Abstract
For two decades diffusion fiber tractography has been used to probe both the spatial extent of white matter pathways and the region to region connectivity of the brain. In both cases, anatomical accuracy of tractography is critical for sound scientific conclusions. Here we assess and validate the algorithms and tractography implementations that have been most widely used - often because of ease of use, algorithm simplicity, or availability offered in open source software. Comparing forty tractography results to a ground truth defined by histological tracers in the primary motor cortex on the same squirrel monkey brains, we assess tract fidelity on the scale of voxels as well as over larger spatial domains or regional connectivity. No algorithms are successful in all metrics, and, in fact, some implementations fail to reconstruct large portions of pathways or identify major points of connectivity. The accuracy is most dependent on reconstruction method and tracking algorithm, as well as the seed region and how this region is utilized. We also note a tremendous variability in the results, even though the same MR images act as inputs to all algorithms. In addition, anatomical accuracy is significantly decreased at increased distances from the seed. An analysis of the spatial errors in tractography reveals that many techniques have trouble properly leaving the gray matter, and many only reveal connectivity to adjacent regions of interest. These results show that the most commonly implemented algorithms have several short-comings and limitations, and choices in implementations lead to very different results. This study should provide guidance for algorithm choices based on study requirements for sensitivity, specificity, or the need to identify particular connections, and should serve as a heuristic for future developments in tractography.
Keywords: Validation, Diffusion magnetic resonance imaging, Histology, Tractography, Dispersion, HARDI, Accuracy
1. Introduction
Diffusion MRI fiber tractography is widely used to probe the structural connectivity of the brain, with a range of applications in both clinical and basic neuroscience [1,2]. However, these techniques are subject to a number of serious pitfalls and limitations which may limit the anatomical accuracy of the reconstructed pathways [3,4]. In addition, the large number of diffusion reconstruction algorithms and tracking strategies that exist are likely to result in different “tracts”, with varying levels of accuracy. As utilization of fiber tractography continually increases, it is necessary to validate these techniques in order to gain insight into the conditions under which they succeed, and more importantly, where they fail.
One approach to validation is through classical tracer injection techniques in animal models, followed by histological analysis to defined the “ground truth” pathways for subsequent comparisons with diffusion tractography. Traditionally, validating the faithfulness of tractography against tracers takes one of two forms. First, some metric of spatial overlap of the tract versus the tracer can be computed, which evaluates the overall layout or spatial extent of the tract. Second, many studies evaluate connectivity measures, disregarding how tracts reach their destinations, with a focus on the strength of the connections between different regions of the brain.
Connection strengths estimated from tractography have been compared with invasive tracer data accumulated in existing atlases or databases, for example the Markov-Kennedy [5] or CoCoMac [6] databases for the macaque, or the Allen Brain Atlas for the mouse [7]. These studies have provided encouraging results, finding moderate to high positive correlations between tractography and connection strengths [8–10], suggesting that the number of reconstructed streamlines is correlated with the strength of connections between brain regions. However, tractography becomes less reliable for longer pathways [10], and results are heavily dependent on decisions made during the tracking process (i.e. the seeding strategy). The use of large-scale tracer databases has the advantage of assessing connectivity of a large number of pathways across many cortical areas, however, they have several disadvantages. Most notably, tracer injection and MRI are typically not employed on the same animal (with few exceptions [11]). Not only can pathway connection strength vary between animals, but variance in brain geometry between injected and scanned animals could lead to mismatches in identifying the location of the injection regions in the subject of interest, together compromising the fidelity of the “ground truth” to which tractography is being compared.
Alternatively, a number of studies have investigated the voxel-wise spatial overlap of histologically-defined white matter trajectories with those from tractography. Validating these measures gives confidence in the ability of tractography to segment specific white matter pathways (with subsequent analysis typically taking some quantitative measure along these pathways). For example, Schmahmann et al. [12] compare one implementation of tractography (diffusion spectrum imaging) to histological tracing and conclude that tractography is able to replicate the major features and geometrical organization of a number of association pathways. Improving upon this, in a series of studies on the macaque brain, Dauget et al. [13,14] register histological sections of labeled fiber tracts in 3D to diffusion tensor imaging (DTI) tractography data. They find a range in spatial agreement, with a range of Dice overlap coefficients (0.2–0.75) dependent on the pathway of interest and various tractography parameters, and note that DTI has difficulties when tracts cross or divide, an issue now referred to as the “crossing fiber” problem.
Building upon these studies, the goal of the present work is to systematically characterize the anatomical accuracy of diffusion fiber tractography – both the spatial extent and tract connections – and to do this on both the scale of individual voxels as well on a larger domain over anatomical regions of interest. To achieve this goal, we utilize the squirrel monkey brain, and compare tractography results directly to registered high-resolution tracer data from the same animal. We aim here to evaluate the algorithms most commonly employed in the literature (all of which are implemented in open-source software packages) in order to reveal the successes and shortcomings of the majority of studies utilizing diffusion tractography to date. In addition to measures of overlap and connectivity for each algorithm, we further assess the effects of user-defined algorithm choices (reconstruction algorithm, seeding strategy, tracking logic), distance from seed point, and effects of probabilistic thresholding on the fidelity of resulting tractograms. The focus of this work is on tractography of the pathways in the motor system. This is because the organization and anatomical connections of this system are well understood [15], and the motor system is a frequent target of tractography as it is particularly relevant for a variety of disabilities or pathologies including stroke [16,17], multiple sclerosis [18,19], Parkinson’s disease [20,21], cerebral palsy [22,23], and tumor removal surgeries [24,25], among others. Herein, we investigate the spatial errors in these tractography algorithms, asking where in the brain these algorithms typically fail, and assess potential reasons for this failure.
2. Methods
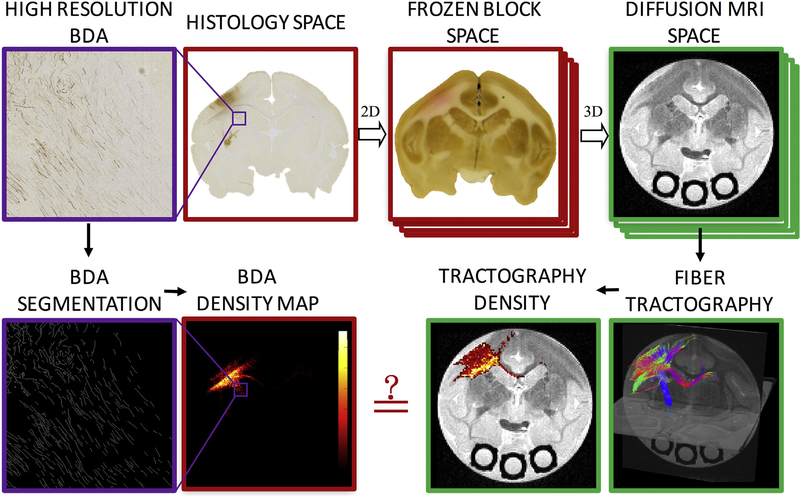
All animal procedures were approved by the Vanderbilt University Animal Care and Use Committee. Fig. 1 shows the methodology pipeline used in this study. Briefly, biotinylated dextran amine (BDA), a histological tracer, was injected into the primary motor cortex (M1) of two squirrel monkey brains. Afterwards, diffusion MRI was acquired on the ex vivo brains and diffusion fiber tractography performed using 40 different algorithms and/or tracking settings. These 40 tractograms resulted in both streamline locations (with the exception of two algorithms) and track density maps, which represent the number of streamlines traversing each voxel. During the brain sectioning digital photographs of the frozen block of the brains were taken to aid in registration of the modalities. Histological sections were processed to visualize BDA and imaged at high resolution. BDA was then segmented from these images in order to create BDA density maps which can be compared directly on a voxel-by-voxel basis to the tractograms and streamline density maps.
Fig. 1.
Methodology pipeline. High resolution BDA micrographs are registered to the corresponding digital photograph of the frozen tissue block, which is registered to the 3D diffusion MRI volume. From the micrograph, BDA is automatically segmented, resulting in a BDA density map. From diffusion MRI, tractography is performed, resulting in tract density maps. Direct, voxel-by-voxel comparisons can now be made between histology and diffusion tractography.
2.1. Tracer injection
For our study we chose BDA, a commonly used neuroanatomical tracer for studying neuronal pathways. BDA is transported in both anterograde and retrograde directions, yielding sensitive and detailed labeling of both axons and terminals, as well as neuronal cell bodies [26]. This tracer relies on axonal transport systems; thus BDA injection was performed prior to ex vivo scanning. Under general anesthesia, BDA (Molecular Probes Inc., Eugene, OR) was injected (as a 10% solution in phosphate buffer) into M1 cortex of the left-hemisphere following the procedures followed in previous studies [11,15]. Pressure injections of BDA were carried out using a 2 μl Hamilton syringe. Eight injections (1 μl/site) were made in order to cover a large M1 region representing the distal forelimb as identified by intracortical micro-stimulation. After each injection, the needle was left in the brain for 5–10 min and then retracted stepwise to avoid leakage of the tracer along the needle track. After surgery, the monkeys were allowed to recover, giving the tracer sufficient time to be transported along axons to all regions connected to the injected M1 cortex.
2.2. Diffusion MRI acquisition
After animal sacrifice, the brains were perfusion fixed with 4% paraformaldehyde preceded by rinse with physiological saline. The brain was removed from the skull and stored in buffered saline overnight. The next day, the brain was scanned on a 9.4 T Varian scanner using a quadrature birdcage volume coil (inner diameter = 63 mm), and immersed in PBS during scanning. Diffusion weighted imaging was performed using a pulsed gradient spin echo multi-shot spinwarp imaging sequence with full brain coverage at 300 μm isotropic resolution (TR = 4.6 s, TE = 42 ms, 32 gradient directions, b ≈ 1000 s/mm2, image matrix = 192 × 128 × 115, 1 b0 image). With a brain volume of approximately 20 cm3, this is roughly equivalent to high resolution protocol of a human brain scanned at ~1.2 mm isotropic. Bore temperature was monitored and maintained at 19–20 °C by circulating air through the bore. Acquisition for a single diffusion-weighted volume took approximately 10 min. The scan time was extended to 50 h in order to facilitate 9 and 10 signal averages, respectively. The b value used in this experiment was lower than is optimal for diffusion studies in fixed tissue [27,28] (approximate mean diffusivity of 0.45 × 10–3 mm2/s [29], about half of that expected in vivo), due to hardware limitations. A low b value decreases the diffusion-related contrast-to-noise ratio (CNR) in the image data (upon which tractography ultimately relies), which has the same effect as higher image noise. To compensate for this shortcoming, we extended the scan time to 50 h, which yielded a CNR comparable to in vivo human studies (equivalent to an in vivo study with mean diffusivity = 0.8 × 10–3 mm2/s and SNR > 50). Thus, although the b-value is lower than optimal, the angular contrast is increased, resulting in voxel-wise reconstructions consistent with expected anatomy. See Supplementary Fig. 1 for example orientation distributions derived from example CSD, QBI, and B&S methods in both single fiber, crossing fiber, and gray matter regions. Because the scan is ex vivo, and only collects a single line of k-space per excitation (as opposed to echo-planar imaging), the data was not pre-processed for motion nor susceptibility induced distortions (effective phase encode bandwidth is infinite).
