Abstract
Enzymes are the ultimate entities responsible for chemical transformations in natural and engineered biosynthetic pathways. However, many natural enzymes suffer from suboptimal functional expression due to poor intrinsic protein stability. Further, stability enhancing mutations often come at the cost of impaired function. Here we demonstrate an automated protein engineering strategy for stabilizing enzymes while retaining catalytic function using deep mutational scanning coupled to multiple-filter based screening and combinatorial mutagenesis. We validated this strategy by improving the functional expression of a Type III polyketide synthase from the Atropa belladonna biosynthetic pathway for tropane alkaloids. The best variant had a total of 8 mutations with over 25-fold improved activity over wild-type in E. coli cell lysates, an improved melting temperature of 11.5 ± 0.6°C, and only minimal reduction in catalytic efficiency. We show that the multiple-filter approach maintains acceptable sensitivity with homology modeling structures up to 4 Å RMS. Our results highlight an automated protein engineering tool for improving the stability and solubility of difficult to express enzymes, which has impact for biotechnological applications.
Keywords: deep mutational scanning, high-throughput screening, enzyme stability, heterologous pathway expression, polyketide synthase, tropane alkaloids
Biomanufacturing is a sustainable alternative to chemical synthetic routes for production of high-value products1. Key factors influencing the rapid advancement of this field include the dramatic increase of available gene coding sequences, reduced cost of synthetic DNA synthesis and assembly2, and improved computational3 and experimental4, 5 tools for engineering biology. Still, generating sufficient end titers and specific productivities to be cost competitive with plant-derived or traditional chemical synthetic routes remains a grand challenge, especially for compounds derived from plant specialized metabolism. For example, extensive engineering efforts led to only microgram per liter titers for reconstitution of opioid biosynthetic pathways6–8 and precursors of monoterpene indole alkaloids9 in yeast. These reported titers are between three and six orders of magnitude too low for supplanting other routes to these chemicals.
The reasons why many plant metabolic pathways yield low titers are multifaceted: intermediate products can build up and be toxic, pathways can be imbalanced, gene expression for pathway members are not optimized, among many other reasons. Nonetheless, at the heart of any pathway are the enzymes responsible for chemical transformation. Consider such a linear pathway of heterologously expressed enzymes. The maximum possible flux (Jmax) for this pathway is given by the product of the turnover number (kcat) and the concentration of active enzyme ([E]active) for the weakest pathway enzyme:
| (1) |
In other words, negligible product flux occurs whenever the concentration of any active enzyme in the pathway approaches zero. In fact, many biomanufacturing platforms have low productivities and titers because one or more pathway enzymes, when overexpressed, have very little activity6, 10–15.
Intrinsic protein biophysics can account for the limited active expression for many of these enzymes. Native proteins are marginally stable, and their native expression levels are often at their solubility limit16. Expression in a different environment can thermodynamically favor the unfolded state or result in aggregation. It follows that stabilization of such poorly expressed enzymes can improve performance of synthetic metabolic pathways. For example, we previously developed a synthetic levoglucosan utilization pathway in E. coli10. Strains harboring the original enzyme, levoglucosan kinase (LGK) from Lipomyces starkeyi, showed weak growth with levoglucosan as the sole carbon source. Strains expressing a thermally stable LGK had 15-fold higher specific growth rates and flux than that of LGK. The catalytic efficiencies for the two enzymes were essentially identical, with the sole difference between the two strains were three point-mutations that increased the melting temperature of the protein by 5.1°C – this increase correlated with an increase in functional enzyme expression. Additionally, the Tang group at UCLA demonstrated that engineering improved solubility and heterologous expression of simvastatin synthase in E. coli increased the productivity of a whole-cell biocatalytic process17.
The above findings have not been extended generally to other biomanufacturing platforms for several reasons. One, modifying active expression by promoter engineering is much easier than by protein engineering. However, in many cases a strong promoter will not drive production of enough active enzyme – this effect is clearly seen by results from Wheeldon and colleagues in metabolic engineering of ester pathways18. Two, the above pathways are examples where the rate-determining step was governed by a single enzyme. Many pathways of interest would have multiple poorly behaved enzymes, making the engineering challenge more difficult. Three, the protein engineering challenge itself is daunting: one has to identify solubility-enhancing mutations; filter away mutations that destroy catalytic activity; and, because the stabilizing effect of any single mutation is often modest, combine many solubility-enhancing mutations at once. This challenge is compounded by the fact that many of the pathway enzymes have neither solved structures or high throughput activity assays, preventing traditional computational design19 and directed evolution approaches, respectively.
To address this challenge, we recently identified stabilizing ‘hits’ using high-throughput screens for stability and solubility in deep mutational scanning experiments20. Existing comprehensive single-mutation functional datasets for two enzymes were compared against datasets generated with the solubility screens. We found a greater than 90% probability of choosing a catalytically neutral mutation by filtering out mutations that were near the active site, not evolutionarily conserved, or buried in the protein core. These encouraging results suggested to us that automated stabilization of proteins using data-driven methods, even in the absence of an activity screen, may be possible. This stabilization method has the following characteristics: (i.) use of deep mutational scanning to identify nearly all mutations that improve soluble expression; (ii.) predictive identification of a subset of these mutations that do not impact catalytic efficiency; (iii.) combine multiple (>5) mutations simultaneously into new designs.
