Abstract
Paired box (Pax) proteins function as regulators of coordinated development in organogenesis by controlling factors such as cell growth and differentiation necessary to organize multiple cell types into a single, cohesive organ. Previous work has suggested that Pax transcription factors may regulate diverse cell types through participation in inductive cell-to-cell signaling, which has not been well explored. Here we show that EGL-38, a Pax2/5/8 ortholog, coordinates differentiation of the C. elegans egg-laying system through separate autonomous and non-autonomous functions synchronized by the EGF pathway. We find that EGL-38 protein is expressed at the correct times to both participate in and respond to the EGF pathway specifying uterine ventral (uv1) cell fate, and that EGL-38 is required for uv1 expression of nlp-2 and nlp-7, which are both markers of and participants in uv1 identity. Additionally, we have separated uv1 cell placement and gene expression as distinct hallmarks of uv1 identity and specification, with different dependencies on EGL-38. The parallels between EGL-38 participation in cell signaling events and previous Pax studies argue that coordination of signaling and response to an inductive pathway may be a common feature of Pax protein function.
1. Introduction
Paired box (Pax) transcription factors are necessary for coordinated development and organogenesis of many mammalian systems, such as thyroid (Pax8), kidney (Pax2/8), central nervous system (Pax2/3/5/6/7/8), B lymphocytes (Pax5), pancreas (Pax4/6), eye (Pax6), and ear (Pax2/8) (Blake and Ziman 2014; Dressler et al. 1990; Lang et al. 2007; Macchia et al. 1998; Magliano, Lauro, and Zannini 2000; Noll 1993; Torres et al. 1995). These transcription factors contribute to the differentiation and maintenance of cell fate, as well as the initiation of cellular programs required for proper cell activity (Mansouri, Chowdhury, and Gruss 1998; Miccadei et al. 2002). It is known that Pax proteins are necessary for initiating complex developmental programs, and previous work has suggested that Pax proteins are capable of coordinating differentiation of diverse cell types due to their involvement in inductive cell-to-cell signaling (Lang et al. 2007; Püschel, Gruss, and Westerfield 1992). For example, research on Pax2, Pax6, and Pax8 has shown these proteins are primarily expressed in cells requiring inductive cell-to-cell signaling for development, arguing that it is likely that Pax2/6/8 are targets of, or participants in, these signaling events (Dressler et al. 1990; Püschel, Gruss, and Westerfield 1992). Involvement in cell-to-cell signaling would provide a clear mechanism for Pax proteins to be able to coordinate cell fate and development among distinct, neighboring cell types; however, as of yet, Pax participation in signaling has not been well characterized in humans or mice.
To investigate the interaction of a Pax protein with signaling, and of Pax coordination of cell fate and developmental programs, we are studying the egl-38 gene in Caenorhabditis elegans. EGL-38 is most similar in sequence to mammalian Pax2/5/8, and similarly possesses the paired-DNA binding domain and octapeptide sequence (Chamberlin et al. 1997). egl-38 functions in the development of the hermaphrodite egg-laying system, hindgut, and male spicule (Chamberlin et al. 1997). In particular, the development of the egg-laying system is ideal for examining the interaction of EGL-38/Pax with an inductive signaling pathway for establishment of cell fate and of execution of cellular programs, as this system is dependent on both EGL-38 and the Epidermal Growth Factor (EGF) pathway for differentiation (Chang, Newman, and Sternberg 1999; Katz et al. 1995)
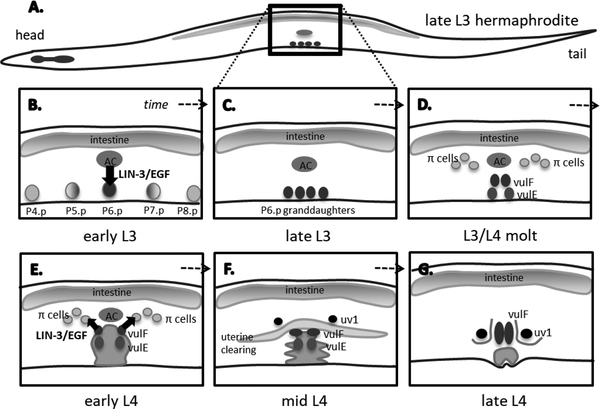
The C. elegans egg-laying system results from coordinated development between vulval cells in the epidermis and mesodermal cells in the somatic gonad (FIGURE 1). First, vulval cell identity is established in response to EGF signaling. The C. elegans EGF signal, LIN-3, is sent from the uterine anchor cell during the L3 larval stage to vulval precursor cells, specifying a cell (P6.p) as the 1° vulval cell (FIGURE 1A) (Hill and Sternberg 1992; Katz et al. 1996). The 1° cell then signals to neighboring cells (to produce the 2° vulval cells), and divides to produce eight cells that form the upper portion of the hermaphrodite vulva. Four of these cells at the apex of the vulva (the vulF cells) then express the lin-3/egf signal gene, which is used for reciprocal EGF signaling back to the uterus during the L4 larval stage (FIGURE 1E) (Chang, Newman, and Sternberg 1999). This reciprocal EGF signal is sent from the vulF cells to a subset of uterine cells surrounding the anchor cell to induce these cells to assume a uterine ventral (uv1) cell identity (Chang, Newman, and Sternberg 1999). The uv1 cells will move into position closer to the vulva to serve as an anchor of the vulva to the uterus, allowing the vulF cells to separate from one another to form the passage through which eggs are laid (FIGURE 1F–G) (Newman and Sternberg 1996). Additionally, the uv1 cells are neurosecretory, and serve an inhibitory role during egg-laying to help control the periodicity and set inactive periods (Banerjee et al. 2017; Collins et al. 2016; Jose et al. 2007).
Figure 1.
(A) A representative schematic of the C. elegans hermaphrodite at the late L3 stage of development. The developing egg-laying system is found on the ventral side of the worm, mid-way down the body. (B-G) A brief representation of egg-laying system development through time. (B) early L3. The LIN-3/EGF signal is sent from the Anchor Cell (AC) to the P6.p cell to specify it as the 1° cell. (C) late L3. P6.p divides, forming 4 granddaughters by late L3. (D) L3/L4 molt. The 1° cell descendants have divided, and (on each side of the animal) there are two vulF and two vulE cells. The π cells have been specified surrounding the AC. (E) early L4. The vulF cells at the apex of the vulva send a reciprocal LIN-3/EGF signal back to the nearest π cells. (F) mid L4. The now-specified uv1 cells move closer to the vulva. (G) late L4. The uv1 cells are in position right next to the vulval cells to serve as an anchor of vulva to uterus.
egl-38 plays a critical role in coordinating the connection between the vulva and the uterus. In an egl-38(lf) mutant, the lin-3/egf signal is not expressed in vulF, and the uv1 cells are not specified (Chang, Newman, and Sternberg 1999; Rajakumar and Chamberlin 2007). Instead, the presumptive uv1 cells retain their original identity and migrate away with their sister cells to fuse with the anchor cell into the uterine seam cell syncytium (Newman, White, and Sternberg 1996). egl-38(lf) mutants also lack vulF cell separation, preventing the laying of eggs (Rajakumar and Chamberlin 2007). Previous work has shown that egl-38 has functions in both the epidermal vulF cells and the mesodermal uv1 cells to contribute to uv1 cell specification and vulF cell separation (Rajakumar and Chamberlin 2007), but its role in coordinating development between these cell types was not clarified.
Here we show how EGL-38 functions in both autonomous (independent of the EGF pathway) and non-autonomous (dependent on the EGF pathway) processes to influence uv1 cell fate and cellular programs necessary for egg-laying function. We have tagged the endogenous egl-38 locus with gfp, which allows for clear examination of EGL-38::GFP localization and timing of expression. We have determined that egl-38 uv1 expression is dependent on the same LIN-3/EGF signal that EGL-38 activates in the vulF cells. Furthermore, our work shows that egl-38 is necessary for the expression in uv1 of two neuropeptide-encoding genes which may have roles in controlling egg-laying and thereby contributing to the functions of differentiated uv1 cells. We have discovered that the uv1 cell traits of positioning above the vulva and of expression of uv1-specific genes are separable activities, with the former being dependent on EGF signaling (and therefore indirectly on EGL-38 in the vulF cell), and the latter requiring direct, autonomous EGL-38 activity within the uv1 cell. EGL-38 coordination of cell fate, development, and its own expression through an inductive cell-to-cell signaling pathway represents a novel Pax function which may represent a concise, direct mechanism for Pax to organize disparate cells into a single, cohesive organ system.
