A total of 150 chicken meat and water samples were cultured, and then, the isolates were identified biochemically by the Vitek® 2 Compact system. Proteomic identification was performed by MALDI‐TOF MS and confirmed by a microchannel fluidics electrophoresis assay. Principal component analysis (PCA) and single‐peak analysis created by MALDI were also used to discriminate the Aeromonas species. The antimicrobial resistance of the A. hydrophila isolates was determined by Vitek® 2 AST cards. In total, 43 samples were positive for Aeromonas and comprised 22 A. hydrophila, 12 Aeromonas caviae (A. caviae), and 9 Aeromonas sobria (A. sobria) isolates. Thirty‐nine out of 43 (90.69%) Aeromonas isolates were identified by the Vitek® 2 Compact system, whereas 100% of the Aeromonas isolates were correctly identified by MALDI‐TOF MS with a score value ≥2.00. PCA successfully separated A. hydrophila, A. caviae and A. sobria isolates into two groups. Single‐peak analysis revealed four discriminating peaks that separated A. hydrophila from A. caviae and A. sobria isolates. The resistance of A. hydrophila to antibiotics was 95.46% for ampicillin, 50% for cefotaxime, 45.45% for norfloxacin and pefloxacin, 36.36% for ceftazidime and ciprofloxacin, 31.81% for ofloxacin and 27.27% for nalidixic acid and tobramycin. In conclusion, chicken meat and water were tainted with Aeromonas spp., with a high occurrence of A. hydrophila. MALDI‐TOF MS is a powerful technique for characterizing aeromonads at the genus and species levels. Future studies should investigate the resistance of A. hydrophila to various antimicrobial agents.
Keywords: Aeromonas spp., antimicrobial resistance, differentiation, microchannel electrophoresis, protein fingerprinting
The sequencing results indicated that A. hydrophila is the most prevalent Aeromonas spp. isolated from food and water. MALDI‐TOF MS is a powerful technique used for identification of Aeromonas at the genus and species‐level. Principal component analysis (PCA) and single‐peak analysis are successful tools to discriminate the Aeromonas spp. VITEK® 2 AST Cards can also use as a detective method of antimicrobial resistance.

1. INTRODUCTION
Bacteria of the genus Aeromonas belong to the family Aeromonadaceae and include nineteen species (Aboyadak, Ali, Goda, Saad, & Salam, 2017; Demarta et al., 2008; Trakhna, Harf‐Monteil, Abdelnour, Maaroufi, & Gadonna‐Widehem, 2009) of gram‐negative, motile, nonlactose fermenting, nonspore forming, facultative anaerobic, and oxidase‐positive organisms. These bacteria can be classified into two large groups according to the host and physiological characteristics (Stratev & Odeyemi, 2016). The first group comprises motile aeromonads, represented by Aeromonas hydrophila (A. hydrophila), which causes various diseases mostly in mammals, including humans. The other group consists of nonmotile species, represented by Aeromonas salmonicida, which causes infections in fish (Bartkova, Kokotovic, Skall, Lorenzen, & Dalsgaard, 2017; Igbinosa, Igumbor, Aghdasi, Tom, & Okoh, 2012). Motile Aeromonas spp. are pathogens that cause foodborne gastroenteritis in humans and extraintestinal infections, such as bacteremia, soft tissue infections, meningitis, endocarditis and osteomyelitis (Alhazmi, 2015), with a high mortality rate in immunocompromised hosts (Gauthier, Vincent, Charette, & Derome, 2017; Igbinosa et al., 2012; Koca & Sarimehmetoglu, 2009; Steinberg & Burd, 2010).
Many people consume chickens daily as a source of animal protein worldwide; hence, hygienic methods of supplying chickens for consumption are critical for public health. Meat can be infected with Aeromonas spp. not only through inadequate processing, cutting and grinding but also by washing carcasses with contaminated water (Ghenghesh, Ahmed, El‐Khalek, Al‐Gendy, & Klena, 2008; Stratev & Odeyemi, 2016). The poor hygienic conditions associated with the processing of raw meat are considered one of the major causes of Aeromonas spp. contamination of meat products (Encinas, Gonzalez, Garcia‐Lopez, & Otero, 1999; Ogu, Madar, Okolo, & Tayubi, 2017; Rajakumar, Ayyasamm, Shanthi, Song, & Lak‐shmanaperumalsamy, 2012). Therefore, the genus Aeromonas has been associated with a wide variety of food and waterborne infections worldwide, particularly in less developed countries due to poor personal hygiene and lack of quality water (Odeyemi & Ahmad, 2017). Most Aeromonas spp. are virulent due to their ability to multiply and produce several toxins in refrigerated conditions (Eley, Geary, & Wilcox, 1993; Kirov, 1993; Humphries & Linscott, 2015; Miyagi, Hirai., & Sano, K., 2016). Because Aeromonas spp. represent commonly isolated pathogens from food as a result of their survival in water and human and animal feces, the threats of foodborne infections with Aeromonas are augmented (Ahmed, Abd El Aal, Ayoub, & Sayed, 2014; Koca & Sarimehmetoglu, 2009).
The most important Aeromonas spp. are A. hydrophila, Aeromonas caviae (A. caviae), and Aeromonas veronii biovar sobria (A. veronii bv. sobria). These organisms are pervasive in water and meat (Encinas et al., 1999; Osman, Aly, Kheader, & Mabrok, 2012; Sharma & Kumar, 2011; Trakhna et al., 2009). A. hydrophila represents the most virulent of these species and produces multifactorial virulence factors, including structural features related to adhesion, cell attack, and escape from the phagocytosis process, and certain extracellular factors, such as aerolysin, which leads to lysis and toxicity of the cells (Abrami, Fivaz, Glauser, Parton, & Goot, 1998; Chopra & Houston, 1999; Citterio & Biavasco, 2015). Nevertheless, some species of Aeromonas were isolated formerly from several food products, and the substantial role of foods of animal origin in the distribution of Aeromonas infections is unclear.
Although biochemical methods, 16S rRNA sequencing and housekeeping genes are considered the standard methods for detecting different Aeromonas spp., they are not widely used due to their cost, labor and time requirements (Chen et al., 2014; Morinaga et al., 2013; Soler et al., 2004; Trakhna et al., 2009). In addition, the exactness of these presently available methods is limited, and the precise and rapid identification of Aeromonas at the species level is still problematic (Benagli et al., 2012; Pérez‐Sancho et al., 2018). From this perspective, to increase the rate of Aeromonas identification at the species level, recent studies have confirmed and recommended that matrix‐assisted laser desorption ionization–time of flight mass spectrometry (MALDI‐TOF MS) as an alternative technique for bacterial identification due to its favorable rapid application (Donohue, Smallwood, Pfaller, Rodgers, & Shoemaker, 2006; Elbehiry, Al‐Dubaib, Marzouk, Osman, & Edrees, 2016; Murray, 2010). This technology is an up‐to‐date approach extensively applied for the identification and discrimination of various microorganisms at the genus and species levels on the basis of MALDI‐TOF mass spectra (Elbehiry et al., 2017; Sandrin, Goldstein, & Schumaker, 2013; Vávrová, Balážová, Sedláček, Tvrzová, & Šedo, 2015).
The development of antimicrobial resistance in various types of bacteria is another significant challenge worldwide (Chugh, 2008; Laith & Najiah, 2013; Li & Webster, 2018). Recently, the antibiotic resistance of Aeromonas spp. has increased because resistance was developed not only in clinical isolates but also in strains isolated from different sources of food products (Alcaide, Blasco, & Esteve, 2010). Throughout the last decade, the distribution of antimicrobial resistance among foodborne pathogens has developed, possibly due to the prolonged administration of medications in the livestock used for human consumption (Adebayo, Majolagbe, Ola, & Ogundiran, 2012; Deng et al., 2016). A previous study conducted by Saavedra et al. (2004) illustrated that the widespread use of various groups of beta‐lactam antibiotics as a method of prophylaxis and treatment of A. hydrophila in humans and animals is considered one of the main causes of the increasing A. hydrophila resistance to amoxicillin, carbenicillin, and ticarcillin. Furthermore, the existence of resistance genes on mobile elements, such as plasmids, transposons and integrons, assists their rapid spread among microorganisms (Romero, Feijoo, & Navarrete, 2012). Similarly, the data on the incidence of antibiotic resistance in Aeromonas spp., particularly in A. hydrophila recovered from chicken meat and water, are sparse. Based on these previously mentioned data, our study was designed to identify various Aeromonas spp. from chicken meat and water samples using MALDI‐TOF MS confirmed by SYBR Green real‐time (RT)‐PCR and microchannel fluidics electrophoresis assays and to study the antimicrobial resistance of A. hydrophila using Vitek 2 Compact AST cards.
