Abstract
Introduction
Cetuximab, a monoclonal antibody to the epidermal growth factor receptor (EGFR), extends survival in combination with standard therapy in head and neck squamous cell carcinoma (HNSCC). However, as effects are modest, and patients experience side effects, a biomarker to predict resistance and personalize therapy is needed. Activation of signaling pathways downstream from receptor tyrosine kinases predicts resistance to such therapies in other cancers. The most common abnormalities downstream from EGFR in HNSCC are in the PI3K pathway, activated via loss of expression of the regulator PTEN, or via PI3K mutation. We studied whether PTEN and/or PI3K abnormalities predict resistance to cetuximab.
Methods
Tumor PTEN and PIK3CA/PI3K p110α were analyzed in samples from subjects treated on two trials of cetuximab-based therapy for patients with metastatic or recurrent HNSCC: E5397, a randomized trial of cisplatin plus placebo versus cisplatin plus cetuximab; and NCI-8070, a randomized trial of cetuximab plus sorafenib versus cetuximab. In situ quantification of PTEN and PI3K p110 α was performed using the AQUA™ method of quantitative immunofluorescence. PI3KCA hot spot mutations were determined with BEAMing.
Results
For E5397, in multivariable analysis, PTEN expressing/PIK3CA WT patients tended to improve PFS with cetuximab compared to placebo (N = 48; HR = 0.54, Wald p = 0.0502). High PTEN expression was significantly associated with superior PFS among patients treated on NCI-8070 (N = 37; HR = 0.35, p = 0.008).
Conclusion
Loss of PTEN expression may be associated with lack of benefit from cetuximab. This analysis is limited by small sample size, and PTEN as a potential predictive biomarker merits validation in larger sample sets.
Keywords: Head and neck cancer, Cetuximab, PTEN, Biomarkers
Introduction
The epidermal growth factor receptor (EGFR) is overexpressed in head and neck squamous cell carcinoma (HNSCC); such overexpression is associated with advanced disease and reduced survival [1,2]. Dysregulation of EGFR signaling has been shown to stimulate tumor cell proliferation, inhibit apoptosis, and promote angiogenesis and metastatic spread [3]. Cetuximab, a chimeric immunoglobulin G1 monoclonal antibody that competitively inhibits ligand binding to the extra-cellular domain of the EGFR receptor, increases response and survival in patients with recurrent and/or metastatic HNSCC when used in combination with platinum-based chemotherapy [4–6]. In combination with radiotherapy, cetuximab also prolongs survival in patients with locally advanced HNSCC, compared to radiotherapy alone [7]. The clinical effects are modest, the agent is costly, and patients experience a variety of side effects. Efficacy has also been demonstrated for tyrosine kinase inhibitors which target either EGFR alone, or which have activity across the HER family: these agents have however not been demonstrated to improve overall survival in unselected populations, and are also associated with toxicity [8–11]. Thus, a biomarker which could predict de novo resistance would be useful in personalizing therapy.
Although no biomarker has been demonstrated to predict resistance to EGFR inhibition in HNSCC, successful strategies to identify sources of resistance to receptor tyrosine kinase (RTK) inhibition in other solid tumors have focused on activation of the signal transduction pathways through which the target RTKs exert their effects. Cetuximab, in addition to its activity in HNSCC, is effective in metastatic colorectal cancer [12]. In colorectal cancer, mutation in the downstream effectors of EGFR signaling, K-ras and B-raf, has been shown to predict cetuximab resistance [13–15]. Testing of these genomic markers is now standard of care in metastatic colorectal cancer, and the 58% of colorectal cancer patients who bear these mutations are not treated with cetuximab or the EGFR-directed monoclonal antibody, panitumumab [16]. Activation of signaling streams downstream to targetable RTKs has been shown to predict resistance in breast cancer as well [17]. Ras mutations are uncommon in HNSCC and translation of these findings to head and neck cancer has lagged [18,19]. Abnormalities in EGFR signaling which are predicted to activate the PI3K/Akt/mTOR pathway have been recognized, and are dominated by PTEN loss of expression/function and by activating mutations in PIK3CA [20]. Phosphorylated Akt content is inversely correlated with nuclear PTEN expression and approximately a third of HNSCC exhibit loss of PTEN expression [21,22]. We reasoned that a signature which reflected aberrancies in PTEN and PIK3CA expected to result in PI3K/Akt/mTOR pathway activation would predict resistance to cetuximab, and wished to test this in tumor samples from patients treated with or without cetuximab.
Methods
We analyzed tumor PTEN and PIK3CA/PI3K p110α in samples from subjects treated on two trials of cetuximab-based therapy for patients with metastatic/recurrent HNSCC.
Study population
The first cohort was drawn from patients enrolled on E5397, a cooperative group, phase III randomized trial of cisplatin plus placebo versus cisplatin plus cetuximab in recurrent or metastatic HNSCC; the results have been previously reported [12]. E5397 enrolled 117 eligible and evaluable patients from June 1999 to June 2001, of whom 57 were randomized to the cetuximab-containing arm. Seventy patients in total received cetuximab after 13 eligible placebo patients received cetuximab on progression. Quantitative immunohistochemistry was used to analyze PTEN expression on specimens from 67 patients from this trial. Additionally, 3 PIK3CA mutations (E542K and E545K in exon 9 and H1047R in exon 20) were determined by BEAMing (Inostics, Heidelberg, Germany) on 52 patients in the E5397 cohort.
The second cohort was drawn from NCI-8070, a phase II randomized trial of cetuximab plus sorafenib versus single agent cetuximab in R/M HNSCC carried out at the Moffitt Cancer Center [23]. Fifty-six patients were enrolled between July 2009 and October 2011, 52 of whom received cetuximab, all at the standard dosing schedule of 400 mg/m2 IV on day 1 followed by 250 mg/m2 IV weekly. Expression of PTEN and of the p110α catalytic subunit of PI3Kwere analyzed with quantitative immunohistochemistry for 37 of these patients, on tumor samples obtained, with Institutional Review Board approval, from the Moffitt Cancer Center biosample repository.
