Abstract
Poorer liver function is positively associated with diabetes in Mendelian randomization (MR) studies. Observationally, adiposity is associated with poorer liver function. To clarify the etiology, we assessed the association of liver enzymes with adiposity observationally and using two-sample MR for validation. In the “Children of 1997” birth cohort, we used multivariable linear regression to assess the associations of alanine transaminase (ALT) and alkaline phosphatase (ALP) at ~17.5 years with body mass index (BMI) (n = 3,458). Using MR, genetic predictors of ALT, ALP and gamma glutamyltransferase (GGT), were applied to genome-wide association studies of BMI (n = 681,275), waist circumference (WC) (n = 224,459) and waist-hip ratio (WHR) (n = 224,459) to obtain unconfounded estimates. Observationally, ALT was positively associated with BMI (0.10 kg/m2 per IU/L, 95% confidence interval (CI) 0.09 to 0.11). ALP was inversely associated with BMI (−0.018 kg/m2 per IU/L, 95% CI −0.024 to −0.012). Using MR, ALT was inversely associated with BMI (−0.14 standard deviation per 100% change in concentration, 95% CI −0.20 to −0.07), but not WC or WHR. ALP and GGT were unrelated to adiposity. Poorer liver function might not cause adiposity; instead higher ALT might reduce BMI, raising the question as to the role of ALT in body composition.
Subject terms: Type 2 diabetes, Risk factors
Introduction
Observationally, poorer liver function, particularly nonalcoholic fatty liver disease (NAFLD), is associated with higher risk of type 2 diabetes mellitus (T2DM)1, but these studies are difficult to interpret because of the difficulty of distinguishing between correlated measures of liver function and the possibility of confounding by poor health causing both poor liver function and T2DM2. Recently, Mendelian randomization (MR) studies, taking advantage of the random allocation of genetic endowment at conception to obtain un-confounded estimates3, have clarified the role of liver function in T2DM. Specifically, these studies suggest that higher alanine aminotransferase (ALT)4,5 or aspartate aminotransferase (AST)5 rather than other measures of liver function, such as glutamyltransferase (GGT)4–6, could play a role in T2DM, although one small MR study found no association of ALT with T2DM7. Adiposity is also a very well-established cause of T2DM8,9. Whether specifically poor hepatocyte function relates to adiposity and contributes to T2DM, and by what mechanism is not entirely clear, although within an evolutionary biology framework we have previously suggested a mechanism via sex hormones4. Circulating levels of endogenous sex hormones are associated with both adiposity10 and fatty liver11. Observationally, poor liver function is associated with obesity12, but these studies are open to confounding by lifestyle, including diet and physical activity, health status, and socioeconomic position (SEP). As such, whether poor liver function is an additional contributor to the obesity epidemic remains uncertain, as experimental evidence is lacking.
To inform this important public health question as to whether liver function plays a role in obesity, we conducted two complimentary analyses with different assumptions and study designs. Observationally, we examined the association of liver function indicated by ALT and alkaline phosphatase (ALP) with adiposity in young people in a setting (Hong Kong) with little clear socio-economic patterning of obesity, so as to reduce confounding by poor health and SEP using Hong Kong’s “Children of 1997” birth cohort13. We also used an MR study to assess the effects of genetically predicted liver enzymes (ALT, ALP, and GGT)14 on adiposity indices, i.e., body mass index (BMI), waist circumference (WC), and waist-hip ratio (WHR), using the Genetic Investigation of ANthropometric Traits (GIANT) consortium15–17, overall and by sex.
Results
Children of 1997
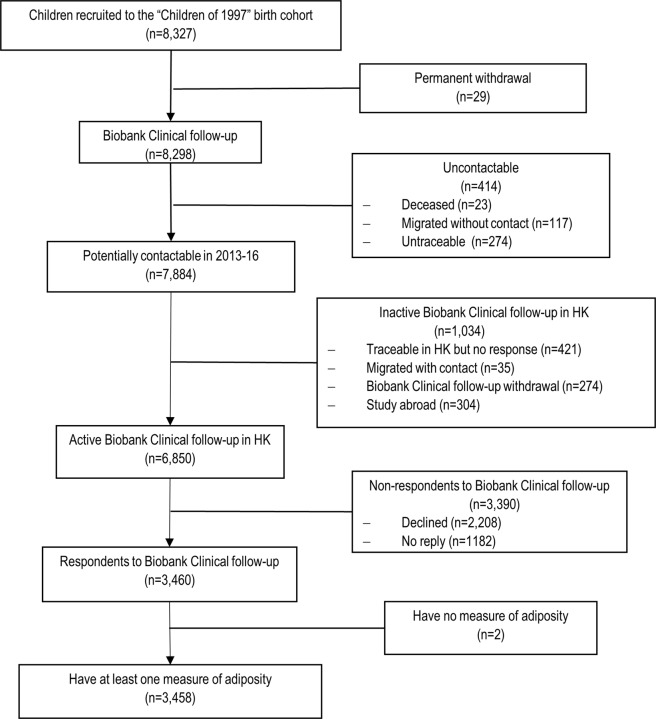
In the “Children of 1997” Biobank Clinical follow-up, 3,460 of 6,850 potentially active follow-up participants took part (51% follow-up). 3,458 had at least one measure of BMI, WC, or WHR, as shown in Fig. 1. The 4,869 participants without adiposity measures were not different from the included participants in terms of sex, second-hand and maternal smoking exposure, and SEP with relatively small Cohen effect sizes (<0.13) (Supplemental Table S1). The mean and standard deviation (SD) of BMI, WC, and WHR were 20.9 kg/m2 (SD 3.5 kg/m2), 72.3 cm (SD 9.2 cm), and 0.77 (SD 0.06). Boys had higher BMI, WC, and WHR than girls. Maternal smoking was associated with larger BMI and WC. SEP had little association with BMI, WC, or WHR (Table 1).
Figure 1.
Flow chart of Hong Kong’s “Children of 1997” birth cohort Biobank Clinical follow-up, Hong Kong, China, 1997 to 2016.
Table 1.
Baseline characteristics by body mass index (BMI), waist circumference (WC), and waist-hip ratio (WHR) among participants in Hong Kong’s “Children of 1997” birth cohort, Hong Kong, China, 1997 to 2016.
