Abstract
The complete genome of the strain Labrenzia sp. PHM005, a free-living producer of a pederin analog 18-O-demethyl pederin, hereinafter labrenzin, has been sequenced. This strain contains two replicons comprising a circular chromosome of 6,167,349 bp and a circular plasmid (named p1BIR) of 19,450 bp. A putative gene cluster responsible for the synthesis of labrenzin (lab cluster) has been identified showing that it encodes a trans-AT mixed type PKS/NRPS biosynthetic pathway that is responsible for the synthesis of pederin and possibly an onnamide analog. The putative boundaries of the lab gene cluster were determined by genetic comparisons with other related strains, suggesting that the cluster consists of a 79-kb region comprising 3 genes encoding multidomain hybrid polyketide synthase/non-ribosomal peptide synthetase (PKS/NRPS) proteins (PKS4, PKS/NRPS13, and PKS/NRPS15), and 16 auxiliary enzymes. Transcriptomic analyses suggest that all the genes of the cluster are expressed in our culture conditions (i.e., in minimal medium in the absence of any specific inducer) at detectable levels. We have developed genetic tools to facilitate the manipulation of this strain and the functional characterization of the cluster genes. We have created a site-directed mutant unable to produce pederin, demonstrating experimentally for the first time the role of the cluster in the synthesis of pederin. This work paves the way to unravel the clues of the biosynthesis of pederin family compounds and opens the door to modify and overproduce these anticancer drugs for industrial and pharmaceutical purposes.
Keywords: pederin, biosynthesis, trans-AT PKS, expression, genetic modification
Introduction
Pederin is a natural polyketide extracted for the first time from insect genus Paederus Curt. (Coleoptera: Staphylinidae) as non-protein insect toxin (Frank and Kanamitsu, 1987). Twenty five million field-collected Paederus fuscipes were used to isolate pure pederin (Pavan and Bo, 1952) and its chemical formula was further determined (Cardani et al., 1965). It has been found that females of genera Paederus use pederin to chemically defend their offspring against predators (Kellner and Dettner, 1996). Pederin displays potent and selective bioactivities that have triggered biomedical interests (Soldati et al., 1966; Wan et al., 2011). It has a huge therapeutic potential because it is shown to be a highly potent anticancer agent (Richter et al., 1997). Closely related cytotoxic compounds like theopederins, mycalamides, and onnamides have been isolated from several genus of marine sponges (Pannell et al., 1988; Sakemi et al., 1988; Fusetani et al., 1992). It has been suggested that these molecules could be synthesized by symbiotic microbes existing in these marine invertebrates (Piel et al., 2004b).
The first insights into the true producers of pederin were obtained by cloning a fraction of the pederin biosynthesis genes (ped cluster) from total DNA of P. fuscipes beetles (Piel, 2002). The partial ped cluster that was initially located on a 54-kb region bordered by transposase pseudogenes encoded a mixed modular polyketide synthase/non-ribosomal peptide synthetase. The ped cluster was distributed among two distinct regions (pedIJK and pedABCDEF) of the genome from an uncultured bacterial symbiont of the insect that exhibited a similarity to genera Pseudomonas (Piel, 2002; Piel et al., 2004a, c). Supporting this finding, a dominant bacterium with the closest relationship to P. aeruginosa had previously been detected in Paederus sabaeus (Kellner, 2002).
An onn gene cluster closely related to ped genes was isolated later on from the metagenome of the marine sponge Theonella swinhoei, which was described as a source of onnamides and theopederins (Piel et al., 2004b). The isolated genes also belonged to a prokaryotic symbiont and the close similarity and complexity of the ped and onn biosynthetic systems suggested that they were derived from a common ancestral gene cluster.
Today, more than 30 members constitute the pederin-like family, containing almost an identical core region and variable polyketide or amino acid termini (Piel et al., 2005). Nonetheless, the gene clusters associated to their biosynthesis have mostly been identified in symbiont metagenomes, such as psymberin (Fisch et al., 2009), diaphorin (Nakabachi et al., 2013), and nosperin (Kampa et al., 2013) with an exception of cusperin (Kust et al., 2018) recently identified in a free-living cyanobacterium.
Moreover, the isolation of labrenzin from the culture of a marine heterotrophic α-Proteobacterium, Labrenzia sp. strain PHM005 isolated from marine sediment was reported (Schleissner et al., 2017; Cañedo et al., 2018). This was the first report of the production of a pederin-like compound by a free-living marine bacterium that could be cultured in the laboratory.
Despite the research carried out on the pederin biosynthetic pathways, direct evidence that such pathways were responsible for the pederin synthesis have not been published yet. Here, we describe the sequencing of the complete genome of Labrenzia sp. PHM005 and the identification of the lab gene cluster responsible for the synthesis of labrenzin. We have been able to manipulate genetically this strain to show experimentally a direct relationship of the lab cluster with the biosynthesis of labrenzin. The genomic and transcriptomic analyses carried out with this cultivable bacterium revealed new insights into the biosynthesis of pederin family compounds.
Materials and Methods
Culture Media and Bacterial Growth Conditions
Culture media used to grow Labrenzia sp. strain PHM005 (provided by PharmaMar) and to produce labrenzin were as follows: (i) Marine Broth (DIFCO 2216) (MB); (ii) Modified marine basal media (MBM) (Baumann and Baumann, 1981) containing NaCl (20 g/L) and 0.2% glucose as carbon source; (iii) MBM supplemented with vitamins (MBM + vit) [B12 (50 μg/L), panthotenic acid (50 μg/L), riboflavin (50 μg/L), pyridoxamine (10 μg/L), biotin (20 μg/L), folic acid (20 μg/L), nicotinic acid (50 μg/L), p-aminobenzoic acid (50 μg/L), thyamine (50 μg/L)]; (iv) MBM supplemented with biotin (20 μg/L) (MBM + BIOTIN); (v) MBM supplemented with thyamine (50 μg/L) (MBM + THYAMINE); (vi) MBM supplemented with biotin (20 μg/L) and thyamine (50 μg/L) (MBM + B + T).
The strain was usually grown overnight in 50 mL falcon tubes in MB at 30°C with shaking at 220 rpm. The overnight culture was washed in 0.85% NaCl solution and diluted to an optical density (OD600) ≈ 0.1 in 20 ml of fresh MBM medium. The OD600 was measured every 3 h during the cultivation.
DNA Extraction, Sequencing and Genome Assembly
The strain was grown in MB for 24 h and the genomic DNA was extracted using Blood & Cell Culture DNA Mini Kit (Qiagen). The genome of a strain PHM005 was sequenced by a PacBio RSII sequencer and was assembled de novo in a single contig. Samples were prepared at PharmaMar and library construction, sequencing, data processing and assembly were performed by Macrogen.
Several sequencing errors were detected in the trans-AT PKS sequences that resulted in frameshifts that render several unexpected stop codons. The errors were corrected by Sanger sequencing of specific PCR amplicons surrounding the sequencing errors (Supplementary Figure S1). Primers used for PCR amplification for Sanger sequencing were designed using Primer3 implemented in the Geneious v.10.0.2 software (Supplementary Table S1). The circular map of a Labrenzia sp. PHM005 genome was generated using a CGView (Stothard and Wishart, 2005).
To determine the presence of plasmids in the strain PHM005 that cannot be detected by the PacBio sequencing method as well as to correct other putative PacBio sequencing errors, the genome of strain PHM005 was re-sequenced using an Illumina MiSeq system with a 300 nt pair-end library at the Complutense University of Madrid. The library was constructed following the manufacturer’s recommendations (Nextera DNA Flex, Illumina). The reads were trimmed and assembled into contigs using CLC Genomics Workbench software package (Quiagen).
