Supplemental Digital Content is available in the text
Keywords: esophagus, genotype guided, inflammation, pharmacogenetics, proton pump inhibitor-nonresponsive eosinophilic esophagitis, proton pump inhibitor-responsive esophageal eosinophilia
ABSTRACT
Objective:
Proton pump inhibitors (PPIs) are an effective treatment for eosinophilic esophagitis (EoE); however, only 30% to 60% of patients respond. Common genetic variants in CYP2C19 and STAT6 associate with PPI plasma concentration and magnitude of inflammatory response, respectively. Our objective was to determine if genetic variation in the genes for CYP2C19 and STAT6 influence differentiation between PPI responsive esophageal eosinophilia versus PPI nonresponsive EoE (PPI-REE, PPI-nonresponsive EoE).
Methods:
Genomic DNA was isolated from 92 esophageal tissue biopsies collected from participants of a prospective clinical trial of high-dose PPI therapy for esophageal eosinophilia in children.
Results:
Of the 92 patients examined, 57 (62%) were PPI-REE and 35 (38%) were PPI-nonresponsive EoE. Forty-six of the 92 patients were further characterized by pH probe monitoring; there was no association between reflux index and carriage of CYP2C19∗17 (P = 0.35). In children who received a PPI dose between ≥1.54 and ≤2.05 mg/kg/day, binary logistic regression modeling showed that carriage of CYP2C19∗17 associated with PPI-nonresponsive EoE (odds ratio (OR) [95% confidence interval (CI)] = 7.71 [1.21, 49.11], P = 0.031). Carriage of STAT6 allelic variant rs1059513 predicts PPI-REE (OR [95% CI] = 6.16 [1.44, 26.4], P = 0.028), whereas carriage of STAT6 rs324011 synergizes with CYP2C19∗17 to predict PPI-nonresponsive EoE (rs324011 OR [95% CI] = 5.56 [1.33, 20.72], P = 0.022; CYP2C19∗17 OR [95% CI] = 8.19[1.42, 50.57], P = 0.023).
Conclusions:
Common variants in CYP2C19 and STAT6 associate with a PPI-nonresponsive EoE outcome of PPI therapy for esophageal eosinophilia suggesting that response rates may be improved by adopting a genotype-guided approach to PPI dosing.
What Is Known/What Is New
What Is Known
Proton pump inhibitors are an effective treatment for esophageal eosinophilia with a variable response rate of 30% to 60%.
Proton pump inhibitor pharmacodynamics are strongly influenced by genetic variation in CYP2C19.
STAT6 genetic variants associate with eosinophilic esophagitis.
What Is New
Carriers of CYP2C19∗17 are more likely to fail proton pump inhibitor therapy for esophageal eosinophilia within a defined dose range.
Different STAT6 genetic variants associate with pre-proton pump inhibitor eosinophil counts and a proton pump inhibitor-responsive esophageal eosinophilia outcome.
STAT6 rs324011 synergizes with CYP2C19∗17 to predict a proton pump inhibitor-nonresponsive eosinophilic esophagitis outcome.
Esophageal eosinophilia patients may benefit from genotype-guided dosing of proton pump inhibitors.
Children treated with proton pump inhibitor (PPI) medications to reduce the inflammation associated with esophageal eosinophilia have initial and sustained response rates of 30% to 60% and 70%, respectively (1,2). The emerging consensus is that PPI medications represent a therapy for eosinophilic esophagitis (EoE) much like dietary elimination and swallowed steroids (3). Whether PPI responsive esophageal eosinophilia (PPI-REE) is, however, mediated by a reduction of esophageal gastric acid exposure or by recently identified anti-inflammatory properties of PPIs, remains controversial (3). Pharmacogenomic factors that influence the outcome of PPI therapy for esophageal eosinophilia remain to be identified.
Individual variability in PPI pharmacokinetics and pharmacodynamics is strongly influenced by genetic variation in CYP2C19(4,5). CYP2C19 variants that confer loss of enzymatic function (LOF, ∗2, ∗3, etc) and lead to a poor metabolizer (PM) phenotype, associate with reduced PPI clearance and increased plasma concentrations of PPI compared to normal metabolizers (NM) (6). Similarly, gain of function (GOF, ∗17) variants have been identified that lead to an extensive metabolizer (EM) phenotype, increased clearance, and decreased plasma concentrations of PPI (6,7). The impact of CYP2C19∗17 GOF allele on PPI-REE in children is not known. In the present study, we hypothesize that carriage of CYP2C19∗17 alleles negatively influences PPI responsiveness in children with EoE. Previous studies have demonstrated that eosinophilic inflammation in EoE is driven by STAT6-dependent local expression of eotaxin-3 (CCL26), and that PPIs can block the chromatin remodeling necessary for STAT6 binding and transcriptional activation of CCL26(8–13). Therefore, we hypothesize that genetic variants of STAT6 may also influence responsiveness to PPI treatment in children with EoE.