2. MATERIALS AND METHODS
2.1. Sample collection
A total of 150 samples, including chicken meat (n = 75) and water (n = 75), were collected from three different sites (Buraidah, Unaizah, and Albukairyah) in the Al‐Qassim region, Saudi Arabia. The samples were collected five times at nearly monthly intervals (in April, May, June, August, and September 2017). Three hundred grams of each chicken meat sample collected from six randomly selected local retail shops, and supermarkets were placed in a separate sterilized plastic bag for the isolation process. One hundred milliliters of each water sample was collected randomly from private drinking water wells, houses, and retailers. The samples collected from houses and retailers were treated first with a sterile sodium thiosulphate solution (13.2 mg/L) to neutralize chlorine and stop its bactericidal action (Massa, Armuzzi, Tosques, Canganella, & Trovatelli, 1999). All samples were kept under ice‐cold conditions, and bacteriological investigations were carried out within 2 hr of collection. All meat and water samples were processed in the Microbiology Laboratory, College of Public Health and Health Informatics, Qassim University for isolation. Isolates were preserved in Cryobank vials at −80°C until the identification process was carried out.
2.2. Isolation of Aeromonas spp.
Thirty grams of each meat specimen was added to 225 ml of alkaline peptone water (pH 8.4 ± 0.2 at 25°C, Sigma‐Aldrich, USA), homogenized in a blender (Stomacher® 400, Thomas Scientific, USA) for 2 minutes and incubated at 30ºC for 18–24 hr. Likewise, 10 ml of each water sample was inoculated in 90 ml of peptone water with 1% NaCl (w/v) at pH 8.6 adjusted with sodium hydroxide and incubated at 30ºC for 18–24 hr. The cultures were streaked onto Aeromonas Isolation Agar (Sigma‐Aldrich) containing 5 mg/L ampicillin, which supports the growth of Aeromonas spp. After incubation of all plates at 28ºC for 24–48 hr, the colonies appeared slightly to deep green. Three to five typical colonies were subcultured onto glutamate starch phenol red agar (Sigma‐Aldrich), and after incubation, the colonies appeared as yellow colonies surrounded by a yellow zone and were identified primarily as Aeromonas spp. if they were gram‐negative, oxidase‐positive and glucose fermenting. The Voges–Proskauer reaction; esculin hydrolysis; lysine decarboxylase; and fermentation of arabinose, salicin, and sorbitol were then carried out to differentiate Aeromonas at the species level (A. hydrophila, A. caviae, and A. veronii bv. sobria) according to the method described by Janda, Abbott, and Carnahan (1995).
2.3. Biochemical analysis of Aeromonas using the Vitek 2 Compact system
The Vitek 2 Compact ((bioMérieux. Marcy l'Etoile, France) Gram‐Negative Identification (GNI) and antibiotic susceptibility testing (AST) cards were used to identify and determine the antibiotic susceptibilities of Aeromonas spp. according to the manufacturer's recommendations. In brief, 3–4 fresh colonies were suspended in sterilized physiological saline (aqueous 0.45% NaCl, pH 4.5 to 7.0) and thoroughly mixed. The Mcfarland turbidity was adjusted in the range from 0.50 to 0.63 using DensiChekTM (BioMe′rieux, France). Five milliliters of this suspension was loaded into Vitek 2 ID‐GNI and AST gram‐negative (AST‐GN04) cards. The Vitek 2 Cassette was finally loaded with cards and suspension tubes into the device. The unknown organisms were compared to the reference strains stored in the Vitek 2 Compact software for proper identification.
2.4. Rapid identification of Aeromonas spp. using MALDI Biotyper
We applied MALDI Biotyper Reference Library for Clinical Applications (MBT‐CA) Database version V.3.3.1.2 (Bruker Daltonik, Bremen, Germany) which has been approved by FDA under Section 510(k) as a powerful method for rapid and precise identification and discrimination of Aeromonas spp. The Proteomic identification was performed according to the ethanol/formic acid extraction method designated by Bruker Corporation. Briefly, a fresh colony of overnight culture, incubated at 28°C for 24 hr, was utilized for each isolate and inoculated onto two spots of the target plate, and every colony was then covered with 1 µl of matrix solution (saturated α‐cyano‐4‐hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid). The microbial spectra were directly produced by applying Compass IVD software, and the identification was directly conducted with a MALDI Biotyper machine.
2.5. Data analysis in MALDI Biotyper
The score value of the unidentified spectrum in the range from zero to three was determined by matching the unknown spectra with the spectra stored in the Bruker library. The accuracy of the strain recognition was detected as designated by the measures of Bruker Daltonik. The device performed the precise detection of species when the log score ranged from 2.3 to 3.0; nevertheless, the species and genus levels were recognized in the range from 2.00 to 2.29 and from 1,700 to 1,999, respectively. Furthermore, a score of 0.00 to 1.69 means that the proof of identity is not reliable. The diverse spectra created by the Microflex LT Compass IVD software were measured in a m/z range from 2,000 to 20,000 Da. To distinguish between Aeromonas spp., mathematical testing of the data sets was generated on the basis of principal component analysis (PCA), and the findings were illustrated in a three‐dimensional (3d) score plot created directly by compass software. According to the MBT‐CA Database, which contains 47 reference Aeromonas spp. and subspecies, the PCA dendrogram setting was utilized for species grouping.
2.6. Molecular identification of Aeromonas spp. using SYBR Green RT‐PCR
2.6.1. DNA extraction
DNA extraction of the field isolates was achieved by QuickGene‐810 (AutoGen, Japan) using the QuickGene DNA tissue kit S (DT‐S), which was applied according to the manufacturer's recommendations. Briefly, 3–5 fresh colonies of each sample grown on soybean casein digest agar were transferred into a sterilized microcentrifuge tube containing 180 µl MDT lysis buffer and 20 µl proteinase K, and the lysate was then centrifuged at 8,000 g for 5 min. The supernatant was transferred to a new Eppendorf tube, and 180 µl LDT buffer was added. Two hundred forty microliters of absolute ethanol (Panreac, Barcelona, Spain) was added, and the tube was properly agitated. The lysate was transferred into the cartridge supplied with the kits and then inserted into the machine. Finally, the concentration and purity of the extracted DNA were determined by the NanoDrop™ 2000 spectrophotometer (Thermo Scientific, MA, USA).
2.6.2. Primers used in the study
The isolates were further confirmed to the species level by 16S rRNA, aerolysin (aerA), polar flagella (Fla), and hemolysin (ASA1) genes analysis. A specific 16S rRNA region was carefully chosen for detecting Aeromonas spp. The primer express software, ver. 2.0 (Applied Biosystems, USA) was used to designate the primers, and their specificity was investigated with the BLAST program (Table 1).
Table 1.
Oligonucleotide primers used to detect A. hydrophila, A. caviae, and A. sobria genes
| Target gene | Oligonucleotide sequence (5’−3’) | GenBank accession number | Size (bp) |
|---|---|---|---|
| 16S rRNA‐F | GGCCTTGCGCGATTGTATAT | DQ455052 | 103 |
| 16S rRNA‐R | GTGGCGGATCATCTTCTCAGA | ||
| AerA‐F | CAAGGCTGATATCTCCTATCCCTATG | AF485770 | 67 |
| AerA‐R | GCCACTCAGGGTCAGGTCAT | AY352352 | |
| Fla‐F | TCCAACCGTYTGACCTC | AF198617 | 608 |
| Fla‐R | GMYTGGTTGCGRATGGT | AF002709 | |
| ASA1‐F | TAA AGG GAA ATA ATG ACG GCG | X65046 | 249 |
| ASA1‐R | GGC TGT AGG TAT CGG TTT TCG |
2.6.3. SYBR Green RT‐PCR assay and electrophoresis for PCR products
SYBR Green RT‐PCR for detection of the A. hydrophila, A. caviae, and A. sobria specific genes was then performed using a 7500 Fast Real‐Time PCR System (Applied Biosystems). Briefly, a 20 µl reaction volume containing 10 µl of Maxima SYBR Green qPCR Master Mix (2×), no ROX (Thermo Scientifics), 1 µl forward primer, 1 µl reverse primer, 1 µl target DNA and 7 µl of RNase/DNase free water was used. All reactions were carried out in duplicate. Regular amplification parameters were carried out as follows: 50°C for 2 min, 95°C for 2 min, followed by 40 amplification cycles, each of which comprised 95°C for 15 s and 60°C for 1 min. Amplification results were expressed by plotting Delta Rn (ΔRn) versus cycle number for detection of the Aeromonas genes. Electrophoresis for PCR products was then carried out using a LabChip GX Touch 24 device (PerkinElmer, USA). DNA 1 K Assay Quick was used for chip and preparation of samples according to the manufacturer's procedures.
