Cytochrome P450 monooxygenases (P450s) are able to introduce oxygen atoms into nonreactive hydrocarbon compounds under mild conditions, thereby offering significant advantages compared to chemical catalysts. Promiscuous P450s with broad substrate specificity and reaction diversity have significant potential for applications in various fields, including synthetic biology. The study of the function, molecular mechanisms, and rational engineering of substrate-promiscuous P450s from microbial sources is important to fulfill this potential. Here, we present a microbial substrate-promiscuous P450, CYP105D7, which can catalyze hydroxylation of steroids. The loss of the bulky side chains of Arg70 and Arg190 in the active site and substrate entrance resulted in an up to 9-fold increase in the substrate conversion rate. These findings will support future rational and semirational engineering of P450s for applications as biocatalysts.
Keywords: cytochrome P450, Streptomyces avermitilis, hydroxylation, steroids, bioconversion, CYP105D7
ABSTRACT
Our previous study showed that CYP105D7, a substrate-promiscuous P450, catalyzes the hydroxylation of 1-deoxypentalenic acid, diclofenac, naringenin, and compactin. In this study, 14 steroid compounds were screened using recombinant Escherichia coli cells harboring genes encoding CYP105D7 and redox partners (Pdx/Pdr, RhFRED, and FdxH/FprD), and the screening identified steroid A-ring 2β- and D-ring 16β-hydroxylation activity. Wild-type CYP105D7 was able to catalyze the hydroxylation of five steroids (testosterone, progesterone, 4-androstene-3,17-dione, adrenosterone, and cortisone) with low (<10%) conversion rates. Structure-guided site-directed mutagenesis of arginine residues around the substrate entrance and active site showed that the R70A and R190A single mutants and an R70A/R190A double mutant exhibited greatly enhanced conversion rates for steroid hydroxylation. For the conversion of testosterone in particular, the R70A/R190A mutant's kcat/Km values increased 1.35-fold and the in vivo conversion rates increased significantly by almost 9-fold with high regio- and stereoselectivity. Molecular docking analysis revealed that when Arg70 and Arg190 were replaced with alanine, the volume of the substrate access and binding pocket increased 1.08-fold, which might facilitate improvement of the hydroxylation efficiency of steroids.
IMPORTANCE Cytochrome P450 monooxygenases (P450s) are able to introduce oxygen atoms into nonreactive hydrocarbon compounds under mild conditions, thereby offering significant advantages compared to chemical catalysts. Promiscuous P450s with broad substrate specificity and reaction diversity have significant potential for applications in various fields, including synthetic biology. The study of the function, molecular mechanisms, and rational engineering of substrate-promiscuous P450s from microbial sources is important to fulfill this potential. Here, we present a microbial substrate-promiscuous P450, CYP105D7, which can catalyze hydroxylation of steroids. The loss of the bulky side chains of Arg70 and Arg190 in the active site and substrate entrance resulted in an up to 9-fold increase in the substrate conversion rate. These findings will support future rational and semirational engineering of P450s for applications as biocatalysts.
INTRODUCTION
P450s form a superfamily of heme-thiolate proteins that catalyze an enormous variety of reactions as part of xenobiotic metabolism, steroid biosynthesis, and natural product biosynthesis (1). P450s are useful for mediating the regio- and stereospecific hydroxylation, epoxidation, demethylation, sulfoxidation, and N-oxidation of diverse substrates (2). The “promiscuity” of enzymes is a particularly promising starting point for the development of versatile biocatalysts through the application of directed laboratory evolution and protein engineering techniques (3). Some P450s can accommodate ambiguous substrates in the same active site. Generally, enzymes catalyze the conversion of a range of similar substrates through identical reaction mechanisms, representing what is referred to as broad substrate specificity (4). For example, CYP154C3 from Streptomyces griseus can catalyze 16α-hydroxylation of a broad range of steroids, which are not considered representative of promiscuity (5). Certain P450s can catalyze the conversion of dissimilar substrates with different binding modes, conferring promiscuous functions to these enzymes (4). A typical example is human CYP3A4, which is able to metabolize approximately half of the drugs in clinical use (6). However, recombinant P450 expression, membrane binding, protein folding, and posttranslational modifications remain obstacles impeding successful application. Microbial P450s are of vital interest as new biocatalysts because of their tremendous diversity and functional versatility and because their heterologous expression occurs more easily than that of mammalian P450s (7).
