The facultative endosymbiont Arsenophonus plays an important role in regulating reproduction through son killing, enemy resistance, and the dietary breadth of its insect hosts. In this study, we found Arsenophonus could alter aphid performance on the amino-acid-deficient diets. Arsenophonus infection increased aphid requirements for the amino acid Phe, but decreased requirements for the Leu. Cotton and cucumber leaves contained drastically different titers of free amino acids Phe and Leu, and aphids living on these two plants were infected with different incidences of Arsenophonus. We hypothesize that host specialization or the host plant range of aphids may be mediated by Arsenophonus.
KEYWORDS: Aphis gossypii, Arsenophonus, amino acid, genotype, host specialization, net reproductive rate
ABSTRACT
Genetic polymorphism and endosymbiont infection are ubiquitous in aphid populations. It has been known that the obligate symbiont Buchnera provides aphids with essential amino acids which cannot be ingested from plant sap. Buchnera often coexists with facultative endosymbionts in aphids. However, it is unclear whether the facultative endosymbionts affect the aphid’s amino acid requirements from diet. In this study, we found that the facultative endosymbiont status in populations of the cotton-melon aphid Aphis gossypii was associated with aphid genotype or host plant. The infection frequency of Arsenophonus in aphids living on cotton was significantly higher than that in aphids on cucumber, and cucumber leaves contained higher titers of free amino acids than cotton leaves, especially amino acids Leu, Arg, Ile, Val, and Phe. The net reproductive rates of five aphid genotypes infected with Arsenophonus were not different on the complete-amino-acid diet, but the values were significantly different among seven Arsenophonus-free aphid genotypes. Moreover, the net reproductive rates of aphids on the amino-acid-deficient diet were significantly affected by Arsenophonus infection and aphid genotype. Arsenophonus infection decreased aphid performance on the Phe-free diet but improved performance on the Leu-free diet and did not affect the performance on the Ile-free or Val-free diet. Arsenophonus infections altered aphid requirements for amino acids that were significantly different in cotton and cucumber leaves, suggesting this endosymbiont would modulate the host specialization of this aphid.
IMPORTANCE The facultative endosymbiont Arsenophonus plays an important role in regulating reproduction through son killing, enemy resistance, and the dietary breadth of its insect hosts. In this study, we found Arsenophonus could alter aphid performance on the amino-acid-deficient diets. Arsenophonus infection increased aphid requirements for the amino acid Phe, but decreased requirements for the Leu. Cotton and cucumber leaves contained drastically different titers of free amino acids Phe and Leu, and aphids living on these two plants were infected with different incidences of Arsenophonus. We hypothesize that host specialization or the host plant range of aphids may be mediated by Arsenophonus.
INTRODUCTION
Aphids require amino acids to support survival and reproduction (1, 2). When Myzus persicae (Sulzer) aphids fed on histidine-, isoleucine-, or methionine-free diets, their body weights decreased (1). The pea aphid Acyrthosiphon pisum (Harris) needs 10 essential amino acids and one nonessential cysteine; individual omission of all these 11 amino acids leads aphids to decreased body weight and even no reproduction on the cysteine-, isoleucine-, methionine- or tryptophan-free diet (3). Amino acids cysteine, methionine, tryptophan, and phenylalanine are essential to the cotton-melon aphid Aphis gossypii Glover, whereas cystine and tyrosine are not required (4, 5). Some clones of Aphis fabae Scopoli displayed lower nymphal survival, nymphal growth rate, or intrinsic rate of population increase on diets omitting histidine, methionine, threonine, or valine (2).
Phloem sap of a plant contains insufficient and imbalanced nutrients for aphids, especially the essential amino acids (6, 7). It has been proved that endosymbionts are involved in the nutrient synthesis of aphids, which complements the insufficient nutrition in plant sap. It is well known that the obligate endosymbiont Buchnera aphidicola supplies aphids with essential amino acids lacking in their phloem sap diet (1, 8–11). Despite massive loss of genes in Buchnera, approximately 10% of the genome encoded biosynthesis of almost all essential amino acids (12–15). In addition, facultative endosymbionts can sometimes have nutritional roles that are not obligate for the host. Serratia symbiotica would compensate for the loss of Buchnera in A. pisum (16). The genome analysis suggested that Hamiltonella defensa might reciprocate with the production of heme, ubiquinone, and pyridoxal-5-phosphate vitamin B6 (17). In whiteflies, it has been found that the facultative endosymbiont Arsenophonus was able to synthesize B vitamins (18). However, it is still unknown whether Arsenophonus is associated with the requirements of aphids for amino acids.
Aphid populations developed various genetic differentiations and formed obvious host specialization (19–21). Community structure of endosymbionts in aphids varies with the genetic backgrounds of hosts (22–25). Aphid populations were often infected with various facultative endosymbionts, such as Serratia, Regiella, Rickettsia, and Spiroplasma (26). In North America, eight species of facultative endosymbionts were detected in the pea aphid and displayed different frequencies between locations and host plants (27). Aphid populations feeding on different host plants were found to be infected with different endosymbionts and belong to different genotypes (27–29). In a previous study, we found that the population genetic structures were different between cotton-melon aphids on cotton and those on cucumber (29), and in this study we also found that the facultative endosymbiont Arsenophonus statuses were different in the aphid populations collected from cotton and cucumber. Arsenophonus has also been found in wild cotton-melon aphid populations (30, 31). Based on genetic differentiation, facultative endosymbionts, and host specialization of cotton-melon aphids, here, we hypothesized that the facultative endosymbiont Arsenophonus and genetic background of aphids were associated with the performance of cotton-melon aphids on different nutrient diets and consequently contribute to the host specialization. Therefore, in this study, we focused on the performance of cotton-melon aphids on diets lacking one of five amino acids, the titers of which in the cotton and cucumber leaves were significantly different, and five Arsenophonus-infected and seven Arsenophonus-free aphid genotypes originally collected from cotton and cucumber were used to address the role of Arsenophonus in mediating the amino acid requirements of aphids. The result will highlight the Arsenophonus-mediated host specialization in aphids.
RESULTS
Relationship between facultative endosymbiont status and genotype of aphids.