METHODS
Study Participants
Study participants were prospectively recruited to the parent study at 2 pediatric hospitals in Madrid, Spain between February 2013 and April 2015 as previously described (1). Briefly, children from 2 to 16 years of age who presented with heartburn, chest pain, food impaction, abdominal pain, vomiting, regurgitation, dysphagia, and feeding difficulties, and also had esophageal eosinophilia (≥15 eos/0.24 mm2, peak value), were enrolled in the primary study. Because CYP2C19 is not fully expressed in the human liver during infancy (14), only children 2 years or older were included in the present study. Following an initial endoscopy with biopsy, participants were treated with PPI (n = 88 esomeprazole, n = 3 lansoprazole, n = 1 omeprazole; twice daily at a target dose of 1 mg/kg/dose, for a total dose of 2 mg/kg/day, up to a maximum dose of 80 mg/day). The mean duration (standard deviation) of PPI therapy was 10.0 (1.4) weeks, with a high of 13.9 and a low of 4.6 weeks. Three patients who received PPI therapy for <8 weeks were included in the cohort (range 4.6–7.7 weeks). A second endoscopy with biopsy was performed while participants were still taking PPI. As reflected in Figure S1 (Supplemental Digital Content 1), the dose range across all 92 participants was 0.46 to 2.4 mg/kg/day. Three patients received <1 mg/kg/day. The variation in PPI dose (mg/kg/day) in the other 89 patients was either due to reaching the maximum daily dose of 80 mg or a result of trying to achieve their target dose while being restricted to prescribing available esomeprazole tablet preparations of 20 and 40 mg. In the present study, there were a total of 92 patients examined (full cohort), of which 46 were randomly selected to receive pH probe monitoring before initiation of PPI (pH probe cohort) and 46 did not (non-pH probe cohort).
Histological Definition of Disease and Response to Proton Pump Inhibitor
Biopsies were performed (at least 2 from the distal esophagus and 2 from the proximal-mid esophagus) according to the guidelines for diagnosis and monitoring of EoE (15,16). All biopsies were targeted to areas with abnormal endoscopic findings if present. Following fixation in 10% buffered formalin and staining with hematoxylin and eosin, eosinophil counts from single high-power microscope fields (hpf) corresponding to an area of 0.24 mm2 were recorded. Esophageal eosinophilia was defined as having a peak eosinophil count of ≥ 15 per hpf in 1 or more esophageal biopsy specimens at baseline. After PPI treatment, PPI-REE was defined as <15 eos/hpf and complete PPI-REE was defined as <5 eos/hpf on all esophageal biopsies obtained during the follow-up upper gastrointestinal endoscopy. PPI-nonresponsive EoE was defined as ≥15 eos/hpf on any of the esophageal biopsies obtained during the follow-up upper gastrointestinal endoscopy.
Genotyping
Genomic DNA was isolated from formalin-fixed paraffin-embedded sections of esophageal biopsy tissue (17) and genotyping reactions were conducted as previously described (17). The STAT6 single-nucleotide polymorphisms (SNPs) interrogated and the TaqMan assays used were rs1059513 (C___7480847_10), rs324015 (C____620398_10), rs3024974 (C__26439023_10), rs841718 (C___7480858_10), rs324011 (C____620399_10), rs167769 (C____620401_20), rs2598483 (C__15984966_10), and rs12368672 (C__31186828_10). The CYP2C19 SNPs interrogated and assays used were as previously described (17). In this study, no carriers of CYP2C19 rs4986893 (∗3, inactive), rs41291556 (∗8, inactive), or rs17884712 (∗9, inactive) were identified; therefore, the LOF phenotype that characterizes PMs is defined in this study as carriers of 1 or 2 copies of rs4244285 (diplotypes ∗1/∗2 + ∗2/∗2), without rs12248560 (∗17). The GOF phenotype that characterizes EMs is defined as carriers of 1 or 2 copies of rs12248560 (diplotypes ∗1/∗17 + ∗17/∗17) without rs4244285 (∗2). Individuals who are ∗1/∗1 are defined as NMs. Diplotype ∗2/∗17 was not assigned to a metabolizer phenotype.
Statistical Analysis
Analyses were conducted in R base version 3.5.1 (2018) (18). A 2-sided Fisher exact test (exact P value) was used for comparison of proportions in count data. A 2-sided Wilcoxon rank-sum test (exact P value) was used to determine whether 2 independent samples were selected from populations having the same distribution. A 2-sided Kolmogorov-Smirnov test was used to test for equality between the empirical distribution functions of 2 samples. Continuous variables were transformed using the powerTransform function of the R package car(19). Negative binomial regression from the R statistical package MASS(20) was employed with auto-optimization of the dispersion parameter to assess relationships between independent variables and count dependent variables. Binary logistic regression was used to assess relationships between independent variables and binary dependent variables. A Bayesian version of binary logistic regression (function bayesglm from package arm(21)) was used when perfect separation of a factor was encountered. The Akaike Information Criterion (AIC) (22) was used to assess relative performance of all models. Plots were produced using function ggplot from the R statistical package ggplot2(23). Probability plots of clinical outcome versus dose of PPI were generated using the sjp.glm function from the R statistical package sjPlot(24). Linkage disequilibrium between genetic markers was determined using the r2fast function of R package GenABEL(25). For comparison, LD for the same variants was determined using rAggr(26) (http://raggr.usc.edu/) within the all European cohort (CEU+FIN+GBR+IBS+TSI) of the 1000 Genomes (27) and HapMap (28) databases. Forest plots were prepared with R package forestplot(29). When differences between values with confidence intervals (CIs) were calculated, the MOVER-D method (30) was used to propagate imprecision. Inflation of type-1 error through multiple testing has been addressed by correction of reported P values using the method of Bonferroni (31).