The CYP105 family plays a crucial role in the degradation of xenobiotics and the biosynthesis of natural products (8). This family comprises at least 17 subfamilies represented in the Streptomyces (9). CYP105A3 from Streptomyces carbophilus and CYP105A1 from S. griseolus have both been successfully applied in industry as biocatalysts. CYP105A3 converts compactin to pravastatin via 6β-hydroxylation, and a mutant (G52S/F89I/P159A/D269E/T323A/E370V/T85F/T119S/V194N/N363Y) showed 29.3-fold-higher biotransformation activity than the wild type after engineering (10, 11). CYP105A1 catalyzes the sequential hydroxylation of vitamin D3 into 1α,25-dihydroxyvitamin D3, and a double mutant (R73V/R84A) yielded 435-fold-higher kcat/Km values than the wild type (12, 13). In our previous studies, CYP105D7 from S. avermitilis was shown to catalyze the hydroxylation of 1-deoxypentalenic acid, diclofenac, flavanone, and compactin in vivo and/or in vitro, and this enzyme exhibited broad substrate specificity that suggested the possibility of industrial applications (9, 14–16).
Steroids represent important natural compounds with a wide range of beneficial properties, including anticancer, anti-inflammatory, immunosuppressive, progestational, antiandrogenic, diuretic, and contraceptive activities (17, 18). The functions of steroids rely on properties related to their chemical structure, including the number, type, and positions of the functional groups. In terms of industrial applications, P450s are key enzymes for the synthesis of highly valuable steroids by stereo- and regioselective hydroxylation. These enzymes are notable biocatalysts in their ability to insert hydroxyl groups into unreactive C-H bonds under mild conditions. In recent years, several microbial P450s have been reported to catalyze the hydroxylation of steroids, for example, the members of the CYP106 family (CYP106A1 and CYP106A2), the CYP154 family (CYP154C3, CYP154C4, CYP154C5, and CYP154C8), the CYP109 family (CYP109B1 and CYP109E1), and the CYP260 family (CYP260A1 and CYP260B1) (5, 18–23).
The catalytic activity of P450s requires one or more redox partners to transfer two electrons from NAD(P)H to the heme iron (7, 24). There are five major redox partner systems. Class I is a three-component system used by most bacterial and mitochondrial P450s and consists of an iron-sulfur ferredoxin (Fdx) and a flavin adenine dinucleotide (FAD)-containing ferredoxin reductase (Fpr) (25). Class II is a two-component system comprising a single cytochrome P450 reductase (CPR) containing FAD/flavin mononucleotide (FMN) (26, 27). Class III and IV are both single-component systems, as exemplified by P450 BM3 from Bacillus megaterium and P450 RhF from Rhodococcus sp. NCIMB 9784. Class V system P450s, such as P450nor from Fusarium oxysporum, transfer electrons directly from NAD(P)H to P450. For practical purposes, one or more surrogate redox partners are often employed during functional characterization or the synthetic application of P450s, such as putidaredoxin reductase (Pdr)/putidaredoxin (Pdx) from Pseudomonas putida or the RhFRED reductase domain from Rhodococcus sp. (15, 27–29). The S. avermitilis genome contains six ferredoxin reductase genes and nine ferredoxin genes (9), and FprD and FdxH have been used as native electron transfer partners for CYP105D7 in the hydroxylation of daidzein (30, 31).
In this study, we screened various steroid compounds for CYP105D7 hydroxylation activity using recombinant Escherichia coli cells harboring CYP105D7-pdx-pdr and identified steroid hydroxylation activity. We also conducted an in vivo biotransformation study using three different P450 redox partners (Pdx/Pdr, RhFRED, and FdxH/FprD) to determine the optimal redox partner. Furthermore, we prepared several single and double mutants of CYP105D7 using site-directed mutagenesis targeted at arginine residues around the entrance and substrate-binding pocket, which resulted in up to 9-fold increases in steroid conversion compared with the wild-type enzyme.
RESULTS
Steroid substrate screening and bioconversion of testosterone by CYP105D7.
