The mechanism underlying bacterial twitching motility is an important research area because it is closely related to social and pathogenic behaviors. The mechanism mediating cell-to-cell perception of twitching motility is largely unknown. Using Lysobacter as a model, we found in this study that the interspecies signal indole caused Lysobacter to exhibit irregular, random twitching motility via activation of gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1. In addition, population-dependent behavior induced by 13-methyltetradecanoic acid, a quorum-sensing signaling molecule designated LeDSF, was involved in twitching motility by indirectly regulating gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC. The results demonstrate that the twitching motility of Lysobacter is regulated by these two signaling molecules, offering novel clues for exploring the mechanisms of twitching motility and population-dependent behaviors of bacteria.
KEYWORDS: interspecies and intraspecies signal indole, interspecies and intraspecies signal LeDSF, Lysobacter enzymogenes, quorum-sensing, twitching motility
ABSTRACT
The twitching motility of bacteria is closely related to environmental adaptability and pathogenic behaviors. Lysobacter is a good genus in which to study twitching motility because of the complex social activities and distinct movement patterns of its members. Regardless, the mechanism that induces twitching motility is largely unknown. In this study, we found that the interspecies signal indole caused Lysobacter to have irregular, random twitching motility with significantly enhanced speed. Deletion of qseC or qseB from the two-component system for indole signaling perception resulted in the disappearance of rapid, random movements and significantly decreased twitching activity. Indole-induced, rapid, random twitching was achieved through upregulation of expression of gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1. In addition, under conditions of extremely low bacterial density, individual Lysobacter cells grew and divided in a stable manner in situ without any movement. The intraspecies quorum-sensing signaling factor 13-methyltetradecanoic acid, designated L. enzymogenes diffusible signaling factor (LeDSF), was essential for Lysobacter to produce twitching motility through indirect regulation of gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC. These results demonstrate that the motility of Lysobacter is induced and regulated by indole and LeDSF, which reveals a novel theory for future studies of the mechanisms of bacterial twitching activities.
IMPORTANCE The mechanism underlying bacterial twitching motility is an important research area because it is closely related to social and pathogenic behaviors. The mechanism mediating cell-to-cell perception of twitching motility is largely unknown. Using Lysobacter as a model, we found in this study that the interspecies signal indole caused Lysobacter to exhibit irregular, random twitching motility via activation of gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1. In addition, population-dependent behavior induced by 13-methyltetradecanoic acid, a quorum-sensing signaling molecule designated LeDSF, was involved in twitching motility by indirectly regulating gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC. The results demonstrate that the twitching motility of Lysobacter is regulated by these two signaling molecules, offering novel clues for exploring the mechanisms of twitching motility and population-dependent behaviors of bacteria.
INTRODUCTION
The genus Lysobacter comprises a group of ubiquitous environmental bacteria that are emerging as a new source of bioactive natural products (1, 2). Lysobacter species exhibit two characteristic features among environmental Gram-negative bacteria: the first is that they are prolific producers of lytic enzymes and antibiotics, and the second is that they exhibit population-dependent behavior, such as twitching motility and quorum-sensing activity. The first of those Lysobacter features has been well studied by several research groups; indeed, multiple species of the genus are known to produce promising antibiotics. For example, lysobactin from Lysobacter sp. ATCC 53042 is a potent cyclic depsipeptide that blocks cell wall biosynthesis in bacteria (3–5), and the WAP-8294A family of Lysobacter enzymogenes OH11 and Lysobacter sp. WAP-8294 exhibits potent activities against the Gram-positive pathogen methicillin-resistant Staphylococcus aureus (MRSA) (6–8). In addition to the WAP-8294A family, L. enzymogenes produces another promising antifungus antibiotic, heat-stable antifungal factor (HSAF), as well as its analogs, which are a family of polycyclic tetramate macrolactams with distinct modes of action and biosynthesis patterns (9–11). Moreover, Lysobacter antibioticus OH13 produces the broad-spectrum antibiotic phenazine (12). In contrast to the signaling pathways that control the biosynthesis of natural products, those that control twitching motility and related population-dependent activities in Lysobacter remain largely unknown.
Twitching motility is conferred by the type IV pilus (Tfp) in many bacteria that exhibit social and pathogenic activities (13–16). Tfp production and assembly are regulated by diverse transcriptional mechanisms. For instance, the Tfp-dependent motility of Myxobacteria is coregulated via distinct regulating pathways by the PilSR and PilS2R2 two-component systems (17). PilB and PilT play are motors of pilus extension and retraction, respectively, in Myxococcus xanthus (18). Furthermore, traction force microscopy was recently employed to show that the force generated during Myxobacteria twitching motility was higher when the cells were in a group than when they were individually isolated (19). PilS-PilR, which is essential for activating the major pilin-encoding gene pilA, has been recognized as an important regulatory pathway for Tfp formation in Pseudomonas aeruginosa (20, 21). In Lysobacter, 14 genes responsible for twitching motility have been identified, including pilA, pilM, pilN, pilO, pilP, pilQ, pilR, pilG and clp (22–24). Among the genes regulating twitching motility, clp, is a global regulator that can control the Tfp-dependent twitching motility of L. enzymogenes OH11 by directly binding to the promoter regions of pilA and pilMNOPQ (25).
Endogenous and xenogenous signaling play essential roles in multiple bacterial physiological and biochemical behaviors (26–30). In one study assessing Lysobacter signaling, a group of diffusible signaling factors (LeDSFs) in L. enzymogenes was identified (31); among them, 13-methyltetradecanoic acid was the major compound and was named L. enzymogenes DSF (LeDSF) in ensuing studies. DSF signaling is mediated by the two-component regulatory system RpfC/RpfG, which is responsible for sensing DSFs and for conveying the signal for subsequent gene expression (32). Although the downstream signal transduction pathway in L. enzymogenes is still unclear, evidence suggests that activated RpfG hydrolyzes cyclic di-GMP of Clp-c-di-GMP, a ubiquitous second messenger that controls numerous cellular processes by regulating intracellular protein effectors and riboswitches. This process leads to release of Clp, which is related to the cAMP receptor protein Crp, and activation of downstream genes (31, 33).
Indole is another type of ubiquitous interkingdom signaling molecule (34, 35). A recently study of ours reported that indole is able to reverse the intrinsic antibiotic resistance of Lysobacter through the two-component regulatory system QseC/QseB. Site-specific mutations in the qseC gene also resulted in antibiotic susceptibility (36). Besides, this antibiotic resistance reversal was facilitated by a novel BtuD-associated importer which can transfer both vitamin B12 and antibiotics (37). So we wanted to know whether other characteristics of L. enzymogenes are also related to indole. At the same time, some studies have reported that indole can affect the motility of some other bacteria, such as the indole-decreased motility of Escherichia coli strains and of ymgA, ymgB and ymgC mutants by 35% to 65% (38); also, exogenous indole can significantly reduce the number of flagella and inhibit the motility of Salmonella enterica serovar strains (39). However, with regard to twitching activity, it is largely unknown how xenogenous small molecules are recognized and transmitted by Lysobacter to regulate twitching activity and the Tfp system. In this study, we found that the interspecies signal indole caused Lysobacter to produce irregular random twitching motility by regulating the gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1. In addition, the intraspecies quorum-sensing signaling factor 13-methyltetradecanoic acid, designated LeDSF, was also essential for Lysobacter to produce twitching motility through indirect regulation of gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC. The synergistic regulation pattern opens a novel approach for studying the mechanism of bacterial twitching activity in the future.
