Abstract
We describe a sibling pair displaying an early infantile-onset, progressive neurodegenerative phenotype, with symptoms of developmental delay and epileptic encephalopathy developing from 12 to 14 months of age. Using whole exome sequencing, compound heterozygous variants were identified in SLC5A6, which encodes the sodium-dependent multivitamin transporter (SMVT) protein. SMVT is an important transporter of the B-group vitamins biotin, pantothenate, and lipoate. The protein is ubiquitously expressed and has major roles in vitamin uptake in the digestive system, as well as transport of these vitamins across the blood–brain barrier. Pathogenicity of the identified variants was demonstrated by impaired biotin uptake of mutant SMVT. Identification of this vitamin transporter as the genetic basis of this disorder guided targeted therapeutic intervention, resulting clinically in improvement of the patient’s neurocognitive and neuromotor function. This is the second report of biallelic mutations in SLC5A6 leading to a neurodegenerative disorder due to impaired biotin, pantothenate and lipoate uptake. The genetic and phenotypic overlap of these cases confirms mutations in SLC5A6 as the genetic cause of this disease phenotype. Recognition of the genetic disorder caused by SLC5A6 mutations is essential for early diagnosis and to facilitate timely intervention by triple vitamin (biotin, pantothenate, and lipoate) replacement therapy.
Subject terms: Medical genomics, Neurodevelopmental disorders, Personalized medicine, Translational research, Metabolic disorders
Introduction
While individually rare, inborn errors of metabolism are over-represented amongst the childhood neurodegenerative diseases, with an increasing number of novel disorders identified.1 These disorders are caused by perturbed transport or processing of essential metabolites and have a profound impact on nervous system development and function. In the majority of cases, this effect is compounded by a lack of effective therapy. However, contemporary elucidation of the molecular abnormalities underpinning these disorders is informing therapeutic development, with early intervention translating to favourable clinical responses in a growing number of cases.2
Highlighting the importance of recognising novel, ultra-rare disorders with the potential for personalised therapeutic intervention is the recently described disorder of sodium-dependent multivitamin transport (SMVT).3,4 The SMVT protein (encoded by SLC5A6) is ubiquitously expressed and plays a major role in the uptake of biotin, pantothenate and lipoate in the digestive system, as well as in transporting these B-group vitamins across the blood–brain barrier. This report showed that pathogenic mutations in SLC5A6 resulted in aberrant cellular uptake of these critical organic cofactors, manifesting clinically in a single patient as a multisystem disease with infantile neurocognitive regression.4
Here we describe a second, non-consanguineous sibling pair, presenting in early infancy with a progressive neurodegenerative phenotype. Whole exome sequencing (WES) revealed compound heterozygous mutations in SLC5A6, with pathogenicity of the identified variants demonstrated by impaired biotin uptake of mutant SMVT. Targeted vitamin replacement therapy was initiated resulting in sustained clinical improvement, providing further evidentiary support for disease causality and expanding the recognised spectrum of inborn diseases of biotin metabolism.4–8
Results
Family presentation
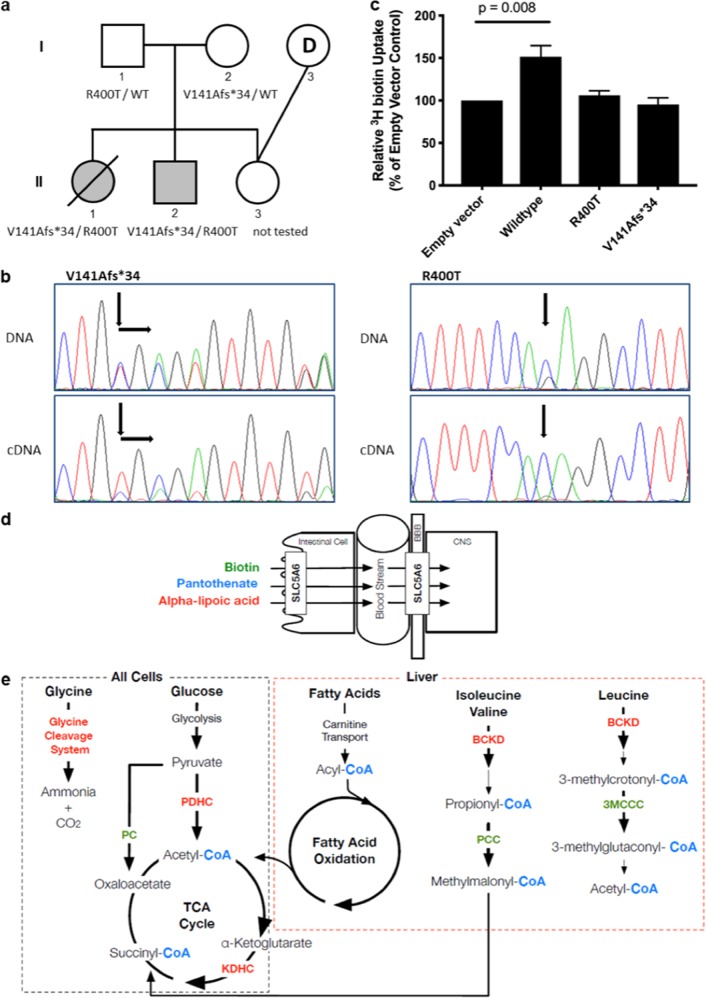
Two siblings (II-1 and II-2) presenting with a profound neurodevelopmental delay during infancy were born to unaffected, unrelated parents (I-1 and I-2). Before a molecular diagnosis was determined for II-1 and II-2, the parents chose an ovum donor (I-3) to reduce the risk of recurrence in their third child (II-3) (Fig. 1a).