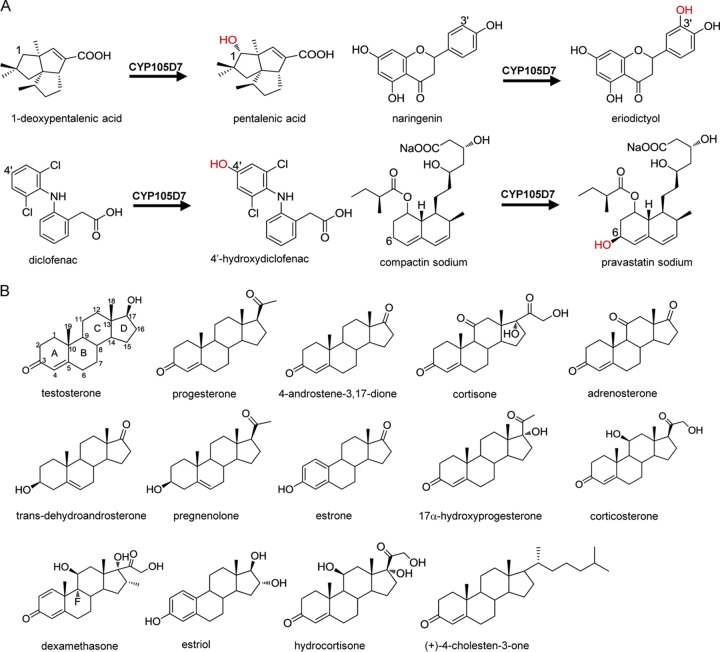
We carried out in vivo steroid substrate screening of CYP105D7 using redox partners Pdx/Pdr. The candidate substrates comprised 14 steroid compounds, including testosterone, progesterone, 4-androstene-3,17-dione, cortisone, adrenosterone, trans-dehydroandrosterone, pregnenolone, estrone, 17α-hydroxyprogesterone, corticosterone, dexamethasone, estriol, hydrocortisone, and (+)-4-cholesten-3-one (Fig. 1). Bioconversion assays revealed that five of these steroid compounds were converted into products. Testosterone was converted by CYP105D7 with a low conversion rate of 6.21%. The product was analyzed by high-performance liquid chromatography-time of flight mass spectrometry (HPLC-TOF-MS), and the data showed that it had an [M+Na]+ molecular ion at m/z 327.1931, which revealed that a hydroxyl group was introduced into testosterone. We isolated the product, and the hydroxyl group was determined to be C-2β based on nuclear magnetic resonance (NMR) (data are available in the supplemental material). Thus, the product of the biotransformation of testosterone was considered 2β-hydroxytestosterone (Fig. 2B). Bioconversion products of another four steroid compounds comprising progesterone (compound 3), 4-androstene-3,17-dione, cortisone, and adrenosterone were analyzed by HPLC-TOF-MS, and the conversion rates were 7.97%, 5.61%, 10.25%, and 7.94%, respectively.
FIG 1.
(A) Structural and conversion information for known substrates of CYP105D7. (B) Steroid compounds used to screen for substrates of CYP105D7.
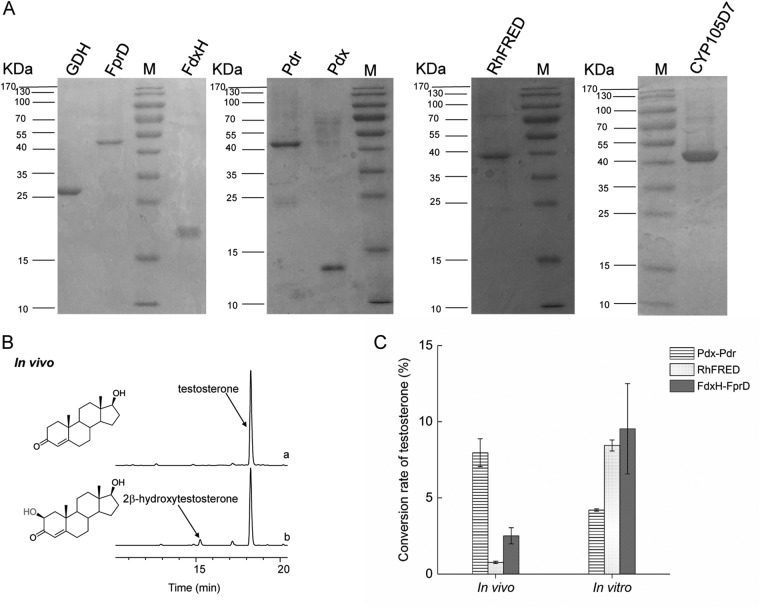
FIG 2.