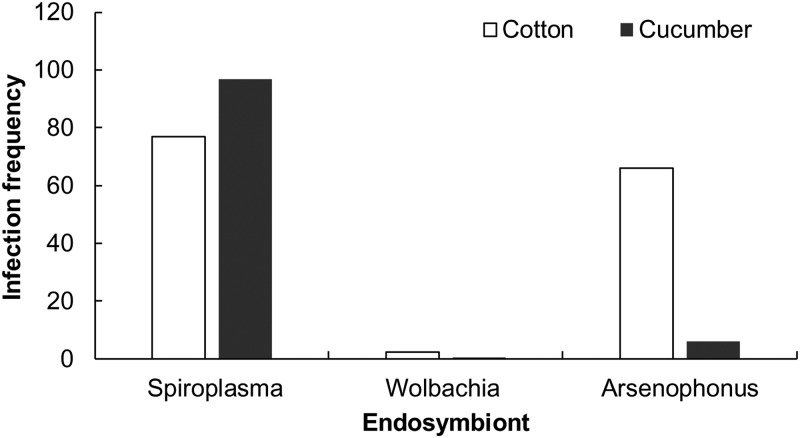
Three facultative endosymbionts, Arsenophonus, Spiroplasma, and Wolbachia, were found in cotton-melon aphids collected from cotton, cucumber, zucchini, and cowpea in Nanjing, China, and their infection frequencies were approximately 44, 87, and 2%, respectively. Approximately 6% of the aphids were not infected with any known facultative endosymbionts (Table 1). The infection statuses of endosymbionts were significantly different among aphid genotypes (χ2 = 668.3, df = 42, P < 0.001). The infection frequencies of Arsenophonus were significantly higher in the aphid genotypes CA9 (81.7%), CA1 (66.7%), and AG5 (49.4%) and were lowest in genotypes CA8 and AG11 (χ2 = 580.3, df = 6, P < 0.001) (Table 1). Aphids collected from cotton and cucumber showed different infection patterns of facultative endosymbionts (χ2 = 163.77, df = 2, P < 0.001) (Fig. 1). The infection frequency of Arsenophonus in aphids on cotton was significantly higher than that of aphids on cucumber (χ2 = 234.38, df = 1, P < 0.001). On the contrary, the infection frequency of Spiroplasma in aphids on cucumber was higher than that of aphids on cotton (χ2 = 50.74, df = 1, P < 0.001). Aphids infected with Wolbachia were infrequent in populations on both cotton and cucumber (Fig. 1).
TABLE 1.
Frequency of cotton-melon aphids belonging to 12 genotypes and infected with facultative endosymbionts
| Genotype | Genotype in this study | Frequency of aphids with endosymbiont infection (%)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ars+ | Spi+ | Wol+ | Ars+ + Spi+ | Ars+ + Wol+ | Spi+ + Wol+ | Ars+ + Spi+ + Wol+ | None | Total | ||
| AG1 | CA2 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.08 | 0.17 |
| AG2 | CA3 | 0.00 | 0.00 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.08 |
| AG3 | CA8 | 0.17 | 7.75 | 0.00 | 0.42 | 0.00 | 0.17 | 0.00 | 0.33 | 8.83 |
| AG4 | 0.00 | 2.92 | 0.00 | 1.08 | 0.00 | 0.00 | 0.00 | 0.08 | 4.08 | |
| AG5 | 0.67 | 2.25 | 0.00 | 2.58 | 0.00 | 0.08 | 0.00 | 1.00 | 6.58 | |
| AG6 | CA4 | 0.25 | 14.67 | 0.00 | 2.17 | 0.00 | 0.42 | 0.08 | 1.25 | 18.83 |
| AG7 | CA9 | 5.08 | 5.25 | 0.00 | 27.75 | 0.17 | 0.00 | 0.83 | 2.33 | 41.42 |
| AG8 | CA1 | 0.33 | 0.92 | 0.08 | 2.08 | 0.00 | 0.00 | 0.08 | 0.25 | 3.75 |
| AG9 | 0.00 | 0.00 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.08 | 0.17 | |
| AG10 | 0.00 | 0.00 | 0.00 | 0.25 | 0.00 | 0.00 | 0.00 | 0.08 | 0.33 | |
| AG11 | 0.00 | 14.17 | 0.00 | 0.25 | 0.00 | 0.17 | 0.08 | 0.83 | 15.50 | |
| AG12 | 0.00 | 0.17 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 | |
| Total | 6.50 | 48.17 | 0.08 | 36.83 | 0.17 | 0.83 | 1.08 | 6.33 | 100 | |
Shown are results from a total of 1,200 aphid samples collected in 2016 in Nanjing, China. Ars+, Arsenophonus infected; Spi+, Spiroplasma infected; Wol+, Wolbachia infected; None, not infected with any of nine endosymbionts.
FIG 1.
Infection frequency (%) of facultative endosymbionts in aphids collected from cotton and cucumber.
Compositions of free amino acids in cotton and cucumber leaves.
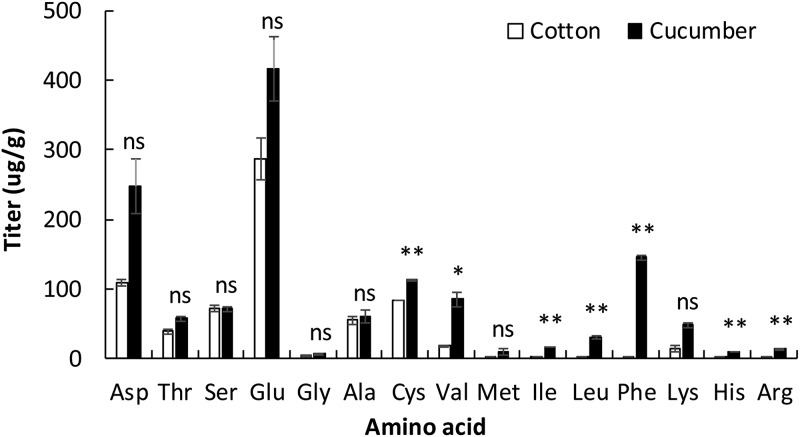
The compositions of free amino acids in cotton and cucumber leaves were not significantly different based on multivariate analysis of variance (MANOVA) for titers of 15 free amino acids (F1, 4 = 29.423, P = 0.137). However, titers of seven free amino acids—cysteine (Cys), valine (Val), isoleucine (Ile), leucine (Leu), phenylalanine (Phe), histidine (His), and arginine (Arg)—were significantly higher in cucumber leaves than in cotton leaves (Fig. 2).
FIG 2.
Titer of 15 free amino acids in cotton and cucumber leaves. * and ** indicate significant differences between cotton and cucumber using the t test followed by the Bonferroni correction at P = 0.05/15 and P = 0.01/15, respectively. ns, no significant differences at P > 0.05/15.
Performance of Arsenophonus-infected and Arsenophonus-free aphids on the complete-amino-acid diet.