Fig. 1.
Biotin, pantothenate and lipoate-dependent metabolic pathways and the effect of identified variants in SLC5A6. a Pedigree of the non-consanguineous family. b Chromatograms from Sanger sequencing of patient DNA compared to cDNA show decreased expression of the V141Afs*34 allele and stable expression of the R400T allele. c Uptake of radiolabelled biotin by HeLa cells transfected with empty vector, wild-type or mutant SMVT expression constructs. Uptake by mutant constructs is decreased compared to wild-type (p = 0.008) and not significantly different to empty vector (p > 0.05). Data show the mean and standard error of the mean (n = 4). d SCL5A6 function and e Enzymes for which the vitamins Biotin (green), Pantothenate (blue) and Alpha-lipoic acid (red) play a role as important cofactors in: the degradation pathways of the amino acids leucine, isoleucine, valine and glycine; glucose energy metabolism; the TCA cycle; and fatty acid oxidation metabolism. All pathways apart from the glycine cleavage system play a fundamental role in cellular energy production. The pathways involved with fatty acid metabolism and branch chain amino acid breakdown occur almost exclusively in the liver. BBB blood–brain barrier, BCKD Branch chain ketoacid dehydrogenase, CNS central nervous system, KDHC ketoglutarate dehydrogenase complex, PC pyruvate carboxylase, PCC propionyl-CoA carboxylase, PDHC pyruvate dehdrogenase complex, 3MCCC 3-methyl crotonyl-CoA carboxylase
Detailed clinical descriptions of both affected siblings can be found in the supplementary information and are summarised in Table 1. In brief, both siblings were born at term without complication following a benign antenatal period. Birth occipitofrontal head circumference (OFC) was at the 50th centile with 34 cm (II-1) and 34.5 cm (II-2), followed by postnatal reduction (2nd–10th centile) in both children. Early developmental progress was age appropriate with a plateau in global development from 14 months (II-1) and 12 months (II-2). II-1 walked independently (15 months), however regressed to crawling (20 months) and non-ambulation (24 months). Her vocabulary of 15–20 words (15 months) retracted to 10 words (23 months), only using ‘mum’ and ‘dad’ by 30 months. II-2 commenced few independent steps at 12 months and used a walking assist device until 5 years and 6 months when he regressed to crawling. His vocabulary of 10 single words by 15 months regressed to non-verbal by 24 months.
Table 1.
Clinical features and outcome following treatment of the three patients with biallelic SLC5A6 mutations causing SMVT deficiency
| Clinical features | This study—patient II-1 | This study—patient II-2 | Subramanian 2017 |
|---|---|---|---|
| Neurological | |||
| Neurocognitive regression | Onset: 14 months |
Onset: 12 months Post treatment: Neurocognitive progressa -walking with assistance -4–6 word vocabulary -improved social interaction/attention -utilising cup and spoon |
Onset: infantile Post treatment: Neurocognitive progressb -walking with assistance -5-word vocabulary -improved social interaction/personality |
| Microcephaly | Yes, relativec | Yes, relativec | Yes |
| Neuro-ophthalmological | Left esotropia |
Nystagmus Dyskinetic saccades Binocular esotropia Post treatment: Nystagmus marked reduction |
Nystagmus Post treatment: Nystagmus resolved |
| Spasticity | No | No | Yes |
| Hyperreflexia | Yes | Yesd | NR |
| Seizures | No |
Yes (well controlled) Post treatment: No significant change Reduced anticonvulsant requirement |
NR |
| Hyperacusis | Yes | Yes (resolved pre-treatment) | NR |
| Peripheral neuropathy | NR |
Mixed demyelinating & axonal sensorimotor polyneuropathy Post treatment: Electrographically resolvede |
NR |
| Neuroimaging (MRI)f |
No cerebral atrophy Right cerebellar haemorrhagic foci T2/FLAIR signal hyperintensity (periventricular & parieto-occipital white matter) |
Cerebral atrophy (progressive) Cerebellar atrophy (progressive) Brainstem (pontine) atrophy Thin corpus callosum T2/FLAIR signal hyperintensity (central segmental tract & peritrigonal regions) Mega cisterna magna 1H-MRS (SVS) [31 and 144 ms; basal ganglia/frontoparietal white matter]: reduced NAA and broad lactate doublets |
Cerebral atrophy Brainstem (pontine) atrophy Thin corpus callosum |
