Abstract
Nonsense-mediated mRNA decay (NMD) is one of the best characterized and most evolutionarily conserved cellular quality control mechanisms. Although NMD was first found to target one-third of mutated, disease-causing mRNAs, it is now known to also target ~10% of unmutated mammalian mRNAs to facilitate appropriate cellular responses — adaptation, differentiation or death — to environmental changes. Mutations in NMD genes in humans are associated with intellectual disability and cancer. In this Review, we discuss how NMD serves multiple purposes in human cells by degrading both mutated mRNAs to protect the integrity of the transcriptome and normal mRNAs to control the quantities of unmutated transcripts.
Eukaryotic cells are subject to a constant battery of environmental insults, which can change the protein-coding potential of their DNA. To maintain the integrity of gene expression, mRNAs are inspected for errors before they have the opportunity to produce deleterious proteins in bulk. One such mechanism, nonsense-mediated mRNA decay (NMD), inspects mRNAs for common errors that result in the introduction of a premature termination codon (PTC) and cleaves and eliminates them from the transcriptome. PTCs are particularly problematic because they can result in the production of nonfunctional and/or dominant-negative proteins.
In this Review, we discuss how key RNA-associated proteins are rearranged to detect and eliminate faulty transcripts that may differ from their wild-type counterparts by as little as a single nucleotide. NMD is exploited by cells also to shape the transcriptome in ways that promote responses to environmental changes, including those that occur during development, differentiation or stress. Originally discovered in humans in the context of a disease (β0-thalassaemia), we also discuss problems that arise when NMD is faulty or transcripts otherwise escape NMD and how these problems may be remedied by therapeutics in the future (Supplementary Box 1).
Key NMD factors
NMD is a translation-dependent mRNA quality control pathway that selectively degrades mRNAs harbouring a PTC or another NMD-activating feature. NMD is an evolutionarily conserved process that is mediated in all tested eukaryotes by the RNA-dependent helicase and ATPase UPF1, which was first discovered in the budding yeast Saccharomyces cerevisiae1. UPF1 binds to mRNAs undergoing translation2,3 that harbour a PTC by interacting with the translation termination complex, which is composed of eukaryotic release factor 1 (eRF1) and eRF3 (REFS4–9) (FIG. 1a). UPF1 also binds promiscuously to any physically accessible transcript, including the ribosome-free 3′ untranslated regions (UTRs) of not only NMD targets but also non-NMD targets in a manner dependent on its ATPase and helicase activities7,10–14.
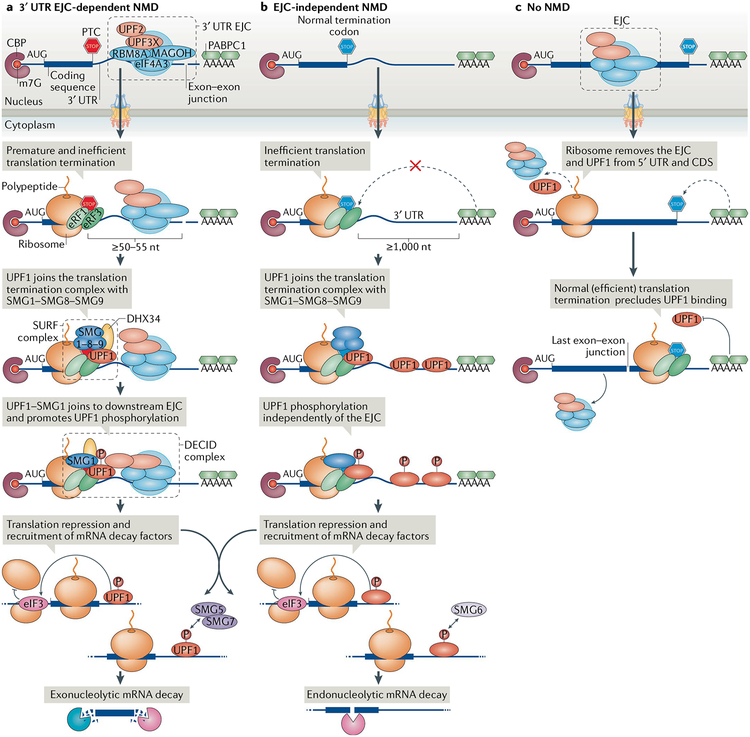
Fig. 1 |. Discriminating between targets and nontargets of nonsense-mediated mRNA decay.
Pre-mRNA splicing is accompanied by deposition of an exon junction complex (EJC) and EJC-associating nonsense-mediated mRNA decay (NMD) factors, such as UPF3X, ~24 nucleotides (nt) upstream of each exon–exon junction. The mRNAs are exported to the cytoplasm and translated. a | 3′ untranslated region (UTR) EJC-mediated NMD. Should translation terminate at a premature termination codon (PTC) that is located ≥50–55 nt upstream of an exon–exon junction, the ribosome will not dislocate the EJC, which is effectively in the 3′ UTR. Because most termination codons are located in the final exon and thus downstream of exon–exon junctions, this situation is abnormal, since an EJC is located downstream of the termination codon. Such translation termination is inefficient, presumably because the EJC interferes with the interaction between polyadenylate-binding protein 1 (PABPC1) and eukaryotic release factor 3 (eRF3). UPF1 and the complex of serine/threonine kinases SMG1–SMG8–SMG9 then join the eRF1–eRF3 translation termination complex and form the SMG1–UPF1–eRFs (SURF) complex. Next, UPF1–SMG1 join the downstream EJC and form the decay-inducing (DECID) complex, where UPF1 is activated by SMG1-mediated phosphorylation (P). UPF1 phosphorylation represses further translation initiation and triggers mRNA decay, which is accomplished by recruiting nucleases, either directly, as in the case of endonucleolytic decay by SMG6, or indirectly (not shown), as in the case of exonucleolytic decay through the SMG5–SMG7 heterodimer. b | EJC-independent NMD. At unusually long, unstructured 3′ UTRs, PABPC1 is too distant from the PTC to efficiently recruit eRF1–eRF3 to initiate translation termination. The presence of UPF1 on the 3′ UTR increases the probability of UPF1 activation by phosphorylation and thus the probability of NMD. c | No NMD. Normally, translating ribosomes remove EJCs and any promiscuously bound UPF1 from the 5′ UTR and from the coding sequence (CDS) but do not travel beyond the stop codon into the 3′ UTR. PABPC1, through its interactions with eRF1–eRF3 or translation initiation factor eIF4G (not shown), is thought to preclude UPF1 from joining the termination complex. CBP, cap-binding protein; eIF4A3, eukaryotic initiation factor 4A3; RBM8A, RNA-binding protein 8A.
UPF1 contains two evolutionarily conserved functional domains: an amino-terminal cysteine- and histidine-rich (CH) domain and a carboxy-terminal RNA helicase domain. UPF1 hydrolyses ATP to translocate 5′-to-3′ along RNAs15–17. The ATPase-dependent helicase activity of UPF1 promotes the decay steps of NMD by translocating along single-stranded RNA and interacting with NMD-activating features, unwinding structured RNA and disassembling messenger ribonucleoproteins to facilitate the completion of mRNA degradation4,17–20. In vitro, the helicase domain of human UPF1 associates with ~10 nucleotides of RNA21,22 and exhibits slow (1.92 bp s−1 for the helicase domain on a structured single-stranded RNA) but highly processive translocating activity18,23 of more than 10 kb (REF18).
The ATPase and helicase activities of UPF1 are stimulated by interactions with two other NMD factors, UPF2 and UPF3X (also known as UPF3B)21,22. UPF1, which forms a closed intramolecular structure through interactions between its terminal domains24, is relaxed to an open configuration when the CH domain interacts in trans with UPF2 (REFS22,25,26). The UPF1–UPF2–UPF3X complex has an evolutionarily conserved role in NMD9,25–31. Of the three NMD factors, UPF1 is at least tenfold more abundant in either S. cerevisiae or human cells than UPF2 or UPF3 (REFS32,33). This suggests that UPF1 has functions that are independent of the other UPF proteins. In vertebrates, there are two UPF3 paralogues, UPF3 (also known as UPF3A) and UPF3X, which differentially function in NMD to support embryogenesis, neurogenesis or gametogenesis29,34–37. UPF3X and UPF3, which is a poor activator of NMD relative to UPF3X, compete for binding to UPF2, indicating that the abundance of UPF3 serves as a molecular rheostat that fine-tunes NMD34,37 (TABLE 1). We note that different pathways of NMD have been described: all require UPF1, but each has different requirements for some other NMD factors38–41.
Table 1 |.
Modulation of nonsense-mediated mRNA decay in mammalian cells
| Stimulus or cause | Mechanism of NMD inhibition | Process promoted | How the process is promoted | Refs |
|---|---|---|---|---|
| Increased ratio of UPF3 to UPF3X | UPF3 represses NMD by competing with UPF3X for UPF2 binding | Spermatogenesis | Upregulation of NMD targets that promote spermatogenesis | 34,37 |
| Hypoxia | NMD suppression by eIF2α phosphorylation | ISR | Upregulation of NMD targets encoding the ISR effectors ATF4, ATF3 and CHOP to promote adaptation to hypoxia | 163 |
| Amino acid deprivation | NMD suppression by eIF2α phosphorylation | ISR | Upregulation of ATF4, ATF3, CHOP and proteins encoding amino acid transporters and proteins that promote autophagy | 160,162 |
| Production of ROS | NMD suppression by eIF2α phosphorylation | ISR | Upregulation of ATF4, ATF3, CHOP and proteins that promote ROS scavenging | 164 |
| Endoplasmic reticulum stress | NMD suppression by eIF2α phosphorylation | ISR, UPR | Upregulation of ATF4, ATF3, CHOP and UPR components | 160,163,167 |
| Chemotherapeutics that severely damage DNA | Caspase-mediated cleavage of UPF1 generates a dominant-negative UPF1 fragment | Apoptosis | Upregulation of NMD targets, including those that encode pro-apoptotic proteins | 168,169 |
| Decreased levels of NMD factors in embryonic stem cells | Transcriptional downregulation of NMD factor genes | Endoderm specification | Upregulation of NMD targets, including those needed for endoderm differentiation | 174 |
| Expression of miR-128, miR-9, miR-124 | miR-128 expression downregulates UPF1, MLN51 and UPF3X; miR-9 and miR-124 expression downregulates UPF3X expression | Neural differentiation | Upregulation of NMD targets, including those promoting neural differentiation | 175,176 |
ATF, activating transcription factor; CHOP, CCAAT-enhancer-binding protein homologous protein; eIF2α, eukaryotic translation initiation factor 2α; ISR, integrated stress response; miR, microRNA; MLN51, metastatic lymph node 51; NMD, nonsense-mediated mRNA decay; ROS, reactive oxygen species; UPR, unfolded protein response.