The PacBio sequencing errors detected in the assembled chromosome were corrected manually after mapping the Illumina reads to PHM005 genome sequenced by PacBio using the Geneious v.10.0.2 software.
The Whole Genome projects have been deposited at GenBank under the accession CP041191 (Labrenzia sp. PHM005) and CP041190 (p1BIR). The DNA sequencing data have been deposited at sequence read archive (SRA) under the accession SRS5035208.
Genomic Analyses
The genome of strain PHM005 was annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). AntiSMASH v. 4.0. was used for a rough secondary metabolites cluster mining and the identification of the individual PKS/NRPS domains. The specificity of NRPS adenylation domains and trans-ATs was predicted based on a combination of tools implemented in antiSMASH 4.0 (Stachelhaus code, NRPS Predictor3, pHMM, SANDPUMA and Minowa).
Whole genome alignment of Labrenzia sp. PHM005, L. alexandrii DFL-11, L. aggregata RMAR6-6, Labrenzia sp. CP4 and Labrenzia sp. VG12 was performed using the progressive Mauve algorithm with default settings from the Geneious v.10.0.2. In addition, each strain was aligned separately to Labrenzia sp. PHM005 to calculate the minimum weight for Locally Collinear Blocks (LCB).
Average nucleotide identity (ANI) was calculated using reciprocal best hits (two-way ANI) between two genomic datasets in an online tool developed at Kostas lab (Rodriguez-R and Konstantinidis, 2016).
To establish gene cluster boundaries, the proximal DNA regions from each strain were aligned using MAFFT with the scoring matrix BLOSUM62 and the gap open penalty set to 1.
The dot plot of the protein alignment of PKSs and theirs Ped homologs was generated based on the score matrix BLOSUM62, window size 10 and threshold 23.
ECH1 protein domains were aligned using Geneious v.10.0.2 default parameters.
Genetic Manipulations
The gene deletion was performed based on the I-SceI endonuclease system using pSEVA312S and pSEVA428S plasmids (Martínez-García and de Lorenzo, 2011; Silva-Rocha et al., 2013). I-SceI endonuclease causes double strand breaks that trigger DNA repair by homologous recombination. Primers PKS4 UP F and PKS4 UP R (5′-ACTA GTCTAGA CGCAGTCGTCCTGATGAGAT-3′ and 5′-ATCC CGAGCTCATGGGAACGTCCAGTATCGC-3′, respectively) were designed to amplify the upstream sequence (574 bp) flanking the deletion region. Primers PKS4 DOWN F and PKS4 DOWN R (5′-ATCCCGAGCTCGGTCGTATTC AACACCCGGT-3′ and 5′-TACCCAAGCTTAGGATCCTGC AAGAAGCCAC-3′, respectively; SacI restriction site is underlined) were designed to amplify the downstream flanking sequence (637 bp). The upstream and downstream sequences were ligated flanking the SacI restriction site and subcloned into the pCRTM-Blunt II-TOPOTM plasmid (Invitrogen) to generate pTOPO-PKS4UP-DOWN plasmid. The pTOPO-PKS4UP-DOWN and suicide plasmid pSEVA312S were both digested with PstI and SpeI. The UP-DOWN fragment released from the pTOPO-PKS4UP-DOWN was further ligated into previously digested pSEVA312S to generate pSEVA312SUP-DOWN and transformed into E. coli cc118λpir. Triparental conjugation with donor E. coli cc118λpir and helper E. coli HB101 harboring the plasmid pRK600 was used to transform Labrenzia sp. PHM005 for the genome integration of the suicide plasmid. The PHM005 transconjugants T1 were selected on the Petri plates with Marine Agar (MA) (Difco 2216) containing as selective antibiotics chloramphenicol (5 μg/mL) and kanamycin (10 μg/mL). Subsequently, pSEVA428S was transformed to PHM005 T1 by triparental conjugation and selected on the MA plates with selective antibiotics chloramphenicol (5 μg/mL), kanamycin (10 μg/mL) and streptomycin (50 μg/mL). The induction of I-SceI endonuclease and the selection of the recombinant PHM005ΔPKS4 on MA, as well as the following curation of the pSEVA428S were performed as described (Martínez-García and de Lorenzo, 2011). The deletion recombinants were screened by PCR-amplification using external primers M1 F and M1R (5′-TCTTGGTGGACGAGACCAGT-3′ and 5′-GGCTTCGTGTGGTGAAATGC-3′, respectively) and the deletion (763 bp) was confirmed by Sanger sequencing.
RNA Extraction and Gene Expression Analysis
Mutant strain PHM005ΔPKS4 was cultured in MBM + vit medium and the total RNA was extracted by High-pure Isolation Kit (Roche) after 24 h. Extracted RNA was additionally treated by DNA-free DNaseTreatment and Removal Reagents (Ambion). The RNA concentration was measured by using a NanoPhotometer (Implen). Reverse transcription of 1 μg of DNA-free RNA per sample was done using the Transcriptor First Strand cDNA Synthesis Kit (Roche) to generate cDNA and RT-PCR semi-quantitative analyses were carried out to check the mRNA levels. The rpoD gene was used as the housekeeping gene. Primers for RT–PCR for Labrenzia sp. PHM005 pederin cluster genes, as well as the housekeeping rpoD gene, were designed using Primer3 implemented by Geneious v.10.0.2 (Supplementary Table S2). Relative mRNA expression was visualized in 1.5% agarose gel. Negative controls contained RNA samples without the reverse transcriptase. The RNA extraction from the strain PHM005 for the transcriptomics analysis was performed in the early exponential phase (12 h) as described previously. Transcriptomics and bioinformatics analyses were carried out by Vertis Biotechnologie AG. The percentage of the mapped reads to annotated genes is 78.4, 80.8, and 77.0% for each replicate sample. The results represent a mean of three replicates.
Extraction of Labrenzin and HPLC-MS Analyses
Strain PHM005 was cultured in media described above in three replicates and samples were extracted at 24 h, 72 h, and 6 days of growth. The mutant PHM005ΔPKS4 was cultured in MBM + vit. Samples were centrifuged and the supernatant was extracted twice with ethyl acetate (1:1, vol:vol) and the two organic fractions were combined and dried on a rotary evaporator. The dry matter was dissolved in methanol and filtered through 0.22-μm polytetrafluoroethylene syringe filters before HPLC-MS analysis. Labrenzin was purified and prepared as a standard by PharmaMar. HPLC-MS was carried out using a mass spectrometry system (Thermo Mod. FinniganTM LXQTM) with on line HPLC Surveyor comprising pump with four separate solvent feeds (Surveyor MS pump Plus), an auto-sampler for multi sample analysis (Surveyor AS Plus), a Photo Diode Array detector (Surveyor PDA Plus) and a mass spectrometer LXQ equipped with linear ion trap. Ionization source used was electrospray ionization (ESI). Separation was performed on a C18 column (ZORBAX Eclipse plus C18, 5 μm, 4.6 × 250 mm, Agilent Technologies, Santa Clara, CA, United States). Solvent A was 100% water and solvent B was 100% acetonitrile. The flow rate was 1 ml min–1 and the gradient (per cent solvent A/B) was t = 2 min, 100% A; t = 5 min, 95% A; t = 25 min, 0% A; t = 27 min, 0% A; t = 30 min, 100% A; t = 35 min, 100% A.