RESULTS
PPI-REE, GERD, PPI Dose, and CYP2C19
The schema for this study is given in Figure S1 (Supplemental Digital Content 1). The baseline characteristics of study patients in the 5 cohorts examined (pH, non-pH, interquartile range (IQR), non-IQR, and full) stratified by clinical outcome, are given in Table S1 (Supplemental Digital Content 2), of the online supplement. Among the pH probe cohort, 32 (70%) were PPI-REE and 14 (30%) were PPI-nonresponsive EoE. Eight of 32 (25%) patients from the PPI-REE group had an elevated reflux index (>4%) from the baseline pH probe study compared to 3 of 14 (21.4%) patients in the PPI-nonresponsive EoE group (P = 1.0). Within the pH probe cohort, the range of PPI doses was 1.18 to 2.33 mg/kg/day. We did not find evidence for an association between reflux index and carriage of CYP2C19∗17 (P = 0.35). The probability of achieving a PPI-REE clinical outcome ranged from 41% to 88% when going from 1.18 to over 2.33 mg/kg/day (Fig. S2 Supplemental Digital Content 3). Binary logistic regression modeling (BLRM) of the association between PPI dose and PPI-REE found that for each unit increase in PPI dose, the odds that a patient would have a PPI-REE outcome tended to increase 7.68-fold (PPI-REE odds ratio (OR) [95% confidence interval (CI)] = 7.68 [0.60, 0.97], P = 0.11, Fig. S2, Supplemental Digital Content 3).
Next we investigated whether CYP2C19∗17 GOF associates with PPI-REE in the pH probe cohort. BLRM of the association between carriage of CYP2C19∗17 GOF and PPI-REE outcome (dominant genetic model for CYP2C19∗17 GOF with race, sex, age, PPI dose, and PPI type included as covariates) found that children who were carriers of CYP2C19∗17 GOF had 8.2-fold better odds of receiving a PPI-nonresponsive EoE diagnosis than children who did not carry CYP2C19∗17 GOF (PPI-REE OR [95% CI] = 0.12 [0.02,0.67], P = 0.02; complete PPI-REE outcome OR [95% CI] = 0.15[0.03, 0.94], P = 0.04; Fig. 1A). Although the pH and non-pH probe cohorts received similar mean (SD) PPI doses (1.79 (1.48) vs 1.83 (1.33) mg/kg/day, P = 0.57), larger proportions of patients receive doses at both the low (<1.54 mg/kg/day) and high (>2.05 mg/kg/day) ends of the concentration range in the non-pH probe cohort relative to the pH probe cohort (Fig. 1B). Specifically, a greater fraction of the pH probe cohort fell within the IQR of doses of the full cohort (67% vs 28%, P = < 0.001, Fig. 1B). Using BLRM (as above), we found that carriage of CYP2C19∗17 GOF was not associated with either PPI-REE or complete PPI-REE outcomes in the non-pH cohort (PPI-REE OR [95% CI] = 1.38 [0.34,5.61], P = 0.65; complete PPI-REE OR [95% CI] = 1.54 [0.37,6.46], P = 0.56, Fig. 1A). In patients who, however, fell within the IQR of the full cohort for PPI dosage, carriers of CYP2C19∗17 GOF had 7.7-fold better odds of failing PPI therapy and receiving a PPI-nonresponsive EoE diagnosis relative to noncarriers (PPI-REE OR [95% CI] = 0.13[0.02,0.83], P = 0.03, Fig. 1A).
FIGURE 1.
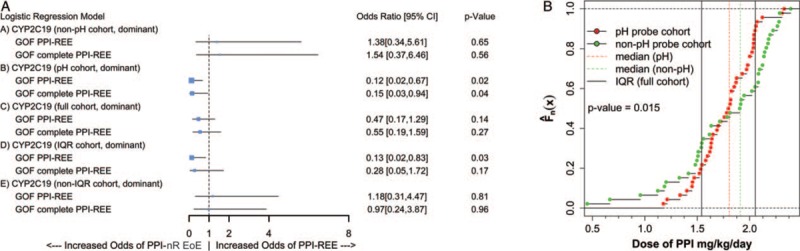
Association of CYP2C19∗17 GOF with outcome of PPI therapy is dose dependent. A, Binary logistic regression modeling of CYP2C19∗17 as predictor of PPI therapy outcome: (A) non-pH, (B) pH, (C) full, (D) IQR cohorts, and (E) non-IQR cohorts. All models include race, sex, age, PPI dose, and PPI type as covariates. B, Plot of the empirical cumulative distribution ( ) of PPI dose for the pH probe (red) and the non-pH probe (green) cohorts. Solid vertical lines indicate the IQR for the full cohort, dashed vertical lines indicate the medians for the pH probe (red), and the non-pH probe (green) cohorts. The 2 distributions are statistically different from each other (Kolmogorov-Smirnov test P value = 0.015). CI = confidence interval; GOF = gain of function; IQR = interquartile range; PPI-REE = proton pump inhibitor-responsive esophageal eosinophilia.