| Electroencephalogram (EEG)g | Not done |
Background slowing (encephalopathy) Epileptiform activity: generalised and multifocal spike-wave (2–3 Hz) Post treatment: Improved background rhythmsh Epileptiform activity reduced |
Normal |
| Histopathology |
Central nervous system: axonal spheroids Peripheral nervous system: undefined thickening Skeletal muscle biopsy: denervation atrophy |
Cutaneous biopsy: membranous cytoplasmic inclusionsi | Skeletal muscle biopsy: normal |
| Gastrointestinal | |||
|
Feeding difficulties/ failure to thrive |
Yes, bulbar dysfunction | Yes, bulbar dysfunction | Yes |
|
Nasogastric tube/ gastrostomy feedingj |
Yes | Yes | Yes |
| GI haemorrhage | Yesk | Yesl | Yesm |
| Other | GORD |
Cyclical vomiting GORD Post treatment: cyclical vomiting improved |
NR |
| Cardiopulmonary | |||
|
Asthma Right heart failuren |
Asthma tracheobronchomalacia ECG: non-specific ST & T-wave changeso |
NR | |
| Other | |||
| Hypogammaglobulinemia | NR |
Yes, Isolated IgG deficiency Post treatment: NR |
Yes, IgG/IgA deficiency Post treatment: Resolved |
| Osteopenia | No | No |
Yes Post treatment: Resolved |
| Digital clubbing | NR | Yesp | NR |
| Treatment | |||
| Nil |
Biotin (intramuscular) 10 mg weekly Dexpanthenol (intramuscular) 250 mg weekly α-lipoic acid (intravenous) 300 mg weekly |
Biotin (oral) 10–30 mg/day Pantothenic acid (oral) 250–500 mg/day α-Lipoic acid (oral) 150–300 mg/day |
|
GORD gastro-oesophageal reflux disease, NR not reported
aReported at +5 years from commencement of therapy
bReported at +2 years, 9 months from commencement of therapy
cRelative postnatal microcephaly (OFC growth retardation)
dInitial hyperreflexia; progressive obtundation of myotatic reflexes presumed secondary to peripheral neuropathy
eConfirmed by serial nerve conduction studies pre/post treatment
fMost recent pre-treatment study [age]: II-1 [1 year, 12 months]; Supplementary Fig. 2A, II-2 [7 years, 5 months]; Supplementary Fig. 2B and C, Subramanian [12 months]
gMost recent pre-treatment study [age]: II-2 [5 years, 5 months], Subramanian [6 months]
hPost-treatment EEG [+4 years, 8 months]
iSupplementary Fig. 3B
jInsertion: II-1 (nasogastric tube [2 years, 7 months]; nil gastrostomy), II-2 (gastrostomy [6 years, 3 months]), Subramanian (nasogastric tube [infancy]; gastrostomy [6 months])
kPerforated duodenal ulcer/duodenal artery
lSuspected Mallory–Weiss tear in context of cyclical vomiting
mReported secondary to thrombocytopenia
nFeatures of right heart failure at autopsy, presumed secondary to pre-mortem cardiopulmonary resuscitation
oNo clinical cardiomegaly and normal screening echocardiogram [8 years, 9 months]; non-specific ST segment and T wave changes present on ECG
pDigital clubbing of uncertain aetiology (note: normal high-resolution chest CT)
Neurological assessment demonstrated progressive truncal ataxia with dyskinetic appendicular movements from 14 months (II-1) and 12 months (II-2). II-2 developed mixed semiology seizures including focal dyscognitive, absence, tonic spasms and generalised convulsive seizures with electrographic features of encephalopathy with generalised and independent multifocal spike-wave discharges (Supplementary Fig. 1A). The seizures proved well controlled on a regimen of levetiracetam, topiramate and vigabatrin. Refractory cyclical vomiting (II-2) was present from 3 years, 5 months of age. As neurophysiological assessment, neuroimaging, biochemical, single-gene testing and mitochondrial genome sequencing did not provide a diagnosis for this family, WES was performed.
At 2 years 7 months, II-1 died secondary to acute gastrointestinal haemorrhage following perforation of a duodenal ulcer. Post-mortem examination revealed a small cerebrum and cerebellum for age (brain weight 977 g), with preservation of overall neural architecture. Cardiomegaly, hepatic congestion and aspiration pneumonia were also observed, likely secondary to pre-mortem cardiopulmonary resuscitation. Histopathological analysis demonstrated a global increase in glial density with reactive gliotic change. Axonal spheroids, up to 100 μm in diameter, were ubiquitously present (Supplementary Fig. 3A). Peripheral nerves demonstrated variable axonal irregularity and patchy denervation atrophy was evident within skeletal muscle.