2.7. Antimicrobial resistance of A. hydrophila using Vitek 2 Compact AST cards
Vitek 2 Compact AST‐GN04 cards (MedexSupply, Passaic, NJ, USA) were used to determine the susceptibility of A. hydrophila to antimicrobial agents. As shown in Table 5, five groups of antibiotics were tested in the present study. All antimicrobial agents were chosen according to these five groups, which can be measured by Vitek 2 system cards (Cockerill et al., 1995). Throughout the evaluation period, A. hydrophila ATCC 35654 was used as a quality control strain and was checked at regular intervals.
Table 5.
Susceptibility percentage for 22 A. hydrophila strains recovered from chicken meat and water samples
| Antimicrobial agent | Conc. (µg) | Vitek 2 Compact system | |||||
|---|---|---|---|---|---|---|---|
| S | I | R | |||||
| No | % | No | % | No | % | ||
| Beta‐lactam penicillins | |||||||
| Ampicillin | 10 | 1 | 4.54 | 0 | 0 | 21 | 95.46 |
| Piperacillin | 100 | 22 | 100 | 0 | 0 | 0 | 0 |
| Ticarcillin | 75 | 22 | 100 | 0 | 0 | 0 | 0 |
| Beta‐lactam/Beta‐lactam inhibitors | |||||||
| Amoxicillin/clavulanic acid | 20/10 | 22 | 100 | 0 | 0 | 0 | 0 |
| Piperacillin/tazobactam | 100/20 | 22 | 100 | 0 | 0 | 0 | 0 |
| Ticarcillin/clavulanic acid | 75/10 | 22 | 100 | 0 | 0 | 0 | 0 |
| Cephalosporins | |||||||
| Cefotaxime | 30 | 11 | 50.00 | 0 | 0 | 11 | 50.00 |
| Ceftazidime | 30 | 14 | 63.63 | 0 | 0 | 8 | 36.36 |
| Cefpdoxime | 10 | 17 | 77.27 | 1 | 4.54 | 4 | 18.18 |
| Cefepime | 30 | 22 | 100 | 0 | 0 | 0 | 0 |
| Cefpirome | 30 | 22 | 100 | 0 | 0 | 0 | 0 |
| Quinolones | |||||||
| Ofloxacin | 5 | 11 | 50.00 | 4 | 18.18 | 7 | 31.81 |
| Norfloxacin | 10 | 12 | 54.54 | 0 | 0 | 10 | 45.45 |
| Pefloxacin | 30 | 12 | 54.54 | 0 | 0 | 10 | 45.45 |
| Nalidixic acid | 30 | 12 | 54.54 | 1 | 4.54 | 9 | 27.27 |
| Ciprofloxacin | 10 | 13 | 59.10 | 3 | 13.63 | 6 | 36.36 |
| Aminoglycosides | |||||||
| Amikacin | 30 | 21 | 95.45 | 0 | 0 | 0 | 4.55 |
| Gentamicin | 10 | 21 | 95.45 | 0 | 0 | 0 | 4.55 |
| Netilmicin | 30 | 22 | 100 | 0 | 0 | 0 | 0 |
| Isepamicin | 30 | 22 | 100 | 0 | 0 | 0 | 0 |
| Tobramycin | 10 | 14 | 63.63 | 2 | 9.10 | 6 | 27.27 |
2.8. Statistical analysis
The data obtained from our study were imported into the Statistical Package for the Social Sciences (SPSS), and all estimations were carried out using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA).
3. RESULTS
3.1. Incidence of Aeromonas spp. in chicken meat and water
The incidence of Aeromonas was examined in 75 chicken meat and another 75 water samples. According to our findings, of 150 chicken meat and water samples, 43 (28.66%) were positive for Aeromonas spp. Out of 75 chicken meat and 75 water samples, 31 (43.33%) and 12 (16%) were positive for Aeromonas spp., respectively. Among the positive samples, 22 (51.16%) A. hydrophila, 12 (27.9%) A. caviae, and 9 (20.93%) A. sobria strains were isolated from both chicken meat and water samples (Table 2). Of the 31 chicken meat samples recognized as tainted with Aeromonas spp., 17 (54.83%) A. hydrophila, 8 (25.8%) A. caviae, and 6 (19.35%) A. sobria strains were isolated. Of the 12 positive water samples for Aeromonas spp., 5 (41.66%), 4 (33.33%), and 3 (25%) were positive for A. hydrophila, A. caviae, and A. sobria, respectively (Table 2).
Table 2.
Frequency of Aeromonas species in positive chicken meat and water samples
| Sample origin | Positive samples | Aeromonas species | ||||||
|---|---|---|---|---|---|---|---|---|
| A. hydrophila | A. caviae | A. sobria | ||||||
| No. | % | No. | % | No. | % | No. | % | |
| Chicken meat | 31 | 72.1 | 17 | 54.83 | 8 | 25.80 | 6 | 19.35 |
| Water | 12 | 27.9 | 5 | 41.66 | 4 | 33.33 | 3 | 25 |
| Total | 43 | 100 | 22 | 51.16 | 12 | 27.90 | 9 | 20.93 |
3.2. Biochemical identification of Aeromonas isolates
The Vitek™ 2 Compact system properly identified 39 of 43 (90.69%) Aeromonas spp., as 21/22 (95.45%) strains of A. hydrophila, 10/12 (83.33%) strains of A. caviae and 8/9 (88.88%) strains of A. sobria (Table 3).
Table 3.
Identification of Aeromonas spp. recovered from chicken meat and water using Vitek™ II Compact ID‐GNI cards
| Aeromonas spp. | No. of tested isolates | Correctly identified | Misidentified | Not identified | |
|---|---|---|---|---|---|
| No. | % | ||||
| A. hydrophila | 22 | 21 | 95.45 | 1 | 0 |
| A. caviae | 12 | 10 | 83.33 | 0 | 2 |
| A. sobria | 9 | 8 | 88.88 | 1 | 0 |
| Total | 43 | 39 | 90.69 | 2 | 2 |
3.3. Accurate identification of Aeromonas spp. using MALDI‐TOF MS
In the current study, 43 Aeromonas isolates were investigated by the Microflex LT device, and the generated spectra were compared with the stored spectra in the Bruker library of Compass software. The precise identification rates for species listed in the Bruker Daltonics Compass 2.0 database by the MALDI Biotyper system were 21/22 (95.45%) for A. hydrophila, 12/12 (100%) for A. caviae, and 9/9 (100%) for A. sobria. In Table 4, we report that 9/22 (40.90%) A. hydrophila, 4/12 (33.33%) A. caviae, and 4/9 (44.44%) A. sobria were correctly identified at the species level, with a score value ranging from 2,300 to 3,000. Moreover, 12/22 (54.54.76%) A. hydrophila, 8/12 (66.66%) A. caviae, and 4/9 (44.44%) A. sobria were also identified at the species level, with a score value ranging from 2,000 to 2,299. However, one isolate of A. hydrophila was identified at the genus level with a score value ranging from 1.7 to 1.99. In contrast, zero isolates were not identified. A current gel view demonstrated the created spectra for all Aeromonas spp. Several spectra were distributed within the range from 2,000 to 11,000 m/z (Figure 1), and the higher peaks were determined between 4,000 and 10,000 m/z (Figure 2).
Table 4.
Score values for 43 Aeromonas species of broiler chicken identified by MALDI Biotyper
| Category | Score range | Identification level | Aeromonas species | |||||
|---|---|---|---|---|---|---|---|---|
| A. hydrophila | A. caviae | A. sobria | ||||||
| No. | % | No. | % | No. | % | |||
| 1 | 2.3–3 | Species | 9/22 | 40.90 | 4/12 | 33.33 | 4/9 | 44.44 |
| 2 | 2–2.29 | Species | 12/22 | 54.54 | 8/12 | 66.66 | 4/9 | 44.44 |
| 3 | 1.7–1.9 | Genus | 1/22 | 4.54 | 0/12 | 0 | 1/9 | 11.11 |
| 4 | 0–1.6 | Not identified | 0/22 | 0 | 0/12 | 0 | 0/9 | 0 |
Figure 1.
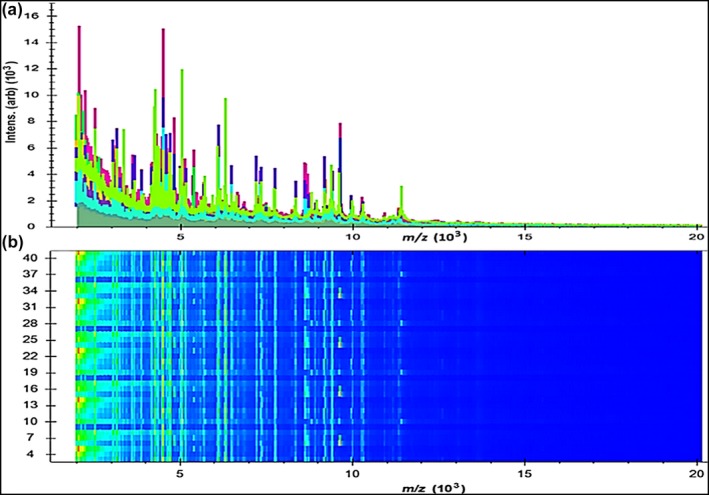
Mass spectrum protein profiles of 43 Aeromonas spp.; (a) Distribution of peaks within the line spectra ranging from 2,000 to 11,000 Da; (b) The gel profile of protein spectra in which the varied color of spots was the gathering of spectra with several contents
Figure 2.