Quantitative determination of immunofluorescence for PTEN expression in the E5397 cohort
PTEN expression was determined by automated quantitative analysis (AQUA) on the PM-2000 (HistoRx, New Haven) as described previously [24]. The PTEN antibody was selected from Cell Signaling Technology (CST) based on published comparison of five optimized antibodies for PTEN detection [25]. Additionally, all but CST exhibited non-specific nuclear staining in negative controls. We derived a cut-point generated in five HNSCC tissue microarrays, consisting of 371 HNSCC as well as 10 positive (small intestine, median AQUA score 2833.2) and 10 negative (breast and colon carcinoma, median AQUA score 205.5) controls. A cut-point of 570 provided 100% specificity, 100% sensitivity, and identified 30% of the HNSCCs as PTEN null, consonant with the literature. Slides were deparaffinized and blocked for endogenous peroxidases; antigen retrieval was performed using EDTA buffer at pH8. All potential non-specific binding sites were blocked using bovine serum albumin. Slides were then incubated overnight with primary antibodies against PTEN (1:100, CST, Cat. 9559) and cytokeratin in Da Vinci Green (Biocare Medical, PD900). Counter-staining was performed with a secondary antibody (Alexa Fluor 555 anti-mouse and Cy5 anti-rabbit), after which slides were cover-slipped with nuclear visualization media (ProLong Gold Antifade with DAPI-Invitrogen, Cat. P36931). Automated image capture was performed with the HistoRx PM-2000 epifluorescent microscope using the AQUAsition software package.
Quantitative determination of immunofluorescence for PTEN expression in the NCI-8070 cohort
PTEN expression was determined using fluorescent IHC and automated quantitative analysis (AQUA) as described above. The concentration of primary antibody was 1:200 (CST, Cat. 9559). Thirty-eight slides from 37 patients were analyzed with two control slides. The AQUA score cut-point was 1177, representing the boundary between the lowest and middle tertiles, consistent with our prior cut-point, to permit translation to the wider dynamic range of the next-generation PM2000 microscopy platform.
Quantitative immunofluorescent determination of PI3 kinase p110α expression in the NCI8070 cohort
PI3 kinase p110α expression was determined by AQUA in 37 analyzable patients, and in a cell line microarray. The slides were incubated with primary antibodies against PI3 kinase p110α (1:100, CST, Cat. 4249) and cytokeratin-diluted in Da Vinci Green (Biocare Medical, PD900) over 24 h. Secondary antibody staining was performed and 2 control slides were stained alongside slides. Slides were cover-slipped with nuclear visualization media, which was ProLong Gold Antifade with DAPI (Invitrogen, Cat. P36931). Automated image capture was performed with the HistoRx PM-2000 using the AQUAsition software package. Images were validated for the presence of tumor (cores showing < 5% tumor area per field were excluded), before scoring and analysis. An AQUA cut-point of 882 was chosen, reflecting the median among the 37 patients on the NCI-8070 trial.
Identification of PIK3CA mutation in the ECOG 5397 cohort by BEAMing
BEAMing (Beads, Emulsification, Amplification, and Magnetics) is a novel reliable technique for identifying mutant DNA [25]. Three hotspot PIK3CA mutations (E542K and E545K in exon 9 and H1047R in exon 20) were determined by BEAMing (Inostics, Heidelberg, Germany) on fifty-four patients in the E5397 cohort.
PIK3CA mutant cell line microarray construction
To generate HNSCC-specific controls, we analyzed PI3K expression in HNSCC cell lines of known PIK3CA mutation status. Cell lines were grown to confluency to reach > 2 × 108 total cell number, then fixed in 10% neutral buffer formalin overnight, and washed repeatedly in PBS and 80% ethanol to remove excess formalin. Cells were centrifuged to form a pellet, embedded in 2% low melting agarose, and stored in 70% ethanol for at least 24 h before being embedded in paraffin for long term storage. 0.6 mm cores from each cell line pellet were placed on the recipient microarray block using a Tissue Microarrayer (Beecher Instrument, Silver Spring, MD) with two-fold redundancy. HSC-2 (purchased from JCRB Cell Bank), SCC61 (a gift from Dr. Ralph Weichselbaum at the University of Chicago) and SCC90 (a gift from Dr. Wendell Yarbrough at Yale Cancer Center) were pelleted, formalin-fixed and paraffin-embedded. The PIK3CA mutational status of these cell lines is as follows: Wild-type: UPCI SCC090 and SCC25; Mutant (substitution missense): SCC61 (E542K), HSC2 (H1047R), and SQ9G (E545Q).
Statistical analyses
For both studies, the primary endpoint was progression-free survival (PFS) and the secondary endpoint was overall survival (OS). PFS is defined as the time between study entry date and date of progression or death from any cause, censored at date of last known alive. OS is defined as the time between study entry date and date of death, censored at date of last known alive. Descriptive statistics were used to characterize patient characteristics and marker expression. Patients with marker data available were dichotomized into low and high expression groups, with cut-points as described. The Wilcoxon rank sum test and the Fisher’s exact test were used to make comparisons between groups when appropriate. Event-time distributions were estimated by Kaplan-Meier estimates. Cox proportional hazards (PH) models were used to estimate hazard ratios (HR) and test for significance for OS and PFS, adjusting for potential covariates when appropriate. All p-values are two-sided. A level of p < 0.05 is considered statistically significant.