) of PPI dose for the pH probe (red) and the non-pH probe (green) cohorts. Solid vertical lines indicate the IQR for the full cohort, dashed vertical lines indicate the medians for the pH probe (red), and the non-pH probe (green) cohorts. The 2 distributions are statistically different from each other (Kolmogorov-Smirnov test P value = 0.015). CI = confidence interval; GOF = gain of function; IQR = interquartile range; PPI-REE = proton pump inhibitor-responsive esophageal eosinophilia.
STAT6, Baseline Esophageal Eosinophilia, and PPI-REE
We selected eight STAT6 variants for analysis of associations with outcome of PPI therapy for EoE based on literature reports of associations with EoE (32,33), allergy (34), asthma (35,36), eczema (37), serum immunoglobulin E (IgE) (38,39), or viral infections (40) (Table S2, Supplemental Digital Content 4). Genotype counts, SNP frequencies, and Hardy-Weinberg equilibrium P values for the STAT6 variants are given in Table S2 (Supplemental Digital Content 4). The strongest associations were found when assuming a recessive genetic model. Of the 8 STAT6 SNPs interrogated, 4 were present at a frequency ≥0.35 in our population, allowing for their analysis using a recessive genetic model (rs841718, rs324011, rs167769, rs12368672; frequency range = 0.36–0.42, Table S2, Supplemental Digital Content 4) (41). Of these 4 SNPs, 3 were in linkage disequilibrium (r2 ≥ 0.8: rs324011, rs167769, and rs12368672; Fig. S3, Supplemental Digital Content 5).
In our initial analysis (1), we determined that pre-PPI eos/hpf tended to be higher in patients who would eventually fail PPI therapy (peak eos/hpf median [95% CI], PPI-REE = 45[32,65] vs PPI-nonresponsive EoE = 83[71,100], P = <0.01, Table S1, Supplemental Digital Content 2). Therefore, we examined the association between STAT6 variants and pre-PPI eos/hpf in the full cohort of 92 patients (Table 1). Carriers of 2 copies of any of the 4 SNPs (rs841718, rs324011, rs167769, rs12368672) tended to have elevated distal pre-PPI eos/hpf relative to individuals who had 0 or 1 copy (range of the median difference in distal eos/hpf [95% CI], = 25[−46,50] to 50[6,85], Table 1). In particular, 2 SNPs were associated with a >1.7-fold increase in distal pre-PPI eos/hpf in individuals who carry 2 copies relative to individuals who carry 0 or 1 copy: rs324011 (PPI-REE rate ratio (RR) [95% CI] = 1.56 [1.06, 2.3], P = 0.048) and rs167769 (PPI-REE RR [95% CI] = 1.66 [1.1,2.51], P = 0.032).
TABLE 1.
Association between STAT6 SNPs and eosinophil counts
| SNP | Outcome* | Model | Counts 0/1/2 | eos/hpf (0)† | eos/hpf (1)† | RR‡ | P§ |
| rs841718 | Distal pre-PPI eos | Recessive | 81/11 | 55[40,67] | 80[10,100] | 1.21[0.79,1.86] | 0.744 |
| rs324011 | Distal pre-PPI eos | Recessive | 79/13 | 50[37,64] | 85[58,100] | 1.56[1.06,2.3] | 0.048 |
| rs167769 | Distal pre-PPI eos | Recessive | 81/11 | 50[37,64] | 100[58,132] | 1.66[1.1,2.51] | 0.032 |
| rs12368672 | Distal pre-PPI eos | Recessive | 80/12 | 50[37,67] | 90[58,100] | 1.54[1.03,2.31] | 0.070 |
| rs12368672 | Δ Peak eos (post-pre) | Dominant | 33/59 | −24.5[−39, −15] | −41.5[−60, −23] | 1.58[1.13,2.2] | 0.043 |
| rs1059513 | Distal post-PPI eos | Additive | 72/18/2 | 10.5[4,17] | 1[0,2] | 0.31[0.13,0.73] | 0.044 |
eos = eosinophils; RR = rate ratio; pre-PPI = baseline before initiation of proton pump inhibitor; post-PPI = following 8 weeks of PPI therapy; hpf = high power field (0.24 mm2).
*Peak value was the highest recorded value from all biopsies in all regions sampled.
†Genetic model coding, recessive: carriage of 0 or 1 copies of the SNP is coded as 0, carriage of 2 copies of the SNP is coded as 1; dominant: carriage of 0 copies of the SNP is coded as 0, carriage of 1 or 2 copies of the SNP is coded as 1; additive carriage of 0 copies of the SNP is coded as 0, carriage of 1 copy of the SNP is coded as 1, carriage of 2 copies of the SNP is coded as 2.