Here we tested this method rigorously using a recently uncovered biosynthetic pathway to tropinone, a common intermediate for nearly all plant tropane alkaloids (TA)21. Specifically, we identified solubility-enhancing mutations in a Type III polyketide synthase from Atropa belladonna (Ab) that expresses very poorly in both bacterial and yeast systems. We then designed new variants with improved in vivo and in vitro stability without appreciably impacting catalytic efficiency. Finally, we developed an automated computational screening process for rapid identification of potential beneficial mutations, which was robust even in the absence of a high-quality structural model. Combined, our results showcase the use of data-driven approaches to improve enzyme stability and provide a new engineering tool for biomanufacturing.
Results and Discussion
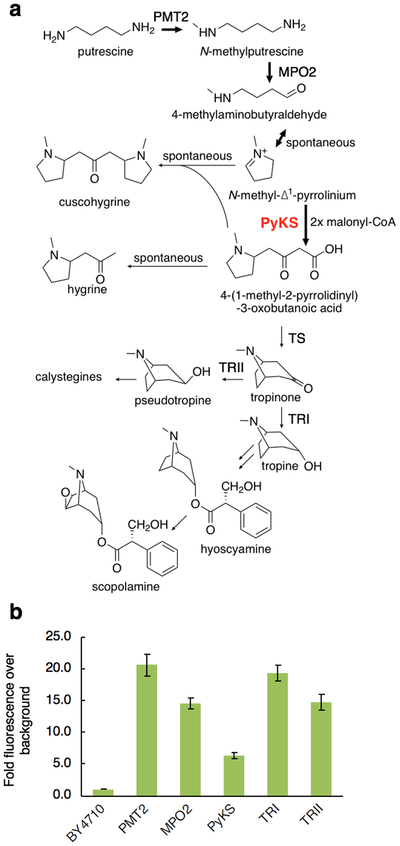
Nearly all TAs, including anticholinergics hyoscyamine and scopolamine, come from the central precursor tropinone (Figure 1a). In Ab, tropinone is derived from the pathway precursor putrescine by four enzymes: Putrescine N-Methyl Transferase (PMT2), N-Methylputrescine Oxidase (MPO2), a Type III PKS Pyrrolidine Ketide Synthase (PyKS), and the cytochrome P450 Tropinone Synthase (TS) / CYP82M3 (Figure 1a).
Figure 1.
The Tropane Alkaloids (TA) pathway from Atropa belladonna (Ab). A.) The conversion of putrescine to N-methylputrescine by PMT2 is the first committed step in TA biosynthesis. The enzyme engineered for improved solubility in this work, PyKS, performs two rounds of ketide synthase (Claisen condensation) activity on N-methyl-Δ1-pyrrolinium with two units of malonyl-CoA to form 4-(1-methyl-2-pyrrolidinyl)-3-oxobutanoic acid. Hyoscyamine and scopolamine are medicinally relevant small molecules. B.) Expression yield of Ab genes in yeast via GFP-tagging confirms poor heterologous expression of PyKS. Fluorescence of S. cerevisiae strain BY4710 cells expressing GFP-tagged Ab genes under galactose induction was quantified using flow cytometry. Error bars represent one standard deviation of at least three independent measurements.
To test our enzyme stabilization method, we chose PyKS as several lines of evidence clearly point to poor functional expression. First, attempts to express and purify PyKS from E. coli for previous biochemical characterization work21 yielded extremely low levels of active protein in the absence of a Glutathione S-Transferase (GST) solubility tag. Essentially all of the protein was insoluble and activity sharply declines at temperatures in excess of 25°C. Second, we quantified the mean fluorescence of GFP-tagged Ab gene products expressed in Saccharomyces cerevisiae BY471022 by flow cytometry and found that the PyKS expressing cells were less fluorescent compared to the other genes, indicating poor soluble expression of GFP-tagged PyKS relative to the other pathway enzymes in yeast (Figure 1b). Third, while PMT2 expression in Nicotiana benthamiana results in high yields of N-methylputrescine that diminishes substantially when MPO2 is co-infiltrated with PMT2, the tropinone yield is ~20-fold less with simultaneous PyKS and TS expression, suggesting that PyKS and/or TS expression is limiting21. Together, these considerations prompted us to engineer increased functional expression of PyKS.
In effort to improve the expression of the PyKS, we sought to use deep mutational scanning coupled to a high-throughput screen for stability and solubility20. We first explored the use of yeast surface display (YSD) coupled to FACS as our previous work utilized this screening platform. The PyKS coding sequence was cloned into the pETConNK backbone20 and expressed in S. cerevisiae EBY100 by galactose induction. We were unable to successfully display the PyKS on the yeast surface (Supporting Figure S1) despite testing several alternate induction temperatures (18–30°C) as well as mutating a potential N-linked glycosylation site at Asn339 to alanine that we hypothesized could disrupt proper folding and display on yeast surface.