2. Results
2.1. EGL-38 protein is expressed during the development of the C. elegans egg-laying system
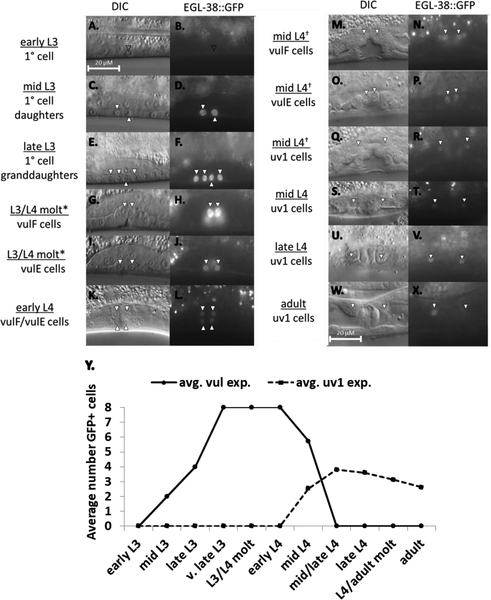
Genetic experiments suggest that egl-38 functions in both the vulva and the uterus of C. elegans (Rajakumar and Chamberlin 2007). Previously, however, egl-38 expression was evaluated only with transcriptional reporter transgenes, which do not inform us about protein expression and localization. Therefore, we utilized CRISPR-mediated genome editing to tag the endogenous locus so that the EGL-38 protein is tagged on the C-terminus with GFP. We find that in this strain, we detect EGL-38::GFP protein fluorescence in all of the expected hermaphrodite tissues, including the vulva, uterus, and hindgut (not shown) and that the protein localizes to the nuclei of cells as predicted for a transcription factor protein. We evaluated protein expression at different time points, beginning in the early L3 stage prior to vulval precursor cell (VPC) division (FIGURE 2). During early L3, we observed that EGL-38::GFP is absent from egg-laying system cells, but is first detected in both P6.px daughters and all four P6.pxx granddaughters following the first and second divisions of the VPCs, respectively (FIGURE 2A–F). After the final VPC divisions, the cells vulF and vulE (derived from P6.p) express EGL-38::GFP; this expression persists through early L4 into mid-L4 (FIGURE 2G–P). At the mid-L4 time point, expression in the uv1 cells can be detected concurrent with vulF and vulE expression (FIGURE 2M–R). EGL-38::GFP expression in vulF and vulE diminishes during mid-L4 so that by the transition to late L4, only uv1 cell expression remains (FIGURE 2S–T). This timing is consistent with the idea that this expression participates in, or is responsive to, the developmental coordination between the two tissues. The uv1 cells continue to express EGL-38::GFP through the adult molt into adulthood (FIGURE 2S–X). Thus EGL-38 protein is expressed in both vulval and uterine cells consistent with the genetic functions of egl-38. EGL-38::GFP expression continues into adulthood in the uv1 cells, but not the vulval cells, indicating that EGL-38 may have a role in the function of mature uv1 cells.
Figure 2.
EGL-38::GFP is expressed during C. elegans egg-laying system development. (A-X) Expression of tagged EGL-38::GFP in the mid-plane view of the developing hermaphrodite egg-laying system. Individual worms were staged and screened for a single time point. Cells of interest are indicated with an arrowhead in both DIC and fluorescent panels. (A-B) Early L3. The P6.p cell lacks expression (empty arrowhead). (C-D) Mid-L3. The two P6.p daughter cells strongly express EGL-38::GFP. (E-F) Late L3. The four P6.p granddaughters all accumulate EGL-38::GFP (third arrowhead rotated to allow visualization of the AC immediately above). (G-J*) L3–L4 molt. Same animal, image plane shifted. vulF (G-H) and vulE (I-J) cell expression. (K-L) Early L4. Presence in vulF and vulE continues. (M-R†) Mid-L4. Same animal, image plane shifted. EGL-38::GFP continues in vulF (M-N) and vulE (O-P) as expression begins in the presumptive uv1 cells prior to positioning (Q-R). (S-T) Mid-L4. By the end of the mid-L4 stage, uv1 expression is apparent while fluorescence in vulF and vulE has diminished to undetectable levels. (U-V) Late L4. uv1 cell expression continues. (W-X) Adult. EGL-38::GFP persists in uv1 cells into young adulthood, but is not as consistent as the worms age. (Y) Time-course trend of EGL-38::GFP expression. Values represent average number of GFP-positive cells, with each time point corresponding to 30 animals. To limit photo bleaching, data were collected from only one side of each worm, but are extrapolated to include cells on the other side of the developing vulva to represent all of the cells of each structure (a full eight vulval cells by late L3, and four uv1 cells by mid-L4).
2.2. EGL-38 expression in uv1 is dependent on EGF signaling
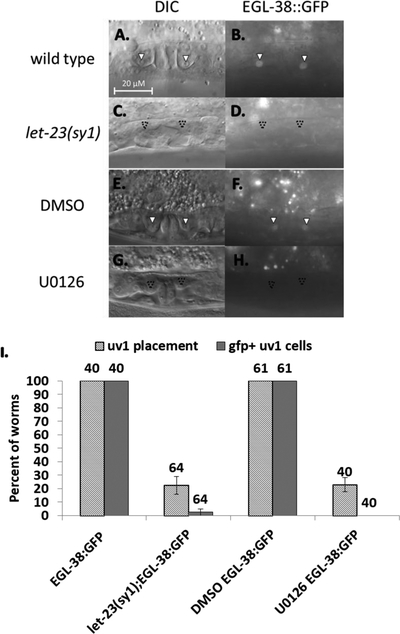
egl-38 mutants exhibit defects in both vulF and uv1 cell specification. In particular, egl-38 mutants are defective in vulF expression of lin-3/egf, which is used to signal to presumptive uv1 cells to promote their development (Chang, Newman, and Sternberg 1999; Rajakumar and Chamberlin 2007). LIN-3/EGF-initiated uv1 cell specification is marked by two features: proper placement of the uv1 cells near the vulva, and expression of uv1-specific genes. If this EGF signaling is disrupted, then the presumptive uv1 cells migrate away from the vulval region and fuse with the uterine seam cell as do the other neighboring uterine (π) cells (Newman and Sternberg 1996; Rajakumar and Chamberlin 2007). We have demonstrated that EGL-38::GFP is expressed in both the signaling and responding cells of this developmental event. To investigate the relationship between EGL-38::GFP expression and EGF signaling, we utilized both genetic disruption and chemical inhibition. For an initial genetic approach, we evaluated EGL-38::GFP expression in animals mutant for the EGF receptor gene, let-23(sy1). As this is a non-null allele, we focused solely on vulvaless let-23(sy1);egl-38::GFP worms, to ensure that vulF cells are absent and unable to communicate with the uterine cells. These worms also lack vulval EGL-38::GFP, as there are no induced vulval cells. We find that placement of presumptive uv1 cells is eliminated in the majority of vulvaless let-23(sy1) worms, and that all lack any uv1 EGL-38::GFP expression (FIGURE 3). Thus expression of EGL-38::GFP in the uv1 cells is dependent on let-23/EGFR activity in the egg-laying system. However, since these animals are both mutant for let-23/EGFR and vulvaless, we sought to more directly test whether EGL-38::GFP is dependent on vulval cell signaling using the MEK inhibitor U0126 (Duncia et al. 1998). Specifically timed application of U0126 allows blocking of the vulF to uv1 EGF signal without affecting the original EGF signal from the anchor cell that triggers the VPCs to divide and form the vulva. We treated late L3 EGL-38::GFP worms with U0126, and find that these animals have morphologically normal vulvas while lacking uv1 cell specification. Similar to the let-23(sy1) mutants, these animals are disrupted for uv1 cell placement, and lack uv1 EGL-38::GFP expression (FIGURE 3). We note that the effect of U0126 is slightly stronger than the effect of let-23(sy1) in this experiment and the ones below. This may reflect the fact that let-23(sy1) is a non-null allele, and retains some activity and that there are non-vulval sources of diffusible lin-3/egf that may compensate for the missing vulF signaling in let-23(sy1) mutants (Aroian et al. 1990; Aroian and Sternberg 1991; Hwang and Sternberg 2004). Nevertheless, in both let-23(sy1) mutants and U0126-treated animals, uv1 cell placement near the vulva was disrupted, and EGL-38::GFP expression was absent. Together, these data argue that EGL-38 expression in uv1 is dependent on EGF signaling from the vulF cells.