2.3. Diffusion MRI fiber tractography
Forty diffusion tractograms were created. Details of each are out-lined in Table 1. All were created using open-source software packages – Diffusion Toolkit and Trackvis [30], FSL [31], DSI Studio [32], and MRtrix [33] – typically using the default or recommended settings and strategies. For example, constraints (FA, curvature, etc.) and tracking choices (step-size, integration, etc.) are set as default or as done on website tutorials, and whole brain tracking is performed prior to the filtering techniques assessed. We note that this is not a compressive list of all software packages or tracking algorithms, but is representative of a number of algorithms and strategies often employed. Table 1 describes any pre-processing (if performed), the reconstruction strategy, software package, tractography method and algorithm, seeding strategy, as well as additional information that may be useful. Detailed descriptions of each method, including software versions, specific commands and implementations, and tracking parameters are given as supplementary information. Reconstruction techniques included DTI [34], Qball Imaging [35], Ball and Sticks (B&S) [36], and Constrained Spherical Deconvolution (CSD) [37]. Tractography included both deterministic and probabilistic methods each with a variety of tract propagation strategies. Seed regions included both the seed defined by the BDA injection region (named “Seed”) (see Section 2.4) as well as the seed dilated 600 μm into the white matter (“Seed600 μm”), a strategy often employed in order to propagate out of the gray matter. Seeds were used both as a literal seed region, meaning that streamline propagation begins in this mask, or used as a region of interest (ROI) “AND” region subsequent to full brain seeding, where streamlines were included if they pass through this region. The number of streamlines generated, or extracted, was generally determined by the software default parameters. After tractography, the number of streamlines traversing each voxel is counted, and saved as track density maps.
Table 1.
Diffusion MRI tractography algorithms.
| Algorithm # | Pre-Processing? | Reconstruction Technique | Software | Tractography Method | Algorithm | Seed | Miscellaneous |
|---|---|---|---|---|---|---|---|
| 1 | None | DTI | Diffusion Toolkit + Trackvis | Deterministic | FACT + Spline filtering | Seed (as ROI) | Diffusion Toolkit default parameters |
| 2 | Seed600um (as ROI) | ||||||
| 3 | Sphere ROI (r=10v) centered on Seed | ||||||
| 4 | None | Qball | Diffusion Toolkit + Trackvis | Deterministic | FACT + Spline filtering | Seed (as ROI) | Diffusion Toolkit default parameters |
| 5 | Seed600um (as ROI) | ||||||
| 6 | Sphere ROI (r=10v) centered on Seed | ||||||
| 7 | None | Ball & Sticks (bedpostx) | FSL | Probablistic | Probtrackx | Seed (as seed) | FSL Default Parameters: No FA Threshold |
| 8 | Seed600um (as seed) | ||||||
| 9 | None | DTI | DSI Studio | Deterministic | DSI Studio default tracking | Seed (as ROI) | 2000 tracks generated |
| 10 | Seed (as seed) | ||||||
| 11 | Seed600 (as ROI) | ||||||
| 12 | Seed600 (as seed) | ||||||
| 13 | Sphere ROI (r=10v) centered on seed | ||||||
| 14 | Sphere seed (r=10v) centered on seed | ||||||
| 15 | None | Qball | DSI Studio | Deterministic | DSI Studio default tracking | Seed (as ROI) | - SH order = 4 - No ODF Sharpening - 2000 tracks generated |
| 16 | Seed (as seed) | ||||||
| 17 | Seed600 (as ROI) | ||||||
| 18 | Seed600 (as seed) | ||||||
| 19 | Sphere ROI (r=10v) centered on seed | ||||||
| 20 | Sphere seed (r=10v) centered on seed | ||||||
| 21 | Response function estimated from highest FA voxels | CSD | MRTrix | Probablistic* | iFOD2 | Seed (as seed) | MRTrix3 Default Parameters - 5000 Tracks generated |
| 22 | Seed600um (as seed) | ||||||
| 23 | Seed (as ROI) | ||||||
| 24 | Seed600 (as ROI) | ||||||
| 25 | Response function estimated from iterative method | CSD | MRTrix | Probablistic* | iFOD2 | Seed (as seed) | MRTrix3 Default Parameters - 5000 Tracks generated |
| 26 | Seed600um (as seed) | ||||||
| 27 | Seed (as ROI) | ||||||
| 28 | Seed600 (as ROI) | ||||||
| 29 | None | DTI | MRTrix | Deterministic | TensorDet | Seed (as seed) | MRTrix3 Default Parameters - 5000 Tracks generated |
| 30 | Seed600um (as seed) | ||||||
| 31 | Seed (as ROI) | ||||||
| 32 | Seed600 (as ROI) | ||||||
| 33 | None | DTI | MRTrix | Probabalistic | TensorProb | Seed (as seed) | MRTrix3 Default Parameters - 5000 Tracks generated |
| 34 | Seed600um (as seed) | ||||||
| 35 | Seed (as ROI) | ||||||
| 36 | Seed600 (as ROI) | ||||||
| 37 | Response function estimated from highest FA voxels | CSD | MRTrix | Probablistic* | SIFT + iFOD2 | Seed (as ROI) | 10 million streamlines SIFTed to 1 million |
| 38 | Seed600um (as ROI) | ||||||
| 39 | Response function estimated from iterative method | CSD | MRTrix | Probablistic* | SIFT + iFOD2 | Seed (as ROI) | 10 million streamlines SIFTed to 1 million |
| 40 | Seed600 (as ROI) |
We note that the MRTrix iFOD2 algorithm uses samples drawn from a fiber orientation distribution to build the streamlines, and aims to reflect the anatomical distribution of fibers rather than statistical uncertainty. Thus, these are not “strictly” probabilistic, but share many similarities and are often described as probabilistic.
2.4. Histology acquisition
Following MRI scanning, the brains were frozen and cut serially on a microtome in the coronal plane at 50 μm thickness. The surface of the frozen tissue block (i.e. the “block-face”) was digitally photographed prior to cutting every third section (i.e. at 150 μm intervals). These block-face images have been shown to produce more robust inter-modality registration results by providing a relatively undistorted intermediate reference space between the histological and MRI data [38].
Sections were divided into six series. Every sixth thin section was processed for BDA histochemistry [26], producing a series of 172 sections (83 of which contained evidence of BDA stain, or connections to M1). Whole-slide Brightfield microscopy was performed using a Leica SCN400 Slide Scanner at 20× magnification, resulting in a maximum in-plane resolution of 0.5 μm/pixel.
2.5. Ground truth M1 connectivity
The “ground truth” connectivity of the injection area was determined by the presence of BDA-labeled axons in our high-resolution histology, which displayed as brown in the digital images. BDA-labeled fibers were segmented and counted following a series of morphological processes: top-hat filtering was performed to correct uneven illumination, global thresholding to extract fibers (segmenting brown [r/g/b = 165/42/42] using the “colorseg” function available on MathWorks File Exchange), and morphological operations to remove non-fiber objects (objects < 11 pixels, empirically chosen) and to remove branch points of overlapping fibers. Histological images were down-sampled to the resolution of the MRI-data (300 μm isotropic), and the number of BDA fibers per voxel was counted, resulting in BDA density maps. These BDA density maps represent the ground truth “strength of connections” to the M1 injection area.
2.6. Registration
In order to make direct comparisons between diffusion MRI tractography and histology, a multi-step registration procedure was utilized. The chosen procedure is similar to the registration framework validated in previous studies [39], which showed that the accuracy of the overall registration was approximately one MRI voxel (~300 μm).
Briefly, the high-resolution Leica image was down-sampled to 128 μm/pixel (down-sample factor of 256), and registered to the down-sampled photograph (256 × 256 pixels at a resolution of 128 μm/pixel) of the corresponding tissue block using a 2D affine transformation followed by a 2D non-rigid transformation, semi-automatically calculated via the Thin-Plate Spline algorithm [40]. Next, all down-sampled block face photographs were assembled into a 3D volume and registered to the 3D diffusion MRI volume (the non-diffusion weighted volume) using a 3D affine transformation followed by a non-rigid transformation automatically calculated via the Adaptive Bases Algorithm [41]. Deformation fields produced by all registration steps were concatenated in order to transfer BDA density maps into MRI space. Immediately following spatial transformation, the Jacobian matrix of the corresponding deformation field was calculated and used to compensate the density change caused by geometric transformations. Finally, direct, voxel-by-voxel comparisons could be made between the diffusion MRI datasets (tractograms) and histology datasets (BDA density maps).
2.7. Anatomical accuracy measures
Measures were calculated which describe the anatomical fidelity of the resulting tractograms, several of which have been previously employed in the validation literature. Here, measures are divided into voxel-wise fidelity metrics, and ROI-based fidelity metrics.
In the following, the BDA defined volume is represented by Bj (j = 1,2, …, m) and tractography volume represented by Ti (i = 1,2, …, n).
2.7.1. Voxel-wise measures
- Bundle Overlap (OL) [42,43]: The proportion of voxels that contain BDA fibers (i.e. voxels in the BDA binary image volume) that are traversed by at least one streamline. The OL describes how well tractography is able to describe the volume occupied by BDA fibers and is defined as:
where |•| denotes cardinality.(1) - Modified Hausdorff Distance, mean value (HDmean): The Hausdorff distance is derived by calculating all the distances from a point in one set (voxels containing streamlines) to the closest point in the other set (voxels containing BDA), and taking the maximum of these distances. The distribution of minimum distances is heavily weighted towards zero, with a small number of voxels that produce large distances. Here, we took the mean of the minimum distances. For example, an HDmean = 5 mm means that the voxels containing streamlines are, on average, within 5 mm from the true BDA pathways.
where sup represents supremum and inf the infimum.(3) - Modified Hausdorff Distance, 90th percentile (HD90): Here, we took the 90th percentile of the minimum distances. In this case, an HD90 = 5 mm means that 90% of the streamlines are within 5 mm from the true BDA pathways.
(4)
All voxel-wise measures were calculated without inclusion of the seed region, as this region should always contain streamlines.
2.7.2. ROI-based measures
A total of 71 regions of interest were defined in MRI-space, as previously described in [29,44,45]. Briefly, 20 Gy matter labels (both cortical and deep gray matter) were defined on a separate histological dataset (of the same brain) based on cytoarchitectural features revealed in Nissl-stained sections. ROIs were manually labeled by an experienced neuroanatomist (author IS), digitized, and transformed to MRI-space using similar registration procedures as above. In addition, 51 white matter labels were created using fiber tractography, manually defining seed points, and refining tracts based on known anatomy. Labels were quality checked by a neuroanatomist (IS), assessing the coarse shape and organization of each tract. These data have been incorporated into the first digital atlas of the squirrel monkey brain [46], and will be released as a web-viewer tool to facilitate navigation through labels, histology, and MRI (manuscript in preparation).
Using these 71 labels, anatomical fidelity metrics of sensitivity, specificity, and accuracy were derived for all tractograms.
Sensitivity – True positive rate; measures the proportion of positives (regions that are occupied by BDA) that are correctly identified as such (using tractography). Sensitivity measures the ability to correctly detect all connections of the M1 region.
Specificity – True negative rate; measures the proportion of negatives (regions that do not contain BDA) that are correctly identified as such (do not contain streamlines). Specificity measures the ability to correctly identify voxels that do not have connections with M1.
Accuracy – The number of correct assessments (both true positives and true negatives) overall assessments. The accuracy measures the proportion of regions that are correctly identified as either connected, or not connected, with the M1 injection region.
All metrics, both voxel-wise and ROI-based are computed for all algorithms. Additionally, effects of reconstruction strategy, tracking algorithm (deterministic vs. probabilistic), seeding strategy, and tracking logic (using the seed region as an ROI vs. using it as a literal seed) are assessed by grouping algorithms and performing the non-parametric Kruskal Wallis test in order to test for statistically significant differences. In addition, we look at the effects of the probabilistic threshold and distance from the seed region on these accuracy measures.