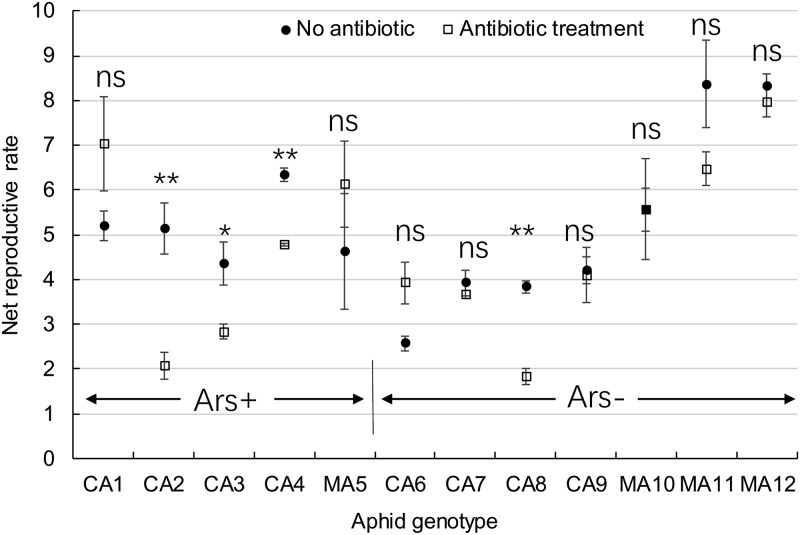
The net reproductive rates of five aphid genotypes infected with Arsenophonus were not significantly different on the complete amino acid diet (F4, 10 = 1.215, P = 0.364), but the values were significantly different among seven genotypes not infected with Arsenophonus, and these aphid genotypes from cucumber (MA10 to MA12) had significantly higher net reproductive rates than those (CA6 to CA9) from cotton (F6, 14 = 14.54, P < 0.001). When all five Arsenophonus-infected genotypes were treated using antibiotics to cure the symbiont, their net reproductive rates on the complete amino acid diet also became different among genotypes (F4, 10 = 10.31, P = 0.001). Elimination of Arsenophonus using antibiotics resulted in the decreased net reproductive rates of the three aphid genotypes CA2, CA3, and CA4, whereas there were no changes of the two genotypes CA1 and MA5 (Fig. 3). The significant differences in the net reproductive rates were still maintained among seven Arsenophonus-free genotypes after antibiotic treatment (F6, 14 = 25.45, P < 0.001), but none of these seven genotypes except for genotype CA8 altered their net reproductive rates as the antibiotic treatment was performed (Fig. 3). Arsenophonus infection affected performance of some aphid genotypes on the complete-amino-acid diet.
FIG 3.
Net reproductive rates (R0s) of 12 genotypes aphids (five Arsenophonus infected [Ars+], and seven Arsenophonus free [Ars−]) on the complete amino acid diet when they were treated or not treated with antibiotics. * and ** indicate significant differences in the net reproductive rates of a genotype between antibiotic treatment and no treatment at P = 0.05 and P = 0.01, respectively. ns, no significant differences.
Relative density of Buchnera in Arsenophonus-infected and Arsenophonus-cured aphids.
The relative density of Buchnera in the MA5 aphids from cucumber hosting Arsenophonus was significantly higher than that in the CA1, CA2, CA3, and CA4 aphids from cotton (F4, 10 = 39.14, P < 0.001). When the facultative symbiont Arsenophonus was eliminated by antibiotics, the relative densities of obligate symbiont Buchnera in the CA1 and MA5 aphids increased, whereas the densities in CA2, CA3, and CA4 aphids remained constant (Fig. 4). The interactions between aphid genotype and Arsenophonus infection affected the Buchnera relative density in aphids (F4, 20 = 4.32, P = 0.011) (Fig. 4).
FIG 4.
Relative density of Buchnera in Arsenophonus-infected (Ars+) and Arsenophonus-cured (Ars−) aphids. * indicates significant difference between infect and cured lineages at P = 0.05. ns, no significant differences.
Performance of Arsenophonus-infected aphids on the amino-acid-deficient diet.
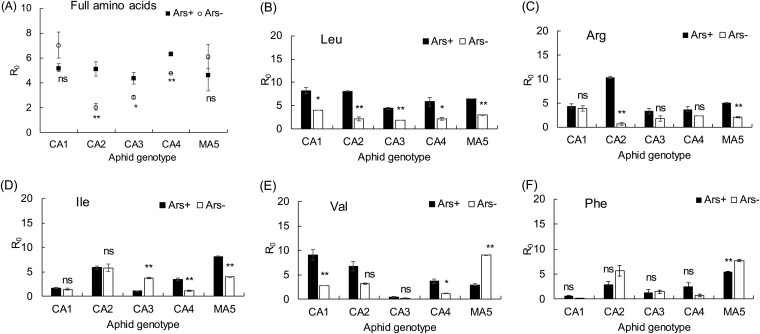
Feeding on the complete-amino-acid diet, CA1 and MA5 aphids did not change their net reproductive rates when their Arsenophonus infections were cured by antibiotics (Fig. 5A). On diets lacking a specific amino acid, however, the cure of Arsenophonus resulted in significant changes in the net reproductive rates of CA1 and MA5 aphids (Fig. 5B to F). Arsenophonus-cured CA1 had a significantly lower net reproductive rate on the diets lacking one of the amino acids Leu and Val than the Arsenophonus-infected CA1 aphids (Fig. 5B and E), whereas they had the same net reproductive rate as the Arsenophonus-infected CA1 on the diets lacking Arg, Ile, or Phe (Fig. 5C, D, and F). The Arsenophonus-cured MA5 aphids also showed a lower net reproductive rate on the diets lacking one of amino acids Leu, Arg, and Ile than the Arsenophonus-infected MA5 aphid (Fig. 5B to D), but they had a higher net reproductive rate on diets lacking Val or Phe than the Arsenophonus-infected one (Fig. 5E and F). Moreover, the net reproductive rates of CA2, CA3, and CA4 were significantly decreased on the complete-amino-acid diet when cured of Arsenophonus (Fig. 5A), but on the diets lacking one of amino acids Leu, Arg, Ile, Val, and Phe, their net reproductive rates would be lower than, equivalent to, or higher than the Arsenophonus-cured aphids, depending on the aphid genotype and amino acid (Fig. 5B to F).
FIG 5.
The net reproductive rates (R0s) of five aphid genotypes on the complete-amino-acid diet (A) or Leu-free (B), Arg-free (C), Ile-free (D), Val-free (E), and Phe-free (F) diets before (Ars+) and after (Ars−) antibiotic treatment. * and ** indicate significant differences between antibiotic treatment and no treatment at P = 0.05 and P = 0.01, respectively. ns, no significant differences.