| Characteristics | BMI (kg/m2) | WC (cm) | WHR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | Mean (SD) | P-value* | No. | % | Mean (SD) | P-value* | No. | % | Mean (SD) | P-value* | |
| BMI (kg/m2) | 3457 | 20.9 (3.5) | — | — | — | — | — | — | — | |||
| WC (cm) | — | — | — | — | 3453 | — | 72.3 (9.2) | — | — | — | — | — |
| WHR | — | — | — | — | — | — | — | — | 3452 | — | 0.77 (0.06) | — |
| Sex | 3457 | — | — | <0.001 | 3453 | — | — | <0.001 | 3452 | — | — | <0.001 |
| Girl | 1716 | 49.6% | 20.7 (3.3) | — | 1713 | 49.6% | 69.7 (7.8) | — | 1712 | 49.6% | 0.75 (0.05) | — |
| Boy | 1741 | 50.4% | 21.2 (3.8) | — | 1740 | 50.4% | 74.9 (9.7) | — | 1740 | 50.4% | 0.79 (0.07) | — |
| Unknown | 0 | — | — | — | 0 | — | — | — | 0 | — | — | — |
| Second-hand and maternal smoking exposure | 3457 | — | — | <0.001 | 3453 | — | — | <0.01 | 3452 | — | — | 0.05 |
| None | 943 | 27.3% | 20.6 (3.1) | — | 943 | 27.3% | 71.6 (8.3) | — | 942 | 27.3% | 0.77 (0.06) | — |
| Prenatal second-hand smoking | 1280 | 37.0% | 20.9 (3.7) | — | 1278 | 37.0% | 72.3 (9.5) | — | 1278 | 37.0% | 0.77 (0.06) | — |
| Postnatal second-hand smoking | 961 | 27.8% | 21.3 (3.8) | — | 959 | 27.8% | 73.0 (9.8) | — | 959 | 27.8% | 0.77 (0.07) | — |
| Maternal smoking | 128 | 3.7% | 21.5 (3.4) | — | 128 | 3.7% | 73.6 (9.4) | — | 128 | 3.7% | 0.78 (0.06) | — |
| Unknown | 145 | 4.2% | 20.6 (3.1) | — | 145 | 4.2% | 71.0 (7.8) | — | 145 | 4.2% | 0.76 (0.06) | — |
| Highest parental education level | 3457 | — | — | 0.71 | 3453 | — | — | 0.39 | 3452 | — | — | 0.29 |
| Grade < =9 | 991 | 28.7% | 21.0 (3.8) | — | 990 | 28.7% | 72.6 (9.8) | — | 990 | 28.7% | 0.77 (0.07) | — |
| Grades 10–11 | 1489 | 43.1% | 20.9 (3.4) | — | 1486 | 43.0% | 72.2 (8.8) | — | 1485 | 43.0% | 0.77 (0.06) | — |
| Grades > =12 | 961 | 27.8% | 20.9 (3.5) | — | 961 | 27.8% | 72.3 (9.3) | — | 961 | 27.8% | 0.77 (0.06) | — |
| Unknown | 16 | 0.5% | 20.8 (2.0) | — | 16 | 0.5% | 69.2 (5.1) | — | 16 | 0.5% | 0.74 (0.04) | — |
| Highest parental occupation | 3457 | — | — | 0.01 | 3453 | — | — | 0.07 | 3452 | — | — | 0.42 |
| I (unskilled) | 99 | 2.9% | 20.7 (3.1) | — | 99 | 2.9% | 71.6 (8.9) | — | 99 | 2.9% | 0.77 (0.06) | — |
| II (semiskilled) | 285 | 8.2% | 21.3 (3.7) | — | 284 | 8.2% | 72.8 (9.3) | — | 284 | 8.2% | 0.77 (0.07) | — |
| III (semiskilled) | 503 | 14.6% | 20.8 (3.5) | — | 503 | 14.6% | 72.2 (9.4) | — | 503 | 14.6% | 0.77 (0.07) | — |
| III (nonmanual skilled) | 879 | 25.4% | 21.0 (3.7) | — | 878 | 25.4% | 72.5 (9.4) | — | 877 | 25.4% | 0.77 (0.06) | — |
| IV (managerial) | 440 | 12.7% | 21.4 (3.8) | — | 440 | 12.7% | 73.3 (10.2) | — | 440 | 12.7% | 0.77 (0.07) | — |
| V (professional) | 797 | 23.1% | 20.7 (3.3) | — | 797 | 23.1% | 71.8 (8.7) | — | 797 | 23.1% | 0.77 (0.06) | — |
| Unknown | 454 | 13.1% | 20.7 (3.4) | — | 452 | 13.1% | 71.6 (8.3) | — | 452 | 13.1% | 0.77 (0.06) | — |
| Household income per head at recruitment | 3457 | — | — | 0.12 | 3453 | — | — | 0.14 | 3452 | — | — | 0.45 |
| First quintile | 572 | 16.5% | 20.8 (3.5) | — | 571 | 16.5% | 71.9 (9.0) | — | 571 | 16.5% | 0.77 (0.07) | — |
| Second quintile | 616 | 17.8% | 21.0 (3.7) | — | 614 | 17.8% | 72.4 (9.9) | — | 614 | 17.8% | 0.77 (0.07) | — |
| Third quintile | 618 | 17.9% | 21.2 (3.7) | — | 617 | 17.9% | 73.2 (9.6) | — | 617 | 17.9% | 0.77 (0.06) | — |
| Fourth quintile | 631 | 18.3% | 20.8 (3.4) | — | 631 | 18.3% | 72.1 (8.8) | — | 631 | 18.3% | 0.77 (0.06) | — |
| Fifth quintile | 646 | 18.7% | 20.8 (3.2) | — | 646 | 18.7% | 72.0 (8.6) | — | 645 | 18.7% | 0.77 (0.07) | — |
| Unknown | 374 | 10.8% | 21.1 (3.7) | — | 374 | 10.8% | 72.2 (9.6) | — | 374 | 10.8% | 0.77 (0.07) | — |
| Type of housing at recruitment | 3457 | — | — | 0.18 | 3453 | — | — | 0.51 | 3452 | — | — | 0.47 |
| Public | 1448 | 41.9% | 21.1 (3.6) | — | 1445 | 41.8% | 72.5 (9.4) | — | 1445 | 41.9% | 0.77 (0.07) | — |
| Subsidized home ownership scheme | 545 | 15.8% | 20.8 (3.6) | — | 544 | 15.8% | 72.0 (9.2) | — | 544 | 15.8% | 0.77 (0.06) | — |
| Private | 1359 | 39.3% | 20.9 (3.4) | — | 1359 | 39.4% | 72.3 (9.1) | — | 1358 | 39.3% | 0.77 (0.06) | — |
| Unknown | 105 | 3.0% | 20.5 (3.0) | — | 105 | 3.0% | 71.2 (7.5) | — | 105 | 3.0% | 0.76 (0.05) | — |
*Two-side P-value from independent t-test or analysis of variance (ANOVA).
Table 2 shows ALT was positively associated with BMI, WC, and WHR adjusted for potential confounders. ALP was negatively associated with BMI, WC, and WHR. The associations of ALP with BMI, WC, and WHR differed by sex, with the inverse associations only evident in boys.
Table 2.
Adjusted associations of liver enzymes (ALT and ALP) with adiposity indices (BMI, WC, and WHR) at ~17.5 years in the Hong Kong’s “Children of 1997” birth cohort, Hong Kong, China.
| Liver enzyme | Outcome | Sex-adjusted | Sex interaction | Boys | Girls | |||
|---|---|---|---|---|---|---|---|---|
| Beta | 95% CI | p-value | Beta | 95% CI | Beta | 95% CI | ||
| ALT (IU/L) | BMI (kg/m2) | 0.10 | 0.09 to 0.11 | 0.19 | 0.10 | 0.09 to 0.12 | 0.09 | 0.07 to 0.11 |
| WC (cm) | 0.25 | 0.23 to 0.28 | 0.04 | 0.27 | 0.24 to 0.30 | 0.21 | 0.17 to 0.26 | |
| WHR | 0.0013 | 0.0012 to 0.0015 | 0.08 | 0.0014 | 0.0012 to 0.0016 | 0.0011 | 0.0008 to 0.0014 | |
| ALP (IU/L) | BMI (kg/m2) | −0.018 | −0.024 to −0.012 | <0.001 | −0.024 | −0.031 to −0.017 | 0.006 | −0.006 to 0.018 |
| WC (cm) | −0.03 | −0.05 to −0.02 | <0.001 | −0.05 | −0.07 to −0.03 | 0.04 | 0.01 to 0.07 | |
| WHR | −0.0002 | −0.0003 to −0.0001 | 0.002 | −0.0002 | −0.0004 to −0.0001 | 0.0001 | −0.0001 to 0.0003 | |
ALT: alanine aminotransferase; ALP: alkaline phosphatase.
