Abstract
We used real-time polymerase chain reaction (PCR) targeting the cdc2 gene and direct fluorescent microscopy examination (DFME) to evaluate the prevalence of Pneumocystis jirovecii among immunocompetent patients without clinical pulmonary infection and immunosuppressed patients evaluated for opportunistic pulmonary infections. Among 102 bronchoalveolar lavage samples collected from immunocompetent patients without infection, none tested positive for P. jirovecii by either DFME or real-time PCR despite the presence of other comorbidities. Among patients with suspected pulmonary infection and tested with either assay, real-time PCR produced a higher number of positive results compared to DFME and increased P. jirovecii detection by 7% when added to DFME-negative samples. Real-time PCR may have increased sensitivity for P. jirovecii detection over DFME and decrease the risk of sample contamination compared to conventional and nested PCR. The use of single-copy gene targets (e.g., cdc2) may lower the rate of “colonization” detection and confer a high predictive value for Pneumocystis pneumonia.
Keywords: P. jirovecii, Pneumocystis pneumonia, Real-time PCR, Direct examination
1. Introduction
Pneumocystis jirovecii is a ubiquitous and low-virulent fungus that can produce significant respiratory disease in the immunocompromised host. Pneumocystis pneumonia (PCP) is well described in patients with immunologically advanced HIV infection but is also encountered in other immunosuppressed patient groups including those with select hematologic malignancies, solid organ and bone marrow transplantations, and those prescribed chronic steroids or other cell-mediated immunity suppressive therapies (Russian and Levine, 2001; Sowden and Carmichael, 2004). P. jirovecii has global distribution, and most people have serologic evidence of infection during early childhood (Meuwissen et al., 1977; Respaldiza et al., 2004; Vargas et al., 2001). PCP, however, is characteristically unusual in the immunocompetent host (Kovacs and Masur, 2009).
The conventional laboratory and current “gold-standard” method to identify P. jirovecii is direct microscopic examination of stained respiratory smears and tissue specimens. Gomori methenamine silver, cresyl echt violet, toluidine blue-O, modified Wright-Giemsa (Diff-Quik), Gram-Weigert, Papanicolaou, calcofluor white, and immunofluorescent stains are commonly used to identify the cystic and trophic forms of the organism (Limper, 1996). While direct microscopic examination provides a fairly specific means to identify the organism, it lacks reliable sensitivity to exclude infection.
Direct microscopy offers a rapid and inexpensive approach for P. jirovecii detection; however, polymerase chain reaction (PCR) has demonstrated a 13–21% higher sensitivity (Arcenas et al., 2006; Caliendo et al., 1998; Cartwright et al., 1994; Eisen et al., 1994) and provides for a more objective interpretation of results. PCR assays are not standardized, however, and vary significantly in their detection technology, nucleic acid target, primer-probe sequences, turnaround time, and capacity to yield a qualitative versus quantitative result. Additional variables affecting organism detection include the type of clinical sample [e.g., sputum versus bronchoalveolar lavage (BAL)], as well as the laboratory experience in performing the testing.
P. jirovecii has occasionally been detected by PCR in respiratory samples from immunocompetent patients without evidence of pulmonary infection. Often labeled airway “colonization”, the prevalence of P. jirovecii infection without disease remains unclear, and concerns for false-positive results generated by some PCR assays complicate the interpretation of previous reports. Conventional and nested PCR platforms incorporate an “open system” of amplicon procurement. Tubes containing amplicon are opened during the detection phase and for subsequent rounds of PCR, raising the potential for sample cross-contamination. Real-time PCR with the Roche LightCycler™ (Roche Applied Sciences, Indianapolis, IN) assay utilizes a “closed-system” with a carousel-based thermal cycler platform and fluorescence detection system. No tubes are opened, minimizing the potential for cross-sample contamination (Arcenas et al., 2006). While there is more experience using open conventional and nested PCR assays, we postulated that positive results generated by real-time PCR may more directly correlate to PCP and could be used in place of direct microscopy.
2. Materials and methods
2.1. Study design
Our study analyzed respiratory samples from 2 separate patient populations by real-time PCR and direct fluorescent microscopy examination (DFME) to detect P. jirovecii. One set of samples came from immunocompetent patients undergoing bronchoscopy for noninfection indications. The other set was collected from immunosuppressed patients evaluated for suspected respiratory tract infections, including PCP. An analysis of the patient’s medical records from our center was performed to determine the presence of clinical PCP if the results of DFME and PCR were discordant. Our objectives were to (a) identify the P. jirovecii rate of detection among immunocompetent patients without clinical PCP by real-time PCR and DFME; (b) compare rates of test positivity on respiratory samples from immunosuppressed patients with suspected pulmonary infection, including PCP; and (c) determine the likelihood of clinical PCP among a subgroup of immunosuppressed patients with discordant assay results. The study was approved by the IRB at our medical center.