Results
PTEN and PIK3CA analysis in ECOG 5397 cohort
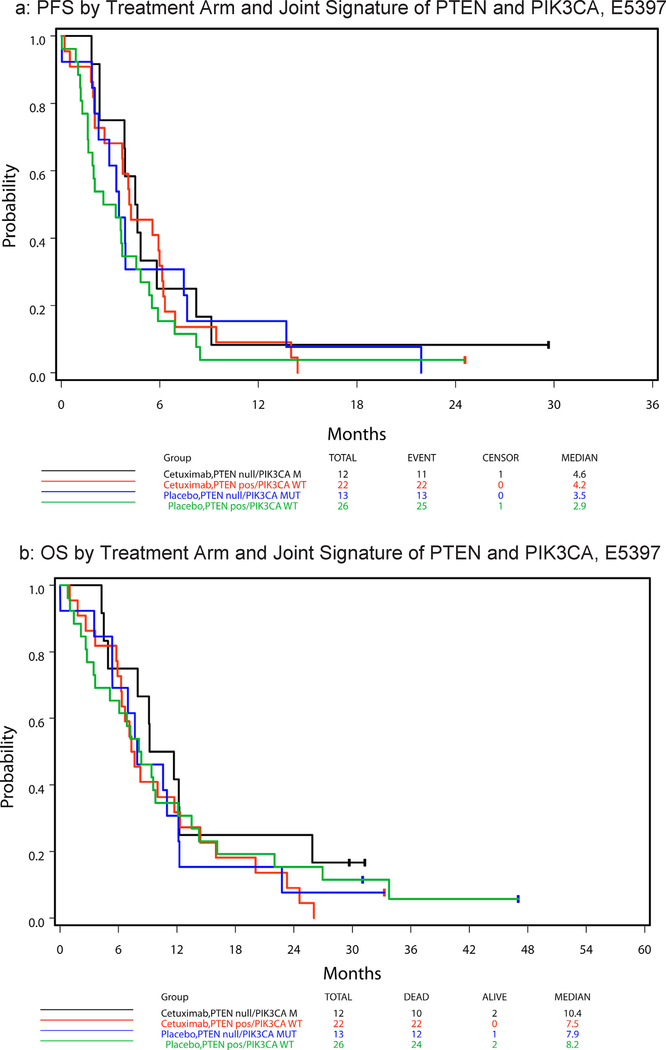
Twenty-three of sixty-seven (34%) tumors were PTEN null and two of fifty-two analyzable tumors (4%) had a PIK3CA mutation at hotspot E542K or E545K (Table 1). Neither tumor with PIK3CA mutation had lost PTEN expression. Table 2 summarizes hazard ratios and median PFS and OS by PTEN status, with respect to treatment arm and across arms. No statistically significant difference in PFS or OS was observed in these samples between patients with PTEN expression and those with PTEN null tumors, either with treatment arms analyzed separately or combined (given no statistically significant 2-way interaction effect of treatment and PTEN status on PFS (p = 0.98) and OS (p = 0.36)). Fig. 2a and b display, respectively, the PFS and OS curves by treatment arm and the joint signature of PTEN and PIK3CA disruption (i.e., PTEN null/PIK3CA mutated and PTEN expressing/PIK3CA WT). There was no statistically significant treatment effect noted in univariate analysis on PFS or OS for either joint gene abnormality in the E5397 cohort. However, in multivariable analysis, PTEN expressing/PIK3CA WT patients had a strong tendency to improve PFS with cetuximab compared to placebo (HR = 0.54 (95% CI 0.29–1.00), Wald p = 0.0502), an effect not observed in the PTEN null/PIK3CA mutant group (Table 3).
Table 1.
PTEN/PIK3CA status by treatment arm, E5397 (among patients with specimens available for marker analysis).
| Treatment Arm |
Total | |||||
|---|---|---|---|---|---|---|
| Cetuximab |
Placebo |
|||||
| N | % | N | % | N | % | |
| PIK3CA* | ||||||
| Mutant | 1 | 4 | 1 | 4 | 2 | 4 |
| Wild-type | 25 | 96 | 25 | 96 | 50 | 96 |
| Unknown | 1 | – | 1 | – | 2 | – |
| PTEN+ | ||||||
| Null | 11 | 35 | 12 | 33 | 23 | 34 |
| Positive | 20 | 65 | 24 | 67 | 44 | 66 |
| PTEN/PIK3CA& | ||||||
| PTEN null/PIK3CA mut | 12 | 35 | 13 | 33 | 25 | 34 |
| PTEN pos/PIK3CA WT | 22 | 65 | 26 | 67 | 48 | 66 |
Fifty four specimens analyzed for PIK3CA.
Sixty-seven specimens analyzed for PTEN.
Seventy-four specimens in total across PIK3Ca and PTEN analysis.
Table 2.
Hazard ratios and median PFS and OS by PTEN status and treatment arm, E5397.
| Event-Time | Arm | PTEN Status* | # of Events/N | Median (Months) | HR (pos/null) (95% CI) | Wald p + |
|---|---|---|---|---|---|---|
| PFS | Cetuximab | Positive | 20/20 | 5.1 (2.6, 6.2) | 1.07 (0.50, 2.30) | 0.86 |
| Null | 10/11 | 4.5 (2.3, 8.2) | ||||
| Placebo | Positive | 23/24 | 3.5 (1.6, 4.8) | 1.12 (0.55, 2.27) | 0.75 | |
| Null | 12/12 | 3.4 (1.9, 7.5) | ||||
| Overall | Positive | 43/44 | 4.1 (2.6, 5.6) | 1.14 (0.68, 1.91) | 0.63 | |
| Null | 22/23 | 3.9 (2.3, 4.8) | ||||
| OS | Cetuximab | Positive | 20/20 | 8.8 (5.9, 14.4) | 1.52 (0.68, 3.41) | 0.31 |
| Null | 9/11 | 9.2 (4.5, 25.9) | ||||
| Placebo | Positive | 22/24 | 8.2 (3.4, 12.2) | 0.91 (0.44, 1.88) | 0.79 | |
| Null | 11/12 | 7.8 (3.5, 12.3) | ||||
| Overall | Positive | 42/44 | 8.2 (6.3, 12.2) | 1.11 (0.65, 1.90) | 0.69 | |
| Null | 20/23 | 9.1 (5.4, 11.7) |
The PTEN status was dichotomized at 570 based on the PTEN AQUA epithelial score.
Univariate Cox PH analysis.
Fig. 2.
E5397 survival by treatment arm and signature.
Table 3.