BMI: body mass index; WC: waist circumference; WHR: waist-hip ratio.
Adjustment: household income, highest parental education, type of housing, highest parental occupation, second-hand and maternal smoking.
Mendelian randomization
Genetic variants
In total, 4 single nucleotide polymorphisms (SNPs) independently predicting ALT, 14 SNPs independently predicting ALP, and 26 SNPs independently predicting GGT at genome-wide significance were obtained (Supplemental Table S2)14. All the palindromic SNPs were aligned based on effect allele frequency (Supplemental Table S3), except for rs2073398 (GGT1, GGTLC2), predicting GGT, which was replaced by rs5751901 (R2 = 0.95) for GIANTUKB. Rs6834314 (HSD17B13, MAPK10) predicting ALT and rs944002 (EXOC3L4) predicting GGT were replaced in the genome-wide association study (GWAS) Anthropometric 2015 Waist by rs13102451 (R2 = 1.00) and rs2297067 (R2 = 0.98). Two SNPs, rs516246 (FUT2) and rs8038465 (CD276) predicting GGT had rather different allele distributions for GGT and adiposity indices in GIANT (GWAS Anthropometric 2015 BMI and the GWAS Anthropometric 2015 Waist)15,16. They were dropped in a sensitivity analysis separately. No proxy SNP (R2 > 0.9) of rs516246 could be found in GIANTUKB. (Supplemental Table S4).
Of the 4 SNPs predicting ALT, rs2954021 (TRIB1) predicted both ALT and ALP, and rs738409 (PNPLA3) is highly associated with NAFLD. Of the 14 SNPs predicting ALP, rs281377 (FUT2) is highly associated with resting metabolic rate; rs579459 is located in the ABO gene. Of the 26 SNPs predicting GGT, rs516246 (FUT2) is associated with obesity-related traits; rs1260326 (GCKR) is associated with Crohn’s disease which might be associated with adiposity (Supplemental Table S2). The F statistics and variance explained (r2) were 15 and 0.001 for ALT, 158 and 0.035 for ALP, and 45 and 0.019 for GGT. As such the MR study had 80% power with 5% alpha to detect a difference of 0.11, 0.02, and 0.02 in BMI effect size for ALT, ALP, and GGT respectively.
Genetically instrumented ALT with BMI, WC, and WHR
Genetically instrumented ALT was negatively associated with BMI using inverse variance weighting (IVW), which was also evident using a weighted median (WM) and after excluding the potentially pleiotropic SNPs. The negative association was more obvious for women. ALT was not associated with WC or WHR using any method with and without potentially pleiotropic SNPs. Few MR-Egger intercepts differed from the null, giving little indication of pleiotropy. Heterogeneity was almost absent after the pleiotropic SNP were excluded (Tables 3–5).
Table 4.
Estimates of the effect of genetically instrumented liver enzymes (ALT, ALP, and GGT) (per 100% change in concentration) on WC (standard deviation) using Mendelian randomization with different methodological approaches with and without potentially pleiotropic SNPs.
| Liver enzyme | Method | SNPs | All | Men | Women | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | 95% CI | MR-Egger | I2 | Beta | 95% CI | MR-Egger | I2 | Beta | 95% CI | MR-Egger | I2 | |||
| Intercept p- value | (p-value) | Intercept p-value | (p-value) | Intercept p-value | (p-value) | |||||||||
| ALT | IVW | 4 | −0.06 | −0.23 to 0.12 | — | 34.5% (0.21) | 0.0001 | −0.35 to 0.35 | — | 66.4% (0.03) | −0.08 | −0.26 to 0.10 | — | 9.3% (0.35) |
| 3 | −0.03 | −0.18 to 0.11 | — | 40.2% (0.19) | 0.08 | −0.13 to 0.29 | — | 0.0% (0.66) | −0.1 | −0.28 to 0.08 | — | 25.8% (0.26) | ||
| 2 | −0.03 | −0.29 to 0.24 | — | — | 0.08 | −0.31 to 0.46 | — | — | −0.06 | −0.39 to 0.27 | — | — | ||
| WM | 4 | −0.06 | −0.22 to 0.10 | — | — | 0.06 | −0.17 to 0.29 | - | — | −0.1 | −0.29 to 0.10 | — | — | |
| 3 | −0.04 | −0.21 to 0.12 | — | — | 0.08 | −0.15 to 0.31 | — | — | −0.12 | −0.32 to 0.08 | — | — | ||
| MR-Egger | 4 | 0.01 | −0.41 to 0.44 | 0.72 | — | 0.35 | −0.34 to 1.05 | 0.26 | — | −0.23 | −0.62 to 0.17 | 0.42 | — | |
| 3 | −0.08 | −0.70 to 0.54 | 0.87 | — | 0.06 | −0.44 to 0.56 | 0.93 | — | −0.2 | −0.86 to 0.46 | 0.75 | — | ||
| ALP | IVW | 14 | −0.02 | −0.16 to 0.11 | — | 72.6% (<0.001) | −0.02 | −0.18 to 0.14 | — | 62.8% (<0.001) | −0.03 | −0.15 to 0.10 | — | 51.0% (0.01) |
| 13 | −0.02 | −0.15 to 0.12 | — | 73.9% (<0.001) | −0.004 | −0.15 to 0.15 | — | 56.5% (0.006) | −0.03 | −0.16 to 0.10 | — | 54.2% (0.01) | ||
| 11 | −0.13 | −0.33 to 0.08 | — | 70.8% (<0.001) | −0.09 | −0.33 to 0.15 | — | 55.7% (0.01) | −0.15 | −0.34 to 0.04 | — | 45.0% (0.05) | ||
| WM | 14 | 0.01 | −0.08 to 0.10 | — | — | 0.002 | −0.12 to 0.12 | — | — | 0.03 | −0.09 to 0.14 | — | — | |
| 13 | 0.02 | −0.07 to 0.10 | — | — | 0.01 | −0.11 to 0.13 | — | — | 0.02 | −0.09 to 0.14 | — | — | ||
| 11 | −0.13 | −0.32 to 0.06 | — | — | −0.19 | −0.43 to 0.06 | — | — | −0.06 | −0.28 to 0.15 | — | — | ||
| MR-Egger | 14 | 0.1 | −0.13 to 0.32 | 0.20 | — | 0.14 | −0.13 to 0.40 | 0.15 | — | 0.06 | −0.16 to 0.28 | 0.33 | — | |
| 13 | 0.09 | −0.14 to 0.32 | 0.27 | — | 0.1 | −0.15 to 0.36 | 0.31 | — | 0.08 | −0.15 to 0.30 | 0.26 | — | ||