Figure 3.
EGL-38::GFP expression is dependent on EGF signaling. (A-B) Wild type late L4. EGL-38::GFP is present in uv1 cells (solid arrowhead). . (C-D) let-23(sy1). uv1 cells are absent in the loss-of-function let-23 background, and EGL-38::GFP expression is also absent in this background (approximate position marked with an empty arrowhead. uv1 positioning can vary between worms). (E-F) DMSO control treatment on wild-type animals. uv1 placement and expression of EGL-38::GFP match wild type in the control treatment. (G-H) U0126 treatment. Blocking the EGF pathway in mid L3 stage results in a lack of uv1 cells, and absent EGL-38::GFP expression. (I) Percent of worms with correctly placed uv1 cells and with expression of EGL-38::GFP in the uv1 cells. Above each bar is shown the number of worms screened. Error bars are shown for the standard error of proportion. For each pairwise comparison of control to mutant/treatment (wild type to let-23(sy1); DMSO to U0126), the difference is significant at a p<0.05 for each pairwise comparison of experiment to control (2-tailed Z-test).
2.3. Expression of neuropeptide genes nlp-2 and nlp-7 in the uv1 cells is dependent on EGF signaling and egl-38
In the vulF cells, a critical function for egl-38 is to promote expression of lin-3/egf; however, EGL-38 targets and functions in the uv1 cells are unknown (Chang, Newman, and Sternberg 1999). The uv1 cells are neurosecretory cells and inhibit egg-laying in response to the passage of eggs out of the uterus (Banerjee et al. 2017; Collins et al. 2016; Jose et al. 2007). To determine if EGL-38 has any direct targets within the uv1 cells, we sought to identify genes which are expressed in the mature uv1 cells. Previous work led us to focus on Neuropeptide-like proteins (nlps), as one such nlp, nlp-7, has been identified as necessary for uv1 inhibition of egg-laying activity (Banerjee et al. 2017). nlps are non-insulin and non-FMRFamide related peptides, with little known about most members of this family (Nathoo et al. 2001). To determine if any additional nlps are expressed in uv1 cells, therefore representing a possible EGL-38 activation target, we tested five genes, nlp-2, 6, 7, 9, and 12. We constructed a reporter with ~2 kb of upstream promoter sequence driving the expression of gfp. Only Pnlp-2::gfp and Pnlp-7::gfp were found to express in uv1 beginning in mid-L4, and persisting into the adult stages. The study which identified nlp-7 involvement in egg-laying also found that the FMRF-like peptide-encoding gene, flp-11, functions with nlp-7 in the uv1 cell. However, we were not able to detect expression of Pflp-11::gfp in uv1 cells using available reporters. Consequently, we focused our studies on nlp-2 and nlp-7.
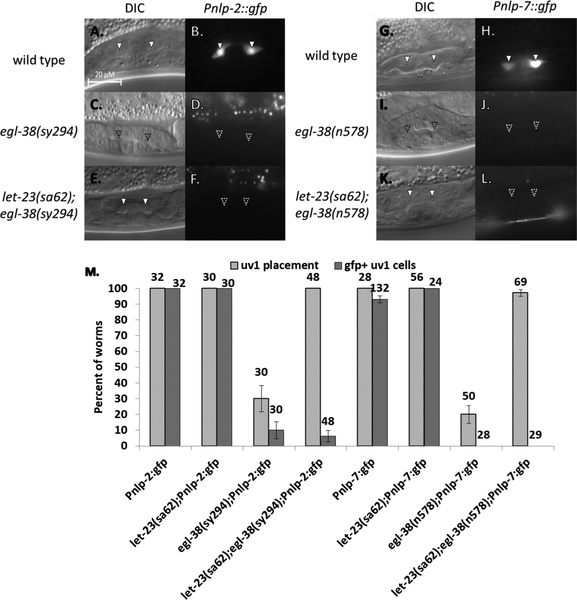
We first sought to determine if Pnlp-2::gfp and Pnlp-7::gfp expression is dependent on uv1 cell specification and if they are markers of mature uv1 cell fate. To do so, we examined the interaction of Pnlp-2::gfp and Pnlp-7:gfp with the EGF pathway by first utilizing the let-23(sy1) allele (FIGURE 4A–Q). In let-23(sy1); Pnlp-2::gfp and let-23(sy1); Pnlp-7::gfp vulvaless animals, expression of the reporter was reduced (to 50% and 25%, respectively). As with EGL-38::GFP, we also examined reporter expression when late L3 animals were treated with U0126 to specifically block signaling from vulF to the uv1 cells. For both Pnlp-2::gfp and Pnlp-7::gfp U0126-treated animals, none of the worms showed reporter expression. Neither reporter exhibits expression in non-uv1 uterine cells in any background, while expression in neuronal cells outside the egg-laying system continued unabated in all altered conditions. Thus, nlp-2 and nlp-7 exhibit a similar dependence as EGL-38 on the EGF pathway for uv1 cell expression. Given that expression from both reporters in the egg-laying system occurs only in the differentiated uv1 cells, it is suggestive that both nlps function in adult uv1 cell activity.
Figure 4.
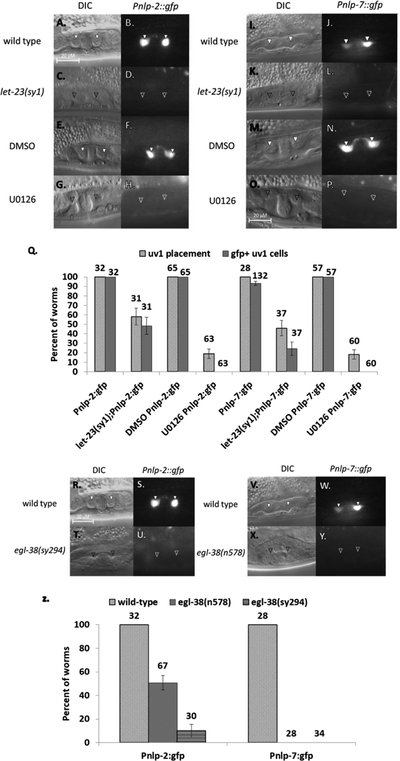
Expression of nlp-2 and nlp-7 in uv1 cells is dependent on EGF signaling and egl-38. (A-H) Pnlp-2::gfp expression. (A-B) Wild type. Pnlp-2::gfp expression in mid-L4 (solid arrowhead). (C-D) let-23(sy1) background. Worms lack uv1 placement and Pnlp-2::gfp expression (approximate position of a normal uv1 cell marked with an empty arrowhead). (E-F) DMSO control treatment. uv1 cell placement and expression matches wild type in late L4. (G-H) U0126 treatment. uv1 cells are absent, as is Pnlp-2::gfp expression. (I-P) Pnlp-7::gfp expression. (I-J) Wild type. Pnlp-7::gfp expression in mid-L4. (K-L) let-23(sy1) background. uv1 cell placement and Pnlp-7::gfp are both absent. (M-N) DMSO control treatment. Worms in late L4 have uv1 placement and expression as in wild type. (O-P) U0126 treatment. uv1 cells and expression are absent. (Q) Percent of worms with correctly placed uv1 cells or with expression of Pnlp-2::gfp or Pnlp-7::gfp in the uv1 cells. Above each bar is shown the number of worms screened. Error bars are shown for the standard error of proportion. For each pairwise comparison of control to mutant/treatment (wild type to let-23(sy1); DMSO to U0126), the difference is significant at a p<0.05 for all pairwise comparisons (2-tailed Z-test). (R-Y) Pnlp-2::gfp and Pnlp-7::gfp expression in the mid-plane of L4 hermaphrodites in wild type or loss-of-function egl-38 backgrounds. (R-U) Pnlp-2::gfp expression. (R-S) Wild type. uv1 cell placement and Pnlp-2::gfp expression in mid/late L4. (T-U) egl-38(sy294) background. Worms lack uv1 cells and expression from the reporter in loss-of-function egl-38. (V-Y) Pnlp-7::gfp expression. (V-W) Wild type. uv1 placement and Pnlp-7::gfp expression in mid/late L4. (X-Y) egl-38(n578) background. uv1 cells and Pnlp-7::gfp expression are both absent in the egl-38 mutant. (Z) Percent of worms with correctly placed uv1 cells or with expression of Pnlp-2::gfp or Pnlp-7::gfp in the uv1 cells in wild type or loss-of-function egl-38 background. Above each bar is shown the number of worms screened. Error bars are shown for the standard error of proportion. For each pairwise comparison of control to mutant (wild type to egl-38(n578 or sy294)), the difference is significant at a p<0.05 for all pairwise comparisons (2-tailed Z-test).