RESULTS
Indole regulates the twitching motility of Lysobacter enzymogenes.
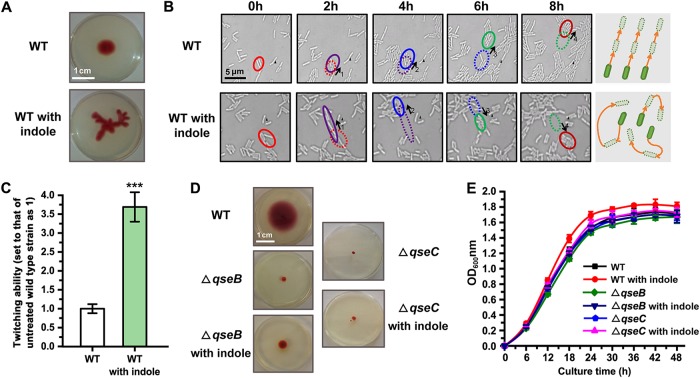
The twitching motility of bacteria is involved in a variety of physiological and sociological behaviors. L. enzymogenes, a type of ubiquitous bacterium that possesses multiple social activities, displays obvious twitching motility and can spread uniformly on semisolid medium without directional selectivity. In this study, we showed that wild-type L. enzymogenes exhibited significantly increased twitching motility when treated with the interspecies signaling molecule indole. Interestingly, the motility pattern of the treated strain became apparently irregular compared with that of the untreated wild-type strain (Fig. 1A). To examine the reason for this phenomenon, we carried out a dynamic imaging assay, the results of which indicated that the twitching motility direction of untreated L. enzymogenes was uniform. As shown in Fig. 1B and Movies S1 and S2 in the supplemental material, a group of bacterial cells moved in the same direction at similar speeds, though a few cells were found to be irregularly rotating. However, in the presence of indole, regular motility was replaced by irregular movements with frequent reorientation. The average speed of the twitching motility of L. enzymogenes was also increased by 3.5-fold under conditions of indole treatment (Fig. 1C). We call this behavior “rapid random twitching” (RRT).
FIG 1.
Indole regulates the twitching motility of Lysobacter enzymogenes. (A) A comparison of the twitching motility patterns of the untreated wild-type (WT) strain and the wild-type strain treated with 0.5 mM indole in motility test medium for 48 h. (B) Dynamic imaging assay under the microscope to detect the twitching motility patterns of the wild-type strain left untreated and/or treated with 0.5 mM indole. The red, purple, blue, green, and dark red circles represent the positions of the bacteria at 0, 2, 4, 6, and 8 h, respectively. 1, motion process at 0 to 2 h; 2, motion process at 2 to 4 h; 3, motion process at 4 to 6 h; 4, motion process at 6 to 8 h. The solid black triangle indicates a fixed position reference. The magnification setting of the microscope was ×640. (C) The speeds of twitching motility of the untreated wild-type strain and the wild-type strain treated with indole were compared. In this experiment, we set the value corresponding to the twitching speed of untreated bacteria to 1. (D) Analysis of twitching ability of the wild-type strain and ΔqseC/B mutants in the presence or absence of 0.5 mM indole. To show the results clearly, the chosen cultivation time was 72 h. The results are representative of biological duplicates. (E) Growth of the wild-type strain and ΔqseC/B mutants. The results are representative of biological duplicates. The error bars represent the standard deviations of results from at least three replicates. OD600nm, optical density at 600 nm. A t test of unpaired unequal variances was performed for testing differences between groups. For statistical analysis, ***, **, and * indicate P < 0.001, P < 0.01, and P < 0.05, respectively.
Our previous studies demonstrated that indole signals can be recognized by the two-component system consisting of QseC, a sensor protein with histidine kinase activity, and QseB, a response regulator that modulates expression of downstream target genes (quorum-sensing E. coli regulators C and B). To determine whether the change in twitching activity described above was driven by the perception of the indole signal, we analyzed the twitching activity of qseC and qseB deletion (ΔqseC/B) mutants. The results showed that indole could no longer induce RRT in ΔqseC or ΔqseB mutants. In addition, the motility of both ΔqseC and ΔqseB mutants was significantly decreased in the presence or absence of indole (Fig. 1D). To exclude the possibility that this result was caused by a change in the mutants’ growth speed, we monitored bacterial growth and found that the mutants grew similarly to the wild-type strain regardless of the presence or absence of indole (Fig. 1E). These results indicate that QseC/QseB-mediated indole signaling is involved in the control of twitching motility in Lysobacter.
Indole regulates twitching motility through the gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1.
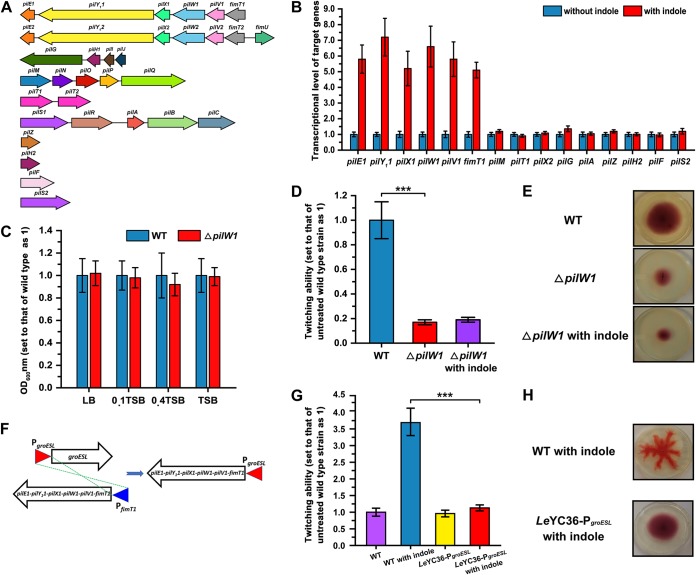
To explore the mechanism of indole-driven RRT activity of Lysobacter, we analyzed the type IV pilin (Tfp) biosynthesis system of L. enzymogenes YC36. Tfp formation relies on 10 gene clusters (or genes) that are distributed in different genomic locations, namely, gene clusters pilE1-pilY11-pilX1-pilW1-pilV1-fimT1, pilE2-pilY12-pilX2-pilW2-pilV2-fimT2-fimU, and pilG-pilH1-pilI-pilJ, gene clusters pilM-pilN-pilO-pilP-pilQ, pilT1-pilT2, and pilS1-pilR-pilA-pilB-pilC, and solo genes pilZ, pilH2, pilF, and pilS2 (Fig. 2A). Among them, indole significantly upregulated gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 by 5-fold to 7-fold compared with untreated L. enzymogenes YC36 as shown by real-time quantitative PCR (qPCR), though the other 9 gene clusters were not affected by indole (Fig. 2B). Consistently, deletion of the qseB gene, which prevents the perception of indole signals, resulted in very weak expression of pilE1-pilY11-pilX1-pilW1-pilV1-fimT1. This result led us to speculate that indole regulates assembly of minor pilins through qseC/B and thus affects twitching motility behavior. To test this hypothesis, we first analyzed whether pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 is necessary or redundant for twitching motility. We knocked out the pilW1 gene because this gene is key for this gene cluster and essential for fiber stability and function (40). According to the results, mutation of pilW1 did not affect the growth of L. enzymogenes (see Fig. S1 in the supplemental material), while the mutant displayed significantly reduced twitching motility (Fig. 2C and D). Regardless of the presence or absence of indole, the twitching activity of the ΔpilW1 mutant was only 20% of that of the wild-type strain (Fig. 2D). In addition, no RRT was observed, even in the presence of indole (Fig. 2E). Moreover, we adopted a double crossover strategy to replace the native promoter of the pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 gene cluster by the promoter of groESL and obtained the mutant LeYC36-PgroESL (Fig. 2F). The groESL promoter was selected because previous studies have shown that groESL is expressed consistently under conventional culture conditions. Our results indicated that the pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 gene cluster was no longer positively regulated by exogenous indole and that the presence of indole did not cause the mutant to exhibit the RRT phenotype (Fig. 2G and H).