| 11 | 0.27 | −0.43 to 0.97 | 0.25 | — | 0.47 | −0.32 to 1.27 | 0.14 | — | 0.12 | −0.53 to 0.78 | 0.39 | — | ||
| GGT | IVW | 26 | −0.01 | −0.05 to 0.03 | — | 14.2% (0.26) | −0.04 | −0.10 to 0.03 | — | 24.8% (0.12) | 0.01 | −0.05 to 0.07 | — | 35.7% (0.04) |
| 23 | 0.003 | −0.04 to 0.04 | — | 0.0% (0.56) | −0.02 | −0.07 to 0.04 | — | 1.4% (0.44) | 0.02 | −0.04 to 0.08 | — | 30.8% (0.08) | ||
| WM | 26 | −0.04 | −0.10 to 0.02 | — | — | −0.01 | −0.09 to 0.06 | — | — | −0.002 | −0.08 to 0.07 | — | — | |
| 23 | −0.04 | −0.09 to 0.02 | — | — | −0.01 | −0.09 to 0.06 | — | — | −0.002 | −0.08 to 0.07 | — | — | ||
| MR-Egger | 26 | 0.05 | −0.05 to 0.14 | 0.18 | — | 0.05 | −0.10 to 0.19 | 0.23 | — | 0.05 | −0.09 to 0.19 | 0.50 | — | |
| 23 | 0.01 | −0.09 to 0.10 | 0.91 | — | 0.02 | −0.12 to 0.16 | 0.57 | — | 0.01 | −0.14 to 0.15 | 0.84 | — | ||
Excluded SNPs predicting ALT: rs2954021 (TRIB1) when SNP = 3, excluded rs738409 (PNPLA3) in addition when SNP = 2; excluded SNPs predicting ALP: rs2954021 (TRIB1), when SNP = 13; excluded rs281377 (FUT2) and rs579459 (ABO) in addition when SNP = 11; excluded SNPs predicting GGT: rs516246 (FUT2), rs1260326 (C2orf16, GCKR), and rs8038465 (CD276) when SNP = 23.
SNP: single nucleotide polymorphism; ALT: alanine aminotransferase; ALP: alkaline phosphatase; GGT: gamma glutamyltransferase; WC: waist circumference; IVW: inverse variance weighted; WM: weighted median.
Table 3.
Estimates of the effect of genetically instrumented liver enzymes (ALT, ALP, and GGT) (per 100% change in concentration) on BMI (standard deviation) using Mendelian randomization with different methodological approaches with and without potentially pleiotropic SNPs.
| Liver enzyme | Method | SNP | Men and women together using All-GIANTUKB | Men using GWAS Anthropometric 2015 BMI | Women using GWAS Anthropometric 2015 BMI | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | 95% CI | MR-Egger | I2 | Beta | 95% CI | MR-Egger | I2 | Beta | 95% CI | MR-Egger | I2 | |||
| Intercept p-value | (p-value) | Intercept p-value | (p-value) | Intercept p-value | (p-value) | |||||||||
| ALT | IVW | 4 | −0.17 | −0.33 to −0.01 | — | 84.6% (<0.001) | −0.13 | −0.39 to 0.13 | — | 60.6% (0.05) | −0.19 | −0.34 to −0.04 | — | 0.0% (0.76) |
| 3 | −0.14 | −0.20 to −0.07 | — | 0.0% (<0.64) | −0.08 | −0.24 to 0.09 | — | 0.0% (0.39) | −0.18 | −0.34 to −0.03 | — | 0.0% (0.59) | ||
| 2 | −0.17 | −0.29 to −0.05 | — | — | −0.24 | −0.56 to 0.07 | — | — | −0.19 | −0.48 to 0.10 | — | — | ||
| WM | 4 | −0.13 | −0.20 to −0.05 | — | — | −0.06 | −0.25 to 0.12 | — | — | −0.19 | −0.36 to −0.02 | — | — | |
| 3 | −0.12 | −0.20 to −0.05 | — | — | −0.05 | −0.23 to 0.13 | — | — | −0.18 | −0.36 to −0.01 | — | — | ||
| MR-Egger | 4 | 0.02 | −0.25 to 0.30 | 0.11 | — | 0.24 | −0.09 to 0.58 | 0.01 | — | −0.16 | −0.48 to 0.15 | 0.85 | — | |
| 3 | −0.09 | −0.25 to 0.06 | 0.54 | — | 0.14 | −0.26 to 0.53 | 0.25 | — | −0.19 | −0.57 to 0.19 | 0.96 | — | ||
| ALP | IVW | 14 | −0.07 | −0.18 to 0.04 | — | 88.5% (<0.001) | 0.03 | −0.10 to 0.17 | — | 60.4% (0.002) | −0.08 | −0.21 to 0.04 | — | 58.7% (0.003) |
| 13 | −0.06 | −0.16 to 0.05 | — | 86.2% (<0.001) | 0.05 | −0.07 to 0.17 | — | 51.6% (0.02) | −0.08 | −0.21 to 0.05 | — | 61.3% (0.002) | ||
| 11 | −0.07 | −0.23 to 0.09 | — | 88.3% (<0.001) | −0.02 | −0.22 to 0.18 | — | 55.7% (0.01) | −0.17 | −0.33 to −0.01 | — | 34.6% (0.12) | ||
| WM | 14 | −0.05 | −0.11 to −0.001 | — | — | 0.07 | −0.03 to 0.18 | — | — | −0.06 | −0.16 to 0.04 | — | — | |
| 13 | −0.05 | −0.10 to −0.001 | — | — | 0.07 | −0.03 to 0.18 | — | — | −0.06 | −0.16 to 0.04 | — | — | ||
| 11 | −0.07 | −0.18 to 0.04 | — | — | −0.12 | −0.33 to 0.09 | — | — | −0.11 | −0.29 to 0.07 | — | — | ||
| MR-Egger | 14 | 0.03 | −0.17 to 0.22 | 0.23 | — | 0.15 | −0.06 to 0.37 | 0.18 | — | −0.04 | −0.26 to 0.18 | 0.63 | — | |
| 13 | −0.001 | −0.19 to 0.19 | 0.50 | — | 0.12 | −0.09 to 0.33 | 0.39 | — | −0.05 | −0.28 to 0.19 | 0.74 | — | ||
| 11 | 0.28 | −0.24 to 0.79 | 0.17 | — | 0.21 | −0.49 to 0.92 | 0.50 | — | 0.01 | −0.54 to 0.56 | 0.51 | — | ||
| GGT* | IVW | 26/25 | 0.04 | −0.01 to 0.08 | — | 85.6% (<0.001) | 0.01 | −0.05 to 0.07 | — | 40.7% (0.02) | 0.001 | −0.06 to 0.07 | — | 56.0% (<0.001) |
| 23/24 | 0.04 | −0.003 to 0.09 | — | 83.2% (<0.001) | 0.02 | −0.03 to 0.07 | — | 12.2% (0.29) | 0.02 | −0.05 to 0.08 | — | 50.4% (0.003) | ||
| WM | 26/25 | 0.03 | 0.01 to 0.06 | — | — | 0.02 | −0.05 to 0.09 | — | — | −0.01 | −0.07 to 0.06 | — | — | |
| 23/24 | 0.03 | 0.01 to 0.06 | — | — | 0.02 | −0.05 to 0.09 | — | — | −0.01 | −0.07 to 0.06 | — | — | ||
| MR-Egger | 26/25 | 0.06 | −0.05 to 0.17 | 0.65 | — | 0.05 | −0.09 to 0.19 | 0.47 | — | 0.04 | −0.11 to 0.19 | 0.56 | — | |
| 23/24 | 0.05 | −0.06 to 0.15 | 0.97 | — | 0.03 | −0.10 to 0.15 | 0.96 | — | −0.01 | −0.16 to 0.14 | 0.67 | — | ||
*Rs516246 (FUT2) predicting GGT is not available in GIANTUKB, 25 SNPs remained.