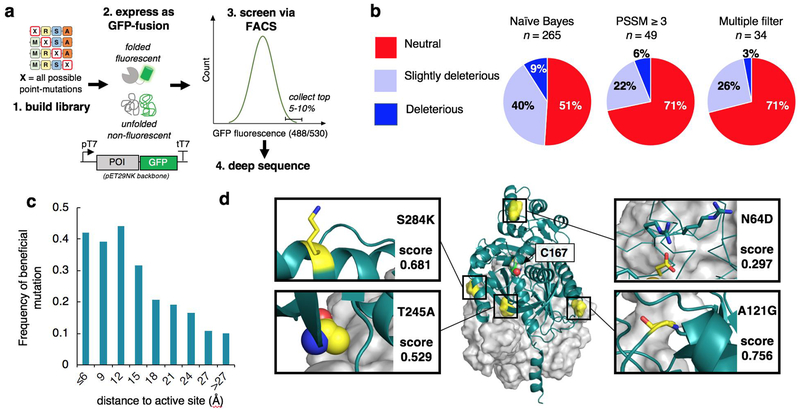
Based on the failure of PyKS to yeast display, we assessed an alternative screen involving fusing a protein of interest to a monomeric GFP variant. Upon expression, folded proteins will permit the folding and subsequent chromophore formation of GFP, while unfolded proteins will be non-fluorescent23. Expression of a protein library can then be screened by fluorescence intensity using FACS (Figure 2a). We first assessed the ability of the screen to identify known stabilizing mutants of the model protein LGK. We fused LGK to fluorescent protein variant mGFPmut324, created a comprehensive single-site saturation mutagenesis library using nicking mutagenesis25, and induced fusion protein expression by IPTG in E. coli BL21 Star (DE3). Individual cells were sorted using FACS and two populations were collected: a gated reference population and the top 5% of cells based on GFP fluorescence intensity. Libraries were harvested, prepared, and deep sequenced in a standardized pipeline26. Detailed statistics for library deep sequencing and FACS sorting are given in Table S1 and Table S2, respectively. The resulting deep sequencing datasets were converted to a solubility score centered about a wild-type score of zero. A solubility score greater than zero indicates that the protein fusion has a higher fluorescence than the wild-type fusion. The per-position scores are provided in Supporting Data S1.
Figure 2.
Deep mutational scanning using GFP-fusion solubility screen and filtering to identify catalytically neutral stabilizing mutations. A.) Method overview. A comprehensive site-saturation library for a protein of interest (POI) is generated using nicking mutagenesis (1). The POI is genetically encoded as an N-terminal fusion to GFP (2), where upon expression folded POIs will permit the folding and subsequent chromophore formation of GFP, while unfolded protein will be non-fluorescent. The amount of folded fusion protein per cell correlates with solubility/stability of the POI variant. The library is expressed in E. coli and screened for GFP fluorescence using FACS (3), and the resulting pre- and post-screening libraries are deep sequenced (4). B.) Results from applying various filtering strategies to the levoglucosan kinase (LGK) GFP-fusion and fitness selections datasets. The best strategy “Multiple filter” includes distance to active site (15 ≥ Å), PSSM (≥ 0), contact number (≤16), and exclusion of mutations to/from proline. C.) The frequency of beneficial (solubility-enhancing) mutations of PyKS identified from the GFP-fusion experiment as a function of distance to catalytic active site. D.) Structural analysis of high solubility score mutations indicates that many improve surface charge characteristics (S284K, N64D), hydrophobic core packing (T245A), and secondary structural elements like loops (A121G). The grey surface representation is the dimer subunit. C167 is the putative catalytic nucleophile.
We evaluated the ability of the solubility deep mutational scans to identify known stabilizing mutations in LGK (Table S3). These mutations were previously shown to rescue enzyme solubility in the context of other destabilizing mutations with an in vitro characterized change in melting temperature (ΔTm) ≥ 1°C in the parental background10. We identified a mutation as stabilizing if its solubility score was above 0.15, which corresponds to a mean fluorescence intensity of 10% above the wild-type sequence. The GFP fusion screens with this threshold identified 9/12 of these mutations (p-value 2.0×10−9). Changing the threshold for identifying solubility-enhancing mutations based on the distribution of synonymous wild-type codons did not alter the significance of the results (Table S4).