FIG 2.
Indole regulates the twitching motility of L. enzymogenes through the gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1. (A) Analysis of type IV pilin (Tfp) biosynthesis and assembly gene clusters of L. enzymogenes YC36. (B) The transcriptional levels of Tfp biosynthesis genes in the presence or absence of 0.5 mM indole. The value corresponding to the relative transcription level of untreated L. enzymogenes YC36 was set as 1. (C) OD600 analysis of the wild-type strain and ΔpilW1 mutant in LB, 0.1 Trypticase soy broth (TSB), 0.4 TSB, and TSB media for 12 h of cultivation. The OD600 of wild-type L. enzymogenes YC36 was set as 1. (D) Analysis of twitching ability of the ΔpilW1 mutant in the presence or absence of 0.5 mM indole. The twitching ability of the untreated wild-type strain was set as 1. (E) Comparison of the twitching motility patterns of the wild-type strain and ΔpilW1 mutant in the presence or absence of 0.5 mM indole in motility test medium. To show the results clearly, we cultivated the strains for 72 h. The results are representative of biological duplicates. (F) Construction of mutant LeYC36-PgroESL. We replaced the promoter of the pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 gene cluster with the promoter of groESL. Sequencing verification of the mutant is provided in the supplemental material. (G) Analysis of twitching ability of the LeYC36-PgroESL mutant in the presence or absence of 0.5 mM indole. The twitching ability of the untreated wild-type strain was set as 1. (H) Comparison of the twitching motility patterns of the wild-type strain and ΔpilW1 mutant in the presence of 0.5 mM indole in motility test medium. To show the results clearly, we cultivated strains for 72 h. The results are representative of biological duplicates. The error bars represent the standard deviations of results from at least three replicates. A t test of unpaired unequal variances was performed for testing differences between groups. For statistical analysis, ***, **, and * indicate P < 0.001, P < 0.01, and P < 0.05, respectively.
Intraspecies LeDSF and interspecies indole signals regulates the twitching motility of Lysobacter.
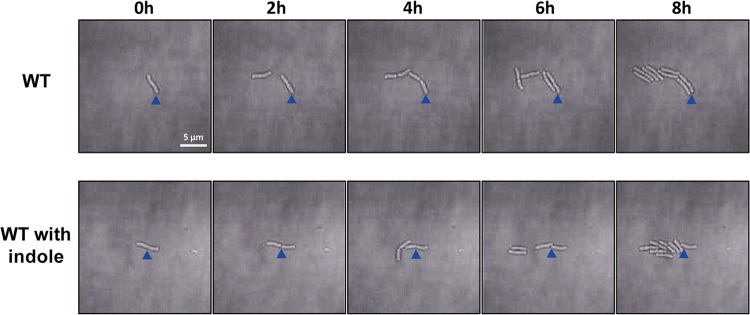
In general, twitching motility is observed with high bacterial density, though relatively few studies of this type of social activity have been performed with low bacterial density or even with individual bacteria. In this study, we observed whether twitching motility appears at low concentrations of L. enzymogenes cells. The results showed that under conditions of extremely low density, individual bacterial cells grew and divided in a stable manner in situ without any movement Moreover, the twitching ability was not enhanced after indole was added (Fig. 3; see also Movies S3 and S4). These results suggested that there are some other substances regulate the twitching ability of L. enzymogenes at low density.
FIG 3.
Dynamic imaging experiments assessing the twitching motility of individual bacterial cells in the absence or presence of 0.5 mM indole. The results showed that the isolated cells did not exhibit twitching motility regardless of the presence of indole. Each solid triangle indicates a fixed position reference. The magnification setting of the microscope was ×640.