Excluded SNPs predicting ALT: rs2954021 (TRIB1) when SNP = 3, excluded rs738409 (PNPLA3) in addition when SNP = 2; excluded SNPs predicting ALP: rs2954021 (TRIB1), when SNP = 13; excluded rs281377 (FUT2) and rs579459 (ABO) in addition when SNP = 11; excluded SNPs predicting GGT: rs516246 (FUT2), rs1260326 (C2orf16, GCKR), and rs8038465 (CD276) when SNP = 23 from GWAS Anthropometric 2015 BMI projects, excluding SNPs: rs516246 (FUT2) and rs1260326 (C2orf16, GCKR) when SNP = 24 from GIANTUKB.
SNP: single nucleotide polymorphism; ALT: alanine aminotransferase; ALP: alkaline phosphatase; GGT: gamma glutamyltransferase; BMI: body mass index; IVW: inverse variance weighted; WM: weighted median.
Table 5.
Estimates of the effect of genetically instrumented liver enzymes (ALT, ALP, and GGT) (per 100% change in concentration) on WHR (standard deviation) using Mendelian randomization with different methodological approaches with and without potentially pleiotropic SNPs.
| Liver enzyme | Method | SNP | All | Men | Women | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | 95% CI | MR-Egger | I2 | Beta | 95% CI | MR-Egger | I2 | Beta | 95% CI | MR-Egger | I2 | |||
| Intercept p-value | (p-value) | Intercept p-value | (p-value) | Intercept p-value | (p-value) | |||||||||
| ALT | IVW | 4 | 0.04 | −0.09 to 0.18 | — | 0.0% (0.73) | 0.09 | −0.13 to 0.30 | — | 10.2% (0.34) | 0.02 | −0.24 to 0.27 | — | 56.2% (0.08) |
| 3 | 0.03 | −0.11 to 0.17 | — | 0.0% (0.81) | 0.14 | −0.08 to 0.35 | — | 0.0% (0.83) | −0.04 | −0.22 to 0.13 | — | 0.0% (0.81) | ||
| 2 | −0.02 | −0.28 to 0.24 | — | — | 0.15 | −0.27 to 0.55 | — | — | −0.12 | −0.45 to 0.20 | — | — | ||
| WM | 4 | 0.05 | −0.10 to 0.20 | — | — | 0.12 | −0.11 to 0.35 | — | — | −0.02 | −0.21 to 0.16 | — | — | |
| 3 | 0.04 | −0.11 to 0.19 | — | — | 0.13 | −0.10 to 0.36 | — | — | −0.03 | −0.21 to 0.16 | — | — | ||
| MR-Egger | 4 | −0.004 | −0.29 to 0.28 | 0.70 | — | 0.29 | −0.16 to 0.74 | 0.31 | — | −0.19 | −0.74 to 0.36 | 0.40 | — | |
| 3 | 0.08 | −0.25 to 0.41 | 0.73 | — | 0.1 | −0.41 to 0.61 | 0.88 | — | 0.06 | −0.35 to 0.47 | 0.59 | — | ||
| ALP | IVW | 14 | −0.03 | −0.16 to 0.10 | — | 73.6% (<0.001) | −0.11 | −0.26 to 0.05 | — | 60.0% (0.002) | 0.04 | −0.11 to 0.19 | — | 66.2% (<0.001) |
| 13 | −0.03 | −0.17 to 0.10 | — | 74.9% (<0.001) | −0.1 | −0.26 to 0.06 | — | 61.4% (0.002) | 0.02 | −0.12 to 0.17 | — | 63.4% (0.001) | ||
| 11 | −0.11 | −0.34 to 0.12 | — | 77.5% (<0.001) | −0.08 | −0.36 to 0.20 | — | 66.7% (<0.001) | −0.11 | −0.32 to 0.11 | — | 57.9% (0.008) | ||
| WM | 14 | 0.02 | −0.06 to 0.10 | — | — | −0.12 | −0.25 to 0.00 | — | — | 0.12 | 0.01 to 0.24 | — | — | |
| 13 | 0.02 | −0.06 to 0.10 | — | — | −0.12 | −0.24 to −0.001 | — | — | 0.12 | 0.01 to 0.23 | — | — | ||
| 11 | −0.002 | −0.17 to 0.17 | — | — | 0.02 | −0.23 to 0.27 | — | — | 0.02 | −0.17 to 0.21 | — | — | ||
| MR-Egger | 14 | 0.08 | −0.14 to 0.30 | 0.23 | — | −0.04 | −0.31 to 0.23 | 0.54 | — | 0.17 | −0.09 to 0.43 | 0.21 | — | |
| 13 | 0.1 | −0.12 to 0.32 | 0.14 | — | −0.06 | −0.34 to 0.23 | 0.69 | — | 0.22 | −0.01 to 0.45 | 0.04 | — | ||
| 11 | 0.35 | −0.42 to 1.13 | 0.22 | — | 0.7 | −0.18 to 1.57 | 0.07 | — | 0.11 | −0.64 to 0.86 | 0.56 | — | ||
| GGT | IVW | 26 | 0.03 | −0.01 to 0.07 | — | 13.6% (0.27) | 0.02 | −0.03 to 0.08 | — | 0.0% (0.88) | 0.04 | −0.02 to 0.10 | — | 37.8% (0.03) |
| 23 | 0.03 | −0.02 to 0.07 | — | 19.8% (0.20) | 0.03 | −0.03 to 0.09 | — | 0.0% (0.85) | 0.03 | −0.03 to 0.10 | — | 37.4% (0.04) | ||
| WM | 26 | −0.01 | −0.06 to 0.05 | — | - | 0.02 | −0.06 to 0.10 | — | — | −0.01 | −0.08 to 0.07 | — | — | |
| 23 | −0.01 | −0.07 to 0.05 | — | — | 0.02 | −0.06 to 0.10 | — | — | −0.02 | −0.09 to 0.06 | — | — | ||
| MR-Egger | 26 | 0.03 | −0.06 to 0.13 | 0.95 | — | 0.08 | −0.05 to 0.21 | 0.31 | — | 0.01 | −0.13 to 0.15 | 0.64 | - | |
| 23 | 0.04 | −0.07 to 0.15 | 0.84 | — | 0.07 | −0.06 to 0.21 | 0.50 | — | 0.03 | −0.12 to 0.18 | 0.94 | — | ||
Excluded SNPs predicting ALT: rs2954021 (TRIB1) when SNP = 3, excluded rs738409 (PNPLA3) in addition when SNP = 2; excluded SNPs predicting ALP: rs2954021 (TRIB1), when SNP = 13; excluded rs281377 (FUT2) and rs579459 (ABO) in addition when SNP = 11; excluded SNPs predicting GGT: rs516246 (FUT2), rs1260326 (C2orf16, GCKR), and rs8038465 (CD276) when SNP = 23.