Survival in CDDP + cetuximab vs. CDDP + placebo in PTEN null/PIK3CA mutant vs. other, E5397.
| PTEN/PIK3CA Status | Time-to-Event Endpoint | Cox Regression Analysis* | # of Events/N | HR (95% CI) (C225 vs Placebo) | Wald P |
|---|---|---|---|---|---|
| PTEN null/PIK3CA mut | PFS | Univariate | 24/25 | 0.76 (0.34–1.70) | 0.50 |
| Multivariable | 23/24 | 0.76 (0.32–1.83) | 0.54 | ||
| OS | Univariate | 22/25 | 0.69 (0.30, 1.60) | 0.39 | |
| Multivariable | 22/24 | 0.39 (0.12–1.26) | 0.11 | ||
| PTEN pos/PIK3CA WT | PFS | Univariate | 47/48 | 0.70 (0.39–1.25) | 0.23 |
| Multivariable | 44/44 | 0.54 (0.29–1.00) | 0.05 | ||
| OS | Univariate | 46/48 | 1.21 (0.67–2.19) | 0.53 | |
| Multivariable | 41/43 | 0.91 (0.45–1.83) | 0.79 |
For PFS, cell differentiation (well/moderately differentiated vs. poorly differentiated) was fitted into the multivariable model. For OS, ECOG PS (0 vs. 1), weight loss in previous 6 months (< 5% vs. ≥5%), and age (< 65 vs. ≥65) were fitted into the multivariable model.
PTEN and PI3K analysis in NC1–8070 cohort
In view of the small sample size and the low yield of PIK3CA mutations in our analysis of E5397, we examined markers of PI3K pathway activation in a second group of patients with metastatic/recurrent HNSCC treated with cetuximab with or without sorafenib. Clinical data were provided for 52 patients on NCI-8070. PTEN and PI3 p110α analysis by AQUA™ were performed in 37 patients who had tumor samples available for testing. Table 4 lists patient characteristics at study entry for all 52 patients and for those with results for PTEN and PI3K expression (n = 37). The two cohorts are similar, with the exception of a statistically significant difference in ethnicity (p = 0.04). Table 5 summarizes median and range statistics for PTEN and PI3K p110α expression, by arm and overall. There is no significant difference in the distribution of the PTEN expression (p = 0.07) or PI3K p110α expression (p = 0.39) between the two arms. No association was observed between PTEN status and PI3K p110α status (PTEN high expression 11/18 vs. 14/19 for PI3K p110α low and high expression, respectively, p = 0.50).
Table 4.
Patient characteristics by PTEN data availability (n = 52), NCI-8070.
| PTEN Data Availability |
Total (n = 52) | P* | |||||
|---|---|---|---|---|---|---|---|
| No (n = 15) |
Yes (n = 37) |
||||||
| N | % | N | % | N | % | ||
| Age (Median, Range) | 59 [26, 74] | 59 [40, 73] | 59 [26, 74] | 0.89 | |||
| Sex | |||||||
| Male | 13 | 87 | 32 | 86 | 45 | 87 | 1.00 |
| Female | 2 | 13 | 5 | 14 | 7 | 13 | |
| Race | |||||||
| White | 13 | 100 | 33 | 92 | 46 | 94 | 1.00 |
| Black | 0 | 0 | 1 | 3 | 1 | 2 | |
| Other | 0 | 0 | 2 | 5 | 2 | 4 | |
| Unknown | 2 | – | 1 | – | 3 | – | |
| Ethnicity | |||||||
| Non-Hispanic | 10 | 71 | 35 | 95 | 45 | 88 | 0.04 |
| Hispanic or Latino | 4 | 29 | 2 | 5 | 6 | 12 | |
| Unknown | 1 | – | 0 | – | 1 | – | |
Wilcoxon rank sum test and Fisher’s exact test when appropriate.
Table 5.
Descriptive statistics of marker expression, NCI-8070.
| Marker | Treatment Arm | N | Minimum | 1st Tertile | Median | 2nd Tertile | Maximum |
|---|---|---|---|---|---|---|---|
| PTEN | Cetuximab | 16 | 481.0 | 1568.4 | 2237.5 | 2857.4 | 5307.1 |