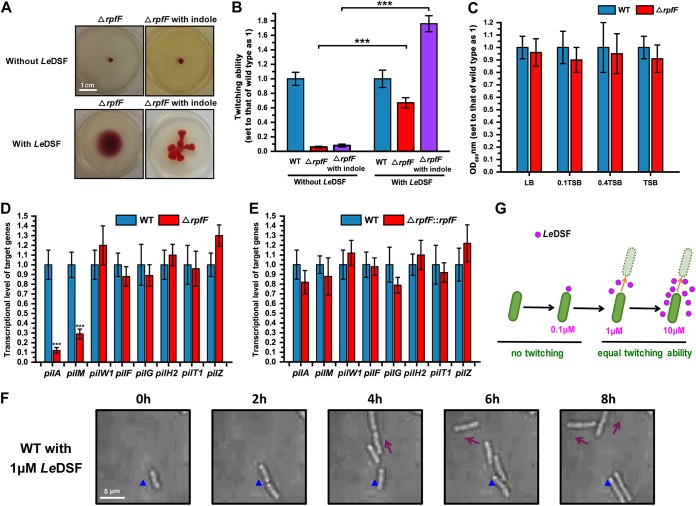
Our findings prompted us to speculate that some quorum-sensing molecules are involved in population-dependent twitching motility. Therefore, we focused our attention on LeDSF, the only intraspecies quorum-sensing molecule identified in the Lysobacter genus so far. Our previous studies have shown that the rpf gene cluster, comprising rpfB, rpfF, rpfG, and rpfC, directs the biosynthesis and sensing of LeDSF and that loss of rpfF causes Lysobacter to lose the ability to produce LeDSF (31). Our results demonstrated that the twitching motility of the ΔrpfF mutant disappeared completely. Indeed, twitching motility and RRT behavior could not be restored, even in the presence of indole (Fig. 4A and B). To exclude the possibility that the observed behavior of the ΔrpfF mutant was caused by decreased growth capacity, we examined the growth of the mutant strain and found that rpfF knockout did not affect growth under conditions of exposure to various culture media (Fig. 4C; see also Fig. S1). When 1 μM exogenous LeDSF was added, the twitching motility of the ΔrpfF strain was restored. Moreover, the exogenous LeDSF-indole combination stimulated RRT formation in the ΔrpfF strain (Fig. 4A and B). Transcriptional analyses showed that deletion of rpfF reduced the expression level of the pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC gene clusters (Fig. 4D) and that expression of the two gene clusters was restored when the ΔrpfF mutant was complemented with ectopically expressed rpfF (Fig. 4E). The pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 gene cluster, which is related to assembly of minor pilins, was not affected by loss of rpfF (Fig. 4D and E). These results indicate that the synthesis and assembly of the Tfp system is regulated not by only one signaling molecule but by at least the interspecies indole and intraspecies LeDSF signaling molecules. Quorum-sensing molecules are produced only when the bacterial population reaches a certain density; low-density bacteria cannot produce LeDSF and therefore cannot generate twitching motility. Thus, we sought to determine whether exogenous LeDSF is able to endow low-density Lysobacter with twitching motility. Subsequently, 1 μM exogenous LeDSF was added to the culture medium, and obvious twitching activity was observed in individual cells, confirming our speculation. Moreover, under conditions in which the amount of LeDSF in the environment was insufficient to stimulate the quorum-sensing activities of the bacteria (0.1 μM), the single cells exhibited no twitching motility (37). Conversely, when LeDSF was present at greater than 1 μM, single Lysobacter cells displayed obvious, regular twitching motility, though an increase in the concentration of LeDSF did not enhance the speed of bacterial movement (Fig. 4F and G; see also Movie S5). This result suggests that when the concentration of the signal molecule LeDSF reaches a certain threshold, it can stimulate the Tfp system.
FIG 4.
Intraspecies LeDSF regulates the twitching motility of L. enzymogenes by indirectly regulating gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC. (A) Comparison of the twitching motility patterns of the ΔrpfF mutant in the presence or absence of 0.5 mM indole or 1 μM LeDSF in motility test medium. For this experiment, we cultivated strains for 48 h. The results are representative of biological duplicates. (B) Analysis of twitching ability of the ΔrpfF mutant in the presence or absence of 0.5 mM indole or 1 μM LeDSF. (C) OD600 analysis of the wild-type strain and ΔrpfF mutant in LB, 0.1 TSB, 0.4 TSB, and undiluted TSB media for 12 h of cultivation. The OD600 of the wild-type strain was set as 1. (D and E) The transcriptional levels of Tfp biosynthesis genes in ΔrpfF (D) and ΔrpfF::rpfF (E) mutants. The relative transcription level of untreated L. enzymogenes YC36 was set as 1. (F) Twitching motility of low-density, individual L. enzymogenes cells induced by exogenous 1 μM LeDSF. The results showed that individual cells exhibit twitching motility in the presence of 1 μM LeDSF. The magnification setting of the microscope was ×640. The solid triangle indicates a fixed position reference. The results are representative of biological duplicates. (G) A schematic diagram of the LeDSF-induced twitching motility of L. enzymogenes YC36. The error bars represent the standard deviations of results from at least three replicates. A t test of unpaired unequal variances was performed for testing differences between groups. For statistical analysis, ***, **, and * indicate P < 0.001, P < 0.01, and P < 0.05, respectively.
DISCUSSION
The molecular mechanisms and physiological functions of the interspecies signal indole in bacteria have been studied by several research groups. For example, early studies have illustrated that SidA, a DNA-binding transcriptional dual regulator, mediates indole signaling (41–43). In antibiotic-resistant bacteria, indole activates expression of several stress response genes and xenobiotic exporter genes, including mdtA, mdtE yceL, cusB, acrD, acrE, and emrK (28, 44). There is ample evidence showing that indole reduces persistence in E. coli (45–47). Furthermore, indole can induce the population-dependent antibiotic resistance of bacteria. A more recent study reported that indole signaling is sensed by Lysobacter through the qseB/C two-component system, thus reversing intrinsic multiantibiotic resistance (37). However, it remains largely unknown whether indole signaling affects other bacterial social behaviors and, if so, what mechanisms are involved.
Twitching motility is one of the major social activities of a wide range of bacteria, including M. xanthus, which exhibits complex social behaviors, and the opportunistic pathogen P. aeruginosa. The pilus is mainly composed of a combination of major pilins and low-abundance minor pilins. We show here that the interspecies signaling molecule indole is sensed by cells and then regulates gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 of L. enzymogenes YC36 through the two-component system QseC/QseB and that intraspecies LeDSF regulates expression of gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC. Why does Lysobacter use both intraspecies and interspecies signals to regulate twitching motility? We hypothesize that this strategy is beneficial to environmental adaptation in bacteria. When there are no intraspecies signals or interspecies signals in the environment, Lysobacter cells are aware that there is no competitive pressure and that they do not need to improve their survival abilities through social behaviors, such as twitching motility. However, when the intraspecies signal LeDSF appears in the environment, Lysobacter can perceive that there is more competition, and it becomes necessary for cells to communicate with each other through regular twitching motility. When the environment has both the intraspecies signal LeDSF and the interspecies signal indole, a given Lysobacter cell can recognize the surrounding Lysobacter cells and other bacterial species, causing Lysobacter to initiate a series of physiological behaviors, including rapid random twitching motility, to adapt to the changing external environment and to invoke competitive behaviors (15, 16).
Minor pilins are important players in the biogenesis and function of Tfp in several bacterial species, including E. coli, M. xanthus, and P. aeruginosa (48–55) In P. aeruginosa, it has been shown that minor pilins are positively regulated by the two-component system FimS-AlgR (40). However, whole-genome sequencing of L. enzymogenes YC36 showed that the strain did not possess the FimS-AlgR two-component system. Thus, how gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 is regulated in Lysobacter is an interesting issue. In this study, we found that the two-component system QseB/C regulates gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1. Nonetheless, it remains unclear whether QseB directly regulates gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 or regulates those genes indirectly by regulating other regulatory genes, such as the global regulatory factor CLP gene. This issue needs to be addressed in a follow-up study.