It is well known that many of stability-enhancing mutations result in enzymes with reduced catalytic efficiency27. Therefore, we asked whether we could predict mutations resulting in neutral or improved catalytic efficiency. To this end, we closely followed filtering methods of our previous work using YSD screens20. Briefly, we compared a previously published single-mutation fitness dataset10 with the GFP fusion dataset. We classified mutations by distance to active site, evolutionary conservation as quantified by a position specific scoring matrix (PSSM), and degree of burial in the protein core measured by contact number. We assessed a strict multiple-filter (PSSM ≥ 0, distance to active site ≥ 15Å, contact number ≤ 16, and no mutations involving a proline), naïve Bayes classification, and a hybrid method combining filtering on PSSM ≥ 3 with Bayes analysis on the remaining filters. Consistent with our previous results using YSD20, the multiple-filter performed best (Figure 2b): for the GFP screening dataset the probability of finding a neutral mutation is 71% with only a 3% chance of uncovering a deleterious mutation. While only 34 LGK mutations (out of >6,000 total) pass this stringent multiple-filter, most proteins can be stabilized sufficiently with approximately 5–15 mutations.
Buoyed by these results, we next sought to apply the validated method to engineer PyKS variants with improved functional expression. The objective was to perform a GFP-fusion deep mutational scan on PyKS (Figure 2a), filter the resulting hits for probability of maintaining catalytic activity with the multiple-filter, and then combine multiple mutations into active designs with improved expression. We performed the deep mutational scanning experimental and analysis pipeline as for LGK in which we sorted a single-site saturation mutagenesis library of PyKS fused to GFP with FACS and deep sequenced the top 8% of the population by fluorescence. Unfortunately, the reference population for the gene tile covering residues 157–234 did not grow after FACS, and so we chose to omit these positions from further analysis.
Deep sequencing the reference population revealed 84.3% coverage of single nonsynonymous (NS) mutations (5107/6060, see Table S5 for complete library statistics). Nonsense mutations had a mean solubility score of −0.653 ± 0.48 (1 s.d.), which was significantly lower than the mean of −0.0561 ± 0.54 for missense mutations (P-value < 0.0001, two-tailed unpaired Student’s T-test). To evaluate the reproducibility of the method, we performed replicate sorting, deep sequencing, and analysis for one gene tile. The Pearson’s correlation coefficient between replicates was found to be 0.72, which is low compared to previous deep mutational scanning experiments (coefficients of 0.8520 and 0.9328 have been previously reported from our lab). As reproducibility generally improves with increasing depth of sequencing coverage, we calculated Pearson’s correlation coefficients for mutations with at least 100 read counts in the reference population and found the coefficient improves to 0.83. Thus, the relatively low depth of coverage in this experiment partially but not completely explains the relatively high variance between replicates. Since we are interested in variants with improved functional expression, we next asked how correlation scales with coverage for variants with a solubility score at or above 0.15. We found that variants with ≥50 or ≥100 average selected read counts had a Pearson’s of 0.84 (n = 247) or 0.90 (n = 193), respectively (Supporting Figure S2). These were deemed reasonable thresholds for reliability of the deep sequencing experiment to identify stabilizing mutations. Full datasets for the PyKS deep mutational scan are provided in Supporting Data S2.
The GFP-fusion experiment identified an astounding 1,115 beneficial missense mutations (solubility score at or above 0.15) with ≥50 selected read counts (19.4% of total tested). To facilitate analysis, we generated a comparative model of PyKS with I-TASSER using default options29 (PDB file for model provided in Supporting Data S3). Hits were spread across the primary and tertiary sequence of PyKS (Figure 2c), with 246 out of 303 tested positions (81.2%) showing at least one mutation with an improved solubility score. These mutations occur at the surface of the enzyme, near the putative active site in the core, as well as at the potential homodimer interface (Figure 2d).
These 1,115 individual hits were sorted using the multiple-filter validated on the LGK dataset, with the adjustment of permitting mutations with a higher contact number (≤24 cutoff instead of ≤16) through if they passed a more stringent PSSM filter cutoff (≥3 instead of ≥0). This adjustment is justified as filtering on PSSM ≥3 alone shows comparable results to the multiple-filter method (Figure 2b). The resulting set of hits post-filter was comprised of 116 mutations at 56 unique positions spread throughout the protein sequence and structure (Figure 3a and Table S6). Notably, there were approximately 3 times more filter-passing hits for PKS than LGK.
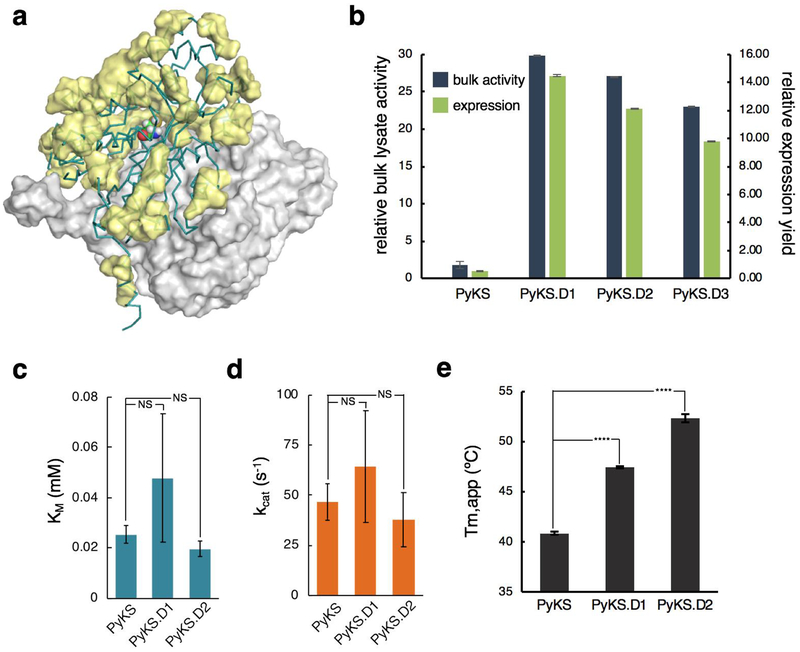
Figure 3.