Treatment outcomes
Upon identification of the pathogenic SLC5A6 mutations, II-2 commenced parenterally administered triple vitamin replacement therapy at 7 years and 1 month of age, with doses of Biotin (10 mg, intramuscular), Dexpanthenol (250 mg, intramuscular) and α-lipoic acid (300 mg, intravenous) given weekly. To date, treatment has improved the overall condition of II-2 (Table 1); he restarted crawling and climbing to a standing position (+1 year, 8 months) and regained use of a walk-assist frame (+3 years, 4 months). He demonstrated improved attention and persistence at tasks (+1 year, 11 months), and from +3 years is more communicative, regaining a limited (4–6 words) vocabulary at +5 years, with improved consistency in following single-step directions. His seizure control remains effective and there has been interval improvement towards normal in background electrographic activity, albeit with persisting epileptiform discharges (Supplementary Fig. 1B). His cyclical vomiting has attenuated in frequency and duration. His nerve conduction studies confirm near total and sustained resolution of peripheral neuropathy from +3 years.
Identification of candidate variants
WES resulted in an average coverage of the exome of 72.21 (II-1) and 88.19 (II-2), with 90.4% (II-1) and 91.8% (II-2) of the targets being covered at least 10× (Supplementary Table 1). Filtering for rare (<1% in population databases), non-synonymous variants resulted in 487 variants for II-1 and 454 variants for II-2. Overlap analysis between the two siblings did not identify variants in OMIM-reported genes known to cause neurodegeneration. Two variants were however identified in SLC5A6, V141Afs*34 (Chr2(GRCh37):g.27,429,780GCA>G; c.422_423delTG), and R400T (Chr2(GRCh37):g.27,426,109C>G; c.1199G>C).
Sanger sequencing of the unaffected parents showed each was a heterozygous carrier of one variant, p.V141Afs*34 (maternal) and p.R400T (paternal) (Fig. 1a), and confirmed the presence, in compound heterozygosity, of both variants in the affected individuals. The unaffected half-sibling was not tested for the paternal allele.
Evaluation of SLC5A6 variants
Qualitative analysis of SLC5A6 wild-type and mutant allele expression was performed by Sanger sequencing of patient cDNA. Expression of the V141Afs*34 truncated allele was decreased compared to wild-type. The R400T allele was stably expressed and was the only allele present at that location in the compound heterozygous, affected children (Fig. 1b).
Biotin uptake assays were initially performed on primary dermal fibroblasts. After growing the cells for 24 h in media supplemented with tritium-labelled biotin, cell lysates were harvested and the amount of radioactivity quantitated by scintillation counting. Whilst decreased biotin uptake was observed in cells derived from the compound heterozygous, affected children relative to their heterozygous, unaffected parents, this difference was not statistically significantly different compared to controls (Supplementary Fig. 4A).
Biotin uptake was then assessed in cultured HeLa cells transiently transfected with expression constructs for wild-type and mutant SLC5A6. Cell lysates were again harvested and quantitated after 24 h of growth in media containing radiolabelled biotin. Expression of wild-type SMVT significantly increased biotin uptake by 1.5-fold over the empty vector negative control (p = 0.008) (Fig. 1c). In contrast, the uptake of biotin by the R400T and V141Afs*34 mutant SMVT proteins was not different from uptake in the empty vector control (Fig. 1c). This result confirmed both variants result in a severe loss of function.
Discussion
Here we provide consolidative evidence implicating disordered SMVT function in an infantile neurodegenerative phenotype. SMVT is a sodium-dependent transporter of biotin, pantothenate and lipoate; organic enzyme co-factors which are integral to fatty acid metabolism and cellular energy production (Fig. 1d, e).9,10
It is notable that prior defined inborn errors of biotin metabolism, holocarboxylase synthetase deficiency (HLCS; MIM#253270) and biotinidase deficiency (BTD; MIM#253260) share phenotypic similarities with the cases described herein. Untreated patients demonstrate progressive cognitive and neuromotor decline, often with prominent seizures, with variable response to biotin replacement therapy.5–7,11 Further clinical overlap is evident amongst inborn mutations affecting pantothenate metabolism, which are implicated as causative of pantothenate kinase-associated neurodegeneration (PKAN/NBIA1, PANK2; MIM#234200) and COASY protein-associated neurodegeneration (CoPAN/NBIA6, COASY; MIM#615643). In addition, neuroaxonal spheroids which were widely distributed throughout the cerebrum of patient II-1 (Supplementary Fig. 3A), are a characteristic neuropathological feature of pantothenate kinase-associated neurodegeneration.12–14 More recently, perturbed lipoic acid biosynthesis has been implicated in the pathological disruption of mitochondrial oxidative metabolism. Features of these disorders are varied but, in many cases, include infantile encephalopathic presentations with severe epilepsy, similar to that reported in the cases of SMVT deficiency here.15–19
While some degree of phenotypic heterogeneity is evident between the described sibling pair and the unrelated index case, the basis of these differences remains uncertain given the small number of cases identified to date.4 It is likely though, that the temporal progression of untreated disease in II-2 has contributed to more severe neurological manifestations. Further delineation of these findings will be informed by identification and longitudinal investigation of additional cases.