‡Median (95% CI) are reported.
§Reported value is from negative binomial regression modeling with eosinophil counts as the dependent variable and genotype counts as the independent variable. Values in bold indicate associations that are significant. A Bonferroni correction has been applied to P values where appropriate.
Next, we tested for associations between the change in post-PPI versus pre-PPI eos/hpf (Δ peak eos/hpf) in individuals who carried 1 or 2 copies of any of the 8 interrogated STAT6 variants (Table 1). We found that carriers of 1 or 2 copies of rs12368672 (frequency = 0.39, Table S2, Supplemental Digital Content 4) have a Δ peak eos/hpf that is 1.7-fold larger than that seen in individuals who do not carry rs12368672 (median difference eos/hpf [95% CI], = −17 [−38,7], RR [95% CI] = 1.58 [1.13, 2.2], P = 0.043, Table 1). Carriers of rs1059513 (frequency = 0.12, Table S2, Supplemental Digital Content 4) have a 10.5-fold lower post-PPI eos/hpf relative to noncarriers (median difference eos/hpf [95% CI], = −9.5 [−16, −3], RR [95% CI] = 0.31 [0.13, 0.73], P = 0.044, Table 1).
Finally, we tested for associations between carriage of STAT6 variants and outcome of PPI therapy (Fig. 2). Given the previous results, we focused our analysis on three of the four linked variants rs324011, rs167769, rs12368672, and on rs1059513. In BLRMs examining the association between STAT6 variant and a PPI-REE outcome (dominant genetic model, covariates as above), individuals who carried 1 or 2 copies of rs1059513 had 6.2-fold better odds of achieving a PPI-REE outcome after PPI therapy than individuals who did not carry rs1059513 (PPI-REE OR [95% CI] = 6.16 [1.44, 26.35], P = 0.028) (Fig. 2). When considering complete PPI-REE, the odds improved to 7-fold more likely for carriers of rs1059513 relative to noncarriers (PPI-REE OR [95% CI] = 7.06 [1.98, 24.9], P < 0.01).
FIGURE 2.
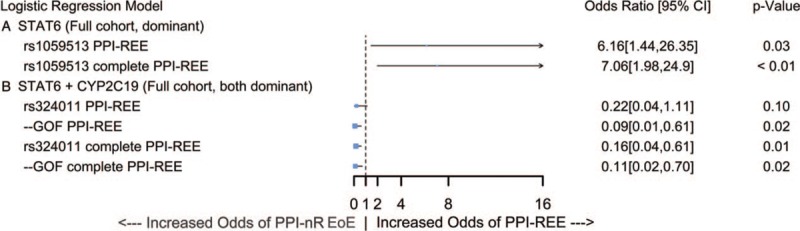
PPI-REE outcome in carriers of STAT6 and CYP2C19∗17 GOF alleles. Binary logistic regression modeling of: (A) STAT6 rs1059513 as predictor of PPI-REE/complete PPI-REE outcome following 8 weeks of PPI therapy in the full cohort and (B) STAT6 and CYP2C19∗17 GOF as co-predictors of PPI-REE in the full cohort. All models include race, sex, age, PPI dose, and PPI type as covariates. CI = confidence interval; GOF = gain of function; PPI-REE = proton pump inhibitor-responsive esophageal eosinophilia.
CYP2C19, STAT6, and PPI-REE
Terms for STAT6 (dominant genetic model), CYP2C19 (dominant genetic model), and their interaction were included in BLRMs with PPI-REE or complete PPI-REE as the outcome (covariates as above, Fig. 2). We found that when rs1059513 and CYP2C19∗17 GOF were modeled as copredictors of outcome, carriers of 1 or 2 copies of rs1059513 had 14.9-fold better odds of responding to PPI therapy and receiving a diagnosis of PPI-REE than individuals who did not carry rs1059513-when considering only individuals who do not carry CYP2C19∗17 GOF (PPI-REE OR [95% CI] = 14.9 [1.87, 119], P = 0.02; complete PPI-REE OR [95% CI] = 7.02 [1.88, 26.3], P < 0.01). When rs324011 and CYP2C19∗17 GOF were modeled as co-predictors of outcome, rs324011 did not significantly predict PPI-REE but did significantly predict complete PPI-REE. Carriers of 1 or 2 copies of rs324011 had 6.10-fold better odds of failing to achieve complete PPI-REE relative to non-carriers (PPI-REE OR [95% CI] = 0.22 [0.04, 1.11], P = 0.10; complete PPI-REE OR [95% CI] = 0.16 [0.04, 0.61], P = 0.01). While the model for rs1059513 did not benefit from the addition of the CYP2C19 GOF term as shown by a positive Δ AIC: 2.11 and a nonsignificant P value for the CYP2C19∗17 GOF term, the model for rs324011 did (ΔAIC complete PPI-REE: −2.94; PPI-REE OR [95% CI] = 0.11 [0.02, 0.70], P = 0.02). The OR of the interaction term between rs324011 and GOF (PPI-REE OR [95% CI] = 8.76 [0.90, 84.8], P = 0.05) implies that the influence of CYP2C19∗17 GOF allele on EoE outcome increases 8.76-fold as rs324011 increases from 0 copies to 1 or 2 copies.
