Few studies pertaining to proteases in ruminant mycoplasmas have been reported. Here, we focus on proteases that are secreted outside the mycoplasma cell using a mass spectrometry approach. The most striking result is the identification, within the Mycoplasma mycoides cluster, of a serine protease that is exclusively detected outside the mycoplasma cells and is responsible for casein digestion. This protease may also be involved in the posttranslational processing of surface proteins, as suggested by analysis of mutants showing a marked reduction in the secretion of extracellular proteins. By analogy, this finding may help increase understanding of the mechanisms underlying this ectodomain shedding in other mycoplasma species. The gene encoding this protease is likely to have been acquired via horizontal gene transfer from Gram-positive bacteria and sortase-associated surface proteases. Whether this protease and the associated ectodomain shedding are related to virulence has yet to be ascertained.
KEYWORDS: Mycoplasma, posttranslational cleavage, exosecretion, proteases
ABSTRACT
Many mycoplasma species are isolated from the ruminant lungs as either saprophytes or true pathogens. These wall-less bacteria possess a minimal genome and reduced metabolic capabilities. Accordingly, they rely heavily on their hosts for the supply of essential metabolites and, notably, peptides. Seven of 13 ruminant lung-associated Mycoplasma (sub)species were shown to possess caseinolytic activity when grown in rich media and assessed with a quantitative fluorescence test. For some species, this activity was detected in spent medium, an indication that proteases were secreted outside the mycoplasma cells. To identify these proteases, we incubated concentrated washed cell pellets in a defined medium and analyzed the supernatants by tandem mass spectrometry. Secreted-protease activity was detected mostly in the species belonging to the Mycoplasma mycoides cluster (MMC) and, to a lesser extent, in Mycoplasma bovirhinis. Analyzing a Mycoplasma mycoides subsp. capri strain, chosen as a model, we identified 35 expressed proteases among 55 predicted coding genes, of which 5 were preferentially found in the supernatant. Serine protease S41, acquired by horizontal gene transfer, was responsible for the caseinolytic activity, as demonstrated by zymography and mutant analysis. In an M. capricolum mutant, inactivation of the S41 protease resulted in marked modification of the expression or secretion of 17 predicted surface-exposed proteins. This is an indication that the S41 protease could have a role in posttranslational cleavage of surface-exposed proteins and ectodomain shedding, whose physiological impacts still need to be explored.
IMPORTANCE Few studies pertaining to proteases in ruminant mycoplasmas have been reported. Here, we focus on proteases that are secreted outside the mycoplasma cell using a mass spectrometry approach. The most striking result is the identification, within the Mycoplasma mycoides cluster, of a serine protease that is exclusively detected outside the mycoplasma cells and is responsible for casein digestion. This protease may also be involved in the posttranslational processing of surface proteins, as suggested by analysis of mutants showing a marked reduction in the secretion of extracellular proteins. By analogy, this finding may help increase understanding of the mechanisms underlying this ectodomain shedding in other mycoplasma species. The gene encoding this protease is likely to have been acquired via horizontal gene transfer from Gram-positive bacteria and sortase-associated surface proteases. Whether this protease and the associated ectodomain shedding are related to virulence has yet to be ascertained.
INTRODUCTION
Bacteria belonging to the Mycoplasma genus can colonize many animal hosts. They are wall-less and have very small genomes, typically around 1,000 kbp, resulting from reductive evolution from low-G+C Firmicutes. Consequently, they depend on their host for the supply of cholesterol, amino acids, nucleotides, etc. They are often host specific, and many are pathogenic. Mycoplasmas colonizing ruminants are a good study model, as they comprise a huge diversity of species and include the type species of the genus, Mycoplasma mycoides subsp. mycoides. M. mycoides subsp. mycoides was the first mycoplasma to be isolated, in 1898 (1), and is the causative agent of contagious bovine pleuropneumonia, a disease notifiable to the World Organization for Animal Health (OIE). Like many other ruminant mycoplasmas, M. mycoides subsp. mycoides shows marked tissue tropism toward the respiratory tract, where it induces severe lesions. It therefore came somewhat as a surprise that no obvious virulence factors were identified when the entire M. mycoides subsp. mycoides genome was sequenced (2).
A decade after the genome was sequenced, Browning et al. illustrated that the complexity of mycoplasma pathogenesis is “predominantly attributable to the immunopathological response of the host to the persistence of these pathogens” (3). This suggested that any gene that is involved in optimal adhesion, efficient nutriment scavenging, immune evasion, or immunomodulation and that is not required for in vitro growth might be involved in virulence (3). In this general picture, H2O2 production was a notable exception, as it corresponds to one of the few cases of production of cytotoxic compounds by mycoplasmas (4). However, H2O2 may not be indispensable for strain virulence (5). Until recently, mycoplasma virulence studies have focused mainly on interactions between the surface of the bacterium and its host. It was clear that mycoplasma immunopathology was linked to an imbalanced immunological response leading to exacerbated inflammation. Extensive work was performed as early as 1971 (6) and recently (7) to attempt to decipher the immune responses of the hosts. However, there is also a body of work focusing on mycoplasma cell-associated pathogenesis. Mycoplasma bovis, for example, is able to display a wide array of variable surface antigens coded by variable surface lipoproteins (Vsps) (8). The in vivo variability of Vsps, together with immunological factors of the host, may contribute to mycoplasma persistence and immunomodulation (9, 10).
More recently, targeted proteolysis of surface antigens, coupled with variable cleavage efficiency, was identified as another mechanism participating in the diversification of surface-exposed antigens (11). In the porcine respiratory pathogen Mycoplasma hyopneumoniae, posttranslational processing is an important mechanism for creating cell surface diversity (12), as 35 surface-associated proteins were shown to be subject to endoproteolytic cleavage. These modifications, affecting adhesins, lipoproteins, or moonlighting proteins, are likely to expand the mycoplasma antigen repertoire of the mycoplasma cell surface. Proteolytic cleavage often enhances the binding of host molecules such as plasminogen, whose conversion into the serine protease plasmin may have an important impact on host tissues and immune effector molecules (13). M. mycoides subsp. mycoides and many other mycoplasma species also express a “mycoplasma immunoglobulin protease,” together with a mycoplasma immunoglobulin binding protein (14). This two-protein system allows the cleavage of host immunoglobulins and may therefore play a key role in immune evasion by mycoplasmas. Proteolysis obviously plays an important role in the natural history of mycoplasma species. This has notably been studied in the porcine pathogen M. hyopneumoniae, in which endoproteases are responsible for ectodomain shedding, with profound impact on host-pathogen interactions. A surface protein, P159, is cleaved, and this processing event generates new surface protein diversity, as well as functional redundancy in glycosaminoglycan and cilium binding (15). In addition, aminopeptidase activity has been evidenced at the surface of this pathogen (16, 17).
Fewer data are available for other lung-pathogenic mycoplasmas. The aim of this study was to determine which proteases may be expressed in mycoplasmas colonizing ruminant lungs and if those proteases may be secreted outside the mycoplasma cell as part of an exoproteome. Naturally, this study was beset with the same difficulties encountered in studying extracellular exopolysaccharides (18), i.e., the need for rich and complex growth media to ensure mycoplasma growth. A similar approach was therefore adopted, based on the incubation of washed, concentrated mycoplasma cells into a defined medium allowing these mycoplasma cells to maintain their metabolism for some time. Secreted exoproteins were then analyzed by phenotypic tests such as by assays of digestion of skimmed milk and fluorescent casein and by casein zymography as well as by tandem mass spectrometry (MS-MS). The main involvement of an extracellular protease was finally confirmed by the use of mutant strains from two species in which the mutation occurred in orthologous genes and for which the exported caseinolytic activity was abolished.
RESULTS
Protease activity of mollicutes found in ruminant lungs.
The global peptidase activity of mollicutes found in the respiratory tract of ruminants was first estimated by a quantitative approach using whole cultures in complex medium and fluorescently labeled casein as a universal substrate. The caseinolytic activity of 28 Mycoplasma and 2 Acholeplasma strains, corresponding to species usually isolated from ruminant lungs (Table 1), was assessed using two independent stationary-phase cultures in modified Hayflick’s medium (m-Hayflick). There was a high heterogeneity of results in comparisons of one species to another, while the results obtained with different strains within a (sub)species were usually homogeneous, with the notable exception of Mycoplasma capricolum subsp. capripneumoniae, M. putrefaciens, M. ovipneumoniae, and M. bovirhinis. The highest levels of caseinolytic activities were measured for members of the Mycoplasma mycoides cluster (MMC), with relative activity (RA) values ranging from 47% to 95%. M. leachii and M. mycoides subsp. capri strains yielded the highest values. M. ovipneumoniae and M. bovirhinis strains presented values within this high range, though certain strains and culture replicates showed lower values. In contrast, M. putrefaciens, which is phylogenetically closely related to the Mycoplasma mycoides cluster, displayed very low RA values, comparable to those of distantly related species such as M. arginini and M. alkalescens (RA, 4% to 29%). All other species provided intermediate values ranging from 23% to 74%. It is noteworthy that M. agalactiae showed lower values (RA, 23% to 30%) than its close relative M. bovis (RA, 41% to 49%). Strains from seven (sub)species, namely, M. capricolum subsp. capripneumoniae, M. capricolum subsp. capricolum, M. mycoides subsp. mycoides, M. mycoides subsp. capri, M. ovipneumoniae, M. bovirhinis, and M. bovis, displaying high overall activity, were selected for analysis of extracellular proteolytic activity. All these species displayed tropism toward ruminant lungs. M. arginini, which showed low overall caseinolytic activity, was selected as a negative control. In spite of their high caseinolytic activity, M. leachii strains were not included in subsequent analyses because current clinical cases are scarce and most often associated with polyarthritis rather than with pneumonia.
TABLE 1.