| Cetuximab + Sorafenib | 21 | 441.7 | 851.8 | 1293.0 | 2091.5 | 6263.3 | |
| Total | 37 | 441.7 | 1177.3 | 1568.4 | 2385.9 | 6263.3 | |
| PI3 p110 | Cetuximab | 16 | 311.4 | 882.1 | 986.5 | 1206.1 | 3101.4 |
| Cetuximab + Sorafenib | 21 | 370.6 | 790.2 | 853.6 | 1049.3 | 2777.1 | |
| Total | 37 | 311.4 | 801.0 | 882.1 | 1142.8 | 3101.4 |
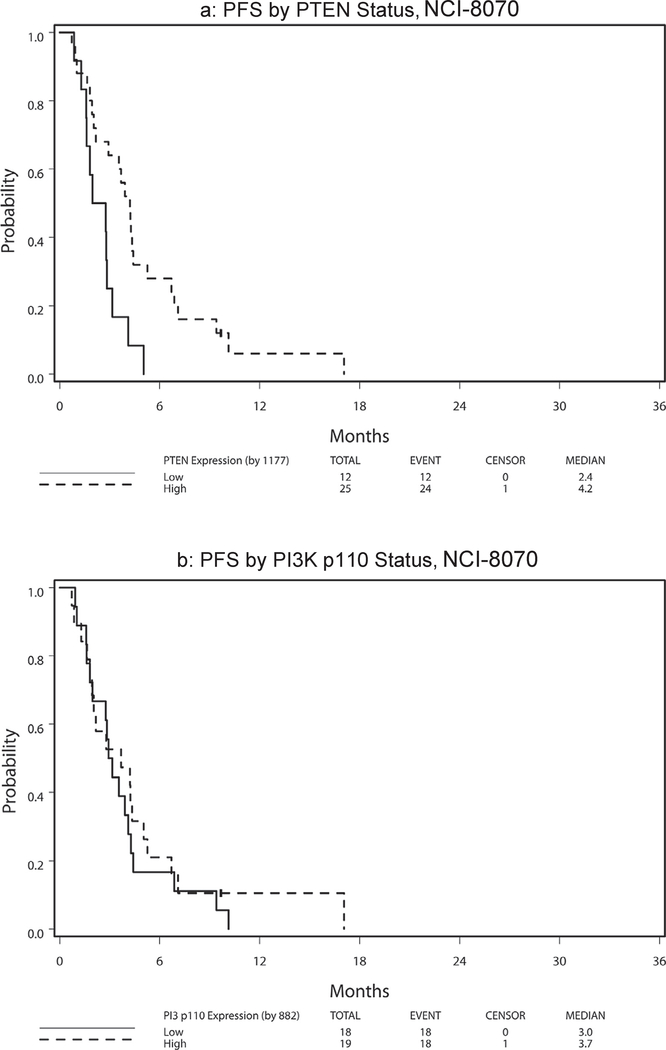
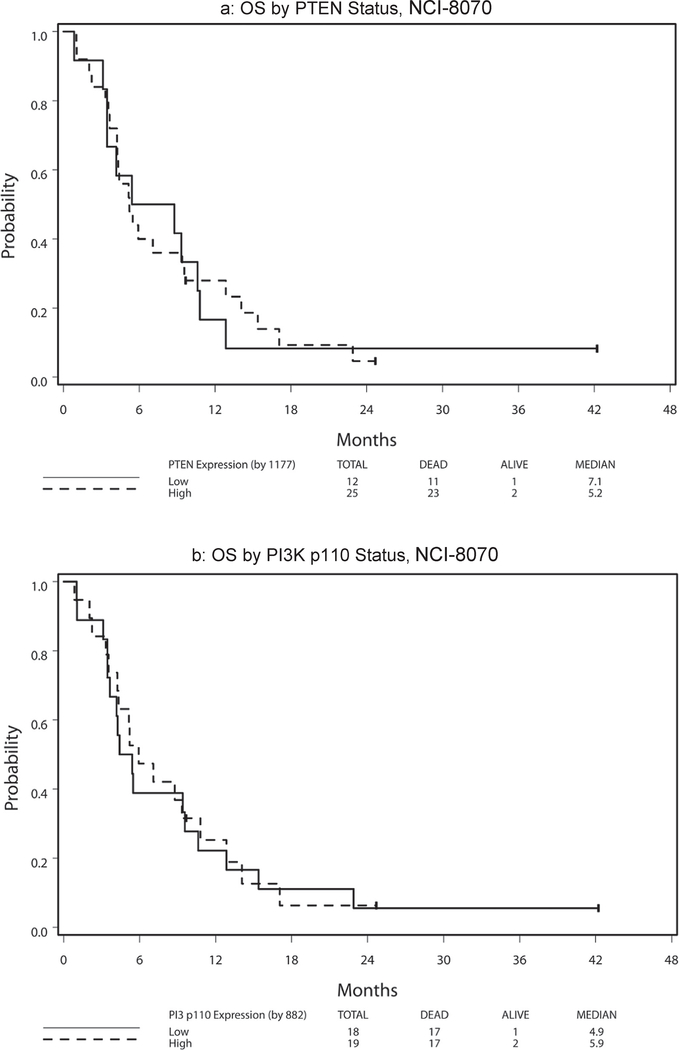
Table 6 presents hazard ratio and median PFS and OS by marker status (high vs. low) with respect to treatment arm and across arms. Because no significant 2-way interaction effect of treatment and marker status and no main treatment effect were noted (all p > 0.11), the following results were analyzed and reported with treatment arms combined. High PTEN expression was significantly associated with superior PFS among patients treated on NCI-8070 (HR = 0.35, p = 0.008; Fig. 3a). This finding remained significant after adjusting for age (< 65 vs. ≥65), sex, race (White vs. Other), and ethnicity (non-Hispanic vs. Hispanic/Latino) (HR = 0.32, 95% CI = (0.14, 0.74), P = 0.008). No PTEN effect was noted for OS (HR = 0.96, p = 0.91; Fig. 4a). The PI3K p110α expression levels revealed no effect on PFS [HR(high/low) = 0.80, p = 0.50; Fig. 3b] or OS [HR(high/low) = 0.94, p = 0.85; Fig. 4b]. There was no statistically significant 2-way interaction effect between PTEN and PI3K p110α on OS (p = 0.96) or on PFS (p = 0.88) (see Fig. 1).
Table 6.
Hazard ratios and median OS and PFS by PTEN and PI3 p110 status, NCI-8070.
| Marker | Event-Time | Arm | PTEN Status1 | # of Events/N | Median (Months) | HR (High vs. Low) (95% CI) | Wald P& |
|---|---|---|---|---|---|---|---|
| PTEN* | PFS | Cetuximab | High | 13/13 | 3.7 (1.9, 6.7) | 0.49 (0.13, 1.85) | 0.29 |
| Low | 3/3 | 2.8 (1.3, 5.0) | |||||
| Cetuximab + Sorafenib | High | 11/12 | 4.2 (0.9, 6.9) | 0.24 (0.08, 0.75) | 0.01 | ||
| Low | 9/9 | 2.0 (0.9, 3.2) | |||||
| Overall | High | 24/25 | 4.2 (2.2, 5.3) | 0.35 (0.16, 0.76) | 0.008 | ||
| Low | 12/12 | 2.4 (1.3, 3.2) | |||||
| OS | Cetuximab | High | 12/13 | 5.5 (3.3, 12.9) | 0.88 (0.24, 3.19) | 0.84 | |
| Low | 3/3 | 8.8 (3.5, 12.9) | |||||
| Cetuximab + Sorafenib | High | 11/12 | 4.8 (1.1, 15.4) | 1.07 (0.43, 2.67) | 0.89 | ||
| Low | 8/9 | 5.4 (0.9, 10.8) | |||||
| Overall | High | 23/25 | 5.2 (4.2, 9.6) | 0.96 (0.47, 1.98) | 0.91 | ||
| Low | 11/12 | 7.1 (3.1, 10.8) | |||||
| PI3 p110+ | PFS | Cetuximab | High | 11/11 | 3.7 (1.6, 5.3) | 2.22 (0.61, 8.20) | 0.23 |
| Low | 5/5 | 3.6 (1.8, 10.1) | |||||
| Cetuximab + Sorafenib | High | 7/8 | 3.5 (0.7, 17.1) | 0.60 (0.22, 1.59) | 0.30 | ||
| Low | 13/13 | 2.9 (1.6, 4.1) | |||||
| Overall | High | 18/19 | 3.7 (1.8, 5.0) | 0.80 (0.41, 1.55) | 0.50 | ||
| Low | 18/18 | 3.0 (1.8, 4.1) | |||||
| OS | Cetuximab | High | 10/11 | 5.2 (2.3, 12.9) | 1.29 (0.44, 3.80) | 0.65 | |
| Low | 5/5 | 0.94 (3.5, 22.9) | |||||
| Cetuximab + Sorafenib | High | 7/8 | 7.6 (0.9, 17.1) | 0.70 (0.27, 1.79) | 0.46 | ||
| Low | 12/13 | 4.3 (3.1, 9.6) | |||||
| Overall | High | 17/19 | 5.9 (3.6, 10.8) | 0.94 (0.48, 1.84) | 0.85 | ||
| Low | 17/18 | 4.9 (3.5, 9.6) |
PTEN low defined as PTEN expression in the first vs. second and third tertile.