DISCUSSION
The present study reports novel associations between common genetic variants of CYP2C19 and STAT6 and PPI-REE in children who received high-dose PPI therapy for pediatric esophageal eosinophilia. Previously, we have shown that carriage of CYP2C19∗17 GOF alleles associates with pH probe acid exposure outcomes (17) in children with gastroesophageal reflux disease. In the present study, clinically significant esophageal acid exposure, however, does not differentiate PPI-REE from PPI-nonresponsive EoE, and we did not find an association between CYP2C19∗17 and pH probe acid exposure outcomes. We did demonstrate that carriers of CYP2C19∗17 GOF have 8.2-fold better odds of failing PPI therapy and receiving a PPI-nonresponsive EoE diagnosis than children who did not carry CYP2C19∗17 GOF (PPI-REE OR [95% CI] = 0.12 [0.02, 0.67], P = 0.02) in a cohort of patients who received pH probe monitoring. Although patients were randomly chosen to receive pH probe monitoring, a classification tree analysis found that most individuals within the pH probe cohort received a PPI dose within the range of ≥1.569 to <2.075 mg/kg/day, which corresponds well with the IQR dose range of ≥1.54 to <2.05 mg/kg/day for the full cohort. Subsequent analysis of the IQR cohort confirmed that carriers of CYP2C19∗17 GOF have 7.7-fold better odds of failing PPI therapy and receiving a PPI-nonresponsive EoE diagnosis relative to noncarriers (PPI-REE OR [95% CI] = 0.13 [0.02, 0.83], P = 0.03). These results suggest that carriage of CYP2C19∗17 GOF may only influence outcome of PPI therapy within a range of PPI doses (including >1.54 and <2.05 mg/kg/day).
In the present study, we show that 2 linked variants of STAT6, rs324011, and rs167769 associate with increased pre-PPI eos/hpf and that carriage rs324011 also predicts failure of PPI therapy in binary logistic regression models that include CYP2C19∗17 GOF variant as a copredictor. Remarkably, we find a significant interaction between CYP2C19∗17 GOF and STAT6 variant rs324011 suggesting that the influence of CYP2C19∗17 GOF on outcome of PPI therapy for EoE increases almost 9-fold in individuals who are carriers of rs324011. We also find that individuals who carry 1 or 2 copies of rs1059513 have 6.2-fold better odds of achieving a PPI-REE outcome following PPI therapy than individuals who did not carry rs1059513 (PPI-REE OR [95% CI] = 6.16 [1.44, 26.35], P = 0.02). In subanalyses we found that these results were robust for the entire intention-to-treat population, which included 4 patients who received PPIs other than esomeprazole, 3 patients who received PPI therapy for <8 weeks, and 5 patients who received ≤1 mg/kg/day PPI (Supplementary Analysis S1 Supplemental Digital Content 6).
One proposed mechanism for the effect of PPI on EoE is reduced esophageal exposure to gastric acid as is seen with gastroesophageal reflux disease. A recent systematic review and meta-analysis of PPI trials for EoE, however, failed to demonstrate a significant trend in response between patients with pathologic versus normal pH probe outcomes (65% vs 45%) (42), which is consistent with our findings in the present study. The effects of CYP2C19∗17 allele appears to be exerted within a specific range of PPI doses, empirically defined in this study by the IQR of the PPI dose range and does not appear to exert influence at the low and high ends of the dose range. This finding is consistent with the possibility that the high PPI dose range employed by this study compensates for carriage of CYP2C19∗17. This general strategy of therapeutic dose adjustment to compensate for variants of drug metabolizing enzymes is the cornerstone of precision medicine (43).
Several studies have shown that PPIs block STAT6 binding to and transcriptional activation of CCL26(12,13), which is an important chemokine that mediates chemotaxis of eosinophils to the esophagus in EoE (44,45). Variants of STAT6 are known to be associated with diseases that are driven by allergic inflammation including allergy (34), asthma (35,36), eczema (37), serum IgE (38,39), or viral infections (40) and food allergies (46), and a recent genome wide association study conducted by Rothenberg et al (32) identified a variant of STAT6 (rs167769) that is strongly associated with EoE. Upon activation of ST2 expressing cells by IL-33, production of IL-13 is increased (47) leading to activation of STAT6 via IL-4R (48). STAT6 upregulates GATA-3, the master regulator of Th2 inflammatory cell differentiation, IgE class switching in B cells, and expression of major histocompatibility complex class II and CD23 (a low-affinity receptor for IgE (FcεRII)), thus increasing antigen presentation and immune reactivity (49–52). Specifically, STAT6 upregulates transcription of CCL26 (eotaxin-3) 53-fold in esophageal eosinophilia relative to levels found in peptic esophagitis (9) and 490-fold over levels found in normal esophageal biopsies (11). We confirm that common genetic variants of STAT6 influence response to PPI therapy for EoE.