Results
Genome Characteristics of Labrenzia sp. PHM005
The complete genome sequence of Labrenzia sp. strain PHM005 revealed two replicons comprising a circular chromosome of 6,167,349 bp (accession CP041191) and a circular plasmid (named p1BIR) of 19,450 bp (accession CP041190) shown in Figure 1. The genome average G + C content is 55%, contains 51 tRNAs and 3 copies of rRNA sequences (5S-23S-16S) (Figure 1). The genome contains 5,988 coding sequences that were classified by Rapid Annotation Subsystem Technology (RAST) Server (Aziz et al., 2008) into 483 subsystems and 26 categories (Supplementary Figure S2). Functional comparison of genome sequences in the RAST database revealed Labrenzia alexandrii (strain DFL-11) (score 548) as the closest neighbor of strain PHM005, followed by Labrenzia aggregata (strain IAM 12614) (score 539), Roseibium sp. (strain TrichSKD4) (score 328), Agrobacterium tumefaciens (strain C58) (score 207) and Sinorhizobium meliloti (strain 5A14) (score 198).
FIGURE 1.
Circular map of the genome of Labrenzia sp. PHM005 and its plasmid p1BIR. (A) From outer to inner circles: The first four circles show the coding sequence (CDS), transfer ribonucleic acid (tRNA), ribosomal ribonucleic acid (rRNA), and open reading frame (ORF). The fifth circle represents the GC content (black). The sixth circle demonstrates the GC skew curve (positive GC skew, green; negative GC skew, violet). The genome position scaled in 500 kbp from base 1 is shown on the inner circle. (B) The circular map of plasmid p1BIR (Geneious version 10.0 created by Biomatters. Available from http://www.geneious.com). Dark green arrows represent genes coding for proteins with homology in other species from NCBI database and light green represents hypothetical proteins.
The genomes of five Labrenzia species, i.e., Labrenzia sp. (strain PHM005) (accession CP041191), L. alexandrii (strain DFL-11) (accession CM011002), L. aggregata (strain RMAR6-6) (accession CP019630), Labrenzia sp. (strain CP4) (accession CP011927) and Labrenzia sp. (strain VG12) (accession CP022529), were analyzed for average nucleotide identity (ANI) to provide intraspecies and interspecies relationships (Supplementary Figure S3). Strain PHM005 appears to be most similar to L. alexandrii (strain DFL-11) although both genomes do not seem to belong to the same species since the ANI value is below 95% (Goris et al., 2007). Nonetheless, only L. aggregata (strain RMAR6-6) and Labrenzia sp. (strain CP4) most likely belong to the same species showing an ANI value of 97.76%.
Plasmid p1BIR (accession CP041190) has a G + C content of 48.5% and contains 17 ORFs most of them coding for putative replication and conjugation related proteins (Figure 1 and Supplementary Table S3). The plasmid carries two putative Abi family proteins described to be involved in bacteriophage abortive infection resistance systems (Garvey et al., 1995). A plasmid copy number of 59 was calculated taking into account the genome and plasmid size and the number of Illumina reads covering them, i. e., 3,820,045 reads and 712,018 reads, respectively (SRA accession SRS5035208).
Analysis of the Secondary Metabolite Gene Clusters in Labrenzia sp. PHM005
The circular genome of Labrenzia sp. PHM005 was set for a secondary metabolite gene cluster mining using an online platform antiSMASH 4.0. In total, 101 gene clusters have been identified: (i) one bacteriocin or other unspecified ribosomally synthesized and post-translationally modified peptide product (RiPP) cluster; (ii) 4 putative fatty acid clusters; (iii) 4 putative saccharide clusters; (iv) 89 putative clusters of unknown type; (v) one putative polyhydroxyalkanoate biosynthetic gene cluster; (vi) one putative mixed PKS/saccharide cluster; (vii) one trans-AT PKS/NRPS cluster. The similarity of these clusters with the known clusters from MIBiG repository is shown in Supplementary Table S4. Additionally, the rest of the clusters were screened for similarity in other microorganisms and we have counted 17 clusters which lie within the order of Rhizobiales sharing from 10 to 32% of cluster genes (Supplementary Table S5).
Proposed Modular Biosynthesis of Labrenzin
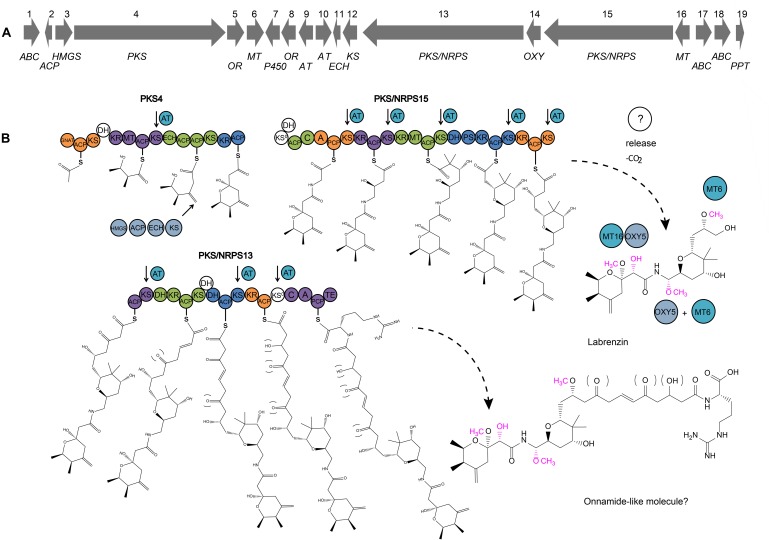
The trans-AT PKS/NRPS cluster, further named lab cluster, consists of a 79-kb region (776,792–855,905 bp) comprising 3 genes encoding multi-domain PKS/NRPS proteins (PKS4, PKS/NRPS13, and PKS/NRPS15), and 16 auxiliary enzymes (Figure 2A).
FIGURE 2.
(A) Map of the lab cluster comprising genes for labrenzin production. (B) Scheme predicting the pederin/onnamide modular biosynthesis in a free-living Alfaproteobacterium Labrenzia sp. PHM005. Specific domains have been identified and analyzed using antiSMASH. Domains (shown in circles) that are part of the same module are represented in the same color, less the putative non-functional domains in white. Module boundaries are defined as described by Vander Wood and Keatinge-Clay (2018). GNAT, GCN5-related N-acetyltransferase domain; ACP, acyl carrier protein domain; KS, ketosynthase domain; KS0, non-elongating ketosynthase domain; KR, ketoreductase domain; MT, methyltransferase domain; ECH, enoyl- CoA hydratase/isomerase domain; A, adenylation domain; AT, trans-acyltransferase; C, condensation domain; DH, dehydratase domain; PS, pyran synthase; PCP, peptidyl carrier protein domain; TE, thioesterase domain; OR, oxidoreductase; MT, methyltransferase. Putative trans-AT docking site associated with its respective KS is indicated by arrows. Two pathways are indicated: the synthesis of the labrenzin and the onnamide A questioned to be the complete molecule of the proposed pathway. Putative functional groups are marked inside brackets and putative modifications by tailoring enzymes are colored in pink.