Combinatorial PyKS designs enable higher enzyme flux via engineered enzyme stability. A.) Positions with filtered solubility-enhancing hits are shown in yellow surface representation on the PyKS model structure. B.) Relative expression yield and bulk lysate activity from E. coli lysates expressing wild-type and stabilized PyKS designs. Cells over-expressing each sample were lysed and assayed for PyKS activity as well as GFP fluorescence intensity (485/507 nm). Measurements are normalized relative to wild-type. Error bars represent one standard deviation of three independent measurements. C-D.) KM and kcat kinetic characterization of untagged (no GFP-fusion) purified wild-type and designed PyKS enzymes. Error bars represent one standard deviation of three independent measurements. NS = not statistically significant. E.) Apparent melting temperatures (Tm,app) of wild-type and designed PyKS enzymes. Error bars represent one standard deviation of three technical replicates.
We first tested the solubility screen and the multiple-filter method before making combinatorial designs. Thus, we produced 6 of these 116 point mutants (V12I, S37A, M115R, A121G, T245A, S284K) along with two mutants with high solubility scores that did not pass the filter (P106A – proline mutation, not evolutionarily conserved, high contact number; A143V – high contact number). These mutants, along with the wild-type sequence, were expressed in E. coli BL21 Star (DE3) and induced with IPTG under standard conditions. Lysates of GFP-fused PyKS mutants were tested for their relative fluorescence intensity per OD600 (expression yield) and PyKS enzymatic activity21 (Table 1). Whereas the 6 mutants that passed the filter had comparable or improved expression yield and activity to wild-type, both A143V and P106A expressed as soluble fusion proteins but had no measurable catalytic activity. These results highlight the importance of employing the binary filter for discriminating desired stabilizing mutations that are catalytically neutral.
Table 1.
Characterization of solubility-enhancing hits identified from PyKS scan. Yellow highlight indicates mutations that fail the multiple-filter method.
| Mutation | Solubility Score | Distance to Active Site (Å) | PSSM score | Contact Number | Relative RFU/OD600‡ | DKA Response Factor* | DKA Est. Conc. (μM) replicate 1** | DKA Est. Conc. (μM) replicate 2** |
|---|---|---|---|---|---|---|---|---|
| WT | 1.00 | 1 | 0.8 | 1 | ||||
| V12I | 0.206 | 39.1 | 3 | 10 | 1.24 | 0.62 ± 0.13 | 0.5 | 0.6 |
| S37A | 0.967 | 23.5 | 3 | 14 | 1.86 | 1.27 ± 0.004 | 1.1 | 1.2 |
| P106A | 0.641 | 19.4 | −2 | 29 | 1.43 | 0.007 ± 0.002 | 0 | 0 |
| M115R | 0.871 | 25.0 | 3 | 19 | 3.41 | 3.39 ± 0.58 | 2.7 | 3.6 |
| A121G | 0.756 | 32.1 | 6 | 13 | 3.21 | 4.30 ± 0.85 | 3.4 | 4.6 |
| A143V | 0.257 | 19.0 | 0 | 29 | 0.47 | 0.025 ± 0.013 | 0 | 0 |
| T245A | 0.529 | 18.5 | 3 | 21 | 2.09 | 1.40 ± 0.28 | 1.4 | 1.1 |
| S284K | 0.681 | 20.9 | 6 | 10 | 1.73 | 1.25 ± 0.48 | 0.9 | 1.6 |
Fluorescent units for GFP relative to OD600 of culture normalized relative to wild-type.
Response factors refer to WT normalization of peak areas for DKA (4-(1-methyl-2-pyrrolidinyl)-3-oxobutanoic acid). These peak areas refer to area under the curve for LC-MS/MS analysis of DKA. Error bars indicate 1 s.d. of technical replicates (n = 2).
Estimated concentration of DKA in reported enzyme assays based on standard curves run within one week of the experiment.