3. Results
3.1. Histological results
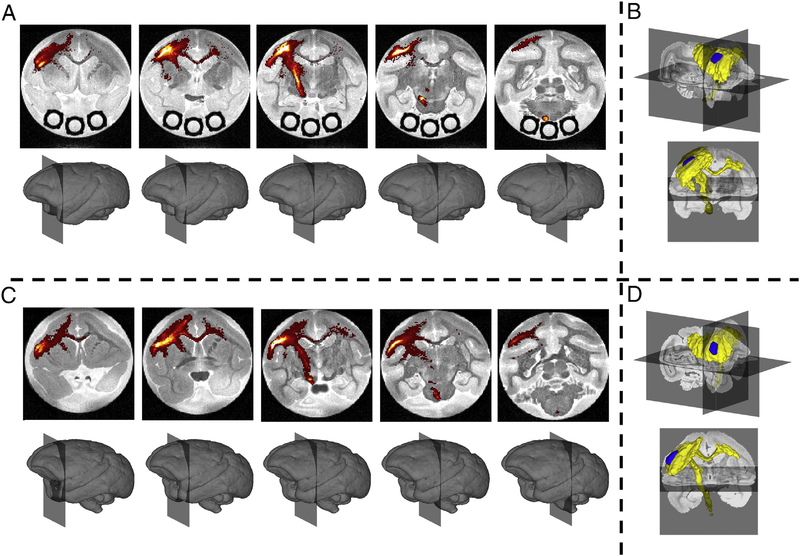
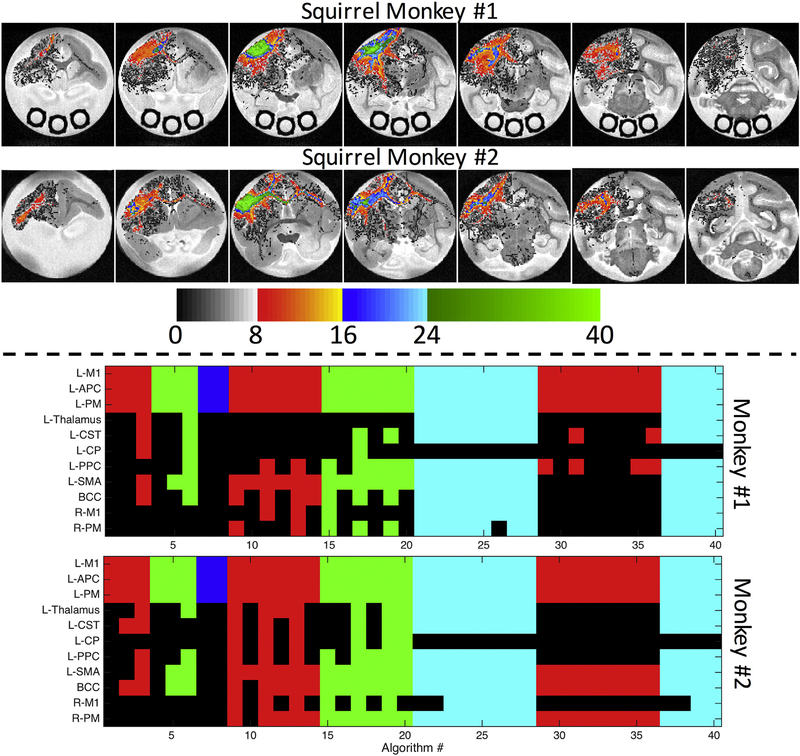
The “ground truth” BDA connectivity of the M1 injection region is shown in Fig. 2, as both BDA density maps overlaid on MRI coronal slices (A,C) and as a binary map (B,D) in a tri-planar view for each subject. Most notably, the highest BDA densities occur in the cortex of the injection region, with dense projection fibers down the corticospinal tract (CST) traversing the genu of the internal capsule (IC) and the cerebral peduncles (CP). As expected based on existing literature, the M1 injection region also has connections with the ipsi-lateral anterior parietal cortex (APC), premotor cortex (PM), ventrolateral thalamus, posterior parietal cortex (PPC), and supplementary motor area (SMA). In addition, fibers coursing through the body of the corpus callosum (BCC) connect with the M1 and PM cortex of the contra-lateral hemisphere. We note that the M1 forelimb representation of monkey #2 was found to be slightly more rostral and ventral to that of monkey #1.
Fig. 2.
Histological Results. BDA density is shown overlaid on the non-diffusion weighted volume for five coronal slices for monkey #1 (A) and monkey #2 (C). A BDA mask is shown as a volume rendering indicating the presence of BDA in a given voxel for monkeys #1 (B) and #2 (D). Injection region is shown in blue, BDA mask is shown in yellow.
3.2. Tractograms
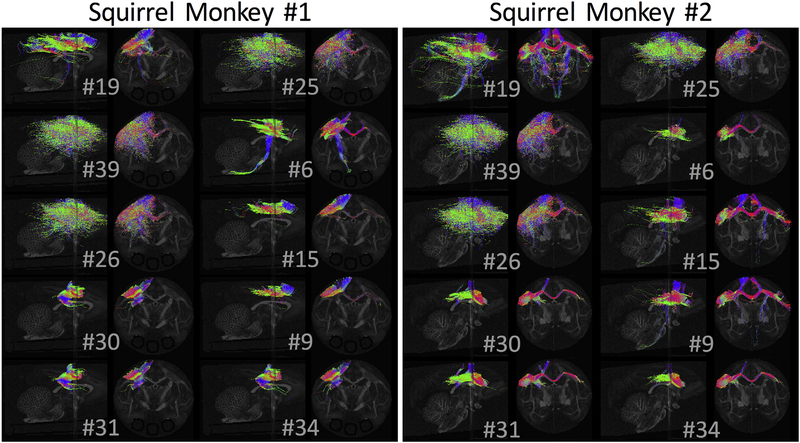
The generated streamlines for 10 randomly selected tractography methods are shown in Fig. 3 for each monkey. Qualitatively, there is tremendous variability in the resulting connectivity profiles, both in spatial extent and the pathways represented. Many algorithms result in very limited connectivity to the seed region, restricted largely to the adjacent gray matter, while other algorithms cover large expanses of the left hemisphere. Notably, few algorithms project to the contra-lateral hemisphere (particularly for monkey #1), and even fewer correctly follow the CST through the CPs.
Fig. 3.
Standard-practice pipelines vary widely in resulting tractography reconstructions. Diffusion Tractograms for streamline-generating algorithms are shown in both coronal and sagittal planes, for 10 randomly selected algorithms.
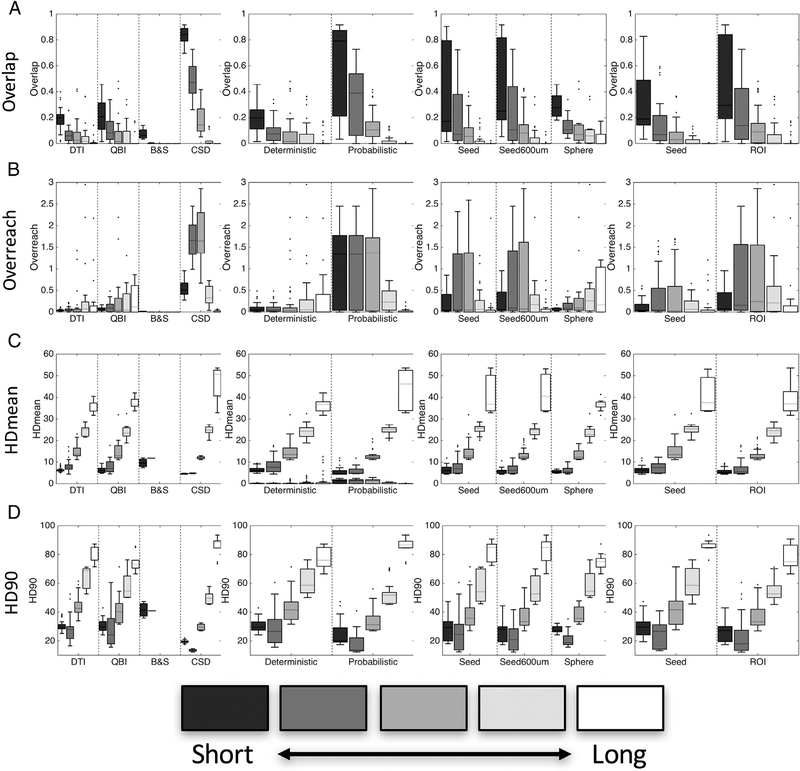
3.3. Voxel-wise anatomical accuracy
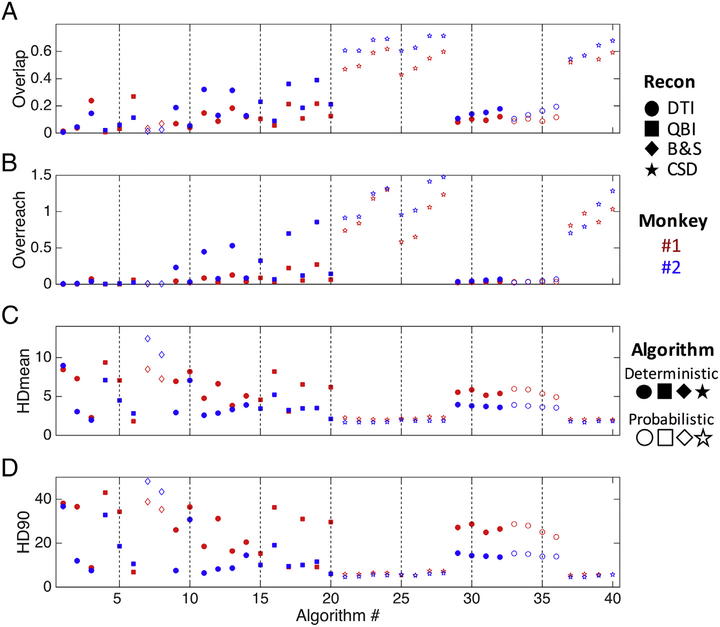
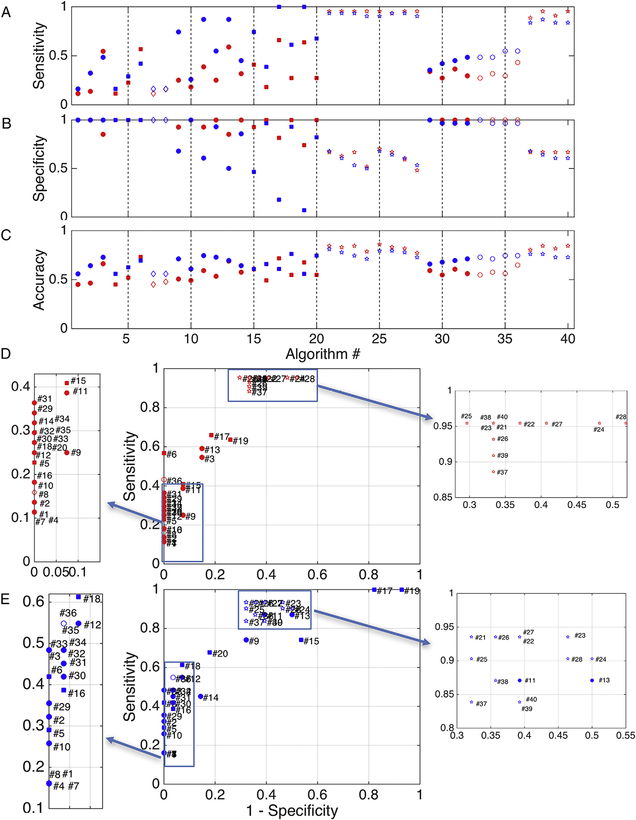
Results for the voxel-wise fidelity metrics are shown for all algorithms in Fig. 4, with marker shape, color, and fill representing differing reconstruction methods, monkey number, and algorithms, respectively. Again, there is large variation in all measures. Importantly, all results show similar trends for both monkeys. Overlap measures of OL (Fig. 4, A) vary from as low as 0.01 up to 0.71 for the most successful algorithms, indicating that the most successful tested methods recover only as much as 70% of the spatial extent of the pathways. CSD methods generally show the highest overlap measures, and utilizing the dilated seed (seed600) usually results in a greater overlap when compared to the corresponding algorithm using the undilated seed region.
Fig. 4.
Voxel-wise anatomical accuracy measures. Values for overlap (A), overreach (B), modified Hausdorff distance [mean] (units of voxels) (C), and modified Hasudorff distance [90th percentile] (D) are shown for all algorithms. Reconstruction methods are designated by symbol shape, subject number by color, and tracking algorithm by the presence (or absence) of shape fill.