The core pederin molecule is constructed following the “assembly line” biosynthesis by modular PKSs/NRPSs and further modified by tailoring enzymes as proposed in the scheme (Figure 2B). The structure of biosynthesis intermediates of labrenzin has been deducted from the specificity of enzymatic domains of each PKS/NRPS module. The domain analysis based on antiSMASH results and the described conserved motifs (El-Sayed et al., 2003) of the three PKS/NRPS are listed in Supplementary Table S6. According to the modular structure predicted by antiSMASH v.4.0., the PKS4 from strain PHM005 consists of a “starter” module 0 with a GCN5-related N-acetyltransferase family (GNAT) domain that catalyzes the decarboxylation of malonyl-CoA to generate acetyl-CoA and its translocation to the ACP unit. After that, three elongating β-ketoacyl synthases (KSs) incorporate the 3 malonyl-CoA building units. Compared to PedI from P. fuscipes symbiont (Piel et al., 2004a), the first module of PKS4 contains an additional dehydratase (DH) domain, most probably inactive, since it lacks the typical conserved motif HXXXGXXXXP (El-Sayed et al., 2003). Also, the presence of only one ECH domain in the third module does not appear to affect the structure of the first pyran ring of the pederin core structure.
The domain architecture of the PKS/NRPS15 from strain PHM005 is almost identical to PedF (Piel, 2002). The singularity of PKS/NRPS15 is the presence of a putative inactive DH domain attached to the first non-elongating KS0, while in PedF there is one putative inactive DH domain in the last module.
According to antiSMASH predictions, the PKS/NRPS13 from strain PHM005, follows the domain order described in PedH, which is hypothesized by Piel (2002) to synthesize the chain containing the terminal arginine residue found in the onnamide analogs (Matsunaga et al., 1992). However, there are a few exceptions. Substrate predictions of PKS/NRPS13 by pHHM and SANDPUMA indicate arginine and alanine, respectively, as candidate amino acids for the chain terminal of the putative onnamide-like molecule, therefore we cannot conclude with accuracy the last amino acid incorporation. PKS/NRPS13 also possesses another, probably inactive DH domain in the third module. The last non-elongating KS lacks the histidine residue and the ACP located downstream lacks the conserved GxDS motif (El-Sayed et al., 2003).
Pederin Cluster Boundaries
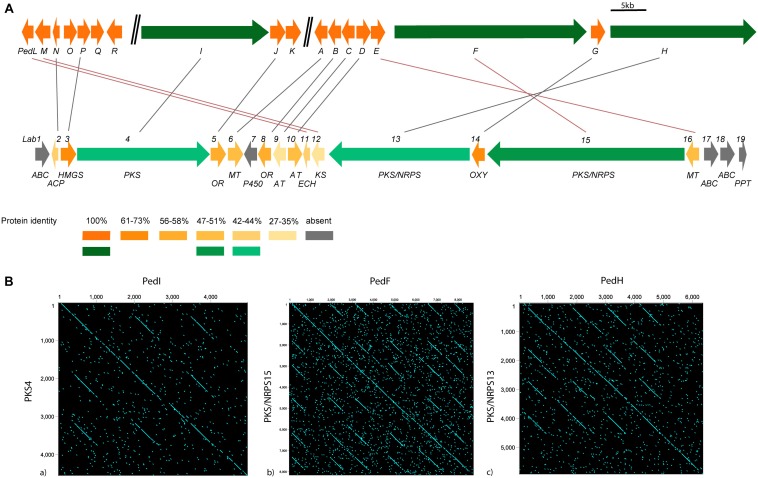
The genome of L. alexandrii (strain DFL-11) was used to estimate the size and the putative boundaries of the lab cluster in the genome of strain PHM005. The alignment of both genomes is shown in Figure 3. This alignment revealed that the lab cluster has been probably inserted within two rDNA regions. The putative cluster limits were further compared with the genomes of 3 other species of Labrenzia, i.e., L. aggregata (strain RMAR6-6) (accession CP019630), Labrenzia sp. (strain CP4) (accession CP011927) and Labrenzia sp. (strain VG12) (accession CP022529). Using the five previously described Labrenzia species we performed a whole genome alignment using a progressive Mauve algorithm from the Geneious version 10.0.2 (Supplementary Figure S4). According to these algorithms, L. alexandrii (strain DFL-11) showed the highest local collinearity number (Supplementary Table S7). Interestingly, the genes located downstream from the lab gene cluster were found in the same orientation in all 5 species. The similarity and the corresponding gene positions in each species are shown in Figure 3B. On the contrary, the genes located upstream the lab cluster were only found in L. alexandrii (strain DFL-11) (gene position at genome between 3,628,890 bp and 3,648,023 bp). Accordingly, to these comparisons, the homologous genes present in at least two genomes were discarded as candidates for the biosynthesis of labrenzin allowing us to fix the putative boundaries.
FIGURE 3.
Representation of the lab gene cluster and its bordering genes distributed among the selected Labrenzia species genomes. (A) MAFFT alignment of Labrenzia alexandrii DFL-11 and Labrenzia sp. PHM005. Coding sequences are represented by arrows: gray, CDS; red, ribosomal DNA; green and orange, lab gene cluster; light blue; identical genes. (B) MAFFT alignment and localization of downstream bordering genes in 5 Labrenzia species.
Comparative Analysis With ped, onn and dip Gene Clusters
The results of the BLAST protein homology analysis indicate the similarity with pederin, onnamide and diaphorin gene clusters (Supplementary Table S8), i.e., other members of the pederin-family biosynthetic clusters. Using as a reference the genes of the pederin cluster of the symbiont from P. fuscipes, the homologous genes and their protein identities of the corresponding genes of Labrenzia sp. PHM005 are visualized in Figures 4A,B. The first observed difference between the two almost identical biosynthetic systems is the gene distribution in their host genomes. In the symbiont bacterium of P. fuscipes, the pederin genes are clustered in three genomic islands flanked by IS elements and transposons (Piel et al., 2004c, 2005). However, the pederin biosynthetic genes of strain PHM005 are clustered together, suggesting its horizontal gene transfer, a very common phenomenon in bacteria as it is hypothesized by Roth and Lawrence (1996).
FIGURE 4.
Comparative analysis of a pederin cluster from a symbiont bacterium of P. fuscipes and the free-living Alphaproteobacterium Labrenzia sp. PHM005. Modified scheme of the pederin cluster as described from a: (A) bacterium symbiont of P. fuscipes (Piel et al., 2004b) (above) and the newly described labrenzin cluster from Labrenzia sp. PHM005 (below). Homologous genes between two clusters are indicated by lines. Protein identity is visualized using colored scale that matches the identity percentage of all the genes from the cluster, using as reference the symbiont bacterium of P. fuscipes genes. PKS and NRPS genes are shown in green and tailoring enzymes in orange and gray. ABC, transporter; ACP, acyl carrier protein; AT, acyltransferase; ECH, enoyl- CoA hydratase/isomerase domain; HMGS, hydroxymethylglutaryl-CoA synthase; KS, ketosynthase; MT, methyltransferase; OR, oxidoreductase; OXY, oxygenase; P450, cytochrome hydroxylase; PPT, phosphopantetheinyl transferase. (B) Graphical representation of the homology using dotplotting between PKS 4, 13, and 15 and PedI, PedH, and PedF, respectively.
The lab gene cluster of strain PHM005 comprises 3 putative ABC transporters, one cytochrome P450 and one 4′-phosphopantetheinyl transferase, which have not been identified in P. fuscipes or T. swinhoei. On the other hand, there are three proteins in the P. fuscipes symbiont, that are absent in Labrenzia sp. PHM005, i.e., a putative PedQ esterase, a PedR regulator and a protein of unknown function named PedK in P. fuscipes or OnnE and OnnF in T. swinhoei (Piel et al., 2004b). The tailoring enzymes in strain PHM005 showed protein identity < 60%, with the exception of the hydroxymethylglutaryl-CoA synthase (HMGS) and the oxygenase (OXY) which have 61 and 73% identity, respectively. It has been demonstrated that β-branching enzymes known as HMGS cassettes introduce alkyl side-chains into β-position and they usually comprise HMGS, ACP, ECH and stand-alone KS modules (Buchholz et al., 2010). Among the tailoring enzymes in lab cluster, there is a presence of HMGS, stand-alone ACP, KS and ECH, two putative acetyl-transferases (AT) and one oxidoreductase (OR).