(A) SDS-PAGE analysis (4.5% to 12.5% acrylamide gradient gel) was performed using Coomassie brilliant blue staining of the following purified His6-tagged proteins (atomic masses): glutamate dehydrogenase (GDH; 30 kDa), FprD (45 kDa), FdxH (17 kDa), Pdr (46 kDa), Pdx (12 kDa), RhFRED (36 kDa), and CYP105D7 (45 kDa). (B) HPLC analysis of the CYP105D7-mediated hydroxylation of testosterone using E. coli BL21(DE3) (no P450) as the negative control and E. coli cells harboring pET11-sav7469-pdx-pdr. (C) Conversion rates of CYP105D7-mediated testosterone bioconversion in vivo (left) and in vitro (right) using the following redox partners: Pdx/Pdr (from Pseudomonas putida), RhFRED (from Rhodococcus sp. NCIMB 9784), and FdxH/FprD (from Streptomyces avermitilis). Values indicate means ± standard deviations (SD) of results from three independent experiments.
Optimal redox partner selection for CYP105D7.
To investigate the bioconversion efficiency of CYP105D7, we reconstituted the self-sufficient CYP105D7-RhFRED fusion system and the CYP105D7-FdxH/FprD coexpression system and compared these with CYP105D7-Pdx/Pdr both in vivo and in vitro. Testosterone was used as the substrate in the conversion assays (Fig. 2C). The Pdx/Pdr system was the most efficient among the three redox partners in vivo. For the in vitro assays, CYP105D7 and the redox partner proteins Pdx, Pdr, RhFRED, FdxH, FprD, and glucose-6-phosphate dehydrogenase (G6P-DH) were expressed and purified to homogeneity by SDS-PAGE (Fig. 2A). Prior to performing the enzyme assays, we optimized the proportions of P450 and the redox partners to 10 μM CYP105D7, 50 μM Pdx, and 10 μM Pdr (or 50 μM FdxH and 10 μM FprD or 50 μM RhFRED). Relative to the in vivo assays, the enzyme assays performed in vitro showed increased conversion rates with the RhFRED and FdxH/FprD systems and decreased conversion rates with Pdx/Pdr. We did not observe any correlation between the in vivo and in vitro results. Thus, Pdx/Pdr was selected as the redox partner for further bioconversion assays.
Targeted amino acid selection and mutagenesis analysis.
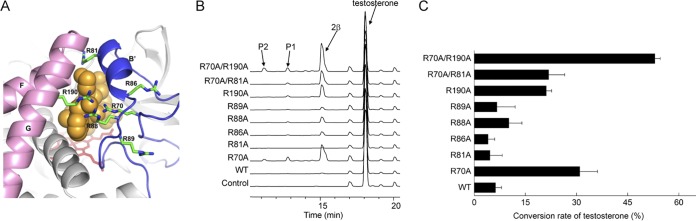
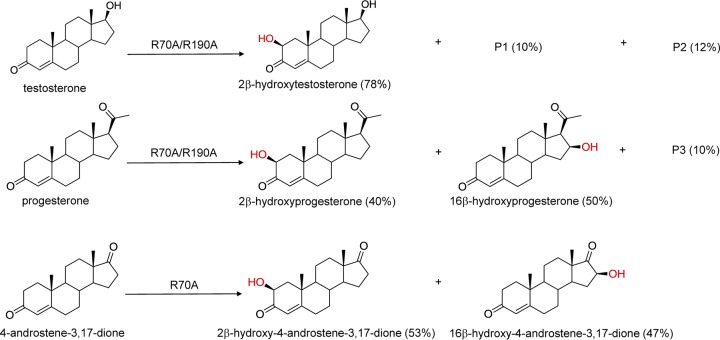
In a previous study, we determined the crystal structure of CYP105D7 in complex with diclofenac (16). The crystal structure of CYP105D7 showed that there were six arginine residues (Arg70, Arg81, Arg86, Arg88, Arg89, and Arg190) surrounding the entrance of the substrate-binding pocket and the active site (Fig. 3A). The three arginine residues Arg70, Arg81, and Arg190 of CYP105D7 are located in positions similar to those of Arg73, Arg84, and Arg193 in CYP105A1, and mutation of these residues in the latter enzyme significantly enhanced its catalytic activity toward the substrate, vitamin D3 (13). Motivated by these findings, we mutated all six of the arginine residues of CYP105D7 to alanine and conducted bioconversion assays. We observed that two single mutants, R70A and R190A, exhibited significantly increased conversion rates of 31% and 21% for testosterone compared with the wild type (Fig. 3B and C). Next, we constructed double mutants, R70A/R81A and R70A/R190A, and observed that the conversion rate of testosterone was significantly increased for R70A/R190A to 53% with high regio- and stereoselectivity (Table 1). Moreover, the R70A/R190A mutant produced another two products, P1 and P2, which gave [M+Na]+ molecular ions at m/z of 327.1945 and 343.1882, respectively. This result indicated that one and two hydroxyl groups were introduced into testosterone. Although we did not determine the structures of P1 and P2, we speculated that P1 and P2 may be 16β-hydroxylation and 2β, 16β-dihydroxylation products on the basis of the bioconversion results of progesterone and 4-androstene-3,17-dione described below.