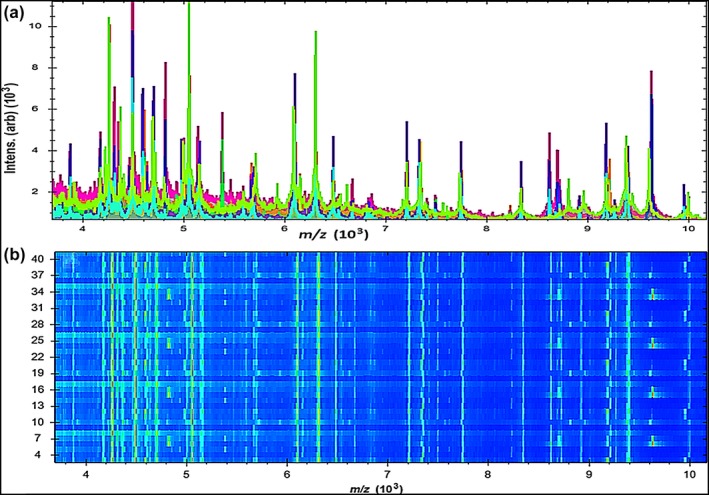
Mass spectral profiles of 43 Aeromonas spp.; (a) higher strength peaks were scattered within the line spectra ranging from 4,000 to 10,000 Da; (b) The gel profile of protein spectra distributed within the same range
Furthermore, a supplementary mathematical tool called PCA was generated in our study by MALDI Biotyper Compass software to explore the degree of similarity and variation in the protein spectra. Numerous protein spectra of the identified isolates were clarified in three‐dimensional (3d) PCA as shown in Figure 3a. Each peak was identified with 3 loading values originating from the calculation of three principal components (PC1, PC2, and PC3). In our analysis, the entire peaks listed in the MALDI Biotyper Compass 2.0 database were analyzed by the PCA tool, which separated A. hydrophila, A. sobria, and A. caviae isolates into two distinctive groups, as shown in the 3d PCA. Nevertheless, two strains of A. hydrophila were found in the A. sobria cluster. The A. caviae strains did not create a distinct group but were localized in the A. hydrophila cluster (Figure 3a). Based on the PCA calculation, the impacts of PC1, PC2, and PC3 on the creation of a profile in a percentage plot of the difference elucidated were nearly 45%, 17%, and 9%, respectively (Figure 3b).
Figure 3.
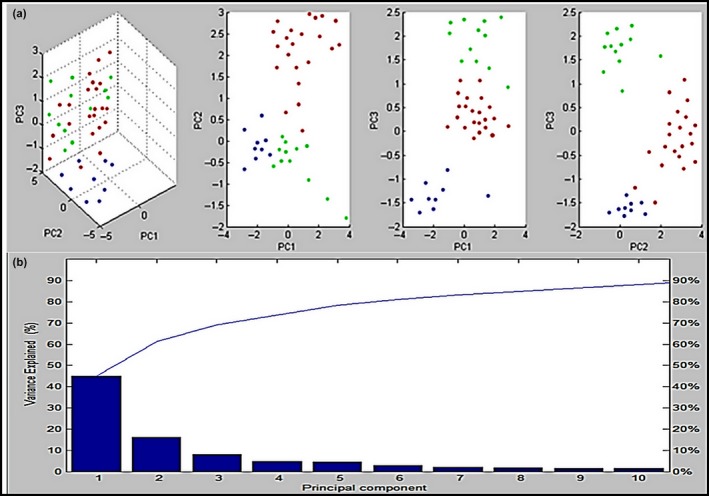
The dimensional image from PCA displays the difference between 43 Aeromonas spp.; (a) the grouping of A. hydrophila (red), A. caviae (green), and A. sobria (blue) in the first three model of PC (PC1, PC2, PC3); (b) the influence of ten principal components to the profiling classification in plot of percentage explained variance of PC. The contributions of PC1, PC2, and PC3 were around 45%, 17%, and 9%, correspondingly
Likewise, we analyzed a single peak for all Aeromonas spp. to explore the distinctive differences in the three Aeromonas spp. Higher peak intensities were detected in A. hydrophila at 3,194 Da, 4,031 Da, 5,383, and 7,611 m/z, whereas they were missed in A. caviae and A. sobria (Figure 4). Otherwise, the averaged spectra of A. sobria isolates exhibited definite peaks at 3,367, 4,351, 7,335, and 9,635 m/z, whereas they were missed in A. hydrophila and A. caviae. Moreover, the higher peak intensity at 7,347 m/z was identified in A. caviae and absent in both A. sobria and A. hydrophila (Figure 5). Analysis of A. hydrophila spectra demonstrated that the 3,194, 4,031, 5,383, and 7,611 m/z peaks were frequently found in 59% (13/22), 54.5% (12/22), 90.9% (20/22), and 95.45% (21/22) of the A. hydrophila strains, respectively. Moreover, the 3,367, 4,351, 7,335, and 9,635 m/z peaks commonly existed in ~67% (6/9), ~89% (8/9), ~78% (7/9), and 100% (9/9), of the A. sobria spectra, respectively.
Figure 4.
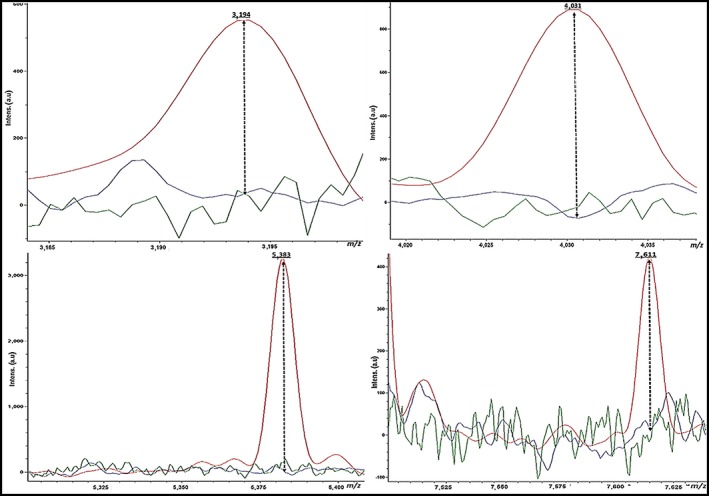
Higher peaks intensity (3,194, 4,030, 5,383, and 7,611 Da) were detected in A. hydrophila (red), whereas they were missed in A. sobria (green) and A. caviae (blue)
Figure 5.
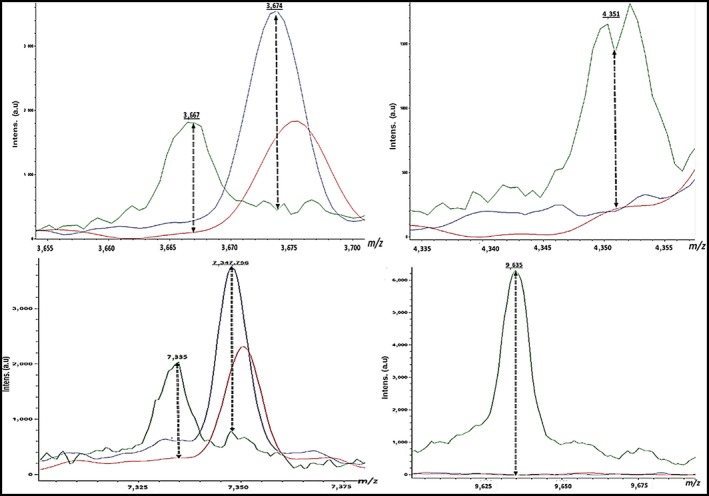
Higher peaks intensity (3,667, 4,351, 7,335 and 9,635 Da) were detected in A. sobria (green), whereas they were missed in A. hydrophila (red) and A. caviae (blue). Moreover, a higher peak intensity (7,347 Da) was detected in A. caviae (blue) while it was missed in A. hydrophila (red) and A. sobria (green)
3.4. Confirmation of the identification of Aeromonas isolates using SYBR Green RT‐PCR
The SYBR Green RT‐PCR technique was then carried out to confirm the MALDI Biotyper results. The primers precisely targeting regions of the A. hydrophila, A. caviae, and A. sobria, 16S rRNA, aerA, fla, and ASA1 genes were designed to identify pathogenic A. hydrophila, A. caviae and A. sobria strains. PCR amplification with these primers yielded amplicons of the expected molecular weights. Amplification of each gene was tested separately, and the size of each expected product was confirmed. The sizes obtained after the LabChip analysis were 107, 67, 608 and 249 bp for the 16S rRNA, aerA, fla, and ASA1 PCR products, respectively. Comparing the results of identification accomplished by MALDI‐TOF Mass Spectrometry and SYBR Green RT‐PCR illustrated an agreement of 100%; therefore, the PCR was succeeded to confirm the results of MALDI.