We next investigated the dependence of nlp-2 and nlp-7 expression on EGL-38 activity in the mature uv1 cells. We crossed the reporter transgenes into two egl-38 loss-of-function backgrounds, egl-38(n578) and egl-38(sy294) (FIGURE 4R–Z). These alleles both affect egg-laying system development (Rajakumar and Chamberlin 2007). Expression from the reporter transgenes for both genes is affected. Pnlp-7::gfp expression is eliminated in both egl-38(n578) and egl-38(sy294) backgrounds while Pnlp-2::gfp exhibited significantly reduced expression. These results demonstrate that both nlp-2 and nlp-7 expression is additionally dependent on egl-38 for expression in the uv1 cells.
2.4. EGFR activation bypasses the requirement for egl-38 to promote uv1 cell placement, but not neuropeptide gene expression.
Our EGL-38::GFP expression results and previous genetic mosaic results indicate that EGL-38 is present and can act in both vulF and uv1 cells (Rajakumar and Chamberlin 2007). However, since egl-38 is required for expression of lin-3/egf in vulF cells, it is possible that expression of nlp-2 and nlp-7 is due indirectly to EGL-38 activity in vulF and not directly to its activity in uv1. To separate these possibilities, we examined the epistatic relationship between let-23/egfr and egl-38 in regard to nlp-2 and nlp-7. Utilizing a let-23(sa62) gain-of-function allele, which remains constitutively active in the absence of the lin-3/egf signal, we created double mutants with egl-38 (FIGURE 5). For the Pnlp-2::gfp strain, we utilized egl-38(sy294), as this mutation had a stronger effect on Pnlp-2::gfp expression, whereas for Pnlp-7::gfp, we employed egl-38(n578).
Figure 5.
Activated let-23/EGFR can bypass the egl-38 defect in uv1 placement, but not nlp gene expression. Pnlp-2::gfp and Pnlp-7::gfp expression in the mid-plane of L4 hermaphrodites in wild type, egl-38(lf), and let-23(gf);egl-38(lf) backgrounds. uv1 cells with accurate placement are indicated with a solid arrowhead; absent uv1 cells are marked at the approximate position with an empty arrowhead. uv1 positioning can vary between worms. (A-F) Pnlp-2::gfp expression. (A-B) Wild type. uv1 cells are correctly placed and expressing Pnlp-2::gfp. (C-D) egl-38(sy294) background. Worms lack uv1 cells and expression. (E-F) let-23(sa62); egl-38(sy294) background. uv1 cell placement has been restored, but Pnlp-2::gfp expression remains absent. (G-L) Pnlp-7::gfp expression. (G-H) Wild type. Worms have uv1 cell placement and expression of reporter. (I-J) egl-38(n578) background. Both uv1 cell placement and Pnlp-7::gfp expression are absent. (K-L) let-23(sa62);egl-38(n578) background. uv1 cell placement has been restored, but Pnlp-7::gfp expression is absent. (M) Percent of worms with correctly placed uv1 cells or with expression of Pnlp-2::gfp or Pnlp-7::gfp in the uv1 cells in wild-type or mutant background. Above each bar is shown the number of worms screened. Error bars are shown for the standard error of proportion. For each pairwise comparison of control to mutant the difference is significant at a p<0.05 for all comparisons (2-tailed Z-test).
For both reporters, let-23(sa62) background predictably resulted in wild-type levels of uv1 cell placement and in reporter expression. There was no over-production of uv1-defined cells or other extraneous cells in the egg-laying system. We constructed double mutants let-23(sa62); egl-38(sy294); Pnlp-2::gfp and let-23(sa62); egl-38(n578); Pnlp-7::gfp. For both strains, the uv1 cell placement defect found in egl-38(lf) worms was rescued by let-23(sa62), whereas the expression of Pnlp-2::gfp and Pnlp-7::gfp was not. Importantly, our data show that placement of the uv1 cells near the vulva and expression of nlp-2 and nlp-7 in the uv1 cells are distinct events with separate dependencies on egl-38 activity. Consequently, we interpret that nlp-2 and nlp-7 expression in the uv1 cells is dependent on egl-38 autonomous activity from within the uv1 cells. Because both nlp-2 and nlp-7 are expressed only in the differentiated uv1 cells, their dependence on EGL-38 indicates that a function for EGL-38 within uv1 is to activate developmental programs necessary for egg-laying.
2.5. nlp-2 is a direct target for EGL-38 in uv1 cells
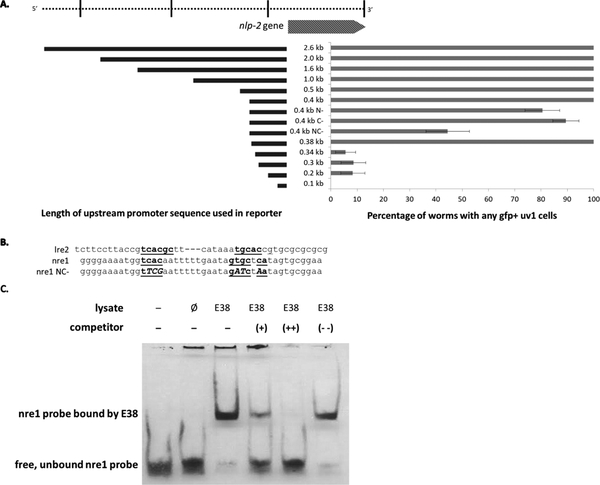
We chose nlp-2 to further examine the relationship of EGL-38 with the uv1 cell neuropeptide genes. The sequence included in the Pnlp-2 reporter consists of around 2.6 kb of upstream sequence. To identify the regulatory element responsible for uv1 cell expression, truncations were introduced beginning at the 5’ upstream end of the original 2.6 kb fragment to create successively smaller reporter genes, and the expression profile of the subsequent reporter was analyzed (FIGURE 6). Due to the abrupt loss of uv1 expression upon deletion, we interpret that a uv1 cell enhancer element exists between 0.34 and 0.38 kb from the start codon of nlp-2. We evaluated this sequence for prospective Pax binding sites using the lre2 sequence from lin-48, a known target for EGL-38 in the hindgut cells (FIGURE 6B) (Johnson et al. 2001). We identified one site with some similarity, which we term nlp-2 response element 1 (nre1). We introduced mutations into this sequence to alter the potential binding sites for the N-, the C-, or both termini of the EGL-38 DNA-binding domain. Only transgenes with both termini mutations exhibited a strong reduction of Pnlp-2::gfp expression, although not to the extent observed when the nre1 enhancer region is deleted (FIGURE 6A). We utilized EMSA to ask whether the DNA binding domain of EGL-38 protein can directly bind the genetically required regulatory sequence, nre1, in vitro. We found that a bacterially expressed Paired Domain of EGL-38 (EGL-38 PD) caused a mobility shift in labelled nre1 probe migration (FIGURE 6C). This shift was specific to lysates expressing the EGL-38 PD, as lysates expressing the empty vector failed to cause a similar shift. Further, the binding of EGL-38 PD to nre1 was competed away in a dose-dependent manner by the known EGL-38 target sequence lre2, but not by a probe with partially overlapping sequence to nre1 (Johnson et al. 2001). Altogether, these data argue that nlp-2 is a direct target for EGL-38 in the uv1 cells.