PI3 p110 low defined as PI3 p110 expression ≤882 and high as > 882, which is the median based on the 37 patients on the Moffitt trial.
Univariate Cox PH analysis.
Fig. 3.
NCI-8070 survival by PTEN status.
Fig. 4.
Survival by PTEN and PI3K status, NCI-8070.
Fig. 1.
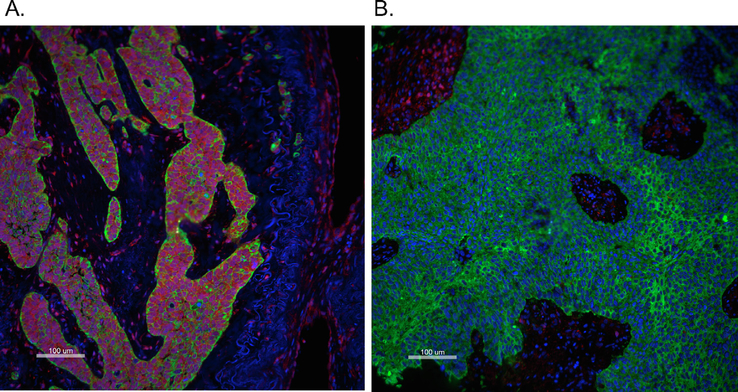
Representative immunofluorescent staining images of PTEN (Red), cytokeratin (Green) and DAPI (blue) showing a specimen with high (A: PTEN in Tumor Mask Normal AQUA score = 9603.22) and low PTEN expression (B: PTEN in Tumor Mask Normal AQUA score = 405.25) based on a cutoff AQUA score = 570. Scale bar: 100 μm (large images). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
E5397 survival by treatment arm and signature.
Discussion
Previous studies have failed to identify EGFR-based biomarkers for cetuximab activity in HNSCC [6,18,26–29]. The work presented here demonstrates for the first time that loss of PTEN expression in HNSCC is associated with absence of benefit from cetuximab. Our initial analysis of PTEN expression in cetuximab-treated or -untreated patients suggested improved outcome in patients with preserved PTEN expression and no mutation of PIK3CA, with an adjusted HR for progression of 0.54 (p = 0.0502). As interpretation of the data from the ECOG trial was limited by sample size and as well as potentially by the age of the tissue samples, we pursued analysis of PTEN expression as a marker of cetuximab resistance or sensitivity in a larger cohort of patients treated with cetuximab (drawn from a multi-institutional trial of cetuximab with or without sorafenib in which sorafenib appeared to have no activity). We analyzed PTEN in the tumor samples and correlated this with PFS on cetuximab therapy; this analysis demonstrated that patients with PTEN high-expressing tumors had an improved PFS in response to cetuximab therapy compared to patients with PTEN low-expressing tumors.
PTEN loss is a relatively common abnormality downstream of EGFR, described in up to 30% of HNSCC, and may be associated with activation of the EGFR pathway in HNSCC [21,30,31]. In a study of HPV-associated oropharynx cancer, PTEN loss (assessed by FISH) was identified in seven out of twenty-one (33%) cases [32]. Chiosea et al. analyzed DNA samples obtained from 252 formalin-fixed paraffin-embedded (FFPE) HNSCC tumor samples using next-generation sequencing. They demonstrated PTEN genomic alterations (PTEN mutation or loss) in 15% of HPV-mediated and 5% of HPV-negative tumors [33]. In another study, protein expression of PTEN, p53, PI3K3, Akt and mTOR (all evaluated by IHC) were investigated according to HPV status (evaluated by ISH) in 65 tonsillar tumors [34]. This study revealed that preserved PTEN (nuclear and cytoplasmic) expression was more frequently observed in HPV-mediated compared to HPV-negative tonsillar SCC cases (P = 0.037), with predominant PTEN distribution in the nucleus. Overall, PTEN expression was lost in 47% of tumors. PTEN was absent in 27% of HPV-associated compared to 57% of HPV-negative tumors. The study also showed a significant correlation between nuclear PTEN expression and disease-free survival (P = 0.27) [34]. Notably, in preclinical models of breast, prostate and non-small cell lung cancer, PTEN loss has been shown to be associated with cetuximab resistance [35].
Genetic abnormalities of the PI3K pathway are also relatively common in HNSCC [35]. It was shown in protein expression correlative studies from the E2303 trial of cetuximab-based induction and chemo-radiotherapy in locally advanced HNSCC that PI3K/AKT pathway activation is associated with inferior PFS and OS and may predict resistance to EGFR-targeted therapy [36]. Previous data suggested PIK3CA mutations in approximately 8% of HNSCC samples [18], but more recent data from TCGA study identified PIK3CA mutations in 21% of HNSCC samples, with 73% of the PIK3CA mutations localized to hotspots that promote activation [20].
The BEAMing methodology we employed to ascertain PIK3CA mutation status was centered on a small number of hotspot mutations which are more frequently detected among HPV-associated HNSCC. This type of HNSCC may have constituted only a small proportion of metastatic/recurrent HNSCCs during the E5397 accrual period E5397, explaining the low frequency with which we detected PIK3CA mutations. We also studied expression of the p110a catalytic subunit of PI3K in an exploratory analysis in this cohort, and this was unrevealing. Although our limited hotspot PIK3CA mutational and PI3K p110a expression analysis did not confirm a role for interrogation of PI3K as part of a cetuximab resistance signature, our sample sizes were small and our methods did not encompass all PIK3CA abnormalities. Our data do not provide definitive evidence that PIK3CA abnormalities do not predict cetuximab resistance; rather, in light of the known prevalence of PIK3CA mutations in HNSCC, further examination of PIK3CA mutation status is merited.