3.5. Antimicrobial resistance of A. hydrophila
Vitek 2 Compact AST‐GN04 cards were used to determine the susceptibility of A. hydrophila to antimicrobial agents (Table 5). Our findings indicated that 51.16%, 27.90%, and 20.93% of bacterial isolates recovered from chicken meat and water samples were A. hydrophila, A. caviae, and A. sobria, respectively. As a result of these findings, we focused on A. hydrophila resistance to various antimicrobial agents using Vitek 2 Compact cards. As shown in Table 5, of 22 A. hydrophila isolates, 21 (95.46%) were resistant to ampicillin (beta‐lactam penicillins), but all strains were sensitive to piperacillin, ticarcillin and beta‐lactam/beta‐lactam inhibitors (amoxicillin/clavulanic acid, piperacillin/tazobactam, ticarcillin/clavulanic acid). A total of 50%, 36.36%, and 18.18% of A. hydrophila isolates were resistant to third‐generation cephalosporins (cefotaxime, ceftazidime, and cefpodoxime), respectively, whereas fourth‐generation cephalosporins (cefepime and cefpirome) showed strong activity against all tested isolates. A total of 31.81%, 45.45%, 45.45%, 27.27%, and 36.36% of A. hydrophila isolates were resistant to the tested quinolones (ofloxacin, norfloxacin, pefloxacin, nalidixic acid, and ciprofloxacin), respectively. In contrast, the susceptibility of A. hydrophila to aminoglycosides was 100% for netilmicin and isepamicin, 95.45% for amikacin and gentamicin and 63.63% for tobramycin.
4. DISCUSSION
The accurate identification of various pathogens is an essential step of diagnosis, and the time‐to‐result obtained is very significant to start the selected treatment as soon as possible. Rapid and accurate analytical tools are necessary for monitoring the food and water safety and screening of any undesirable pathogens, which may cause noteworthy health hazards upon consumption. Aeromonas spp. are known to cause various infections in humans. Because its developing significance as an emerging pathogen isolated from food and water, it is imperative to combat this bacterium (Praveen, Debnath, Shekhar, Dalai, & Ganguly, 2016).
As culture‐ and biochemical‐based identification of different microorganisms are difficult and time‐consuming, MALDI‐TOF MS was significantly used here for the early identification and discrimination of various pathogens from environmental samples by introducing a simple, rapid, precise, and low‐cost identification method compared to other methods (Elbehiry et al., 2016; Singhal, Kumar, Kanaujia, & Virdi, 2015; van Belkum, Welker, Pincus, Charrier, & Girard, 2017). Recently, MALDI‐TOF MS has been revealed to be an important method for the rapid identification of bacterial threats that might contaminate drinking water and food products (Singhal et al., 2015).
In our study, the identification rates for Aeromonas isolates were 21/22 (95.45%) for A. hydrophila, 12/12 (100%) for A. caviae and 9/9 (100%) for A. sobria. These findings prove that the mass spectral data generated by MALDI Biotyper Compass 2.0 Software for all isolates were satisfactory to differentiate between the genus Aeromonas at the species and strain levels. The higher level of precise identification compared to that of the former studies might be due to the updated Compass 2.0 database utilized in our study (Lo et al., 2015; Seng et al., 2009). Similar results were obtained by Donohue et al. (2007), who used the m/z signature of the recognized Aeromonas reference isolates to allocate species of unidentified environmental isolates. They reported that MALDI‐TOF MS quickly and precisely categorized fourteen species and four subspecies of Aeromonas, including A. hydrophila, A. caviae, A. jandaei, and A. veronii bv. sobria, which were the most clinically significant species of the genus Aeromonas.
A previous study of Aeromonas isolates was also conducted by Donohue et al. (2006), who indicated that the signals created by MALDI‐TOF MS Compass 2.0 Software after analysis of protein spectra might be utilized as specific biomarkers for the successful identification and discrimination of Aeromonas at the species and below the species level. Another study was carried out by Chen et al. (2014) for proteomic identification of 217 Aeromonas strains using cluster analysis of spectra created by MALDI‐TOF MS. They reported that the Aeromonas strains were precisely identified as 96.7% S (A. dhakensis), 90% A. hydrophila, 96.7% A. veronii, and 100% A. caviae. Böhme et al. (2011) utilized MALDI‐TOF MS successfully in the accurate identification of 26 species of seafood spoilage and pathogenic gram‐negative bacteria, including A. hydrophila, Acinetobacter baumannii, Pseudomonas spp., and Enterobacter spp. Therefore, proteomic identification has been illustrated to be a powerful tool for species identification. Another MS study was evaluated by Lamy, Kodjo, Laurent, and CoIBVH Group (2011) for proteomic identification of aeromonads. They found that the genus‐level precision was detected at 100% compared with rpoB gene sequencing, which makes this system one of the most reliable and rapid techniques for the identification of various microorganisms.
The main benefit of MALDI‐TOF technology for routine diagnosis is the precise identification of various pathogens that, by traditional techniques, are frequently categorized to the genus or even genus‐group level (Elbehiry et al., 2017; García, Allende, Legarraga, Huilcaman, & Solari, 2012; McElvania TeKippe & Burnham, 2014; Porte et al., 2017). With MALDI‐TOF MS, these microorganisms have recently been identified without the high costs and significant time span related to multiple biochemical tests and/or 16S rRNA analysis (McElvania TeKippe & Burnham, 2014). Our assessment established that the MALDI‐TOF technology quickly and exactly identified nearly all Aeromonas isolates tested at the species level with a score value ≥2.00. However, one isolate of A. hydrophila in our study was identified at the genus level with a score value ranging from 1.7 to 1.99. Species identification by MALDI‐TOF MS is still not always reached as a result of small quantities of material, weak protein signals, and inadequate representation in the stored Compass 2.0 software (Bizzini et al.., 2011; Carrasco et al., 2016; Croxatto, Prod´hom, G., & Greub, G., 2012; Lau et al., 2014). Similar results were obtained by Benagli et al. (2012), who tested 741 clinical and environmental Aeromonas isolates using MALDI‐TOF MS and found that 93% of these strains were positively identified with a score value ≥2.00. In addition, we used MALDI‐TOF MS to discriminate between A. hydrophila, A. caviae, and A. sobria using PCA analysis and single‐peak analysis. PCA analysis successfully separated A. hydrophila, A. caviae, and A. sobria isolates into two groups. Single‐peak analysis revealed four discriminating peaks that separated A. hydrophila from A. sobria and A. caviae isolates. Han (2010) indicated that PCA is a commonly utilized calculation tool to extract, show and rank the difference within a data set. The main aim of PCA is to decrease the dimensionality of a data set, concurrently recollecting the information present in the data (Shao et al., 2012).
Likewise, the SYBR Green RT‐PCR established here was effectively used to confirm the identification of A. hydrophila, A. caviae, and A. sobria from chicken meat and water samples. For 16S rRNA, aerA, fla, and ASA1 gene detection, a good correlation between MALDI‐TOF MS and PCR analysis was found regardless of the origin of the isolates (meat or water). In addition to the LabChip preparation time (20 min), the PCR product can be shown with a microchannel fluidic apparatus in five min. After protein analysis, a molecular technique was used to identify the unconfirmed Aeromonas isolates in approximately 2.5 hr (Persson, Al‐Shuweli, Yapici, Jensen, & Olsen, 2015; Trakhna et al., 2009).
The distribution of drug resistance among A. hydrophila was also evaluated in our study, as previous surveys showed the development of this pathogen as one of the major opportunistic human pathogens (Laith & Najiah, 2013; Rey et al., 2009). The Vitek 2 Compact GN cards were used in the current study to detect the degree of resistance for 22 A. hydrophila against various antimicrobial agents commonly used for gram‐negative bacteria. Out of the 22 A. hydrophila isolates, 17 isolates were obtained from 75 chicken meat samples and 5 isolates were isolated from the 75 water samples. There was no significant difference between source of isolates in relation to their susceptibilities to various antibiotics.