Due to the increase of Buchnera density in CA1 and MA5 aphids after the elimination of Arsenophonus (Fig. 4), the aphid genotypes CA2, CA3, and CA4 with the same densities of Buchnera before and after antibiotic treatment were selected to reanalyze the role of Arsenophonus in the amino acid requirements of aphids by MANOVA. The result showed that both the aphid genotype and antibiotic treatment significantly affected the net reproductive rates of both the Arsenophonus-infected and Arsenophonus-free aphids when data from all diets lacking Leu, Arg, Ile, Val, and Phe were pooled (Table 2), but on each of the amino-acid-deficient diets, the effects of antibiotic treatment on Arsenophonus-infected aphids were not quite the same as those on the Arsenophonus-free aphids (Table 3). Antibiotic treatment resulted in the significant change in net reproductive rates of both the Arsenophonus-infected and Arsenophonus-free aphids on the diet lacking Arg (Table 3 and Fig. 5C). On the contrary, antibiotic treatment did not lead to a significant change of the net reproductive rates in both the Arsenophonus-infected and Arsenophonus-free aphids on the diet lacking Ile or Val when data from all aphid genotypes were pooled (Table 3), because there were different effects among aphid genotypes (Fig. 5D and E). Overall, Arsenophonus infection did not affect the requirements of aphids for Ile and Val (Table 3). However, on the diets lacking Leu or Phe, antibiotic treatment produced a drastic change in the net reproductive rates of the Arsenophonus-infected aphids (Table 3 and Fig. 5B and F), but not the Arsenophonus-free aphids (Table 3). Arsenophonus infection altered the amino acid requirements of aphids for Leu and Phe. Arsenophonus infection improved the performance of aphids on the Leu-deficient diet (Fig. 5B), but it decreased the performance on the Phe-deficient diet (Fig. 5F).
TABLE 2.
Effects of aphid genotype and antibiotic treatment on the net reproductive rate of Arsenophonus-infected and -free aphids analyzed by MANOVA
| Aphid typea | Effect | Roy's greatest root | F | df |
P value | |
|---|---|---|---|---|---|---|
| Numerator | Denominator | |||||
| Ars+ | Genotype | 36.192 | 57.907 | 5 | 8 | <0.001 |
| Antibiotic | 10.971 | 15.360 | 5 | 7 | 0.001 | |
| Ars− | Genotype | 32.260 | 145.169 | 6 | 27 | <0.001 |
| Antibiotics | 4.486 | 20.637 | 5 | 23 | <0.001 | |
Ars+, Arsenophonus infected; Ars−, Arsenophonus free.
TABLE 3.
Effects of antibiotic treatment on the net reproductive rate of Arsenophonus-infected and -free aphids on diet lacking one of the amino acids Leu, Arg, Ile, Val, and Phe as analyzed by MANOVAa
| Aphid typeb | Amino acid | Type III SS | df | F | P value |
|---|---|---|---|---|---|
| Ars+ | Leu | 13.508 | 1 | 20.889 | 0.0008 |
| Arg | 6.581 | 1 | 16.344 | 0.0019 | |
| Ile | 0.041 | 1 | 0.0894 | 0.7705 | |
| Val | 9.187E−05 | 1 | 0.0003 | 0.9871 | |
| Phe | 9.213 | 1 | 14.012 | 0.0032 | |
| Ars− | Leu | 0.287072 | 1 | 0.547157 | 0.4659 |
| Arg | 24.21308 | 1 | 85.63252 | <0.0001 | |
| Ile | 0.8 | 1 | 1.091195 | 0.3055 | |
| Val | 3.548563 | 1 | 6.432599 | 0.0173 | |
| Phe | 0.360469 | 1 | 0.590888 | 0.4487 |
Bold text indicates the significant effect on this amino acid requirement of aphids at P < 0.01. SS, sum of squares.
Ars+, Arsenophonus infected; Ars−, Arsenophonus free.
DISCUSSION
Genetic background mainly affects nutrient requirements of insects. The amino acid requirements of two biotypes (C and J) in pea aphids were different; biotype C needed arginine, leucine, lysine, and tryptophan, but biotype J needed phenylalanine, threonine, and valine (32). Six clones in pea aphids displayed different adult masses on diets lacking individual essential amino acids (23). The result in this study showed that the net reproductive rate of the Arsenophonus-free A. gossypii on the complete-amino-acid diet varied with their genotypes or the host plants on which aphids were originally collected, and the net reproductive rates of aphid genotypes from cucumber were significantly higher than that from cotton. The dominant aphid genotypes on different host plants were different (19, 29, 33), and contents of free amino acids in cotton and cucumber leaves were also different (34). In this study, we found at least seven amino acids (Leu, Arg, Ile, Val, Phe, His, and Cys) were different in titer between cotton and cucumber leaves, and the titers in cucumber leaves were generally higher than that in cotton leaves. A. gossypii aphids on cotton do not use cucumber and vice versa (21, 33–35). These results imply that the amino acid nutrition in a specific host plant might only meet the demand for some specific genotypes in aphid populations. Different aphid genotypes in Sitobion avenae (Fabricius) exhibited differential preference and performance for different barley genotypes (36). Those aphid genotypes depending closely on a specific nutrient or host plant genotype may find it easy to form host specialization. The nutrition requirements of aphids are associated with their genetic background.
However, in this study we also found that five aphid genotypes infected with Arsenophonus had the same net reproductive rates on the complete-amino-acid diet, but on the amino acid-deficient diet, the Arsenophonus-infected and Arsenophonus-cured aphids exhibited different levels of performance. This implied that besides genetic background, the nutrient requirements of aphids were also affected by facultative endosymbionts. There is a close nutritional relation between endosymbionts and their hosts. It has been well known that the obligate endosymbiont Buchnera supplies necessary nutrients for its insect hosts (1, 8, 12–14). On the other hand, a few studies also indicated or implied that some species of facultative endosymbionts, such as S. symbiotica, H. defensa, and Regiella insecticola, were involved in tryptophan biosynthesis or affected the aphids’ performance on a low-amino-acid diet (23, 37, 38). In this study, we found that the facultative symbiont Arsenophonus improved aphid performance on the Leu-free diet, but decreased aphid performance on the Phe-free diet, and it did not affect aphid performance on the Ile- or Val-free diet. This result suggests that Arsenophonus infections can alter amino acid requirements of aphids. Our study provides evidence of a direct correlation between Arsenophonus and amino acid requirements in aphids.
