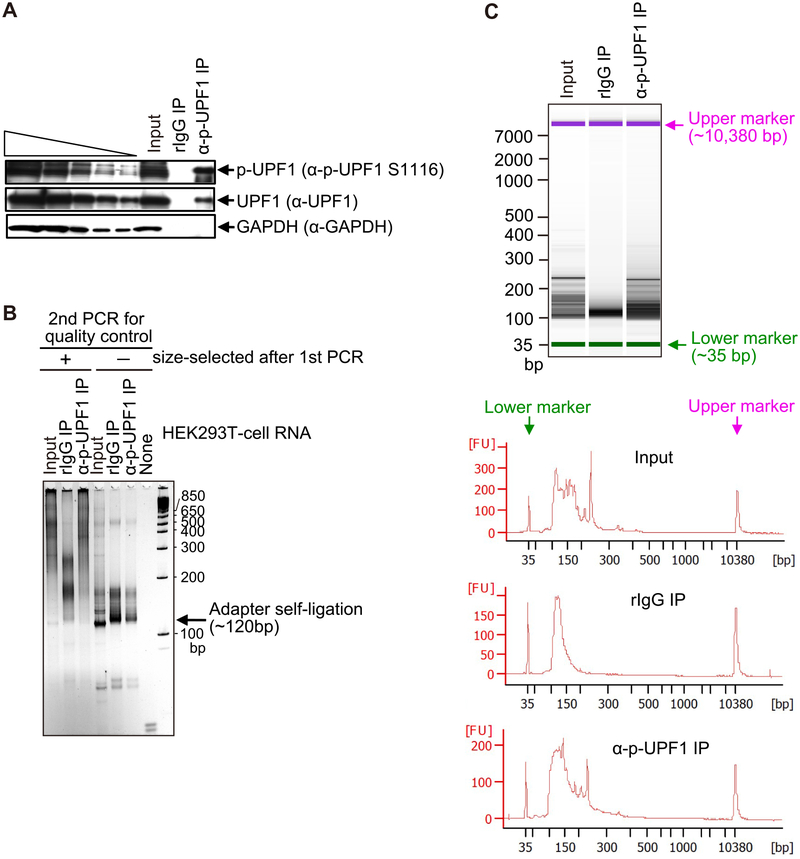
Fig. 2.
Quality check of IP specificity and first and second PCR-sample sizes and amounts used for NMD-DegSeq. (A) Western blot of lysates of HEK293T cells using the specified antibodies prior to (Input) or after IP using anti(α)-p-UFP1 S1116 antibody (i.e. (α)-p-UFPl) or, as a negative control, rabbit (r)IgG. The left-most five lanes are 3-fold dilutions of Input sample (i.e. before IP). (B) SYBR Gold staining after the second PCR-amplification for the purpose of quality control. Analyses are of RNA samples prior to (Input); after (+) IP using anti(α)-p-UFP1 S1116 antibody or, as a negative control, rabbit (r)IgG; or no RNA (None). These RNAs were subjected to the first PCR and, subsequently, as second PCR, after which PCR products were (+) or were not (−) size-selected (the latter has Adapter self-ligation products that should be eliminated prior to library construction). (C) Agilent Bioanalyzer 2100 analyses using capillary electrophoresis (upper) and tracing (lower) of fluorescent samples after the first PCR. Results confirm adequate quality and quantity of samples before (Input) and after IP so that index PCR can be performed to generate samples for Illumina sequencing. FU, fluorescence units; bp, base pairs.