Despite the fact that biotin, pantothenate and lipoate feed into numerous biochemical pathways (Fig. 1d, e), biochemical analyses of metabolites in both our patients and the previously reported patient did not detect any abnormalities.4 This is suggestive of a redundancy in B-group vitamin transport within the gastrointestinal tract, in contrast to the critical role of SMVT in transport across the blood–brain barrier, which may explain the neurological features but lack of biochemical abnormalities.20 It is similarly interesting that neuroimaging features and pathological findings specific to the basal ganglia, commonly observed in disorders of biotin, pantothenate and lipoate metabolism, were not evident in our patients or the previously described individual.4
The mutations we identified in SLC5A6, V141Afs*34 (29 alleles, 0.01%) and R400T (2 alleles, 0.0008%), have both been observed at extremely low frequency in the population database gnomAD, but never in homozygosity.21 The V141Afs*34 variant is predicted to result in a truncated protein product, removing many of the functionally important domains (Supplementary Fig. 4B).9,22,23 This truncation was supported by the cDNA experiment (Fig. 1b), which also suggested nonsense-mediated decay by displaying reduced expression of this allele. The R400T variant is located on an intracellular loop of SMVT, close to a transmembrane domain (Supplementary Fig. 4B). Multiple species alignment shows the R400 residue is moderately conserved, retained in 25 of 39 vertebrate species (Supplementary Fig. 4C), with the lysine residue present in other species having similar chemical properties to arginine. The molecular basis of impaired SMVT function due to this missense mutation is unclear.
SMVT deficiency is extremely rare, with only one similar case being reported to date (Table 1).4 In order to identify additional patients, we contacted researchers with cohorts of patients without a genetic diagnosis for suspected mitochondrial and neurological disease, with existing WES data, as well as sharing the genotypic and phenotypic information of patient II-2 on MatchMaker Exchange.24 However, no additional patients with homozygous or compound heterozygous variants in SLC5A6 were identified.
The paucity of clinically affected individuals due to pathogenic variants in SLC5A6 may be explained by the critical role of biotin, pantothenate and lipoate in numerous metabolic pathways. The SMVT protein is solely responsible for intestinal biotin uptake and has been shown to be responsible for 89% of biotin transport across the blood–brain barrier.9,20,25 It is therefore likely that carrying biallelic amorphic alleles, resulting in no functional SMVT protein, is lethal. This is supported by the absence of homozygous loss-of-function variants in gnomAD. Conversely, individuals with two hypomorphic alleles may have sufficient residual SMVT function to remain clinically asymptomatic. This is similar to that observed in other metabolic disorders, such as biotinidase deficiency, where patients carrying two alleles with residual function are clinically well.6 Both the earlier described patient and our patients carry a single amorphic (null) allele in combination with a hypomorphic allele, likely resulting in sufficient transporter activity to support life, but inadequate to meet biological demands after birth.4
Complete SLC5A6 knockout mice have not been reported. Only one-third of conditional (intestine-specific) SLC5A6 knockout mice were viable and display growth retardation, decreased bone density and histological abnormalities of the gastrointestinal tract, overlapping with phenotypic features (e.g. gastrointestinal bleeding and osteopenia) present in the human patients reported to date.4,25 While the mechanisms underlying these varied clinical characteristics remain uncertain, it is notable that the phenotype of the conditional knockout mouse could also be rescued by over-supplementation of biotin and pantothenic acid.26
For patient II-2, triple replacement therapy was initiated upon molecular diagnosis of SMVT deficiency at 7 years of age. The case described in the earlier report commenced treatment at 19 months of age, which prevented further neurodegeneration and significantly improved clinical outcomes in this child.
This is the second report of biallelic SLC5A6 mutations underpinning infantile neurodegenerative disease with subsequent therapeutic response. This result reinforces the importance of critical cellular cofactors in nervous system function and development, and emphasises the potential for significant clinical benefit, at the individual patient level, that can result from access to exploratory molecular analysis. Introduction of neonatal or even preconceptual molecular screening would allow identification of an increasing number of rare and ultra-rare novel disorders, where early intervention is supported by therapeutic benefit. In the case of patients with SLC5A6 mutations, this provides the opportunity to dramatically attenuate or perhaps even ameliorate neurodegeneration when identified early within the disease course.
Methods
Patients
Written informed consent to perform WES was obtained under institutionally approved molecular diagnostic consent protocols (SA Pathology), in accordance with the Declaration of Helsinki. Experimental treatment regime was approved by the Women’s and Children’s Health Network Drug and Therapeutics Committee (30 September 2014), and access obtained following approval under the Australian Government Therapeutic Goods Administration Special Access Scheme.