We next selected 21 mutations at 19 positions to include in combinatorial libraries, which were constructed using multi-site nicking mutagenesis25 (primer sequences listed in Table S7). BL21 Star (DE3) cells expressing the combinatorial libraries were cultured, induced with IPTG and screened by a combination of FACS and visible plate screening. Given the large theoretical size of the combinatorial library (2.1×106), FACS enrichment prior to visual plate screening enabled us to discard the bulk of “failures” and thus only screen the top variants for fluorescence intensity by eye. There were three clear hits from this fluorescence-based screening, which we named PyKS.D1, PyKS.D2, and PyKS.D3 (full amino acid and nucleotide sequences are listed in Supporting Text S1). These designs had 8–11 total mutations with 6 in common: PyKS.D1 (V12I, S37A, N64E, M115R, A121G, L235V, T245A, S284K, I301V, S318E, D366E), PyKS.D2 (V12I, S37A, N64D, M115R, A121G, T245A, G357A, D366E), and PyKS.D3 (V12I, S37A, N64E, N90M, M115R, A121G, T245A, D366E). Based on the homology structure, the shared mutations generally appear to alter surface charge characteristics, core packing, loop flexibility, or dimeric interface contacts (Figure 2d). For example, N64E/D introduces a negative charge to a patch on the surface that is otherwise positive, while T245A lies at the dimeric interface where it presumably strengthens the protein-protein interaction. Lastly, A121G likely improves loop flexibility.
We performed lysate relative expression and activity measurements exactly as performed for the point mutants. Compared with wild-type, all three designs had a greater than 10-fold improvement in relative expression, with PyKS.D1 showing a 27.1 ± 0.14-fold improvement in relative activity (Figure 3b). When lysate activity is normalized to gene expression, PyKS.D1 observed a 1.89 ± 0.027-fold improvement over wild-type. This higher functional expression is not the result of increased mRNA expression as all variants had statistically insignificant or slightly decreased mRNA expression relative to PyKS wild-type (Supporting Figure S3). Thus, we conclude that most of the relative lysate activity improvement can be attributed to improved gene expression from improved protein solubility and/or stabilization.
To confirm that these designs were not dependent on being in the GFP fusion context, we produced and purified recombinant PyKS, PyKS.D1, and PyKS.D2 and evaluated their activity on malonyl-CoA and starter unit N-methyl-Δ1-pyrrolinium cation as substrates (Supporting Figure S4). PyKS is an efficient enzyme, with a kcat of 47 s−1 and approx. 20 μM KM for both substrates. Both PyKS.D1 and PyKS.D2 had similar activity to PyKS (Figure 3c and 3d): PyKS.D1 has a marginally higher average kcat and PyKS.D2 has a slightly lower average kcat, but neither is significantly different than PyKS (p-value = 0.29 and 0.16, respectively, Figure 3d). There was also no statistically significant difference in Michaelis constant for either substrate (Figure 3c). We also assessed the apparent melting temperatures (Tm,app) using a well-established dye-shift thermal assay30 (Supporting Figure S5). While PyKS had a Tm,app of 40.9±0.2°C, both PyKS.D1 (47.5±0.1°C) and PyKS.D2 (52.4±0.4°C) showed statistically significant (p < 0.0001 for both) increases in melting temperatures (Figure 3e). It is important that we are not claiming that all thermally stabilizing mutations improve functional expression. Improvements to functional expression result from several effects including aggregation propensity, thermodynamic stability, and kinetic folding and oligomerization rates; these effects are only somewhat correlated with thermal stability. However, these results confirm that our designs that result in higher function expression in vivo also have higher thermal stability in vitro while maintaining statistically similar catalytic parameters to wild-type PyKS.
Finally, to facilitate automatic generation of a list of mutations to pass into a combinatorial library, we wrote a custom python script for PyRosetta31. Given an input enzyme sequence and structure, this script evaluates the four different components of the filter, returning a positive (passed filter) or negative (failed filter result) value for every possible point-mutation in the protein sequence. This script has been integrated into the Rosetta Macromolecular software package and is available on GitHub (User: raisanoshin). Details for using the script can be found in Supporting Text S2.
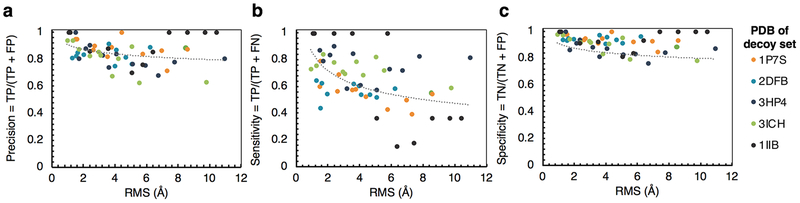
The multiple-filter includes terms that do not require structural information (PSSM, mutations involving proline) as well as terms that do (distance to active site, contact number). A final key question to assess the general utility of our method is how precise, sensitive, and specific the multiple-filtering method is for enzymes with approximated 3D structures generated with homology modeling. While not all enzymes will have experimentally determined structures, almost all enzyme classes (e.g. polyketide synthases, P450s, glycosyl hydrolases, terpene synthases, etc.) have at least one solved structure. In these cases, comparative models can be calculated with accuracies of approximately 4–5 Å RMSD or better using current comparative modeling computational software packages32.