Target choice and activation of NMD
NMD selectively degrades mRNAs harbouring a PTC (FIG. 1a) or another NMD-activating feature, such as an unusually long 3′ UTR (FIG. 1b). One long-standing question is how cells discriminate target mRNAs from nontarget mRNAs. Recent transcriptome-wide analyses indicate that UPF1 exists on both NMD targets and mRNAs that are not targets10,12,14,42. This contradicts an old model of NMD, in which UPF1 is recruited only to NMD targets. Instead, at least two different mechanisms have been proposed for target discrimination: 3′ UTR exon junction complex (EJC)-dependent NMD and 3′ UTR EJC-independent NMD; the latter is also known as long 3′ UTR-mediated NMD or EJC-independent NMD. Generally, a 3′ UTR EJC mediates the most efficient NMD.
3′ UTR EJC-dependent NMD.
Splicing involves the deposition of a large protein complex called the EJC approximately 20–24 nucleotides upstream of most (~80%) spliced exon–exon junctions without any sequence preference43–46. Transcriptome-wide analyses estimate that ~30–35% of pre-mRNA splicing events in human and mouse cells routinely introduce PTCs that are predicted to trigger NMD47–49. It is also reported that ~50% of human transcripts contain at least one upstream open reading frame (uORF) that ends with a PTC that can trigger NMD when the uORF is used50,51. The core of the nuclear EJC is composed of eukaryotic initiation factor 4A3 (eIF4A3), which is a helicase that anchors the EJC to the RNA, RNA-binding protein 8A (RBM8A; the human homologue of Y14) and MAGOH52. This core is joined by other proteins, including UPF3X and RNA-binding protein with serine-rich domain 1 (RNPS1)45,46,52. Notably, EJCs are compositionally heterogeneous: besides containing either UPF3X or UPF3, other components can undergo a switch, for example, RNPS1 with metastatic lymph node 51 (MLN51; also known as CASC3) after export to the cytoplasm46,52. Variations in EJC composition regulate its roles in alternative splicing53–55, mRNA export43,56 and mRNA translation57–59. When positioned downstream of a termination codon, the best-characterized role of the EJC is the targeting of mRNAs for and strongly activating NMD41,60,61 (FIG. 1a).
During translation, proteins associated with 5′ UTRs or with coding regions are generally removed by processive translocation of, respectively, 40S or 80S ribosomes (FIG. 1c). As active ribosomes do not transit through 3′ UTRs but dissociate once translation terminates, proteins associated with 3′ UTRs remain associated with the mRNA62,63. This distinction forms the basis of the ‘50–55 nucleotide rule’: NMD occurs if a PTC is located ≥50–55 nucleotides upstream of an exon–exon junction64,65 (FIG. 1a). In these cases, the leading edge of the terminating ribosome falls short of physically removing the EJC. After the first round of translation terminates, UPF3X in complex with UPF2, at what is effectively a 3′ UTR EJC22,66–68, recruits UPF1 from the termination complex to the EJC so that it stimulates UPF1 helicase activity21,22. Subsequently, the serine/threonine kinase SMG1 phosphorylates UPF1 (REFS6,12,69) (see below). The deposition of EJCs and their differential association with auxiliary factors are crucial modulators of NMD. For example, EJC-associating factors such as RNPS1 (REFS61,70), MLN51 (REFS41,70), SRSF138,71,72, CWC22 (REFS73–75) and ICE1 (REF76) interact with core EJC proteins and can influence the sensitivity of bound transcripts to NMD.
SMG1 is a phosphatidylinositol 3-kinase-related kinase that preferentially phosphorylates serine and threonine residues upstream of glutamine residues (S/TQ motifs), which are enriched in the amino terminus and carboxy terminus of UPF1 (REFS69,77,78). Translation termination at normal termination codons involves eRF1 and eRF3 binding to the A site of the 80S ribosome, where they promote the release of the nascent peptide. During translation termination of the type that triggers NMD, both UPF1 and SMG1 are recruited to the eRF1-eRF3 translation termination complex to form the SMG1–UPF1–eRFs (SURF) complex6. SURF promotes UPF1 phosphorylation by SMG1 through the interaction with, possibly through bridging, the downstream 3′ UTR EJC to form the decay-inducing (DECID) complex6 (FIG. 1a). In humans, until the SMG1–UPF1 complex joins the EJC79–82, SMG1 is in a complex (SMG1c) with SMG8 and SMG9, and its kinase activity is inhibited, likely because, once UPF1 is phosphorylated, the bound mRNA is committed to degradation6,12,78,83,84. Phosphorylation is likely preceded by the action of the RNA helicase DEAH box polypeptide 34 (DHX34), which associates with the SMG1c complex85. In the context of EJC-mediated NMD, DHX34 promotes the association of UPF1 with EJC-bound UPF2 to form the DECID complex. One model of 3′ UTR EJC-mediated NMD posits that the helicase activity of UPF1 reels in the RNA intervening between the PTC and the EJC20. In the case of normal translation termination, in which the termination codon is located in the final exon or is upstream of but sufficiently close to a 3′ UTR EJC such that the terminating ribosome removes the EJC, no EJC remains to join UPF1, and thus, NMD is not triggered.
Recently, another layer of complexity has been added to the role of translation termination in NMD following the observation that UPF3X directly interacts with eRF3 in vitro86, thereby raising the possibility that the interaction of UPF1 with eRF1-eRF3 is indirect rather than direct5,8. At least in vitro, UPF3X was found to inhibit translation by decreasing the efficiency of peptidyl-tRNA hydrolysis, which fits with a model proposed for S. cerevisiae and extended to humans in which aberrant translation termination allows time for NMD to ensue87. The most extreme interpretation of the in vitro data88 is that, because UPF1 is not present, UPF1 is not involved at all in termination but instead is needed to promote target mRNA cleavage and remodelling of the 3′-cleavage fragment (see below). These observations do not seem to fit with current models of NMD, and potential caveats apply given that these in vitro assays used limiting amounts of eRF1 and eRF3, so that termination was inefficient, and saturating amounts of UPF3X. A more recent study, also performed in vitro, reported that UPF1 has no function in ribosome elongation, translation termination or ribosome recycling89.
NMD generally occurs in association with the nucleus, during or immediately after nuclear export of the mRNA90–92 and while the mRNA is bound at its 5′-cap by the cap-binding protein (CBP) heterodimer CBP20-CBP80 (REFS60,93). Thus, the first round of translation and 3′ UTR EJC-mediated NMD often take place efficiently and quickly once an mRNA can be accessed by cytoplasmic ribosomes94–96. It is during this ‘pioneer round’ of translation, which is defined as the translation of CBP20-CBP80-bound mRNA, that 3′ UTR EJC-mediated NMD largely occurs97. Notably, mRNAs can be bound during this round by more than one ribosome, depending on the frequency of translation initiation and the size of the ORF, as evident from polysome profiling98, biochemical analyses7 and the finding that subsequent translation initiation events on an NMD target need to be prevented for the decay steps of NMD to occur59. During NMD, CBP80 interacts with and chaperones UPF1 together with SMG1 to the terminating ribosome to promote SURF formation99 and subsequently to promote UPF1 and SMG1 binding to the EJC99.
Once CBP20–CBP80 has been replaced by the steady-state cytoplasmic CBP eIF4E at the 5′ caps of NMD targets, 3′ UTR EJC-mediated NMD is largely over — mRNAs become relatively insensitive to NMD and manifest half-lives equal to those of their PTC-free counterparts90–92,96. This is because CBP8099 and the EJCs, both of which promote NMD, have been largely removed before or concurrently with eIF4E binding100. Although eIF4E-bound NMD targets may also be subject to 3′ UTR EJC-mediated NMD101,102, what fraction of eIF4E-bound mRNAs relative to CBP20–CBP80-bound mRNAs is targeted for NMD remains unknown. Notably, like NMD in S. cerevisiae103, long 3′ UTR-mediated NMD (EJC-independent NMD) continues beyond the pioneer round of translation, thus targeting both CBP20–CBP80-bound and eIF4E-bound mRNAs60.
EJC-independent NMD.
Transcripts without a 3′ UTR EJC may also be targeted for NMD, although the molecular mechanism of this process is less well understood than it is for 3′ UTR EJC-dependent targets (FIG. 1b). In S. cerevisiae, although only a small proportion of transcripts (<5%) are subject to pre-mRNA splicing104,105, ~50% of transcripts are routinely targeted for NMD owing to transcriptional and translational errors106,107. Thus, most NMD is not due to the presence of an EJC. Likewise, in human cells, some NMD targets do not follow the ≥50–55 nucleotide rule and are targeted for decay in an EJC-independent manner8,39,108–111. For these targets, cells may perceive translation termination as faulty and recruit UPF1 because one or more proteins that promote NMD outcompete proteins that stimulate translation termination8,87,112. Studies in both S. cerevisiae and human cells demonstrate that shortening the physical distance between a termination codon and the poly(A) tail of an mRNA reduces not only UPF1 binding to the mRNA7,10,11 but also the sensitivity of the mRNA to NMD8,112,113. Normally, binding of the mostly cytoplasmic114 polyadenylate-binding protein 1 (PABPC1) to the poly(A) tail stimulates recruitment of eRF1 and eRF3 to the terminating ribosome115 to promote timely and efficient translation termination. In line with this, tethering PABPC1 between a PTC and the poly(A) tail stabilizes PTC-containing mRNAs8,112,113, possibly by mimicking a shorter 3′ UTR and thus promoting proper translation termination over NMD. Likewise, tethering PABPC1-interacting partners such as eRF3 or translation initiation factor eIF4G between a PTC and the poly(A) tail suppresses NMD116,117. There is molecular evidence that SURF complex formation, and specifically the eRF3–UPF1 interaction, competes with the eRF3–PABPC1 interaction118,119, suggesting that a cellular balance between NMD-promoting proteins or complexes and NMD antagonists can determine the efficiency of mRNA targeting for NMD.
Interestingly, mRNAs can assume a ‘loop’ configuration, in which simultaneously poly(A)-bound PABPC1 and cap-bound CBP80 (REF.120) or eIF4E121 directly interact with eIF4G. This closed-loop configuration, by placing poly(A)-bound PABPC1 in close proximity to a PTC residing close to the 5′ cap, may inhibit NMD despite the great distance between the PTC and the poly(A) tail in a linear configuration119,120,122. Alternatively, NMD resistance could result from translation reinitiation downstream of the PTC, which represents a different type of competition between translation and DECID complex formation123,124. An additional indication that a closed-loop configuration does not generally influence the efficiency of NMD is that, in S. cerevisiae, NMD is more efficient the closer a PTC is to the 5′ cap125.