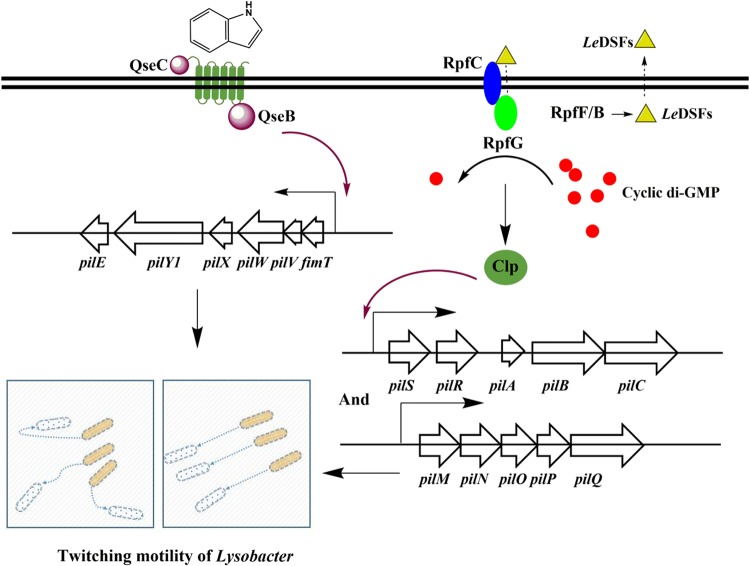
Compared with regulation of indole-induced gene cluster pilE1-pilY11-pilX1-pilW1-pilV1-fimT1, regulation of gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC has been clarified through this study and previously published research. For example, the Qian group showed that the Clp global regulator has a dual role in controlling twitching motility in L. enzymogenes OH11 (22). Clp directly controls transcription of pilA and pilM-pilN-pilO-pilP-pilQ by binding to their promoter regions. This finding suggests that LeDSF does not directly regulate major pilin biosynthesis and assembly genes but modulates them indirectly through RpfC and RpfG, leading to a reduction in cyclic di-GMP levels. The absence of rpfF leads to the absence of LeDSF and to elevations in the cellular level of cyclic di-GMP (56); thus, inactivated Clp can no longer activate the pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC gene clusters (Fig. 5). Overall, the abundance of Clp directly controls gene clusters pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC.
FIG 5.
Schematic diagram of the synergistic regulation of the twitching motility of L. enzymogenes by the intraspecies signal LeDSF and the interspecies signal indole. Reduction in cyclic di-GMP levels through activation of RpfG allows Clp to bind to the promoters of the pilM-pilN-pilO-pilP-pilQ and pilS1-pilR-pilA-pilB-pilC gene clusters. Whether QseB directly regulates expression of pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 is unknown.
The twitching motility of L. enzymogenes is closely associated with the attachment and colonization of fungal mycelia (57). Thus, Tfp formation is recognized as a potential antifungal strategy employed by Lysobacter. As Lysobacter species also produce multiple bioactive natural products and the regulatory mechanism remains largely unexplored, it will be interesting to explore whether indole/LeDSF-induced twitching motility also plays a role in regulating antibiotic production in Lysobacter.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general methods.
Lysobacter strains were grown on lysogeny broth medium (LB) or 40%-strength tryptic soy broth (TSB; Sigma) (1/10) at 30°C (58). E. coli DH5α was grown in LB medium and used as the host for DNA manipulation. When required, the following antibiotics were used: gentamicin, 40 mg/ml; ampicillin, 100 mg/ml; kanamycin, 50 mg/ml. The concentration of indole in all experiments was 0.5 mM. Genomic DNA of L. enzymogenes was prepared as previously described (59). Plasmid construction and related DNA fragment extractions were performed following the instructions provided with kits purchased from Omega (plasmid minikit I and gel extraction kit; Omega, USA). Restriction enzymes and other molecular biology materials were purchased from TaKaRa (TaKaRa Bio Group, Japan). PCR primers were synthesized by Tsingke Biological Technology Company (Qingdao, China). The bacterial strains and plasmids used in this study are described in Table 1. The primers used are described in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source (reference) |
|---|---|---|
| Strains | ||
| L. enzymogenes YC36 | Wild-type strain | This study |
| Escherichia coli DH5α | Competent cells that can be used for DNA manipulation | This study |
| Escherichia coli S17-1 | RP4-2Tc::Mu-Kn::Tn7 pro hsdR recA; host for required plasmids; conjugal donor | This study |
| Escherichia coli ΔpilW | pilW gene deletion mutant strain | This study |
| Escherichia coli ΔpilW::pilW | pilW complementary strain of the ΔpilW mutant | This study |
| Escherichia coli ΔrpfF | rpfF gene deletion strain | Laboratory of L. Du (31) |
| Escherichia coli ΔrpfF::rpfF | rpfF complementary strain of the ΔrpfF mutant | This study |
| Escherichia coli ΔqseB | qseB gene deletion strain | Laboratory of L. Du (36) |
| Escherichia coli ΔqseC | qseC gene deletion strain | Laboratory of L. Du (36) |
| Plasmids | ||
| pEX18 | Gmr; oriT+ sacB+, gene replacement vector with MCS from pUC18a | This study |
| pEX18-T | Used for gene deletion; pEX18 carrying the orf gene from L. enzymogenes | This study |
| pHmgA-P | Contains promoter of HSAF and selection marker (used for target gene complementation) | Laboratory of L. Du (71) |
| pHmgA-P-G | The complementary gene was linked to pHmgA-P and formed pHmgA-P-G; this plasmid was used for expression of the target gene | This study |
Gmr, gentamycin resistance.
TABLE 2.