This study had several limitations including small sample size, variation in PPI dose and length of therapy, lack of pH measurement in a large portion of the cohort, lack of a validated questionnaire for symptom assessment, and the potential for additional genetic variants identified in previous genome wide association study studies (32,33) to act as confounders and influence clinical outcome of PPI therapy.
CONCLUSIONS
In conclusion, the effect of PPI medications in pediatric EoE appears to be through a dose-dependent mechanism associated with CYP2C19∗17 that does not correlate with esophageal gastric acid exposure as measured by pH probe monitoring. Genetic variants of STAT6 associate with pre-PPI eos/hpf (rs324011, rs167769, and rs12368672), PPI-REE (rs1059513), and interact with CYP2C19∗17 to increase the odds of PPI-nonresponsive EoE (rs324011). Taken together, our results suggest that genetic variants in CYP2C19 and STAT6 are important factors that influence the pharmacogenetics/genomics of PPI therapy in EoE. Furthermore, our data support an anti-inflammatory mechanism for PPI efficacy in EoE. Pediatric EoE patients may benefit from future genotype-guided personalized PPI therapy.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
This work was funded by a grant from the Nemours Foundation; Anonymous Foundation Grant.
The authors report no conflicts of interest.
REFERENCES
- 1.Gutierrez-Junquera C, Fernandez-Fernandez S, Cilleruelo ML, et al. High prevalence of response to proton-pump inhibitor treatment in children with esophageal eosinophilia. J Pediatr Gastroenterol Nutr 2016; 62:704–710. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez-Junquera C, Fernandez-Fernandez S, Cilleruelo ML, et al. Long-term treatment with proton pump inhibitors is effective in children with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2018; 67:210–216. [DOI] [PubMed] [Google Scholar]
- 3.Dellon ES, Liacouras CA, Molina-Infante J, et al. Updated international consensus diagnostic criteria for eosinophilic esophagitis: proceedings of the AGREE conference. Gastroenterology 2018; 155:1022.e10–1033.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lima JJ, Franciosi JP. Pharmacogenomic testing: the case for CYP2C19 proton pump inhibitor gene-drug pairs. Pharmacogenomics 2014; 15:1405–1416. [DOI] [PubMed] [Google Scholar]
- 5.Franciosi JP, Mougey EB, Williams A, et al. Association between CYP2C19 extensive metabolizer phenotype and childhood anti-reflux surgery following failed proton pump inhibitor medication treatment. Eur J Pediatr 2018; 177:69–77. [DOI] [PubMed] [Google Scholar]
- 6.Furuta T, Shirai N, Sugimoto M, et al. Pharmacogenomics of proton pump inhibitors. Pharmacogenomics 2004; 5:181–202. [DOI] [PubMed] [Google Scholar]
- 7.Lima JJ, Lang JE, Mougey EB, et al. Association of CYP2C19 polymorphisms and lansoprazole-associated respiratory adverse effects in children. J Pediatr 2013; 163:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JY, Zhang X, Nguyen N, et al. Proton pump inhibitors decrease eotaxin-3 expression in the proximal esophagus of children with esophageal eosinophilia. PLoS One 2014; 9:e101391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest 2006; 116:536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeck J, Woisetschlager M. Activation of eotaxin-3/CCLl26 gene expression in human dermal fibroblasts is mediated by STAT6. J Immunol 2001; 167:3216–3222. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharya B, Carlsten J, Sabo E, et al. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol 2007; 38:1744–1753. [DOI] [PubMed] [Google Scholar]
- 12.Cheng E, Zhang X, Huo X, et al. Omeprazole blocks eotaxin-3 expression by oesophageal squamous cells from patients with eosinophilic oesophagitis and GORD. Gut 2013; 62:824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Cheng E, Huo X, et al. Omeprazole blocks STAT6 binding to the eotaxin-3 promoter in eosinophilic esophagitis cells. PLoS One 2012; 7:e50037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koukouritaki SB, Manro JR, Marsh SA, et al. Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 2004; 308:965–974. [DOI] [PubMed] [Google Scholar]
- 15.Dellon ES, Gonsalves N, Hirano I, et al. ACG clinical guideline: evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE). Am J Gastroenterol 2013; 108:679–692. quiz 693. [DOI] [PubMed] [Google Scholar]
- 16.Lucendo AJ, Molina-Infante J, Arias A, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United European Gastroenterol J 2017; 5:335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franciosi JP, Mougey EB, Williams A, et al. Association between CYP2C19∗17 alleles and pH probe testing outcomes in children with symptomatic gastroesophageal reflux. J Clin Pharmacol 2018; 58:89–96. [DOI] [PubMed] [Google Scholar]
- 18.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/, 2018. [Google Scholar]
- 19.Weisberg S, Fox J. An {R} Companion to Applied Regression. 2nd edThousand Oaks, CA: Sage; 2011. [Google Scholar]
- 20.Venables WN, Ripley BD. Modern Applied Statistics With S. New York: Springer; 2002. [Google Scholar]
- 21.Gelman A, Su Y-S. arm: Data Analysis Using Regression and Multilevel/Hierarchical Models. R package version 1.10-1, https://CRAN.R-project.org/package=arm, 2018. [Google Scholar]