In mammals, EJC-independent NMD is often observed at endogenous nonmutated transcripts with longer (>1 kb) 3′ UTRs8, even though they do not undergo translation termination at a PTC. Given the positive correlation between 3′ UTR length and UPF1 binding7,10,11, EJC-independent NMD is often termed long 3′ UTR-mediated NMD. In yeast, most mRNAs (>90%) harbouring a longer-than-average 3′ UTR are targeted by NMD126. However, in mammalian cells, both NMD and 3′ UTR structures are more complicated. Transcriptome-wide analyses measuring mRNA abundance and half-life indicate that 3′-UTR length and susceptibility to NMD do not strongly correlate in either human or mouse cells, confounding predictions of NMD susceptibility11,127. This could be because mammalian mRNA 3′ UTRs contain various cis-acting regulatory elements that directly or indirectly inhibit NMD128. For example, 3′ UTR binding by NMD-inhibitory factors such as the cytidine deaminase APOBEC1 complex, which converts a CAA glutamine codon into a UAA termination codon, polypyrimidine tract-binding protein 1 (PTBP1) or hnRNPL can inhibit NMD of cellular, viral or both types of transcript provided that APOBEC1, PTBP1 or hnRNPL binds immediately downstream of the termination codon to shield the 3′ UTR from binding UPF1 and prevent recognition of the mRNA as an NMD target129–131 (see below).
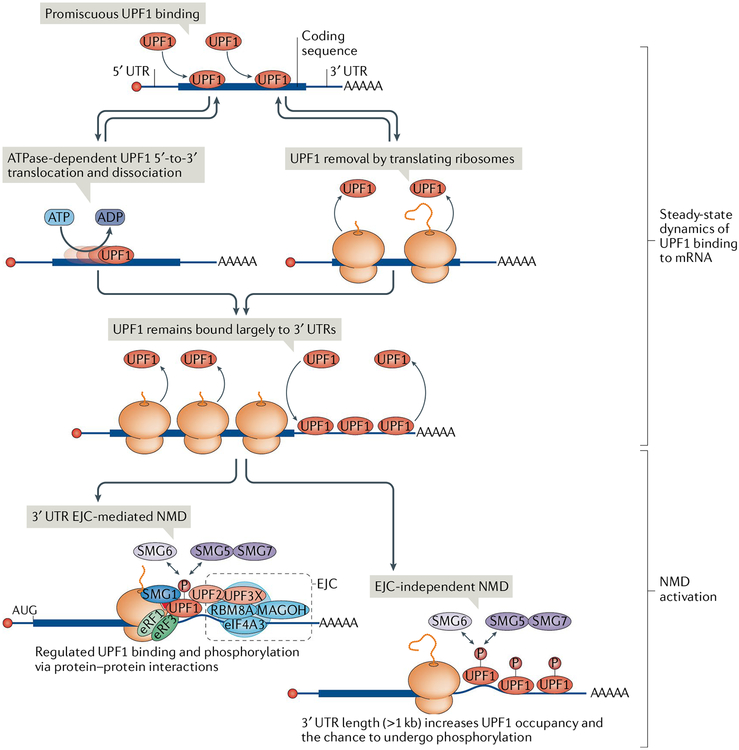
Transcriptome-wide analysis in human cells of phosphorylated UPF1, which is the activated form of UPF1, indicates it is enriched not only on 3′ UTR EJC-mediated NMD targets but also on the 3′ UTRs of EJC-independent NMD targets12,132. How is bound UPF1 phosphorylated without forming the DECID complex (DECID cannot form because a downstream EJC is not present)? Recent biochemical and biophysical studies indicate that UPF1 binding is promiscuous: it is present on most cellular mRNAs, but UPF1 dissociates from nontarget mRNAs in a manner dependent on its ATPase and helicase activities12,13 (FIG. 2). Presumably, a long 3′ UTR increases the probability of UPF1 occupancy and thereby the chance that occupied UPF1 will undergo phosphorylation. Furthermore, UPF1 is frequently observed bound to G-rich sequences11 and GC-rich sequences132 that are predicted to form stable secondary structures. Data indicate that these structures stall UPF1 translocation and thus increase the chance of UPF1 phosphorylation and recruitment of other decay-inducing factors132. That NMD can be activated independently of a 3′ UTR EJC is also supported by experiments that artificially tethered UPF1 to 3′ UTRs, resulting in mRNA decay29,60. Furthermore, a 3′ UTR EJC is dispensable for subsets of NMD targets, as indicated by the finding that UPF1 can trigger mRNA decay when it is recruited to 3′ UTRs by the double-stranded RNA-binding protein staufen, thereby triggering staufen-mediated mRNA decay133, or when it is recruited by stem-loop-binding protein, thereby triggering the decay of cell-cycle-regulated histone mRNAs134. Notably, unlike 3′ UTR EJC-dependent NMD, which is activated during the pioneer round of translation, these types of decay and EJC-independent NMD can also be activated later, during the steady-state rounds of translation60,135,136.
Fig. 2 |. UPF1 binding to mRNA and activation of nonsense-mediated mRNA decay.
Cellular UPF1 is largely nonphosphorylated and promiscuously binds to and dissociates from accessible RNA, including mRNAs, using its ATP-dependent 5′-to-3′ RNA helicase activity. UPF1 bound to the mRNA 5′ untranslated region (5′ UTR) and coding sequence is also actively removed by translocating ribosomes, and as a result, UPF1 is relatively enriched on the mRNA 3′ UTR. UPF1 binding to mRNA can activate nonsense-mediated mRNA decay (NMD) through different mechanisms. A 3′ UTR exon junction complex (EJC) increases UPF1 phosphorylation (P) through protein-mediated interactions, whereas a long 3′ UTR (>1 kb) can increase UPF1 occupancy on the 3′ UTR and thus the probability of UPF1 phosphorylation. In both cases, NMD is activated by serine/threonine kinase SMG1-meditated UPF1 phosphorylation, which in turn results in translation repression and recruitment of mRNA decay factors such as SMG5–SMG7 and SMG6. eIF4A3, eukaryotic initiation factor 4A3; eRF, eukaryotic release factor; RBM8A, RNA-binding protein 8A.
Degradation of the NMD target
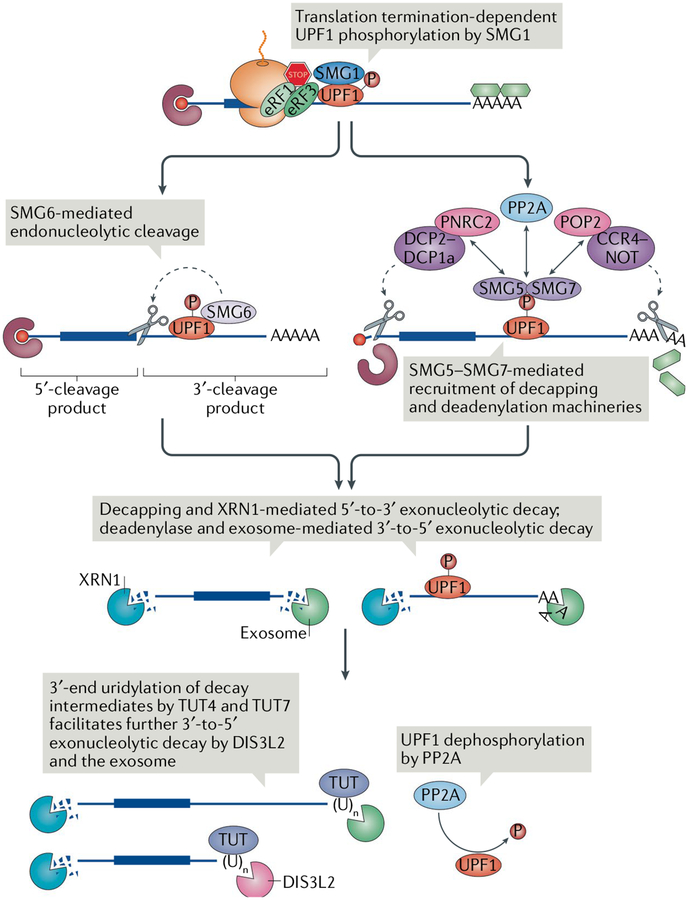
UPF1 phosphorylation by SMG1 is a prerequisite for and represents a commitment step in NMD12,45,59 (FIG. 3). Phosphorylation of UPF1 at multiple residues within its amino-terminal and carboxy-terminal regions prevents further rounds of translation initiation by promoting its interaction with eIF3 and is crucial for mRNA decay59, serving as a platform for recruitment of RNA degradation enzymes6,12,78,83,84.
Fig. 3 |. Degradation of the nonsense-mediated mRNA decay targets.
Following triggering of nonsense-mediated mRNA decay (NMD) by translation termination through serine/threonine kinase SMG1-mediated UPF1 phosphorylation (P), phosphorylated UPF1 recruits either the endonuclease SMG6 or the SMG5–SMG7 complex. SMG6 cleaves the mRNA near the premature termination codon (PTC), whereas SMG5–SMG7 recruits the deadenylation complex CCR4–NOT through its subunit POP2 and the decapping complex mRNA-decapping enzyme 2 (DCP2)–DCP1a through its subunit PNRC2. DCP2–DCP1a can also be recruited by CCR4–NOT (not shown). SMG5–SMG7 also recruits protein phosphatase 2A (PP2A), which dephosphorylates UPF1. NMD intermediates from either pathway are degraded 5′-to-3′ by the exonuclease XRN1 and 3′-to-5′ by the exosome or the exosome-free 3′-to-5′ exonuclease DIS3-like exonuclease 2 (DIS3L2). Terminal uridylyltransferase 4 (TUT4) and TUT7 can append nontemplated uridines at the 3′ ends; the uridylated decay intermediates are favoured for degradation by DIS3L2. eRF, eukaryotic release factor.
RNA degradation can proceed through several pathways. Phosphorylated UPF1 can directly recruit SMG6 (also known as EST1A), an endonuclease with a PilT amino-terminal domain, which cleaves target mRNA in the vicinity of the termination codon27,137–139 (FIG. 3). This generates a 5′-cleavage product that is likely degraded 3′-to-5′ not only by the exosome140 but also by DIS3-like exonuclease 2 (DIS3L2)141 independently of the exosome142 and a 3′-cleavage product bearing UPF1 and, if present, one or more EJ Cs. The 3′-cleavage product must be stripped of its protein components by the helicase activity of UPF1 to provide access to the 5′-to-3′ exoribonuclease XRN1 (REF19) (FIG. 3). The RNA helicase Moloney leukaemia virus 10 (MOV10) was also reported to facilitate mRNA decay by remodelling the 3′ UTR42.