Another difference regarding the ped cluster is the number of methyl-transferases (MT). Ped cluster described in the beetle symbiont comprises 3 MTs (PedA, PedE and PedO) while lab cluster in PHM005 strain has only two (MT6 and MT16). In comparison with pederin (4 O-methyl groups), labrenzin has only 3 O-methyl groups (see Figure 5A). Interestingly, labrenzin lacks the methylation at the terminal C18-OH group. Previous in vitro experimental analysis established that PedO is responsible for the methylation of the C18-OH group in pederin and it was hypothesized that PedA or PedE could have a dual function (Zimmermann et al., 2009). According to BLAST identity (Supplementary Table S8), MT16 shows 51% identity to PedE, while MT6 shows 47% and 43% identity to PedA and PedO, respectively. OnnG and OnnH from the onnamide cluster are homologous to PedA and PedE, respectively, but there is no Onn homolog for PedO (Piel et al., 2004b). Another close pederin analog, diaphorin, has only one O-methyl group because diaphorin gene cluster lacks the PedA and PedO orthologs (Nakabachi et al., 2013). Nonetheless, the BLAST identity between DipM and PedE is 60%.
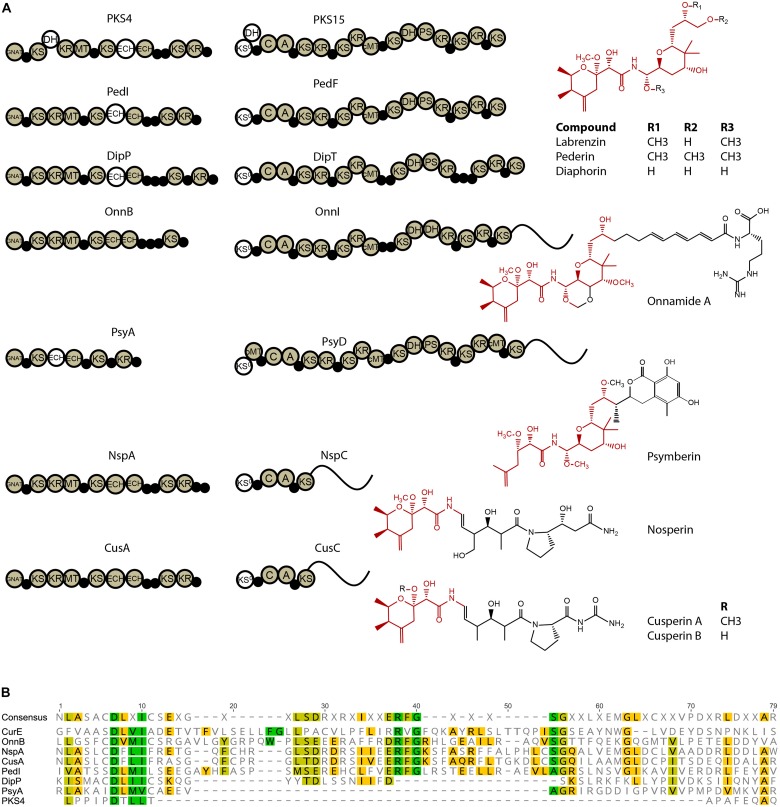
FIGURE 5.
(A) Modular scheme and domain comparison of PKS/NRPSs responsible for the biosynthesis of pederin family compounds isolated from organisms of different ecological niches: labrenzin, Labrenzia sp. PHM005; pederin, P. fuscipes symbiont; diaphorin, Profftella armatura; onnamide A, T. swinhoei symbiont; psymberin, Psammocinia sp. symbiont; nosperin, Nostoc sp.; cusperin, Cuspidothrix issatschenkoi (top to bottom). Common core structure of the respective polyketides is colored red. (B) Alignment (aa) of putative ECH1 domains from the first PKS in onnamide A, nosperin, cusperin, pederin, diaphorin, psymberin and labrenzin (top to bottom).
According to these comparisons and BLAST identities with PedA, PedE, and PedO, we suggest that MT16 and MT6 are functional analogs of PedE and PedA, respectively. In this case, MT16 might have a dual function as PedE and the lack of PedO analog results in the absence of methylation at the terminal C18-OH group.
The three PKS/NRPS genes show less than 50% identity, but conserving the modular repeats of the enzymes as visualized by dot-plot (Figure 4B).
PKS/NRPS Domain Comparison Between the Known Pederin Family Compounds
Apart from pederin and onnamide gene clusters mentioned above, other biosynthetic gene clusters of compounds from the pederin family, such as those described for the biosynthesis of diaphorin, psymberin, nosperin and cusperin were inspected for the PKS/NRPS domain architecture using the antiSMASH v.4.0 software (Figure 5A). The starting PKS responsible for the synthesis of the first pyran ring of pederin is conserved in all these compounds, except in the psymberin starting PKS which lacks the second module resulting in the absence of three C atoms. There is also a difference in the number of the tandem ACP domains found in the third module of the starting PKS.
In addition, the third module comprises two enoyl-CoA hydratase (ECH1 and ECH2) domains in OnnB, while the rest of the starting PKSs contain a single ECH domain based on antiSMASH analysis. The ECH domains can exist as separate enzymes, as a part of the HMGS-cassette, or as embedded domains in the PKSs, like ECH2 in curacin A (Liu et al., 2006). It is hypothesized that the ECH domains are responsible for the exomethylene group formation in the pederin pyran structure (Piel et al., 2004c). The amino acid sequence of the putative ECH1 domains in all 7 PKSs were aligned using as reference the characterized ECH1 functional domain (CurE) from the curacin A biosynthetic pathway (Liu et al., 2006) (Figure 5B). The OnnB, NspA, CusA, and PedI maintain the conserved amino acid motifs, while DipP, PsyA, and PKS4 have deleted the regions containing the conserved motifs of ECH1, suggesting that they might not be functional and that only one functional ECH domain is enough to generate the corresponding compounds.
Lab Cluster Expression Analysis, Functional Gene Analysis and Labrenzin Production
We have determined that Labrenzia sp. PHM005 can grow in the rich medium marine broth (MB) and the defined medium Marine Basal medium (MBM) containing glucose as carbon source and supplemented with vitamins as described in Experimental procedures (Figure 6). Labrenzin was detected by mass spectrometry in both media within the first 24 h of growth (Figure 6).
FIGURE 6.
Growth curves and extracted ion chromatograms of culture extracts. (A) Strain is grown in Marine Broth (MB) and Marine Basal Medium (MBM) supplemented with different vitamin mixtures as indicated in the legend. (B) MS chromatograms showing the labrenzin extracted from different media after 24 h of cultivation (from up to down): MB; MBM; MBM + VIT; MBM + B + T; MBM + Biotin; MBM + Thyamine; standard.