The algorithms with the largest overlap generally also have higher over-reach (Fig. 4, B), sometimes with more false-positive voxels than the total number of voxels within the BDA volume itself (OR > 1). However, most algorithms have very small, or no, OR.
HDmean for all methods lies between 1.7 and 12.4 voxels (Fig. 4, C), meaning that using the implemented methods, voxels from tractography, on average, are between 1.7 and 12.4 voxels away from those of the BDA volume, or vice-versa (note this is a symmetric measure). Generally, the methods that on average differ the least from BDA are those implementing CSD as a reconstruction method. HD90 (Fig. 3, D) shows trends very similar to that of HDmean, with 90% of tractography within 48 voxels from BDA for the worst case, and within 5 voxels for the best case.
3.4. ROI-based anatomical accuracy
Fig. 5 displays the results of the ROI-based tractography fidelity measures of sensitivity (A), specificity (B), and accuracy (C). Similar to voxel-wise results, there is a wide range of performance across algorithms. Specifically, many CSD methods are able to detect a majority of the connections to M1 (high sensitivity), but lack in specificity. Conversely, nearly all other methods are highly specific, rarely indicating false positive voxels. Taken together, there is still some variation in overall ROI-based accuracy, ranging between 0.45 (algorithm #1, monkey #1) and 0.85 (algorithm #25, monkey #1). ROC curves (Fig. 5, D and E) reiterate that even on the broader scale of larger ROI domains, there is always a tradeoff in sensitivity and specificity, with a majority of methods at the two extremes, and interestingly, the DTI algorithms that use a spherical ROI (centered on the injection region) as the seed (indicated by green circles) lie in between. All results (both voxel- and ROI-based) are given in tabular form in Supplementary Table 1.
Fig. 5.
ROI-based anatomical accuracy measures. Values for sensitivity (A), specificity (B), and accuracy (C) are shown for each algorithm, along with ROC plots of sensitivity vs. 1 – specificity for animal #1 (D) and animal #2 (E). Shapes, color, and fill are the same as in Fig. 4.
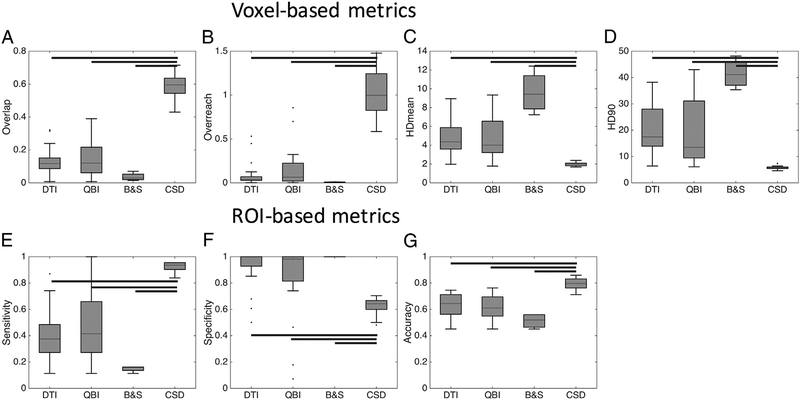
3.5. Reconstruction strategy
We next assessed the effects of the reconstruction strategy on both the voxel-based and ROI-based accuracy measures, and results are shown in Fig. 6. In agreement with qualitative observations above, CSD has statistically significantly greater OL, and OR (Fig. 6, A,B), and significantly smaller HDmean and HD90 than all other methods (Fig. 6, C,D). The B&S method has significantly greater BTO than QBI and DTI, and no differences in any other metric. Similarly, CSD has significantly greater sensitivity, lower specificity, and greater accuracy than all other reconstruction techniques (Fig. 6, E–G).
Fig. 6.
Reconstruction strategy affects track anatomical accuracy measures. Algorithms were grouped by reconstruction strategy, and statistically significant differences are indicated by solid bars – results are shown for both monkeys.
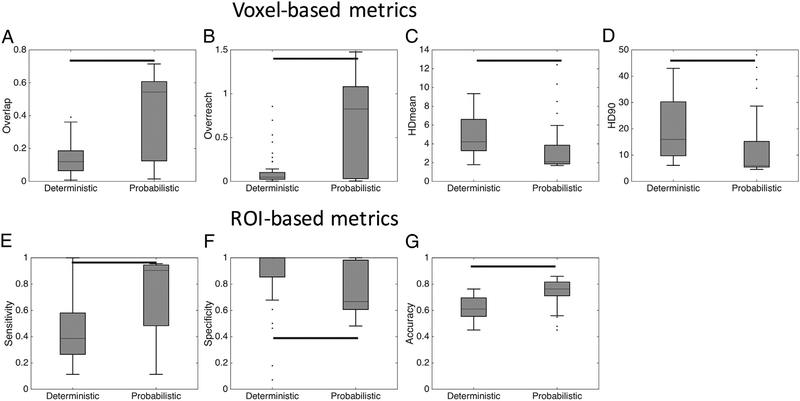
3.6. Tracking algorithm
Differences between deterministic and probabilistic algorithms were also statistically significant, as shown in Fig. 7. For voxel-wise metrics, probabilistic algorithms indicate greater overlap measures and smaller HD distances (Fig. 7, A,C,D), but a greater OR (Fig. 7, B). For ROI-based measures, probabilistic algorithms indicate greater sensitivity, reduced specificity, and an overall greater accuracy (Fig. 7, E–G) than deterministic algorithms.
Fig. 7.
Tracking algorithm affects track anatomical accuracy measures. Algorithms were grouped by tracking strategy (deterministic and probabilistic), and statistically significant differences are indicated by solid bars – results are shown for both monkeys.
3.7. Seeding strategy
No statistically significant differences were found between the use of the three different seeds: the injection region (“seed”), the injection region dilated into the white matter 600 μm (“seed600”) and a sphere centered on the seed (“sphere”) (Supplementary Fig. 2). While not statistically significant, there are some general trends that dilating into white matter slightly increases the median overlap values (OL) as well as the ROI-based sensitivity (Supplementary Fig. 2).
3.8. Seeding logic and inclusion criteria
The use of the injection region as either a seed or as an ROI after whole brain seeding only had a statistically significant effect on the HDmean (Supplementary Fig. 3), where the use as an ROI has a significantly decreased HDmean. No other measure was statistically significant. However, the use of the injection region as an ROI had an increased OL and a decreased HD90, yet this came with an increased OR. For ROI metrics, the use as an ROI had an increased sensitivity and accuracy, but decreased specificity (Supplementary Fig. 3). Again, none of these reached statistical significance.
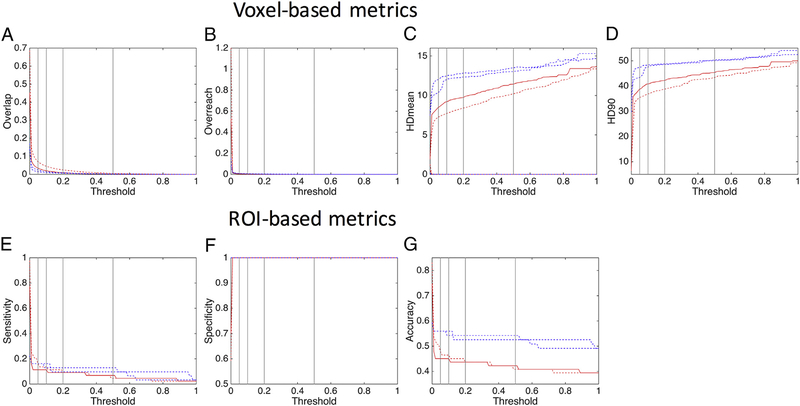
3.9. Probabilistic threshold
For probabilistic algorithms, an “uncertainty” threshold is commonly chosen, usually 5%–10% of the maximum number of streamlines in a voxel, with voxels containing less than this threshold usually disregarded. Here, we assess the effects of the threshold on algorithms #7 and #8, which differ in only the seed used (#8 utilized the dilated seed) (Fig. 8). The OL (Fig. 8, A) is quickly reduced with an increasing threshold, reaching values of 0.03 and 0.07 (for seed and seed600) at a threshold of 5%. The OR (Fig. 8, B) is almost eliminated entirely at a threshold of just 1%. Interestingly, the both HDmean and HD90 (Fig. 8, C, D) are very low with no threshold (due to the increased OL), and increase with increasing thresholds. For ROIs (Fig. 8, E–G), the thresholds between 0% and 5% have the largest change in metrics. A threshold of 5% yields sensitivity, specificity, and accuracy values of 0.11, 1.0, and 0.45 (for monkey #1) and 0.16, 1.0, 0.55 (for monkey #2), while a threshold of 0% results in a sensitivity of 0.97 (#1) and 0.77 (#2), specificity of 0.59 (#1) and 0.61 (#2), and accuracy of 0.83 (#1) and 0.77 (#2). There is comparatively little change in these metrics after 2% or 3% thresholds.
Fig. 8.
Probabilistic threshold affects track anatomical accuracy measures. Analysis is performed on algorithm #7 and #8 for subject #1 (red) and #2 (blue) which differ only in seed (#8 uses a dilated seed). Vertical lines represent thresholds at 5%, 10%, 20%, and 50% of the maximum number of streamlines in a voxel.
3.10. Track lengths and distance from seed
Table 2 shows the number of tracks generated for each algorithm, along with the mean, median, maximum, and standard deviation of tract lengths (in units of mm). Most algorithms were limited by the software defined default maximum number of streamlines, or simply by the number of tracks passing through the seed ROI. We highlight the three algorithms with the lowest (blue) and highest (green) lengths for each category. For reference, the squirrel monkey brain is approximately 35 mm across (left to right), 30 mm in height (at the level of the injected region), and 50 mm in length (anterior to posterior). Some of the standard methods implemented track with maximum lengths of only 10–15 mm, with average lengths on the order of 1 mm, clearly not able to cover the spatial extent of the true connections.
Table 2.
Streamline lengths (in mm). The number of tracks, and mean, median, maximum, and standard deviation of streamline lengths are shown for all algorithms. Blue and green boxes highlight the three minimum and maximums of select categories, respectively.