Primers used in this study
| Purpose | Primer | Sequence |
|---|---|---|
| qPCR assay | pilE-up | 5′-AGGCCAAGGCCGATCTGGC-3′ |
| pilE-down | 5′-GATGGTCAGGTTCTTGCAC-3′ | |
| pilY1-up | 5′-AACGGCAGCGTGCTGACCT-3′ | |
| pilY1-down | 5′-AGCTCTTCCTTCAGGCGCG-3′ | |
| pilX-up | 5′-ACGCATGTCGACCAATTAC-3′ | |
| pilX-down | 5′-TGCCGACCTTGATGCTGGA-3′ | |
| pilW-up | 5′-CTGACCAATCCGGCCGGCT-3′ | |
| pilW-down | 5′-CAGCACGACGACGTCGCTG-3′ | |
| pilV-up | 5′-AACTACCGCACCCGCGCCA-3′ | |
| pilV-down | 5′-GCTGCGGCGGACGATCGC-3′ | |
| fimT-up | 5′-TGGCCCTGCCCAGTTTCAC-3′ | |
| fimT-down | 5′-CACGCTCTCGGTGCGGC-3′ | |
| pilM-up | 5′-CGGCATGCCGGCCGATCT-3′ | |
| pilM-down | 5′-AGATCGGCCGGCATGCCG-3′ | |
| pilT-up | 5′-GGTCGAAGACCCGATCGA-3′ | |
| pilT-down | 5′-GATGCGGTCCACGGTCT-3′ | |
| pilX2-up | 5′-GAACTACGTGCCGTATCCGATC-3′ | |
| pilX2-down | 5′-GTCATGCGCCGCTACGGAGCG-3′ | |
| pilG-up | 5′-ATCGACGGATTCGAGGCGT-3′ | |
| pilG-down | 5′-GCGGATCGCGTCGAGGAGC-3′ | |
| pilA-up | 5′-TCGCCGAGTCGGCCAACGA-3′ | |
| pilA-down | 5′-GCGGCAGGACGACGGGAT-3′ | |
| pilZ-up | 5′-TACATGCCGTTCCTGAAGTA-3′ | |
| pilZ-down | 5′-TCAACGTGCCCGCCAGCAG-3′ | |
| pilH2-up | 5′-AGAAGACGGCGCCGCCGGC-3′ | |
| pilH2-down | 5′-AGGGCAGCAACTGGCCGAT-3′ | |
| pilF-up | 5′-ACGATTTCAAGAACGACCA-3′ | |
| pilF-down | 5′-GTTGAGCGCCGCGCCGCGT-3′ | |
| pilS2-up | 5′-CGCAGCCTGGCCGGCCAC-3′ | |
| pilS2-down | 5′-AGCCGGCGATGAAGCGCT-3′ | |
| Construction of vectors for gene disruption | pilW-up | 5′-TGGGGTACCACCGTTCCGACCG-3′ |
| pilW-down | 5′-TAGGAATTCAACTGGTCGCGGGCA-3′ | |
| Construction of vectors for gene in-frame deletion | pilW-1-up | 5′-CGGGATCCACGCCGATGCCGAGCAACA-3′ |
| pilW-1-down | 5′-GCTCTAGACGCCACGGATCGCTCCGT-3′ | |
| pilW-2-up | 5′-GCTCTAGAATCTTGCCGTTGTAGTAG-3′ | |
| pilW-2-down | 5′-CCCAAGCTTTGCGCAAGTTCGAACTGT-3′ | |
| Mutant verification | pilW-V-up | 5′-CCTATGGTTGCGATGGCTCA-3′ |
| pilW-V-down | 5′-GCAAGGCGATTAAGTTGGGTA-3′ | |
| Gene complementation | rpfF-C-up | 5′-CGGGATCCATGAGCACCATCGAAA-3′ |
| rpfF-C-down | 5′-CCGCTCGAGTTACGCGGCCACGGC-3′ | |
| Construction of mutant LeYC36-PgroESLa | PfimT1-1-up | 5′-CGGAATTCGTCGCACAGCAGCCAGGG-3′ |
| PfimT1-1-down | 5′-GGGGTACCGCTTATGCGAATCAAGAC-3′ | |
| PgroESL-1-up | 5′-GGGGTACCGAAGGCCTTCCTCAGCCG-3′ | |
| PgroESL-1-down | 5′-CGGGATCCGGCGACCTCTGTAAGTAAT-3′ | |
| PfimT1-2-up | 5′-CGGGATCCATGAGCAGGCGGCGAACTG-3′ | |
| PfimT1-2-down | 5′-CCCAAGCTTGAAATCGCGCTTGCCGCCT-3′ | |
We replaced the promoter of the pilE1-pilY11-pilX1-pilW1-pilV1-fimT1 gene cluster with the promoter of groESL.
Twitching motility test for L. enzymogenes.
The motility test medium contained 1% peptone, 0.3% beef extract, 0.5% NaCl, and 0.4% agar. To facilitate visual observation of the motility of L. enzymogenes, 2,3,5-triphenyltetrazolium chloride (1% TTC; 5 ml) was added to the medium (1 liter) as a color indicator (60). As the bacterial cells grew, the colorless TTC was converted into reddish TPF by the reductase activities of the bacterial cells. Each of the strains was inoculated at the center of the plate using a sterile toothpick to penetrate the 3-mm-thick solid motility test medium, and the plates were placed in a 28°C incubator for 2 to 3 days. An indirect measurement assay was performed to calculate the twitching speed of the bacteria. Bacterial movement can be observed by continuous photography under a microscope for 30 h. The bacteria were randomly selected for twitching speed calculation according to their movement distance at a specific time point under the microscope. In this experiment, we set the value representing the twitching speed of untreated bacteria to 1.
RNA extraction and qPCR.
Wild-type and mutant strains of L. enzymogenes YC36 were grown in 100 ml 40% TSB medium for 36 h. A 3-ml aliquot of cells was transferred to sterile tubes and centrifuged for 5 min at 15,000 × g. TRIzol solution was used to isolate total RNA from the cells following the manufacturer’s instructions. A PrimeScript reverse transcription reagent kit was used with a genomic DNA (gDNA) eraser kit (TaKaRa Biotechnology) to remove DNA and to reverse transcribe the mRNA. qPCR was then carried out in a total reaction volume of 20 μl containing 200 nM primers, 0.5 μl of diluted cDNA template (0.5 μl of RNA sample was used as a negative template to ensure that DNA contamination was removed), 10 μl of EvaGreen 2× qPCR Master Mix and 8.5 μl of RNase-free double-distilled water (ddH2O). 16S rRNA was used as the reference gene. qPCR was performed using a StepOne qPCR system (AB Applied Biosystems). The qPCR program was designed as described previously (61). All experiments were repeated three times.
Bioinformatics analyses and gene information.
Genomic DNA of our Lysobacter enzymogenes YC36 strain (GenBank accession number CP040656) was extracted according to a procedure previously described by Moore et al. (62), and the DNA G+C content was determined by whole-genome sequencing. The genome sequencing of strain L. enzymogenes YC36 was performed on an Illumina HiSeq platform (Illumina, USA). SOAPdenovo assembler software was applied to assemble these reads (63) (http://sourceforge.net/projects/soapdenovo2/files/SOAPdenovo2/). Putative coding sequences (CDSs) were identified by Glimmer 3.0 (64). RNAmmer (65) and tRNAscan (66) were used to predict rRNAs and tRNAs, respectively, and small RNA (sRNA) was identified using the Rfam database (67). The annotation and bioinformatics analyses of this genome were carried out by the use of the RAST server (68) (http://rast.nmpdr.org/) and EMBOSS (The European Molecular Biology Open Software Suite) (http://emboss.open-bio.org/).The annotated sequences of the twitching-motility-related genes were checked by BLAST with the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The primers for qPCR and the gene manipulation assays were designed using Primer Premier 5 (69). ENDscript 2 software was used to compare the target proteins (70). The primers used for qPCR are listed in Table 2. Gene information and GenBank accession numbers of the Tfp assembly system of L. enzymogenes YC36 are listed in Table S1 in the supplemental material.
Twitching-motility detection under a microscope.
All images were captured under an inverted microscope. A FCS2 flow cell system (Bioptechs) was used to record time-lapse images. The cells were cultured, collected, washed three times with 40% TSB medium, and imaged on a gel pad that contained 2% low-melting-temperature agarose. L. enzymogenes cells were observed at 30°C. An indirect measurement assay was performed to calculate the twitching speed of the bacteria. Bacterial movement can be observed by continuous photography under a microscope for 30 h. The bacteria were randomly selected for twitching speed calculation according to their movement distance (measured in micrometers) at a specific time point under the microscope. The magnification setting of the microscope was ×640.
Generation of gene deletion, promoter replacement, and gene complemented mutants.