Genetic studies
WES was performed on genomic DNA samples from II-1 and II-2 at the Centre for Cancer Biology ACRF Genomics Facility. Exonic sequences were enriched using the SeqCap EZ Human Exome Library v3.0 kit (Roche NimbleGen) and libraries sequenced as 100 bp paired-end reads on the HiSeq 2000 platform (Illumina).
Read alignment to the UCSC human genome assembly hg19 was performed with the Burrows–Wheeler Aligner (v.0.6.2). Picard was used to mark duplicates and GATK (v2.7-2) was used for local realignment around indels and to recalibrate quality scores. SNV’s and small insertions and deletions were detected using GATK’s Unified Genotyper (v2.7-2).
To rule out sample identity errors, SNP calls from WES data were compared to calls from the Infinium CytoSNP-850K BeadChip assay (Illumina), performed according to the manufacturer’s protocol at the Centre for Cancer Biology ACRF Genomics Facility on genomic DNA samples.
Variant filtering and prioritisation
WES results were first investigated for variants in OMIM genes known to be causative of neurodegeneration, but no likely pathogenic variants were identified. Results were then filtered to highlight potentially causative variants under three modes of inheritance: autosomal recessive with homozygous or compound heterozygous variants, and autosomal dominant due to parental mosaicism. Variants were only considered if they were present in both affected children, were predicted to cause a functionally important effect (SNPEFF effect moderate or high) and were rare in the general population (ExAC MAF <1% for AR or <0.1% for AD).21 Variants were further prioritised by only considering those predicted to be pathogenic by in-silico programmes (CADD score >20) and in conserved regions (GERP score >2). Variants present in concordant zygosity in our in-house genomic variant database were excluded (Supplementary Table 1).
Candidate variants from WES were confirmed and co-segregation assessed by Sanger sequencing of PCR-amplified genomic DNA from affected and unaffected family members.
Gene expression analysis
Total RNA was extracted from dermal fibroblasts (I-1, I-2, II-1 and II-2) using the RNeasy Mini Kit (Qiagen). Reverse transcription was performed using the SuperScript III Reverse Transcriptase kit (Invitrogen), as per the manufacturer’s instructions, with 150 ng of Random Primers (Promega) and 1 μg of total RNA. RNA complimentary to cDNA was removed using 1 μL of RNase H (Invitrogen). Gene expression was assessed by Sanger sequencing of PCR-amplified cDNA.
Cell culture
HeLa cell cultures were maintained in Dulbecco’s modified Eagle medium (DMEM) with 20 mM HEPES, supplemented with 10% foetal bovine serum (Sigma-Aldrich) and 1% penicillin–streptomycin–glutamine (Gibco). Cells were split every 3–4 days at a ratio of 1:2. HeLa cells were purchased from the American Type Culture Collection (ATCC, Reference CCL-2). Primary dermal fibroblast cultures were established following skin biopsy of I-1, I-2, II-1 and II-2 at the Women’s and Children’s Hospital, Adelaide. Cultured fibroblasts were maintained using basal Eagle’s medium 1 × (BME) (Gibco) with 10% foetal bovine serum, 1% l-glutamine, 200 mM and 1% penicillin–streptomycin (all from Sigma-Aldrich). Cells were split every 7–10 days at a ratio of 1:2.
Generation of expression constructs
Wild-type SLC5A6 expression clones were obtained from OriGene (RC204865). Mutant alleles were created by site-directed mutagenesis, following the manufacturer’s protocol for the Quikchange II XL Site-Directed Mutagenesis Kit (Stratagene). Products were incubated with the DpnIrestriction enzyme to digest the parental (non-mutated) plasmid dsDNA. Wild-type, mutagenized and pCMV6-entry empty vector clones were transformed into XL-10 Gold Ultracompetent Cells (Agilent Technologies) and incubated overnight. Single colonies were grown overnight in liquid culture and clones purified using the QIAfilter Plasmid Midi Kit (QIAGEN). The entire SLC5A6 insert in wild-type and mutant clones was verified by Sanger sequencing in both directions.
Transfection of HeLa cells
HeLa cells were transfected using the Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s protocol. Cells were seeded in 12-well plates (2 × 105 cells) and incubated overnight. The following day, cells were transfected using 0.25 and 1 μg, respectively, of empty-vector, wild-type or mutant expression construct.