Caseinolytic activity in mycoplasma cultures from species that can be isolated from ruminant lungsa
| Phylogenetic group |
Species and subspecies | Main host |
Disease/clinical signs | Strain | Isolation yr |
Country of origin or source (reference) |
% overall caseinolytic activityb |
|
|---|---|---|---|---|---|---|---|---|
| Assay 1 | Assay 2 | |||||||
| Spiroplasma | Mycoplasma capricolum subsp. capripneumoniae | Goat | Contagious caprine pleuropneumonia | 16125 | 2016 | United Arab Emirates | 85 | 63 |
| 9231 Abomsa | 1982 | Ethiopia | 47 | 58 | ||||
| Mycoplasma capricolum subsp. capricolum | Goat | Contagious agalactia | CkT | 1955 | United States | 82 | 90 | |
| Ck-mut | 2010 | Allam et al. (22) | 40 | 45 | ||||
| 94157 | 1994 | Ethiopia | 87 | 79 | ||||
| Mycoplasma leachii | Cattle | Pneumonia | PG50T | 1963 | Australia | 86 | 91 | |
| ML06049 | 2005 | Nigeria | 95 | 95 | ||||
| Mycoplasma mycoides subsp. mycoides | Cattle | Contagious bovine pleuropneumonia | Gladysdale | 1953 | Australia | 64 | 71 | |
| 16113 | 2016 | Namibia | 64 | 72 | ||||
| Rita | 1987 | Cameroon | 60 | 68 | ||||
| Rita-mut | 2006 | Unpublished data from CIRAD | 22 | 26 | ||||
| PG1T | <1931 | NK | 48 | 59 | ||||
| Mycoplasma mycoides subsp. capri | Goat | Contagious agalactia | GM12 | 1983 | United States | 92 | 89 | |
| 95010 | 1995 | France | 75 | 75 | ||||
| Mycoplasma putrefaciens | Goat | Contagious agalactia | KS1T | ∼1954 | United States | 12 | 4 | |
| 9231b | 1992 | France | 11 | 29 | ||||
| Hominis | Mycoplasma alkalescens | Cattle | Pneumonia arthritis mastitis otitis | PG51T | 1961 | Australia | 7 | 4 |
| 5561 | 2005 | Nigeria | 8 | 5 | ||||
| Mycoplasma arginini | Ruminants | Opportunistic | Pontaumur | ∼1970 | France | 20 | 17 | |
| Tizi ouzou | ∼1970 | Algeria | 13 | 12 | ||||
| Mycoplasma ovipneumoniae | Sheep | Pneumonia conjunctivitis mastitis | 9139-2/90 | 1991 | Ethiopia | 31 | 35 | |
| Y98T | ∼1971 | Australia | 63 | 59 | ||||
| 14811 | 2007 | France | 74 | 74 | ||||
| Mycoplasma agalactiae | Goat | Contagious agalactiae | PG2T | 1952 | Spain | 24 | 30 | |
| 94093-5633 | 1991 | France | 30 | 23 | ||||
| Mycoplasma bovis | Cattle | Pneumonia mastitis | PG45T | 1962 | United States | 49 | 41 | |
| Oger2 | 1975 | France | 45 | 45 | ||||
| M. bovirhinis | Cattle | Commensal | MV5 | ∼1970 | France | 49 | 69 | |
| PG43 | 1965 | England | 57 | 32 | ||||
| F11513 | 2017 | France | 58 | 53 | ||||
| Phytoplasma/acholeplasma | Acholeplasma laidlawii | Environment | Commensal | PG9T | <1963 | NK | 30 | 31 |
| PG8 | <1963 | NK | 27 | 30 | ||||
A superscript “T” indicates the type strain. NK, not known. Data corresponding to species belonging to the Mycoplasma mycoides cluster are indicated with boldface. Mutant strain data are boldface and underlined.
Overall caseinolytic activities are expressed as percent activity based on the logarithmic values corresponding to relative fluorescence unit (RFU) data measured in whole cultures in modified Hayflick’s medium.
Assessment of extracellular proteolytic activity.
The extracellular caseinolytic activity of a strain from each of the 8 selected (sub)species was assessed on milk plates, where proteases diffusing in the agar generate a translucent ring around the culture (Fig. 1). A wide area of milk digestion was observed for M. mycoides subsp. capri and M. capricolum subsp. capricolum strains, although the appearance differed between the two species. For M. mycoides subsp. capri strains, there was a sharp zone where the milk was completely digested and a wider zone with diffuse digestion. For all other strains, the digestion zone was diffuse with various diameters. M. capricolum subsp. capripneumoniae strains displayed a conspicuous zone of digestion despite the cultures on solid media being barely visible, while M. mycoides subsp. mycoides and M. bovirhinis strains induced only weak digestion. M. bovis and M. ovipneumoniae strains, as well as the negative-control strain of M. arginini, did not display any detectable activity (data not shown). This phenotypic test clearly suggests that M. mycoides subsp. capri, M. capricolum subsp. capricolum, M. capricolum subsp. capripneumoniae, and, to a much lesser extent, M. mycoides subsp. mycoides and M. bovirhinis, are all able to produce extracellular caseinolytic proteases released from the cell into the environment whereas M. ovipneumoniae and M. bovis are not. The extracellular caseinolytic activity of these strains was then confirmed by performing quantitative analysis using the fluorescently labeled casein assay in culture supernatants. The assay was first applied to cell pellets and supernatants from stationary-phase cultures in m-Hayflick medium (Table 2). The supernatants were filtered through 0.1-μm pores, and no living mycoplasma cells were found in the sample. All strains belonging to the Mycoplasma mycoides cluster (Table 1) exhibited higher caseinolytic activities in the supernatant than in the cell pellet. The opposite was found for the four other strains tested, including M. bovirhinis, which showed higher activities in the pellet. It is noteworthy that for M. ovipneumoniae and M. arginini, the activity was exclusively detected in the cell pellet. Under these experimental conditions, only the Mycoplasma mycoides cluster strains seemed to secrete proteases into the culture supernatant.
FIG 1.
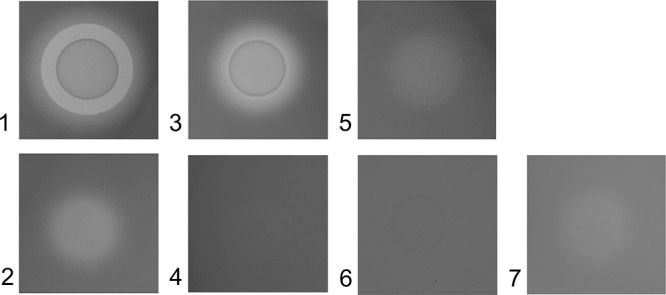
Casein digestion on an agar plate (modified Hayflick’s medium) supplemented with 0.4% (wt/vol) milk. An area of casein digestion was observed for strains producing extracellular caseinolytic proteases. The strongest activity was observed for M. mycoides subsp. capri strain 95010 (panel 1), while a digested zone was evidenced for M. capricolum subsp. capripneumoniae Abomsa, although no conspicuous growth was observed for that fastidious strain (panel 2). The activity observed for M. capricolum subsp. capricolum Ck strain (panel 3) was completely abolished in the Ck-mut strain (panel 4) although its culture was clearly visible on the agar surface. Lower activity was evidenced for M. mycoides subsp. mycoides Rita (panel 5), and no activity was recorded for the mutated Rita-mut strain (panel 6). Slight activity was recorded for M. bovirhinis MV5 (panel 7).
TABLE 2.
Caseinolytic activities of selected mycoplasma strains in modified Hayflick’s or Opti-MEM medium, in pellet or supernatant
| Phylogenetic group | Species and subspecies |
Straina | Modified Hayflick’s mediumb |
Opti-MEM mediumc |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % overall caseinolytic activityd |
% supernatant activityd |
Cellular activityd | Casein digestion on agar platese |
Overall caseinolytic activityd |
Supernatant activityd |
Cellular activityd |
T0 titerf |
T16 titerf |
||||
| Assay 1 |
Assay 2 |
|||||||||||
| Spiroplasma (Mycoplasma mycoides cluster) | M. capricolum subsp. capripneumoniae | 9231 Abomsa | 47 | 58 | 46 | 22 | + | 38 | 29 | −3 | 9.3 | 9.3 |
| M. capricolum subsp. capricolum | CkT | 82 | 90 | 85 | 43 | + | 90 | 69 | 24 | 9.9 | 10.0 | |
| Ck-mut | 40 | 45 | 13 | 19 | − | 25 | 13 | −1 | 9.0 | 9.3 | ||
| 94157 | 87 | 79 | 59 | 41 | ++ | 94 | 83 | 30 | 9.3 | 9.6 | ||
| M. mycoides subsp. mycoides | 16113 | 64 | 72 | 54 | 26 | + | 27 | 1 | 2 | 8.7 | 9.8 | |
| 8740-Rita | 60 | 68 | 56 | 30 | + | 11 | 3 | −1 | 8.8 | 8.8 | ||
| Rita-mut | 22 | 26 | 5 | 24 | − | 7 | −8 | 6 | 8.5 | 8.7 | ||
| M. mycoides subsp. capri | 95010 | 75 | 75 | 77 | 54 | ++ | 99 | 97 | 23 | 9.3 | 9.3 | |
| Hominis | M. arginini | Tizi Ouzou | 13 | 12 | 2 | 29 | − | 13 | −3 | 9 | 9.8 | 5.6 |
| M. ovipneumoniae | Y98T | 63 | 59 | 0 | 26 | − | 40 | −5 | 9 | 8.9 | 6.7 | |
| M. bovis | Oger2 | 45 | 45 | 9 | 48 | − | 25 | 3 | 2 | 8.9 | 7.6 | |
| M. bovirhinis | MV5 | 49 | 69 | 29 | 76 | + | 50 | 49 | 20 | 9.6 | 9.6 | |
Strains chosen for extracellular activity research. Mutant strain names are in bold and underlined. Ck: California Kid.
Culture in modified Hayflick’s medium. Mutant data are in bold and underlined.
Culture in supplemented Opti-MEM medium. Mutant data are in bold and underlined.
Activity data are expressed in relative activity percentages.
Casein digestion on milk agar plates was evaluated by measurement of the translucent area around bacterial spots and expressed in arbitrary units based on translucent area observations. −, no translucent area; +, blurred and small translucent area; ++, sharp and large translucent area; see Fig. S2.
Titer (in log10 CFU per milliliter) at 0 h (T0) and after 16 h of incubation (T16) in supplemented Opti-MEM medium.
Extracellular caseinolytic activity in defined medium.
Studies aiming at the characterization of extracellular components of mycoplasma cultures require the use of chemically defined media devoid of the complex supplements such as horse serum and yeast extract that are necessary for mycoplasma growth (18, 19). Here, a medium devoid of uncharacterized peptides, Opti-MEM, might facilitate extracellular protease identification. This medium was able to maintain mycoplasma viability during 16 h for the majority of the species tested, except for M. arginini, M. ovipneumoniae, and M. bovis (Table 2). Within the Mycoplasma mycoides cluster, M. capricolum subsp. capricolum and M. mycoides subsp. capri still exhibited a very high level of caseinolytic activity in Opti-MEM supernatant, while the activity of M. capricolum subsp. capripneumoniae declined and that of M. mycoides subsp. mycoides completely disappeared. This was not due to a loss of viability, as the titers in cell suspensions remained stable until supernatant collection. In contrast to the observations made in rich medium, the caseinolytic activity of M. bovirhinis was higher in the supernatant after incubation in a chemically defined, serum-free medium. As in complex medium, none or negligible caseinolytic activity was detected for M. arginini, M. bovis, and M. ovipneumoniae supernatants in Opti-MEM (Table 2). However, the titers of viable cells in the Opti-MEM suspensions dropped sharply for those three species.