To construct vectors for gene deletion in L. enzymogenes YC36, upstream and downstream fragments were amplified using the primer pairs listed in Table 2. Genomic DNA was used as the PCR template. The upstream and downstream fragments of each gene were cloned into pEX18 to generate pEX18-G. The resulting plasmids were transferred into L. enzymogenes via conjugation according to a method described previously (59). Confirmed colonies were subjected to a second round of crossover to produce the gene deletion mutants. Target colonies were selected by PCR and verified by sequencing. Similarly, the promoter replacement experiment was performed using a double-crossover strategy and the same vector as described above. The only difference was the addition of PgroELS promoter sequences between the upstream and downstream homologous arms. (Please see Table 2 for primer information.) pHmgA-P was used for a gene complement assay, with the complete gene sequence amplified and linked to the vector to generate pHmgA-P-G. pHmgA-P-G plasmids were transferred into L. enzymogenes by conjugation according to a previously described method (71).
Accession number(s).
Newly determined sequence data were deposited in GenBank under accession numbers MK967260 to MK967276.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (no. 31870023, 31571970, and 41506160), the Fundamental Research Funds for the Central Universities (no. 201941009), the Young Elite Scientists Sponsorship Program by CAST (no. YESS20160009), and the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (no. 2018SDKJ0406-4).
Y.W. and Y.H. conceived the project. Y.W., L.D., and Y.H. designed the experiments. T.F., Y.H., B.L., Z.L., Y.Y., X.L., and Q.S. carried out experiments. Y.W., T.F., and Y.H. analyzed the data. Y.W. wrote the manuscript draft. L.D. and T.F. revised the manuscript. All of us read and approved the submission for publication.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01742-19.
REFERENCES
- 1.Xie Y, Wright S, Shen Y, Du L. 2012. Bioactive natural products from Lysobacter. Nat Prod Rep 29:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panthee S, Hamamoto H, Paudel A, Sekimizu K. 2016. Lysobacter species: a potential source of novel antibiotics. Arch Microbiol 198:839–837. doi: 10.1007/s00203-016-1278-5. [DOI] [PubMed] [Google Scholar]
- 3.Bonner DP, O’Sullivan J, Tanaka SK, Clark JM, Whitney RR. 1988. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. II. Biological properties. J Antibiot 41:1745–1751. doi: 10.7164/antibiotics.41.1745. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Rokni-Zadeh H, De VM, Ghequire MG, Sinnaeve D, Xie GL, Rozenski J, Madder A, Martins JC, De Mot R. 2013. The antimicrobial compound xantholysin defines a new group of Pseudomonas cyclic lipopeptides. PLoS One 8:e62946. doi: 10.1371/journal.pone.0062946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee W, Schaefer K, Qiao Y, Srisuknimit V, Steinmetz H, Müller R, Kahne D, Walker S. 2016. The mechanism of action of lysobactin. J Am Chem Soc 138:100–103. doi: 10.1021/jacs.5b11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato A, Nakaya S, Ohashi Y, Hirata H, Fujii K, Harada KI. 1997. WAP-8294A2, a novel anti-MRSA antibiotic produced by Lysobacter sp. J Am Chem Soc 119:6680–6681. doi: 10.1021/ja970895o. [DOI] [Google Scholar]
- 7.Zhang W, Li Y, Qian G, Wang Y, Chen H, Li YZ, Liu F, Shen Y, Du L. 2011. Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob Agents Chemother 55:5581–5589. doi: 10.1128/AAC.05370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Olson AS, Su W, Dussault PH, Du L. 2015. Fatty acyl incorporation in the biosynthesis of WAP-8294A, a group of potent anti-MRSA cyclic lipodepsipeptides. RSC Adv 5:105753–105759. doi: 10.1039/c5ra20784c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. 2007. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother 51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. 2011. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc 133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lou L, Chen H, Cerny RL, Li Y, Shen Y, Du L. 2012. Unusual activities of the thioesterase domain for the biosynthesis of the polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes C3. Biochemistry 51:4–6. doi: 10.1021/bi2015025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Qian G, Ye Y, Wright S, Chen H, Shen Y, Liu F, Du L. 2016. Heterocyclic aromatic N-oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org Lett 18:2495–2498. doi: 10.1021/acs.orglett.6b01089. [DOI] [PubMed] [Google Scholar]
- 13.Chang YW, Kjær A, Ortega DR, Kovacikova G, Sutherland JA, Rettberg LA, Taylor RK, Jensen GJ. 2017. Architecture of the Vibrio cholerae toxin-coregulated pilus machine revealed by electron cryotomography. Nat Microbiol 2:16269. doi: 10.1038/nmicrobiol.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma G, Burrows LL, Singer M. 2018. Diversity and evolution of myxobacterial type IV pilus systems. Front Microbiol 9:1630. doi: 10.3389/fmicb.2018.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grohmann E, Christie PJ, Waksman G, Backert S. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol 107:455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hospenthal MK, Costa TRD, Waksman G. 2017. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol 15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 17.Bretl DJ, Müller S, Ladd KM, Atkinson SN, Kirby JR. 2016. Type IV-pili dependent motility is co-regulated by PilSR and PilS2R2 two-component systems via distinct pathways in Myxococcus xanthus. Mol Microbiol 102:37–53. doi: 10.1111/mmi.13445. [DOI] [PubMed] [Google Scholar]
- 18.Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J Bacteriol 190:2411–2421. doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabass B, Koch MD, Liu G, Stone HA, Shaevitz JW. 2017. Force generation by groups of migrating bacteria. Proc Natl Acad Sci U S A 114:7266–7271. doi: 10.1073/pnas.1621469114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilmury SLN, Burrows LL. 2018. The Pseudomonas aeruginosa PilSR two-component system regulates both twitching and swimming motilities. mBio 9:e01310-18. doi: 10.1128/mBio.01310-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehl-Fie TE, Porsch EA, Miller SE, St Geme JW. 2009. Expression of Kingella kingae type IV pili is regulated by σ54, PilS, and PilR. J Bacteriol 191:4976–4986. doi: 10.1128/JB.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Zhao Y, Zhang J, Zhao Y, Shen Y, Su Z, Xu G, Du L, Huffman JM, Venturi V, Qian G, Liu F. 2014. Transcriptomic analysis reveals new regulatory roles of Clp signaling in secondary metabolite biosynthesis and surface motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol 98:9009–9020. doi: 10.1007/s00253-014-6072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Qian G, Chen Y, Du L, Liu F, Yuen GY. 2015. PilG is involved in the regulation of twitching motility and antifungal antibiotic biosynthesis in the biological control agent Lysobacter enzymogenes. Phytopathology 105:1318–1324. doi: 10.1094/PHYTO-12-14-0361-R. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Xia J, Su Z, Xu G, Gomelsky M, Qian G, Liu F. 2017. Lysobacter PilR, the regulator of type IV pilus synthesis, controls antifungal antibiotic production via a cyclic di-GMP pathway. Appl Environ Microbiol 83:e03397-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Shen D, Odhiambo BO, Dan X, Han S, Chou SH, Qian G. 2018. Two direct gene targets contribute to Clp-dependent regulation of type IV pilus-mediated twitching motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol 102:7509–7519. doi: 10.1007/s00253-018-9196-x. [DOI] [PubMed] [Google Scholar]