Figure 6.
nlp-2 is a direct target for EGL-38. (A) Percent of worms with expression of Pnlp-2::gfp or its deletion derivatives. A full-length reporter (shown at top) was constructed and then deletions taken from the reporter to isolate a uv1 cell enhancer region. On the left is shown the length of upstream promoter sequence which was included in the reporters. Worms were screened in mid-late L4 for uv1 expression of the deletion reporter. Deletion of between 0.34 and 0.38 kb upstream from the start codon significantly reduces reporter expression. We define this 40 bp region as the uv1 cell enhancer for nlp-2. Point mutations in this enhancer made to the sequences hypothesized to bind the N- and both termini (NC-) of EGL-38 significantly decrease reporter expression. Residual reporter fluorescence in the mutant construct may be due to the degenerate Pax DNA-binding domain consensus sequence (Czerny, Schaffner, and Busslinger 1993), or failure to identify the key nucleotides. Data are recovered from more than 40 worms per reporter; error bars are shown for the standard error of proportion. For each pairwise comparison of control to mutant the difference is significant at a p<0.05 (comparing 0.4 kb N-, 0. 4 kb NC-, 0.34 kb, 0.3 kb, 0.2 kb, and 0.1 kb to wild type). (B) Comparison of the Pax binding enhancer regions from lin-48 (lre2; Johnson et al., 2001) and nlp-2 (nre1; this work). The mutations introduced in the 0.4 kb reporter are indicated under nre1 (NC-). (C) EMSA showing the EGL-38 DNA-binding Paired domain interaction with the nre1 probe containing the nlp-2 uv1 cell enhancer. In lane 1, no lysate is added. Lane 2 is lysate from bacteria bearing the empty vector. In lane 3, lysate from bacteria expressing the paired domain of EGL-38 (indicated as E38) can bind to the nre1 probe and shift mobility. In lanes 4 and 5, this binding is competed away by increasing amounts of unlabeled lre2, a sequence known to bind specifically to EGL-38. In lane 6, an unlabeled negative probe that corresponds to nlp-2 sequences that partially overlap lre2 is unable to compete away E38 binding to nre1.
3. Discussion
egl-38 has previously been shown to participate in the development of vulval and uterine cells necessary for egg-laying in C. elegans (Chamberlin et al. 1997; Chang, Newman, and Sternberg 1999; Rajakumar and Chamberlin 2007). However, past work focused on its functions in the vulF cells. Here, we have explored more in depth the interaction of egl-38 with the signaling end of the EGF pathway (non-autonomous functions) and the activity of egl-38 within the responding uv1 cells for execution of cellular programs (autonomous functions). EGL-38::GFP protein is present in both the vulF and uv1 cells at developmental times when it can be functionally responsible for, and responsive to, the EGF pathway. In fact, EGL-38::GFP expression in uv1 is dependent on the EGF pathway, and egl-38 is also required for expression of the neuropeptide genes nlp-2 and nlp-7, which have a similar dependence on the EGF pathway as EGL-38. In addition, activated LET-23/EGFR can bypass the requirement for egl-38 in uv1 cell placement, but not in promoting Pnlp-2::gfp and Pnlp-7::gfp expression, showing that uv1 cell positioning and gene expression are two unique, separate aspects of uv1 cell fate. Ultimately, EGL-38 activity in uv1 is coordinated to EGL-38 activity in vulF through the EGF signaling pathway. EGL-38 activates lin-3/egf in the vulF cells; EGF signaling from vulF directs the expression of EGL-38 in the uv1 cells; EGL-38 in uv1 activates transcription of nlp-2 and nlp-7 to mediate cellular functions.
Consistent with an active role for egl-38 in uv1 cells, our results argue that nlp-2 is a direct target for EGL-38, as EGL-38 is capable of binding in vitro to a uv1 cell specific enhancer of nlp-2. We find that nlp-7 expression likewise exhibits dependence on egl-38, although a direct interaction has not been established. nlp-2 and nlp-7 likely both act during egg-laying. The uv1 cells are generally inhibitory, releasing neuropeptides to control the periodicity of egg-laying following mechano-sensory deformation (Alkema et al. 2005; Collins et al. 2016; Jose et al. 2007). uv1 cells synapse with the hermaphrodite specific neurons; the lack of a direct junction indicates that uv1 activity must be through processes such as neuropeptide vesicle release (Banerjee et al. 2017). Consistent with this idea, nlp-7 (with flp-11) has been shown genetically to inhibit egg-laying (Banerjee et al. 2017). Thus we find that EGL-38 has uv1 cell targets important for execution of a differentiated fate, in addition to its roles in coordinating development of egg-laying system cells.
Pax proteins orthologous to EGL-38 are necessary components of cellular development and differentiation for numerous systems, such as the kidney (Pax2/8), thyroid (Pax8), and B cells (Pax5) (Dressler et al. 1990; Lang et al. 2007; Macchia et al. 1998; Magliano, Lauro, and Zannini 2000; Torres et al. 1995). In most organs, such as the kidneys, Pax protein expression is significantly downregulated in the differentiated tissue (Blake and Ziman 2014; Czerny, Schaffner, and Busslinger 1993; Dressler et al. 1990; Lang et al. 2007; Rothenpieler and Dressler 1993; Terzić et al. 1998). However, in some systems, such as the thyroid, Pax activity is necessary after cell specification for expression of terminally differentiated genes required for cellular function. Pax8 is necessary for establishment of the thyroid follicular cells, as well as for expression of thyroid genes including thyroglobulin and thyroperoxidase (Tg and TPO) (Magliano, Lauro, and Zannini 2000; Mansouri, Chowdhury, and Gruss 1998; Miccadei et al. 2002). Interestingly, a study found that in thyroid-derived cell lines, re-introduction of Pax8 resulted in expression of endogenous targets without fully rescuing the differentiated follicular cell phenotype, indicating that while Pax8 is necessary for both processes, only expression of terminally differentiated targets directly involves Pax8 (Fabbro et al. 1994; Magliano, Lauro, and Zannini 2000). Similarly, we have shown in C. elegans that EGL-38 is present both in developing cells of the vulva and in the differentiating uv1 cells, but that EGL-38 also persists in uv1 cells into adulthood. EGL-38 protein becomes abruptly undetectable in the vulF and vulE cells at a time point following expression of the lin-3/egf signal in early/mid-L4 larval stage. In the uv1 cells, expression tapers off more gradually as the worms are in the adult stage, presumably accommodating the EGL-38 role in activating nlp-2 and nlp-7, which are involved in the adult egg-laying function. Both the thyroid and the uv1 cells are neurosecretory organs with important physiological signaling functions in the body (Collins et al. 2016; Jose et al. 2007). Thus a common theme is that Pax2/5/8 activity participates in both the differentiation and function of these neurosecretory organs.