Biotin uptake assay
Biotin uptake studies were performed using primary dermal fibroblasts (I-1, I-2, II-1 and II-2), and HeLa cells transfected with empty-vector pCMV6 or constructs expressing wild-type SLC5A6 or mutant protein.27 Dermal fibroblasts were seeded in 12-well plates (2.5 × 105 cells) in basal Eagle’s medium 1 × (BME) (Gibco) with 20% foetal bovine serum (Sigma-Aldrich) and 1% l-glutamine. 24 h after seeding, cells were incubated in Krebs–Ringer buffer (133 mM NaCl, 4.93 mM KCl, 1.23 mM MgSO4, 0.85 mM CaCl2, 5 mM glucose, 5 mM glutamine, 10 mM HEPES and 10 mM MES, pH 7.4) containing 20 nM 3H biotin (American Radiolabeled Chemicals). Biotin uptake was measured after 24 h by washing cells twice with 1 ml of ice-cold Krebs–Ringer buffer then lysed in 0.2 ml of 1 M NaOH. Radioactive content in the cell lysates was quantitated using a scintillation counter with OptiPhase Supermix scintillation cocktail (Perkin Elmer). Protein content in the lysates was measured using a Bradford protein assay kit. Uptake assays in transfected HeLa cells were performed as above except cells were incubated in Krebs–Ringer buffer containing 80 nM 3H biotin for 24 h post-transfection. Each transfection and uptake assay were performed in triplicate, and the results calculated as cpm/mg of lysate. The data from four discrete assays (n = 4) was combined for statistical analysis. A two-tailed Student’s t-test was performed comparing the empty vector and expression constructs.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper.
Supplementary information
Acknowledgements
We would like to thank the patients and their family for their participation in this study. We also thank all staff of the Genetics and Molecular Pathology Research Laboratory, the staff of the Centre for Cancer Biology ACRF Genomics Facility and the Women’s and Children’s Hospital Neurology ward. We would also like to acknowledge the contribution of the following laboratories in reviewing the exome data of their undiagnosed patients; Susan Hayflick (Oregon Health and Science University), Manju Kurian (University College London), Kym Boycott (FORGE Canada) and Fowzan Alkuraya (King Faisal Specialist Hospital and Research Centre). This research was supported by the Australian Genomic Health Alliance NHMRC Targeted Call for Research into Preparing Australia for the Genomics Revolution in Healthcare (GNT1113531), Therapeutic Innovation Australia, the Australian Cancer Research Foundation (to H.S.S.); and NHMRC (APP1068885) (to G.W.B. and S.W.P.). Additional support provided by Cancer Council SA’s Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health, and NHMRC Fellowship (APP1023059) (to H.S.S.); the Australian Government Research Training Programme Scholarship and the Australian Genomics Health Alliance & NHMRC (GNT1113531) (to A.B.B.); and The Hospital Research Foundation Fellowship (to P.A.).
Author contributions
A.B.B., P.A., N.J.S. and D.B. drafted the manuscript. J.F. and A.W.S. processed WES data, and A.B.B. and K.S.K. performed data analysis. A.B.B. and S.W.P. performed functional analyses. A.B.B., C.N.H., K.S.K., S.W.P., P.A., D.A.M., G.W.B., D.B., N.J.S. and H.S.S. contributed to interpretation and discussion of results. J.L. performed surgical pathology investigations. N.J.S., D.B., D.A.M., J.M.F. provided clinical care for the patients with N.J.S. overseeing therapy. H.S.S. and N.J.S. conceived and supervised the study, contributing equally, and should be considered co-senior authors. All authors read and approved the manuscript.
Data availability
Sequence data has been deposited at the European Genome-phenome archive, which is hosted by the European Bioinformatics Institute, under accession #EGAS00001003861. All unique materials and datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nicholas J. Smith, Hamish S. Scott
Contributor Information
Nicholas J. Smith, Email: nicholas.smith@sa.gov.au
Hamish S. Scott, Email: hamish.scott@sa.gov.au
Supplementary information
Supplementary information is available for this paper at 10.1038/s41525-019-0103-x.
References
- 1.Pierre G. Neurodegenerative disorders and metabolic disease. Arch. Dis. Child. 2013;98:618–624. doi: 10.1136/archdischild-2012-302840. [DOI] [PubMed] [Google Scholar]
- 2.Patay Z, Blaser SI, Poretti A, Huisman TA. Neurometabolic diseases of childhood. Pediatr. Radiol. 2015;45(Suppl. 3):S473–S484. doi: 10.1007/s00247-015-3279-y. [DOI] [PubMed] [Google Scholar]
- 3.Richter T, et al. Rare disease terminology and definitions—a systematic global review: report of the ISPOR Rare Disease Special Interest Group. Value Health. 2015;18:906–914. doi: 10.1016/j.jval.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian VS, Constantinescu AR, Benke PJ, Said HM. Mutations in SLC5A6 associated with brain, immune, bone, and intestinal dysfunction in a young child. Hum. Genet. 2017;136:253–261. doi: 10.1007/s00439-016-1751-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgartner, M. R. & Suormala, T. Biotin-responsive Disorders (Springer, Berlin, Heidelberg, 2012).