Facultative endosymbionts may mediate dietary breadth in polyphagous aphids (28, 39, 40). Arsenophonus promoted the specialization of Aphis craccivora Koch on locust trees, and the Arsenophonus-free locust-tree-origin clones performed no better on locust trees than alfalfa-origin clones (39). Aphis glycines Matsumura soybean aphids infected with Arsenophonus performed better on a resistant soybean than their paired uninfected isolines (40). In this study, we further found that Arsenophonus affected the performance or fitness of cotton-melon aphids on the amino-acid-deficient diet. The absence of amino acid biosynthetic capabilities was found in the facultative endosymbionts. A previous study based on genome sequence showed that endosymbiont Arsenophonus presented no genes to metabolize histidine, arginine, and proline, but retained the conserved ABC transporters for proline, arginine, and methionine, suggesting that the endosymbiont might increase uptake of amino acids from its environment (41). Although Arsenophonus has no complete capacity to biosynthesize essential amino acids, some lineages or clades of this endosymbiont retain a part of enzymes in the biosynthetic pathways of isoleucine, valine (Val), phenylalanine (Phe), lysine, tryptophan, leucine (Leu), threonine, and arginine (Arg) (18). In this study, we found that Arsenophonus altered the performance of aphids on an amino-acid-deficient diet—especially on the Phe-free or Leu-free diet. Moreover, the effect of Arsenophonus on aphid performance on amino-acid-deficient diets was dependent on the genotype of aphids. These results suggest that the Arsenophonus may be associated with the nutrition metabolism of the cotton-melon aphids. Based on the result that Arsenophonus infection could alter Buchnera density in some aphid genotypes (for example, CA1 and MA5), we speculated that Arsenophonus might assist the obligate symbiont Buchnera to biosynthesize amino acids and then alter aphid performance on amino-acid-deficient diets. In addition, different strains of Arsenophonus might play different roles in determining host fitness; for example, the S-type Arsenophonus increased the susceptibility of its host brown planthopper Nilaparvata lugens to insecticides, but the N-type did not (42). The strains of Arsenophonus in aphids belonging to different genotypes might be different. Therefore, the effect of Arsenophonus on the performance of aphids varied with aphid genotype.
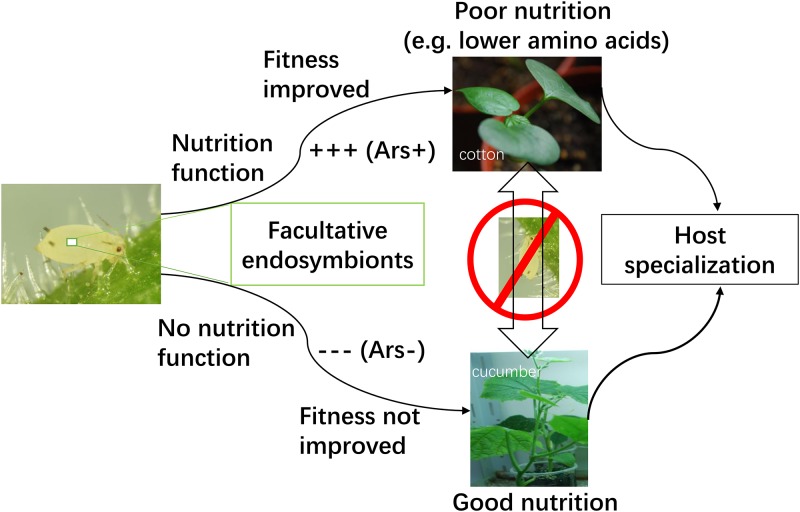
The A. gossypii aphids on cotton were infected with Arsenophonus more frequently than on cucumber, and the free-amino-acid titers in cotton leaves were generally lower than that in cucumber leaves (for example, Phe and Leu). Arsenophonus infection was also found in A. gossypii populations on cotton in Cameroon and northern China, but not found on melon in France (30, 43). The host-specialized biotypes in A. gossypii populations on cotton and cucurbits are ubiquitous in nature (19, 21, 33, 35). Arsenophonus infection in aphids feeding on cotton may compensate for the shortage of amino acids in this plant, due to the nutrition function of endosymbionts (Fig. 6). The aphids infected with a specific endosymbiont can use host plants with “poor” nutrition, but endosymbiont-free aphids cannot and only use host plants with “good” nutrition (abundant amino acids). Therefore, the endosymbiont-mediated host specialization will be promoted (Fig. 6). We speculated that the facultative endosymbiont Arsenophonus may play a role in promoting host specialization in A. gossypii.
FIG 6.
A mode of the facultative endosymbiont-mediated host specialization in aphids.
As a facultative endosymbiont, the effects of Arsenophonus on host fitness were not consistently evident (40). Arsenophonus-reduced or -expanded dietary breadth in A. craccivora was dependent on host genotype (39). In this study, the effects of Arsenophonus on amino acid requirements of aphids were also inconsistent in different genotypes and for different amino acids. We considered that Arsenophonus was still a double-edged endosymbiont in aphids, with both positive and negative effects on its host. This phenomenon was also found in another facultative endosymbiont, Regiella, in the grain aphid S. avenae (44). The function in nutrient regulation of Arsenophonus will promote the coevolution of the facultative endosymbiont and aphid. Therefore, the mechanism of Arsenophonus to alter aphid performance on the amino-acid-deficient diet is worth studying.
MATERIALS AND METHODS
Cotton-melon aphids.
The cotton-melon aphids, Aphis gossypii Glover, were collected from cotton and cucumber fields in Nanjing, China. Aphids from cotton were reared on cotton leaves, and those from cucumber were fed on cucumber leaves under conditions of 14 h of light and 10 h of darkness at 25°C. In order to explore the relationship between endosymbiont status and aphid genotype, 1,200 cotton-melon aphids from cotton, cucumber, zucchini, and cowpea (300 aphids per host plant) were collected from August to October of 2016 in Nanjing, China, and the infection frequencies of nine facultative endosymbionts and genotype of each of the aphids were examined by PCR.
PCR detection of aphid genotype and endosymbiont infection.
All the aphids used in this study were genotyped by PCR using six microsatellite locus primers (45), and the analysis detected the presence of nine facultative endosymbionts, Arsenophonus, Hamiltonella, Regiella, Rickettsia, Rickettsiella, Serratia, Spiroplasma, Wolbachia, and X-type, using the diagnostic PCR method (28, 46–48). Only three facultative endosymbionts (Arsenophonus, Spiroplasma, and Wolbachia) were found in the cotton-melon aphids. Based on the endosymbiont status, a total of 12 genotype lineages of aphid were set up. Five genotype lineages (four from cotton [CA1 to -4] and one from cucumber [MA5]) were only infected with one species of facultative endosymbionts Arsenophonus (here called Arsenophonus-infected strains), and the other seven lineages (four from cotton [CA6 to -9] and three from cucumber [MA10 to -12]) were not infected with any known facultative endosymbionts (called Arsenophonus-free strains). All aphid lineages were infected with the obligate endosymbiont Buchnera. Aphid lineages were reared using leaves as their original host plant in petri dishes (diameter of 90 mm and height of 15 mm). The leaf in a dish was replaced with a fresh one every 3 to 4 days.
Arsenophonus elimination.
Selective elimination of Arsenophonus in aphids was conducted using antibiotics (400 μg/ml ampicillin, 200 μg/ml cephalosporin, and 200 μg/ml gentamicin) (49) added into the artificial diet (Table 4) containing 20 amino acids, vitamins, minerals, and sucrose (1, 50). Newborn nymphs were fed on the antibiotic diet for 6 days and then removed onto leaves of their original host plants. When antibiotic-treated aphids generated offspring, the mother and part of the offspring were chosen to examine the presence of Arsenophonus by PCR. These offspring from a mother whose Arsenophonus endosymbiont was cured were maintained on leaves in dishes as the Arsenophonus-cured lineage. All the 12 genotype aphid lineages infected or not infected with Arsenophonus were treated by antibiotics by the same method. These Arsenophonus-cured lineages were used for experiments after 20 generations had been maintained on leaves without antibiotics. Before experiments, these aphids were examined again in three successive generations in order to confirm the absence of Arsenophonus.
