The aim of the current study was to determine the genomic sequence of parvovirus strain GX-Tu-PV-1, which was isolated from a turkey in Guangxi Province, South China. The analysis showed that the genome sequence of GX-Tu-PV-1 was 81.3% to ∼99.3% similar to those of other turkey parvoviruses (TuPVs) and 79.8% to ∼92.1% related to chicken parvovirus (ChPV). This study will help in understanding the epidemiology and molecular characteristics of parvovirus in turkeys.
ABSTRACT
The aim of the current study was to determine the genomic sequence of parvovirus strain GX-Tu-PV-1, which was isolated from a turkey in Guangxi Province, South China. The analysis showed that the genome sequence of GX-Tu-PV-1 was 81.3% to ∼99.3% similar to those of other turkey parvoviruses (TuPVs) and 79.8% to ∼92.1% related to chicken parvovirus (ChPV). This study will help in understanding the epidemiology and molecular characteristics of parvovirus in turkeys.
ANNOUNCEMENT
Parvovirus (family Parvoviridae) spp. that infect vertebrate hosts make up the subfamily Parvovirinae, while those that infect arthropods make up the subfamily Densovirinae (1–4). According to the latest classification by the International Committee on Taxonomy of Viruses (https://talk.ictvonline.org/taxonomy/), the Parvovirinae subfamily is divided into eight genera (5–8) (Amdoparvovirus, Aveparvovirus, Bocaparvovirus, Copiparvovirus, Dependoparvovirus, Erythroparvovirus, Protoparvovirus, and Tetraparvovirus). Chicken parvovirus (ChPV) and turkey parvovirus (TuPV) are classified as members of the new genus Aveparvovirus (Galliform aveparvovirus 1).
In this study, viral DNA was extracted from tracheal and cloacal swabs from 12 turkeys using the EasyPure viral DNA/RNA kit (Transgen, Beijing, China). TuPV was identified by PCR, using primers designed to target the conserved region (nonstructural [NS] gene, 561 bp) first (1). The two positive samples were confirmed by partial sequencing of the nonstructural 1 (NS1) gene using primers NSF and NSR (Table 1). BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis revealed 98.0% to ∼100% nucleotide (nt) sequence identity to the TuPV 1030 strain (GenBank accession number KM598418). Based on an alignment of the sequences of three TuPV isolates deposited in GenBank, the specific pairs of primers (Table 1) were then designed to amplify the genome of one positive TuPV sample named GX-Tu-PV-1. The gel-electrophoresed PCR products were purified by using the AxyPrep DNA gel extraction kit (Hangzhou, China), followed by cloning into the pMD-19T vector (TaKaRa, Japan). Sanger sequencing was performed on a DNA analyzer (Invitrogen, Guangzhou, China). The undetermined 5′ and 3′ terminal fragments were amplified using the 5′ rapid amplification of cDNA ends (RACE) system kit v.2 (TaKaRa) and reverse transcription-PCR with oligo(dT) primers, respectively. The amplified fragments were cloned into the pMD-19T vector (TaKaRa), and 4 and 8 clones of the 5′ and 3′ terminal regions, respectively, were sequenced. The sequences were obtained by assembling overlapping contigs, followed by editing using the EditSeq and MegAlign programs of DNAStar 7.0 green (DNAStar, Madison, WI).
TABLE 1.