- 6.Pomponio RJ, Reynolds TR, Cole H, Buck GA, Wolf B. Mutational hotspot in the human biotinidase gene causes profound biotinidase deficiency. Nat. Genet. 1995;11:96–98. doi: 10.1038/ng0995-96. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y, et al. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat. Genet. 1994;8:122–128. doi: 10.1038/ng1094-122. [DOI] [PubMed] [Google Scholar]
- 8.Bailey LM, et al. Reduced half-life of holocarboxylase synthetase from patients with severe multiple carboxylase deficiency. Hum. Mutat. 2008;29:E47–E57. doi: 10.1002/humu.20766. [DOI] [PubMed] [Google Scholar]
- 9.Ghosal A, Said HM. Structure-function activity of the human sodium-dependent multivitamin transporter: role of His115 and His254. Am. J. Physiol. Cell Physiol. 2011;300:C97–C104. doi: 10.1152/ajpcell.00398.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quick M, Shi L. The sodium/multivitamin transporter: a multipotent system with therapeutic implications. Vitam. Horm. 2015;98:63–100. doi: 10.1016/bs.vh.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zempleni J, Hassan YI, Wijeratne SS. Biotin and biotinidase deficiency. Expert Rev. Endocrinol. Metab. 2008;3:715–724. doi: 10.1586/17446651.3.6.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurian MA, Hayflick SJ. Pantothenate kinase-associated neurodegeneration (PKAN) and PLA2G6-associated neurodegeneration (PLAN): review of two major neurodegeneration with brain iron accumulation (NBIA) phenotypes. Int. Rev. Neurobiol. 2013;110:49–71. doi: 10.1016/B978-0-12-410502-7.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruer MC, et al. Novel histopathologic findings in molecularly-confirmed pantothenate kinase-associated neurodegeneration. Brain. 2011;134:947–958. doi: 10.1093/brain/awr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malandrini A, et al. Ultrastructure and immunoreactivity of dystrophic axons indicate a different pathogenesis of Hallervorden–Spatz disease and infantile neuroaxonal dystrophy. Virchows Arch. 1995;427:415–421. doi: 10.1007/BF00199391. [DOI] [PubMed] [Google Scholar]
- 15.Solmonson A, DeBerardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 2018;293:7522–7530. doi: 10.1074/jbc.TM117.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soreze Y, et al. Mutations in human lipoyltransferase gene LIPT1 cause a Leigh disease with secondary deficiency for pyruvate and alpha-ketoglutarate dehydrogenase. Orphanet J. Rare Dis. 2013;8:192. doi: 10.1186/1750-1172-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habarou F, et al. Biallelic mutations in LIPT2 cause a mitochondrial lipoylation defect associated with severe neonatal encephalopathy. Am. J. Hum. Genet. 2017;101:283–290. doi: 10.1016/j.ajhg.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayr JA, Feichtinger RG, Tort F, Ribes A, Sperl W. Lipoic acid biosynthesis defects. J. Inherit. Metab. Dis. 2014;37:553–563. doi: 10.1007/s10545-014-9705-8. [DOI] [PubMed] [Google Scholar]
- 19.Mayr JA, et al. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am. J. Hum. Genet. 2011;89:792–797. doi: 10.1016/j.ajhg.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida Y, et al. Major involvement of Na(+)-dependent multivitamin transporter (SLC5A6/SMVT) in uptake of biotin and pantothenic acid by human brain capillary endothelial cells. J. Neurochem. 2015;134:97–112. doi: 10.1111/jnc.13092. [DOI] [PubMed] [Google Scholar]
- 21.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosal A, Subramanian VS, Said HM. Role of the putative N-glycosylation and PKC-phosphorylation sites of the human sodium-dependent multivitamin transporter (hSMVT) in function and regulation. Biochim. Biophys. Acta. 2011;1808:2073–2080. doi: 10.1016/j.bbamem.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nabokina SM, Subramanian VS, Said HM. Association of PDZ-containing protein PDZD11 with the human sodium-dependent multivitamin transporter. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G561–G567. doi: 10.1152/ajpgi.00530.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philippakis AA, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosal A, Lambrecht N, Subramanya SB, Kapadia R, Said HM. Conditional knockout of the Slc5a6 gene in mouse intestine impairs biotin absorption. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;304:G64–G71. doi: 10.1152/ajpgi.00379.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabui S, et al. Biotin and pantothenic acid oversupplementation to conditional SLC5A6 KO mice prevents the development of intestinal mucosal abnormalities and growth defects. Am. J. Physiol. Cell Physiol. 2018;315:C73–C79. doi: 10.1152/ajpcell.00319.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reidling JC, Lambrecht N, Kassir M, Said HM. Impaired intestinal vitamin B1 (thiamin) uptake in thiamin transporter-2-deficient mice. Gastroenterology. 2010;138:1802–1809. doi: 10.1053/j.gastro.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data has been deposited at the European Genome-phenome archive, which is hosted by the European Bioinformatics Institute, under accession #EGAS00001003861. All unique materials and datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.