| Algorithm # | #Tracks | Mean Length | Median Length | Max Length | St.D. Lengths |
|---|---|---|---|---|---|
| 1 | 216 | 2.3 | 1.2 | 15.8 | 2.5 |
| 2 | 360 | 4.2 | 3.1 | 16.1 | 3.6 |
| 3 | 8340 | 6.8 | 5.1 | 54.5 | 7.0 |
| 4 | 435 | 1.0 | 0.7 | 10.4 | 1.0 |
| 5 | 602 | 1.8 | 0.9 | 14.6 | 2.2 |
| 6 | 11872 | 3.8 | 1.8 | 39.6 | 4.5 |
| 7 | 9645000 | N/A | N/A | N/A | N/A |
| 8 | 11795000 | N/A | N/A | N/A | N/A |
| 9 | 2000 | 7.8 | 4.2 | 46.2 | 7.3 |
| 10 | 2000 | 4.0 | 1.6 | 26.2 | 5.3 |
| 11 | 2000 | 12.2 | 12.0 | 63.5 | 8.6 |
| 12 | 2000 | 6.5 | 2.0 | 31.5 | 7.3 |
| 13 | 2000 | 13.6 | 13.7 | 63.5 | 9.0 |
| 14 | 2000 | 7.4 | 3.0 | 37.2 | 7.6 |
| 15 | 2000 | 12.2 | 14.1 | 66.2 | 8.1 |
| 16 | 2000 | 7.0 | 3.6 | 25.4 | 6.4 |
| 17 | 2000 | 18.5 | 18.8 | 101.9 | 12.5 |
| 18 | 2000 | 9.2 | 5.2 | 34.9 | 8.3 |
| 19 | 2000 | 19.3 | 19.5 | 101.9 | 12.7 |
| 20 | 2000 | 9.6 | 5.8 | 39.6 | 8.7 |
| 21 | 5000 | 16.0 | 15.1 | 29.9 | 9.8 |
| 22 | 5000 | 17.2 | 16.8 | 29.9 | 9.8 |
| 23 | 4876 | 21.2 | 23.4 | 29.9 | 8.9 |
| 24 | 5000 | 22.2 | 25.3 | 29.9 | 8.5 |
| 25 | 5000 | 10.9 | 8.1 | 29.9 | 8.4 |
| 26 | 5000 | 11.8 | 9.5 | 29.9 | 8.6 |
| 27 | 4136 | 17.4 | 17.3 | 29.9 | 9.3 |
| 28 | 5000 | 18.0 | 18.0 | 29.9 | 9.1 |
| 29 | 5000 | 6.2 | 5.0 | 25.7 | 4.6 |
| 30 | 5000 | 6.9 | 5.8 | 25.7 | 4.5 |
| 31 | 2092 | 8.8 | 9.2 | 25.6 | 4.8 |
| 32 | 2568 | 9.2 | 10.0 | 26.1 | 4.6 |
| 33 | 5000 | 4.4 | 3.1 | 24.4 | 3.2 |
| 34 | 5000 | 4.6 | 3.5 | 26.0 | 3.1 |
| 35 | 1348 | 6.3 | 5.4 | 19.7 | 3.8 |
| 36 | 1734 | 6.5 | 5.6 | 25.9 | 3.9 |
| 37 | 6098 | 13.2 | 10.6 | 29.9 | 9.7 |
| 38 | 6921 | 14.1 | 12.0 | 29.9 | 9.8 |
| 39 | 8756 | 9.7 | 6.4 | 29.9 | 8.2 |
| 40 | 9833 | 10.5 | 7.3 | 29.9 | 8.6 |
We next examine the anatomical accuracy measures as distance from the seed varies. This was done by binning results (BDA and tractography) into bins based on a Euclidean distance from the center of the seed, and calculating metrics for each case. Fig. 9 shows the results of these experiments, with algorithms again separated based on reconstruction method, tracking method, seed region, and seeding logic. Intuitively, the overlap measures (Fig. 9, A) decrease with increase distance from seed, for all tracking parameters. In nearly all cases, the overlap is zero by the 4th “bin”, a distance of 14.7 mm (see figure legend). The OR values (Fig. 9, B) show interesting trends, with most overreach occurring in the medium-distance range (a range still in the ipsi-lateral hemisphere). This is because there is very little over-reach for small distances, and few streamlines propagate longer distances (see Table 2) thereby contributing only little to OR. For the HD distances (Fig. 9, C, D), in all cases the maximum distance between BDA and streamlines increases for the pathways further from the seeds. If streamlines do propagate greater distances, they do not do so accurately.
Fig. 9.
Track length (distance from seed) affects track anatomical accuracy measures. Measures are binned across equidistant intervals (in mm) of < 4.9, 4.9–9.8, 9.8–14.7, 14.7–19.6, and > 19.6 – results are shown for both monkeys.
3.11. Pathway representations
To better understand which pathways are more (or less) represented by standard algorithms on both voxel and ROI scales, we visualized the number of algorithms that pass-through a given voxel (Fig. 10, top), as well as those that indicate connections to a given ROI (Fig. 10, bottom). Besides the seed region (M1), the voxels most represented are in the cortical areas just anterior (PM cortex) and posterior (PPC) to the injection region, in addition to the superficial white matter immediately below the cortex (Fig. 10, top) for both monkeys. Few algorithms overlap in the corpus callosum or overlap in the CST, and fewer still have any overlapping voxels in the contra-lateral hemisphere.
Fig. 10.
Algorithms vary in estimating both spatial extent and connectivity. The number of algorithms reaching given voxels (top) are shown overlaid on select coronal slices. On the scale of ROIs (bottom), if an algorithm has at least one streamline reach a region known to be occupied by BDA, the index displays as a color (as opposed to black for “no connection”). Colors indicate the reconstruction method used (red: DTI; green: Qball; blue: B&S; cyan: CSD). Regions on the vertical axis are listed in order of BDA densities derived from histology (i.e. most BDA occurs in M1).
On the scale of ROIs, Fig. 10 (bottom) shows the BDA connections ranked from highest to lowest, with a colored index (colored based on reconstruction method) if the algorithm reaches these regions. All algorithms indicate connections with APC and PM regions. Few reach the thalamus, and those that do are largely grouped between #21–28 and #37–40, employing CSD. Similarly, only a few reach the CST (only 3 and 5 DTI methods for monkey #1 and #2, respectively) and fewer extend through to the CP. The ipsi-lateral cortical areas (PPC and SMA) are largely represented for most methods, while the contralateral connections are few, and again mostly dependent on the reconstruction method or software package. Monkey #2 was slightly more successful in identifying contralateral connections than tracking on Monkey #1.
3.12. Spatial errors in tractography
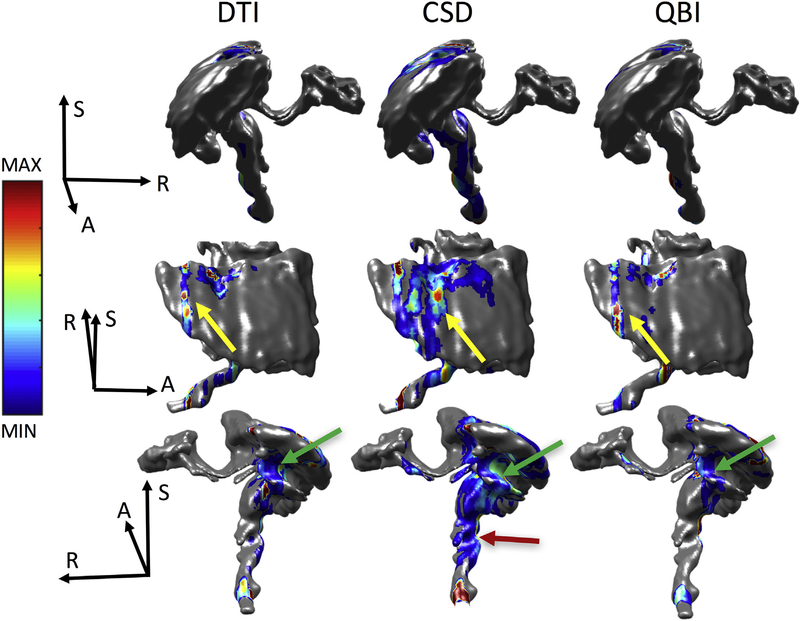
We next ask two questions, where do errors occur? And what do these “error voxels” have in common? Following every individual streamline, we record where it first exits the BDA mask, meaning it no longer coincides with the M1 pathway. Fig. 11 shows three views each of DTI, CSD, and QBI reconstructions (concatenation of all algorithms using that reconstruction) highlighting where errors occur for monkey #1 (Monkey #2 results are shown in supplementary Fig. 4). Qualitatively, all three methods show hot spots of errors posterior to the injection region, in the gray matter, in the PPC (yellow arrow). In addition, all show a grouping of errors projecting anteriorly into the sulcal fundi (inferior to the injection region) instead of following the U-fibers along the gyral stalk (green arrow). Finally, the CSD methods, of which many project down the CST, show evidence of prematurely exiting this pathway anteriorly (red arrow).
Fig. 11.
Where tractography goes wrong. Density maps overlaid on the BDA fiber mask indicating where tractography first exits the BDA mask are shown for DTI (left), CSD (middle), and QBI (right), in three different orientations. Note that for the B&S algorithm, streamline outputs are not given, so we cannot query where error occurs. Results for subject #2 are given as Supplementary Fig. 3. Error density maps are scaled individually from maximum to minimum error (see colorbar) where gray indicates no streamline error.
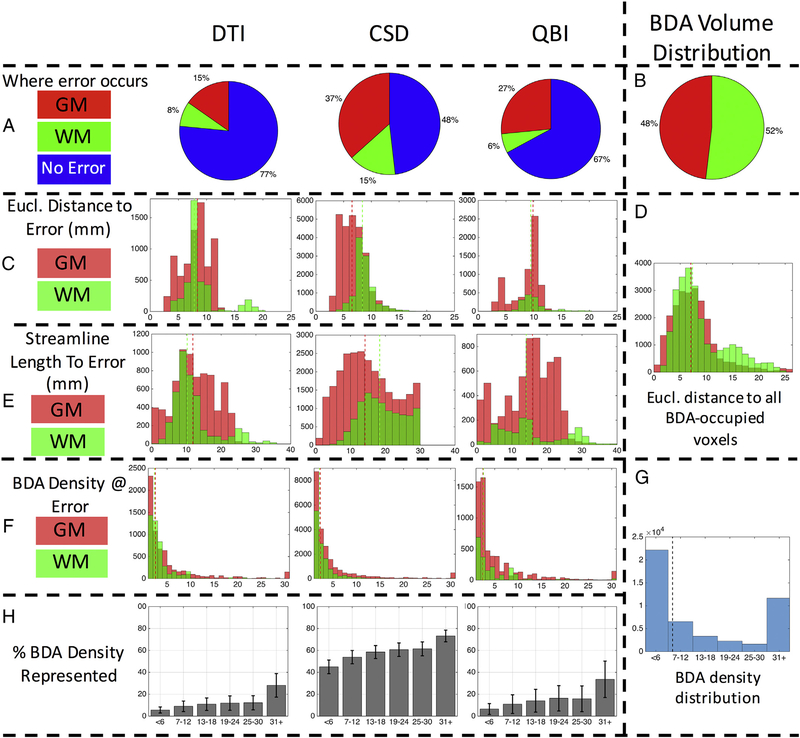
We quantify several measures at these error locations (Fig. 12 and Supplementary Fig. 5 for monkey #1 and #2, respectively). For all reconstruction methods, we find that most errors actually occur in gray matter regions (Fig. 12, A), whereas the BDA is distributed approximately equally between white and gray matter (Fig. 12, B). In agreement with lower OR and increased specificity, a large majority of streamlines for DTI and QBI never “go wrong”, never leaving the true pathways volume. When streamlines do leave the BDA mask, most errors occur < 7–10 mm away from the seed region (Fig. 12, C) in both white and gray matter. To give a reference for distance, the distances from the seed to all voxels occupied by BDA is shown in Fig. 12, D, with median distances of about 7 mm. The Euclidean distance to the error is always less than the actual length of the streamlines themselves (Fig. 12, E), which generally propagate between 10 and 20 mm before they become unreliable. We note that MRTrix default tracking parameters have a hard cut-off of maximum streamline length of 100 times the voxel size, thus all of the tracking implementations utilizing CSD reconstructions (all done in MRTrix) have a maximum streamline length of 30 mm.
Fig. 12.
Several sources contribute to tractography error. Pie plots show where error occurs (A) as well as BDA volume distribution in white and gray matter (B). The Euclidean distance to error voxels is shown for white and gray matter (C) as well as the distance to all voxels occupied by BDA (D). The streamline length to the error is shown for white and gray matter (E). Finally, the BDA density right before an error occurs is shown (F), as well as the overall BDA density distribution (G), and the percent of BDA density represented for each algorithm (H). Results for subject #2 are given as Supplementary Fig. 3.
A look at the overall distribution of BDA density in BDA-positive voxels (Fig. 12, G) shows that a majority of voxels have a very low density, with prevalence decreasing quickly as density increases. Quantifying the BDA density along a streamline’s last correct step (Fig. 12, F), we find that a majority occur in regions of very low BDA (< 2 BDA fibers per voxel) and very rarely do streamlines deviate from pathways with strong connections to M1. In addition, the voxels with higher BDA densities are consistently more represented by tractography than those with lower densities (Fig. 12, H).