Alternatively to recruiting SMG6, phosphorylated UPF1 can directly recruit the SMG5–SMG7 heterodimer, which in turn recruits the CCR4–NOT deadenylation complex through an interaction between SMG7 and the POP2 (also known as CNOT8) subunit143–145 (FIG. 3). CCR4–NOT then removes the poly(A) tail and recruits the decapping complex member mRNA-decapping enzyme 2 (DCP2)144,145. Phosphorylated UPF1 and SMG5 can additionally directly recruit the decapping complex and the decapping enhancer PNRC2 (REFS45,146–149) (FIG. 3). The decapped and/or deadenylated mRNA is then subject to 5′-to-3′ and/or 3′-to-5′ decay, respectively. In S. cerevisiae, a complex of Upf1, Dcp1, Dcp2, Nmd4 and Ebs1 has been identified, of which the latter two factors may be similar to SMG5 and SMG7 (REF.150), pointing to possible overall similarities in NMD components between yeast and humans. In Caenorhabditis elegans, NMD has been shown to interact with components of nonstop RNA decay, which degrades mRNA lacking a stop codon, to complete mRNA degradation and ribosome recycling151. Conceivably, this coupling of processes may also apply to mammals.
The SMG6 and SMG5–SMG7 pathways seem to be partially redundant because, at least in human HeLa cells, they act on the same cohort of transcripts143,152. Nevertheless, the extent to which a particular transcript is targeted by one decay pathway over the other may vary in different cell types and even different sources of the same cell type owing to differences in passaging conditions139,141. It should also be noted that deadenylation and exosome-mediated decay of the 3′ ends of decay intermediates are accompanied by the addition of nontemplated nucleotides, which are largely uridines appended by terminal uridylyltransferase 4 (TUT4) and TUT7 and removed by DIS3L2 (REF141) (FIG. 3). Whereas the addition of non-uridine residues to decay intermediate 3′ ends inhibits decay, addition of uridine promotes 3′-to-5′ decay by DIS3L2 (REF141). Following degradation, phosphorylated UPF1 is dephosphorylated by the protein phosphatase 2A (PP2A) holoenzyme, which is recruited by SMG5–SMG712,83,153,154 (FIG. 3).
There has been some controversy regarding the sub-cellular location of the decay steps of NMD. As NMD factors colocalize with cytoplasmic processing body (P-body) components, NMD was proposed to occur in P-bodies, which are indeed cytoplasmic compartments where decapping and 5′-to-3′ degradation machineries are enriched155. However, genetic evidence strongly refutes that P-bodies are necessary for NMD. For example, depletion of Ge-1, which is a protein required for P-body maintenance, has little impact on NMD156,157. Recent evidence from mammalian cells demonstrates that, as in S. cerevisiae158, the decay steps of NMD are initiated while the mRNA target is bound by ribosomes141. Additionally, as in S. cerevisiae158, mammalian-cell NMD decay intermediates have not been detected in ribosome-free fractions, in which P-bodies should reside141. As in the case of RNAi in fruitfly cells, where polysome-free, partially degraded mRNAs are sent to P-bodies156, P-body formation may involve late steps of mRNA decay rather than represent the site where NMD is initiated.
Physiological roles of NMD
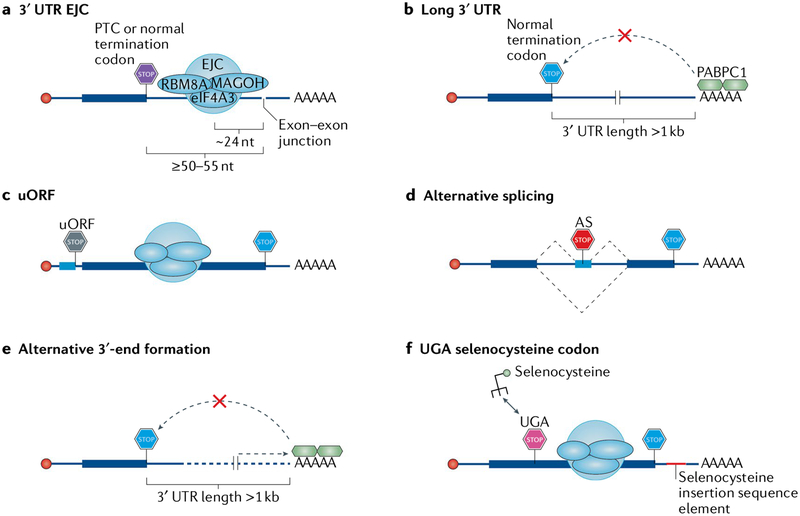
In addition to its quality control function, which usually involves mRNA degradation, NMD also controls the abundance of ~10% of the cellular transcriptome. Features that explain why at least some transcripts are recognized by the NMD machinery (FIG. 4a) include an unusually long (>1 kb) 3′ UTR (FIG. 4b); an uORF with a termination codon that resides ≥50–55 nucleotides upstream of an exon–exon junction (FIG. 4c); regulated alternative splicing that introduces a PTC (FIG. 4d); regulated alternative 3′-end formation that results in normal termination codons triggering 3′ UTR EJC-dependent NMD (if an alternative poly(A) site is chosen that positions a splice site properly downstream of the termination codon) or EJC-independent NMD (FIG. 4e); or a UGA codon encoding selenocysteine that is recognized as a PTC when selenocysteine concentrations are low, and selenocysteine cannot be efficiently incorporated (FIG. 4 f). Furthermore, although some nonmutated endogenous transcripts may not have identifiable features that mark them as NMD targets, disruption of cellular NMD through depletion of an NMD factor or treatment with a small-molecule NMD inhibitor prolongs their half-lives, indicating that they are NMD targets. Thus, it is not surprising that NMD has the capacity to co-regulate the abundance of entire groups of genes47,159–162. Furthermore, as a post-transcriptional mechanism, NMD can be used to support speedy cellular responses to various stimuli. These features are used by cells, for example, both during development and when stressed, or are circumvented by infecting viruses (Supplementary Box 1).
Fig. 4 |. Features of cellular mRNAs that activate nonsense-mediated mRNA decay.
a | An exon–exon junction located ≥50–55 nucleotides (nt) downstream of either a premature termination codon (PTC) or a normal termination codon, with an exon junction complex (EJC) deposited ~24 nt upstream of the junction so that it is far enough from the termination codon and cannot be removed by the terminating ribosome. b | A long (more than ~1 kb), unstructured 3′ untranslated region (3′ UTR), which places the nonsense-mediated mRNA decay (NMD) suppressor polyadenylate-binding protein 1 (PABPC1) too far from the termination codon. c | An upstream open reading frame (uORF) whose termination codon is interpreted as a PTC because an EJC lies ≥50–55 nt downstream of it. d | Alternative splicing (AS) that generates a termination codon that can function as a PTC with any of the above-described features. Alternative splicing may also convert a normal termination codon into one that triggers NMD. e | Alternative 3′-end formation that generates either a 3′ UTR EJC (not shown) or a long 3′ UTR. f | In the few selenoprotein-encoding mRNAs, a UGA codon encoding selenocysteine in cis to a selenocysteine insertion element in a 3′ UTR. The selenocysteine insertion element normally directs the incorporation of selenocysteine, but when cellular selenocysteine concentrations are low, the process becomes inefficient and the UGA codon may be read as a termination codon, which, if properly placed, leads to NMD. eIF4A3, eukaryotic initiation factor 4A3; RBM8A, RNA-binding protein 8A.
Adaptation to stress and environment.
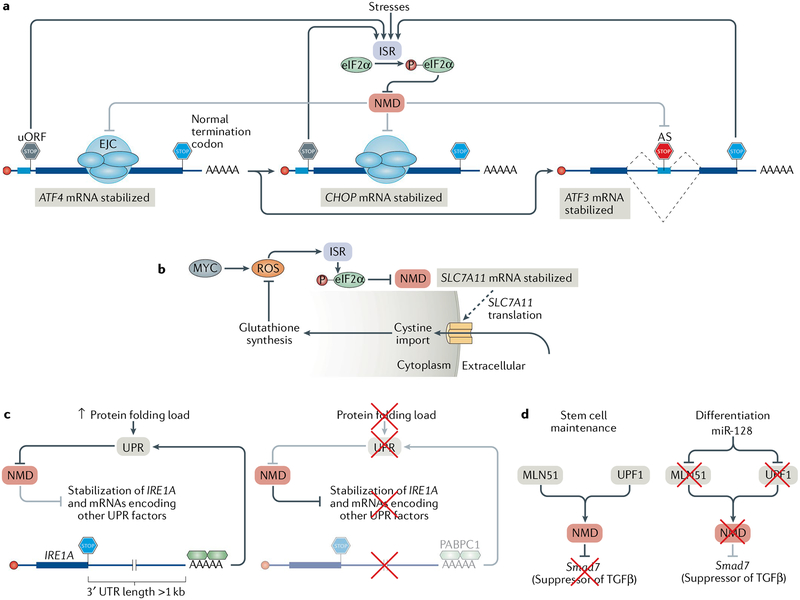
A number of stresses including hypoxia163, amino acid deprivation160 and the production of reactive oxygen species (ROS)164 can blunt NMD (TABLE 1). These stresses lead to activation of the integrated stress response (ISR) with the aim of re-stablishing homeostasis165 (FIG. 5a). The ISR induces eukaryotic translation initiation factor 2α (eIF2α) phosphorylation, which decreases protein synthesis, in general, but promotes both the stability and translation of the mRNA encoding activating transcription factor 4 (ATF4). ATF4, in turn, upregulates the expression of the transcription factors CCAAT-enhancer-binding protein homologous protein (CHOP; also known as DDIT3) and ATF3, which together orchestrate the production of effectors of the ISR. ATF4, CHOP and ATF3 are all encoded by natural NMD targets, thereby explaining how cells use NMD to keep the ISR at bay until stress of sufficient magnitude promotes ISR activation and NMD suppression.
Fig. 5 |. Physiological roles of nonsense-mediated mRNA decay.
a | Various extracellular stresses can induce the integrated stress response (ISR). During the ISR, eukaryotic initiation factor 2α (eIF2α) is phosphorylated (P), leading to the suppression of nonsense-mediated mRNA decay (NMD). NMD suppression enables the expression of NMD targets, including those encoding the transcription factors activating transcription factor 4 (ATF4), CCAAT-enhancer-binding protein homologous protein (CHOP) and ATF3, which coordinate the expression of proteins that alleviate the stresses. Following resolution of the ISR, NMD is resumed and suppresses the expression of ATF4 and CHOP, because their mRNAs contain an upstream open reading frame (uORF), and of ATF3, because of a premature termination codon introduced by alternative splicing (AS), thereby ensuring that the ISR is only active during stress. b | Cells that constitutively express the oncogene MYC produce toxic reactive oxygen species (ROS), which activate the ISR and cause eIF2α phosphorylation and NMD suppression, thereby stabilizing SLC7A11 mRNA. SLC7A11 encodes a transporter that imports cystine into the cytoplasm, which is used in the synthesis of the ROS scavenging molecule glutathione. c | Cells with increases in protein-folding demand in the endoplasmic reticulum (ER) induce the unfolded protein response (UPR) in order to increase the folding capacity of the ER (left). The UPR inhibits NMD, and as a result, NMD targets, such as the IRE1A mRNA, which has an unusually long 3′ untranslated region (3′ UTR), are stabilized. IRE1α together with the products of many other natural NMD targets then coordinates the UPR. When the protein-folding capacity of the ER matches demand (right), NMD is active and degrades the IRE1A mRNA and other mRNAs encoding key UPR regulators, thereby ensuring that innocuous stresses do not activate the UPR. d | NMD maintains the pluripotency of mouse neural stem cells by targeting Smad7 mRNA, which encodes a negative regulator of TGFβ signalling. During neural differentiation, the brain-specific microRNA miR-128 is expressed and inhibits the exon junction complex (EJC) component metastatic lymph node 51 (MLN51) and UPF1. This suppresses NMD, enables the production of SMAD7 and facilitates differentiation.