Identification of genes potentially coding for proteases in Mycoplasma mycoides subsp. capri and Mycoplasma bovirhinis species and tandem mass spectrometry analysis.
As the greatest caseinolytic activity in culture supernatants was found in M. mycoides subsp. capri strains, the genome of strain 95010 was chosen as a model for the Mycoplasma mycoides cluster. Within this genome, 55 genes are predicted to potentially code for proteases (Table 3). Thirty-six were retrieved from the MEROPS database, while 19 additional genes were identified through MaGe data mining. In the closely related subspecies M. mycoides subsp. mycoides, this number was reduced to 36, as orthologues were either absent or present in the form of pseudogenes (see Table S1B in the supplemental material).
TABLE 3.
Predicted protease-coding genes and tandem mass spectrometry detection of proteases for M. mycoides subsp. capri (strain 95010)a
| Protein accession no. |
Ratio × 10,000 (spectral count) |
Fold change |
Mnemonic | Gene name |
Annotation (MEROPS and MaGe) | |
|---|---|---|---|---|---|---|
| Pellet (26,893) |
Supernatant (11,964) |
|||||
| CBW53764.1 | 50 | 2 | 0.0 | MLC_0360 | ftsH | FtsH-2 peptidase |
| CBW53797.1 | 0 | 0 | MLC_0690 | Family S9 unassigned peptidases | ||
| CBW53831.1 | 1 | 1 | MLC_1030 | Subfamily S41A nonpeptidase homologues | ||
| CBW53842.1 | 1 | 0 | MLC_1140 | Family S9 unassigned peptidases | ||
| CBW53849.1 | 42 | 14 | 0.3 | MLC_1210 | pyrG | CTP synthetase |
| CBW53854.1 | 4 | 0 | 0.0 | MLC_1260 | Subfamily S8A unassigned peptidases | |
| CBW53874.1 | 141 | 317 | 0.2 | MLC_1460 | pepA | Family M17 unassigned peptidases |
| CBW53913.1 | 56 | 37 | 0.6 | MLC_1850 | pepF | Oligopeptidase F |
| CBW53915.1 | 84 | 195 | 2.3 | MLC_1870 | pepA | Family M17 unassigned peptidases |
| CBW53985.1 | 0 | 397 | 397.0 | MLC_2570 | Subfamily S41A nonpeptidase homologues | |
| CBW54055.1 | 0 | 38 | 38.4 | MLC_3270 | Subfamily S41A nonpeptidase homologues | |
| CBW54056.1 | 3 | 53 | 13.2 | MLC_3280 | Subfamily S41A nonpeptidase homologues | |
| CBW54067.1 | 45 | 165 | 3.6 | MLC_3390 | pepQ | Subfamily M24B unassigned peptidases |
| CBW54074.1 | 0 | 0 | MLC_3460 | Subfamily C1A unassigned peptidases | ||
| CBW54082.1 | 0 | 2 | MLC_3540 | lip1 | Family S33 unassigned peptidases | |
| CBW54162.1 | 49 | 16 | 0.3 | MLC_4340 | lon | Lon-A peptidase |
| CBW54168.1 | 5 | 16 | 2.7 | MLC_4400 | Family C56 nonpeptidase homologues | |
| CBW54169.1 | 0 | 0 | MLC_4410 | abc | Family C39 unassigned peptidases | |
| CBW54212.1 | 40 | 18 | 0.5 | MLC_4840 | pepO | Family M13 unassigned peptidases |
| CBW54233.1 | 50 | 29 | 0.6 | MLC_5050 | lip2 | Family S33 unassigned peptidases |
| CBW54234.1 | 15 | 0 | 0.0 | MLC_5060 | lip2 | Family S33 unassigned peptidases |
| CBW54235.1 | 14 | 16 | 1.1 | MLC_5070 | lip3 | Family S33 unassigned peptidases |
| CBW54242.1 | 12 | 3 | 0.2 | MLC_5140 | nagA | Family M38 nonpeptidase homologues |
| CBW54257.1 | 0 | 0 | MLC_5290 | Subfamily C1A unassigned peptidases | ||
| CBW54260.1 | 35 | 66 | 1.8 | MLC_5320 | pepV | Peptidase V |
| CBW54267.1 | 0 | 0 | MLC_5390 | Family C108 unassigned peptidases | ||
| CBW54280.1 | 0 | 0 | MLC_5520 | lspA | Family A8 nonpeptidase homologues | |
| CBW54282.1 | 1 | 0 | MLC_5540 | pepD | Subfamily S9C unassigned peptidases | |
| CBW54326.1 | 2 | 0 | MLC_5980 | Subfamily S8A unassigned peptidases | ||
| CBW54404.1 | 6 | 21 | 2.9 | MLC_6750 | map | Subfamily M24A unassigned peptidases |
| CBW54445.1 | 0 | 0 | MLC_7150 | Family M79 unassigned peptidases | ||
| CBW54490.1 | 7 | 0 | 0.0 | MLC_7600 | pldB | Family S33 unassigned peptidases |
| CBW54520.1 | 6 | 14 | 1.9 | MLC_7900 | Esterase EstB | |
| CBW54631.1 | 0 | 0 | MLC_9010 | Subfamily M23B nonpeptidase homologues | ||
| CBW54632.1 | 0 | 1 | MLC_9020 | Subfamily M23B nonpeptidase homologues | ||
| CBW54642.1 | 3 | 0 | 0.0 | MLC_9120 | Subfamily S8A unassigned peptidases | |
| CBW53777.1 | 22 | 29 | 1.3 | MLC_0490 | Putative peptidase DUF31 | |
| CBW53798.1 | 3 | 0 | 0.0 | MLC_0700 | O-Sialoglycoprotein endopeptidase | |
| CBW53816.1 | 0 | 0 | MLC_0880 | pepQ | Proline dipeptidase | |
| CBW53878.1 | 0 | 0 | MLC_1500 | Papain-like cysteine peptidase superfamily | ||
| CBW53944.1 | 2 | 1 | MLC_2160 | Peptidase_M78 | ||
| CBW53958.1 | 3 | 3 | 0.8 | MLC_2300 | Inactive homologue of metal-dependent proteases | |
| CBW53994.1 | 1 | 10 | 5.8 | MLC_2660 | Putative peptidase DUF31 | |
| CBW54029.1 | 0 | 0 | MLC_3010 | Peptidase_M78 | ||
| CBW54081.1 | 1 | 8 | 4.8 | MLC_3530 | Papain-like cysteine peptidase superfamily | |
| CBW54164.1 | 1 | 1 | MLC_4360 | Papain-like cysteine peptidase superfamily | ||
| CBW54165.1 | 12 | 12 | 0.9 | MLC_4370 | Papain-like cysteine peptidase superfamily | |
| CBW54170.1 | 1 | 0 | MLC_4420 | Metalloprotease catalytic domain superfamily, predicted | ||
| CBW54304.1 | 30 | 10 | 0.3 | MLC_5760 | clpB | ATP-dependent Clp protease ATP binding subunit |
| CBW54318.1 | 13 | 11 | 0.8 | MLC_5900 | Putative peptidase DUF31; Mycoplasma IgG protease | |
| CBW54320.1 | 18 | 44 | 2.4 | MLC_5920 | Putative peptidase DUF31; Mycoplasma IgG protease | |
| CBW54322.1 | 4 | 0 | 0.0 | MLC_5940 | Putative peptidase DUF31; Mycoplasma IgG protease | |
| CBW54324.1 | 7 | 14 | 1.8 | MLC_5960 | Putative peptidase DUF31; Mycoplasma IgG protease | |
| CBW54539.1 | 1 | 1 | MLC_8090 | Putative peptidase DUF31 | ||
| CBW54606.1 | 0 | 0 | MLC_8760 | Zinc metalloprotease | ||
Mnemonics of genes which were found to be consistently expressed in either the pellet or the supernatant (proportion, >0.001) are indicated with boldface. Proteases whose proportion was significantly higher in the supernatant are indicated with boldface and underlining.
Similarly, the number of protease-coding genes was drastically reduced for M. capricolum subsp. capripneumoniae (n = 35) compared to M. capricolum subsp. capricolum (n = 44) (Table S1A). Only 39 putative protease genes were predicted in the M. bovirhinis genome (Table S1C). Thus far, our experimental approach was based on phenotypic detection of protease activity, more specifically, of caseinolytic activity. For the sake of completeness, we also developed a general proteomic approach consisting of the characterization of all secreted proteases detected in Opti-MEM supernatants. For this purpose, mycoplasma cell suspensions were incubated as described above in Opti-MEM for 16 h before being harvested as two separated fractions, i.e., the cell pellet and the supernatant. Both fractions were subjected to MS-MS analysis. Of the 55 putative proteases from M. mycoides subsp. capri 95010 that were identified after genome mining, 35 were detected by tandem mass spectrometry analysis either in the pellet or in the supernatant (Table 3), whereas 18 of the 39 predicted were detected in M. bovirhinis MV5 samples (Table S1C).
FtsH, which is a membrane-bound, cytoplasm-oriented, energy-dependent AAA-positive (AAA+) protease (20), was used as a control. This protease is a universally conserved protein with well-established localization in the cytoplasm, presenting two hydrophobic domains anchoring it to the membrane. Its ratio within the pellet samples was quite stable, whichever mycoplasma strain was studied, with a mean of 6.7 × 10−3 (n = 15; minimum = 4.5 × 10−3; maximum = 8.5 × 10−3) and a supernatant/pellet fold change level which ranged from 0.04 to 0.7, an indication that FtsH was detected mostly in the cell pellet. This preliminary analysis confirmed that our procedure for sample preparation did not induce a noticeable level of release of membrane fragments in the supernatant attributable due to cell lysis or vesicle formation (21). In contrast, a number of proteases were highly expressed and detected both in the pellet and in the supernatant (0.5-fold to 2.5-fold change). This was notably the case for endopeptidases such as PepA (1.2 to 2.9), PepF (0.6 to 2.1), and PepV (1.8 to 3.3).
Proteases located preferentially in the supernatant.
To detect which proteases were significantly overrepresented in the supernatant, a supernatant-versus-pellet fold change distribution analysis was performed for each of the strains. This allowed detection of which fold change values could be considered “outlier” values compared to the “normal curve” values of the distribution (see Fig. S1 in the supplemental material). Most supernatant/pellet fold change values were located within the first two intervals (0 to 0.5 and 0.5 to 1), which corresponded to proteins detected mostly in the pellet sample. For values above 1, the curve had a negative exponential shape up to a fold change value of 5, which was considered the outlier limit for that example. The curves for all other strains had similar shapes, but the upper limits differed slightly (data not shown). This analysis allowed the detection of proteases that were preferentially overrepresented in supernatant samples from M. mycoides subsp. capri 95010 (Table 3) as well as the other members of the Mycoplasma mycoides cluster and M. bovirhinis (Table S1C).