An interesting facet of Pax2/5/8 gene function is that their role in cellular differentiation, combined with temporal and spatial expression patterns, indicates that participating in inductive cell-to-cell signaling may be a common theme for proteins in this group (Dressler et al. 1990; Püschel, Gruss, and Westerfield 1992). For example, in HeLa, CaSki, and renal proximal tubule cell cultures, PAX2 expression was increased by exposure to EGF (de Graaff et al. 2012; Liu et al. 1997). Similarly, in both eutopic and ectopic tissue of endometriosis patients, the expression of PAX2 and EGFR was tightly correlated, indicating that PAX2 expression may be dependent on EGFR activity (de Graaff et al. 2012). Aberrant expression of PAX proteins in cancer tissue is concomitant with high levels of EGF and hallmarks of increased EGF activity, such as the maintenance of tumor growth, proliferation, and survivability (Brand et al. 2011; Herbst 2004; Konecny et al. 2009; Muratovska et al. 2003; J. Wang et al. 2018; Q. Wang et al. 2008). Knockdown of PAX2 in endometrial cancer lines significantly decreases cancer cell viability (Jia et al. 2016; L.-P. Zhang et al. 2011). The relationship of these PAX2/5/8 proteins with inductive signaling pathways has not yet been examined during mammalian development, given the lethal or significant loss-of-function phenotypes that result from loss of either the Pax2/5/8 gene or of signaling activity. However, our study has observed parallels between Pax2/5/8 and the EGF pathway that were highlighted by these adult disease state and cell culture experiments. Therefore, coordination of signaling and responding within a developing organ may be a common developmental function for Pax transcription factors.
4. Experimental Procedures
4.1. Genetic strains and worm culture
Strains were cultured under standard conditions for C. elegans (Brenner 1974; Stiernagle 2006). All experiments were performed at 20°C unless otherwise noted. The reference wild type C. elegans strain is N2; additional C. elegans strains used were:
CM2762 egl-38(gu253[egl-38::gfp])
CM2238 unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp)
CM2694 unc-119(e2498); guEx1554 (Pnlp-7:gfp)
NY2040 unc-119(e2498); ynIs40(Pflp-11:gfp)
CM2763 let-23(sy1); egl-38(gu253[egl-38::gfp])
CM2766 let-23(sy1); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp)
CM2767 let-23(sy1); unc-119(e2498); guEx1554 (Pnlp-7:gfp)
CM2760 egl-38(sy294); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp)
CM2768 egl-38(n578); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp)
CM2427 let-23(sa62); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp)
CM2444 let-23(sa62); unc-119(e2498); egl-38(sy294); guEx1372 (Pnlp-2(0.4kb)::gfp)
CM2769 egl-38(sy294); unc-119(e2498); guEx1554 (Pnlp-7:gfp)
CM2770 egl-38(n578); unc-119(e2498); guEx1554 (Pnlp-7:gfp)
CM2740 let-23(sa62); unc-119(e2498); guEx1554 (Pnlp-7:gfp)
CM2739 let-23(sa62); unc-119(e2498); egl-38(n578); guEx1554 (Pnlp-7:gfp)
4.2. Generating tagged EGL-38:GFP
We tagged the endogenous egl-38 locus with gfp sequences at the 3’ end using methods of (Dickinson and Goldstein 2016). Briefly, we used synthesized sgRNA and purified Cas9 (Prior et al. 2017) combined with a repair template that introduces gfp sequences and selection markers removed via self-excising cassette (Dickinson and Goldstein 2016). A 20-bp guide RNA targeting the desired region of genomic egl-38 was chosen using the MIT CRISPR design tool (Zhang lab, MIT, http://crispr.mit.edu). The guide RNA was ordered and harnessed to the Synthego EZ Scaffold, providing a 100-mer guide RNA (Synthego, Inc.). PCR-generated homology arms, with a 3-point mutated PAM sequence (primers are in Supplemental File 1) were assembled into the GFPŜEĈ3xFlag vector pDD282 (NEBuilder HiFi DNA Assembly Cloning Kit E5520S) and confirmed by PCR and sequencing (Dickinson et al. 2013). Injection mixes contained 50 ng/μL repair template, 20 ng/μL myo-2::mCherry (pCFJ90), 300 mM KCl, and 20mM HEPES, mixed with 5 μM guide RNA and 5 μM Cas9 enzyme (Frøkjaer-Jensen et al. 2008; Prior et al. 2017). This mix was injected into the gonads of N2 adult worms; three days later, 250 μg/mL hygromycin was added to each plate to select for transformed F1 offspring. On days six-eight surviving Roller, non-myo-2::mCherry expressing F1 adults were singled out and allowed to self-cross. Multiple offspring of Rollers were selected and allowed to self-cross, and plates with a homozygous parent were identified. These animals were heat-shocked to remove the self-excising cassette, and screened for wild-type movement and GFP expression. Correct insertion of the GFP was confirmed by PCR and sequencing with flanking genomic primers. The tagged strain is viable, fertile, and appears wild type.
4.3. Generating nlp-2 and nlp-7 reporters and deletion transgenes
Reporter strains for nlp-2 and nlp-7 were constructed using pDD95.69 from the Fire Vector kit (Addgene.org). For nlp-2, 2.6 kB of upstream genomic promoter sequence was amplified using a 5’ SphI-tagged primer and a 3’ SalI-tagged primer; the 2.6 kb amplicon includes only the start codon from the coding sequence. Similarly, for nlp-7, 2.7 kb of upstream promoter (including the start codon) was amplified using a 5’ Sal-tagged primer and a 3’ MscI-tagged primer. The amplicons of both genes were ligated into the promoter-less pPD95.69 plasmid and transformed into DH5α bacteria. nlp-2 deletion clones were generated by designing upstream primers tagged with SphI that were more proximal to the translational start site, and paired with the same 3’ primer. Site-directed mutagenesis of the nlp-2 promoter sequence in pRJ124 (0.4 kb) were performed using standard reagents, according to the Stratagene protocol (Stratagene Quikchange II Site-Directed Mutagenesis Kit). Positive clones were verified by restriction enzyme digest and sequencing. Injection mixes containing 75 ng/μL of confirmed plasmid and 15 ng/ μL of the rescue plasmid unc-119(pTJ1043) were injected into RH10 (unc-119(e2498)) adult hermaphrodites. non-Unc F1 offspring were selected and allowed to self to identify transgenic lines. At least three lines were obtained and screened for fluorescence. The reported data represent an individual line with the most consistent expression pattern.
4.4. Microscopy
Morphological and fluorescent phenotypes were evaluated using a Zeiss Axioplan microscope, under 100x magnification. Hermaphrodite animals were observed at mid-L3 larval stage through adult, depending on the experiment. Larval stage was defined on the basis of the vulval morphology in the mid-plane. Comparably-staged adult animals were evaluated by selecting L4 animals to fresh plates, and evaluating them 24 hours later. Images were taken at auto-calculated DIC and epi-fluorescent exposures, varying from 0.5–0.8 ms and 1.2–1.9 ms respectively. Worms were immobilized on agar pads consisting of 3.5% noble agar in water with a 10 mM sodium azide solution. Screening for vulval cell and uv1 phenotypes involved locating the vulval or uv1 cells in the uppermost plane of the worm; expression in uv1 cells on the side where the worm was laying was not evaluated due to variability in the data due to photo-bleaching of the GFP.
4.5. U0126 experiments
U0126 was dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 10mM. U0126 and DMSO control plates were made by adding 15 μM of U0126 or DMSO to the top of 5 mL NGM in 35 mm petri plates, drying overnight, and spotting with OP50 bacteria. Gravid worms were floated from plates using M9, dissolved with a bleach solution, and the remaining eggs were rinsed three times with M9. The eggs were hatched overnight (~18 hours) with rotation. Synchronized L1 larvae were plated onto standard NGM plates and placed at 20°C. Thirty-two hours later, the L3 larvae were removed and plated to the U0126 or DMSO plates, and returned to 20°C. Screening was performed 8 hours later, when larvae are mid-L4.