Combination strategies targeting the PI3K pathway can be a potential therapeutic option to overcome resistance and enhance the efficacy of EGFR inhibition in HNSCC. Trials of the EGFR kinase inhibitors dacomitinib and afatinib demonstrate shorter PFS when PTEN is lost [37,38]. RAS mutations are rare in HNSCC. A recent study analyzed the activating RAS mutations in tumors from cetuximab-naive HNSCC patients by NGS and compared this with liquid biopsies taken during and after cetuximab/platinum/5-fluorouracil treatment [39].
The majority of cetuximab-naïve tumors did not carry RAS mutations, with the exception of 4.3% with HRAS mutations. Liquid biopsies revealed acquired KRAS, NRAS or HRAS mutations in more than one-third of patients after cetuximab exposure. Almost half of patients with on-treatment disease progression showed acquired RAS mutations, while no RAS mutations were found in the non-progressive subset of patients, indicating that acquisition of RAS mutant clones correlated significantly with clinical resistance to EGFR inhibition.
Cetuximab is an IgG1 monoclonal antibody, and its potential role in immune modulation has been intensively studied. Cetuximab has been shown to augment dendritic cell (DC)-mediated phagocytosis of colon carcinoma cells [40]. Cetuximab-induced DC maturation, likely mediated by effects on NK cells, influences tumor antigen presentation [41]. Most recently, broad effects of cetuximab on T cell repertoire, as measured by T cell receptor sequence diversity, have been demonstrated by Kansy, et al. and correlate with therapeutic response [42]. Cetuximab effects on immune cells are likely independent of direct signaling mediated effects on tumor cells, and the relative contributions of these mechanisms likely differ across patients and across the duration of therapy. Investigators are currently exploring the combination of cetuximab with immune checkpoint inhibition [NCT01860430], and further characterization of resistance arising from abnormalities in the signaling pathway will be necessary to fully interpret such studies.
Since constitutive activation of the EGFR signaling stream appears to lead to resistance across tumor types and across activating mutations, it is unsurprising that this principle is now borne out in HNSCC. We believe that our findings justify analysis of PTEN loss in larger cetuximab-based randomized trials. Further development of a highly reproducible quantitative assay to appropriately identify this population is needed. We recommend that in future studies with cetuximab and possibly other EGFR inhibitors, PTEN be considered as a predictive biomarker. Validation of our findings would support stratification by PTEN status in future trials of EGFR or HER family inhibitors in HNSCC.
Acknowledgements
This study was coordinated by the ECOG-ACRIN Cancer Research Group (Peter J. O’Dwyer, MD and Mitchell D. Schnall, MD, PhD, Group Co-Chairs) and supported by the National Cancer Institute of the National Institutes of Health under the following award numbers: CA180820, CA180794, CA180826, CA180847, CA180802. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. NCI-8070 was supported by consortium N01-CM-62208 to Moffit Cancer Center.
Footnotes
Conflict of interest
N Eze, MD: None.
J-W Lee, PhD: None.
D-H Yang, PhD: None.
F Zhu, PhD: None.
V Neumeister, PhD:
T Sandoval-Schaefer, PhD: None.
R Mehra, MD: None.
J A Ridge, MD, PhD: None.
A Forastiere, MD: None.
C H Chung, MD: Research funding, Lilly; Honoraria, Bristol Myers Squibb and Ignyta.
B Burtness, MD: Research funding: Bristol Myers Squibb and Imclone Systems; Honoraria: Bristol Myers Squibb and Imclone Systems.
References
- [1].Dassonville O, Formento JL, Francoual M, Ramaioli A, Santini J, Schneider M, et al. Expression of epidermal growth factor receptor and survival in upper aerodigestive tract cancer. J Clin Oncol 1993;11:1873–8. [DOI] [PubMed] [Google Scholar]
- [2].Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res 2002;62:7350–6. [PubMed] [Google Scholar]
- [3].Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res 2003;284:31–53. [DOI] [PubMed] [Google Scholar]
- [4].Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med 2008;359:1116–27. [DOI] [PubMed] [Google Scholar]
- [5].Goldstein NI, Prewett M, Zuklys K, Rockwell P, Mendelsohn J. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res 1995;1:1311–8. [PubMed] [Google Scholar]
- [6].Burtness B, Goldwasser MA, Flood W, Mattar B, Forastiere AA. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. J Clin Oncol 2005;23:8646–54. [DOI] [PubMed] [Google Scholar]
- [7].Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med 2006;354:567–78. [DOI] [PubMed] [Google Scholar]
- [8].Machiels JP, Haddad RI, Fayette J, Licitra LF, Tahara M, Vermorken JB, et al. Afatinib versus methotrexate as second-line treatment in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck progressing on or after platinum-based therapy (LUX-Head & Neck 1): an open-label, randomised phase 3 trial. Lancet Oncol 2015;16:583–94. [DOI] [PubMed] [Google Scholar]
- [9].Martins RG, Parvathaneni U, Bauman JE, Sharma AK, Raez LE, Papagikos MA, et al. Cisplatin and radiotherapy with or without erlotinib in locally advanced squamous cell carcinoma of the head and neck: a randomized phase II trial. J Clin Oncol 2013;31:1415–21. [DOI] [PubMed] [Google Scholar]
- [10].Stewart JS, Cohen EE, Licitra L, Van Herpen CM, Khorprasert C, Soulieres D, et al. Phase III study of gefitinib compared with intravenous methotrexate for recurrent squamous cell carcinoma of the head and neck [corrected]. J Clin Oncol 2009;27:1864–71. [DOI] [PubMed] [Google Scholar]
- [11].Argiris A, Ghebremichael M, Gilbert J, Lee JW, Sachidanandam K, Kolesar JM, et al. Phase III randomized, placebo-controlled trial of docetaxel with or without gefitinib in recurrent or metastatic head and neck cancer: an eastern cooperative oncology group trial. J Clin Oncol 2013;31:1405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337–45. [DOI] [PubMed] [Google Scholar]
- [13].Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol 2015;26:13–21. [DOI] [PubMed] [Google Scholar]