The most striking one was a serine protease (GenBank accession no. CBW53985.1 and its orthologues, namely, MLC_2570, MCAP_0240, and MCCP01_0297), which was detected almost exclusively in the supernatants (fold change, 397) and not in the pellets from all species of Mycoplasma mycoides cluster strains, with the exception of M. mycoides subsp. mycoides, in which it was not detected. Another one (CBW54056.1; MLC_3280), a predicted lipoprotein with a S41 protease domain, was also detected in 4 of 6 Mycoplasma mycoides cluster strains, but with lower fold change values (2 to 13). Various other proteases were overrepresented in the supernatant of Mycoplasma mycoides cluster strains. For M. capricolum subsp. capricolum type strain California kid (Ck), two S41 proteases were detected, namely, MCAP_0240, which is orthologous to M. mycoides subsp. capri MLC_2570, and MCAP_0329, which is orthologous to CBW54056.1 (MLC_3280) Mycoplasma mycoides subsp. capri strain 95010. The latter was equally extensively detected in the M. capricolum subsp. capricolum Ck mutant, suggesting that MCAP_0329 is not involved in the extracellular caseinolytic activity of M. capricolum subsp. capricolum. The four proteases that were overrepresented in the supernatant of M. bovirhinis strain MV5 displayed a DUF31 domain and were analogues of the Mycoplasma IgG protease (MIP) evidenced in M. mycoides subsp. capri (14). These proteases were also detected in the M. mycoides subsp. capri 95010 strain, both in the pellet and in the supernatant, but they were not found overrepresented in the supernatant of Mycoplasma mycoides cluster strains.
These results fully confirm the data obtained by phenotypic detection, which resulted in the identification of the two main proteases, corresponding to genes MLC_2570 and MBVR141_0224, in M. mycoides subsp. capri and M. bovirhinis, respectively. We were not able to detect any overrepresented proteases in M. mycoides subsp. mycoides strains (Table S1B). Some proteases were indeed detected in the supernatant but with fold change values that could not be considered to represent deviations from the standard distribution. In contrast, all the other subspecies and strains of the Mycoplasma mycoides cluster displayed proteases preferentially secreted into the supernatant.
Identification of caseinolytic proteases by zymography.
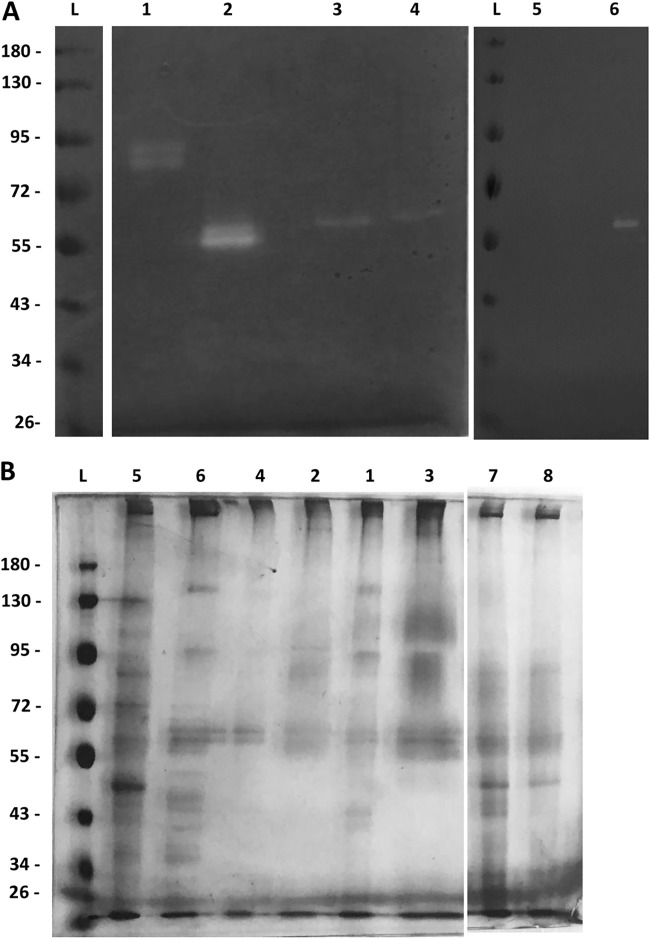
Culture supernatants obtained after incubation in Opti-MEM were concentrated by lyophilization and analyzed by casein zymography (Fig. 2). A single band of digestion representing an estimated molecular weight of 55 kDa was observed for M. capricolum subsp. capricolum and M. capricolum subsp. capripneumoniae, while two bands were observed for M. mycoides subsp. capri, with one also estimated at 55 kDa and the other estimated at 52 kDa. For M. mycoides subsp. mycoides Rita, one of the zymograms showed a faint band at 55 kDa (data not shown) but the band was not reproducible and thus was not taken into account for further analysis. Tandem mass spectrometry detected specific peptides from orthologues of MLC_2570 peptidase S41 in all the excised bands (Table 4) from strains of the Mycoplasma mycoides cluster. For M. mycoides subsp. capri, MLC_2570 represented the majority of spectral counts for both the 55-kDa and 52-kDa bands, while few peptides corresponding to other proteases were also detected (MLC_1460 and MLC_3270). There was a marked difference between the actual size of the zymography band, 55 kDa, and the predicted molecular weight of MLC_2570, 74.5 kDa, which suggests proteolytic cleavage. The estimated molecular weight of a protein encompassing all amino acids between the detected N-terminal and C-terminal peptides is 53.5 kDa (Fig. 3). This indicates that the actual cleavage sites may be located very close to the peptides detected at the extremities. For M. capricolum subsp. capripneumoniae, MCCP01_0297 was the only protease detected in the excised band, and few peptides were detected for it. In the case of M. capricolum subsp. capricolum, the majority of the peptides detected in the sliced 55-kDa band corresponded to another S41 protease, MCAP_0329. However, MCAP_0240, the orthologue of MLC_2570, was also detected. As for M. bovirhinis MV5, the two bands of 85 and 80 kDa contained specific peptides corresponding to MBVR141_0224, a putative DUF31 peptidase.
FIG 2.
Detection of caseinolytic activity by zymography. Supernatant from mycoplasmas was incubated in supplemented Opti-MEM. (A) Casein zymogram. (B) SDS-PAGE. Lanes L, Ladder PageRuler (top to bottom, 180, 130, 100, 70, 55, 40, 35, and 25 kDa); lane 1, M. bovirhinis MV5; lanes 2, M. mycoides subsp. capri 95010; lanes 3, M. capricolum subsp. capripneumoniae Abomsa; lane, 4, M. capricolum subsp. capricolum 94157; lanes 5, M. capricolum subsp. capricolum Ck-mut; lanes 6, M. capricolum subsp. capricolum Ck; lane 7, M. mycoides subsp. mycoides Rita; lane 8, M. mycoides subsp. mycoides Rita-mut. The discolored bands, showing caseinolytic activity, were cut and then analyzed by tandem mass spectrometry. The zymography picture (panel A) has been spliced to remove the lanes with samples which did not yield any digested bands (M. arginini, M. bovis, M. ovipneumoniae, and M. mycoides subsp. mycoides).
TABLE 4.
Proteins identified by tandem mass spectrometry from caseinolytic zymogram bandsa
| Strain | Mnemonic | Protein accession no. |
Annotation | MW (kDa) | Spectral count |
|||
|---|---|---|---|---|---|---|---|---|
| 55 kDa | 52 kDa | 85 kDa | 80 kDa | |||||
| M. mycoides subsp. capri 95010 | MLC_2570 | CBW53985.1 | CHP, predicted TMB protein and tail specific protease | 74.5 | 35 | 56 | ||
| CBW54005.1 | Pyruvate kinase | 53.6 | 9 | 5 | ||||
| CBW54529.1 | ATP synthase alpha chain | 58.1 | 6 | 2 | ||||
| MLC_1460 | CBW53874.1 | Leucyl aminopeptidase | 49.8 | 0 | 4 | |||
| MLC_3270 | CBW54055.1 | CHP predicted transmembrane protein, peptidase S41 | 78.3 | 4 | 0 | |||
| CBW54480.1 | Phosphoglucomutase | 63.8 | 2 | 0 | ||||
| M. capricolum subsp. capricolum Ckid | MCAP_0329 | ABC01488.1 | lpp, C-terminal processing peptidase family S41 | 71.7 | 15 | |||
| MCAP_0240 | ABC01466.1 | Membrane protein, peptidase S41 | 75.5 | 7 | ||||
| ABC01270.1 | Pyruvate kinase | 53.6 | 4 | |||||
| ABC01646.1 | Membrane protein, putative | 206.8 | 4 | |||||
| MCAP_0328 | ABC01790.1 | Membrane protein, peptidase S41 | 78.9 | 4 | ||||
| ABC01474.1 | Arginine deiminase | 46.6 | 3 | |||||
| ABC01292.1 | ATP synthase F1, alpha subunit | 58.0 | 2 | |||||
| M. capricolum subsp. capripneumoniae Abomsa | CDZ17831.1 | lpp, ABC transporter substrate-binding protein | 60.1 | 12 | ||||
| CDZ18073.1 | Dihydrolipoyllysine-residue acetyltransferase | 46.8 | 10 | |||||
| CDZ17832.1 | ABC transporter, ATP binding component | 59.4 | 8 | |||||
| CDZ17891.1 | ATP synthase (subunit alpha, component F1) | 58.1 | 4 | |||||
| MCCP01_0297 | CDZ18087.1 | Putative conserved membrane protein, peptidase S41 | 75.5 | 4 | ||||
| M. bovirhinis MV5 | MBVR141_0224 | BBA22185.1 | HP, lpp, putative peptidase (DUF31) | 104.0 | 16 | 70 | ||
| BBA22123.1 | Hypothetical protein | 87.7 | 68 | 47 | ||||
| BBA22285.1 | Hypothetical protein | 87.2 | 34 | 0 | ||||
| BBA22436.1 | Hypothetical protein | 98.5 | 31 | 0 | ||||
| BBA22541.1 | Surface protein | 80.9 | 26 | 28 | ||||
| BBA22061.1 | Hypothetical protein | 106.0 | 15 | 6 | ||||
| BBA22174.1 | Elongation factor G | 77.0 | 11 | 0 | ||||
| BBA22209.1 | Phosphoketolase | 91.5 | 7 | 0 | ||||
| MBVR141_0761 | BBA22491.1 | HP, lpp, putative peptidase (DUF31) | 93.9 | 5 | 5 | |||
| MBVR141_0284 | BBA22211.1 | HP, putative peptidase (DUF31) | 88.1 | 4 | 0 | |||
| BBA22472.1 | Membrane protein | 90.2 | 4 | 1 | ||||
| BBA22169.1 | Hypothetical protein | 120.0 | 2 | 2 | ||||
Protein accession numbers with boldface correspond to predicted proteases. Mnemonic designations with boldface and underlining correspond to the deduced gene coding for the caseinolytic protease. CHP, conserved hypothetical protein; DUF, domain of unknown function; HP, hypothetical protein; lpp, lipoprotein; TMB, transmembrane region; MW, molecular weight.