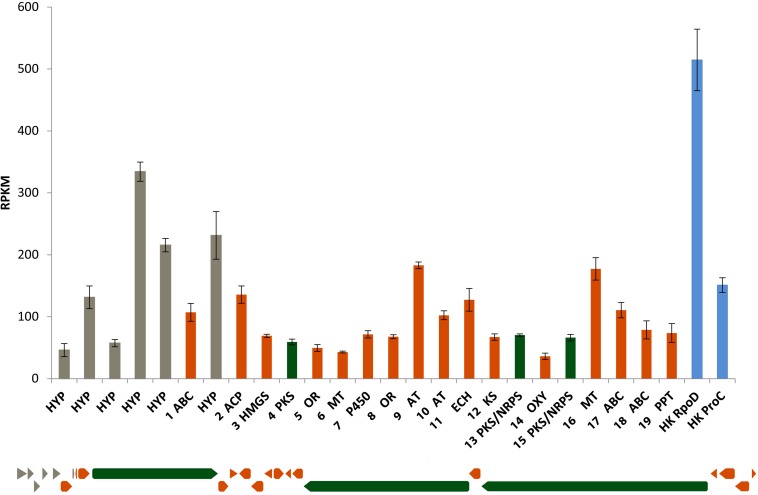
The MBM + vit medium was chosen for growing the bacterium to carry out the transcriptomics analysis. Figure 7 shows the expression levels of lab cluster genes and the housekeeping genes in the early exponential phase. All the lab cluster genes are expressed suggesting that these genes are activated during the exponential phase of growth and none of the lab cluster genes is completely silenced.
FIGURE 7.
Lab cluster gene expression levels in early exponential phase cultured in MBM + vit medium. The order of the bars follows the order of the genes (from left to right). The results represent the mean of three replicates. Blue bars represent housekeeping genes RpoD and ProC.
To investigate the association between the described gene cluster and the production of labrenzin we performed a gene knockout by homologous recombination where the sequence comprising the putative promoter region of the gene encoding PKS4 and the region encoding the GNAT domain of the first module of PKS4 was deleted to generate a non-producing mutant (ΔPKS4) (see Experimental procedures section).
The analyses of the culture extracts of the wild type and mutant strain are shown in Figures 8A–C. The MS ionization pattern of labrenzin, as purified standard or present in the culture extracts, is shown in the Supplementary Figure S5. As expected, the mutant strain was unable to produce labrenzin. The gene expression analysis performed with the mutant strain suggested that the deletion has produced a polar effect on the expression of the genes coding for putative oxidoreductase (5) and methyl-transferase (6) that are located downstream of the gene encoding PKS4, suggesting that the three genes are transcribed as an operon (Figure 8D).
FIGURE 8.
(A) Extracted ion chromatograms of the supernatant extracts obtained after 72 h of cultivation of Labrenzia sp. PHM005 in MBM + vit medium: (A) labrenzin standard; (B) wild type; (C) ΔPKS4. (D) RT-PCR analysis of the mRNA expression of lab gene cluster in the PHM005ΔPKS4 mutant cultured in MBM + vit medium. The numbers represent genes from the cluster. Each control lane contains a sample lacking a reverse transcriptase.
Discussion
The potent and selective bioactivities of pederin family compounds have triggered their pharmaceutical interest during the last years mainly as drug candidates for cancer therapy (Wan et al., 2011). The production of pederin analogs had been ascribed to non-cultivable endosymbiont bacteria until the recent discovery of a pederin analogs produced by the free-living bacterium Labrenzia sp. PHM005 (Schleissner et al., 2017). These compounds were only obtained after extraction from insects or marine organisms (Helfrich and Piel, 2016), and by chemical synthesis through a multi-step process due to its complex structure (Wu et al., 2011). The pederin and onnamide biosynthetic clusters described so far have been reconstructed using partial sequences obtained from metagenomics analyses and it had not been possible to fix precisely the boundaries of these clusters as well as to carry out regulatory and expression analyses. The discovery of the labrenzin produced by the cultivable strain Labrenzia sp. PHM005 has opened a new technical window not only to understand its biosynthesis and regulation, but also to apply biotechnological tools for industrial purposes and for developing large-scale production of these anti-cancer agents. In this sense, we have developed genetic tools to carry out metabolic engineering including the possibility of using for the first time in these genera replicative plasmids such as pSEVA428S and others pSEVA plasmids with different selection markers and replicative origins (data not shown). Therefore, this work has laid the foundations and expanded the tools to explore this organism.
Based on in silico genomic approaches we have identified and described a trans-AT PKS/NRPS cluster that is responsible for the synthesis of labrenzin a pederin analog in Labrenzia sp. PHM005. This lab cluster consists of three main PKSs/NPRSs genes complemented by several tailoring genes that might be responsible not only for the synthesis of labrenzin but also for the synthesis of an onnamide analog. Interestingly, the partially published sequence of the homologous onnamide gene cluster (onn cluster) found in the metagenome of the sponge T. swinhoei contains all the PKS genes in a single region (Piel et al., 2004b) like in the strain PHM005. It has been postulated that the diverse onnamide and pederin analogs produced by the sponge might be generated by the same onn cluster by using alternative synthetic mechanisms (Piel et al., 2004b). Since the lab cluster of strain PHM005 also contains the PKS/NRPS protein assigned to the biosynthesis of onnamide (Piel, 2002), it can be classified as a pederin and onnamide gene cluster. However, we have not detected onnamide analogs under our cultivation conditions so far.
Our data suggest that all the genes encoding the trans-AT PKS/NRPS of the cluster are expressed during the growth of Labrenzia sp. PHM005 and thus, it could be assumed that an onnamide analog should be synthesized. However, we cannot discard the possibility of an oxidative cleavage by OXY, a putative monooxygenase homologous to PedG, releasing the pederin intermediate as discussed previously (Piel, 2010). The putative monooxygenase is hypothesized to either cleave off the intermediate stalled at the terminus module of PedF, homologous to PKS15 in lab cluster, or process the already synthetized onnamide-like precursor. Furthermore, recent in vitro studies on SNAC and ACP derivatives demonstrated that PedC has proofreading activity in PKS systems and is able to hydrolase thioesters of different chain lengths (Dimitrova et al., 2017). The existence of the second AT in the lab cluster, homologous to PedC, newly denominated as acyl hydrolase (AH) by Dimitrova and co-workers could be another possible candidate for an intermediate release.
The lab cluster of Labrenzia sp. PHM005 contains only two MTs in contrast with the three MTs present in the cluster described by Piel (2002). This observation is consistent with the fact that labrenzin lacks the methylation at the terminal C18-OH and reinforces the hypothesis of Zimmermann et al. (2009), which suggested that PedO was responsible for the methylation of C18-OH and at least one of the two MTs, PedA or PedE, should have a dual function to be able to methylate two different OH groups. The functionality of tailoring enzymes of lab cluster is currently under study in our laboratory.
The biosynthesis of secondary metabolites is influenced by a wide variety of environmental and physiological signals, presumably reflecting the range of conditions that trigger their production in nature. Our data show that all genes from lab cluster are transcribed in our laboratory conditions at a detectable level with the sole exception of the gene encoding the HMGS enzyme. This apparent constitutive expression of the cluster could be explained with the absence of any specific regulator within the cluster or even closed to the cluster, in contrast with the finding in the ped cluster described in the P. fuscipes symbiont that codes for a putative regulator PedR belonging to the LysR family (Piel et al., 2004b).
The genetic manipulation of Labrenzia sp. PHM005 strain has been our first milestone toward the unraveling the biosynthesis of pederin and pederin-like compounds. We constructed a mutant strain deleting the promoter region and a GNAT module of the PKS4. The resulting strain was unable to produce pederin demonstrating for the first time that the lab cluster was certainly involved in the pederin biosynthesis. The genetic manipulation of Labrenzia sp. PHM005 opens the possibility of studying the pathway at a molecular level, not only to unravel the biosynthetic mechanisms for pederin synthesis but also to generate new pederin family analogs for pharmaceutical purposes.