2.1.1. Prospective P. jirovecii testing of immunocompetent patients without PCP undergoing bronchoscopy
2.1.1.1. Patient enrollment criteria for bronchoscopy and data collection.
The inclusion and exclusion criteria to define immunocompetence and other factors for study enrollment of adult patients are listed in Table 1. All patients enrolled were 18 years or older and could not have a defined immunosuppressive condition before bronchoscopy. Enrolled patients were required to have a clinically indicated bronchoscopy scheduled for a noninfection indication and have no symptoms of a respiratory tract infection or prior diagnosis of PCP. With patient written consent, a BAL was added to each patient’s bronchoscopic procedure.
Table 1.
Inclusion and exclusion criteria for immunocompetent patients undergoing BAL
| Inclusion criteria |
|
| Exclusion criteria |
|
All patients were tested for HIV infection at the time of bronchoscopy by a serum chemiluminescence immunoassay (CIA) (VITROS Anti-HIV 1+2, Ortho Clinical Diagnostics Inc., Raritan, NJ) and excluded if they were either HIV-positive or had received an antimicrobial with anti-Pneumocystis activity within the past year.
2.1.1.2. Bronchoscopy with BAL.
Whenever possible, the BAL specimen was obtained from the unaffected region of the lung as indicated for bronchoscopy. Ten aliquots (10 mL each) of normal saline for a total of 100 mL were instilled with the bronchoscope wedged in the fourth-order bronchus to sample the distal airspaces of the radiographically normal lung. Typically, the right middle lobe or the lingula was lavaged. The BAL fluid was recovered by immediate return suction. The recovered BAL specimens were separated into cellular and fluid components by centrifugation at 600 × g for 10 min, as described previously (Limper et al., 1993). All BAL components were stored at −20 °C until PCR testing.
2.1.1.3. Laboratory methods
2.1.1.3.0. Direct examination for P. jirovecii.
A 100-μL aliquot of the BAL fluid was concentrated using a cytospin procedure. The slide was stained with a KOH and calcofluor white solution and analyzed by fluorescent microscopy.
2.1.1.3.0. LightCycler PCR testing.
Real-time PCR detection of P. jirovecii was performed using the LightCycler™ 2.0 platform PCR as previously described (Arcenas et al., 2006) with slight modifications to the extraction procedure. In brief, 500 μL of the raw BAL and 100 μL proteinase K (Roche Applied Sciences) were pipetted into a 1.5-mL tube containing 0.1-mm silica glass beads and 2.4-mm zirconia beads (Biospec Products). Samples were then incubated at 55 °C for 15 min on a Thermo-mixer (Eppendorf) at 1400 rpm and subsequently placed on a 95 °C heat block for 5 min. To facilitate complete lysis and nucleic acid liberation, samples were placed on a Disruptor Genie (Scientific Industries, Bohemia, NY) for 2 min and then centrifuged for 5–10 s at 20 800 × g (14 000 rpm) to collect the sample at the bottom of the tube. Two hundred microliters of the prepared lysate was pipetted into a sample tube for MagNa Pure Compact extraction (Roche Applied Sciences) utilizing the Nucleic Acid Isolation Kit (small volume), Total NA Plasma program, and a final elution volume of 100 μL. Following extraction, 5 μL of sample and 15 μL PCR master-mix were tested on the LightCycler PCR platform to amplify a 166-bp region of the cdc2 gene of P. jirovecii. The PCR platform utilizes a melting curve analysis to determine positive and negative results. Assay performance characteristics with the cdc2 real-time PCR were previously established (Arcenas et al., 2006).
2.1.2. Retrospective review of P. jirovecii test results submitted from immunosuppressed patients with suspected pulmonary infection
We performed a retrospective review of all patients tested at our center by DFME and/or real-time PCR for P. jirovecii between 2005 and 2007. All samples were obtained from patients with presumptive immunosuppressive conditions and submitted by providers suspecting pulmonary infection. Patient samples submitted from outside our center had specific P. jirovecii testing requests while those from within our center mostly had P. jirovecii testing done as part of an “immunocompromised host” panel of preselected tests. Samples included sputum, BAL fluid, bronchial washings, tracheal secretions, and direct lung tissue.
Test results were analyzed from all respiratory samples as well from a BAL fluid subgroup. Patients with both negative DFME and PCR results were considered to not have either P. jirovecii infection or colonization. The medical records of patients at our medical center that tested negative by DFME but positive by PCR were reviewed to determine the clinical likelihood for PCP.