FIG 3.
Identification of peptides detected by tandem mass spectrometry for the predicted S41 protease (CBW53985.1, MLC_2570) of M. mycoides subsp. capri strain 95010. The N-terminal and C-terminal transmembrane regions are boxed. The predicted S41 superfamily domain is represented in bold with a boxed serine active site. The specific peptides detected by tandem mass spectrometry in the concentrated supernatant are underlined.
Assessment of the extracellular protease activity of predicted S41 peptidases by analysis of mutant strains.
To confirm the activity of MLC_2570 orthologues, two mutant strains with transposon insertions in these orthologues were studied. One was an M. capricolum subsp. capricolum strain already described (22), with an insertion in MCAP_0240 (Ck-mut), and the other an M. mycoides subsp. mycoides mutant previously obtained at CIRAD, with an insertion in MSC_0281 (Rita-mut). These insertions had a drastic effect on caseinolytic activities. Grown in rich m-Hayflick medium, activity of mutant strains was negligible or nonexistent in culture supernatants, but it was still detectable in the cell pellets (Table 2). When incubated in a defined medium, Ck-mut lost its caseinolytic activity, both in the pellet and in the supernatant, while the parental strain displayed an increased level of activity, mostly in the supernatant. The results obtained with Rita-mut were less marked, as the parental strain also failed to show caseinolytic activity after incubation in Opti-MEM. Concordant results were obtained on milk agar plates, where M. capricolum subsp. capricolum and M. mycoides subsp. mycoides mutants completely lost their milk digestion properties, while cultures were clearly visible (Fig. 1). Finally, the digested band of 55 kDa that was observed with M. capricolum subsp. capricolum Ck in a zymogram performed with the concentrated supernatant was not observed with Ck-mut (Fig. 2). These analyses confirmed the role of secreted S41 peptidase MLC_2570 orthologues in casein degradation. Analysis of concentrated supernatants from two independent replicates of M. capricolum subsp. capricolum Ck cultures revealed that peptides from 20 predicted surface-exposed proteins, lipoproteins, or proteins bearing N-terminal or C-terminal predicted transmembrane regions were detected at significantly higher levels in the supernatant than in the cell pellet. The results obtained with three Ck-mut cultures showed that the extracellular secretion profile of 17 of these 20 proteins was significantly altered (Table 5). Their extracellular secretion was heavily reduced or even abolished, and 4 of them could no longer be detected either in the supernatant or in the cell pellet. The expression and extracellular secretion levels of 3 of these 20 proteins was unaltered.
TABLE 5.
List of proteins whose extracellular secretion was modified in the Ck-mut strain compared to the original Ck straina
| Category | Protein accession no. |
Spectral count for strain (titer [log/ml]): |
Mnemonic | Location | Annotation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ck (9.4) |
Ck1 (9.7) |
Ck-mut1 (9.1) |
Ck-mut5 (8.9) |
Ck-mut50 (8.7) |
||||||||||
| P | S | P | S | P | S | P | S | P | S | |||||
| Gene translation abolished | ABC01099.1 | 1 | 47 | 3 | 72 | 0 | 1 | 0 | 1 | 0 | 0 | MCAP_0843 | lpp | Transglutaminase |
| ABC01278.1 | 0 | 44 | 0 | 70 | 0 | 0 | 0 | 0 | 0 | 0 | MCAP_0860 | 1 TMB | Hypothetical protein | |
| ABC01319.1 | 0 | 25 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | MCAP_0863 | —b | DUF2570 | |
| ABC01807.1 | 0 | 100 | 11 | 146 | 0 | 0 | 0 | 0 | 0 | 0 | MCAP_0864 | 1 TMB | Topoisomerase? | |
| Gene translation and protein exosecretion reduced | ABC01143.1 | 0 | 50 | 0 | 47 | 4 | 8 | 3 | 4 | 5 | 0 | MCAP_0351 | 1 TMB | IgG-blocking virulence domain |
| ABC01224.1 | 0 | 56 | 0 | 38 | 3 | 19 | 3 | 9 | 1 | 4 | MCAP_0513 | lpp | Transglutaminase-like superfamily | |
| ABC01376.1 | 0 | 157 | 2 | 271 | 0 | 18 | 0 | 9 | 0 | 0 | MCAP_0862 | 1 TMB | DUF342 | |
| ABC01488.1 | 0 | 43 | 0 | 35 | 0 | 7 | 0 | 3 | 0 | 0 | MCAP_0329 | 1 TMB | Peptidase S41 | |
| ABC01574.1 | 8 | 161 | 9 | 209 | 0 | 6 | 0 | 0 | 0 | 0 | MCAP_0861 | 1 TMB | DUF342 | |
| ABC01698.1 | 4 | 106 | 10 | 80 | 30 | 43 | 19 | 18 | 17 | 5 | MCAP_0514 | lpp? | Transglutaminase-like superfamily | |
| ABC01774.1 | 0 | 69 | 3 | 57 | 7 | 8 | 5 | 3 | 3 | 0 | MCAP_0349 | 1 TMB | IgG-blocking virulence domain | |
| ABC01864.1 | 0 | 16 | 0 | 11 | 4 | 4 | 1 | 0 | 0 | 0 | MCAP_0345 | 1 TMB | IgG-blocking virulence domain | |
| Protein exosecretion reduced or abolished | ABC01302.1 | 34 | 187 | 20 | 118 | 81 | 36 | 109 | 29 | 172 | 6 | MCAP_0115 | 1 TMB | RecF/RecN/SMC N-terminal domain |
| ABC01413.1 | 15 | 75 | 16 | 65 | 32 | 44 | 34 | 17 | 32 | 3 | MCAP_0720 | lpp | lppQ | |
| ABC01444.1 | 12 | 59 | 13 | 38 | 22 | 11 | 25 | 2 | 20 | 0 | MCAP_0348 | lpp | Peptidase | |
| ABC01466.1 | 24 | 156 | 68 | 214 | 54 | 28 | 63 | 29 | 76 | 20 | MCAP_0240 | 2 TMB | Peptidase S41 | |
| ABC01836.1 | 3 | 26 | 5 | 18 | 23 | 0 | 27 | 0 | 16 | 0 | MCAP_0019 | 1 TMB | Secreted thousand residue frequently tandem | |
| Gene translation and protein exosecretion unmodified | ABC01469.1 | 0 | 23 | 0 | 19 | 1 | 22 | 7 | 39 | 4 | 17 | MCAP_0607 | lpp | Transglutaminase-like superfamily |
| ABC01669.1 | 0 | 13 | 0 | 15 | 0 | 28 | 0 | 11 | 0 | 3 | MCAP_0399 | 2 TMB | Hypothetical protein | |
| ABC01832.1 | 0 | 16 | 1 | 22 | 3 | 26 | 3 | 11 | 1 | 2 | MCAP_0401 | 2 TMB | Hypothetical protein | |
DUF, domain of unknown function; lpp, lipoprotein; P, pellet; S, supernatant; TMB, transmembrane region.
This protein is predicted to be 273 aa long in the Ck genome. However, orthologous genes in M. capricolum subsp. capricolum genomes are longer and contain one TMB.
In silico analysis of serine protease CBW53985.1 (MLC_2570).
Protein CBW53985.1 was by far the most overrepresented protease in the supernatants, especially in M. mycoides subsp. capri strains. It is 651 amino acids (aa) long and possesses two predicted transmembrane domains located at its N and C extremities (positions 7 to 29 and 627 to 646, respectively) and a predicted signal peptidase I (SPI) site at positions 29 to 30. BLASTP analysis revealed a “C-terminal processing peptidase family S41; peptidase family S41” domain spanning positions 383 to 543 with a high level of probability (2.7 e−23), with an active-site serine at position 477 (Fig. 3). High (>78%) levels of identity were observed with orthologous genes in mycoplasmas of the Mycoplasma mycoides cluster (M. mycoides subsp. mycoides, M. capricolum subsp. capricolum, M. capricolum subsp. capripneumoniae, and M. leachii) but also in M. feriruminatoris (82.6% identity). Two other proteases of M. mycoides subsp. capri were detected by BLASTP analysis with lower levels of identity (49.1% and 44.5%, respectively). Both (namely, CBW54055.1 [MLC_3270] and CBW53831.1 [MLC_1030]) had greater sizes (679 aa and 770 aa, respectively) but very similar structures, including an SPI site at the N-terminal extremity and an S41 domain and a transmembrane fragment at the C-terminal extremity. In addition, another M. mycoides subsp. capri 95010 protein displayed an S41 domain (CBW54056.1; MLC_3280), was predicted to be a lipoprotein, and did not display any C-terminal transmembrane fragment but did yield lower identity values by BLASTP. Similar mycoplasmal S41 serine proteases were detected by performing a BLASTP analysis that excluded the Mycoplasma mycoides cluster genomes, notably in M. agalactiae, M. alkalescens, M. auris, M. bovis, M. putrefaciens, and M. salivarium (see the supplemental material). More distantly related S41 bacterial serine proteases were also detected by BLAST analyses of the conserved S41 domain against the nonredundant database (excluding mycoplasmas). Four S41 serine proteases from Ruminococcus flavefaciens (WP_009982655.1, WP_028518726.1, WP_082325677.1, and WP_080693401.1) were detected and had very similar features. Interestingly, a typical LPXTG sortase-associated cell wall anchor domain was located shortly before the C-terminal transmembrane fragment in all four S41 Ruminococcus peptidases.