FIG 3.
(A) Conserved arginine residues (R70, R81, R86, R88, R89, and R190) are positioned around the substrate access area of CYP105D7. The BC loop and FG helices of CYP105D7–diclofenac are shown in blue and magenta, respectively. Ligand molecules (DIF-1 and DIF-2) are shown as orange spheres, and arginine residues are shown as green sticks. (B) HPLC chromatograms of the biotransformation of testosterone using E. coli cells harboring the pET11-sav7469-pdx-pdr wild type and its variants. (C) Conversion rates (percent) of testosterone hydroxylation yields in vivo with the CYP105D7 wild type and the corresponding mutants (R70A, R81A, R86A, R88A, R89A, R190A, R70A/R81A, and R70A/R190A) with redox partners Pdx/Pdr. Conversion rates were calculated based on the substrate concentrations, which were determined by calibration with an external standard. Values indicate means ± SD of results from three independent experiments.
TABLE 1.
Conversion rate of steroid oxidation and product selectivity of CYP105D7 and its mutants from bioconversion assay
| Steroid and strain/mutant | Conversion rate (%) | Product selectivity (%) |
||||
|---|---|---|---|---|---|---|
| 2β | 16β | P1 | P2 | P3 | ||
| Testosterone | ||||||
| WT | 6 | 100 | ||||
| R70A | 31 | 80 | 14 | 6 | ||
| R81A | 5 | 100 | ||||
| R86A | 4 | 88 | 12 | |||
| R88A | 10 | 87 | 10 | 3 | ||
| R89A | 7 | 95 | 5 | |||
| R190A | 21 | 91 | 9 | |||
| R70A/R81A | 22 | 92 | 7 | 1 | ||
| R70A/R190A | 53 | 78 | 10 | 12 | ||
| Progesterone | ||||||
| WT | 8 | 67 | 33 | |||
| R70A | 17 | 30 | 60 | 10 | ||
| R81A | 5 | 100 | ||||
| R86A | 11 | 71 | 29 | |||
| R88A | 10 | 68 | 32 | |||
| R89A | 7 | 31 | 69 | |||
| R190A | 19 | 76 | 22 | 2 | ||
| R70A/R81A | 26 | 41 | 41 | 18 | ||
| R70AR/190A | 39 | 40 | 50 | 10 | ||
| 4-Androstene-3,17-dione | ||||||
| WT | 6 | 89 | 11 | |||
| R70A | 31 | 53 | 47 | |||
| R81A | 1 | 100 | ||||
| R86A | 5 | 75 | 25 | |||
| R88A | 7 | 75 | 25 | |||
| R89A | 2 | 85 | 15 | |||
| R190A | 16 | 82 | 18 | |||
| R70A/R81A | 8 | 39 | 61 | |||
| R70AR/190A | 24 | 61 | 39 | |||
Kinetics parameters of the CYP105D7 hydroxylation of testosterone.
The wild type and mutants of CYP105D7 were expressed and purified, and the CO-bound reduced-difference spectra of these CYP105D7 proteins each showed a peak at about 450 nm, which is characteristic of functional P450 enzymes (see Fig. S2 in the supplemental material). We then determined the kinetic parameters of the CYP105D7 hydroxylation of testosterone using redox partners Pdx/Pdr (with a P450/Pdx/Pdr ratio of 1:5:1), and the results are summarized in Table 2. We obtained testosterone conversion rates (Vmax) of 17.33 ± 7.213, 17.9 ± 8.581, 8.714 ± 2.757, and 16.99 ± 303.1 (μM min−1), with Km values of 355.60 ± 189.80, 390.10 ± 235.40, 165.00 ± 81.65, and 303.10 ± 206.6 (μM), respectively (Fig. S3). Using the CYP105D7 wild type as the benchmark, the R70A/R190A mutant had the highest kcat/Km ratio among the mutants, with an increase of 1.35-fold.
TABLE 2.