3. Results
3.1. P. jirovecii testing of BAL fluid from immunocompetent patients without infection
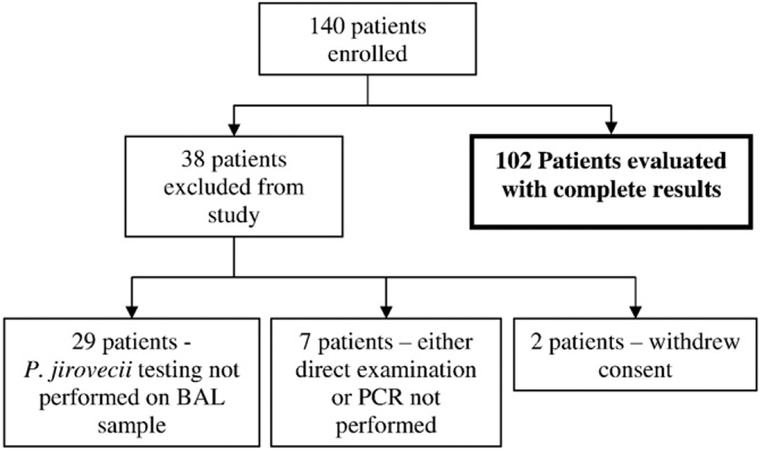
A total of 140 patients were enrolled from March 2005 through May 2007. Thirty-eight patients were subsequently excluded for the reasons listed in Fig. 1. There were 102 remaining patients who met the study enrollment criteria and had complete Pneumocystis and HIV testing performed.
Fig. 1.
Reasons for subsequent patient exclusions from study.
Table 2 lists the indications for bronchoscopy and subsequent diagnoses of enrolled patients. The most common indications for bronchoscopy included mediastinal and/or hilar lymphadenopathy (38%), pulmonary mass or nodules (36%), and chronic infiltrates seen on chest x-ray or CT scan (20%).
Table 2.
Bronchoscopy indications and subsequent diagnoses of enrolled immunocompetent patients
| No. of patients (%) | |
|---|---|
| Indication for bronchoscopy: | |
| Mediastinal/hilar lymphadenopathy | 39 (38%) |
| Pulmonary mass or nodules | 37 (36%) |
| Chronic infiltrate | 20 (20%) |
| Endobronchial lesion/stricture | 3 (3%) |
| Hemoptysis | 2 (2%) |
| Tracheomalacia | 1 (1%) |
| Total | 102 |
| Post-bronchoscopy diagnosis: | |
| Primary or metastatic pulmonary lung cancer | 52 (51%) |
| Indeterminate diagnosis or unknown | 28 (27%) |
| Sarcoidosis | 14 (14%) |
| Nonspecific interstitial lung disease or Interstitial lung fibrosis | 5 (5%) |
| Aseptic organizing pneumonia | 2 (2%) |
| Wegener granulomatosis | 1 (1%) |
| Total | 102 |
| Other: | |
| Presence of documented COPDa | 26 (25%) |
| Active or recentb smoker | 41 (40%) |
Documentation of COPD obtained from pre-bronchoscopy evaluation in medical record; authors suspect prevalence of COPD in study population to be higher than documented.
Recent smoker defined as smoking within the previous 10 years of bronchoscopy.
The most common post-bronchoscopy diagnoses included primary or metastatic lung cancers (51%), indeterminate or unknown pulmonary diagnoses (27%), pulmonary sarcoidosis (14%), and nonspecific interstitial lung disease or pulmonary fibrosis (5%). Twenty-five percent of enrolled patients had documented COPD and 40% had a current or recent history of smoking.
Of these 102 patients undergoing bronchoscopy and BAL for noninfectious indications, all 102 samples (100%) tested negative for P. jirovecii by both DFME and real-time PCR. There were no discordant results between the 2 assays on any of the enrolled patients.
3.2. P. jirovecii testing of respiratory samples from immunosuppressed patients evaluated for pulmonary infection
From January 2005 through May 2007, a total of 3271 respiratory samples from patients evaluated for pulmonary infection including PCP were tested in our laboratory by DFME and/or real-time PCR (Table 3). The respiratory samples included BAL fluid (n = 1422), bronchial washings (n = 634), tracheal secretions (n = 45), direct lung tissue (n = 156), induced sputum (n = 812), and other miscellaneous (nonspecified) sources (n = 202). DFME was the only Pneumocystis test performed on 2145 patients, and 44 (2%) of these were positive. Real-time PCR testing was the only Pneumocystis test performed on 734 patients, and 101 (14%) were positive. A subgroup of 392 patient respiratory samples that tested negative by DFME subsequently had PCR added. From this subgroup, 363 (93%) tested negative by both assays, but 29 (7%) tested positive for P. jirovecii by real-time PCR alone.
Table 3.