Our findings revealed that 95.46% of A. hydrophila demonstrated a strong resistance to ampicillin among all the tested beta‐lactam penicillins. This finding was similar to previous studies conducted by Ramalivhana, Obi, and Moyo (2009) and Laith and Najiah (2013), who evaluated the susceptibility of different antimicrobial agents against A. hydrophila recovered from water and stool samples. They reported that 100% of isolates were resistant to ampicillin. Moreover, previous studies reported 100% Aeromonas resistance rates to ampicillin (Aoki, Egusa, Ogata, & Watanabe, 1971; Igbinosa, 2014; Rall et al., 1998). Most A. hydrophila isolates have intrinsic or chromosomally mediated resistance to ampicillin (Ghenghesh, El‐Mohammady, Levin, Zorgani, & Tawil, 2013; Rall et al., 1998).
In contrast, of the total A. hydrophila isolates tested in the current study, no resistance was detected against beta‐lactam/beta‐lactam inhibitors (amoxicillin/clavulanic acid, piperacillin/tazobactam and ticarcillin/clavulanic acid). A previous study was carried out by Awan, Maqbool, Bari, and Krovacek (2009), who evaluated the activity of ß‐lactam antibiotics against 20 A. hydrophila strains. They reported that A. hydrophila showed a high degree of resistance to ampicillin and cephaloridine, with the highest susceptibility to amoxicillin/clavulanic acid. However, some isolates were sensitive to third‐generation cephalosporins. As reported by Stratev and Odeyemi (2016), most A. hydrophila strains isolated from meat and meat products are resistant to a broad range of antimicrobial drugs.
Fourth‐generation cephalosporins, including cefepime and cefpirome, exhibited higher activity against all strains compared to third‐generation cephalosporins (cefotaxime, ceftazidime, and cefpodoxime), although some isolates were sensitive to these third‐generation cephalosporins, indicating that A. hydrophila has a variable susceptibility against cephalosporins. Similar results were obtained by Morita, Watanabe, Kurata, and Kanamori (1994) and Igbinosa (2014). Moreover, we observed that the aminoglycosides (gentamicin, amikacin, netilmicin, and isepamicin) showed excellent activity against all A. hydrophila strains. A similar report was observed by Awan et al. (2009), Dallal, Yazdi, and Avadisians (2012) and Igbinosa (2014), who found that Aeromonas spp. recovered from various food samples revealed sensitivity to gentamicin.
According to Alcaide et al. (2010), the resistance of Aeromonas spp. to various antibiotics has been augmented because emerging resistance has been established not only in clinical isolates but also in Aeromonas spp. recovered from water and food. Another study described by Adebayo et al. (2012) revealed that the frequency of bacterial resistance to different antibiotics in food and food products has increased throughout the last few years, potentially due to their extensive use in livestock raised for human feeding.
5. CONCLUSIONS
Current research illustrates a high frequency of possibly virulent A. hydrophila among Aeromonas spp. in chicken meat and water samples. Through this study, we confirmed that MALDI‐TOF MS used for the identification of Aeromonas isolates is a powerful, cost‐effective, and accurate method and was able to distinguish A. hydrophila, A. caviae, and A. sobria strains based on PCA and single‐peak analysis. RT‐PCR and microchannel fluidics electrophoresis assays can be used as a confirmatory diagnostic method for MALDI‐TOF MS. Future studies will address the application of this method for the direct identification and differentiation of Aeromonas spp. in food or water samples. Moreover, our findings demonstrate that the A. hydrophila strains in the sample had developed antibiotic resistance. Consequently, the progression of resistance may be predictable; accordingly, the number of effective antibiotics is declining. Because A. hydrophila may threaten human health, the transmission of resistance may have bad impacts for humans.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHORS CONTRIBUTION
AE, EM, and EEA designed the study. AE, EM, EEA, AA, and MH performed all the experiments, and AE, MA and EM analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
ETHICS STATEMENT
Permission has been received from the owners of shops and retailers.
DATA ACCESSIBILITY
All data generated or analyzed during this study are included in this published article.
ACKNOWLEDGEMENTS
This research was made possible with the support of Qassim University, Kingdom of Saudi Arabia.
Elbehiry A, Marzouk E, Abdeen E, et al. Proteomic characterization and discrimination of Aeromonas species recovered from meat and water samples with a spotlight on the antimicrobial resistance of Aeromonas hydrophila . MicrobiologyOpen. 2019;8:e782 10.1002/mbo3.782
REFERENCES
- Aboyadak, I. M. , Ali, N. G. , Goda, A. M. , Saad, W. , & Salam, A. M. E. (2017). Non‐selectivity of R‐S media for Aeromonas hydrophila and TCBS media for Vibrio species isolated from Diseased Oreochromis niloticus . Journal of Aquaculture Research and Development, 8, 496–500. 10.4172/2155-9546.1000496 [DOI] [Google Scholar]
- Abrami, L. , Fivaz, M. , Glauser, P. E. , Parton, R. P. , & van der Goot, F. G. (1998). A pore‐forming toxin interacts with a GPI‐anchored protein and causes vacuolation of the endoplasmic reticulum. Journal of Cell Biology, 140, 525–540. 10.1083/jcb.140.3.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adebayo, E. A. , Majolagbe, O. N. , Ola, I. O. , & Ogundiran, M. A. (2012). Antibiotic resistance pattern of isolated bacterial from salads. Journal of Research in Biology, 2, 136–142. [Google Scholar]
- Ahmed, N. I. , Abd El Aal, S. F. A. , Ayoub, M. A. , & El Sayed, M. S. (2014). Enumeration and characterization of Aeromonas spp. isolated from milk and some dairy products in Sharkia Governorate Egypt. Alexandria Journal of Veterinary Sciences, 40, 52–64. [Google Scholar]
- Alcaide, E. , Blasco, M. D. , & Esteve, C. (2010). Mechanisms of quinolone resistance in Aeromonas species isolated from humans, water and eels. Research in Microbiology, 161, 40–45. 10.1016/j.resmic.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Alhazmi, M. I. (2015). Isolation of Aeromonas spp. from food products: Emerging Aeromonas infections and their significance in public health. Journal of AOAC International, 98, 927–929. [DOI] [PubMed] [Google Scholar]
- Aoki, T. , Egusa, S. , Ogata, Y. , & Watanabe, T. (1971). Detection of resistance factors in fish pathogen Aeromonas liquefaciens . Journal of General Microbiology, 65, 3439 10.1099/00221287-65-3-343 [DOI] [PubMed] [Google Scholar]
- Awan, M. B. , Maqbool, A. , Bari, A. , & Krovacek, K. (2009). Antibiotic susceptibility profile of Aeromonas spp. isolates from food in Abu Dhabi, United Arab Emirates. New Microbiologica, 32, 17–23. [PubMed] [Google Scholar]
- Bartkova, S. , Kokotovic, B. , Skall, H. F. , Lorenzen, N. , & Dalsgaard, I. (2017). Detection and quantification of Aeromonas salmonicida in fish tissue by real‐time PCR. Journal of Fish Diseases, 40, 231–242. [DOI] [PubMed] [Google Scholar]
- Benagli, C. , Demarta, A. , Caminada, A. , Ziegler, D. , Petrini, O. , & Tonolla, M. A. (2012). Rapid MALDI‐TOF MS identification database at genospecies level for clinical and environmental Aeromonas strains. PLoS ONE, 7, e48441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzini, A. , Jaton, K. , Romo, D. , Bille, J. , Prod’hom, G. , & Greub, G. (2011). Matrix‐assisted laser desorption ionization‐time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult‐to‐identify bacterial strains. Journal of Clinical Microbiology, 49, 693–696. 10.1128/JCM.01463-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme, K. , Fernández‐No, I. C. , Barros‐Velázquez, J. , Gallardo, J. M. , Cañas, B. , & Calo‐Mata, P. (2011). Rapid species identification of seafood spoilage and pathogenic Gram‐positive bacteria by MALDI‐TOF mass fingerprinting. Electrophoresis, 32, 2951–2965. 10.1002/elps.201100217 [DOI] [PubMed] [Google Scholar]