During amino acid starvation, NMD suppression leads to the upregulation of a suite of genes, some of which produce mRNAs that have NMD-inducing features and/or encode proteins involved in maintaining amino acid homeostasis160. During starvation, NMD suppression also leads to the induction of autophagy, partly through stabilizing the ATF4 mRNA162. During autophagy, cells engulf their proteins (and other macromolecules) in double-membraned vesicles called autophagosomes in order to recycle the proteins back into amino acids162.
Cells cope with the damaging production of ROS by producing ROS scavenger molecules such as glutathione. When the oncoprotein MYC is constitutively active, ROS are generated, eIF2α is phosphorylated and NMD is suppressed164. This results in the stabilization of mRNA encoding the cystine/glutamate exchanger SLC7A11, which facilitates the import of cystine into cells to produce glutathione and reduce ROS levels (FIG. 5b).
A well-studied cellular stress is endoplasmic reticulum (ER) stress, which occurs when the capacity to fold secretory proteins in the ER is insufficient and leads to the accumulation of unfolded or misfolded proteins. ER stress is detected by a suite of sensor proteins in the ER, including PRKR-like endoplasmic reticulum kinase (PERK; also known as eIF2α kinase 3), IRE1α and ATF6, which monitor the abundance of chaperone proteins relative to their client proteins166. PERK phosphorylates eIF2α, thereby decreasing protein synthesis and thus the protein-folding load and activating ATF4 synthesis166. IRE1α autoactivation of its nuclease activity is required for the noncanonical splicing of an intron from the cytoplasmic mRNA XBP1, thereby producing a transcription factor that increases chaperone production166. Activation of plasma-membrane-bound ATF6 results in its cleavage and the release of an ATF6 fragment that functions as a transcription factor, thereby also boosting chaperone production166. The transcriptional output of these sensors collectively amounts to the unfolded protein response (UPR), which is aimed at restoring ER homeostasis. NMD is suppressed during ER stress, leading to the stabilization of a number of transcripts encoding UPR factors such as ATF4, ATF3, CHOP, ATF6, FSD1L, HERP, IRE1α, PERK, PRDG1, TNRC1 and TRAF2 (REFS160,163,167). As in the case of the ISR, NMD is involved in setting the threshold of UPR activation: low-level stresses should not activate the UPR, whereas moderate stresses should temporally activate the UPR. IRE1α, which is a master regulator of the UPR, is encoded by an mRNA with a long 3′ UTR that is subjected to NMD such that NMD normally suppresses the UPR167 (FIG. 5c). However, once the UPR is activated, eIF2α phosphorylation by PERK suppresses NMD. This feedback loop ensures that the UPR is not activated prematurely because NMD inhibits the UPR; when the UPR is activated, UPR-mediated production of its effectors proceeds fully because the UPR inhibits NMD, and the UPR resolves after the stress ceases, because NMD is resumed.
NMD is also involved in the cellular decision to undergo apoptosis when exposed to stress levels that preclude cellular adaptation. For example, challenging cancer cells with concentrations of chemotherapeutics known to elicit apoptosis results in suppression of NMD168,169. When apoptosis is activated, caspases (the proteases that mediate apoptosis) cleave part of UPF1 to yield a dominant-negative protein that reduces the efficiency of NMD. As a consequence, a number of apoptotic genes that produce NMD targets are upregulated.
Development and differentiation.
NMD is conserved throughout evolution, from yeast to humans. Although not essential in S. cerevisiae, knockout of UPF1 (REF170), UPF2 (REF161), SMG1 (REF171) or SMG6 (REF172) is embryonic lethal in mice. For example, SMG6-knockout embryonic stem cells (ESCs) can be generated172, but these cells fail to differentiate in vitro and in vivo. The failure to differentiate is mediated by high levels of MYC, which is encoded by a natural NMD target and promotes stemness in ESCs. This differentiation defect was also seen following the depletion of UPF1, UPF2, SMG1 or SMG5. The inability of ESCs to differentiate when NMD is compromised could explain why NMD factors are essential in mammalian development. In Drosophila melanogaster, NMD is also essential for viability through its control of a single transcript encoding growth arrest and DNA damage 45 (Gadd45)173. Gadd45 promotes apoptosis by upregulating the mitogen-activated protein kinase signalling pathway.
During cell differentiation, NMD must be tuned in order to promote the proper timing and expression of NMD targets (TABLE 1). In humans, differentiation of the endoderm layer is accompanied by strong downregulation of many NMD factors174, and enforced expression of UPF1 or UPF3X inhibits endoderm-directed differentiation. Another example of NMD downregulation during differentiation is the brain-specific microRNA, miR-128, which is expressed during neural differentiation and targets the mRNAs encoding UPF1 and the EJC component MLN51 (REF175), stabilizing a number of NMD targets involved in promoting neural differentiation176 (FIG. 5d). NMD supports the maintenance of mouse neural stem and progenitor cells by promoting TGFβ signalling through destabilization of mRNAs encoding negative regulators of TGFβ signalling, such as Smad7 (REF176). Thus, in order to differentiate, NMD must be downregulated through transcriptional upregulation of miR-128. Additionally, miR-128 downregulates Upf3x, as do miR-124 and miR-9 (REF176). NMD is also important for haematopoiesis. Conditional ablation of mouse UPF2 in the haematopoietic compartment led to complete loss of all stem and progenitor cells, whereas ablation in mature cells had less effect, suggesting that NMD functions during the developmental transitions of haematopoiesis161.
Not all developmental transitions involve the tuning of NMD activity. Some pathways exploit NMD to prune out of the transcriptome mRNAs involved in differentiation by ensuring that NMD-inducing features are included in those transcripts. During the production of granulocytes, progenitor cells called promyelocytes differentiate to mature granulocytes. Through intron retention, a battery of transcripts that must be destroyed to permit granulocyte differentiation is targeted for NMD177. Genes such as that encoding the nuclear membrane protein lamin B1 are expressed with a retained intron containing a PTC and are thus targeted for NMD, thereby permitting transition to a granulocyte nuclear morphology.
Proper positioning of commissural neurons relative to the spinal cord midline of symmetry in the central nervous system is crucial to neural development. ROBO3, which is a key receptor in commissural neurons, receives guidance cues in the form of SLIT proteins secreted from the midline. The mouse Robo3 mRNA has two isoforms: Robo3.1 and Robo3.2, the latter of which contains an intron that subjects it to NMD. Before midline crossing, Robo3.2 is expressed but translationally repressed, thereby shielding it from NMD. However, upon midline crossing, translation repression is relieved owing to signals from the floorplate at the ventral midline, and Robo3.2 is subjected to NMD. This has two, coupled effects: ROBO3.2 protein is produced, which is followed by quick degradation of the mRNA, thereby ensuring only a temporary burst of protein expression. This is important because ROBO3.2 mediates repulsion from the midline of post-crossing commissural neurons. Neurons with compromised NMD — in vivo using UPF2 conditional-knockout embryos and in vitro using spinal cord explants electroporated with a dominant-negative form of UPF1 — are mislocalized, exhibiting over-repulsion from the midline178.
Eukaryotic cells have other ways to leverage NMD to control gene expression through the modulation of mRNA abundance. Programmed ribosomal frameshifts (PRFs) at repeat sequences able to cause ribosomes to ‘slip’ from one translational reading frame to another were originally discovered to expand the limited coding capacity of virus genomes and ensure proper viral-protein stoichiometries. In eukaryotes, −1 PRF (ribosome shifting 1 nucleotide in the 5′ direction) sequences were computationally identified in up to 10% of genes, where they can cause premature translation termination and transcript destruction through NMD179. The activity of −1 PRF sequences is regulated, as exemplified by the −1 PRF in the mRNA encoding C-C chemokine receptor type 5 (CCR5), which is also an HIV-1 co-receptor179. The −1 PRF on the CCR5 mRNA is stimulated by the formation of a pseudoknot, which is stabilized by direct binding of miR-1224 and generates a PTC, ultimately resulting in NMD limiting the amount of CCR5 protein produced. Depletion of AGO1 showed that other interleukin receptors can be controlled by NMD through miRNAs, thereby explaining how PRF can be used in a sequence-specific manner. This mechanism, which demonstrates the interface of NMD with other regulatory mechanisms, may fine-tune cytokine responses in an effort to prevent a runaway immune response.
NMD-related human disorders
NMD is crucial in humans, and its breakdown may result in disease. The underlying causes of these disorders include expression of transcripts that escape NMD, expression of important transcripts that are mistakenly targeted for NMD and even mutations in the NMD machinery.
NMD modulates inherited diseases.
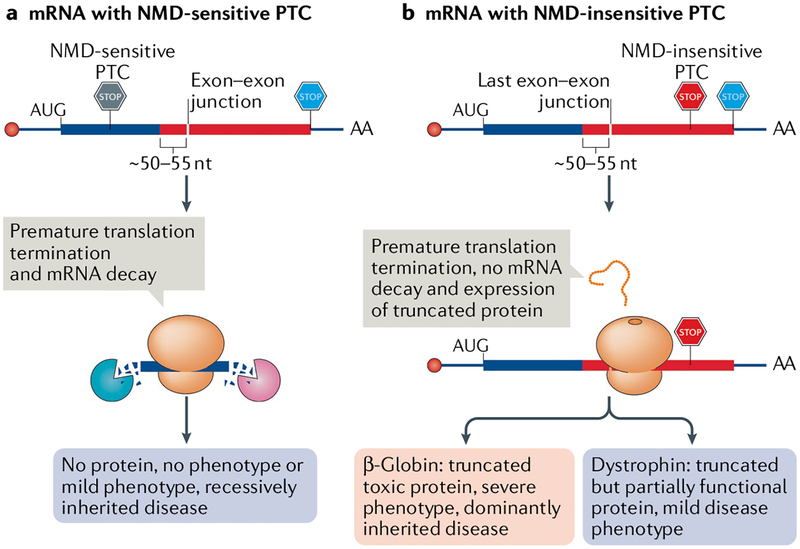
NMD may have evolved to prevent the production of truncated (owing to a PTC) toxic dominant-negative proteins, as illustrated by the ≥50–55 nucleotide rule and its effects on human β-globin mRNA. Individuals with one wild-type β-globin allele and one allele bearing an NMD-sensitive PTC are asymptomatic (FIG. 6a). Individuals with both alleles bearing an NMD-sensitive PTC have severe anaemia owing to the lack of functional β-globin protein. By contrast, in a dominantly inherited form of β-thalassaemia, individuals with one wild-type allele and one allele with an NMD-insensitive PTC produce a truncated dominant-negative β-globin protein92. This dominant-negative protein renders any haemoglobin complexes it forms with α-globin nonfunctional, resulting in ineffective erythropoiesis even though 50% of the produced β-globin protein is normal (FIG. 6b). Another example is of mutations in the SOX10 gene that fail to trigger NMD and produce toxic dominant-negative peptides that lead to a severe combination of diseases, including peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome and Hirschprung disease. However, individuals with mutations that trigger NMD and degradation of SOX10 have mild symptoms of Waardenburg and Hirschsprung disease180. Many other examples that illustrate this protective function of NMD have been documented (see REFS181,182).