4. Discussion
Diffusion MRI tractography is the only non-invasive method that offers the ability to map the structural connectivity of the human brain, and its application has been widely adopted in both small and large-scale studies over the last two decades in order to improve our understanding of normal brain development as well as complex brain disorders. However, the application of these methods is arguably racing ahead of our ability to understand the data and its limitations. It is critical that these methods result in anatomically accurate reconstructions, both in reconstructing major fiber bundles and in quantifying region-to-region connectivity, not only to ensure that sound conclusions are reached on an individual basis, but also for comparative studies across subjects, time, or even across differing diffusion tractography implementations. Here, we aim to answer the question “how reliable are the most commonly used methods?”. In addition, we attempt to determine the most common pitfalls and successes of these algorithms. We accomplish this by performing both diffusion MRI and histological tracing in the same brain. Because of this, we can probe not only brain connectivity, but spatial overlap on the scale of individual MRI voxels.
There are several main takeaways from this study. First, we find a sensitivity and specificity tradeoff in both voxel-wise measures of spatial overlap and in region-to-region measures of tractography accuracy. None of the standard practice algorithms was consistently successful at identifying true positive connections AND true negative connections. With the large number of commonly implemented pipelines investigated, this study helps emphasize the differences between tracking methods. We find a large variation in the tractography reconstructions, both visually and quantitatively. The anatomical accuracy of the reconstructed pathways is dependent on parameter and algorithm choices. For example, the results are most dependent on the voxel-wise reconstruction method used, whether the algorithm is deterministic or probabilistic in nature, and the tract threshold used in analysis. In general, for all algorithms and implementations, the accuracy decreases for increasing distances from the seed. Finally, an analysis of spatial errors indicates that many errors occur in the cortex, errors occur when the true fiber density is low, and results of long-range connectivity (for example to the contra-lateral hemisphere) should be interpreted with caution.
4.1. Anatomical accuracy
Most of the methods implemented in this study do not fully cover the spatial extent of the true fiber pathways connected to M1, as shown by low OL values (Fig. 4). This is particularly true for DTI (recovering just 20% or less of the true bundles), which was, and arguably still is, the most commonly implemented reconstruction algorithm used as a basis for tractography. The algorithms that are able to cover large portions of the true fiber pathways were typically those implemented using CSD reconstruction with probabilistic tractography, however, these suffered from large overreach, sometimes covering twice the spatial extent of the true pathways. Discouragingly, measures of distances between histology and tractography show that streamlines and tracer are, on average, separated by between 2 and 10 voxels (HDmean, Fig. 4), however this number is largely influenced by tracer on the contra-lateral hemisphere. In summary, none of the commonly utilized methods tested were consistently successful at accurately delineating the spatial profile of the true pathways.
While voxel-wise spatial overlap is important, many studies are interested in region-to-region connectivity or general track shapes where voxel-by-voxel accuracy may not be critical. Towards this end, we calculated sensitivity, specificity, and accuracy of these methods to identify the presence of connections to various white and gray matter regions of interest. Much like OL and OR, the sensitivity and specificity varied dramatically depending on algorithm and tracking choices. Notably many algorithms lay at the two extremes of the ROC curve (Fig. 5) with either very high sensitivity (typically CSD implementations with probabilistic tractography), or high specificity (all other algorithms). It is important to point out that neither the reconstruction method nor tracking method (probabilistic versus deterministic) accounts for the sensitivity/specificity tradeoff alone. For example, both deterministic and probabilistic methods cover a wide range of sensitivity/specificity with different reconstruction methods, and vice-versa, a given reconstruction method can span the range of accuracy measures (dependent on tracking method and modulated by other tracking parameters). We note that all CSD methods tested were probabilistic, as this was commonly done in both literature and existing software packages. Interestingly, three of the four algorithms that did not lie at the extremes used DTI with a spherical seed (rather than the pre-defined seed). The high specificity for most algorithms, however, is due to the failure to propagate longer distances (see Discussion, Errors in Propagation, and Discussion, Streamline length), resulting in zero false positive connections.
4.2. Choosing algorithms
Connections of the primary motor cortex are particularly relevant for a variety of disorders, for pre-operative planning, and for basic neuroscience of the healthy brain. In addition to overall accuracy of the reconstructed pathways, we probe which M1 connections are most (or least) represented using different tractography techniques. These results could lend insight into the algorithm of choice if a researcher or clinician is interested in a specific connection. For example, one may be interested in the thalamic connections for deep brain stimulation in patients with Parkinson’s disease or essential tremor [47,48], the contra-lateral motor connections through the corpus callosum in patients with epilepsy [49,50], or the general corticospinal tract delineation for tumor removal surgery [25,51].
In addition, the sensitivity and specificity analysis should help to choose a tracking and reconstruction strategy based on study requirements. An exploratory study of any and all potential connections of M1 could choose an implementation with high sensitivity. Our results would suggest choosing CSD (Fig. 6), using a seed extended into the WM, using a probabilistic method (with a low threshold, if utilized), and using a ROI as an inclusion mask rather than as a seed. On the other hand, if a high specificity is required, an algorithm could be chosen that has an increased specificity (Fig. 5), but has adequate overlap (Fig. 4) and/or meets the requirements for connecting to regions of interest relevant to the study (Fig. 10). These results are only specific to the M1 of the squirrel monkey brain (see Discussion, Limitations), but general trends are expected to be the same for tractography in other specimens or differing tracts, although likely with different absolute values.
4.3. Variability
A surprising result of our study was the large variability in the results, given that all streamlines were generated using the same data set (same b-value, number of diffusion weighted directions, resolution, SNR) and the most basic of algorithms (Fig. 3). This variability can be attributed not only to differences in the parameters we tested – reconstruction method, which seed was used, how the seed was used, and algorithm – but also likely due to the minor variations in implementations from differing software packages. This could include variations in termination index (FA, curvature thresholds), step sizes used, smoothing, inclusion criteria (maximum and minimum streamline lengths), interpolation methods, number of tracts generated, and number of tracts kept for analysis. Analysis of these would make the parameter space intractable, so we’ve chosen to implement each algorithm using the default parameters (or those recommended in existing tutorials) which represent a majority of the usage.
This variability highlights the importance of using the same tracking parameters for a given study. Although we are not aware of comparisons of populations using different tractography methods within a study (i.e., the use of CSD for healthy controls and DTI for the diseased population), it certainly complicates comparisons across studies. Reported findings, statistical differences in indices and locations of these differences, using one technique are almost certainly going to differ when using a different implementation. This study also highlights the importance of seeds, and how small variability in the seed penetration into white matter can lead to large variability in resulting tractography, a factor that could be hard to control given individual differences in brain geometries and size, even if analysis is performed in a common space.
4.4. Probabilistic threshold
A common strategy following probabilistic tractography is to threshold the streamline count to a certain percent of its maximum value (typically 5%), as the voxels containing few streamlines are considered to have a higher uncertainty of connection to the seed region [52]. Our results shown that the biggest geometrical changes in tractography occur between thresholds of approximately 2–3%, beyond which the overreach is reduced and specificity increased, but at the cost of dramatically reduced overlap and sensitivity. Similar to parameter and tracking considerations, this threshold could be tuned based on the requirements of the specific study (see Discussion, Choosing algorithms). And again, there is no clear optimal threshold for probabilistic tractography, with several tradeoffs in anatomical accuracy that must be considered.
4.5. Common and complex algorithms
This study should not be viewed as a “ranking” of algorithms, but rather as a survey and assessment of the methods that are currently most implemented in the literature, and have been for the last decade. For example, many of the algorithms and implementations still used DTI, which has limitations that have been well known in the diffusion community for quite some time [3,53], as well as very basic FACT [54] propagation of streamlines. However, the use of these algorithms is still prevalent, largely because of their availability and ease of implementation in open source software packages, which require only diffusion images and parameters (b-values and b-vectors) to create beautiful tractograms with little to no user intervention. Some of the methods implemented do include more advanced high angular resolution reconstruction algorithms (i.e. CSD, QBI, B&S), complex decisions made for tracking (i.e. probabilistic methods), or even streamline filtering strategies that match fiber densities to the diffusion signal (SIFT [55]), with some significant improvements in many fidelity measures. However, while these address the problem of fibers crossing within an MRI-voxel, they still lack some combination of specificity or sensitivity in all cases.
These algorithms should serve as a benchmark against which future algorithms can be compared. Towards this end, the data available here (as well as a phantom dataset and a macaque dataset published in a different study [56]) are being made available for a diffusion tractography challenge, hosted by IEEE International Symposium on Biomedical Engineering (ISBI 2018). Researchers are free to present their own submissions (even after challenge completion) and tract accuracy parameters will be automatically calculated (https://my.vanderbilt.edu/votem/).
Based on the current results, there is significant room for improvement in tractography. New algorithms and analysis methods are continually proposed and published, and we expect that as the algorithms are validated and the fidelity is shown to improve, these methods will be more commonly employed. Future development should include algorithms that incorporation prior anatomical knowledge [57], use appropriate inclusion/exclusion criteria (dependent on tract) [58], or corrections for length and known tracking biases [59–61].
4.6. Length
As expected, and in agreement with previous theoretical and experimental studies [61–63], nearly all fidelity metrics worsen as the distance from the seed (or streamline length) increases. At the most extreme distances, tractography holds almost no predictive value. While some algorithms were limited in that they could not propagate out of the gray matter (see Discussion, Spatial errors), the ones that did show evidence of streamlines to the other hemisphere did not do so accurately (on the scale of voxels).
4.7. Spatial errors
We assess not only tractography accuracy measures, but also probe where tractography goes wrong. We found that for most methods, the first instance of streamline error actually occurs in the gray matter (Fig. 11). This can be appreciated qualitatively in Fig. 3 (and quantitatively in Fig. 10) where many streamlines show connections to only adjacent cortical regions anteriorly (PM) and posteriorly (APC). In many cases (particularly DTI), tracts cannot properly extend into the white matter, due to a combination of complex gray matter fiber orientations that lead to ambiguous orientation estimates, as well as crossing white matter systems adjacent to the cortex [11]. At first sight, the distances to the first discrepancy between tractography and tracer was surprisingly short, with an average distance of 7–10 mm. However, the streamline lengths to the first errors were on average 10–20 mm, corresponding to 33–66 voxel-lengths, and anywhere from ~66 (10 mm/.15 mm) to as many as ~660 (20 mm/.03 mm) steps, depending on step size (ranging from .03 to .15 mm).
4.8. In relation to previous validation studies
Several validation studies have aimed to determine the successes and limitations of fiber tractography. Specifically, many of these metrics are similarly evaluated on the FiberCup [64] dataset using Tractometer [42]. The FiberCup is a physical phantom meant to represent a coronal slice of the brain, with crossing, curving, and fanning fiber structures. Although the focus was on connectivity metrics, the results of the Tractometer study suggest a much more positive outlook, with many algorithms reconstructing 92% or more of the true fiber bundles [42], whereas many of our algorithms failed to identify regions not immediately adjacent to the seed regions. This could be due to the increased complexity expected in the squirrel monkey tissue, which is expected to be more similar to the complexity seen in the in vivo human brain.
As an alternative, many studies utilize histological tracers for validation. Qualitative comparisons have shown good agreement with histological tracing in the monkey brain [12], or against human cadaver samples [65]. The sensitivity and specificity of tractography in detecting pathways has also been systematically explored in the monkey [10,56,66] and mouse [9], suggesting a moderate to good accuracy in identifying connections and their pathways. However, these all rely on collections of histological data from a brain different than the one studied with tractography, and thus cannot access voxel-wise metrics. Comparisons with our results show that tractography performs much better when estimating connectivity between relatively larger regions of interest rather than fine details on the scale of individual voxels. Alternatively, some studies utilize MR-visible tracers as the ground truth [67,68], also validating some common implementations of tractography (including similar software packages). These studies yielded similar voxel-wise results, although the methods are quite different (MR-visible tracers avoid the complexity of registration with histology; BDA provides higher spatial resolution connectivity maps). Regardless, all studies illustrate a repeated theme – a strong dependence of the results on reconstruction model and tractography settings, and that increased sensitivity comes at the cost of decreased specificity, thus optimizing tracking parameters is important. Because optimal settings for different pathways are likely to vary (due to geometry, location, and complexity), the methodology described in this manuscript should be repeated for several pathways of interest to the neuroscience or neurosurgery communities.