SNP: single nucleotide polymorphism; ALT: alanine aminotransferase; ALP: alkaline phosphatase; GGT: gamma glutamyltransferase; WHR: waist-hip ratio; IVW: inverse variance weighted; WM: weighted median.
Genetically instrumented ALP and GGT with BMI, WC, and WHR
Genetically instrumented ALP and GGT were not clearly associated with BMI, WC, or WHR using any method with and without potentially pleiotropic SNPs. Overall, there was no evidence of pleiotropy based on the null values of the MR-Egger intercepts. Heterogeneity was most evident for ALP (Tables 3–5).
Discussion
This novel study used two different approaches, an observational study and an MR study, with different data sources, assumptions, and different unrelated sources of bias to assess the role of liver enzymes in adiposity. We found the clearest evidence for ALT being inversely associated with BMI, perhaps particularly among women.
We used an observational design to assess the association of liver function, indicated by liver enzymes, with adiposity indices in young people aged 17.5 years and an MR design in adults. However, limitations exist in both study designs. First, liver enzymes represent different aspects of liver function: ALT is a marker of hepatocyte integrity, which is relatively more specific for liver pathology than other indices. ALP and GGT are markers of cholestasis. ALP is not liver specific and also originates from other tissues especially from bone. As such, ALT, ALP, and GGT may not completely or only represent liver function18. Second, the two study designs may not be completely comparable. Specifically, the observational study pertains to Chinese, while the MR study mainly pertains to people of European ancestry because a suitably large GWAS of Chinese people is not publicly available. However, causes are usually consistent although not relevant in all contexts19. Finally, the two study designs have contrasting limitations.
The conventional observational study is open to residual confounding by factors such as diet, lifestyle, and physical activity, which are hard to measure precisely and eliminate, although smoking is rare, alcohol use is low, and adiposity is not strongly associated with SEP in Hong Kong20,21, which may reduce confounding. However, it is difficult to disentangle correlated factors reliably in such observational studies. ALT was also lower than 10 IU/L (n = 254) for 7.3% of the participants in “Children of 1997” and was fixed at 5 IU/L, which was unlikely to affect the estimates, because it was only below the limit of detection for a relatively small proportion of observations. Follow-up in “Children of 1997” was incomplete, however, no major differences were found between the participants with and without adiposity indices (Supplemental Table S1). As such, selection bias from loss-to-follow-up is unlikely.
MR studies are more robust to confounding than conventional observational studies but have strong assumptions. Specifically MR studies rely on the assumptions that the genetic instruments predict the exposure reliably, are independent of confounders of the exposure-outcome association, and are only associated with the outcome via the exposure. The F statistics were all larger than 10, which reduces the risk of weak instrument bias. Pleiotropic effects are possible, but estimates were similar after excluding potentially pleiotropic SNPs, such as rs738409 (PNPLA3) predicting ALT and MR-Egger did not provide statistical evidence of pleiotropy. Although some of the I2 were large, after excluding potential pleiotropic SNPs, in most cases, the I2 became smaller. Estimates for ALP showed some heterogeneity although the MR-Egger regression did not show directional pleiotropy. The GWAS for liver enzymes overlapped slightly (~17%) with the GWAS of adiposity indices from the GIANT consortium but is unlikely to cause bias. We assessed sex differences on the assumption that genetic predictors of liver function are similar for women and men, which we could not test empirically. Finally, MR provides an estimate of the effect of life time exposure rather than indicating the exact size of the corresponding intervention, as such it indicates an etiological pathway.
Observationally, the positive associations of ALT with BMI, WC, and WHR are consistent with most of the previous observational studies in both adolescents and adults22–26. The negative associations of ALP with adiposity are consistent with a previous study among Australian adolescents27, but not with all studies28, although few such studies have been conducted. However, some estimates differed between the observational and MR designs, probably because of the difficulty of distinguishing correlated measures of liver function, the possibility of confounding, and/or observational studies not reflecting life-long effects.
To our knowledge, only one small MR study has assessed the association of liver function with adiposity, and found no association of ALT with BMI7. One possible explanation for ALT potentially reducing BMI, but not WHR or WC, is that ALT, acting via sex hormones, reduces muscle mass rather than or as well as fat mass. ALT reducing muscle mass would be consistent with ALT increasing the risk of diabetes3, because low muscle mass is a potential cause of diabetes29.
Overall, this study suggests that ALT reduces BMI. To further clarify the role of liver function in metabolic conditions whether ALT reduces specifically muscle mass, and thereby causes diabetes should be investigated, because it would mean that muscle mass could be an attractive target of intervention to prevent diabetes.
Methods
The “Children of 1997” birth cohort
The “Children of 1997” birth cohort is a population-representative Chinese birth cohort (n = 8,327) which recruited 88% of all births in Hong Kong in April and May 199730. The study was originally established to assess the effects of second-hand smoke exposure and breastfeeding on health services utilization in the first 18 months of life. Recruitment was conducted at the first postnatal visit to the Maternal and Child Health Centers in Hong Kong. Parents of all newborns were encouraged to attend to obtain free preventive care and vaccinations for their child. Parental and infant characteristics were obtained from a self-administered questionnaire in Chinese at recruitment and subsequent routine visits. In 2007, contact was re-established followed by three postal/telephone questionnaire surveys. From 2013 to 2016 a Biobank Clinical follow-up at 16–18 years was conducted, when liver enzymes were assessed. As a compromise between cost and comprehensiveness, liver enzymes were assessed from plasma ALT and plasma ALP analyzed using the Roche Cobas C8000 System, a discrete photometric chemistry analyzer, with International Federation of Clinical Chemistry standardized method with pyridoxal phosphate and substrates of L-alanine and 2-oxoglutarate for ALT, and an optimized substrate concentration and 2-amino-2-methyl-1-propanol as buffer plus the cations magnesium and zinc for ALP. These analyses were conducted at an accredited laboratory serving a teaching hospital in Hong Kong. Height, weight, and waist and hip circumference were measured using standard protocols.
Children of 1997
Exposure - liver enzymes
Liver function at ~17.5 years was assessed from plasma ALT (IU/L) and plasma ALP (IU/L).
Outcome - Adiposity
Adiposity was assessed from BMI (kg/m2), WC (cm), and WHR, which represent different aspects of adiposity. Although these are not completely normally distributed, we present them in natural units for ease of interpretation, given interpretation was similar using a gamma distribution.