- Carrasco, G. , Caballero, J. , Garrido, N. , Valdezate, S. , Cantón, R. , & Sáez‐Nieto, J. (2016). Shortcomings of the commercial MALDI‐TOF MS database and use of MLSA as an arbiter in the identification of Nocardia species. Frontiers in Microbiology, 7, 542 10.3389/fmicb.2016.00542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. L. , Wu, C. J. , Chen, C. S. , Tsai, P. J. , Tang, H. J. , & Ko, W. C. (2014). A comparative study of clinical Aeromonas dhakensis and Aeromonas hydrophila isolates in southern Taiwan: A. dhakensis is more predominant and virulent. Clinical Microbiology and Infection, 20, 428–434. 10.1111/1469-0691.12456 [DOI] [PubMed] [Google Scholar]
- Chopra, A. K. , & Houston, C. W. (1999). Enterotoxins in Aeromonas‐associated gastroenteritis. Microbes and Infection, 1, 1129–1137. 10.1016/S1286-4579(99)00202-6 [DOI] [PubMed] [Google Scholar]
- Chugh, T. D. (2008). Emerging and re‐emerging bacterial diseases in India. Journal of Biosciences, 33, 549–555. 10.1007/s12038-008-0073-0 [DOI] [PubMed] [Google Scholar]
- Citterio, B. , & Biavasco, F. (2015). Aeromonas hydrophila virulence. Virulence, 6, 417–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill, F. R. , Patel, J. B. , Alder, J. , Bradford, P. A. , Dudley, M. N. , Eliopoulos, G. M. , … Zimmer, B. L. (2013). Performance standards for antimicrobial susceptibility testing; twenty‐third informational supplement, M100–S23, Vol. (1). Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Croxatto, A. , Prod’hom, G. , & Greub, G. (2012). Applications of MALDI‐TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiology Reviews, 36, 380–407. 10.1111/j.1574-6976.2011.00298.x [DOI] [PubMed] [Google Scholar]
- Dallal, M. M. S. , Yazdi, M. K. S. , & Avadisians, S. (2012). Study of prevalence and antibiotic resistance in Aeromonas species isolated from minced meat and chicken samples in Iran. African Journal of Microbiology Research, 6, 460–464. [Google Scholar]
- Demarta, A. , Küpfer, M. , Riegel, P. , Harf‐Monteil, C. , Tonolla, M. , Peduzzi, R. , … Martínez‐Murcia, A. (2008). Aeromonas tecta sp. nov., isolated from clinical and environmental sources. Systematic and Applied Microbiology, 31, 278–286. 10.1016/j.syapm.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Deng, Y. , Wu, Y. , Jiang, L. , Tan, A. , Zhang, R. , & Luo, L. (2016). Multi‐drug resistance mediated by class 1 integrons in Aeromonas isolated from farmed fresh water animals. Frontiers in Microbiology, 7, 935–942. 10.3389/fmicb.2016.00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue, M. J. , Best, J. M. , Smallwood, A. W. , Kostich, M. , Rodgers, M. , & Shoemaker, J. A. (2007). Differentiation of Aeromonas isolated from drinking water distribution systems using matrix‐assisted laser desorption/ionization mass spectrometry. Analytical Chemistry, 79, 1939–1946. [DOI] [PubMed] [Google Scholar]
- Donohue, M. J. , Smallwood, A. W. , Pfaller, S. , Rodgers, M. , & Shoemaker, J. A. (2006). The development of a matrix‐assisted laser desorption/ionization mass spectrometry‐based method for the protein fingerprinting and identification of Aeromonas species using whole cells. Journal of Microbiological Methods, 65, 380–389. 10.1016/j.mimet.2005.08.005 [DOI] [PubMed] [Google Scholar]
- Elbehiry, A. , Al‐Dubaib, M. , Marzouk, E. , Osman, S. , & Edrees, H. (2016). Performance of MALDI biotyper compared with Vitek™ 2 compact system for fast identification and discrimination of Staphylococcus species isolated from bovine mastitis. Microbiologyopen, 5, 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbehiry, A. , Marzouk, E. , Hamada, M. , Al‐Dubaib, M. , Alyamani, E. , Moussa, I. M. , … Hemeg, H. A. (2017). Application of MALDI‐TOF MS fingerprinting as a quick tool for the identification and clustering of foodborne pathogens isolated from food products. New Microbiologica, 40, 269–278. [PubMed] [Google Scholar]
- Eley, A. , Geary, I. , & Wilcox, M. H. (1993). Growth of Aeromonas spp. at 4°C and related toxin production. Letters in Applied Microbiology, 16, 36–39. 10.1111/j.1472-765X.1993.tb01367.x [DOI] [Google Scholar]
- Encinas, J. , Gonzalez, C. , Garcia‐Lopez, M. L. , & Otero, A. (1999). Numbers and species of motile Aeromonads during the manufacture of naturally contaminated Spanish fermented sausages (Longaniza and Chorizo). Journal of Food Protection, 62, 1045–1049. 10.4315/0362-028X-62.9.1045 [DOI] [PubMed] [Google Scholar]
- García, P. , Allende, F. , Legarraga, P. , Huilcaman, M. , & Solari, S. (2012). Bacterial identification based on protein mass spectrometry: A new insight at the microbiology of the 21st century. Revista Chilena De Infectologia, 29, 263–272. [DOI] [PubMed] [Google Scholar]
- Gauthier, F. , Vincent, A. T. , Charette, S. J. , & Derome, N. (2017). Strong genomic and phenotypic heterogeneity in the Aeromonas sobria species complex. Frontiers in Microbiology, 8, 2434–2448. 10.3389/fmicb.2017.02434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghenghesh, K. S. , Ahmed, S. F. , El‐Khalek, R. A. , Al‐Gendy, A. , & Klena, J. (2008). Aeromonas‐associated infections in developing countries. Journal of Infection in Developing Countries, 2, 81–98. 10.3855/T2.2.81 [DOI] [PubMed] [Google Scholar]
- Ghenghesh, K. S. , El‐Mohammady, H. , Levin, H. Y. , Zorgani, A. , & Tawil, K. (2013). Antimicrobial resistance profile of Aeromonas species isolated from Libya. Libyan Journal Medicine, 8, 21234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H. (2010). Nonnegative principal component analysis for mass spectral serum profiles and biomarker discovery. BMC Bioinformatics, 11, S1 10.1186/1471-2105-11-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries, R. M. , & Linscott, A. G. (2015). Laboratory diagnosis of bacterial gastroenteritis. Reviews, 28, 3–31. 10.1128/CMR.00073-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa, I. H. (2014). Antibiogram profiling and pathogenic status of Aeromonas species recovered from Chicken. Saudi Journal of Biological Sciences, 21, 481–485. 10.1016/j.sjbs.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa, I. , Igumbor, E. , Aghdasi, F. , Tom, M. , & Okoh, A. (2012). Emerging Aeromonas species infections and their significance in public health. The Scientific World Journal, 2012, 625023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J. M. , Abbott, S. L. , & Carnahan, A. M. (1995). Aeromonas and Plesiomonas In Murray P. R., Baron E. J., Pfaller M. A., Tenover F. C., & Yolken R. H. (Eds.), Manual of clinical microbiology (6th ed., pp. 507–515). Washington, DC: American Society for Microbiology. [Google Scholar]
- Kirov, S. M. (1993). The public health significance of Aeromonas spp. in foods. International Journal of Food Microbiology, 20, 179–198. [DOI] [PubMed] [Google Scholar]
- Koca, C. , & Sarimehmetoglu, B. (2009). Isolation and identification of motile Aeromonas spp. in turkey meat. Kafkas Universitesi Veteriner Fakultesi Dergisi, 6, 95–98. [Google Scholar]
- Laith, A. R. , & Najiah, M. (2013). Aeromonas hydrophila: Antimicrobial susceptibility and histopathology of isolates from diseased catfish, Clarias gariepinus (Burchell). Journal of Aquaculture Research and Development, 5, 2–7. [Google Scholar]
- Lamy, B. , Kodjo, A. , Laurent, F. & CoIBVH Group (2011). Identification of Aeromonas isolates by matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry. Diagnostic Microbiology and Infectious Disease, 71, 1–5. [DOI] [PubMed] [Google Scholar]
- Lau, S. K. P. , Tang, B. S. F. , Teng, J. L. L. , Chan, T.‐M. , Curreem, S. O. T. , Fan, R. Y. Y. , … Woo, P. C. Y. (2014). Matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry for the identification of clinically significant bacteria that are difficult to identify in clinical laboratories. Journal of Clinical Pathology, 67, 361–366. [DOI] [PubMed] [Google Scholar]
- Li, B. , & Webster, T. J. (2018). Bacteria antibiotic resistance: New challenges and opportunities for implant‐associated orthopaedic infections. Journal of Orthopaedic Research, 36, 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, C. I. , Fall, B. , Sambe‐Ba, B. , Diawara, S. , Gueye, M. W. , Mediannikov, O. , … Fenollar, F. (2015). MALDI‐TOF mass spectrometry: A powerful tool for clinical microbiology at hospital principal de Dakar, Senegal (west Africa). PLoS ONE, 10, e0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa, S. , Armuzzi, R. , Tosques, M. , Canganella, F. , & Trovatelli, L. D. (1999). Note: Susceptibility to chlorine of Aeromonas hydrophila strains. Journal of Applied Microbiology, 86, 168–173. 10.1046/j.1365-2672.1999.00592.x [DOI] [PubMed] [Google Scholar]