Results of real-time PCR and DFME on respiratory samples from patients evaluated for pulmonary infection, including PCP, from January 2005 through May 2007a
| Description | Number of tests |
|---|---|
| Testing performed from all respiratory sourcesb | 3271 |
| DFME only: | 2145 total |
| Negative | 2101 (98%) |
| Positive | 44 (2%) |
| Real-time PCR only: | 734 total |
| Negative | 633 (86%) |
| Positive | 101 (14%) |
| Both tests done on same samplec: | 392 total |
| DFME negative, PCR negative | 363 (93%) |
| DFME negative, PCR positive | 29 (7%) |
| Testing performed from BAL fluid only | 1422 |
| DFME only: | 934 total |
| Negative | 910 (97%) |
| Positive | 24 (3%) |
| Real-time PCR only: | 156 total |
| Negative | 140 (90%) |
| Positive | 16 (10%) |
| Both tests done on same samplec: | 332 total |
| DFME negative, PCR negative | 311 (94%) |
| DFME negative, PCR positive | 21 (6%) |
Patient samples collected from both Mayo Clinic Rochester and Mayo Medical Laboratories.
Respiratory sources include sputum, tracheal secretions, BAL, and bronchial washings.
These samples all tested negative by DFME and subsequently had PCR added.
Among the 1422 (43%) BAL samples tested, DFME was the only Pneumocystis test performed on 934 samples, and 24 (3%) were positive. Real-time PCR testing was the only Pneumocystis test performed on 156 patient samples, and 16 (10%) were positive. A subgroup of 332 patient BAL fluid samples that tested negative by DFME subsequently had PCR added. From this subgroup, 311 (94%) of the samples tested negative by both assays, but 21 (6%) samples tested positive for P. jirovecii by real-time PCR alone.
Five percent of all respiratory samples (174 of 3271) and 4% of the BAL subgroup samples (61 of 1422) tested positive for P. jirovecii using either method. Of the samples tested by either DFME or PCR, there proportionally were more positive results by real-time PCR testing than by DFME (14% versus 2%, respectively); however, direct comparisons between both groups could not be made as most samples were not tested by both methods and the medical records were not available for all patients to confirm clinical disease.
Twenty-seven of the 29 respiratory samples with a negative DFME and subsequent positive PCR result came from patients evaluated at our medical center, and the medical records of these patients were reviewed. Table 4 lists the patient immunosuppressive conditions and the types of specimens tested. Hematologic malignancy/condition was the most common underlying medical condition (n = 9; 33%), and BAL was the most common type of sample tested (n = 18; 67%). Among these 27 patients with discordance test results, 21 (78%) had definitive or probable PCP (P. jirovecii determined to be the primary cause of pulmonary disease based on the positive PCR test result with corresponding clinical and radiologic findings, the absence of an alternative diagnosis, and the clinical decision to treat for PCP) (Table 5). Four patients (15%) had possible PCP (not able to clinically confirm or exclude P. jirovecii contribution to disease, but patients were treated for PCP). Two patients (7%) had an unlikely diagnosis of PCP (P. jirovecii determined not the cause of pulmonary syndrome, an alternative diagnosis was made, no PCP treatment given and patients remained clinically stable or improved). Given the variable immunosuppressive medical conditions and associated therapies, objective immunologic comparisons between the patients could not accurately be determined.
Table 4.
Patient conditions and sample sources among discordant negative DFME and positive PCR resultsa
| Patient no. n = 27 (%) | |
|---|---|
| Primary patient medical condition | |
| Hematologic malignancy and other hematologic disordersb | 9 (33%) |
| Chronic lung disease with immunosuppressive drug therapyc | 5 (19%) |
| Solid organ malignancyd | 4 (15%) |
| Active connective tissue diseasee | 4 (15%) |
| Kidney/pancreas transplantation | 3 (11%) |
| HIV infection/AIDS | 2 (7%) |
| Specimen source | |
| BAL | 18 (67%) |
| Induced sputum | 6 (22%) |
| Tracheal secretions | 2 (7%) |
| Lung tissue/biopsy | 1 (4%) |
Patients and samples from Mayo Clinic Rochester, MN. Note 2 of the 29 total patient samples with discordant test results from Table 3 are omitted as they were not patients at the Mayo Clinic and their records could not be reviewed.
Hematologic malignancies and other hematologic disorders: chronic lymphocytic leukemia (n = 3), multiple myeloma (n = 2), non-Hodgkin lymphoma (n = 1), Hodgkin disease (n = 1), acute myelogenous leukemia (n = 1), and aplastic anemia (n = 1).
Chronic lung disease with immunosuppressive therapy: interstitial lung disease (n = 2), bronchiolitis obliterans (n = 1), steroid-dependent asthma (n = 1) and idiopathic constrictive bronchiolitis (n = 1).
Solid organ malignancies: invasive breast carcinoma (n = 2), glioblastoma multiforme (n = 1), and recurrent pontine glioma (n = 1).
Active connective tissue diseases: Goodpasture disease (n = 2), rheumatoid arthritis (n = 1), and polymyalgia rheumatica (n = 1).
Table 5.