To assess the accuracy of using our PyRosetta script to generate an accurate list of filter-passing mutations on enzymes where a predicted structural model is used as input, we used decoy sets of varying RMSD for six different enzymes with their corresponding experimentally-determined structures. We compared the results of running the script on each decoy with results using the solved structure, generating true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN) for each mutation in the set of all possible mutations. We then calculated precision, sensitivity, and specificity as a function of model RMS shown in Figures 4a–c. For this specific application, the most important criterion is precision, as incorporating too many false positives into designs would disrupt catalytic efficiency of the enzyme. Here, the precision is 0.8 or better up to 4 Å RMS (Figure 4a). Beyond 4 Å RMS the precision vacillates between 0.6 and 1.0. Also important is the sensitivity of the method because we typically end up with 30–50 mutations that pass the filter. For RMS values until 5 Å the sensitivity remains above 0.4 (Figure 4b), which still allows recovery of enough mutations to fix poorly expressed enzymes. The least important metric is specificity, which we include for completeness. Here the specificity remains above 0.8 for RMS lower than 5 Å (Figure 4c).
Figure 4.
Automated generation of filter-passing mutations with a PyRosetta script is robust for use on predicted structural models. A-C.) Precision (a), sensitivity (b), and specificity (c) were calculated by comparing the results of running our PyRosetta mutation filtering script on five different PDB crystal structures and their corresponding decoy sets (see methods).
Concluding remarks
In this work, we performed a high-throughput screen for stability and solubility to test thousands of mutations on a protein sequence. Combinations of those mutations using a stringent multiple-filter led to Type III PyKS designs with enhanced functional expression in E. coli. There are several important takeaways from this project.
First, this work provides validation for the filtering method previously developed by Klesmith et al.20. Results from the point-mutation analysis indicate that although certain mutations provide stabilizing effects, if the position is highly conserved in nature a mutation is likely to be deleterious for function. Indeed, proline 106 is a canonical example of this stability/function trade-off. P106 lies in the middle of a helix, where prolines are generally disfavored, and the solubility screen indicates that several other residues at this position improve overall stability of the protein. However, the PSSM indicates that proline is highly conserved and thus important to catalytic function. The P106A variant increased the solubility of the PyKS-GFP fusion but almost completely ablated activity.
Second, our integrated method has now been validated on 3 different enzymes (PyKS, LGK, TEM-1 BLA)20 and thus ready for general deployment towards different biotechnological applications. A potential strategy moving forward would be to generate structural models for enzymes of interest, run the filtering script to obtain a list of passing mutations, test only the subset, and then combine hits into new designs. While deep sequencing driven protein science enables the generation of previously unthinkable amounts of mutational data33, testing hundreds versus thousands of mutations needed for a comprehensive scan of a gene is certainly more practical and economical. We anticipate several potential platforms would benefit from enzyme stabilization.
Finally, this PyKS is part of a recently described tropane alkaloid pathway. While beyond the scope of the present work, our immediate next steps are to develop biomanufacturing platforms for tropane alkaloids. Our intent is to test the hypothesis that stabilized PyKS variants (and potentially other enzymes) can improve pathway yields, titers, and productivities.
Supplementary Material
Acknowledgements
We thank Prof. Frank DiMaio for donation of the enzyme decoy sets. This work was supported by Michigan State University funds, by NSF CBET Career Award #1254238 to T.A.W., NIH T32 Biotechnology Training Grant (Award # T32-GM110523) to E.E.W, and NSF award numbers IOS-1546617 and MCB-1714093 to C.S.B. C.S.B. is supported in part by Michigan AgBioResearch and through USDA National Institute of Food and Agriculture, Hatch project number MICL02552.
Footnotes
Supporting Information
A Supporting Information PDF file containing Materials and Methods, Supporting Figures, Tables, and Text is included. Supporting Data S1 and S2 contain full LGK and PyKS GFP-fusion datasets, respectively. Data S3 contains a protein data bank file of the PyKS homology model used in this work.
Data Availability
Raw sequencing reads have been deposited in the Sequencing Read Archive. All plasmids described in this work are available upon request. All scripts have been integrated into the Rosetta macromolecular software package, which is freely available to registered academic users.