- 22.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control 1974; 19:716–723. [Google Scholar]
- 23.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, http://ggplot2.org, 2016. [Google Scholar]
- 24.Lüdecke D. sjPlot: Data Visualization for Statistics in Social Science. doi: 10.5281/zenodo.1308157, R package version 2.5.0, https://CRAN.R-project.org/package=sjPlot, 2018. [Google Scholar]
- 25.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL : an R library for genome-wide association analysis, Bioinformatics, Volume 23, Issue 10, 15 May 2007, Pages 1294–1296, 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263–265. [DOI] [PubMed] [Google Scholar]
- 27.Auton A, Brooks LD, et al. 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International HapMap Consortium. The international HapMap project. Nature 2003; 426:789–796. [DOI] [PubMed] [Google Scholar]
- 29.Gordon M, Lumley T. forestplot: Advanced Forest Plot Using ’grid’ Graphics. R package version 1.7.2. https://CRAN.R-project.org/package=forestplot, 2017. [Google Scholar]
- 30.Newcombe RG. Excel spreadsheet to obtain MOVER-D (MOVER for a difference) confidence interval 2016. [Google Scholar]
- 31.Dunn OJ. Estimation of the means for dependent variables. Ann Math Stat 1958; 29:1095–1111. [Google Scholar]
- 32.Rothenberg ME, Spergel JM, Sherrill JD, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet 2010; 42:289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleiman PM, Wang ML, Cianferoni A, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun 2014; 5:5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Ginkel CD, Pettersson ME, Dubois AEJ, et al. Association of STAT6 gene variants with food allergy diagnosed by double-blind placebo-controlled food challenges. Allergy 2018; 73:1337–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian X, Gao Y, Ye X, et al. Association of STAT6 variants with asthma risk: a systematic review and meta-analysis. Hum Immunol 2014; 75:847–853. [DOI] [PubMed] [Google Scholar]
- 36.Duetsch G, Illig T, Loesgen S, et al. STAT6 as an asthma candidate gene: polymorphism-screening, association and haplotype analysis in a Caucasian sib-pair study. Hum Mol Genet 2002; 11:613–621. [DOI] [PubMed] [Google Scholar]
- 37.Ziyab AH, Davies GA, Ewart S, et al. Interactive effect of STAT6 and IL13 gene polymorphisms on eczema status: results from a longitudinal and a cross-sectional study. BMC Med Genet 2013; 14:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amoako-Sakyi D, Adukpo S, Kusi KA, et al. A STAT6 intronic single-nucleotide polymorphism is associated with clinical malaria in Ghanaian children. Genet Epigenet 2016; 8:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schedel M, Frei R, Bieli C, et al. An IgE-associated polymorphism in STAT6 alters NF-kappaB binding, STAT6 promoter activity, and mRNA expression. J Allergy Clin Immunol 2009; 124:583–589. 589.e1-6. [DOI] [PubMed] [Google Scholar]
- 40.Howell MD, Gao P, Kim BE, et al. The signal transducer and activator of transcription 6 gene (STAT6) increases the propensity of patients with atopic dermatitis toward disseminated viral skin infections. J Allergy Clin Immunol 2011; 128:1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform 2012; 10:117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucendo AJ, Arias A, Molina-Infante J. Efficacy of proton pump inhibitor drugs for inducing clinical and histologic remission in patients with symptomatic esophageal eosinophilia: a systematic review and meta-analysis. Clin Gastroenterol Hepatol 2016; 14:13.e1–22.e1. [DOI] [PubMed] [Google Scholar]
- 43.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 2012; 92:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology 2003; 125:1419–1427. [DOI] [PubMed] [Google Scholar]
- 45.Niranjan R, Rayapudi M, Mishra A, et al. Pathogenesis of allergen-induced eosinophilic esophagitis is independent of interleukin (IL)-13. Immunol Cell Biol 2013; 91:408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishnamurthy P, Kaplan MH. STAT6 and PARP family members in the development of T cell-dependent allergic inflammation. Immune Netw 2016; 16:201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitz J, Owyang A, Oldham E, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 2005; 23:479–490. [DOI] [PubMed] [Google Scholar]
- 48.Pesu M, Takaluoma K, Aittomaki S, et al. Interleukin-4-induced transcriptional activation by stat6 involves multiple serine/threonine kinase pathways and serine phosphorylation of stat6. Blood 2000; 95:494–502. [PubMed] [Google Scholar]
- 49.Tindemans I, Serafini N, Di Santo JP, et al. GATA-3 function in innate and adaptive immunity. Immunity 2014; 41:191–206. [DOI] [PubMed] [Google Scholar]
- 50.Takeda K, Tanaka T, Shi W, et al. Essential role of Stat6 in IL-4 signalling. Nature 1996; 380:627–630. [DOI] [PubMed] [Google Scholar]
- 51.Shimoda K, Van Deursen J, Sangster MY, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature 1996; 380:630–633. [DOI] [PubMed] [Google Scholar]
- 52.Kaplan MH, Schindler U, Smiley ST, et al. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 1996; 4:313–319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