Analysis of discordant DFME and PCR results by specimen type
| Clinical likelihood for PCP in patients with discordant P. jirovecii test results |
Sputum (n = 6) | Tracheal secretions (n = 2) | Lung tissue (n = 1) | BAL (n = 18) | Total (n = 27) |
|---|---|---|---|---|---|
| Definite or probableb | 4 (67%) | 2 (100%) | 1 (100%) | 14 (78%) | 21 (78%) |
| Possiblec | 1 (17%) | 0 (0%) | 0 (0%) | 3 (17%) | 4 (15%) |
| Unlikelyd | 1 (17%) | 0 (0%) | 0 (0%) | 1 (6%) | 2 (7%) |
| 100% | 100% | 100% | 100% | 100% |
Discordant samples defined as those with initial negative DFME and subsequent positive PCR.
Definite or probable PCP—P. jirovecii determined to be the primary cause of pulmonary disease based on the positive PCR test result with corresponding clinical and radiologic findings, the absence of an alternative diagnosis, and the decision to treat for PCP.
Possible PCP—not able to clinically confirm or exclude P. jirovecii contribution to pulmonary disease, but patients were treated for PCP.
Unlikely PCP—P. jirovecii determined not to cause the pulmonary syndrome. Patients did not receive treatment for PCP and remained stable or improved.
To determine whether there is a correlation between PCR cycle threshold (Ct) values and smear positivity, we retrospectively reviewed the Ct values from patients tested in 2010 by both PCR and direct smear examination (n = 212; data not shown). This analysis indicated that there was a trend toward lower Ct values (higher concentration of target DNA) and concordant positive smear results. The number of specimens with both smear and PCR results, however, was too small (n = 15) to allow for an accurate correlation to be made between Ct and smear positivity.
4. Discussion
The actual prevalence of P. jirovecii colonization or subclinical infection in immunocompetent patients remains unclear as study results have been conflicting. Early studies using conventional PCR assays were able to detect P. jirovecii from induced sputa, oropharyngeal washes, and BAL of patients with clinically suspected PCP but not from smaller numbers of immunocompetent patients with alternative diagnoses (Oz and Hughe, 2000; Wakefield et al., 1990, 1993). Some clinical laboratories subsequently use nested PCR to improve assay sensitivity for organism detection and found variable positivity rates (0–20%) for P. jirovecii in sputum and oropharyngeal wash samples from immunocompetent patients, including some with chronic lung disease (Medrano et al., 2005; Nevez et al., 1999; Sing et al., 2001; Vargas et al., 2010). Other studies using nested PCR on BAL samples from immunocompetent patients with chronic lung disease and/or steroid use but without clinical PCP have shown an 18–20% detection rate of P. jirovecii (Maskell et al., 2003; Nevez et al., 1997; Sing et al., 1999).
Khan et al. compared immunofluorescence staining to both conventional and nested PCR from BAL and sputum samples of immunosuppressed patients evaluated for PCP and found a progressively higher percentage of positive test results (4.3%, 23.9%, and 45.6%, respectively) by these test methods (Khan et al., 1999). Of interest, the highest percentage of positive tests (29%) from patients without PCP in this study also came from nested PCR testing. Olsson et al. (2001) found a higher sensitivity by nested PCR compared to immunofluorescence staining for PCP diagnoses in non-HIV immunosuppressed patients but a much lower specificity (59% and 97%, respectively) from BAL and sputum samples. Interestingly, Ponce et al. (2010) identified P. jirovecii using nested PCR directly from lung tissue in over 60% of immunocompetent patients tested during autopsy, raising the question whether subclinical or self-limiting P. jirovecii infections are more common that previously thought.
Concerns for sample contamination using nested PCR have led to the recent development of real-time PCR for the detection of P. jirovecii. Alvarez-Martinez et al. (2006) compared nested and real-time PCR assays using primers for the dihydropteroate synthase gene from HIV-positive patients with PCP confirmed by direct examination and found comparative sensitivities of 94% for both PCR assays but a higher specificity for real-time PCR (96% versus 81%). The higher false-positive rate for nested PCR (19%) compared to real-time PCR (4%) was postulated to be a consequence of a higher likelihood for sample contamination using nested PCR. Huggett et al. (2008) compared real-time PCR using primers for the heat shock protein 70 gene with conventional PCR using primers for mitochondrial large subunit rRNA gene (mtLSUrRNA) in HIV-infected patients and also found comparative sensitivity for P. jirovecii detection between both assays (97–98%) but a higher specificity for clinical PCP with real-time PCR testing (96% versus 68%). However, Robberts et al. (2007) compared 9 PCR assays using a variety of primers/targets in both nested and (non-cdc2) real-time PCR assays and found a slightly higher sensitivity and specificity using mtLSUrRNA nested PCR. Thus, the performance interpretation among different PCR assays remains challenging.