Determination of steady-state kinetic parameters for testosterone hydroxylation of CYP105D7 and its mutants (R70A, R190A, and R70A/R190A)
| Strain/mutant | Km (μM) | kcat (min−1) |
Vmax (μM min−1) |
kcat/Km (min−1 M−1) |
Ratio kcat/Kma |
|---|---|---|---|---|---|
| WT | 355.60 ± 189.80 | 0.15 ± 0.06 | 17.33 ± 7.213 | 431.66 | 1 |
| R70A | 390.10 ± 235.40 | 0.19 ± 0.09 | 17.90 ± 8.581 | 494.74 | 1.15 |
| R190A | 165.00 ± 81.65 | 0.06 ± 0.02 | 8.71 ± 2.757 | 338.06 | 0.78 |
| R70A/R190A | 303.10 ± 206.6 | 0.18 ± 0.09 | 16.99 ± 8.726 | 581.99 | 1.35 |
The values are expressed relative to the kcat/Km value determined for the wild type.
Bioconversion of progesterone and 4-androstene-3,17-dione by CYP105D7.
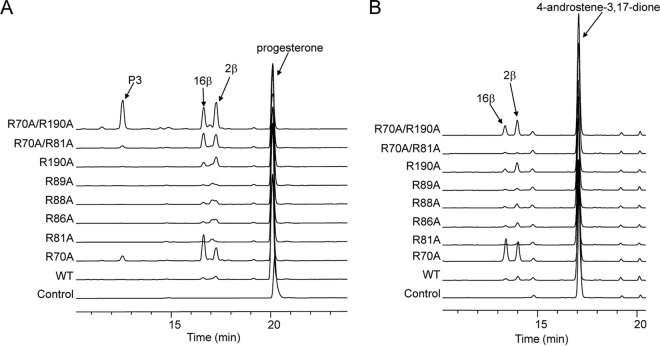
To gain further insights into the substrate specificity and conversion activity of the CYP105D7 mutants, we carried out bioconversion assays using progesterone and 4-androstene-3,17-dione and the wild-type and mutant forms of CYP105D7 (Fig. 4). In the bioconversion of progesterone, R70A, R190A, R70A/R81A, and R70A/R190A showed increased conversion rates compared with the wild type, with R70A/R190A exhibiting the highest conversion rate of 39% (Table 1). This result was quite similar to that calculated for the conversion of testosterone. In the conversion of 4-androstene-3,17-dione, however, R70A, R190A, and R70A/R190A showed visibly increased conversion rates, with the R70A single mutant showing the highest rate of 31%. We further isolated products from the R70A/R190A double mutant and determined the structure of each by HPLC-TOF-MS and NMR. The products of the bioconversion of progesterone were 2β-hydroxyprogesterone and 16β-hydroxyprogesterone. The presence of another, unknown product, P3, showed a [M+Na]+ molecular ion at a m/z of 369.2035, indicating that it contained two hydroxyl groups. We speculate that P3 may be 2β,16β-dihydroxyprogesterone on the basis of the results of a previous study on the hydroxylation of progesterone performed using engineered P450 BM3 (32). In addition, the products of the bioconversion of 4-androstene-3,17-dione were 2β-hydroxy- and 16β-hydroxy-4-androstene-3,17-dione (Fig. 4). These results revealed that CYP105D7 hydroxylation of the two steroids occurred at the A-ring 2β and D-ring 16β positions (Fig. 5). Although we did not determine the products of cortisone and adrenosterone bioconversion, these products also would likely have a hydroxyl group at the position of C-2β based on HPLC-TOF-MS analysis (data not shown). Interestingly, R70A changes the regioselectivity of substrates; for example, in the conversion of progesterone, R70A delivers a product ratio of 2β/16β/other of 30:60:10 compared with the wild-type product ratio of 67:33:0. These results suggest that Arg70 plays an important role in substrate regioselectivity.
FIG 4.
HPLC analysis of the biotransformation of progesterone (A) and 4-androstene-3,17-dione (B) using E. coli cells harboring the pET11-sav7469-pdx-pdr wild type and its variants (R70A, R81A, R86A, R88A, R89A, R190A, R70A/R81A, and R70A/R190A).
FIG 5.
Steroid hydroxylation by CYP105D7. Testosterone was hydroxylated at the 2β position. Progesterone and 4-androstene-3,17-dione were each hydroxylated at the 2β and 16β positions.
Bioconversion of naringenin and diclofenac using the CYP105D7 mutants and comparison with that of steroids.