4.6. Protein Expression and EMSA
The EGL-38 Paired DNA-binding domain was expressed using a pET23a vector in E. coli BL21 cells (strain MC702) (G. Zhang et al. 2005). Expression and lysis were performed as previously described, with empty-vector cultures (strain MC706) as a negative control (Fitzsimmons et al. 2001). In summary, 400 μL of bacterial culture were added to 40 mL LB with 50 μg/mL Carbenicillin. This was grown at 37°C to an OD600 of ~4, before IPTG was added (final concentration 1mM) and cultures shook at room temperature overnight. Cells were pelleted and re-suspended in 1 mL cold buffer Z (25mM HEPES, pH 7.0; 100mM Kcl; 12.5mM MgCl2; 20% glycerol; 0.1% NP-40; 1mM DTT). Cells were sonicated, lysed, and the supernatants stored on ice for use in EMSA. EMSA was carried out as described (Roche DIG Gel Shift Kit, 2nd Generation, #3353591910). 50 mM MgCl2 was included in the binding (Johnson et al. 2001). Protein lysates were diluted 1:50 in Buffer Z, and incubated with 2 ng labelled probes, and 50x/250x un-labelled competitors. Samples were separated on a 6% non-denaturing gel, with 0.5x TBE buffer, transferred to nylon membrane in 0.5x TBE, and performed detection following the DIG Gel Shift protocol.
Supplementary Material
KEY RESOURCES TABLE
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| none | ||
| Bacterial and Virus Strains | ||
| OP50 E. coli | Caenorhabditis Genetics Center | Wormbase ID: OP50 |
| Biological Samples | ||
| none | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| none | ||
| Critical Commercial Assays | ||
| none | ||
| Deposited Data | ||
| none | ||
| Experimental Models: Cell Lines | ||
| none | ||
| Experimental Models: Organisms/Strains | ||
| CM2762 egl-38(gu253[egl-38::gfp]) | This paper | N/A |
| CM2238 unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp) | This paper | N/A |
| CM2694 unc-119(e2498); guEx1554 (Pnlp-7:gfp) | This paper | N/A |
| NY2040 unc-119(e2498); ynIs40(Pflp-11:gfp) | Caenorhabditis Genetics Center | Wormbase ID: NY2040 |
| CM2763 let-23(sy1); egl-38(gu253[egl-38::gfp]) | This paper | N/A |
| CM2766 let-23(sy1); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp) | This paper | N/A |
| CM2767 let-23(sy1); unc-119(e2498); guEx1554 (Pnlp-7:gfp) | This paper | N/A |
| CM2760 egl-38(sy294); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp) | This paper | N/A |
| CM2768 egl-38(n578); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp) | This paper | N/A |
| CM2427 let-23(sa62); unc-119(e2498); guEx1372 (Pnlp-2(0.4kb)::gfp) | This paper | N/A |
| CM2444 let-23(sa62); unc-119(e2498); egl-38(sy294); guEx1372 (Pnlp-2(0.4kb)::gfp) | This paper | N/A |
| CM2769 egl-38(sy294); unc-119(e2498); guEx1554 (Pnlp-7:gfp) | This paper | N/A |
| CM2770 egl-38(n578); unc-119(e2498); guEx1554 (Pnlp-7:gfp) | This paper | N/A |
| CM2740 let-23(sa62); unc-119(e2498); guEx1554 (Pnlp-7:gfp) | This paper | N/A |
| CM2739 let-23(sa62); unc-119(e2498); egl-38(n578); guEx1554 (Pnlp-7:gfp) | This paper | N/A |
| Oligonucleotides | ||
| Primers are listed in Supplemental Table 1 | This paper | N/A |
| Recombinant DNA | ||
| pDD282 | Dickinson et al., 2015 | Addgene.org/66823/ |
| pDD95.69 | Fire Lab Vector Kit, 1999 | Addgene.org/1654 |
| pRJ124 | This paper | N/A |
| Software and Algorithms | ||
| none | ||
| Other | ||
| none | ||
Highlights:
EGL-38 exhibits dynamic expression during C. elegans egg-laying system development
EGF signaling promotes expression of EGL-38 and nlp genes in uterine uv1 cells
EGL-38 promotes uv1 cell fate with cell-autonomous and non-autonomous functions
The neuropeptide gene, nlp-2, is a direct target for EGL-38 in uv1 cells
Acknowledgements
We thank Adriana Dawes, David Ignacio, Maxine Ignacio, and Kristen Navarro for critical reading of the manuscript. We also thank Ahmed Abokor, Abdulrahman Jama, and Benjamin Kaumeyer for assistance in creating plasmids. Some C. elegans strains were supplied by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Some plasmids were provided by Addgene.org.
Funding
This work was supported by the National Science Foundation (DMS-1361251). Allison Webb Chasser was a Cellular, Molecular and Biochemical Sciences Program Graduate Fellow (5T32GM086252) and Pelotonia Cancer Research Graduate Fellow.
References
- Alkema Mark J., Hunter-Ensor Melissa, Ringstad Niels, and Horvitz H. Robert. 2005. “Tyramine Functions Independently of Octopamine in the Caenorhabditis Elegans Nervous System.” Neuron 46(2): 247–60. [DOI] [PubMed] [Google Scholar]
- Aroian et al. 1990. “The Let-23 Gene Necessary for Caenorhabditis Elegans Vulval Induction Encodes a Tyrosine Kinase of the EGF Receptor Subfamily.” Nature 348(6303): 693. [DOI] [PubMed] [Google Scholar]
- Aroian, and Sternberg PW. 1991. “Multiple Functions of Let-23, a Caenorhabditis Elegans Receptor Tyrosine Kinase Gene Required for Vulval Induction.” Genetics 128(2): 251–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee Navonil et al. 2017. “Local Neuropeptide Signaling Modulates Serotonergic Transmission to Shape the Temporal Organization of C. Elegans Egg-Laying Behavior.” PLOS Genetics 13(4): e1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, and Ziman MR. 2014. “Pax Genes: Regulators of Lineage Specification and Progenitor Cell Maintenance.” Development 141(4): 737–51. [DOI] [PubMed] [Google Scholar]
- Brand Toni M., Iida Mari, Li Chunrong, and Wheeler Deric L.. 2011. “The Nuclear Epidermal Growth Factor Receptor Signaling Network and Its Role in Cancer.” Discovery Medicine 12(66): 419–32. [PMC free article] [PubMed] [Google Scholar]
- Brenner S 1974. “The Genetics of Caenorhabditis Elegans.” Genetics 77(1): 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin HM et al. 1997. “The PAX Gene Egl-38 Mediates Developmental Patterning in Caenorhabditis Elegans.” Development 124(20): 3919–28. [DOI] [PubMed] [Google Scholar]
- Chang Chieh, Newman Anna P., and Sternberg Paul W.. 1999. “Reciprocal EGF Signaling Back to the Uterus from the Induced C. Elegans Vulva Coordinates Morphogenesis of Epithelia.” Current Biology 9(5): 237–46. [DOI] [PubMed] [Google Scholar]
- Collins Kevin M et al. 2016. “Activity of the C. Elegans Egg-Laying Behavior Circuit Is Controlled by Competing Activation and Feedback Inhibition.” eLife 5 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5142809/ (March 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Schaffner G, and Busslinger M. 1993. “DNA Sequence Recognition by Pax Proteins: Bipartite Structure of the Paired Domain and Its Binding Site.” Genes & Development 7(10): 2048–61. [DOI] [PubMed] [Google Scholar]
- Dickinson Daniel J., and Goldstein Bob. 2016. “CRISPR-Based Methods for Caenorhabditis Elegans Genome Engineering.” Genetics 202(3): 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson Daniel J., Ward Jordan D., Reiner David J., and Goldstein Bob. 2013. “Engineering the Caenorhabditis Elegans Genome Using Cas9-Triggered Homologous Recombination.” Nature Methods 10(10): 1028–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR et al. 1990. “Pax2, a New Murine Paired-Box-Containing Gene and Its Expression in the Developing Excretory System.” Development 109(4): 787–95. [DOI] [PubMed] [Google Scholar]
- Duncia John V. et al. 1998. “MEK Inhibitors: The Chemistry and Biological Activity of U0126, Its Analogs, and Cyclization Products.” Bioorganic & Medicinal Chemistry Letters 8(20): 2839–44. [DOI] [PubMed] [Google Scholar]