DISCUSSION
Proteases are among the largest families of metabolic enzymes. They operate by a variety of mechanisms and are vital to many aspects of bacterial cell life and pathogenicity (23), such as proteolysis and digestion, cellular respiration, energy storage, transcription, and response to the environment. Proteolytic activity in mycoplasmas was first observed by Longley using inspissated goat or sheep serum for the growth of an organism causing pleuropneumonia in goats (24). This finding prompted the development of methods to evaluate the proteolytic activities of mycoplasmas (25). Only a few Mycoplasma species displayed such activity, including M. arthritidis, which digested gelatin (26), M. bovirhinis, which digested casein, and M. mycoides, which digested gelatin, coagulated serum, and casein (27). M. mycoides subsp. capri strain 95010 is no exception, as 55 protease-coding genes were predicted in its genome consisting of 962 coding DNA sequences (CDS). This subspecies can be considered a model for the mycoplasmas of the Mycoplasma mycoides cluster. It is highly pathogenic and can induce lesions in a variety of organs, but it can also be found as a saprophyte in the ears of normal goats (28, 29). This proves its ability to survive in diverse environments of its caprine host, despite possessing a very small genome of 1.15 megabases.
Within these 55 proteases, one, MLC_2570, appeared to be prominent. This protease was detected solely in the supernatant and not in the cell pellet, and it was shown to be caseinolytic. Orthologs of this gene were detected within the Mycoplasma mycoides cluster sensu stricto but not in closely related mycoplasmas found in ruminants such as M. cottewii or M. yeatsii nor in the rest of the members of the Spiroplasma group of species found in insects or plants. Accordingly, this gene belongs to the group of genes that were most probably acquired via horizontal gene transfer (HGT) by the species of the Mycoplasma mycoides cluster to enable their becoming successful ruminant pathogens (30). The most closely related BLASTP bidirectional best hits outside the Mycoplasma mycoides cluster were observed with M. auris. Best hits were also detected with M. alkalescens and other ruminant pathogens such as M. agalactiae and M. bovis. In such cases, the reciprocal best hits were different protease genes (MLC_1030 and MLC_3270), which seemed to indicate that the HGT involved a number of protease genes, highlighting the importance of these enzymes for evolutionary convergence toward ruminant colonization within divergent mycoplasma lineages. The origin of the HGT may be hinted at by examination of the best hits outside mycoplasma genomes, which corresponded to S41 peptidases of Ruminococcus species (WP_093044261.1). They share the global architecture of MLC_2570, with a similar length, an N-terminal SPI domain, an S41 family motif, and a C-terminal transmembrane domain. These Ruminococcus proteases possess a typical sortase LPXTG motif shortly upstream from the C-terminal transmembrane domain. These sortase motifs are typical of the Gram-positive proteins that are expressed at the cell surface through a covalent linkage to their cell wall (31, 32). It is tempting to draw a parallel with a possible secretion mechanism occurring in mycoplasmas of the Mycoplasma mycoides cluster. However, there are still some clues missing to explain the extracellular secretion of the protease. Neither a signal peptidase I nor a sortase-coding gene has been identified in the Mycoplasma mycoides cluster genomes (33). An alternative cleavage process, based on endoproteolysis, may be involved here. Analyzing an M. capricolum subsp. capricolum Ck mutant lacking the MLC_2570 orthologue, it was not only the S41 protease whose exosecretion in the spent medium was altered but also most of the other proteins that were seen to be overrepresented in the supernatant of the parental Ck strain.
These results suggest that this S41 protease could be involved in the posttranslational processing of many mycoplasma surface-exposed proteins. Further work is needed to determine the cleavage site of the protease and verify which surface proteins are candidate substrates. In mycoplasmas, the proteolysis of surface-exposed proteins seems to be common. This is the case for the MALP-404 lipoprotein of M. fermentans (34), with the release of a soluble lipoprotein fragment and the alteration of the surface phenotype leading to a shorter membrane-anchored fragment acting as a Toll-like receptor macrophage-activating lipopeptide. This study had clearly demonstrated that the release of the MALP-404 fragment entailed extracellular processing, considered a new mechanism for the “secretion” of hydrophilic proteins in mycoplasmas. This mechanism is widely distributed in mycoplasmas, and it has been extensively studied in M. hyopneumoniae (35), where endoproteases are responsible for ectodomain shedding (36), with notable consequences with respect to adhesion and plasminogen activation. The posttranslational processing of adhesion is not limited to mycoplasmas found in animals, as it is also present in the plant pathogen Spiroplasma citri, where it affects adhesion-related proteins (S. citri ARP [ScARP]) (11).
Studying medium-secreted proteins is difficult in mycoplasmas, which need complex and rich media for their growth. This is the reason why we adopted the same strategy that allowed us to characterize secreted polysaccharides (37). Washed mycoplasma cells were incubated into a defined medium, Opti-MEM, which contains only a very limited amount of proteins consisting of growth factors such as insulin and transferrin. This should allow the mycoplasma to maintain some metabolic activities while obviously not enabling its multiplication. This strategy proved efficient, as medium-exported proteins were clearly detected and could be identified by tandem mass spectrometry. However, since this medium did not allow the multiplication of the mycoplasma, its metabolism may have been modified by this stress through a classical “stringent response,” as seen in other bacteria (38). The RelA/SpoT homologue superfamily, which is involved in the regulation of (p)ppGpp alarmone during stress, may not be present in some mycoplasma species (39). However, it is present in the Mycoplasma mycoides cluster and specific spectral counts of RelA were detected in the mycoplasma pellets of all studied strains. In the case of M. mycoides subsp. capri, the secretion of S41 peptidase in the medium is certainly not specific to the incubation into a defined medium, as a high level of caseinolytic activity was also detected in the highly enriched m-Hayflick spent medium. For some species, such as M. mycoides subsp. mycoides, and, to a lesser extent, M. capricolum subsp. capripneumoniae, the caseinolytic activity was more pronounced in rich medium than in Opti-MEM medium.
This raises the issue of the ability of the various mycoplasmas to adapt to stress, a field which is emerging now for these organisms (40, 41). A lower level of adaptability for M. mycoides subsp. mycoides and M. capricolum subsp. capripneumoniae would not be surprising, as these species possess degenerate genomes compared to the close relatives from which they emerged (42, 43). Their reduced gene repertoire may be associated with a restricted ecological niche, i.e., the ruminant lungs, and with an inability to cope with stress encountered in other body compartments. The genes coding for the S41 peptidases are still functional in M. capricolum subsp. capripneumoniae, despite its degenerated genome. This was very noticeable on milk agar, where casein digestion was clearly visible in spite of colony growth being barely noticeable. Expressing and exporting a protease must therefore bring a fitness advantage to these fastidious bacteria. As mycoplasmas are dependent on the supply of peptides for their metabolism, protein digestion by S41 protease could be a first step before further degradations by peptidases (Pep A-F-O-Q-V) and uptake by the oligopeptide ABC transporters. The in vivo relevance of these events, notably in terms of virulence, has yet to be evaluated. The release of peptides and active proteases could well represent major events in the pathological process by disrupting the delicate environment of the lung alveoli. Concomitantly, the antigenic variation that it involves on the cell surface may have profound effects on the interactions with the host innate and adaptive immune responses.
MATERIALS AND METHODS
Mycoplasma strains and culture conditions.
At least 2 strains from each of 13 mollicutes (sub)species that can be isolated from ruminant lungs were analyzed in this study (Table 1). The MSC_0281::pMT85/2res transposon mutant of M. mycoides subsp. mycoides strain 8740 Rita (Rita-mut), with an insertion in the orthologue of M. mycoides subsp. mycoides PG1T MSC_0281, was selected from a mutant bank produced at CIRAD. The procedures for transformation and identification of pMT85/2res transposon insertion site sequences have been previously described (44). The ctpA::Tn4001t mutant of the California Kid type strain, or CkT (Ck-mut), was kindly provided by M. Brown (Department of Infectious Diseases and Pathology, College of Veterinary Medicine, University of Florida). This mutant was generated by random mutagenesis, with the Tn4001t transposon being inserted in the ctpA (MCAP_0240) gene (22). All other strains were cultured in either M-Hayflick medium (44) or commercialized pleuropneumonia-like organism (PPLO) modified broth (Indicia, France). Opti-MEM GlutaMAX medium (Gibco), depleted of proteins except for minute amounts of insulin and transferrin, was used to characterize the extracellular proteases. This defined medium was supplemented with 0.4% (wt/vol) pyruvic acid, 0.02% (wt/vol) DNA (herring sperm) (to maintain cell viability), and 0.1% (wt/vol) ampicillin, and the reaction mixture was subjected to sterilization by passage through a 0.1-μm-pore-size filter unit (Millex low protein binding Durapore, polyvinylidene difluoride [PVDF]). Mycoplasma cultures were incubated at 37°C and 5% CO2 in a humid atmosphere. Culture titers were evaluated by performing six 10-fold dilutions in m-Hayflick and plating 10-μl aliquots of the last three dilutions on agar plates to quantify viable bacteria by colony counting.
Determination of global caseinolytic activity.
The proteolytic activity of mycoplasmas was investigated using casein as the substrate. The overall caseinolytic activity of the mycoplasma strains was assessed by measuring the degradation of fluorescent casein using a protease detection kit (Jena Bioscience PP404S). The microtiter plate operation protocol was used following the manufacturer’s instructions, and fluorescence was measured using an Enspire 2300 fluorimeter (Perkin Elmer), with excitation at 490 nm, emission at 525 nm, 100 flashes, and a top measurement height of 9.5 mm. With the exception of Fig. S2, where the original relative fluorescence units (RFU) were retained, each fluorescence measurement was normalized using the kit’s positive control and the corresponding medium negative control, according to the following formula (with relative activity [RA] expressed as a percentage): RA = 100 × {[(log RFU sample) − (log RFU medium)]/[(log RFU kit_positive_control) − (log RFU medium)]}.
This approach was validated using strains belonging to three different Mycoplasma species, namely, M. mycoides subsp. capri (M. mycoides subsp. capri) strain 95010, M. mycoides subsp. mycoides strain Gladysdale, and M. bovis strain L2. The caseinolytic activity was measured in serial 10-fold dilutions, which were then incubated. The last tube showing some turbidity was considered to be in the exponential phase of growth, while the preceding tubes were at later stages of growth, up to the stationary phase within the first tube of the series. The fluorescence was shown to be stable and at its maximum in the stationary phase for all three species (Fig. S2A). Under our experimental conditions, the choice of medium used for protease detection (Indicia versus m-Hayflick) had no significant impact on the overall activity of M. mycoides subsp. capri 95010, although m-Hayflick yielded a lower background level (Fig. S2B). Finally, the robustness of this assay was demonstrated by analyzing three independent cultures of the same strains, which provided reproducible results (Fig. S2C).