- 26.Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatterjee A, Cook LC, Shu CC, Chen Y, Manias DA, Ramkrishna D, Dunny GM, Hu WS. 2013. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci U S A 110:7086–7090. doi: 10.1073/pnas.1212256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsek MR, Greenberg EP. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci U S A 97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Y, Wang Y, Tombosa S, Wright S, Huffman J, Yuen G, Qian G, Liu F, Shen Y, Du L. 2015. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biotechnol 99:801–811. doi: 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian G, Wang Y, Liu Y, Xu F, He YW, Du L, Venturi V, Fan J, Hu B, Liu F. 2013. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl Environ Microbiol 79:6604–6616. doi: 10.1128/AEM.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan RP, An S, Allan JH, Mccarthy Y, Dow JM. 2015. The DSF family of cell-cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog 11:e1004986. doi: 10.1371/journal.ppat.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bansal T, Alaniz RC, Wood TK, Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A 107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. 2013. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8:e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Y, Wang Y, Yu Y, Chen H, Shen Y, Du L. 2017. Indole-induced reversion of intrinsic multiantibiotic resistance in Lysobacter enzymogenes. Appl Environ Microbiol 83:e00995-17. doi: 10.1128/AEM.00995-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Tian T, Zhang J, Jin X, Yue H, Zhang XH, Du L, Bai F. 2019. Indole reverses intrinsic antibiotic resistance by activating a novel dual-function importer. mBio 10:e00676-19. doi: 10.1128/mBio.00676-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Page R, García-Contreras R, Palermino J-M, Zhang X-S, Doshi O, Wood TK, Peti W. 2007. Structure and function of the E. coli protein YmgB: a protein critical for biofilm formation and acid-resistance. J Mol Biol 373:11–26. doi: 10.1016/j.jmb.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikaido E, Giraud E, Baucheron S, Yamasaki S, Wiedemann A, Okamoto K, Takagi T, Yamaguchi A, Cloeckaert A, Nishino K. 2012. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog 4:5. doi: 10.1186/1757-4749-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marko VA, Kilmury SLN, MacNeil LT, Burrows LL. 2018. Pseudomonas aeruginosa type IV minor pilins and PilY1 regulate virulence by modulating FimS-AlgR activity. PLoS Pathog 14:e1007074. doi: 10.1371/journal.ppat.1007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J, Jayaraman A, Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J, Maeda T, Hong SH, Wood TK. 2009. Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl Environ Microbiol 75:1703–1716. doi: 10.1128/AEM.02081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J, Zhang XS, Hegde M, Bentley WE, Jayaraman A, Wood TK. 2008. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J 2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- 44.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y, Kwan BW, Osbourne DO, Benedik MJ, Wood TK. 2015. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ Microbiol 17:1275–1285. doi: 10.1111/1462-2920.12567. [DOI] [PubMed] [Google Scholar]
- 46.Kwan BW, Osbourne DO, Hu Y, Benedik MJ, Wood TK. 2015. Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol Bioeng 112:588–600. doi: 10.1002/bit.25456. [DOI] [PubMed] [Google Scholar]
- 47.Lee JH, Kim YG, Gwon G, Wood TK, Lee J. 2016. Halogenated indoles eradicate bacterial persister cells and biofilms. AMB Express 6:123. doi: 10.1186/s13568-016-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alm RA, Hallinan JP, Watson AA, Mattick JS. 1996. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol 22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 49.Alm RA, Mattick JS. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol 16:485–496. doi: 10.1111/j.1365-2958.1995.tb02413.x. [DOI] [PubMed] [Google Scholar]
- 50.Alm RA, Mattick JS. 1996. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol 178:3809–3817. doi: 10.1128/jb.178.13.3809-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell MA, Darztns A. 1994. The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol Microbiol 13:973–985. doi: 10.1111/j.1365-2958.1994.tb00489.x. [DOI] [PubMed] [Google Scholar]
- 52.Ramer SW, Schoolnik GK, Wu CY, Hwang J, Schmidt SA, Bieber D. 2002. The type IV pilus assembly complex: biogenic interactions among the bundle-forming pilus proteins of enteropathogenic Escherichia coli. J Bacteriol 184:3457–3465. doi: 10.1128/JB.184.13.3457-3465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winther-Larsen HC, Wolfgang M, Dunham S, van Putten JP, Dorward D, Løvold C, Aas FE, Koomey M. 2005. A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol Microbiol 56:903–917. doi: 10.1111/j.1365-2958.2005.04591.x. [DOI] [PubMed] [Google Scholar]
- 54.Carbonnelle E, Helaine S, Nassif X, Pelicic V. 2006. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol 61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- 55.Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Søgaard-Andersen L, Jensen GJ. 2016. Architecture of the type IVA pilus machine. Science 110:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryan RP. 2013. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology 159:1286–1297. doi: 10.1099/mic.0.068189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia J, Chen J, Chen Y, Qian G, Liu F. 2018. Type IV pilus biogenesis genes and their roles in biofilm formation in the biological control agent Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol 102:833–846. doi: 10.1007/s00253-017-8619-4. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Qian G, Li Y, Wang Y, Wang Y, Wright S, Li Y, Shen Y, Liu F, Du L. 2013. Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS One 8:e66633. doi: 10.1371/journal.pone.0066633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi DY, Reedy RM, Palumbo JD, Zhou JM, Yuen GY. 2005. A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl Environ Microbiol 71:261–269. doi: 10.1128/AEM.71.1.261-269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. 2009. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol 73:1020–1031. doi: 10.1111/j.1365-2958.2009.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Li H, Cui X, Zhang XH. 2017. A novel stress response mechanism, triggered by indole, involved in quorum quenching enzyme MomL and iron-sulfur cluster in Muricauda olearia Th120. Sci Rep 7:4252. doi: 10.1038/s41598-017-04606-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore ERB, Arnscheidt A, Krüger A, Strömpl C, Mau M. 1999. Simplified protocols for the preparation of genomic DNA from bacterial cultures, chapter 1.6.1, p 1–15. In Akkermans ADL, van Elsas JD, de Bruijn FJ (ed), Molecular microbial ecology manual. Kluwer Academic Press, Dordrecht, the Netherlands. [Google Scholar]
- 63.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res 37:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh VK, Mangalam AK, Dwivedi S, Naik S. 1998. Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques 24:318–319. doi: 10.2144/98242pf02. [DOI] [PubMed] [Google Scholar]
- 70.Xavier R, Patrice G. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Qian G, Liu F, Li YZ, Shen Y, Du L. 2013. Facile method for site-specific gene integration in Lysobacter enzymogenes for yield improvement of the anti-MRSA antibiotics WAP-8294A and the antifungal antibiotic HSAF. ACS Synth Biol 2:670–678. doi: 10.1021/sb4000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.