- [14].Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer 2015;51:587–94. [DOI] [PubMed] [Google Scholar]
- [15].Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol 2015;33:692–700. [DOI] [PubMed] [Google Scholar]
- [16].Guren TK, Thomsen M, Kure EH, Sorbye H, Glimelius B, Pfeiffer P, et al. Cetuximab in treatment of metastatic colorectal cancer: final survival analyses and extended RAS data from the NORDIC-VII study. Br J Cancer 2017;116:1271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sanchez V, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci U S A 2013;110:14372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011;333:1154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Comprehensive genomic characterization of head and neck squamous cell carcinomas. Cancer Genome Atlas Network. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Squarize CH, Castilho RM, Abrahao AC, Molinolo A, Lingen MW, Gutkind JS. PTEN deficiency contributes to the development and progression of head and neck cancer. Neoplasia 2013;15:461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee JI, Soria JC, Hassan KA, El-Naggar AK, Tang X, Liu DD, et al. Loss of PTEN expression as a prognostic marker for tongue cancer. Arch Otolaryngol Head Neck Surg 2001;127:1441–5. [DOI] [PubMed] [Google Scholar]
- [23].Gilbert J, Schell MJ, Zhao X, Murphy B, Tanvetyanon T, Leon ME, et al. A randomized phase II efficacy and correlative studies of cetuximab with or without sorafenib in recurrent and/or metastatic head and neck squamous cell carcinoma. Oral Oncol 2015;51:376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 2002;8:1323–7. [DOI] [PubMed] [Google Scholar]
- [25].Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res 2007;67:11111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol 2015;12:11–26. [DOI] [PubMed] [Google Scholar]
- [27].Vermorken JB, Psyrri A, Mesia R, Peyrade F, Beier F, de Blas B, et al. Impact of tumor HPV status on outcome in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck receiving chemotherapy with or without cetuximab: retrospective analysis of the phase III EXTREME trial. Ann Oncol 2014;25:801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rosenthal DI, Harari PM, Giralt J, Bell D, Raben D, Liu J, et al. Association of human papillomavirus and p16 status with outcomes in the IMCL-9815 phase III registration trial for patients with locoregionally advanced oropharyngeal squamous cell carcinoma of the head and neck treated with radiotherapy with or without cetuximab. J Clin Oncol 2016;34:1300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Licitra L, Mesia R, Rivera F, Remenar E, Hitt R, Erfan J, et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann Oncol 2011;22:1078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 2009;9:550–62. [DOI] [PubMed] [Google Scholar]
- [31].Shao X, Tandon R, Samara G, Kanki H, Yano H, Close LG, et al. Mutational analysis of the PTEN gene in head and neck squamous cell carcinoma. Int J Cancer 1998;77:684–8. [DOI] [PubMed] [Google Scholar]
- [32].Sangale Z, Prass C, Carlson A, Tikishvili E, Degrado J, Lanchbury J, et al. A robust immunohistochemical assay for detecting PTEN expression in human tumors. Applied Immunohistochem Mol Morphol: AIMM 2011;19:173–83. [DOI] [PubMed] [Google Scholar]
- [33].Chiosea SI, Grandis JR, Lui VW, Diergaarde B, Maxwell JH, Ferris RL, et al. PIK3CA, HRAS and PTEN in human papillomavirus positive oropharyngeal squamous cell carcinoma. BMC Cancer 2013;13:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, Richmon J, et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann Oncol 2015;26:1216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Holsinger FC, Piha-Paul SA, Janku F, Hong DS, Atkins JT, Tsimberidou AM, et al. Biomarker-directed therapy of squamous carcinomas of the head and neck: targeting PI3K/PTEN/mTOR pathway. J Clin Oncol 2013;31:e137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Psyrri A, Lee JW, Pectasides E, Vassilakopoulou M, Kosmidis EK, Burtness BA, et al. Prognostic biomarkers in phase II trial of cetuximab-containing induction and chemoradiation in resectable HNSCC: eastern cooperative oncology group E2303. Clin Cancer Res 2014;20:3023–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hoon CS, Chan-Kwon J, Sung WH, Jin-Hyoung K, Yeon-Sil K, Min-Sik K. Divergence of P53, PTEN, PI3K, Akt and mTOR expression in tonsillar cancer. Head Neck 2015;37:636–43. [DOI] [PubMed] [Google Scholar]
- [38].Kim HS, Kwon HJ, Jung I, Yun MR, Ahn M-J, Kang BW, et al. Phase II clinical and exploratory biomarker study of dacomitinib in patients with recurrent and/or metastatic squamous cell carcinoma of head and neck. Clin Cancer Res 2015;21:544–52. [DOI] [PubMed] [Google Scholar]
- [39].Cohen EEW, Licitra LF, Burtness B, Fayette J, Gauler T, Clement PM, et al. Biomarkers predict enhanced clinical outcomes with afatinib versus methotrexate in patients with second-line recurrent and/or metastatic head and neck cancer. Ann Oncol 2017;28:2526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Correale P, Botta C, Cusi MG, Del Vecchio MT, De Santi MM, Gori Savellini G, et al. Cetuximab ± chemotherapy enhances dendritic cell-mediated phagocytosis of colon cancer cells and ignites a highly efficient colon cancer antigen-specific cytotoxic T-cell response in vitro. Int J Cancer 2012;130:1577–89. [DOI] [PubMed] [Google Scholar]
- [41].Srivastava RM, Trivedi S, Concha-Benavente F, Gibson SP, Reeder C, Ferrone S, et al. CD137 stimulation enhances cetuximab-induced natural killer: dendritic cell priming of antitumor T-cell immunity in patients with head and neck cancer. Clin Cancer Res 2017;23:707–16. 10.1158/1078-0432.CCR-16-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kansy BA, Shayan G, Jie HB, Gibson SP, Lei YL, Brandau S, et al. T cell receptor richness in peripheral blood increases after cetuximab therapy and correlates with therapeutic response. Oncoimmunology 2018;2018(7):e1494112 10.1080/2162402X.2018.1494112.eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]