Mendelian randomization
Genetic associations with liver enzymes
SNPs associated with plasma log transformed ALT, ALP, and GGT at genome-wide significance (p-value < 5 × 10−8) adjusted for age and sex were obtained from the largest available GWAS of plasma levels of liver enzymes comprising 61,089 adults (~86% European, mean age 52.8 years, 50.6% women)14. For SNPs in linkage disequilibrium (R2 > 0.01), we retained SNPs with the lowest p-value using the Clumping function of MR-Base (TwoSampleMR) R package, based on the 1000 Genomes catalog31. Whether any of the selected SNPs was related to adiposity directly rather than through liver enzymes (pleiotropic effects) was assessed from their known phenotypes obtained from comprehensive curated genotype to phenotype cross-references, i.e., Ensembl (http://www.ensembl.org/index.html) and the GWAS Catalog (https://www.ebi.ac.uk/gwas/). We also identified SNPs from highly pleiotropic genes, such as ABO and GCKR, whose full functionality is not yet clearly understood.
Genetic associations with adiposity
Overall genetic associations with BMI (SD units) were obtained from 2018 GIANT and UK Biobank meta-analysis (GIANTUKB) (n = 681,275)17, a meta-analysis of the GIANT GWAS Anthropometric 2015 BMI15 (mean age 56.0 years, 53.8% women, 95% European) with a newly conducted GWAS of UK Biobank (100% European). The sample overlap is negligible between these two GWAS17. Sex-specific genetic associations with BMI were from the GIANT GWAS Anthropometric 2015 BMI15 (n = 339,224, mean age 56.0 years, 53.8% women, 95% European). Overall and sex-specific genetic associations with WC (SD units) and WHR (SD units) were obtained from the GIANT GWAS Anthropometric 2015 Waist16 (n = 224,459, mean age 54.5 years, 54.6% women, 63.6% European). The GIANTUKB adjusted for age, sex, 10 principal components, recruitment centre, and genotyping batches17. The GIANT (GWAS Anthropometric 2015 BMI and the GWAS Anthropometric 2015 Waist) adjusted for age, age-squared, study-specific covariates in a linear model15,16.
Statistical analyses
In the “Children of 1997” birth cohort, baseline characteristics of cohort participants who were included and excluded were compared using Cohen effect sizes32. Cohen effect sizes indicate the magnitude of the difference independent of sample size. They are usually categorized as 0.10 for small, 0.30 for medium, and 0.50 for large when considering categorical variables32. The associations of adiposity indices with potential confounders were assessed using independent t-test or analysis of variance (ANOVA).
We assessed the associations of liver enzymes with adiposity indices adjusted for potential confounders, i.e., household income, highest parental education, type of housing, highest parental occupation, second-hand and maternal smoking, and sex, using multivariable linear regression. We also assessed whether associations differed by sex from the relevant interaction terms.
In the Mendelian randomization study, the strength of the genetic instruments was indicated by the F-statistic33. A higher F-statistic indicates lower risk of weak instrument bias33. We aligned SNPs for exposure and outcome on allele and effect allele frequency to ensure all SNPs, in particular palindromic SNPs, were aligned correctly. SNPs that could not be unequivocally aligned were replaced by proxies or dropped. SNPs predicting liver enzymes that were not available for adiposity indices were replaced by highly correlated proxies (R2 > 0.9). Potential proxy SNPs were obtained from the GWAS14 and their correlations with other SNPs were obtained using LDlink34,35.
Unconfounded estimates of the effects of liver enzymes on adiposity indices overall and by sex were obtained by meta-analyzing SNP-specific Wald estimates (SNP-outcome association divided by SNP-exposure association) using IVW with random effects for 4+ SNPs, which assumes that balanced pleiotropy, and with fixed effects for 3 SNPs or fewer. We repeated the analysis excluding pleiotropic SNPs that might be associated with the relevant outcome directly rather than via liver enzymes. As sensitivity analyses, WM and MR-Egger regression were used. The WM may generate correct estimates when >50% of weight is contributed by valid SNPs36. MR-Egger generates correct estimates even when all the SNPs are invalid instruments as long as the instrument strength independent of direct effect assumption is satisfied37. A non-null intercept from MR-Egger indicates potential directional pleiotropy and invalid IVW estimates36. Heterogeneity was assessed using the I2 statistic37. Power calculations were performed using the approximation that the sample size for an Mendelian randomization equates to that of the same regression analysis with the sample size divided by the r2 for genetic variant on exposure38.
All statistical analyses were conducted using R version 3.4.2 (R Foundation for Statistical Computing, Vienna, Austria). The R package MendelianRandomization39 was used to generate the estimates.
Ethics approval and informed consent
Ethical approval for the study, including comprehensive health related analyses, were obtained from Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (HKU/HA HKW IRB). Informed written consent was obtained from the parents/guardians or participant if 18 years or older before participation in the Biobank Clinical Follow-up.
The MR study only uses published or publicly-available data. No original data were collected for the MR study. Ethical approval for each of the studies included in the investigation can be found in the original publications (including informed consent from each participant).
Supplementary information
Acknowledgements
The authors thank colleagues at the Student Health Service and Family Health Service of the Department of Health for their assistance and collaboration. They also thank late Dr. Connie O for coordinating the project and all the fieldwork for the initial study in 1997–1998. Data on measures of adiposity (2018 GIANT and UK Biobank meta-analysis, GWAS anthropometric 2015 BMI, GWAS anthropometric 2015 waist) have been contributed by the Genetic Investigation of ANthropometric Traits (GIANT) consortium and have been download from https://portals.broadinstitute.org/collaboration/giant/index.php/Main_Page. This work is a substudy of the “Children of 1997” birth cohort which was initially supported by the Health Care and Promotion Fund, Health and Welfare Bureau, Government of the Hong Kong SAR [HCPF grant 216106] and reestablished in 2005 with support from the Health and Health Services Research Fund, Government of the Hong Kong SAR, [HHSRF grant 03040771]; the Research Fund for the Control of Infectious Diseases in Hong Kong, the Government of Hong Kong SAR [RFCID grant 04050172]; the University Research Committee Strategic Research Theme (SRT) of Public Health, the University of Hong Kong. The Biobank Clinical follow-up was partly supported by the WYNG Foundation.
Author contributions
J.X. Liu conducted the literature review, the analysis and drafted the manuscript. C.M. Schooling conceptualized ideas, designed and directed the analytic strategy and supervised the study from conception to completion, with assistance from S.L. Au Yeung, G.M. Leung. M.K. Kwok, J.Y.Y. Leung, S.L. Lin, and L.L. Hui had full access to all the data in the study and took responsibility for the integrity of the data. All the authors contributed to the interpretation of the data, critically revising the paper and approval of the final version.
Data availability
Data are available upon request from the “Children of 1997” data access committee: aprmay97@hku.hk. The volume and complexity of the data collected preclude public data deposition, because the participants could be identifiable from such extensive data which would comprise participant privacy. Data of the MR study are publicly available summary data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-52489-8.