TABLE 4.
Basic components of the complete-amino-acid diet for feeding aphids
| Essential amino acid | Dose (mg/100 ml) | Nonessential amino acid |
Dose (mg/100 ml) | Vitamin | Dose (mg/100 ml) | Mineral and sucrose | Dose (mg/100 ml) |
|---|---|---|---|---|---|---|---|
| Phenylalanine | 40 | Glutamate | 140 | Calcium pantothenate | 5 | Potassium dihydrogen phosphate | 500 |
| Methionine | 80 | Glutamine | 150 | Inositol | 50 | Magnesium chloride | 200 |
| Arginine | 270 | Aspartic acid | 140 | Ascorbic acid | 100 | Manganese chloride | 0.26 |
| Lysine | 120 | Asparagine | 550 | Choline chloride | 50 | Cupric chloride | 0.14 |
| Tryptophan | 80 | Cysteine | 40 | Biotin | 0.1 | Zinc chloride | 0.28 |
| Threonine | 140 | Alanine | 200 | Pyridoxine hydrochloride | 2.5 | Ferric citrate | 0.67 |
| Valine | 80 | Glycine | 80 | Folic acid | 0.5 | Sucrose | 15,000 |
| Isoleucine | 80 | Tyrosine | 40 | Thiamine hydrochloride | 2.5 | Deionized water | 100,000 |
| Leucine | 80 | Proline | 80 | Nicotinic acid | 10 | ||
| Histidine | 80 | Serine | 80 |
Examination of Buchnera density.
The relative densities of Buchnera in aphids were measured by quantitative PCR (qPCR) based on the relative copy number of the Buchnera groEL gene to that of the ef1α gene of the aphid. The densities of Buchnera in the same genotype of aphids infected with Arsenophonus and eliminated from this symbiont by antibiotics were quantified before these genotypes were used in experiments. Total aphid genomic DNA was extracted from five 5-day-old aphids by the method of Zhang et al. (25). The qPCR was performed using SYBR Premix Ex Taq (TaKaRa) in an ABI 7500 real-time PCR system (Thermo Fisher Scientific). The forward primer GroEL-F (5′-GCCATCCAAAGCCGTATTAGTCA-3′) and the reverse primer GroEL-R (5′-AGTACCGCAACACCACCAGATA-3′) amplified a 116-bp fragment of the groEL gene with 107% efficiency. The forward primer ef1α-F (5′-TCACCATCATTGACGCACCTG-3′) and the reverse primer ef1α-R (5′-CCAGTACCAGCAGCAACGATAAG-3′) amplified a 103-bp fragment of the ef1α gene with 108% efficiency. The qPCR conditions were 95°C for 0.5 min, followed by 40 cycles of 95°C for 5 s, and 60°C for 34 s. All the melting curves had a unique peak. Standard curves (log concentration of DNA on the x axis and PCR cycle number on the y axis) for the groEL and ef1α genes were set up based on the method of Zhang et al. (25), and the gene copy number was calculated using the method described by Whelan et al. (51). The copy number of genes and the relative density of Buchnera in five Arsenophonus-infected lineages (CA1 to -4 and MA5) and their Arsenophonus-cured lineages reared for 20 generations were examined. The examination was performed for three biological replicates in each aphid lineage.
Detection of free amino acids in cotton and cucumber leaves.
The free amino acids in seedling cotton and cucumber with five leaves were examined. Fresh leaves (0.2 g) were cut into pieces and ground well on ice with 1 ml 0.02 N hydrochloric acid solution. The grinding apparatus was washed using 0.02 N hydrochloric acid solution three times, and the solution mixture was filtered. The total extracted solution was adjusted to 10 ml using 0.02 N hydrochloric acid solution. The leaf extraction was maintained at 5°C for 24 h. Leaf extraction (0.8 ml) was transferred into a 2-ml microcentrifuge tube, and then 0.8 ml 4% aqueous sulfosalicylic acid was added, and the tube was shaken well. After 15 min, the mixture was centrifuged at 10,000 rpm at 4°C for 15 min, and the supernatant liquid was filtered with a 0.22-μm-pore aqueous-phase filter. One milliliter of filtrate was used to detect the concentrations of 16 free amino acids in a Hitachi Automatic Amino-Acid Analyzer L-8900 (Hitachi, Japan). Three replications were performed for each of the host plant leaves.
Aphid performance on the diet lacking an amino acid.
We used the artificial diet described by Mittler (1) and Sun and Li (50) for rearing aphids (Table 4). The composition of amino acids in the diet was manipulated to assay the performance of aphids on a diet lacking a specific amino acid. The diet includes all 20 essential and nonessential amino acids and is considered the complete amino acid diet. One hundred fifty microliters of artificial diet was placed between two layers of thin Parafilm, which was fixed to one end of a glass tube (30 mm in diameter and 35 mm in height). Ten apterous adult aphids were transferred onto the Parafilm in the tube, and then the other end of the tube was covered with one layer of Parafilm to prevent escape of aphids.
Ten newborn nymphs in a glass tube were left and considered the original cohort for the population life table. The survival and reproduction of the cohort were surveyed daily. When the original cohort reproduced, all offspring were recorded and then removed. The survey lasted until the death of the original cohort. During the survey, the artificial diet was replaced every 2 days. Life tables of 12 aphid genotypes were carried out on each artificial diet individually lacking phenylalanine (Phe), arginine (Arg), valine (Val), isoleucine (Ile), or leucine (Leu), and those amino acids were significantly different in titers between cotton and cucumber leaves as measured as described above. The aphids reared on the complete-amino-acid diet were the control. The experiments of the life table were conducted for all the Arsenophonus-infected, -free, and -cured aphid strains, and each strain experiment was replicated three times.
Data analyses.