Our results are largely in agreement with a series of studies performed on a high quality, high resolution, ex vivo macaque brain [56,69], where region-to-region connectivity of tractography is compared to existing histological tracer studies. The authors demonstrate that tractography with high sensitivity will likely show low specificity, and vice-versa [56]. They conclude that anatomical accuracy of tractography is fundamentally limited, even with exceptional data quality. In addition to the sensitivity/specificity tradeoff, our results indicate that a large number of commonly used algorithms (both past and present) lack anatomical accuracy in both voxel-wise overlap and region-to-region connectivity. In the macaque study, Reveley et al. [69] find that accurate tracking is dependent on the ability to follow the correct fiber trajectory through the white matter/gray matter boundary, which is complicated by superficial white matter systems immediately adjacent to the cortex. These white matter systems are also the likely cause of failure of many algorithms to properly propagate out of the cortex.
4.9. Limitations and future work
This study has several potential limitations. The first is the sample size. Significant resources are required to perform tracer injections, scan the brain for an extended period of time, performed histological reactions, and register individual slices to block face images, all before quantitative comparisons can be made. Because of this, we are compiling all data and making all resources (both registered histology and MRI) available not only for the ISBI 2018 challenge, but also on a website containing the first digital atlas of the squirrel monkey brain (publication under review). At this point, we have only assessed a single injection site in two monkey brains. Thus, these results are only specific to the connections with the primary motor cortex. Although the trends are likely similar across different pathways, further evaluations are needed to establish the accuracy of other tracts.
In addition, a major limitation of this study is the MRI protocol, and several sources of error are likely related to the sub-optimal acquisition. The acquisition in this study is much closer to what would be expected clinically, rather than in the research environment. The number of diffusion directions is relatively low for high angular resolution diffusion imaging (although on the higher end for tensor-based studies) and the b-value is lower than optimal for ex vivo imaging [28], both of which could be sources of variation not related to the algorithms themselves. However, the relevant orientation contrast-to-noise ratio between parallel and perpendicular diffusivities is increased through multiple averages, facilitating orientation distribution reconstructions. For example, Supplementary Fig. 1 shows coherent single fiber populations in the corpus callosum, crossing fibers where it crosses with the corona radiata, and gray matter distributions largely perpendicular to the white matter/gray matter boundary – with fairly consistent results across multiple reconstruction methods. Thus, the reconstructions are largely successful, giving expected results, despite the limited acquisition. In addition, the FA is preserved ex vivo (Supplementary Fig. 1), with anisotropy very similar to that seen in vivo (and in the human) [29] in both white and gray matter, justifying the decision to not alter or investigate the default FA thresholds in various software implementations. Future work will include more monkeys, and multiple injection sites, as well as different acquisition techniques (more time efficient acquisition) and different acquisition schemes with multiple diffusion weightings or more diffusion gradient directions, for both in vivo and ex vivo brains.
With histology, there are several potential sources of error. First, it is possible that not all axons in the injection region were labeled equally, which could contribute to false negative tractography results. Errors in Image processing and detection of BDA-labeled fibers could also be a source of false positive or false negative BDA in our ground truth dataset. We have taken several precautions to ensure that the tracer gets deposited along the entire length of the axons (so that stain intensity does not vary with distance), and covers the entire M1 region of interest. To ensure as much tracer uptake as possible, we made eight injections covering large portions of the M1 cortex of interest. We also waited several weeks between injection and sacrifice to minimize false negative BDA labels (i.e. to ensure BDA was transported along the entire axon) and verified that BDA was visible in axon terminals in the cortex. In addition, there is potential geometric mismatch that is not corrected through registration. We expect the accuracy to only be on the order of the size of the MR voxels themselves [39].
It is important to point out that this study simply asks “is tractography able to identify pathways and connections associated with a well-defined cortical region (the injection region in M1)”. It does not ask how well tractography extracts a single pathway, instead, M1 shares connectivity with a number of distinct white matter bundles. If a clinician or scientist were trying to extract a known bundle, they may use prior knowledge to place seeds, ROIs, or exclusion regions in order to isolate the intended bundle. For example, to extract the corticospinal tract, it is common to seed from the whole brain, and isolate streamlines that pass through multiple ROIs, typically in the cerebral peduncles and either the superior internal capsule or a gray matter region in the motor cortex. It would be of interest to determine how well algorithms with manual ROI placement are able to extract specific pathways associated with M1 (for example, contralateral connections, thalamic connections, corticospinal projections).
In this study, we do not investigate all possible combinations of parameter choices, instead focusing on reconstruction method, tracking logic, threshold value, and seeding strategy. It would be of interest to also determine the effects of FA, angular threshold, and step size, as has been done on mouse models [70] to systematically study the trends and variation in measures. Instead, we chose several commonly implemented pipelines, with common parameter choices (scaled to the squirrel monkey brain, when necessary), to look at the overall accuracy of the standard of practice techniques.
Finally, the definition of “where tractography goes wrong” needs to be clearly stated. We chose this to mean “where tractography first doesn’t match” the BDA pathways, because at this point tractography is clearly not correct. However, it may – and likely does - go wrong before this point, for example by stepping onto a crossing fiber well inside the BDA mask, possibly due to orientation mismatch or the inability to identify all fiber populations present in a voxel. For this reason, assessing the ability of reconstruction algorithms to correctly describe the distribution of neuronal fibers is also an important step in the validation process [71,72].
5. Conclusions
Diffusion tractography has seen widespread use for investigating the structural connectivity of the human brain. Despite known limitations of common methods, and a large number of advanced algorithms and reconstruction methods, most studies still implement common, open-source tractography methodologies. We found that none of these standard-practice algorithms is consistently successful at recovering the spatial extent of fiber pathways, or revealing region-to-region connectivity. The anatomical accuracy of results is dependent on parameter and algorithm choices, and accuracy decreases at increased streamline lengths. Finally, error analysis indicates that tractography in many cases is not able to leave the gray matter, and is not successful at recovering low density fiber pathways.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award numbers RO1 NS058639 and S10 RR17799. Whole slide imaging was performed in the Digital Histology Shared Resource at Vanderbilt University Medical Center (www.mc.vanderbilt.edu/dhsr).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mri.2018.09.004.
References
- [1].Catani M, Thiebaut de Schotten M. Atlas of human brain connections. 2015.
- [2].Johansen-Berg H, Behrens TE. Just pretty pictures? What diffusion tractography can add in clinical neuroscience. Curr Opin Neurol 2006;19(4):379–85. 10.1097/01.wco.0000236618.82086.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jones DK, Cercignani M. Twenty-five pitfalls in the analysis of diffusion MRI data. NMR Biomed 2010;23(7):803–20. 10.1002/nbm.1543. [DOI] [PubMed] [Google Scholar]
- [4].Jones DK, Knosche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. NeuroImage 2013;73:239–54. Epub 2012/08/01 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- [5].Markov NT, Ercsey-Ravasz MM, Ribeiro Gomes AR, Lamy C, Magrou L, Vezoli J, et al. A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb Cortex 2014;24(1):17–36. 10.1093/cercor/bhs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stephan KE, Kamper L, Bozkurt A, Burns GA, Young MP, Kotter R. Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac). Philos Trans R Soc Lond Ser B Biol Sci 2001;356(1412):1159–86. 10.1098/rstb.2001.0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, et al. A mesoscale connectome of the mouse brain. Nature 2014;508(7495):207–14. 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van den Heuvel MP, de Reus MA, Feldman Barrett L, Scholtens LH, Coopmans FM, Schmidt R, et al. Comparison of diffusion tractography and tract-tracing measures of connectivity strength in rhesus macaque connectome. Hum Brain Mapp 2015;36(8):3064–75. 10.1002/hbm.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Calabrese E, Badea A, Cofer G, Qi Y, Johnson GA. A diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cereb Cortex 2015;25(11):4628–37. 10.1093/cercor/bhv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Donahue CJ, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Behrens TE, Dyrby TB, et al. Using diffusion tractography to predict cortical connection strength and distance: a quantitative comparison with tracers in the monkey. J Neurosci 2016;36(25):6758–70. 10.1523/JNEUROSCI.0493-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gao Y, Choe AS, Stepniewska I, Li X, Avison MJ, Anderson AW. Validation of DTI tractography-based measures of primary motor area connectivity in the squirrel monkey brain. PLoS One 2013;8(10):e75065 Epub 2013/10/08 10.1371/journal.pone.0075065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schmahmann JD, Pandya DN, Wang R, Dai G, D’Arceuil HE, de Crespigny AJ, et al. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain 2007;130(Pt 3):630–53. 10.1093/brain/awl359. [DOI] [PubMed] [Google Scholar]
- [13].Dauguet J, Peled S, Berezovskii V, Delzescaux T, Warfield SK, Born R, et al. Comparison of fiber tracts derived from in-vivo DTI tractography with 3D histological neural tract tracer reconstruction on a macaque brain. NeuroImage 2007;37(2):530–8. 10.1016/j.neuroimage.2007.04.067. [DOI] [PubMed] [Google Scholar]
- [14].Dauguet J, Peled S, Berezovskii V, Delzescaux T, Warfield SK, Born R, et al. 3D histological reconstruction of fiber tracts and direct comparison with diffusion tensor MRI tractography. Med Image Comput Comput Assist Interv 2006;9(Pt 1):109–16. [DOI] [PubMed] [Google Scholar]
- [15].Stepniewska I, Preuss TM, Kaas JH. Architectonics, somatotopic organization, and ipsilateral cortical connections of the primary motor area (M1) of owl monkeys. J Comp Neurol 1993;330(2):238–71. 10.1002/cne.903300207. [DOI] [PubMed] [Google Scholar]
- [16].Moller M, Frandsen J, Andersen G, Gjedde A, Vestergaard-Poulsen P, Ostergaard L. Dynamic changes in corticospinal tracts after stroke detected by fibretracking. J Neurol Neurosurg Psychiatry 2007;78(6):587–92. 10.1136/jnnp.2006.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ward NS, Newton JM, Swayne OB, Lee L, Thompson AJ, Greenwood RJ, et al. Motor system activation after subcortical stroke depends on corticospinal system integrity. Brain 2006;129(Pt 3):809–19. 10.1093/brain/awl002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hubbard EA, Wetter NC, Sutton BP, Pilutti LA, Motl RW. Diffusion tensor imaging of the corticospinal tract and walking performance in multiple sclerosis. J Neurol Sci 2016;363:225–31. 10.1016/j.jns.2016.02.044. [DOI] [PubMed] [Google Scholar]