In our study, we used both cdc2 real-time PCR and DFME and did not detect P. jirovecii from any of 102 enrolled HIV-negative immunocompetent patients who underwent bronchoscopy for noninfection indications. Although considered immunocompetent by the exclusion criteria listed in Table 1, most of our patients did have significant immunomodulatory comorbidities identified after bronchoscopy and/or structural lung disease affiliated with a higher potential for P. jirovecii detection. Despite these comorbidities, and coupled with a relative comparative sensitivity of real-time PCR to other PCR assays, the absence of any positive P. jirovecii test result from this patient group raises the question whether prevalence of P. jirovecii airway colonization is lower or perhaps more variable depending on specific patient populations than previously reported (Nevez et al., 2006). Our findings contrast those of Ponce et al.; however, differences in the PCR methodologies and primers used, clinical samples tested, and select patient demographics (e.g., prevalence of disease in different geographic locations) may also have affected the results.
Patient immunocompetence can be a relative term affected by numerous underlying medical conditions and subsequent therapies. The enrollment criteria for immunocompetent patients by many studies is heterogeneous, and we tried to more clearly articulate and exclude patients with immunosuppressive conditions and treatments that have been associated with PCP and other opportunistic infections. It is possible our strict pre-bronchoscopy enrollment criteria could have led to comparatively lower rates of P. jirovecii detection compared to other published reports.
Among patients evaluated for pulmonary infection including PCP, a higher proportion of respiratory samples tested positive for P. jirovecii by real-time PCR than by DFME; however, most patient samples were tested by only one assay. Our laboratory does not routinely perform both PCR and DFME on patient samples; however, P. jirovecii detection was increased by 7% when PCR testing was added to DFME-negative samples. Although we were not able to compare real-time PCR with DFME on all samples, Arcenas et al. (2006) did directly compare rates of P. jirovecii detection specifically using these 2 assays on BAL fluid and found all P. jirovecii-positive samples by DFME were also positive by cdc2 real-time PCR with an additional 3% positive by PCR alone. No samples were positive for P. jirovecii by DFME and negative by real-time PCR. The cdc2 target gene for PCR amplification in both the study of Arcenas and our study was selected for its highly conserved sequence and essential role in the cell division cycle. Although disagreement between DFME and real-time PCR is low, our findings as well as those of Arcenas et al. support the heightened sensitivity of PCR for P. jirovecii compared to DFME.
Among the subgroup of 27 patients evaluated in our medical center with confirmed immunosuppressive conditions and discordant test results (negative DFME and subsequent positive real-time PCR), 93% of patients were treated for PCP and 84% of these treated patients were considered to have definite or probable disease as documented in their medical records. Thus, in the vast majority of these patients, a positive PCR result was closely correlated with clinical PCP. These findings, coupled with the decreased risk of sample contamination compared to conventional and nested PCR, enable single-copy gene target (e.g., cdc2) to lower the rate of detecting organism “colonization” and confer a high predictive value for clinical PCP.
P. jirovecii can still occasionally be identified by PCR from patients without clinical PCP. Whether this scenario represents airway colonization or subclinical infection remains unclear and may depend on additional patient factors. Among HIV-negative patients evaluated by Nevez et al. (1999) who were found to have P. jirovecii without PCP, all had immunomodulating medical conditions, and there was a higher likelihood for CD4 cell counts to be under 400 cell/μL or CD4 cell/CD8 cell ratio of less than 1. Calderon et al. (2007) identified P. jirovecii without PCP in HIV-negative patients with COPD and found a higher level of proinflammatory cytokines (including IL-8, IL-6, and TNF-α) compared to patients with COPD testing negative for P. jirovecii. Higher rates of P. jirovecii detection without PCP have also been identified in infants with bronchiolitis and sudden infant death syndrome (Morris et al., 2004b, 2008). Collectively, these reports suggest P. jirovecii may still augment pulmonary inflammation and structural lung damage without definitive PCP (Calderon et al., 2007; Morris et al., 2004a).
There are a few limitations to our study that should be considered. Although not the primary objective of this study, most samples from our immunosuppressed patient group did not have both PCR and DFME testing performed and, thus, direct assay comparisons could not be made. The cdc2 gene of P. jirovecii is present as a single copy, which likely reduces sensitivity for P. jirovecii detection compared to other PCR assays using multicopy genes. Although it has been demonstrated that detection of the cdc2 gene of P. jirovecii remains more sensitive than DFME (Arcenas et al., 2006), the sensitivity of the cdc2 gene detection has not been directly compared to other PCR assays using multicopy genes. Additionally, it remains difficult to directly compare different PCR assays because of variability in gene targets, PCR primers, and PCR platform combinations (Robberts et al., 2007). Most studies evaluating P. jirovecii testing platforms use sputum, BAL, and other respiratory fluids. Additional studies comparing different PCR assays and directly using lung tissue are clearly needed, and the determination of whether a Ct cutoff value can be established that correlates with disease severity is an interesting area for future research.
5. Conclusion
The absence of any positive real-time PCR results for P. jirovecii in our immunocompetent patient group and the high correlation of discordant positive PCR and negative DFME results with clinical PCP in our immunosuppressed patient group suggest that real-time PCR does have increased sensitivity and objectivity over DFME. Furthermore, cdc2 real-time PCR may have a strong predictive value for PCP by not amplifying lower numbers of organism that could clinically correspond to colonization. Whether P. jirovecii colonization accurately reflects subclinical pulmonary infection remains an area for further study.