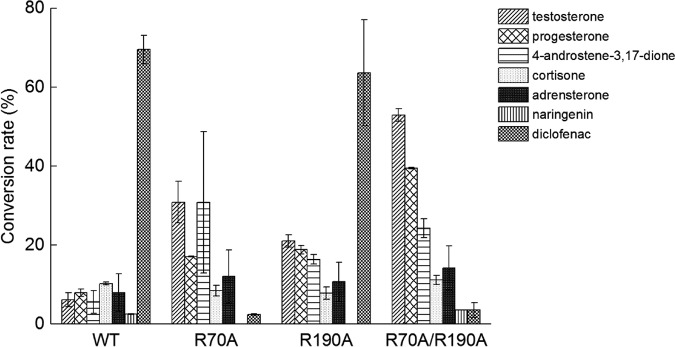
To investigate the efficacy of the bioconversion of other compounds, we next conducted bioconversion assays of diclofenac and naringenin. These two compounds were also hydroxylated by CYP105D7. The three mutants R70A, R190A, and R70A/R190A were analyzed in the conversion assays and the results compared to those obtained with steroids (Fig. 6). Interestingly, the wild type and the R190A mutant exhibited high conversion rates for diclofenac of almost 70%, while the R70A and R70A/R190A mutants showed markedly lower conversion rates. This result confirmed that Arg70 plays a crucial role in the binding of diclofenac. For naringenin, there was no significant difference between the conversion activities of the three mutants and that of the wild type, which suggested that these arginine residues do not affect the binding and hydroxylation of naringenin. For the steroid substrates, the R70A/R190A double mutant showed increased conversion at different levels for each of the four steroid compounds, i.e., testosterone, progesterone, 4-androstene-3,17-dione, and adrenosterone, but the level of conversion of cortisone was lower than that seen with the wild type. This result revealed that Ala70 and Ala190 appear to promote the binding and hydroxylation of relatively hydrophobic substrates.
FIG 6.
Rates of conversion of testosterone, progesterone, 4-androstene-3,17-dione, cortisone, adrenosterone, naringenin, and diclofenac by wild-type CYP105D7 and its mutants (R70A, R190A, and R70A/R190A). Conversion rates were calculated based on the substrate concentrations, which were determined by calibration with an external standard. Values indicate means ± SD of results from three independent experiments.
Homology modeling and molecular docking analysis of R70A/R190A mutant.
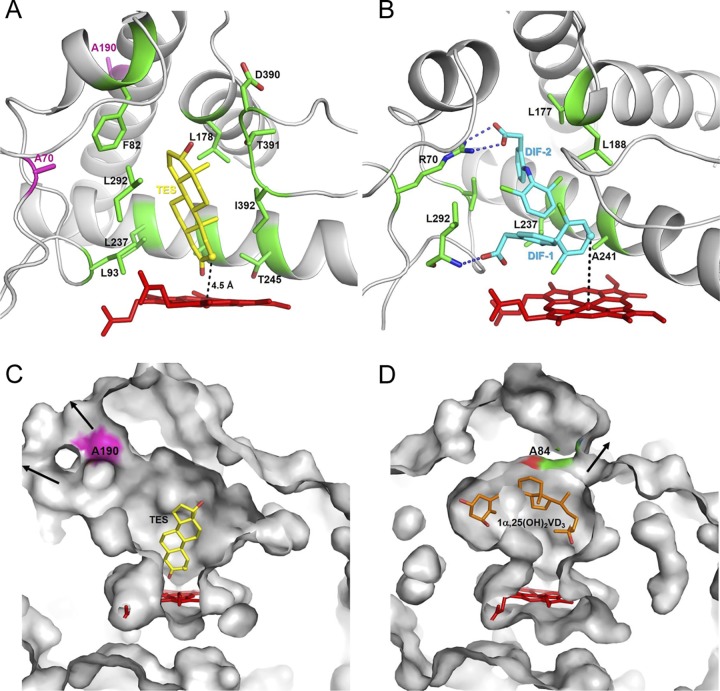
To investigate the possible conformation of steroid substrates within the active site, docking analysis was carried out using testosterone and the R70A/R190A mutant. The structure of R70A/R190A was built using homology modeling with SWISS-MODEL. The crystal structure of CYP105D7 in complex with diclofenac (PDB identifier [ID] 4UBS) was determined in a previous study (16) and used as the template here (Fig. 7B). Panels A and C of Fig. 7 shows the most energetically permissible docking conformation. The C-2 position of testosterone is proximate to the heme group, and the distance to the heme iron is 4.5 Å. The testosterone molecule is surrounded by and interacts with several hydrophobic residues, such as Phe82, Leu292, Leu237, Leu93, and IIe392 (Fig. 7A). The volume of the substrate access and binding pocket was calculated using the CASTp 3.0 server with a probe radius of 1.4 Å (33). The pocket volume of R70A/R190A was 2,080 Å3, which represented a 1.08-fold increase compared with wild-type value of 1,925 Å3 (Fig. 7C). Furthermore, Arg190 in the G helix located at the substrate entrance created a narrow channel that appears to play a crucial role in substrate recognition (Fig. 7C).