References
- 1.Hazlehurst JM, Woods C, Marjot T, Cobbold JF, Tomlinson JW. Non-alcoholic fatty liver disease and diabetes. Metabolism: clinical and experimental. 2016;65:1096–1108. doi: 10.1016/j.metabol.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu L, et al. Liver enzymes and incident diabetes in China: a prospective analysis of 10 764 participants in the Guangzhou Biobank Cohort Study. Journal of epidemiology and community health. 2015;69:1040–1044. doi: 10.1136/jech-2015-205518. [DOI] [PubMed] [Google Scholar]
- 3.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statistics in medicine. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Au Yeung SL, Lin SL, Leung GM, Schooling CM. Liver Enzymes and Risk of Ischemic Heart Disease and Type 2 Diabetes Mellitus: A Mendelian Randomization Study. Scientific reports. 2016;6:38813. doi: 10.1038/srep38813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Silva, N. M. G. et al. Liver Function and Risk of Type 2 Diabetes: Bidirectional Mendelian Randomization Study. Diabetes, db181048, 10.2337/db18-1048 (2019). [DOI] [PMC free article] [PubMed]
- 6.Nano J, et al. Gamma-glutamyltransferase levels, prediabetes and type 2 diabetes: a Mendelian randomization study. International journal of epidemiology. 2017;46:1400–1409. doi: 10.1093/ije/dyx006. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, et al. Mendelian randomization estimates of alanine aminotransferase with cardiovascular disease: Guangzhou Biobank Cohort study. Human molecular genetics. 2017;26:430–437. doi: 10.1093/hmg/ddw396. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow DI. Mendelian Randomization and Type 2 Diabetes. Cardiovascular Drugs and Therapy. 2016;30:51–57. doi: 10.1007/s10557-016-6638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitlock G, et al. Body-mass index and cause-specific mortality in 900 000 adults: collaborative analyses of 57 prospective studies. Lancet (London, England) 2009;373:1083–1096. doi: 10.1016/s0140-6736(09)60318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mongraw-Chaffin ML, et al. Association Between Sex Hormones and Adiposity: Qualitative Differences in Women and Men in the Multi-Ethnic Study of Atherosclerosis. The Journal of clinical endocrinology and metabolism. 2015;100:E596–E600. doi: 10.1210/jc.2014-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazo M, et al. (MESA AC 291) The Association of Endogenous Sex Hormones with Liver Fat - Multi-Ethnic Study of Atherosclerosis (MESA) Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2015;13:1686–1693.e1682. doi: 10.1016/j.cgh.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinella ME. Nonalcoholic fatty liver disease: a systematic review. Jama. 2015;313:2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 13.Schooling CM, Yau C, Cowling BJ, Lam TH, Leung GM. Socio-economic disparities of childhood Body Mass Index in a newly developed population: evidence from Hong Kong’s ‘Children of 1997’ birth cohort. Archives of disease in childhood. 2010;95:437–443. doi: 10.1136/adc.2009.168542. [DOI] [PubMed] [Google Scholar]
- 14.Chambers JC, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nature genetics. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yengo L, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Human molecular genetics. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giannini EG, Testa R, Savarino V. Liver enzyme alteration: a guide for clinicians. CMAJ: Canadian Medical Association journal = journal de l’Association medicale canadienne. 2005;172:367–379. doi: 10.1503/cmaj.1040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez P, Subramanian SV, Schooling CM. Effect measure modification conceptualized using selection diagrams as medication by mechanisms of varying population-level relevance. J Clin Epidemiol. 2019 doi: 10.1016/j.jclinepi.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Au WM, et al. Alcohol Drinking and Pro-drinking Practices in Parents of Hong Kong Adolescents. Alcohol and Alcoholism. 2014;49:668–674. doi: 10.1093/alcalc/agu063. [DOI] [PubMed] [Google Scholar]
- 21.HKSAR, H. K. C. a. S. D. o. (ed. Hong Kong: Census and Statistics Department of HKSAR) (2018).
- 22.Anderson EL, et al. The Prevalence of Non-Alcoholic Fatty Liver Disease in Children and Adolescents: A Systematic Review and Meta-Analysis. PloS one. 2015;10:e0140908. doi: 10.1371/journal.pone.0140908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L, et al. Liver enzymes as mediators of association between obesity and diabetes: the Guangzhou Biobank Cohort Study. Annals of epidemiology. 2017;27:204–207. doi: 10.1016/j.annepidem.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Labayen I, et al. Liver enzymes and clustering cardiometabolic risk factors in European adolescents: the HELENA study. Pediatric obesity. 2015;10:361–370. doi: 10.1111/ijpo.273. [DOI] [PubMed] [Google Scholar]
- 25.Fall T, et al. Age- and sex-specific causal effects of adiposity on cardiovascular risk factors. Diabetes. 2015;64:1841–1852. doi: 10.2337/db14-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu XY, et al. Higher waist-to-height ratio and waist circumference are predictive of metabolic syndrome and elevated serum alanine aminotransferase in adolescents and young adults in mainland China. Public health. 2012;126:135–142. doi: 10.1016/j.puhe.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Booth ML, et al. The population prevalence of adverse concentrations and associations with adiposity of liver tests among Australian adolescents. Journal of paediatrics and child health. 2008;44:686–691. doi: 10.1111/j.1440-1754.2008.01407.x. [DOI] [PubMed] [Google Scholar]
- 28.Nannipieri M, et al. Liver enzymes, the metabolic syndrome, and incident diabetes: the Mexico City diabetes study. Diabetes care. 2005;28:1757–1762. doi: 10.2337/diacare.28.7.1757. [DOI] [PubMed] [Google Scholar]
- 29.Hou WW, Tse MA, Lam TH, Leung GM, Schooling CM. Adolescent testosterone, muscle mass and glucose metabolism: evidence from the ‘Children of 1997’ birth cohort in Hong Kong. Diabetic medicine: a journal of the British Diabetic Association. 2015;32:505–512. doi: 10.1111/dme.12602. [DOI] [PubMed] [Google Scholar]
- 30.Schooling CM, Hui LL, Ho LM, Lam TH, Leung GM. Cohort profile: ‘children of 1997’: a Hong Kong Chinese birth cohort. Int J Epidemiol. 2012;41:611–620. doi: 10.1093/ije/dyq243. [DOI] [PubMed] [Google Scholar]
- 31.Hemani G, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. 2016 doi: 10.1101/078972. [DOI] [Google Scholar]
- 32.Cohen, J. Statistical power analysis for the behavioral sciences. (Academic Press, 1977).
- 33.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two‐sample Mendelian randomization. Genetic Epidemiology. 2016;40:597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics (Oxford, England) 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machiela MJ, Chanock SJ. LDassoc: an online tool for interactively exploring genome-wide association study results and prioritizing variants for functional investigation. Bioinformatics (Oxford, England) 2017 doi: 10.1093/bioinformatics/btx561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgess, S., Bowden, J., Fall, T., Ingelsson, E. & Thompson, S. G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology (Cambridge, Mass.) (2016). [DOI] [PMC free article] [PubMed]
- 38.Freeman G, Cowling BJ, Schooling CM. Power and sample size calculations for Mendelian randomization studies using one genetic instrument. Int J Epidemiol. 2013;42:1157–1163. doi: 10.1093/ije/dyt110. [DOI] [PubMed] [Google Scholar]
- 39.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. International journal of epidemiology. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request from the “Children of 1997” data access committee: aprmay97@hku.hk. The volume and complexity of the data collected preclude public data deposition, because the participants could be identifiable from such extensive data which would comprise participant privacy. Data of the MR study are publicly available summary data.