The fluorescent casein test was also used to evaluate whether the activity was present in the culture supernatant or associated with the mycoplasma pellet. For this purpose, 1-ml volumes of stationary-phase cultures were centrifuged at 12,000 × g for 20 min at 4°C. The supernatants were filtered through a 0.1-μm-pore-size filter unit (Millex low protein binding Durapore, PVDF), while the pellet was resuspended in the original culture volume (i.e., 1 ml) of phosphate-buffered saline (PBS; 10 mM phosphate, 150 mM NaCl, pH 7.8). The absence of residual viable mycoplasmas was confirmed by plating 10-μl volumes of the filtered supernatant onto agar medium. The caseinolytic activity was measured as described above for the two fractions. The same method was applied for the determination of global caseinolytic activity in exoproteome extracts. For this purpose, mycoplasmas grown in m-Hayflick until the late exponential phase of growth were centrifuged 20 min at 12,000 × g, washed twice in nonsupplemented Opti-MEM medium, and concentrated thrice in supplemented Opti-MEM. After incubation for 16 h in supplemented Opti-MEM, the cultures were centrifuged at 12,000 × g for 20 min at 4°C. The pellets were resuspended in the same volume of PBS, and the supernatants were filtered through 0.1-μm-pore-size filter units. The caseinolytic activities of the 16-h cultures incubated in Opti-MEM, as well as the supernatant and pellet corresponding to each culture, were determined as indicated above. Viability losses during Opti-MEM incubation were evaluated by titration of the washed culture transferred into Opti-MEM medium (time zero [T0] titer) and of the same culture after the 16-h incubation period.
Assessment of proteolytic activity on milk agar.
Milk agar plates were prepared by adding 0.4% (wt/vol) dried skimmed milk powder to m-Hayflick agar medium. Mycoplasma stationary-phase cultures (7 μl) were plated and incubated for 60 h. Casein degradation was then assessed against a dark background by evaluating the presence of a translucent area around the culture, indicating casein degradation (−, no translucent area; +, blurred and small translucent area; ++, sharp and large translucent area).
Identification of M. mycoides subsp. capri 95010 and M. bovirhinis MV5 genes coding for putative protease motifs.
The table listing the proteases from Mycoplasma mycoides was retrieved from the MEROPS peptidase database (https://www.ebi.ac.uk/merops/cgi-bin/speccards?sp=sp003189;type=peptidase). As this list included peptidases that were identified in three subspecies or serotypes, namely, M. mycoides subsp. mycoides, M. mycoides subsp. capri serotype LC, and M. mycoides subsp. capri serotype capri (locus tags MSC, MLC, and MMCAP1, respectively), the corresponding genes in the genome of M. mycoides subsp. capri LC strain 95010 (NC_015431) were identified and duplicates discarded. This resulted in a table with 36 entries. However, additional genes bearing putative protease motifs could also be identified in genome annotations. To retrieve them, a query on the MaGe website was performed using the “search by keywords” tool and using “Protease OR peptidase” as the query (https://www.genoscope.cns.fr/agc/microscope/mage/viewer.php). This led to the identification of 19 additional genes potentially coding for proteins with peptidase activity. In addition, the orthologous genes found in the Mycoplasma mycoides cluster species were identified using the MaGe interface. As M. bovirhinis genomes were not integrated in the MEROPS database, any proteins that may be potential peptidases were retrieved by searching using “peptidase OR protease” in the genome of strain HAZ 141_2 (accession AP018135).
Tandem mass spectrometry analysis of the exoproteome.
Mycoplasma cultures, incubated 16 h in supplemented Opti-MEM as described above, were centrifuged at 12,000 × g for 20 min at 4°C. The cell pellets were resuspended in PBS and standardized to obtain 1 mg/ml of proteins, and the culture supernatants were filtered through 0.1-μm-pore-size filters as described above. The Opti-MEM filtered supernatants were freeze-dried (2.5 ml per vial). The freeze-dried supernatants were first reconstituted with 120 μl of sterile MilliQ water and then supplemented with 120 μl of 2× Laemmli buffer. Samples were incubated for 5 min at 99°C and then subjected to SDS-PAGE on a 4% to 12% NuPage gel (Invitrogen) with MES (morpholineethanesulfonic acid) buffer (Invitrogen) for a short electrophoretic migration, as described previously (45). The whole-protein content from each well was extracted as a single polyacrylamide band, processed for in-gel digestion, and subjected to proteolysis with trypsin (Roche) using 0.01% ProteaseMAX surfactant (Promega) for 1 h at 50°C. The resulting peptide fractions were analyzed with a Q-Exactive HF tandem mass spectrometer (Thermo) coupled with an UltiMate 3000 liquid chromatography (LC) system (Dionex-LC Packings) and operated in data-dependent mode as previously described (46). Peptides were analyzed along a 90-min gradient of acetonitrile with scan cycles initiated by a full scan of peptide ions in the Orbitrap analyzer, followed by high-energy collisional dissociation and MS/MS scans of the 20 most abundant precursor ions with 2+ or 3+ charges only. Full-scan mass spectra were acquired from m/z 350 to 1,800 at a resolution of 60,000. Ion selection for MS/MS fragmentation and measurement was performed by applying dynamic exclusion for 10 s. MS/MS spectra were assigned to peptide sequences by the MASCOT Daemon 2.6.0 search engine (Matrix Science) searching against the corresponding annotated theoretical proteome database (M. mycoides subsp. capri [GenBank accession no. FQ377874.1], M. mycoides subsp. mycoides [CP002107.1], M. capricolum subsp. capricolum [CP000123.1], M. capricolum subsp. capripneumoniae [LM995445.1], and M. bovirhinis [AP018135.1]) with the following parameters: full-trypsin specificity, maximum of two missed cleavages, mass tolerances of 5 ppm on the parent ion and 0.02 Da on the MS/MS, carboxyamidomethylated cysteine (+57.0215) as a fixed modification, and oxidized methionine (+15.9949) and deamidation of asparagine and glutamine (+0.9848) as variable modifications. Only peptide matches presenting a MASCOT peptide score with a P value of less than 0.05 were retained. Proteins were considered identified when at least two peptides were assigned. For each protein, peptide-to-spectrum assignments were summed (spectral counts) and compared per conditions.
Method for identification of putative proteases enriched in supernatants versus cell pellets.
The number of detected spectral counts was recorded for each of the putative proteases in both fractions. As the total amount of spectral counts could differ from one sample to another, ratios were calculated by dividing the detected spectral count for each protein by the total number of spectral counts detected in the sample. This ratio yielded estimated proportions of the protein in the samples that could be used for comparisons of one sample to another. A secondary ratio, or supernatant/pellet fold change value, was then calculated by dividing the ratio obtained for one protein in the supernatant by that obtained in the pellet. The distribution of fold change values was evaluated by performing a frequency curve analysis (Microsoft Excel), with a range of 0 to 33 and intervals of 0.5 to detect outlier values.
Casein zymography.
To identify which proteases were able to digest casein, concentrated supernatants and mycoplasma cell pellets were first analyzed by SDS-PAGE (7.5% acrylamide) followed by silver staining. Zymogram preparations were performed on 160-by-180-mm, 1-mm-thick gels. Running gels contained 7.5% (wt/vol) acrylamide–1.5 M Tris (pH 8.8) buffer–0.1% (wt/vol) SDS–ammonium persulfate, supplemented with 0.1% (wt/vol) casein and tetramethylethylenediamine (TEMED) before polymerization. Stacking gels contained a mixture of 3.75% (wt/vol) acrylamide, 0.5 M Tris (pH 6.8), 0.1% (wt/vol) SDS, and ammonium persulfate, supplemented with TEMED before polymerization. A 10-μg volume of each sample was loaded into each well of SDS-PAGE and zymogram gels, avoiding a boiling step. The migration conditions were 70 V (constant) through the stacking gel and 25 mA (constant) through the running gel (all at 4°C) and were maintained until the migration front reached the bottom of the gel. The gel was washed twice for 30 min in washing buffer (2.5% [vol/vol] Triton X-100, 50 mM Tris-HCl [pH 7.5], 5 mM CaCl2, 1 μM ZnCl2) and incubated for 24 h in an incubation buffer that was similar to washing buffer except that the concentration of Triton X-100 was 1% instead of 2.5% (vol/vol). The gel was then stained with Bio-Safe staining solution (Bio-Rad) following the manufacturer’s instructions. Active casein proteases, revealed as translucent bands, were collected for further identification analysis by mass spectrometry. The cell pellets and concentrated supernatants were prepared in a similar way for mass spectrometry analysis.
Analysis of serine protease CBW53985.1 (MLC_2570).
The search for signal peptidase motifs was performed online with the SignalP v5.0 server for Gram-positive bacteria (http://www.cbs.dtu.dk/services/SignalP/), while transmembrane regions were detected using the TMHMM server, v2.0 (http://www.cbs.dtu.dk/services/TMHMM/). BLASTP analysis was performed using the whole protein CBW53985.1 as the query through the Genoscope MaGe interface with available genomes and then through the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the conserved S41 region of CBW53985.1 on the nonredundant database and excluding mycoplasmas (Mycoplasmatales; TaxID: 2085).
Supplementary Material
ACKNOWLEDGMENTS
We are greatly indebted to Mary Brown of the University of Florida College of Veterinary Medicine, FL, USA, who supplied us with the M. capricolum subsp. capricolum mutant (ctpA::Tn4001t; Ck-mut).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01439-19.