Acknowledgments
The authors wish to thank Dr David Maldonado for his significant contributions to this project.
Financial disclosures: No external or commercial financial support was used for this study.
Footnotes
Conflict of interest: None of the authors (J.W.W., A.H.L., T.E.G., T.K., N.L.W., and M.J.B.) of this article have a conflict of interest with any product or company listed in this article.
References
- Alvarez-Martinez MJ, Miro JM, Valls ME, Moreno A, Rivas PV, Sole M, Benito N, Domingo P, Munoz C, Rivera E, Zar HJ, Wissmann G, Diehl AR, Prolla JC, de Anta MT, Gatell JM, Wilson PE, Meshnick SR (2006) Sensitivity and specificity of nested and real-time PCR for the detection of Pneumocystis jiroveci in clinical specimens. Diagn Microbiol Infect Dis 56:153–160. [DOI] [PubMed] [Google Scholar]
- Arcenas RC, Uhl JR, Buckwalter SP, Limper AH, Crino D, Roberts GD, Wengenack NL (2006) A real-time polymerase chain reaction assay for detection of Pneumocystis from bronchoalveolar lavage fluid. Diagn Microbiol Infect Dis 54:169–175. [DOI] [PubMed] [Google Scholar]
- Calderon EJ, Rivero L, Respaldiza N, Morilla R, Montes-Cano MA, Friaza V, Munoz-Lobato F, Varela JM, Medrano FJ, Horra Cde L (2007) Systemic inflammation in patients with chronic obstructive pulmonary disease who are colonized with Pneumocystis jiroveci. Clin Infect Dis 45:e17–e19. [DOI] [PubMed] [Google Scholar]
- Caliendo AM, Hewitt PL, Allega JM, Keen A, Ruoff KL, Ferraro MJ (1998) Performance of a PCR assay for detection of Pneumocystis carinii from respiratory specimens. J Clin Microbiol 36:979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright CP, Nelson NA, Gill VJ (1994) Development and evaluation of a rapid and simple procedure for detection of Pneumocystis carinii by PCR. J Clin Microbiol 32:1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen D, Ross BC, Fairbairn J, Warren RJ, Baird RW, Dwyer B (1994) Comparison of Pneumocystis carinii detection by toluidine blue O staining, direct immunofluorescence and DNA amplification in sputum specimens from HIV positive patients. Pathology 26:198–200. [DOI] [PubMed] [Google Scholar]
- Huggett JF, Taylor MS, Kocjan G, Evans HE, Morris-Jones S, Gant V, Novak T, Costello AM, Zumla A, Miller RF (2008) Development and evaluation of a real-time PCR assay for detection of Pneumocystis jirovecii DNA in bronchoalveolar lavage fluid of HIV-infected patients. Thorax 63:154–159. [DOI] [PubMed] [Google Scholar]
- Khan MA, Farrag N, Butcher P (1999) Diagnosis of Pneumocystis carinii pneumonia: immunofluorescence staining, simple PCR or nPCR. J Infect 39:77–80. [DOI] [PubMed] [Google Scholar]
- Kovacs JA, Masur H (2009) Evolving health effects of Pneumocystis: one hundred years of progress in diagnosis and treatment. JAMA 301:2578–2585. [DOI] [PubMed] [Google Scholar]
- Limper AH (1996) Diagnosis of Pneumocystis carinii pneumonia: does use of only bronchoalveolar lavage suffice? Mayo Clin Proc 71:1121–1123. [DOI] [PubMed] [Google Scholar]
- Limper AH, Specks U, Brutinel WM, Martin WJ II, Rohrbach MS (1993) Interlobar variation in the recovery of bronchoalveolar lavage fluid, cell populations, and angiotensin-converting enzyme in normal volunteers. J Lab Clin Med 121:785–791. [PubMed] [Google Scholar]
- Maskell NA, Waine DJ, Lindley A, Pepperell JC, Wakefield AE, Miller RF, Davies RJ (2003) Asymptomatic carriage of Pneumocystis jiroveci in subjects undergoing bronchoscopy: a prospective study. Thorax 58:594–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano FJ, Montes-Cano M, Conde M, de la Horra C, Respaldiza N, Gasch A, Perez-Lozano MJ, Varela JM, Calderon EJ (2005) Pneumocystis jirovecii in general population. Emerg Infect Dis 11:245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen JH, Tauber I, Leeuwenberg AD, Beckers PJ, Sieben M (1977) Parasitologic and serologic observations of infection with Pneumocystis in humans. J Infect Dis 136:43–49. [DOI] [PubMed] [Google Scholar]