Fig. 6 |. Involvement of nonsense-mediated mRNA decay in disease.
Frameshift or nonsense mutations often introduce a nonsense-mediated mRNA decay (NMD)-sensitive or an NMD-insensitive premature termination codon (PTC). a | mRNAs harbouring an NMD-sensitive PTC that is located ≥50–55 nucleotides (nt) upstream of the last exon–exon junction (PTCs located in the thick blue bar) efficiently undergo NMD, which in turn prevents the production of truncated, potentially toxic proteins. As a consequence, these mutations are recessively inherited and cause either no or only a mild disease in heterozygous individuals. b | mRNAs harbouring an NMD-insensitive PTC, which is generally located ≤50–55 nt upstream of the last exon–exon junction (not shown) or in the last exon (located in the thick red bar), fail to trigger NMD and produce truncated proteins. These truncated proteins could be stable and toxic, as in the case of some β-globin gene mutations (pink). Such mutations can be dominant and patients show a severe disease phenotype, even with one normal allele. However, in some diseases, as illustrated by Becker muscular dystrophy (purple), the truncated protein shows some residual activity, thereby causing milder symptoms than seen in Duchenne muscular dystrophy, in which NMD-sensitive PTCs eliminate dystrophin protein expression.
The protective function of NMD may become a burden when the truncated proteins are not toxic. In fact, some truncated proteins have the potential to lessen the severity of disease. For example, most PTCs in the dystrophin gene trigger NMD, which results in Duchenne muscular dystrophy. However, when a PTC is located in a region that does not trigger NMD, a truncated polypeptide is produced that partially rescues the phenotype, instead causing the milder Becker muscular dystrophy183 (FIG. 6b). Again, other examples of this scenario exist (see REF182). Thus, whether NMD has a protective role or a disruptive role depends on the position of the PTC and, of course, on the physiological role of the protein.
Mutations in UPF3X cause intellectual disability in humans184,185. By sequencing genes on the X chromosomes of families with X-linked mental retardation, mutations in the UPF3X gene (so called because it is located on the X chromosome26) were identified in three families184 — two with Lujan–Fryns syndrome186 and one with FG syndrome187. UPF3X mutations have also been identified in other families with X-linked intellectual disabilities, and some of these families additionally manifest schizophrenia and autism spectrum disorder188–191. Heterozygous deletions in a region encompassing the UPF2 gene have been identified in 11 individuals with intellectual disabilities, and the changes in the transcriptome of these individuals correlate with transcriptomic changes in the individuals with intellectual disabilities, suggesting that NMD is important for brain development and function192. Furthermore, deletions in a region of chromosome 1 that includes the EJC protein RBM8A have been linked to intellectual disabilities and brain defects193.
Mouse models have recapitulated the patient-derived data. Upf3x-knockout mice manifest selective learning defects and a major defect in pre-pulse inhibition, which is common in individuals with schizophrenia194. Neurons from these mice have maturation defects, and their neural stem cells have differentiation defects. Rbm8a-haploinsufficient mice are characterized by neural defects including microcephaly, which is also seen in humans bearing the aforementioned deletion in chromosome 1 (REF195). NMD has a role in timely differentiation, growth and maturation of human neuronal cells176,194,196–199, indicating that these are the defects underlying the phenotypes of individuals with mutations in NMD factors.
NMD in cancer.
A range of tumour types express key tumour-suppressor mRNAs with NMD-inducing mutations. These include mutations in the BRCA1 (REF.200), BRCA2 (REF.201), TP53 (REF.202), Rb203 and WT1 (REF.204) genes. Tumour-suppressor genes have a greater propensity to exhibit NMD-inducing features than do oncogenes205, and degradation of these mRNAs by NMD may protect normal cells from toxic effects of the encoded truncated proteins204.
As NMD-inducing mutations exhibit a high incidence of heterozygosity205,206, how might NMD promote the transformation of cells that retain functional alleles of tumour suppressors? Analysis of matching genomic and transcriptomic data from human cancers revealed that tumours can select for acquired NMD-sensitive PTCs in combination with deletions in the other allele or for NMD-sensitive PTCs in haploinsufficient tumour-suppressor genes to eliminate gene function206,207. Alternatively, tumours may select for NMD-insensitive PTCs that generate dominant-negative proteins.
Although it appears that some tumours leverage an intact NMD system to suppress tumour suppressors, other tumours contain disabling mutations in NMD factors. For example, somatic mutations in the UPF1 gene that disrupt UPF1 function are detected in pancreatic adenosquamous carcinomas208. In lung inflammatory myofibroblastic tumours, UPF1 is likewise mutated and allows the upregulation of the NMD target that encodes NF-κB-inducing kinase (also known as MAP3K14), which promotes chemokine production that typifies this tumour type209. Reduction in NMD activity can also affect clinical outcomes: UPF1 silencing in some hepatocellular carcinomas seems to correlate with poorer prognosis210. Interestingly, in some hypermutable colorectal cancers with microsatellite instability owing to mutations in DNA mismatch repair, UPF1, UPF2, SMG1, SMG6 and SMG7 are overexpressed relative to their microsatellite stable tumour counterparts211. This may enable the microsatellite instability tumours to survive the creation of many different PTC-bearing transcripts because they are degraded by NMD. Indeed, experimental suppression of NMD led to re-expression of many PTC-bearing transcripts and to decreased tumour growth in a mouse xenograft model211. Targeted silencing of NMD was also shown to promote the expression of tumour neo-antigens to decrease tumour volumes212.
Finally, the tumour microenvironment induces a variety of stresses, such as hypoxia and ER stress, to which tumour cells must adapt in order to proliferate. One way to adapt to the tumour microenvironment is to tune NMD activity (see above). NMD is decreased during tumour formation in order to overcome microenvironment-related stresses213. For example, both control-siRNA-treated and UPF1-siRNA-treated tumour cells injected into nude mice grow tumours, but cells with enforced UPF1 expression grow much smaller tumours, presumably because they cannot overcome the stress barriers to tumour development.
Concluding remarks
Transcriptome-wide studies have revealed that NMD not only functions in the destruction of mRNAs harbouring PTCs but also globally shapes the transcriptome and defines the fate of different cell types. Our understanding of NMD has now progressed far beyond the original term, ‘nonsense’-mediated mRNA decay. As NMD activity varies by cell type and tissue type, future studies of the full inventory of NMD targets in various cells and tissues may help to reveal what additional roles NMD has in physiology. As the vast majority of transcripts are bound by UPF1 and a considerable fraction of these transcripts are subject NMD, it is not surprising that NMD modifies the phenotypes of different genetic disorders in humans. A complete understanding of the molecular mechanism of NMD will facilitate the development of therapies based on invoking PTC-based NMD or alternatively inhibiting NMD despite the presence of a PTC (Supplementary Box 1).
Supplementary Material
Dominant-negative proteins.
Mutated proteins that can antagonize the function of the wild-type protein, often because the proteins are part of a macromolecular complex, which is rendered defective by the presence of the mutated protein.
β0-thalassaemia.
Thalassaemia is a blood disorder causing anaemia. In the severe form of β0-thalassaemia, no β-globin protein is detectable in peripheral blood.
NMD-activating feature.
An mRNA feature that increases the probability of the mRNA undergoing nonsense-mediated mRNA decay. Examples include an exon–exon junction complex deposited as a consequence of splicing more than ~30–35 nucleotides downstream of a termination codon, an unusually long (>1 kb) 3′ untranslated region or a selenocysteine codon that is interpreted as a stop codon.
Upstream open reading frame.
(uORF). A short ORF in the 5′ end of mRNA (upstream of the main ORF) that can regulate the translation of the main ORF.
A site.
The amino-acyl site on the ribosome is where charged tRNA molecules (with the exception of the translation-initiating charged tRNA) bind during protein synthesis.
Peptidyl-tRNA hydrolysis.
A process that occurs during translation when a water molecule attacks the bond between the nascent peptide and the tRNA molecule in the ribosome, thereby releasing the completed polypeptide.
Staufen-mediated mRNA decay.
An mRNA decay pathway in which the staufen protein recruits UPF1 to an mRNA 3′ untranslated region, causing translation-dependent destabilization of the mRNA.
Exosome.
A large protein complex that degrades mRNAs through its 3′-to-5′ exoribonuclease activities.
Selenocysteine.
An amino acid that is inserted into mRNA bearing a selenocysteine insertion sequence that directs its incorporation at UGA codons, which otherwise would be recognized as termination codons.
Intron retention.
Occurs when an intron fails to be excised out of a pre-mRNA during alternative splicing, giving rise to a transcript with a premature termination codon.
Programmed ribosomal frameshifts.
(PRFs). During translation, incidents of ribosome ‘slippage’ and adoption of a new reading frame.
Pseudoknot.
A tertiary RNA structure formed by base pairing between the loop of a stem–loop structure and nearby ribonucleotides. It is extremely difficult for helicases to unwind this structure.
Waardenburg syndrome.
A disease manifesting defects in tissues derived from cells in the neural crest lineage (neurocristopathy). Individuals with Waardenburg syndrome have defects in hair, skin and eye pigmentation and may suffer from hearing loss.
Hirschprung disease.
A congenital malady in which nerve cells are missing from the end of the bowel, thereby causing problems with passing stool.
Lujan–Fryns syndrome.
An X-linked disorder causing mild to moderate intellectual disability, facial dysmorphism and arms and legs that are abnormally long and slender.
FG syndrome.
An X-linked disorder characterized by intellectual disability, poor muscle tone and macrocephaly.
Tumour neo-antigens.
Peptides absent from normal cells that are produced by tumour-mutated genes that are presented to and activate the immune system.