The relationship between endosymbiont status and genotype of aphids collected from cotton, cucumber, zucchini, and cowpea represented samples from more than 40 aphids, and the results were analyzed by Pearson’s chi-square test. Effects of aphid genotype and Arsenophonus infection on the relative density of the obligate endosymbiont Buchnera in aphids were analyzed using the GLM model, and differences between Arsenophonus-infected and Arsenophonus-cured aphid lineages were analyzed using Student's t test. The titers of 15 free amino acids in cotton and cucumber leaves were analyzed by MANOVA in GLM to highlight the differences in host plants in amino acid contents, and the mean concentrations of each free amino acid between cotton and cucumber leaves were compared using the t test followed by the Bonferroni correction (P < 0.05/15 = 0.003). The life table parameters were calculated as net reproductive rate (R0) = ∑lxmx, in which lx is the proportion of individuals in the initial cohort alive at age x days, and mx is the mean number of progeny produced per mother aphid alive on day x. The R0s of seven aphid genotypes not infected with Arsenophonus (CA6 to -9 and MA10 to -12) and five genotypes infected with Arsenophonus (CA1 to -4 and MA5) on the complete diet were analyzed by ANOVA followed by the post hoc Tukey’s test to distinguish the differences among different genotypes. The differences in R0 between the Arsenophonus-infected and Arsenophonus-cured lineages were analyzed using Student's t test. The effects of aphid genotype and antibiotic treatment on the R0 of Arsenophonus-infected and Arsenophonus-free aphids feeding on five amino-acid-deficient diets were analyzed using MANOVA in GLM, the R0 on the completed diet was considered a covariant, and the R0s on each amino-acid-deficient diet were considered multiple dependent variables. In order to eliminate the effect of Buchnera density on R0, only data from three Arsenophonus-infected aphid genotypes CA2, CA3, and CA4 were used in this MANOVA, because these three genotypes had the same density of Buchnera before and after antibiotic treatment, and CA1 and MA5 were ruled out. All data analyses were performed using SPSS 25.0.
ACKNOWLEDGMENTS
We thank Wen-Jie Cao and Lin Ma for collecting aphid samples. We thank the two anonymous reviewers for constructive comments and language corrections.
This work was supported by the National Natural Science Foundation of China (grant no. 31672034), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant no. KYCX17_0575). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that we have no competing interests.
REFERENCES
- 1.Mittler TE. 1971. Dietary amino acid requirements of the aphid Myzus persicae affected by antibiotic uptake. J Nutr 101:1023–1028. doi: 10.1093/jn/101.8.1023. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson TL, Douglas AE. 2003. Phloem amino acids and the host plant range of the polyphagous aphid, Aphis fabae. Entomol Exp Appl 106:103–113. doi: 10.1046/j.1570-7458.2003.00014.x. [DOI] [Google Scholar]
- 3.Retnakaran A, Beck SD. 1968. Amino acid requirements and sulfur amino acid metabolism in the pea aphid, Acrythosiphon pisum (Harris). Comp Biochem Physiol 24:611–619. doi: 10.1016/0010-406x(68)91013-x. [DOI] [PubMed] [Google Scholar]
- 4.Turner RB. 1971. Dietary amino acid requirements of the cotton aphid, Aphis gossypii, the sulphur-containing amino acids. J Insect Physiol 17:2451–2456. doi: 10.1016/0022-1910(71)90092-8. [DOI] [Google Scholar]
- 5.Turner RB. 1977. Quantitative requirements for tyrosine, phenylalanine and tryptophan by the cotton aphid, Aphis gossypii (Glover). Comp Biochem Physiol A Physiol 56:203–205. doi: 10.1016/0300-9629(77)90185-2. [DOI] [PubMed] [Google Scholar]
- 6.Sandström J, Pettersson J. 1994. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J Insect Physiol 40:947–955. doi: 10.1016/0022-1910(94)90133-3. [DOI] [Google Scholar]
- 7.Sandstrom J, Moran NA. 1999. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl 91:203–210. doi: 10.1046/j.1570-7458.1999.00485.x. [DOI] [Google Scholar]
- 8.Douglas AE, Prosser WA. 1992. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J Insect Physiol 38:565–568. doi: 10.1016/0022-1910(92)90107-O. [DOI] [Google Scholar]
- 9.Febvay G, Liadouze I, Guillaud J, Bonnot G. 1995. Analysis of energetic amino acid metabolism in Acyrthosiphon pisum: a multidimensional approach to amino acid metabolism in aphids. Arch Insect Biochem Physiol 29:45–69. doi: 10.1002/arch.940290106. [DOI] [Google Scholar]
- 10.Sasaki T, Ishikawa H. 1995. Production of essential amino acids from glutamate by mycetocyte symbiont of the pea aphid, Acyrthosiphon pisum. J Insect Physiol 41:41–46. doi: 10.1016/0022-1910(94)00080-Z. [DOI] [Google Scholar]
- 11.Sasaki T, Hayashi H, Ishikawa H. 1991. Growth and reproduction of the symbiotic and aposymbiotic pea aphids, Acyrthosiphon pisum maintained on artificial diets. J Insect Physiol 37:749–756. doi: 10.1016/0022-1910(91)90109-D. [DOI] [Google Scholar]
- 12.Moran NA, McLaughlin HJ, Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 13.Pérez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- 14.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 15.Gosalbes MJ, Lamelas A, Moya A, Latorre A. 2008. The striking case of tryptophan provision in the cedar aphid Cinara cedri. J Bacteriol 190:6026–6029. doi: 10.1128/JB.00525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga R, Tsuchida T, Fukatsu T. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc Biol Sci 270:2543–2550. doi: 10.1098/rspb.2003.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A 106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos-Garcia D, Juravel K, Freilich S, Zchori-Fein E, Latorre A, Moya A, Morin S, Silva FJ. 2018. To B or not to B: comparative genomics suggests Arsenophonus as a source of B vitamins in whiteflies. Front Microbiol 9:2254. doi: 10.3389/fmicb.2018.02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charaabi K, Carletto J, Chavigny P, Marrakchi M, Makni M, Vanlerberghe-Masutti F. 2008. Genotypic diversity of the cotton-melon aphid Aphis gossypii (Glover) in Tunisia is structured by host plants. Bull Entomol Res 98:333–341. doi: 10.1017/S0007485307005585. [DOI] [PubMed] [Google Scholar]
- 20.Michel AP, Zhang W, Mian M. 2010. Genetic diversity and differentiation among laboratory and field populations of the soybean aphid, Aphis glycines. Bull Entomol Res 100:727–734. doi: 10.1017/S000748531000012X. [DOI] [PubMed] [Google Scholar]
- 21.Satar S, Kersting U, Yokomi R. 2013. Presence of two host races of Aphis gossypii Glover (Hemiptera: Aphididae) collected in Turkey. Ann Appl Biol 162:41–49. doi: 10.1111/j.1744-7348.2012.00578.x. [DOI] [Google Scholar]