REFERENCES
- 1.Nocard E, Roux E, Borrel A, Salimbeni AT, Dujardin-Beaumetz E. 1898. Le microbe de la péripneumonie. Ann Inst Pasteur (Paris) 12:240–262. [Google Scholar]
- 2.Westberg J, Persson A, Holmberg A, Goesmann A, Lundeberg J, Johansson KE, Pettersson B, Uhlen M. 2004. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP). Genome Res 14:221–227. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Browning GF, Noormohammadi AH, Markham PF. 2014. Identification and characterization of virulence genes in mycoplasmas, p 77–90. In Browning GF, Citti C (ed), Mollicutes, molecular biology and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 4.Pilo P, Frey J, Vilei EM. 2007. Molecular mechanisms of pathogenicity of Mycoplasma mycoides subsp. mycoides SC. Vet J 174:513–521. doi: 10.1016/j.tvjl.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szczepanek SM, Boccaccio M, Pflaum K, Liao X, Geary SJ. 2014. Hydrogen peroxide production from glycerol metabolism is dispensable for virulence of Mycoplasma gallisepticum in the tracheas of chickens. Infect Immun 82:4915–4920. doi: 10.1128/IAI.02208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clyde WA., Jr. 1971. Immunopathology of experimental Mycoplasma pneumoniae disease. Infect Immun 4:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones HP, Tabor L, Sun X, Woolard MD, Simecka JW. 2002. Depletion of CD8+ T cells exacerbates CD4+ Th cell-associated inflammatory lesions during murine mycoplasma respiratory disease. J Immunol 168:3493–3501. doi: 10.4049/jimmunol.168.7.3493. [DOI] [PubMed] [Google Scholar]
- 8.Rosengarten R, Behrens A, Stetefeld A, Heller M, Ahrens M, Sachse K, Yogev D, Kirchhoff H. 1994. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect Immun 62:5066–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchenau I, Poumarat F, Grand DL, Linkner H, Rosengarten R, Hewicker-Trautwein M. 2010. Expression of Mycoplasma bovis variable surface membrane proteins in the respiratory tract of calves after experimental infection with a clonal variant of Mycoplasma bovis type strain PG45. Res Vet Sci 89:223–229. doi: 10.1016/j.rvsc.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Christodoulides A, Gupta N, Yacoubian V, Maithel N, Parker J, Kelesidis T. 2018. The role of lipoproteins in mycoplasma-mediated immunomodulation. Front Microbiol 9:1682. doi: 10.3389/fmicb.2018.01682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubrana M-P, Guéguéniat J, Bertin C, Duret S, Arricau-Bouvery N, Claverol S, Lartigue C, Blanchard A, Renaudin J, Béven L. 2017. Proteolytic post-translational processing of adhesins in a pathogenic bacterium. J Mol Biol 429:1889–1902. doi: 10.1016/j.jmb.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Tacchi JL, Raymond BBA, Haynes PA, Berry IJ, Widjaja M, Bogema DR, Woolley LK, Jenkins C, Minion FC, Padula MP, Djordjevic SP. 2016. Post-translational processing targets functionally diverse proteins in Mycoplasma hyopneumoniae. Open Biol 6:150210. doi: 10.1098/rsob.150210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raymond BBA, Djordjevic S. 2015. Exploitation of plasmin(ogen) by bacterial pathogens of veterinary significance. Vet Microbiol 178:1–13. doi: 10.1016/j.vetmic.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Arfi Y, Minder L, Di Primo C, Le Roy A, Ebel C, Coquet L, Claverol S, Vashee S, Jores J, Blanchard A, Sirand-Pugnet P. 2016. MIB-MIP is a mycoplasma system that captures and cleaves immunoglobulin G. Proc Natl Acad Sci U S A 113:5406–5411. doi: 10.1073/pnas.1600546113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raymond BBA, Tacchi JL, Jarocki VM, Minion FC, Padula MP, Djordjevic SP. 2013. P159 from Mycoplasma hyopneumoniae binds porcine cilia and heparin and is cleaved in a manner akin to ectodomain shedding. J Proteome Res 12:5891–5903. doi: 10.1021/pr400903s. [DOI] [PubMed] [Google Scholar]
- 16.Tacchi JL, Raymond BBA, Jarocki VM, Berry IJ, Padula MP, Djordjevic SP. 2014. Cilium adhesin P216 (MHJ_0493) is a target of ectodomain shedding and aminopeptidase activity on the surface of Mycoplasma hyopneumoniae. J Proteome Res 13:2920–2930. doi: 10.1021/pr500087c. [DOI] [PubMed] [Google Scholar]
- 17.Jarocki VM, Santos J, Tacchi JL, Raymond BBA, Deutscher AT, Jenkins C, Padula MP, Djordjevic SP. 2015. MHJ_0461 is a multifunctional leucine aminopeptidase on the surface of Mycoplasma hyopneumoniae. Open Biol 5:140175. doi: 10.1098/rsob.140175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertin C, Pau-Roblot C, Courtois J, Manso-Silván L, Thiaucourt F, Tardy F, Le Grand D, Poumarat F, Gaurivaud P. 2013. Characterization of free exopolysaccharides secreted by Mycoplasma mycoides subsp. mycoides. PLoS One 8:e68373. doi: 10.1371/journal.pone.0068373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan DS, Daubenspeck JM, Laube AH, Renfrow MB, Dybvig K. 2013. O-linked protein glycosylation in mycoplasma. Mol Microbiol 9:1046–1053. doi: 10.1111/mmi.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalbey RE, Wang P, van Dijl JM. 2012. Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev 76:311–330. doi: 10.1128/MMBR.05019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaurivaud P, Ganter S, Villard A, Manso-Silvan L, Chevret D, Boulé C, Monnet V, Tardy F. 2018. Mycoplasmas are no exception to extracellular vesicles release: revisiting old concepts. PLoS One 13:e0208160. doi: 10.1371/journal.pone.0208160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allam AB, Reyes L, Assad-Garcia N, Glass JI, Brown MB. 2010. Enhancement of targeted homologous recombination in Mycoplasma mycoides subsp. capri by inclusion of heterologous recA. Appl Environ Microbiol 76:6951–6954. doi: 10.1128/AEM.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Culp E, Wright GD. 2017. Bacterial proteases, untapped antimicrobial drug targets. J Antibiot (Tokyo) 70:366. doi: 10.1038/ja.2016.138. [DOI] [PubMed] [Google Scholar]
- 24.Longley EO. 1951. Contagious caprine pleuropneumonia (a study of the disease in Nigeria), vol 7 Colonial Research Publication, Stationary Office, London, United Kingdom. [Google Scholar]
- 25.Freundt EA. 1983. Proteolytic activity, p 367–371. In Razin S, Tully JG (ed), Methods in mycoplasmology, vol 1 Academic Press, New York, NY. [Google Scholar]
- 26.Woolcock PR, Czekalowski JW, Hall DA. 1973. Studies on proteolytic activities of mycoplasmas: the preparation and properties of gelatinolytic enzymes from strains of Mycoplasma arthritidis. J Gen Microbiol 78:23–32. doi: 10.1099/00221287-78-1-23. [DOI] [PubMed] [Google Scholar]
- 27.Razin S, Freundt EA. 1984. The mycoplasmas, p 740–793. In Krieg NR, Holt JG (ed), Bergey’s manual of systematic bacteriology, vol I Williams and Wilkins, Baltimore, MD. [Google Scholar]
- 28.Cottew GS. 1985. Mycoplasma in ears. Aust Vet J 62:420. doi: 10.1111/j.1751-0813.1985.tb14125.x. [DOI] [PubMed] [Google Scholar]
- 29.Tardy F, Mercier P, Solsona M, Saras E, Poumarat F. 2007. Mycoplasma mycoides subsp. mycoides biotype large colony isolates from healthy and diseased goats: prevalence and typing. Vet Microbiol 121:268–277. doi: 10.1016/j.vetmic.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Lo W-S, Gasparich GE, Kuo C-H. 2018. Convergent evolution among ruminant-pathogenic Mycoplasma involved extensive gene content changes. Genome Biol Evol 10:2130–2139. doi: 10.1093/gbe/evy172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ton-That H, Marraffini LA, Schneewind O. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta 1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Schneewind O, Missiakas D. 2019. Sortases, surface proteins, and their roles in Staphylococcus aureus disease and vaccine development. Microbiol Spectr 7:PSIB-0004-2018. doi: 10.1128/microbiolspec.PSIB-0004-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adeel M, Bum KS. 2019. A comprehensive in silico analysis of sortase superfamily. J Microbiol 57:431–443. [DOI] [PubMed] [Google Scholar]
- 34.Davis KL, Wise KS. 2002. Site-specific proteolysis of the MALP-404 lipoprotein determines the release of a soluble selective lipoprotein-associated motif-containing fragment and alteration of the surface phenotype of Mycoplasma fermentans. Infect Immun 70:1129–1135. doi: 10.1128/iai.70.3.1129-1135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry IJ, Jarocki VM, Tacchi JL, Raymond BBA, Widjaja M, Padula MP, Djordjevic SP. 2017. N-terminomics identifies widespread endoproteolysis and novel methionine excision in a genome-reduced bacterial pathogen. Sci Rep 7:11063. doi: 10.1038/s41598-017-11296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarocki VM, Tacchi JL, Djordjevic SP. 2015. Non-proteolytic functions of microbial proteases increase pathological complexity. Proteomics 15:1075–1088. doi: 10.1002/pmic.201400386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertin C, Pau-Roblot C, Courtois J, Manso-Silván L, Tardy F, Poumarat F, Citti C, Sirand-Pugnet P, Gaurivaud P, Thiaucourt F. 2015. Highly dynamic genomic loci drive the synthesis of two types of capsular or secreted polysaccharides within the Mycoplasma mycoides cluster. Appl Environ Microbiol 81:676–687. doi: 10.1128/AEM.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pulschen AA, Sastre DE, Machinandiarena F, Crotta Asis A, Albanesi D, de Mendoza D, Gueiros-Filho FJ. 2017. The stringent response plays a key role in Bacillus subtilis survival of fatty acid starvation. Mol Microbiol 103:698–712. doi: 10.1111/mmi.13582. [DOI] [PubMed] [Google Scholar]
- 39.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galperin MY. 2018. What bacteria want. Environ Microbiol 20:4221–4229. doi: 10.1111/1462-2920.14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beier LS, Siqueira FM, Schrank IS. 2018. Evaluation of growth and gene expression of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis in defined medium. Mol Biol Rep 45:2469–2479. doi: 10.1007/s11033-018-4413-3. [DOI] [PubMed] [Google Scholar]
- 42.Thiaucourt F, Manso-Silvan L, Woubit S, Barbe V, Vacherie B, Jacob D, Breton M, Dupuy V, Lomenech AM, Blanchard A, Sirand-Pugnet P. 2011. Mycoplasma mycoides, from “mycoides Small Colony” to “capri”. A microevolutionary perspective. BMC Genomics 12:114. doi: 10.1186/1471-2164-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupuy V, Thiaucourt F. 2014. Complete genome sequence of Mycoplasma capricolum subsp. capripneumoniae strain 9231-Abomsa. Genome Announc 2:e01067-14. doi: 10.1128/genomeA.01067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnefois T, Vernerey M-S, Rodrigues V, Totté P, Puech C, Ripoll C, Thiaucourt F, Manso-Silván L. 2016. Development of fluorescence expression tools to study host-mycoplasma interactions and validation in two distant mycoplasma clades. J Biotechnol 236:35–44. doi: 10.1016/j.jbiotec.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Hartmann EM, Allain F, Gaillard J-C, Pible O, Armengaud J. 2014. Taking the shortcut for high-throughput shotgun proteomic analysis of bacteria, p 275–285. In Vergunst AC, O’Callaghan D (ed), Host-bacteria interactions: methods and protocols. Springer New York, New York, NY. [DOI] [PubMed] [Google Scholar]
- 46.Klein G, Mathé C, Biola-Clier M, Devineau S, Drouineau E, Hatem E, Marichal L, Alonso B, Gaillard J-C, Lagniel G, Armengaud J, Carrière M, Chédin S, Boulard Y, Pin S, Renault J-P, Aude J-C, Labarre J. 2016. RNA-binding proteins are a major target of silica nanoparticles in cell extracts. Nanotoxicology 10:1555–1564. doi: 10.1080/17435390.2016.1244299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.