- [19].Bergsland N, Lagana MM, Tavazzi E, Caffini M, Tortorella P, Baglio F, et al. Corticospinal tract integrity is related to primary motor cortex thinning in relapsing-remitting multiple sclerosis. Mult Scler 2015;21(14):1771–80. 10.1177/1352458515576985. [DOI] [PubMed] [Google Scholar]
- [20].Lu MK, Chen CM, Duann JR, Ziemann U, Chen JC, Chiou SM, et al. Investigation of motor cortical plasticity and Corticospinal tract diffusion tensor imaging in patients with Parkinson’s disease and essential tremor. PLoS One 2016;11(9):e0162265 10.1371/journal.pone.0162265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mahlknecht P, Akram H, Georgiev D, Tripoliti E, Candelario J, Zacharia A, et al. Pyramidal tract activation due to subthalamic deep brain stimulation in Parkinson’s disease. Mov Disord 2017;32(8):1174–82. 10.1002/mds.27042. [DOI] [PubMed] [Google Scholar]
- [22].Kuczynski AM, Dukelow SP, Hodge JA, Carlson HL, Lebel C, Semrau JA, et al. Corticospinal tract diffusion properties and robotic visually guided reaching in children with hemiparetic cerebral palsy. Hum Brain Mapp 2017. 10.1002/hbm.23904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gupta D, Barachant A, Gordon AM, Ferre C, Kuo HC, Carmel JB, et al. Effect of sensory and motor connectivity on hand function in pediatric hemiplegia. Ann Neurol 2017;82(5):766–80. 10.1002/ana.25080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Clark CA, Barrick TR, Murphy MM, Bell BA. White matter fiber tracking in patients with space-occupying lesions of the brain: a new technique for neurosurgical planning? NeuroImage 2003;20(3):1601–8. [DOI] [PubMed] [Google Scholar]
- [25].Mikuni N, Okada T, Nishida N, Taki J, Enatsu R, Ikeda A, et al. Comparison between motor evoked potential recording and fiber tracking for estimating pyramidal tracts near brain tumors. J Neurosurg 2007;106(1):128–33. 10.3171/jns.2007.106.1.128. [DOI] [PubMed] [Google Scholar]
- [26].Reiner A, Veenman CL, Medina L, Jiao Y, Del Mar N, Honig MG. Pathway tracing using biotinylated dextran amines. J Neurosci Methods 2000;103(1):23–37. [DOI] [PubMed] [Google Scholar]
- [27].Miller KL, Stagg CJ, Douaud G, Jbabdi S, Smith SM, Behrens TE, et al. Diffusion imaging of whole, post-mortem human brains on a clinical MRI scanner. NeuroImage 2011;57(1):167–81. Epub 2011/04/09 10.1016/j.neuroimage.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dyrby TB, Baare WF, Alexander DC, Jelsing J, Garde E, Sogaard LV. An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion-weighted imaging datasets. Hum Brain Mapp 2011;32(4):544–63. Epub 2010/10/15 10.1002/hbm.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schilling K, Gao Y, Stepniewska I, Choe AS, Landman BA, Anderson AW. Reproducibility and variation of diffusion measures in the squirrel monkey brain, in vivo and ex vivo. Magn Reson Imaging 2017;35:29–38. 10.1016/j.mri.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang R, Benner T, Sorensen AG, Wedeen VJ, editors. Diffusion toolkit: a software package for diffusion imaging data processing and tractography. International Society for Magnetic Resonance in Medicine (ISMRM); 2007. [Google Scholar]
- [31].Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage 2012;62(2):782–90. 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- [32].Yeh FC, Verstynen TD, Wang Y, Fernandez-Miranda JC, Tseng WY. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS One 2013;8(11):e80713 10.1371/journal.pone.0080713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tournier JD, Calamante F, Connelly A. MRtrix: diffusion tractography in crossing fiber regions. Int J Imaging Syst Technol 2012;22(1):53–66. 10.1002/ima.22005. [DOI] [Google Scholar]
- [34].Basser PJ, Mattiello J, Lebihan D. MR diffusion tensor spectroscopy and imaging. Biophys J 1994;66(1):259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tuch DS. Q-ball imaging. Magn Reson Med 2004;52(6):1358–72. Epub 2004/11/25 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- [36].Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? NeuroImage 2007;34(1):144–55. 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. NeuroImage 2007;35(4):1459–72. 10.1016/j.neuroimage.2007.02.016. [DOI] [PubMed] [Google Scholar]
- [38].Toga AW, Ambach KL, Schluender S. High-resolution anatomy from in situ human brain. NeuroImage 1994;1(4):334–44. Epub 1994/11/01 10.1006/nimg.1994.1018. [DOI] [PubMed] [Google Scholar]
- [39].Choe AS, Gao Y, Li X, Compton KB, Stepniewska I, Anderson AW. Accuracy of image registration between MRI and light microscopy in the ex vivo brain. Magn Reson Imaging 2011;29(5):683–92. Epub 2011/05/07 10.1016/j.mri.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bookstein FL. Principal warps-thin-plate splines and the decomposition of deformations. IEEE Trans Pattern Anal Mach Intell 1989;11(6):567–85. 10.1109/34.24792. [DOI] [Google Scholar]
- [41].Rohde GK, Aldroubi A, Dawant BM. The adaptive bases algorithm for intensity-based nonrigid image registration. IEEE Trans Med Imaging 2003;22(11):1470–9. Epub 2003/11/11 10.1109/tmi.2003.819299. [DOI] [PubMed] [Google Scholar]
- [42].Cote MA, Girard G, Bore A, Garyfallidis E, Houde JC, Descoteaux M. Tractometer: towards validation of tractography pipelines. Med Image Anal 2013;17(7):844–57. 10.1016/j.media.2013.03.009. [DOI] [PubMed] [Google Scholar]
- [43].Maier-Hein KH, Neher PF, Houde JC, Cote MA, Garyfallidis E, Zhong J, et al. The challenge of mapping the human connectome based on diffusion tractography. Nat Commun 2017;8(1):1349 10.1038/s41467-017-01285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gao Y, Parvathaneni P, Schilling K, Zu Z, Choe A, Stepniewska I, et al. A 3D high resolution ex vivo white matter atlas of the common squirrel monkey (Saimiri sciureus) based on diffusion tensor imaging. Proceedings of the SPIE Medical Imaging Conference; February; San Diego, California 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].A brain MRI atlas of the common squirrel monkey, Saimiri sciureus In: Gao Y, Khare SP, Panda S, Choe AS, Stepniewska I, Li X, editors. Proc SPIE Int Soc Opt Eng. 2014. March 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schilling KG, Gao Y, Stepniewska I, Wu TL, Wang F, Landman BA, et al. The VALiDATe29 MRI based multi-channel atlas of the squirrel monkey brain. Neuroinformatics 2017. 10.1007/s12021-017-9334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Frank MJ, Samanta J, Moustafa AA, Sherman SJ. Hold your horses: impulsivity, deep brain stimulation, and medication in Parkinsonism. Science 2007;318(5854):1309–12. 10.1126/science.1146157. [DOI] [PubMed] [Google Scholar]
- [48].Kahn E, D’Haese PF, Dawant B, Allen L, Kao C, Charles PD, et al. Deep brain stimulation in early stage Parkinson’s disease: operative experience from a prospective randomised clinical trial. J Neurol Neurosurg Psychiatry 2012;83(2):164–70. 10.1136/jnnp-2011-300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ferbert A, Priori A, Rothwell JC, Day BL, Colebatch JG, Marsden CD. Interhemispheric inhibition of the human motor cortex. J Physiol 1992;453:525–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Concha L, Gross DW, Wheatley BM, Beaulieu C. Diffusion tensor imaging of time-dependent axonal and myelin degradation after corpus callosotomy in epilepsy patients. NeuroImage 2006;32(3):1090–9. 10.1016/j.neuroimage.2006.04.187. [DOI] [PubMed] [Google Scholar]
- [51].Chen Z, Tie Y, Olubiyi O, Zhang F, Mehrtash A, Rigolo L, et al. Corticospinal tract modeling for neurosurgical planning by tracking through regions of peritumoral edema and crossing fibers using two-tensor unscented Kalman filter tractography. Int J Comput Assist Radiol Surg 2016;11(8):1475–86. 10.1007/s11548-015-1344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jeurissen B, Descoteaux M, Mori S, Leemans A. Diffusion MRI fiber tractography of the brain. NMR Biomed 2017. 10.1002/nbm.3785. [DOI] [PubMed] [Google Scholar]
- [53].Alexander DC, Seunarine KK. Mathematics of crossing fibers In: Jones DK, editor. Diffusion MRI: theory, methods, and application. Oxford. New York: Oxford University Press; 2010. p. 451–64. [Google Scholar]
- [54].Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 1999;45(2):265–9. Epub 1999/02/16. [DOI] [PubMed] [Google Scholar]
- [55].Smith RE, Tournier JD, Calamante F, Connelly A. SIFT: spherical-deconvolution informed filtering of tractograms. NeuroImage 2013;67:298–312. 10.1016/j.neuroimage.2012.11.049. [DOI] [PubMed] [Google Scholar]
- [56].Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A 2014;111(46):16574–9. Epub 2014/11/05 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cottaar M, Bastiani M, Chen C, Dikranian K, Van Essen DC, Behrens TE, editors. Fibers crossing the white/gray matter boundary: a semi-global, histology-informed dMRI model. Singapore: ISMRM Proceedings; 2016. [Google Scholar]
- [58].Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage 2007;36(3):630–44. 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yeh CH, Smith RE, Liang X, Calamante F, Connelly A. Correction for diffusion MRI fibre tracking biases: the consequences for structural connectomic metrics. NeuroImage 2016. 10.1016/j.neuroimage.2016.05.047. [DOI] [PubMed] [Google Scholar]
- [60].Schilling K, Gao Y, Janve V, Stepniewska I, Landman BA, Anderson AW. Confirmation of a gyral bias in diffusion MRI fiber tractography. Hum Brain Mapp 2017. 10.1002/hbm.23936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liptrot MG, Sidaros K, Dyrby TB. Addressing the path-length-dependency confound in white matter tract segmentation. PLoS One 2014;9(5):e96247 10.1371/journal.pone.0096247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Anderson AW. Theoretical analysis of the effects of noise on diffusion tensor imaging. Magn Reson Med 2001;46(6):1174–88. Epub 2001/12/18. [DOI] [PubMed] [Google Scholar]
- [63].Lazar M, Alexander AL. An error analysis of white matter tractography methods: synthetic diffusion tensor field simulations. NeuroImage 2003;20(2):1140–53. [DOI] [PubMed] [Google Scholar]
- [64].Fillard P, Descoteaux M, Goh A, Gouttard S, Jeurissen B, Malcolm J, et al. Quantitative evaluation of 10 tractography algorithms on a realistic diffusion MR phantom. NeuroImage 2011;56(1):220–34. 10.1016/j.neuroimage.2011.01.032. [DOI] [PubMed] [Google Scholar]
- [65].Zemmoura I, Serres B, Andersson F, Barantin L, Tauber C, Filipiak I, et al. FIBRASCAN: a novel method for 3D white matter tract reconstruction in MR space from cadaveric dissection. NeuroImage 2014;103:106–18. 10.1016/j.neuroimage.2014.09.016. [DOI] [PubMed] [Google Scholar]
- [66].Azadbakht H, Parkes LM, Haroon HA, Augath M, Logothetis NK, de Crespigny A, et al. Validation of high-resolution tractography against in vivo tracing in the macaque visual cortex. Cereb Cortex 2015;25(11):4299–309. 10.1093/cercor/bhu326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Knosche TR, Anwander A, Liptrot M, Dyrby TB. Validation of tractography: comparison with manganese tracing. Hum Brain Mapp 2015;36(10):4116–34. 10.1002/hbm.22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dyrby TB, Sogaard LV, Parker GJ, Alexander DC, Lind NM, Baare WF, et al. Validation of in vitro probabilistic tractography. NeuroImage 2007;37(4):1267–77. 10.1016/j.neuroimage.2007.06.022. [DOI] [PubMed] [Google Scholar]
- [69].Reveley C, Seth AK, Pierpaoli C, Silva AC, Yu D, Saunders RC, et al. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion MR tractography. Proc Natl Acad Sci U S A 2015;112(21):E2820–8. 10.1073/pnas.1418198112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Aydogan DB, Jacobs R, Dulawa S, Thompson SL, Francois MC, Toga AW, et al. When tractography meets tracer injections: a systematic study of trends and variation sources of diffusion-based connectivity. Brain Struct Funct 2018;223(6):2841–58. 10.1007/s00429-018-1663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schilling K, Janve V, Gao Y, Stepniewska I, Landman BA, Anderson AW. Comparison of 3D orientation distribution functions measured with confocal microscopy and diffusion MRI. NeuroImage 2016;129:185–97. 10.1016/j.neuroimage.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schilling KG, Janve V, Gao Y, Stepniewska I, Landman BA, Anderson AW. Histological validation of diffusion MRI fiber orientation distributions and dispersion. NeuroImage 2018;165:200–21. 10.1016/j.neuroimage.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.