- 22.Vogel KJ, Moran NA. 2011. Effect of host genotype on symbiont titer in the aphid-Buchnera symbiosis. Insects 2:423–434. doi: 10.3390/insects2030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel KJ, Moran NA. 2011. Sources of variation in dietary requirements in an obligate nutritional symbiosis. Proc Biol Sci 278:115–121. doi: 10.1098/rspb.2010.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong RA, Moran NA. 2016. Intraspecific genetic variation in hosts affects regulation of obligate heritable symbionts. Proc Natl Acad Sci U S A 113:13114–13119. doi: 10.1073/pnas.1610749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YC, Cao WJ, Zhong LR, Godfray HCJ, Liu XD. 2016. Host plant determines the population size of an obligate symbiont (Buchnera aphidicola) in aphids. Appl Environ Microbiol 82:2336–2346. doi: 10.1128/AEM.04131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T. 2002. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol Ecol 11:2123–2135. doi: 10.1046/j.1365-294X.2002.01606.x. [DOI] [PubMed] [Google Scholar]
- 27.Russell JA, Weldon S, Smith AH, Kim KL, Hu Y, Łukasik P, Doll S, Anastopoulos I, Novin M, Oliver KM. 2013. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol Ecol 22:2045–2059. doi: 10.1111/mec.12211. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchida T, Koga R, Fukatsu T. 2004. Host plant specialization governed by facultative symbiont. Science 303:1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- 29.Liu XD, Xu TT, Lei HX. 2017. Refuges and host shift pathways of host-specialized aphids Aphis gossypii. Sci Rep 7:2008. doi: 10.1038/s41598-017-02248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Zhang S, Luo JY, Wang CY, Lv LM, Cui JJ. 2016. Bacterial communities of the cotton aphid Aphis gossypii associated with Bt-cotton in northern China. Sci Rep 6:22958. doi: 10.1038/srep22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayoubi A, Talebi AA, Fathipour Y, Mehrabadi M. 2018. Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae). Insect Sci 27:86–98. doi: 10.1111/1744-7917.12603. [DOI] [PubMed] [Google Scholar]
- 32.Srivastava PN, Gao Y, Levesque J, Auclair JL. 1985. Differences in amino acid requirements between two biotypes of the pea aphid, Acyrthosiphon pisum. Can J Zool 63:603–606. doi: 10.1139/z85-087. [DOI] [Google Scholar]
- 33.Wang L, Zhang S, Luo JY, Wang CY, Lv LM, Zhu XZ, Li CH, Cui JJ. 2016. Identification of Aphis gossypii Glover (Hemiptera: Aphididae) biotypes from different host plants in north China. PLoS One 11:e0146345. doi: 10.1371/journal.pone.0146345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Zhang S, Luo J, Lv L, Wang C, Cui J. 2015. Host biotypes of cotton aphid Aphis gossypii Glover and preliminary analysis of the formation mechanism in Anyang region of China. Cotton Sci 27:372–378. [Google Scholar]
- 35.Liu XD, Zhang LJ, Zhang XX, Zhai BP. 2002. Studies on cotton aphid Aphis gossypii selectivity to host and its host-type. Acta Ecol Sin 22:1281–1285. [Google Scholar]
- 36.Zytynska SE, Preziosi RF. 2011. Genetic interactions influence host preference and performance in a plant-insect system. Evol Ecol 25:1321–1333. doi: 10.1007/s10682-011-9493-7. [DOI] [Google Scholar]
- 37.Douglas AE, Francois C, Minto LB. 2006. Facultative ‘secondary’ bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol Entomol 31:262–269. doi: 10.1111/j.1365-3032.2006.00516.x. [DOI] [Google Scholar]
- 38.Chandler SM, Wilkinson TL, Douglas AE. 2008. Impact of plant nutrients on the relationship between a herbivorous insect and its symbiotic bacteria. Proc Biol Sci 275:565–570. doi: 10.1098/rspb.2007.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner SM, Martinez AJ, Ruan Y-M, Kim KL, Lenhart PA, Dehnel AC, Oliver KM, White JA. 2015. Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol 29:1402–1410. doi: 10.1111/1365-2435.12459. [DOI] [Google Scholar]
- 40.Wulff JA, White JA. 2015. The endosymbiont Arsenophonus provides a general benefit to soybean aphid (Hemiptera: Aphididae) regardless of host plant resistance (Rag). Environ Entomol 44:574–581. doi: 10.1093/ee/nvv031. [DOI] [PubMed] [Google Scholar]
- 41.Darby AC, Choi J-H, Wilkes T, Hughes MA, Werren JH, Hurst GDD, Colbournet JK. 2010. Characteristics of the genome of Arsenophonus nasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol Biol 19(Suppl 1):75–89. doi: 10.1111/j.1365-2583.2009.00950.x. [DOI] [PubMed] [Google Scholar]
- 42.Pan R, Chen M, Yue L, Xing K, Li T, Kang K, Liang Z, Yuan L, Zhang W. 2018. A distinct strain of Arsenophonus symbiont decreases insecticide resistance in its insect host. PLoS Genet 14:e1007725. doi: 10.1371/journal.pgen.1007725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carletto J, Gueguen G, Fleury F, Vanlerberghe-Masutti F. 2008. Screening the bacterial endosymbiotic community of sap-feeding insects by terminal-restriction fragment length polymorphism analysis. Entomol Exp Appl 129:228–234. doi: 10.1111/j.1570-7458.2008.00760.x. [DOI] [Google Scholar]
- 44.Liu XD, Lei HX, Chen FF. 2019. Infection pattern and negative effects of a facultative endosymbiont on its insect host are environment-dependent. Sci Rep 9:4013. doi: 10.1038/s41598-019-40607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanlerberghe-Masutti F, Chavigny P, Fuller SJ. 1999. Characterization of microsatellite loci in the aphid species Aphis gossypii Glover. Mol Ecol 8:693–695. doi: 10.1046/j.1365-294x.1999.00876.x. [DOI] [PubMed] [Google Scholar]
- 46.Fukatsu T, Ishikawa H. 1996. Phylogenetic position of yeast-like symbiont of Hamiltonaphis styraci (Homoptera, Aphididae) based on 18S rDNA sequence. Insect Biochem Mol Biol 26:383–388. doi: 10.1016/0965-1748(95)00105-0. [DOI] [PubMed] [Google Scholar]
- 47.Fukatsu T, Tsuchida T, Nikoh N, Koga R. 2001. Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol 67:1284–1291. doi: 10.1128/AEM.67.3.1284-1291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandström JP, Russell JA, White JP, Moran NA. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228. doi: 10.1046/j.1365-294x.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 49.Wulff JA, Buckman KA, Wu K, Heimpel G, White JA. 2013. The endosymbiont Arsenophonus is widespread in soybean aphid, Aphis glycines, but does not provide protection from parasitoids or a fungal pathogen. PLoS One 8:e62145. doi: 10.1371/journal.pone.0062145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun ZJ, Li ZX. 2017. Host plants and obligate endosymbionts are not the sources for biosynthesis of the aphid alarm pheromone. Sci Rep 7:6041–6048. doi: 10.1038/s41598-017-06465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whelan JA, Russell NB, Whelan MA. 2003. A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods 278:261–269. doi: 10.1016/S0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]