- Morris A, Lundgren JD, Masur H, Walzer PD, Hanson DL, Frederick T, Huang L, Beard CB, Kaplan JE (2004a) Current epidemiology of Pneumocystis pneumonia. Emerg Infect Dis 10:1713–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A, Sciurba FC, Lebedeva IP, Githaiga A, Elliott WM, Hogg JC, Huang L, Norris KA (2004b) Association of chronic obstructive pulmonary disease severity and Pneumocystis colonization. Am J Respir Crit Care Med 170:408–413. [DOI] [PubMed] [Google Scholar]
- Morris A, Wei K, Afshar K, Huang L (2008) Epidemiology and clinical significance of pneumocystis colonization. J Infect Dis 197:10–17. [DOI] [PubMed] [Google Scholar]
- Nevez G, Jounieaux V, Linas MD, Guyot K, Leophonte P, Massip P, Schmit JL, Seguela JP, Camus D, Dei-Cas E, Raccurt C, Mazars E (1997) High frequency of Pneumocystis carinii sp.f. hominis colonization in HIV-negative patients. J Eukaryot Microbiol 44:36S. [DOI] [PubMed] [Google Scholar]
- Nevez G, Magois E, Duwat H, Gouilleux V, Jounieaux V, Totet A (2006) Apparent absence of Pneumocystis jirovecii in healthy subjects. Clin Infect Dis 42:e99–e101. [DOI] [PubMed] [Google Scholar]
- Nevez G, Raccurt C, Vincent P, Jounieaux V, Dei-Cas E (1999) Pulmonary colonization with Pneumocystis carinii in human immunodeficiency virus-negative patients: assessing risk with blood CD4+ T cell counts. Clin Infect Dis 29:1331–1332. [DOI] [PubMed] [Google Scholar]
- Olsson M, Stralin K, Holmberg H (2001) Clinical significance of nested polymerase chain reaction and immunofluorescence for detection of Pneumocystis carinii pneumonia. Clin Microbiol Infect 7:492–497. [DOI] [PubMed] [Google Scholar]
- Oz HS, Hughes WT (2000) Search for Pneumocystis carinii DNA in upper and lower respiratory tract of humans. Diagn Microbiol Infect Dis 37:161–164. [DOI] [PubMed] [Google Scholar]
- Ponce CA, Gallo M, Bustamante R, Vargas SL (2010) Pneumocystis colonization is highly prevalent in the autopsied lungs of the general population. Clin Infect Dis 50:347–353. [DOI] [PubMed] [Google Scholar]
- Respaldiza N, Medrano FJ, Medrano AC, Varela JM, de la Horra C, Montes-Cano M, Ferrer S, Wichmann I, Gargallo-Viola D, Calderon EJ (2004) High seroprevalence of Pneumocystis infection in Spanish children. Clin Microbiol Infect 10:1029–1031. [DOI] [PubMed] [Google Scholar]
- Robberts FJ, Liebowitz LD, Chalkley LJ (2007) Polymerase chain reaction detection of Pneumocystis jiroveci: evaluation of 9 assays. Diagn Microbiol Infect Dis 58:385–392. [DOI] [PubMed] [Google Scholar]
- Russian DA, Levine SJ (2001) Pneumocystis carinii pneumonia in patients without HIV infection. Am J Med Sci 321:56–65. [DOI] [PubMed] [Google Scholar]
- Sing A, Geiger AM, Hogardt M, Heesemann J (2001) Pneumocystis carinii carriage among cystic fibrosis patients, as detected by nested PCR. J Clin Microbiol 39:2717–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A, Roggenkamp A, Autenrieth IB, Heesemann J (1999) Pneumocystis carinii carriage in immunocompetent patients with primary pulmonary disorders as detected by single or nested PCR. J Clin Microbiol 37:3409–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowden E, Carmichael AJ (2004) Autoimmune inflammatory disorders, systemic corticosteroids and pneumocystis pneumonia: a strategy for prevention. BMC Infect Dis 4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas SL, Hughes WT, Santolaya ME, Ulloa AV, Ponce CA, Cabrera CE, Cumsille F, Gigliotti F (2001) Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin Infect Dis 32:855–861. [DOI] [PubMed] [Google Scholar]
- Vargas SL, Pizarro P, Lopez-Vieyra M, Neira-Aviles P, Bustamante R, Ponce CA (2010) Pneumocystis colonization in older adults and diagnostic yield of single versus paired noninvasive respiratory sampling. Clin Infect Dis 50:e19–e21. [DOI] [PubMed] [Google Scholar]
- Wakefield AE, Miller RF, Guiver LA, Hopkin JM (1993) Oropharyngeal samples for detection of Pneumocystis carinii by DNA amplification. Q J Med 86:401–406. [PubMed] [Google Scholar]
- Wakefield AE, Pixley FJ, Banerji S, Sinclair K, Miller RF, Moxon ER, Hopkin JM (1990) Detection of Pneumocystis carinii with DNA amplification. Lancet 336:451–453. [DOI] [PubMed] [Google Scholar]