Primers used for PCR amplification of the turkey parvovirus genome
| Primera | Sequence (5′–3′)b | Nucleotide positionsc |
|---|---|---|
| F1-1 | CTGCTGAGCTGGTAAGATGG | 395–414 |
| R1-2 | TCTTCCCGACTGACTAGATT | 724–743 |
| R1-3 | CCCCCATGATACATTTTGCT | 1751–1770 |
| F2-1 | TTCTAATAACGATATCACTCAAGTTTC | 1841–1867 |
| R1-4 | AACCAGTATAGGTGGGTTCC | 2192–2211 |
| R1-1 | TTTGCGCTTGCGGTGAAGTCTGGCTCG | 2375–2401 |
| F3-1 | CAAGCCGCCATTGTGTTTGT | 3575–3594 |
| R2-2 | GTATTGKGTYTGGTTTTCAG | 3659–3678 |
| R2-1 | AAGTCWAKRTAATTCCATGG | 3694–3713 |
| R3-2 | GTCCCTGTCAAGTCATTAGAG | 3858–3878 |
| R3-1 | TTAATTGGTYYKCGGYRCSCG | 5005–5025 |
| NSF | TTCTAATAACGATATCACTCAAGTTTC | 1841–1867 |
| NSR | TTTGCGCTTGCGGTGAAGTCTGGCTCG | 2375–2401 |
Primers F1-1/R1-1, F1-1/R1-2, F1-1/R1-3, and F1-1/R1-4 were used to amplify the first fragment, F2-1/R1-1 and F2-1/R2-2 were used to amplify the second fragment, and F3-1/R3-1 and F3-1/R3-2 were used to amplify the third fragment of the turkey parvovirus (GX-Tu-PV-1) genome. Primers NSF to NSR were used to amplify the partial NS sequence.
The sequences of the primers were designed according to the sequences of three other known ChPV/TuPV strains (GenBank accession numbers GU214704, GU214706, and NC_024454).
The positions of primers located in the genome are shown according to the U.S. TuPV isolate (TuPV 1030).
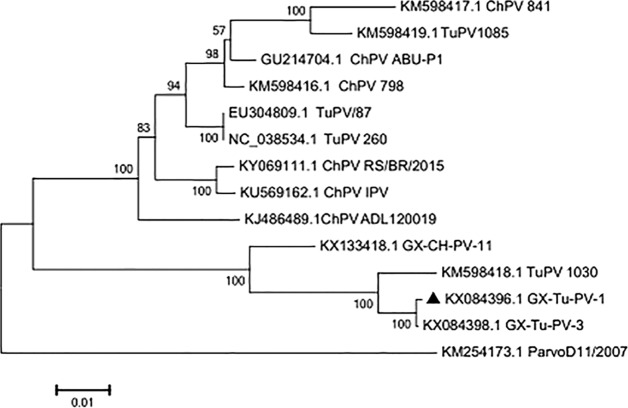
The DNA sequence of the obtained isolate was 4,642 bp long, with an A+T content of 57.1% and G+C content of 42.9%. The entire genome of GX-Tu-PV-1 consists of three open reading frames (ORFs). ORF1 and ORF2 encode a nonstructural (NS) protein, which is involved in viral replication, and the major capsid proteins (VP1 and VP2), respectively. ORF3 encodes a putative protein, NP1, of which the function is unclear (9–13). The genetic diversity of GX-Tu-PV-1 was explored using phylogenetic analyses using ClustalW (http://www.clustal.org/clustal2/) and MEGA 7.0 (14). It showed that the genome sequence of GX-Tu-PV-1 was 81.3% to ∼99.3% related to TuPVs (GenBank accession numbers NC_038534, EU304809, KM598418, KM598419, and KX084398) and 79.8% to ∼92.1% related to ChPV strains (GenBank accession numbers KM598417, GU214704, KM598416, KY069111, KU569162, KJ486489, KX133418, and KM254173) (Fig. 1).
FIG 1.
Neighbor-joining (NJ) tree of the genomic sequences of GX-Tu-PV-1 and 13 other ChPV/TuPV isolates. GenBank accession numbers follow the names of the ChPV/TuPV strains. The numbers near the branches indicate the confidence level calculated by bootstrapping (n = 1,000). The length of each pair of branches represents the distance between sequence pairs. The scale bar represents 0.01-nt substitutions per site. ▲, GX-Tu-PV-1.
The sequence data of the GX-Tu-PV-1 strain will facilitate research on the epidemiology and evolutionary biology of parvoviruses in China.
Data availability.
The genome sequence of GX-Tu-PV-1 was deposited in GenBank under the accession number KX084396.