References
- 1.Paddon CJ, and Keasling JD (2014) Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development, Nat. Rev. Microbiol 12, 355. [DOI] [PubMed] [Google Scholar]
- 2.Kosuri S, and Church GM (2014) Large-scale de novo DNA synthesis: technologies and applications, Nat. Methods 11, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farasat I, Kushwaha M, Collens J, Easterbrook M, Guido M, and Salis HM (2014) Efficient search, mapping, and optimization of multi‐protein genetic systems in diverse bacteria, Mol. Sys. Biol 10, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee ME, DeLoache WC, Cervantes B, and Dueber JE (2015) A highly characterized yeast toolkit for modular, multipart assembly, ACS Synth. Biol 4, 975–986. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz CM, Hussain MS, Blenner M, and Wheeldon I (2016) Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR–Cas9-mediated genome editing in Yarrowia lipolytica, ACS Synth. Biol 5, 356–359. [DOI] [PubMed] [Google Scholar]
- 6.Galanie S, Thodey K, Trenchard IJ, Interrante MF, and Smolke CD (2015) Complete biosynthesis of opioids in yeast, Science 349, 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, and Smolke CD (2016) Engineering biosynthesis of the anticancer alkaloid noscapine in yeast, Nat. Comm 7, 12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLoache WC, Russ ZN, Narcross L, Gonzales AM, Martin VJ, and Dueber JE (2015) An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose, Nat. Chem. Biol 11, 465. [DOI] [PubMed] [Google Scholar]
- 9.Brown S, Clastre M, Courdavault V, and O’Connor SE (2015) De novo production of the plant-derived alkaloid strictosidine in yeast, Proc. Natl. Acad. Sci. U.S.A 112, 3205–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klesmith JR, Bacik J-P, Michalczyk R, and Whitehead TA (2015) Comprehensive sequence-flux mapping of a levoglucosan utilization pathway in E. coli, ACS Synth. Biol 4, 1235–1243. [DOI] [PubMed] [Google Scholar]
- 11.DeLoache WC, Russ ZN, and Dueber JE (2016) Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways, Nat. Comm 7, 11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrenworth A, Sarria S, and Peralta-Yahya P (2015) Pterin-dependent mono-oxidation for the microbial synthesis of a modified monoterpene indole alkaloid, ACS Synth. Biol 4, 1295–1307. [DOI] [PubMed] [Google Scholar]
- 13.Lau W, and Sattely ES (2015) Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone, Science 349, 1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiue E, Brockman IM, and Prather KL (2015) Improving product yields on D‐glucose in Escherichia coli via knockout of pgi and zwf and feeding of supplemental carbon sources, Biotechnol. Bioeng 112, 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, and Keasling JD (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass, Nature 463, 559. [DOI] [PubMed] [Google Scholar]
- 16.Dobson CM, and Vendruscolo M (2017) “Life on the Edge”: The Relationship Between Cell Abundances and Physical Properties of Proteins, Annu. Rev. Biophys 46. [Google Scholar]
- 17.Xie X, Pashkov I, Gao X, Guerrero JL, Yeates TO, and Tang Y (2009) Rational improvement of simvastatin synthase solubility in Escherichia coli leads to higher whole-cell biocatalytic activity, Biotechnol. Bioeng 102, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Lin JL, Palomec L, and Wheeldon I (2015) Microbial host selection affects intracellular localization and activity of alcohol-O-acetyltransferase, Microb. Cell. Fact 14, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenzweig A, Goldsmith M, Hill SE, Gertman O, Laurino P, Ashani Y, Dym O, Unger T, Albeck S, and Prilusky J (2016) Automated structure-and sequence-based design of proteins for high bacterial expression and stability, Mol. Cell 63, 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klesmith JR, Bacik J-P, Wrenbeck EE, Michalczyk R, and Whitehead TA (2017) Trade-offs between enzyme fitness and solubility illuminated by deep mutational scanning, Proc. Natl. Acad. Sci. U.S.A 114, 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedewitz MA, Jones AD, D’Auria JC, and Barry CS (2018) Tropinone synthesis via an atypical polyketide synthase and P450-mediated cyclization, Nat. Comm 9, 5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, and Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR‐mediated gene disruption and other applications, Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- 23.Waldo GS, Standish BM, Berendzen J, and Terwilliger TC (1999) Rapid protein-folding assay using green fluorescent protein, Nat. Biotech 17, 691–695. [DOI] [PubMed] [Google Scholar]
- 24.Bienick MS, Young KW, Klesmith JR, Detwiler EE, Tomek KJ, and Whitehead TA (2014) The interrelationship between promoter strength, gene expression, and growth rate, PLoS One 9, e109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrenbeck EE, Klesmith JR, Stapleton JA, Adeniran A, Tyo KE, and Whitehead TA (2016) Plasmid-based one-pot saturation mutagenesis, Nat. Methods 13, 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowalsky CA, Klesmith JR, Stapleton JA, Kelly V, Reichkitzer N, and Whitehead TA (2015) High-resolution sequence-function mapping of full-length proteins, PLoS One 10, e0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokuriki N, and Tawfik DS (2009) Stability effects of mutations and protein evolvability, Curr. Opin. Struct. Biol 19, 596–604. [DOI] [PubMed] [Google Scholar]
- 28.Wrenbeck EE, Azouz LR, and Whitehead TA (2017) Single-mutation fitness landscapes for an enzyme on multiple substrates reveal specificity is globally encoded, Nat. Comm 8, 15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y (2008) I-TASSER server for protein 3D structure prediction, BMC Bioinf 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavinder JJ, Hari SB, Sullivan BJ, and Magliery TJ (2009) High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering, J. Am. Chem. Soc 131, 3794–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhury S, Lyskov S, and Gray JJ (2010) PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta, Bioinformatics 26, 689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan R, Xu D, Yang J, Walker S, and Zhang Y (2013) A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction, Sci. Rep 3, 2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrenbeck EE, Faber MS, and Whitehead TA (2017) Deep sequencing methods for protein engineering and design, Curr. Opin. Struct. Biol 45, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.