- McElvania TeKippe, E. , & Burnham, C. A. (2014). Evaluation of the Bruker Biotyper and VITEK MS MALDI‐TOF MS systems for the identification of unusual and/or difficult‐to‐identify microorganisms isolated from clinical specimens. European Journal of Clinical Microbiology & Infectious Diseases, 33, 2163–2171. 10.1007/s10096-014-2183-y [DOI] [PubMed] [Google Scholar]
- Miyagi, K. , Hirai, I. , & Sano, K. (2016). Distribution of Aeromonas species in environmental water used in daily life in Okinawa Prefecture, Japan. Environmental Health and Preventive Medicine, 21, 287–294. 10.1007/s12199-016-0528-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga, Y. , Yanagihara, K. , Eugenin, F. L. , Beaz‐Hidalgo, R. , Kohno, S. , & Figueras Salvat, M. J. (2013). Identification error of Aeromonas aquariorum: A causative agent of septicemia. Diagnostic Microbiology and Infectious Disease, 76, 106–109. 10.1016/j.diagmicrobio.2013.01.019 [DOI] [PubMed] [Google Scholar]
- Morita, K. , Watanabe, N. , Kurata, S. , & Kanamori, M. (1994). b‐Lactam resistance of motile Aeromonas isolates from clinical and environmental sources. Antimicrobial Agents and Chemotherapy, 38, 353–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, P. R. (2010). Matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry: Usefulness for taxonomy and epidemiology. Clinical Microbiology and Infection, 16, 1626–1630. 10.1111/j.1469-0691.2010.03364.x [DOI] [PubMed] [Google Scholar]
- Odeyemi, O. A. , & Ahmad, A. (2017). Population dynamics, antibiotic resistance and biofilm formation of Aeromonas and Vibrio species isolated from aquatic sources in Northern Malaysia. Microbial Pathogenesis, 103, 178–185. [DOI] [PubMed] [Google Scholar]
- Ogu, G. I. , Madar, I. H. , Okolo, J. C. , & Tayubi, I. A. (2017). Exposure assessment of chicken meat to heavy metals and bacterial contaminations in Warri metropolis, Nigeria. International Journal of Scientific Innovations, 1, 1–14. [Google Scholar]
- Osman, K. , Aly, M. , Kheader, A. , & Mabrok, K. (2012). Molecular detection of the Aeromonas virulence aerolysin gene in retail meats from different animal sources in Egypt. World Journal of Microbiology and Biotechnology, 28, 1863–1870. 10.1007/s11274-011-0915-z [DOI] [PubMed] [Google Scholar]
- Pérez‐Sancho, M. , Cerdá, I. , Fernández‐Bravo, A. , Domínguez, L. , Figueras, M. J. , Fernández‐Garayzába, J. F. , & Vela, A. I. (2018). Limited performance of MALDI‐TOF for identification of fish Aeromonas isolates at species level. Journal of Fish Diseases, 2018(41), 1485–1493. [DOI] [PubMed] [Google Scholar]
- Persson, S. , Al‐Shuweli, S. , Yapici, S. , Jensen, J. N. , & Olsen, K. E. P. (2015). Identification of clinical Aeromonas species by rpoB and gyrB sequencing and development of a multiplex PCR method for detection of Aeromonas hydrophila, A. caviae, A. veronii, and A. media . Journal of Clinical Microbiology, 53, 653–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte, L. , García, P. , Braun, S. , Ulloa, M. T. , Lafourcade, M. , Montaña, A. , … Weitzel, T. (2017). Head‐to‐head comparison of Microflex LT and Vitek MS systems for routine identification of microorganisms by MALDI‐TOF mass spectrometry in Chile. PLoS ONE, 12, e0177929 10.1371/journal.pone.0177929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen, P. K. , Debnath, C. , Shekhar, S. , Dalai, N. , & Ganguly, S. (2016). Incidence of Aeromonas spp. infection in fish and chicken meat and its related public health hazards: A review. Veterinary World, 9, 6–11. 10.14202/vetworld.2016.6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar, S. , Ayyasamm, P. M. , Shanthi, K. , Song, Y. C. , & Lak‐shmanaperumalsamy, P. (2012). Incidence survival and antibiotic resistance of Aeromonas hydrophila isolated from lamb and chicken meat retail outlets. Journal of Current Perspectives in Applied Microbiology, 1, 53–63. [Google Scholar]
- Rall, V. L. M. , Iaria, S. T. , Heidtman, S. , Pimenta, F. C. , Gamba, R. C. , & Pedroso, D. M. M. (1998). Aeromonas species isolated from Pintado fish (Pseudoplatystoma sp. ): Virulence factors and drug susceptibility. Revista De Microbiologia, 29, 2227. [Google Scholar]
- Ramalivhana, J. N. , Obi, C. L. , & Moyo, S. R. (2009). Antimicrobial susceptibility testing of Aeromonas hydrophila isolated from Limpopo Province, South Africa using VITEK 2 system, micro scan walk away, disk diffusion and E‐test method. African Journal of Microbiological Research, 3, 903–913. [Google Scholar]
- Rey, G. , Fouillet, A. , Bessemoulin, P. , Frayssinet, P. , Dufour, A. , Jougla, E. , & Hémon, D. (2009). Heat exposure and socio‐economic vulnerability as synergistic factors in heat‐wave‐related mortality. European Journal of Epidemiology, 24, 495–502. 10.1007/s10654-009-9374-3 [DOI] [PubMed] [Google Scholar]
- Romero, J. , Feijoo, C. G. , & Navarrete, P. (2012). Antibiotics in aquaculture– Use, abuse and alternatives, health and environment in aquaculture In Carvalho E. D., Silva R. J., & David G. S. (Eds.), Health and environment in aquaculture (pp. 159–198). Santiago, Chile: : InTech. [Google Scholar]
- Saavedra, M. J. , Guedes‐Novais, S. , Alves, A. , Rema, P. , Tacão, M. , Correia, A. , & Martínez‐Murcia, A. (2004). Resistance to beta‐lactam antibiotics in Aeromonas hydrophila isolated from rainbow trout (Oncorhynchus mykiss). International Microbiology, 7, 207–211. [PubMed] [Google Scholar]
- Sandrin, T. R. , Goldstein, J. E. , & Schumaker, S. (2013). MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrometry Reviews, 32, 88–217. 10.1002/mas.21359 [DOI] [PubMed] [Google Scholar]
- Seng, P. , Drancourt, M. , Gouriet, F. , La Scola, B. , Fournier, P. , Rolain, J. M. , & Raoult, D. (2009). Ongoing revolution in bacteriology: Routine identification of bacteria by matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry. Clinical Infectious Diseases, 49, 543–551. 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- Shao, C. , Tian, Y. , Dong, Z. , Gao, J. , Gao, Y. , Jia, X. , … Zhang, X. (2012). The use of principal component analysis in MALDI‐TOF MS: A powerful tool for establishing a mini‐optimized proteomic profile. American Journal of Biomedical Sciences, 4, 85–101. 10.5099/aj120100085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, I. , & Kumar, A. (2011). Occurrence of enterotoxigenic Aeromonas species in foods of animal origin in North East India. European Review for Medical and Pharmacological Sciences, 15, 883–887. [PubMed] [Google Scholar]
- Singhal, N. , Kumar, M. , Kanaujia, P. K. , & Virdi, J. S. (2015). MALDI‐TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Frontiers in Microbiology, 5, 791–806. 10.3389/fmicb.2015.00791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler, L. , Yáñez, M. A. , Chacon, M. R. , Aguilera‐Arreola, M. G. , Catalán, V. , Figueras, M. J. , & Martínez‐Murcia, A. J. (2004). Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. International Journal of Systematic and Evolutionary Microbiology, 54, 1511–1519. 10.1099/ijs.0.03048-0 [DOI] [PubMed] [Google Scholar]
- Steinberg, J. , & Burd, E. M. (2010). Other gram‐negative and gram‐variable bacilli In Mandell G. L., Bennett J. E., & Dolin R. (Eds.), Mandell Douglas, Bennett’s principles and practice of infectious diseases, 7th ed. (pp. 465–478). New York, NY: Churchill Livingstone p. [Google Scholar]
- Stratev, D. , & Odeyemi, O. A. (2016). Antimicrobial resistance of Aeromonas hydrophila isolated from different food sources: A mini‐review. Journal of Infection and Public Health, 9, 535–544. 10.1016/j.jiph.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Trakhna, F. , Harf‐Monteil, C. , Abdelnour, A. , Maaroufi, A. , & Gadonna‐Widehem, P. (2009). Rapid Aeromonas hydrophila identification by TaqMan PCR assay: Comparison with a phenotypic method. Letters in Applied Microbiology, 49, 186–190. [DOI] [PubMed] [Google Scholar]
- van Belkum, A. , Welker, M. , Pincus, D. , Charrier, J. , & Girard, V. (2017). Matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry in clinical microbiology: What are the current issues? Annals of Laboratory Medicine, 37, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vávrová, A. , Balážová, T. , Sedláček, I. , Tvrzová, L. , & Šedo, O. (2015). Evaluation of the MALDI‐TOF MS profiling for identification of newly described Aeromonas spp. Folia Microbiologica (Praha), 60, 375–383. 10.1007/s12223-014-0369-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