ACKNOWLEDGMENTS
This research project was funded by the Guangxi Science Base and Talents Special Program (grant AD17195083), the Guangxi Science Great Special Program (grant AA17204057), the Guangxi BaGui Scholars Program Foundation (grant 2019-79), and the National Ten-Thousand Talents Program of China (grant W02060083).
We declare no conflicts of interest, and we are responsible for the content and writing of the article.
REFERENCES
- 1.Feng B, Xie ZX, Deng XW, Xie LJ, Xie ZQ, Huang L, Fan Q, Luo SS, Huang JL, Zhang YF, Zeng TT, Wang S, Wang LY. 2016. Genetic and phylogenetic analysis of a novel parvovirus isolated from chickens in Guangxi, China. Arch Virol 161:3285–3289. doi: 10.1007/s00705-016-2999-0. [DOI] [PubMed] [Google Scholar]
- 2.Tsai H, Tseng C, Chang P, Mei K, Wang S. 2004. Genetic variation of viral protein 1 genes of field strains of waterfowl parvoviruses and their attenuated derivatives. Avian Dis 48:512–521. doi: 10.1637/7172. [DOI] [PubMed] [Google Scholar]
- 3.Zsak L, Strother KO, Day JM. 2009. Development of a polymerase chain reaction procedure for detection of chicken and turkey parvoviruses. Avian Dis 53:83–88. doi: 10.1637/8464-090308-Reg.1. [DOI] [PubMed] [Google Scholar]
- 4.Murgia AMV, Rauf A, Tang Y, Gingerich E, Lee C, Saif YM. 2012. Prevalence of parvoviruses in commercial turkey flocks. Avian Dis 56:744–749. doi: 10.1637/10076-020812-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 5.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotmore SF, Tattersall P. 2014. Parvoviruses: small does not mean simple. Annu Rev Virol 1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 7.Kapgate SS, Kumanan K, Vijayarani K, Barbuddhe SB. 2018. Avian parvovirus: classification, phylogeny, pathogenesis and diagnosis. Avian Pathol 47:536–545. doi: 10.1080/03079457.2018.1517938. [DOI] [PubMed] [Google Scholar]
- 8.Torre DDL, Nuñez LFN, Puga B, Parra SHS, Astolfi-Ferreira CS, Ferreira AJP. 2018. Molecular diagnostic of chicken parvovirus (ChPV) affecting broiler flocks in Ecuador. Braz J Poult Sci 20:643–125. doi: 10.1590/1806-9061-2018-0730. [DOI] [Google Scholar]
- 9.Day JM, Zsak L. 2013. Recent progress in the characterization of avian enteric viruses. Avian Dis 57:573–580. doi: 10.1637/10390-092712-Review.1. [DOI] [PubMed] [Google Scholar]
- 10.Bidin M, Lojkic I, Bidin Z, Tiljar M, Majnaric D. 2011. Identification and phylogenetic diversity of parvovirus circulating in commercial chicken and turkey flocks in Croatia. Avian Dis 55:693–696. doi: 10.1637/9746-032811-Reg.1. [DOI] [PubMed] [Google Scholar]
- 11.Zsak L, Cha RM, Li F, Day JM. 2015. Host specificity and phylogenetic relationships of chicken and turkey parvoviruses. Avian Dis 59:157–161. doi: 10.1637/10939-092414-ResNote. [DOI] [PubMed] [Google Scholar]
- 12.Domanska-Blicharz K, Jacukowicz A, Lisowska A, Minta Z. 2012. Genetic characterization of parvoviruses circulating in turkey and chicken flocks in Poland. Arch Virol 157:2425–2430. doi: 10.1007/s00705-012-1446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nuñez L, Santander-Parra S, Chaible L, De la Torre D, Buim M, Murakami A, Zaidan Dagli M, Astolfi-Ferreira C, Piantino Ferreira A. 2018. Development of a sensitive real-time fast-qPCR based on SYBR green for detection and quantification of chicken parvovirus (ChPV). Vet Sci 5:69. doi: 10.3390/vetsci5030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Stecher G, Tamura K. 2016. Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence of GX-Tu-PV-1 was deposited in GenBank under the accession number KX084396.