Acknowledgements
The authors thank X. Rambout and H. Cho for critically reading the manuscript and R. Green for helpful conversations. The authors apologize to colleagues whose work was not referenced owing to space limitations. Work on nonsense-mediated mRNA decay in the Maquat laboratory is supported by the National Institutes of Health R01 grant GM059614. T.K. was partially supported by a Schmitt Program in lntegrative Neuroscience from the University of Rochester Medical Center.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41580-019-0126-2.
References
- 1.Leeds P, Peltz SW, Jacobson A & Culbertson MR The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5, 2303–2314 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Atkin AL, Altamura N, Leeds P & Culbertson MR The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell 6, 611–625 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pal M, Ishigaki Y, Nagy E & Maquat LE Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7, 5–15 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czaplinski K et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12, 1665–1677 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanov PV, Gehring NH, Kunz JB, Hentze MW & Kulozik AE Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27, 736–747 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kashima I et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20, 355–367 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurosaki T & Maquat LE Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc. Natl Acad. Sci. USA 110, 3357–3362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh G, Rebbapragada I & Lykke-Andersen J A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLOS Biol 6, e111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Czaplinski K, Rao Y & Peltz SW The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20, 880–890 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogg JR & Goff SP Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143, 379–389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurt JA, Robertson AD & Burge CB Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23, 1636–1650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurosaki T et al. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 28, 1900–1916 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SR, Pratt GA, Martinez FJ, Yeo GW & Lykke-Andersen J Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol. Cell 59, 413–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zund D, Gruber AR, Zavolan M & Muhlemann O Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat. Struct. Mol. Biol 20, 936–943 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharya A et al. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6, 1226–1235 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czaplinski K, Weng Y, Hagan KW & Peltz SW Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1, 610–623 (1995). [PMC free article] [PubMed] [Google Scholar]
- 17.Weng Y, Czaplinski K & Peltz SW Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol 16, 5477–5490 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiorini F, Bagchi D, Le Hir H & Croquette V Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat. Commun 6, 7581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franks TM, Singh G & Lykke-Andersen J Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143, 938–950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shigeoka T, Kato S, Kawaichi M & Ishida Y Evidence that the Upf1-related molecular motor scans the 3′-UTR to ensure mRNA integrity. Nucleic Acids Res 40, 6887–6897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti S et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 41, 693–703 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Chamieh H, Ballut L, Bonneau F & Le Hir H NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol 15, 85–93 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Kanaan J et al. UPF1-like helicase grip on nucleic acids dictates processivity. Nat. Commun 9, 3752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiorini F, Boudvillain M & Le Hir H Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res 41, 2404–2415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He F, Brown AH & Jacobson A Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol 17, 1580–1594 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serin G, Gersappe A, Black JD, Aronoff R & Maquat LE Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol 21, 209–223 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P & Izaurralde E Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 22, 3960–3970 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerenyi Z et al. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27, 1585–1595 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lykke-Andersen J, Shu MD & Steitz JA Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Mendell JT, Medghalchi SM, Lake RG, Noensie EN & Dietz HC Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol 20, 8944–8957 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pulak R & Anderson P mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 7, 1885–1897 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Maderazo AB, He F, Mangus DA & Jacobson A Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol 20, 4591–4603 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maquat LE & Serin G Nonsense-mediated mRNA decay: insights into mechanism from the cellular abundance of human Upf1, Upf2, Upf3, and Upf3X proteins. Cold Spring Harb. Symp. Quant. Biol 66, 313–320 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Chan WK et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat. Struct. Mol. Biol 16, 747–753 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE & Gehring NH Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12, 1015–1022 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou CH, Shum EY & Wilkinson MF RNA degradation drives stem cell differentiation. EMBO J 34, 1606–1608 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shum EY et al. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell 165, 382–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aznarez I et al. Mechanism of nonsense-mediated mRNA decay stimulation by splicing factor SRSF1. Cell Rep 23, 2186–2198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buhler M, Steiner S, Mohn F, Paillusson A & Muhlemann O EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol 13, 462–464 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Chan WK et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J 26, 1820–1830 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gehring NH et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20, 65–75 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Gregersen LH et al. M0V10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol. Cell 54, 573–585 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Le Hir H, Gatfield D, Izaurralde E & Moore MJ The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20, 4987–4997 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Hir H, Izaurralde E, Maquat LE & Moore MJ The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19, 6860–6869 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lejeune F, Li X & Maquat LE Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12, 675–687 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Singh G et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151, 915–916 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Lewis BP, Green RE & Brenner SE Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA 100, 189–192 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Q et al. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev 20, 153–158 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weischenfeldt J et al. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol 13, R35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calvo SE, Pagliarini DJ & Mootha VK Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA 106, 7507–7512 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skarshewski A et al. uPEPperoni: an online tool for upstream open reading frame location and analysis of transcript conservation. BMC Bioinformatics 15, 36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mabin JW et al. The exon junction complex undergoes a compositional switch that alters mRNP structure and nonsense-mediated mRNA decay activity. Cell Rep 25, 2431–2446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashton-Beaucage D et al. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143, 251–262 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Roignant JY & Treisman JE Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143, 238–250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Murigneux V & Le Hir H Transcriptome-wide modulation of splicing by the exon junction complex. Genome Biol 15, 551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt U, Richter K, Berger AB & Lichter P In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains. J. Cell Biol 172, 373–381 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chazal PE et al. EJC core component MLN51 interacts with eIF3 and activates translation. Proc. Natl Acad. Sci. USA 110, 5903–5908 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diem MD, Chan CC, Younis I & Dreyfuss G PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol 14, 1173–1179 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Isken O et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133, 314–327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hosoda N, Kim YK, Lejeune F & Maquat LE CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol 12, 893–901 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Lykke-Andersen J, Shu MD & Steitz JA Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293, 1836–1839 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Dostie J & Dreyfuss G Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol 12, 1060–1067 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Sato H & Maquat LE Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev 23, 2537–2550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagy E & Maquat LE A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci 23, 198–199 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Thermann R et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17, 3484–3494 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchwald G et al. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc. Natl Acad. Sci. USA 107, 10050–10055 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gehring NH, Neu-Yilik G, Schell T, Hentze MW & Kulozik AE Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11, 939–949 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Kim VN, Kataoka N & Dreyfuss G Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293, 1832–1836 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Yamashita A, Ohnishi T, Kashima I, Taya Y & Ohno S Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15, 2215–2228 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Viegas MH, Gehring NH, Breit S, Hentze MW & Kulozik AE The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res 35, 4542–4551 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sato H, Hosoda N & Maquat LE Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell 29, 255–262 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Zhang Z & Krainer AR Involvement of SR proteins in mRNA surveillance. Mol. Cell 16, 597–607 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Alexandrov A, Colognori D, Shu MD & Steitz JA Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc. Natl Acad. Sci. USA 109, 21313–21318 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barbosa I et al. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat. Struct. Mol. Biol 19, 983–990 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Steckelberg AL, Altmueller J, Dieterich C & Gehring NH CWC22-dependent pre-mRNA splicing and eIF4A3 binding enables global deposition of exon junction complexes. Nucleic Acids Res 43, 4687–4700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baird TD et al. ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. eLife 7, e33178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chakrabarti S, Bonneau F, Schussler S, Eppinger E & Conti E Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res 42, 9447–9460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durand S, Franks TM & Lykke-Andersen J Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat. Commun 7, 12434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arias-Palomo E et al. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev 25, 153–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deniaud A et al. A network of SMG-8, SMG-9 and SMG-1 C-terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Res 43, 7600–7611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melero R et al. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat. Commun 7, 10585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamashita A et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23, 1091–1105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ohnishi T et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12, 1187–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 84.Okada-Katsuhata Y et al. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40, 1251–1266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hug N & Cáceres JF The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep 8, 1845–1856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neu-Yilik G et al. Dual function of UPF3B in early and late translation termination. EMBO J 36, 2968–2986 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He F & Jacobson A Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet 49, 339–366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao Z & Wilkinson M An RNA decay factor wears a new coat: UPF3B modulates translation termination. FI000Res 6, 2159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schuller AP, Zinshteyn B, Enam SU & Green R Directed hydroxyl radical probing reveals Upf1 binding to the 80S ribosomal E site rRNA at the L1 stalk. Nucleic Acids Res 46, 2060–2073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belgrader P, Cheng J, Zhou X, Stephenson LS & Maquat LE Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol. Cell. Biol 14, 8219–8228 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng J & Maquat LE Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol. Cell. Biol 13, 1892–1902 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kugler W, Enssle J, Hentze MW & Kulozik AE Nuclear degradation of nonsense mutated beta-globin mRNA: a post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of beta-thalassemia? Nucleic Acids Res 23, 413–418 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ishigaki Y, Li X, Serin G & Maquat LE Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 (2001). [DOI] [PubMed] [Google Scholar]
- 94.Halstead JM et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 347, 1367–1671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Popp MW & Maquat LE A TRICK’n way to see the pioneer round of translation. Science 347, 1316–1317 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Trcek T, Sato H, Singer RH & Maquat LE Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev 27, 541–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maquat LE, Tarn WY & Isken O The pioneer round of translation: features and functions. Cell 142, 368–374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chiu SY, Lejeune F, Ranganathan AC & Maquat LE The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev 18, 745–754 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hwang J, Sato H, Tang Y, Matsuda D & Maquat LE UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 39, 396–409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lejeune F, Ishigaki Y, Li X & Maquat LE The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J 21, 3536–3545 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Durand S & Lykke-Andersen J Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol 20, 702–709 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Rufener SC & Muhlemann O eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol 20, 710–717 (2013). [DOI] [PubMed] [Google Scholar]