- Fabbro Dora et al. 1994. “Expression of Thyroid-Specific Transcription Factors TTF-1 and PAX-8 in Human Thyroid Neoplasms.” Cancer Research 54(17): 4744–49. [PubMed] [Google Scholar]
- Fitzsimmons D et al. 2001. “Highly Conserved Amino Acids in Pax and Ets Proteins Are Required for DNA Binding and Ternary Complex Assembly.” Nucleic Acids Research 29(20): 4154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen Christian et al. 2008. “Single-Copy Insertion of Transgenes in Caenorhabditis Elegans.” Nature Genetics 40(11): 1375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaff AA et al. 2012. “Paired-Box Gene 2 Is down-Regulated in Endometriosis and Correlates with Low Epidermal Growth Factor Receptor Expression.” Human Reproduction (Oxford, England) 27(6): 1676–84. [DOI] [PubMed] [Google Scholar]
- Herbst Roy S. 2004. “Review of Epidermal Growth Factor Receptor Biology.” International Journal of Radiation Oncology, Biology, Physics 59(2 Suppl): 21–26. [DOI] [PubMed] [Google Scholar]
- Hill RJ, and Sternberg PW. 1992. “The Gene Lin-3 Encodes an Inductive Signal for Vulval Development in C. Elegans.” Nature 358(6386): 470–76. [DOI] [PubMed] [Google Scholar]
- Hwang Byung Joon, and Sternberg Paul W.. 2004. “A Cell-Specific Enhancer That Specifies Lin-3 Expression in the C. Elegans Anchor Cell for Vulval Development.” Development 131(1): 143–51. [DOI] [PubMed] [Google Scholar]
- Jia Nan et al. 2016. “DNA Methylation Promotes Paired Box 2 Expression via Myeloid Zinc Finger 1 in Endometrial Cancer.” Oncotarget 7(51): 84785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Andrew D., Fitzsimmons Daniel, Hagman James, and Chamberlin Helen M.. 2001. “EGL-38 Pax Regulates the Ovo-Related Gene Lin-48 during Caenorhabditis Elegans Organ Development.” Development 128(15): 2857–65. [DOI] [PubMed] [Google Scholar]
- Jose Antony M., Bany I. Amy, Chase Daniel L., and Koelle Michael R.. 2007. “A Specific Subset of Transient Receptor Potential Vanilloid-Type Channel Subunits in Caenorhabditis Elegans Endocrine Cells Function as Mixed Heteromers to Promote Neurotransmitter Release.” Genetics 175(1): 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz WS et al. 1996. “A Point Mutation in the Extracellular Domain Activates LET-23, the Caenorhabditis Elegans Epidermal Growth Factor Receptor Homolog.” Molecular and Cellular Biology 16(2): 529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz WS, Hill RJ, Clandinin TR, and Sternberg PW. 1995. “Different Levels of the C. Elegans Growth Factor LIN-3 Promote Distinct Vulval Precursor Fates.” Cell 82(2): 297–307. [DOI] [PubMed] [Google Scholar]
- Konecny GE et al. 2009. “HER2 Gene Amplification and EGFR Expression in a Large Cohort of Surgically Staged Patients with Nonendometrioid (Type II) Endometrial Cancer.” British Journal of Cancer 100(1): 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Deborah et al. 2007. “PAX Genes: Roles in Development, Pathophysiology, and Cancer.” Biochemical Pharmacology 73(1): 1–14. [DOI] [PubMed] [Google Scholar]
- Liu S, Cieslinski DA, Funke AJ, and Humes HD. 1997. “Transforming Growth Factor-Beta 1 Regulates the Expression of Pax-2, a Developmental Control Gene, in Renal Tubule Cells.” Experimental Nephrology 5(4): 295–300. [PubMed] [Google Scholar]
- Macchia PE et al. 1998. “PAX8 Mutations Associated with Congenital Hypothyroidism Caused by Thyroid Dysgenesis.” Nature Genetics 19(1): 83–86. [DOI] [PubMed] [Google Scholar]
- Magliano Marina Pasca di, Lauro Roberto Di, and Zannini Mariastella. 2000. “Pax8 Has a Key Role in Thyroid Cell Differentiation.” Proceedings of the National Academy of Sciences 97(24): 13144–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri Ahmed, Chowdhury Kamal, and Gruss Peter. 1998. “Follicular Cells of the Thyroid Gland Require Pax8 Gene Function.” Nature Genetics 19(1): 87. [DOI] [PubMed] [Google Scholar]
- Miccadei Stefania et al. 2002. “The Synergistic Activity of Thyroid Transcription Factor 1 and Pax 8 Relies on the Promoter/Enhancer Interplay.” Molecular Endocrinology 16(4): 837–46. [DOI] [PubMed] [Google Scholar]
- Muratovska Aleksandra et al. 2003. “Paired-Box Genes Are Frequently Expressed in Cancer and Often Required for Cancer Cell Survival.” Oncogene 22(39): 7989–97. [DOI] [PubMed] [Google Scholar]
- Nathoo Arif N., Moeller Rachael A., Westlund Beth A., and Hart Anne C.. 2001. “Identification of Neuropeptide-like Protein Gene Families in Caenorhabditis Elegans and Other Species.” Proceedings of the National Academy of Sciences 98(24): 14000–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, and Sternberg PW. 1996. “Coordinated Morphogenesis of Epithelia during Development of the Caenorhabditis Elegans Uterine-Vulval Connection.” Proceedings of the National Academy of Sciences 93(18): 9329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, White JG, and Sternberg PW. 1996. “Morphogenesis of the C. Elegans Hermaphrodite Uterus.” Development 122(11): 3617–26. [DOI] [PubMed] [Google Scholar]
- Noll Markus. 1993. “Evolution and Role of Pax Genes.” Current Opinion in Genetics & Development 3(4): 595–605. [DOI] [PubMed] [Google Scholar]
- Prior Harriet, Jawad Ali K., MacConnachie Lauren, and Beg Asim A.. 2017. “Highly Efficient, Rapid and Co-CRISPR-Independent Genome Editing in Caenorhabditis Elegans.” G3 (Bethesda, Md.) 7(11): 3693–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Püschel AW, Gruss P, and Westerfield M. 1992. “Sequence and Expression Pattern of Pax-6 Are Highly Conserved between Zebrafish and Mice.” Development (Cambridge, England) 114(3): 643–51. [DOI] [PubMed] [Google Scholar]
- Rajakumar Vandana, and Chamberlin Helen M.. 2007. “The Pax2/5/8 Gene Egl-38 Coordinates Organogenesis of the C. Elegans Egg-Laying System.” Developmental Biology 301(1): 240–53. [DOI] [PubMed] [Google Scholar]
- Rothenpieler UW, and Dressler GR. 1993. “Pax-2 Is Required for Mesenchyme-to-Epithelium Conversion during Kidney Development.” Development 119(3): 711–20. [DOI] [PubMed] [Google Scholar]
- Stiernagle Theresa. 2006. “Maintenance of C. Elegans.” WormBook. http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html (April 5, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzić J, Muller C, Gajović S, and Saraga-Babić M. 1998. “Expression of PAX2 Gene during Human Development.” The International Journal of Developmental Biology 42(5): 701–7. [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, and Gruss P. 1995. “Pax-2 Controls Multiple Steps of Urogenital Development.” Development 121(12): 4057–65. [DOI] [PubMed] [Google Scholar]
- Wang Jieyu et al. 2018. “Paired Box 2 Promotes Progression of Endometrial Cancer via Regulating Cell Cycle Pathway.” Journal of Cancer 9(20): 3743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Qiuyu et al. 2008. “Pax Genes in Embryogenesis and Oncogenesis.” Journal of Cellular and Molecular Medicine 12(6a): 2281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Guojuan et al. 2005. “Alteration of the DNA Binding Domain Disrupts Distinct Functions of the C. Elegans Pax Protein EGL-38.” Mechanisms of Development 122(7–8): 887–99. [DOI] [PubMed] [Google Scholar]
- Zhang Li-Ping et al. 2011. “RNA Interference of Pax2 Inhibits Growth of Transplanted Human Endometrial Cancer Cells in Nude Mice.” Chinese Journal of Cancer 30(6): 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.