There are remarkable challenges ahead in understanding the regulatory cascades that link environmental and developmental signals to pleiotropic and ultimately pathway-specific regulatory genes for secondary metabolism. The availability of the entire genome sequence of Labrenzia sp. PHM005 and the possibility to develop transcriptome analyses will definitively provide some clues to fully exploit the industrial potential of this strain.
Conclusion
The identification of the lab gene cluster responsible for the synthesis of labrenzin in the cultivable bacterium Labrenzia sp. PHM005 together with the development of the metabolic engineering tools described in this work will facilitate not only the understanding of the mechanisms involved in the biosynthesis of pederin and onnamide analogs that are extremely intriguing, but also the production by fermentation at industrial scale of these compounds that can be very useful to develop a new family of drugs based on one of the most active anticancer molecules described so far.
Data Availability Statement
The Whole Genome projects have been deposited at GenBank under the accession CP041191 (Labrenzia sp. PHM005) and CP041190 (p1BIR). The DNA sequencing data have been deposited at sequence read archive (SRA) under the accession SRS5035208.
Author Contributions
FC and JG conceived and designed the study. DK performed the experiments and analyzed the data. BG and CS participated in the experiment design. LC developed the HPLC-MS method and prepared the standard. PR extracted the DNA for PacBio sequencing. DK and JG wrote the manuscript. BG and CS contributed to preparing the final version of the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest
FC, LC, and PR were employed by the company PharmaMar S.A. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Virginia Rivero, laboratory colleague at the Centro de Investigaciones Biológicas (CSIC) for the technical support with LC–MS experiments. The technical work of Ana Valencia is greatly appreciated.
Footnotes
Funding. The present research was financed in part by a grant from the EU Horizon 2020 Project INMARE 634486 and by the Ministry of Economy and Competitiveness of Spain under the program RETOS-COLABORACIÓN with the project number RTC-2016-4892-1 (DESPOL). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02561/full#supplementary-material
Correction of a parted pederin family gene cluster sequence by Sanger sequencing.
The genome of Labrenzia sp. PHM005 annotated using the RAST. The green part in the bar chart at the leftmost position corresponds to the percentage of genes assigned to different subsystems. The pie chart demonstrates the percentage of each subsystem in Labrenzia sp. PHM005. The gene counts of the each subsystem is shown on the right.
Average nucleotide identity (ANI) comparison shows interspecies and intraspecies relationships of five strains in the Labrenzia genera. Identity scale: Green = 100%; orange > 95%; yellow ≥ 80%; gray < 80%.
Whole genome alignment using by progressive Mauve algorithm (Genious version 10.0.2), from up to the bottom: L. aggregata RMAR-6, Labrenzia sp. CP4, Labrenzia sp. VG12, Labrenzia sp. PHM005, L. alexandrii DFL-11.
Specific MS ionization pattern of labrenzin in culture extracts of the wild type PHM005 and the PHM005ΔPKS4 mutant after 72 h of cultivation in MBM + vit medium. (A) Pure labrenzin (standard) fragmentation patern at RT = 15,72 min. (B) Labrenzin fragmentation patern at RT = 15,72 min extracted from the wild type strain culture. (C) Background fragmentation patern at RT = 15,72 min extracted from the PHM005ΔPKS4 mutant strain culture.
Primers used for the cluster sequence correction.
Primers used in qPCR.
Coding sequences of plasmid p1BIR.
List of clusters homologous to those identified in Labrenzia sp. PHM005 by antiSMASH 4.0.
Homologous gene clusters in other microorganisms according to ClusterBlast by antiSMASH 4.0.
Domain specificity of the three PKS and mixed PKS/NRPS.
Four Labrenzia genomes aligned to Labrenzia sp. PHM005 using progressive Mauve algorithm individually.
BLAST homology of the Labrenzia sp. PHM005 trans- AT PKS gene cluster.
References
- Aziz R. K., Bartels D., Best A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Baumann L. (1981). The Prokaryotes: A Handbook on Habitats, Isolation and Identification of Bacteria. Berlin: Springer. [Google Scholar]
- Buchholz T. J., Rath C. M., Lopanik N. B., Gardner N. P., Håkansson K., Sherman D. H. (2010). Polyketide β-branching in bryostatin biosynthesis: identification of surrogate acetyl-ACP donors for BryR, an HMG-ACP synthase. Chem. Biol. 17 1092–1100. 10.1016/j.chembiol.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañedo M., de la calle F., Rodriguez P., Schleissner C., Zuñiga P. (2018). Anticancer compounds. Pub. No.: WO 2018167270; International Application No.: PCT/EP2018056665. [Google Scholar]
- Cardani C., Ghiringhelli D., Mondelli R., Quilico A. (1965). The structure of Pederin. Tetrahedron Lett. 6 2537–2545. 10.1016/s0040-4039(01)84020-x [DOI] [Google Scholar]
- Dimitrova N., Zamudio J. R., Jong R. M., Soukup D., Resnick R., Sarma K., et al. (2017). Polyketide proofreading by an acyltransferase-like enzyme. PLoS One 32:178059. 10.1371/journal.pone.0178059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed A. K., Hothersall J., Cooper S. M., Stephens E., Simpson T. J., Thomas C. M. (2003). Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10 419–430. 10.1016/s1074-5521(03)00091-7 [DOI] [PubMed] [Google Scholar]
- Fisch K. M., Gurgui C., Heycke N., van der Sar S. A., Anderson S. A., Webb V. L., et al. (2009). Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 5 494–501. 10.1038/nchembio.176 [DOI] [PubMed] [Google Scholar]
- Frank J. H., Kanamitsu K. (1987). Paederus, sensu lato (Coleoptera: Staphylinidae): natural history and medical importance. J. Med. Entomol. 24 155–191. 10.1093/jmedent/24.2.155 [DOI] [PubMed] [Google Scholar]
- Fusetani N., Sugawara T., Matsunaga S. (1992). Theopederins A-E, potent antitumor metabolites from a marine Sponge, Theonella sp. J. Org. Chem. 57 3828–3832. 10.1021/jo00040a021 [DOI] [Google Scholar]
- Garvey P., Fitzgerald G. F., Hill C. (1995). Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61 4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Helfrich E. J. N., Piel J. (2016). Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 33 231–316. 10.1039/c5np00125k [DOI] [PubMed] [Google Scholar]
- Kampa A., Gagunashvili A. N., Gulder T. A. M., Morinaka B. I., Daolio C., Godejohann M., et al. (2013). Metagenomic natural product discovery in lichen provides evidence for a family of biosynthetic pathways in diverse symbioses. Proc. Natl. Acad. Sci. U.S.A. 110 E3129–E3137. 10.1073/pnas.1305867110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner R. L. L. (2002). Molecular identification of an endosymbiotic bacterium associated with pederin biosynthesis in Paederus sabaeus (Coleoptera: Staphylinidae). Insect Biochem. Mol. Biol. 32 389–395. 10.1016/S0965-1748(01)00115-1 [DOI] [PubMed] [Google Scholar]
- Kellner R. L. L., Dettner K. (1996). Differential efficacy of toxic pederin in deterring potential arthropod predators of Paederus (Coleoptera: Staphylinidae) offspring. Oecologia 107 293–300. 10.1007/BF00328445 [DOI] [PubMed] [Google Scholar]
- Kust A., Mareš J., Jokela J., Urajová P., Hájek J., Saurav K., et al. (2018). Discovery of a pederin family compound in a nonsymbiotic bloom-forming Cyanobacterium. ACS Chem. Biol. 13 1123–1129. 10.1021/acschembio.7b01048 [DOI] [PubMed] [Google Scholar]