- 103.Gao Q, Das B, Sherman F & Maquat LE Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc. Natl Acad. Sci. USA 102, 4258–4263 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lopez PJ & Seraphin B Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA 5, 1135–1137 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Spingola M, Grate L, Haussler D & Ares M Jr. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5, 221–234 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malabat C, Feuerbach F, Ma L, Saveanu C & Jacquier A Quality control of transcription start site selection by nonsense-mediated-mRNA decay. eLife 4, e06722 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Celik A, He F & Jacobson A NMD monitors translational fidelity 24/7. Curr. Genet 63, 1007–1010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang J, Sun X, Qian Y, LaDuca JP & Maquat LE At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol 18, 5272–5283 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J, Sun X, Qian Y & Maquat LE Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4, 801–815 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsuda D, Hosoda N, Kim YK & Maquat LE Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat. Struct. Mol. Biol 14, 974–979 (2007). [DOI] [PubMed] [Google Scholar]
- 111.Wang J, Gudikote JP, Olivas OR & Wilkinson MF Boundary-independent polar nonsense-mediated decay. EMBO Rep 3, 274–279 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eberle AB, Stalder L, Mathys H, Orozco RZ & Muhlemann O Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLOS Biol 6, e92 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Amrani N et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118 (2004). [DOI] [PubMed] [Google Scholar]
- 114.Hosoda N, Lejeune F & Maquat LE Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol 26, 3085–3097 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ivanov A et al. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44, 7766–7776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fatscher T, Boehm V, Weiche B & Gehring NH The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA 20, 1579–1592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Joncourt R, Eberle AB, Rufener SC & Muhlemann O Eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay by two genetically separable mechanisms. PLOS ONE 9, e104391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kervestin S, Li C, Buckingham R & Jacobson A Testing the faux-UTR model for NMD: analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie 94, 1560–1571 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Silva AL, Ribeiro P, Inacio A, Liebhaber SA & Romao L Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14, 563–576 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lejeune F, Ranganathan AC & Maquat LE elF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol 11, 992–1000 (2004). [DOI] [PubMed] [Google Scholar]
- 121.Mangus DA, Evans MC & Jacobson A Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4, 223 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peixeiro I et al. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res 40, 1160–1173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Neu-Yilik G et al. Mechanism of escape from nonsense-mediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA 17, 843–854 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J & Maquat LE Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J 16, 826–833 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baker KE & Parker R Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin. Cell Biol 16, 293–299 (2004). [DOI] [PubMed] [Google Scholar]
- 126.Kebaara BW & Atkin AL Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res 37, 2771–2778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tani H et al. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9, 1370–1379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Toma KG, Rebbapragada I, Durand S & Lykke-Andersen J ldentification of elements in human long 3′ UTRs that inhibit nonsense-mediated decay. RNA 21, 887–897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chester A et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J 22, 3971–3982 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ge Z, Quek BL, Beemon KL & Hogg JR Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. eLife 5, e11155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kishor A, Ge Z & Hogg JR hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma. EMBO J 38, e99128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Imamachi N, Salam KA, Suzuki Y & Akimitsu NA GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. GenomeRes 27, 407–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Park E & Maquat LE Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 4, 423–435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Marzluff WF & Koreski KP Birth and death of histone mRNAs. Trends Genet 33, 745–759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim YK, Furic L, Desgroseillers L & Maquat LE Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120, 195–208 (2005). [DOI] [PubMed] [Google Scholar]
- 136.Choe J, Ahn SH & Kim YK The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Res 42, 9334–9349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Eberle AB, Lykke-Andersen S, Muhlemann O & Jensen TH SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol 16, 49–55 (2009). [DOI] [PubMed] [Google Scholar]
- 138.Huntzinger E, Kashima I, Fauser M, Sauliere J & Izaurralde E SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14, 2609–2617 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lykke-Andersen S et al. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev 28, 2498–2517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schmid M & Jensen TH The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci 33, 501–510 (2008). [DOI] [PubMed] [Google Scholar]
- 141.Kurosaki T, Miyoshi K, Myers JR & Maquat LE NMD-degradome sequencing reveals ribosome-bound intermediates with 3′-end non-templated nucleotides. Nat. Struct. Mol. Biol 25, 940–950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Malecki M et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32, 1842–1854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Loh B, Jonas S & Izaurralde E The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27, 2125–2138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Unterholzner L & Izaurralde E SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16, 587–596 (2004). [DOI] [PubMed] [Google Scholar]
- 145.Yamashita A et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol 12, 1054–1063 (2005). [DOI] [PubMed] [Google Scholar]
- 146.Cho H et al. SMG5-PNRC2 is functionally dominant compared with SMG5-SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res 41, 1319–1328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cho H, Kim KM & Kim YK Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell 33, 75–86 (2009). [DOI] [PubMed] [Google Scholar]
- 148.Lai T et al. Structural basis of the PNRC2-mediated link between mrna surveillance and decapping. Structure 20, 2025–2037 (2012). [DOI] [PubMed] [Google Scholar]
- 149.Lykke-Andersen J Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol 22, 8114–8121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dehecq M et al. Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes. EMBO J 37, e99278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Arribere JA & Fire AZ Nonsense mRNA suppression via nonstop decay. eLife 7, e33292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Colombo M, Karousis ED, Bourquin J, Bruggmann R & Muhlemann O Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 23, 189–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Anders KR, Grimson A & Anderson P SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22, 641–650 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chiu SY, Serin G, Ohara O & Maquat LE Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9, 77–87 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fillman C & Lykke-Andersen J RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol 17, 326–331 (2005). [DOI] [PubMed] [Google Scholar]
- 156.Eulalio A, Behm-Ansmant I, Schweizer D & Izaurralde E P-Body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol 27, 3970–3981 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stalder L & Muhlemann O Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 15, 1265–1273 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hu W, Petzold C, Coller J & Baker KE Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol 17, 244–247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Green RE et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 19 (Suppl. 1), i118–i121 (2003). [DOI] [PubMed] [Google Scholar]
- 160.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F & Dietz HC Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet 36, 1073–1078 (2004). [DOI] [PubMed] [Google Scholar]
- 161.Weischenfeldt J et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22, 1381–1396 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wengrod J et al. Inhibition of nonsense-mediated RNA decay activates autophagy. Mol. Cell. Biol 33, 2128–2135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gardner LB Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol 28, 3729–3741 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang D, Wengrod J & Gardner LB Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J. Biol. Chem 286, 40038–40043 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pakos-Zebrucka K et al. The integrated stress response. EMBO Rep 17, 1374–1395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Goetz AE & Wilkinson M Stress and the nonsense-mediated RNA decay pathway. Cell. Mol. Life Sci 74, 3509–3531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Karam R et al. The unfolded protein response is shaped by the NMD pathway. EMBO Rep 16, 599–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jia J et al. Caspases shutdown nonsense-mediated mRNA decay during apoptosis. Cell Death Differ 22, 1754–1763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Popp MW & Maquat LE Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat. Commun 6, 6632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Medghalchi SM et al. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet 10, 99–105 (2001). [DOI] [PubMed] [Google Scholar]
- 171.McIlwain DR et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl Acad. Sci. USA 107, 12186–12191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li T et al. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J 34, 1630–1647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nelson JO, Moore KA, Chapin A, Hollien J & Metzstein MM Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. eLife 5, e12876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lou CH et al. Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Rep 6, 844–857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bruno IG et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 42, 500–510 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lou CH et al. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep 6, 748–764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wong JJ et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154, 583–595 (2013). [DOI] [PubMed] [Google Scholar]
- 178.Colak D, Ji SJ, Porse BT & Jaffrey SR Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell 153, 1252–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Belew AT et al. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature 512, 265–269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Inoue K et al. Translation of SOX10 3′ untranslated region causes a complex severe neurocristopathy by generation of a deleterious functional domain. Hum. Mol. Genet 16, 3037–3046 (2007). [DOI] [PubMed] [Google Scholar]
- 181.Bhuvanagiri M et al. 5-Azacytidine inhibits nonsense-mediated decay in a MYC-dependent fashion. EMBO Mol. Med 6, 1593–1609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Miller JN & Pearce DA Nonsense-mediated decay in genetic disease: friend or foe? Mutat. Res. Rev. Mutat. Res 762, 52–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kerr TP, Sewry CA, Robb SA & Roberts RG Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum. Genet 109, 402–407 (2001). [DOI] [PubMed] [Google Scholar]
- 184.Tarpey PS et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat. Genet 39, 1127–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jaffrey SR & Wilkinson MF Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat. Rev. Neurosci 19, 715–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fryns JP & Buttiens M X-Linked mental retardation with marfanoid habitus. Am. J. Med. Genet 28, 267–274 (1987). [DOI] [PubMed] [Google Scholar]
- 187.Opitz JM & Kaveggia EG Studies of malformation syndromes of man 33: the FG syndrome. An X-linked recessive syndrome of multiple congenital anomalies and mental retardation. Z. Kinderheilkd 117, 1–18 (1974). [DOI] [PubMed] [Google Scholar]
- 188.Addington AM et al. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol. Psychiatry 16, 238–239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Laumonnier F et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol. Psychiatry 15, 767–776 (2010). [DOI] [PubMed] [Google Scholar]
- 190.Lynch SA et al. Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur. J. Med. Genet 55, 476–479 (2012). [DOI] [PubMed] [Google Scholar]
- 191.Xu X et al. Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin. Genet 83, 560–564 (2013). [DOI] [PubMed] [Google Scholar]
- 192.Nguyen LS et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum. Mol. Genet 22, 1816–1825 (2013). [DOI] [PubMed] [Google Scholar]
- 193.Brunetti-Pierri N et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet 40, 1466–1471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huang L et al. A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol. Psychiatry 23, 1773–1786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mao H et al. Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J. Neurosci 35, 7003–7018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Alrahbeni T et al. Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol. Brain 8, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jolly LA, Homan CC, Jacob R, Barry S & Gecz J The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum. Mol. Genet 22, 4673–4687 (2013). [DOI] [PubMed] [Google Scholar]
- 198.Long AA et al. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J. Cell Sci 123, 3303–3315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nguyen LS et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol. Psychiatry 17, 1103–1115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM & Mazoyer S The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum. Mol. Genet 11, 2805–2814 (2002). [DOI] [PubMed] [Google Scholar]
- 201.Ware MD et al. Does nonsense-mediated mRNA decay explain the ovarian cancer cluster region of the BRCA2 gene? Oncogene 25, 323–328 (2006). [DOI] [PubMed] [Google Scholar]
- 202.Anczukow O et al. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum. Mutat 29, 65–73 (2008). [DOI] [PubMed] [Google Scholar]
- 203.Pinyol M et al. Inactivation of RB1 in mantle-cell lymphoma detected by nonsense-mediated mRNA decay pathway inhibition and microarray analysis. Blood 109, 5422–5429 (2007). [DOI] [PubMed] [Google Scholar]
- 204.Reddy JC et al. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J. Biol. Chem 270, 10878–10884 (1995). [DOI] [PubMed] [Google Scholar]
- 205.Mort M, Ivanov D, Cooper DN & Chuzhanova NA A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat 29, 1037–1047 (2008). [DOI] [PubMed] [Google Scholar]
- 206.Hu Z, Yau C & Ahmed AA A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay. Nat. Commun 8, 15943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lindeboom RG, Supek F & Lehner B The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet 48, 1112–1118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu C et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat. Med 20, 596–598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lu J et al. The nonsense-mediated RNA decay pathway is disrupted in inflammatory myofibroblastic tumors. J. Clin. Invest 126, 3058–3062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chang L et al. The human RNA surveillance factor UPF1 regulates tumorigenesis by targeting Smad7 in hepatocellular carcinoma. J. Exp. Clin. Cancer Res 35, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bokhari A et al. Targeting nonsense-mediated mRNA decay in colorectal cancers with microsatellite instability. Oncogenesis 7, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pastor F, Kolonias D, Giangrande PH & Gilboa E lnduction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465, 227–230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang D et al. lnhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol. Cell. Biol 31, 3670–3680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.