- Liu H., Gerwick W. H., Sherman D. H., Jia J., Håkansson K., Gu L. (2006). Metabolic coupling of dehydration and decarboxylation in the curacin a pathway: functional identification of a mechanistically diverse enzyme pair. J. Am. Chem. Soc. 128 9014–9015. 10.1021/ja0626382 [DOI] [PubMed] [Google Scholar]
- Martínez-García E., de Lorenzo V. (2011). Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13 2702–2716. 10.1111/j.1462-2920.2011.02538.x [DOI] [PubMed] [Google Scholar]
- Matsunaga S., Fusetani N., Nakao Y. (1992). Eight new cytotoxic metabolites closely related to onnamide A from two marine sponges of the genus Theonella. Tetrahedron 48 8369–8376. 10.1016/S0040-4020(01)86585-6 [DOI] [Google Scholar]
- Nakabachi A., Ueoka R., Oshima K., Teta R., Mangoni A., Gurgui M., et al. (2013). Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 23 1478–1484. 10.1016/j.cub.2013.06.027 [DOI] [PubMed] [Google Scholar]
- Pannell L. K., Perry N. B., Blunt J. W., Munro M. H. (1988). Mycalamide a, an antiviral compound from a New Zealand sponge of the genus Mycale. J. Am. Chem. Soc. 110 4850–4851. 10.1021/ja00222a067 1797440 [DOI] [Google Scholar]
- Pavan M., Bo G. (1952). Ricerche sulla differenziabilita, natura e attivita del principio tossico di Paederus fuscipes Curt.(Col. Staph.). Mem. Soc. Entomol. Ital. 31 67–82. [Google Scholar]
- Piel J. (2002). A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci U.S.A. 99 14002–14007. 10.1073/pnas.222481399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J. (2010). Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 27 996–1047. 10.1039/c5np00125k [DOI] [PubMed] [Google Scholar]
- Piel J., Butzke D., Fusetani N., Hui D., Platzer M., Wen G., et al. (2005). Exploring the chemistry of uncultivated bacterial symbionts: antitumor polyketides of the Pederin family. J. Nat. Prod. 68 472–479. 10.1021/np049612d [DOI] [PubMed] [Google Scholar]
- Piel J., Hui D., Fusetani N., Matsunaga S. (2004a). Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ. Microbiol. 6 921–927. 10.1111/j.1462-2920.2004.00531.x [DOI] [PubMed] [Google Scholar]
- Piel J., Hui D., Wen G., Butzke D., Platzer M., Matsunaga S., et al. (2004b). Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. PNAS 101 16222–16227. 10.1073/pnas.0405976101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J., Wen G., Platzer M., Hui D. (2004c). Unprecedented diversity of catalytic domains in the first four modules of the putative pederin polyketide synthase. Chembiochem 5 93–98. 10.1002/cbic.200300782 [DOI] [PubMed] [Google Scholar]
- Richter A., Kocienski P., Raubo P., Davies D. (1997). The in vitro biological activities of synthetic 18-O-methyl mycalamide B, 10-epi-18-O-methyl mycalamide B and pederin. Anticancer Drug Des. 12 217–227. [PubMed] [Google Scholar]
- Rodriguez-R L. M., Konstantinidis K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 4:e1900v1 10.7287/peerj.preprints.1900v1 [DOI] [Google Scholar]
- Roth J. R., Lawrence J. G. (1996). Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143 1843–1860. 10.1261/rna.130506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakemi S., Ichiba T., Kohmoto S., Saucy G., Higa T. (1988). Isolation and structure elucidation of onnamide A, a new bioactive metabolite of a marine sponge. Theonella sp. J. Am. Chem. Soc. 110 4851–4853. 10.1021/ja00222a068 [DOI] [Google Scholar]
- Schleissner C., Cañedo L. M., Rodríguez P., Crespo C., Zúñiga P., Peñalver A., et al. (2017). Bacterial production of a pederin analogue by a free-living marine alphaproteobacterium. J. Nat. Prod. 80 2170–2173. 10.1021/acs.jnatprod.7b00408 [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R., Martínez-García E., Calles B., Chavarría M., Arce-Rodríguez A., de Las Heras A., et al. (2013). The standard european vector architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41 666–675. 10.1093/nar/gks1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati M., Fioretti A., Ghione M. (1966). Cytotoxicity of pederin and some of its derivatives on cultured mammalian cells. Experientia 22 176–178. 10.1007/BF01897720 [DOI] [PubMed] [Google Scholar]
- Stothard P., Wishart D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21 537–539. 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- Vander Wood D. A., Keatinge-Clay A. T. (2018). The modules of trans-acyltransferase assembly lines redefined with a central acyl carrier protein. Proteins Struct. Funct. Bioinform. 86 664–675. 10.1002/prot.25493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S., Wu F., Rech J. C., Green M. E., Balachandran R., Horne W. S., et al. (2011). Total synthesis and biological evaluation of pederin, psymberin, and highly potent analogs. J. Am. Chem. Soc. 133 16668–16679. 10.1021/ja207331m [DOI] [PubMed] [Google Scholar]
- Wu F., Green M. E., Floreancig P. E. (2011). Total synthesis of pederin and analogues. Angew. Chemi. Int. Ed. Engl. 50 1131–1134. 10.1002/anie.201006438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann K., Engeser M., Blunt J. W., Munro M. H. G., Piel J. (2009). Pederin-type pathways of uncultivated bacterial symbionts: analysis ofO-methyltransferases and generation of a biosynthetic hybrid. J. Am. Chem. Soc. 131 2780–2781. 10.1021/ja808889k [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correction of a parted pederin family gene cluster sequence by Sanger sequencing.
The genome of Labrenzia sp. PHM005 annotated using the RAST. The green part in the bar chart at the leftmost position corresponds to the percentage of genes assigned to different subsystems. The pie chart demonstrates the percentage of each subsystem in Labrenzia sp. PHM005. The gene counts of the each subsystem is shown on the right.
Average nucleotide identity (ANI) comparison shows interspecies and intraspecies relationships of five strains in the Labrenzia genera. Identity scale: Green = 100%; orange > 95%; yellow ≥ 80%; gray < 80%.
Whole genome alignment using by progressive Mauve algorithm (Genious version 10.0.2), from up to the bottom: L. aggregata RMAR-6, Labrenzia sp. CP4, Labrenzia sp. VG12, Labrenzia sp. PHM005, L. alexandrii DFL-11.
Specific MS ionization pattern of labrenzin in culture extracts of the wild type PHM005 and the PHM005ΔPKS4 mutant after 72 h of cultivation in MBM + vit medium. (A) Pure labrenzin (standard) fragmentation patern at RT = 15,72 min. (B) Labrenzin fragmentation patern at RT = 15,72 min extracted from the wild type strain culture. (C) Background fragmentation patern at RT = 15,72 min extracted from the PHM005ΔPKS4 mutant strain culture.
Primers used for the cluster sequence correction.
Primers used in qPCR.
Coding sequences of plasmid p1BIR.
List of clusters homologous to those identified in Labrenzia sp. PHM005 by antiSMASH 4.0.
Homologous gene clusters in other microorganisms according to ClusterBlast by antiSMASH 4.0.
Domain specificity of the three PKS and mixed PKS/NRPS.
Four Labrenzia genomes aligned to Labrenzia sp. PHM005 using progressive Mauve algorithm individually.
BLAST homology of the Labrenzia sp. PHM005 trans- AT PKS gene cluster.
Data Availability Statement
The Whole Genome projects have been deposited at GenBank under the accession CP041191 (Labrenzia sp. PHM005) and CP041190 (p1BIR). The DNA sequencing data have been deposited at sequence read archive (SRA) under the accession SRS5035208.