FIG 7.
Docking model compared to the crystal structure with different substrates bound. (A) Docking model of CYP105D7-R70A/R190A binding with testosterone. The two mutation sites at A70 and A190 are represented with magenta sticks, and testosterone is represented with yellow sticks. The C-2 position of testosterone, shown as a sphere, is proximate to the heme group, with a distance of 4.5 Å. The residues interacting with the testosterone molecule are F82, L93, L178, L237, T245, L292, D390, T391, and I392. (B) Crystal structure of wild-type CYP105D7 bound with diclofenac. Amino acid residues recognizing DIF-1 and DIF-2 are shown with green sticks, and R70 and L292 are hydrogen bonded with DIF-2 and DIF-1, respectively. The hydroxylation target position, C-4′ of DIF-1, is shown as a sphere. (C) Substrate entrance and active site comparison of the docking model of CYP105D7-R70A/R190A bound with testosterone. (D) Crystal structure of CYP105A1-R84A (PDB ID 2ZBZ) from Streptomyces griseolus bound with the product 1α,25(OH)2VD3. In panels C and D, two figures, representing the ligand, heme, and mutation sites at A190 and A84, are represented with differently colored sticks.
DISCUSSION
In this study, we demonstrated that substrate-promiscuous P450 CYP105D7 was able to catalyze the A-ring 2β- and D-ring 16β-hydroxylation of various steroid molecules, and we significantly improved this conversion rate through rational engineering. Only a few native bacterial P450s have been reported thus far that are capable of catalyzing the stereospecific hydroxylation of steroids, especially at the 2β and 16β positions. For instance, CYP109E1 from Bacillus megaterium is able to hydroxylate testosterone and produce one main product, 16β‐hydroxytestosterone (69%), and one minor product, androstenedione (14%) (23). Agematu et al. conducted testosterone hydroxylation screening using bacterial P450s and revealed that CYP105D4 from Streptomyces lividans and CYP105D5 from S. coelicolor A3(2) hydroxylated testosterone at the 2β position with low conversion rates (34). Although there is little enzymatic and bioconversion information available, the substrate specificity of CYP105D7 seems to be similar to that of CYP105D4 and CYP105D5. Interestingly, the three P450s have high amino acid sequence identity of 73% and sequencing analysis has revealed that they also all contain the six conserved arginine residues. These shared features likely underlie their similar levels of substrate specificity and selectivity. Furthermore, P450 BM3 (CYP102A1) mutants, obtained by direct evolution, are able to catalyze the 2β- and 15β-hydroxylation of testosterone and the 2β- and 16β-hydroxylation of progesterone with high selectivity for either of the regioisomers (32). The substrate selectivity of CYP105D7 seems to resemble that of P450 BM3 mutants, especially for conversion of progesterone. Here, we speculated that P2 and P3 represent the 2β-, 16β-dihydroxylation products referred to above.
In this study, we carried out site-directed mutagenesis targeted at six arginine residues (Arg70, Arg81, Arg86, Arg88, Arg89, and Arg190) surrounding the substrate entrance and active site, and the mutants R70A and R70A/R190A greatly enhanced the conversion rate of steroid hydroxylation. Panel B of Fig. 7 shows that the carboxyl group of the distal diclofenac (DIF-2) bound to Arg70 via a direct salt bridge, which contributed to substrate stability. Interestingly, replacement of Arg70 with alanine resulted in a marked decrease in diclofenac bioconversion and revealed the importance of Arg70 in diclofenac binding and hydroxylation. However, in the bioconversion of steroids, mutation of Arg70 to alanine appears to increase the hydrophobicity and volume of the substrate-binding pocket. Furthermore, loss of the bulky side chain of Arg190 generated a larger substrate entrance that may aid substrate access and product release (Fig. 7C). Interestingly, the role of Arg190 in CYP105D7 is similar to that of Arg84 in CYP105A1 from S. griseolus (Fig. 7D). Although the substrate access channels of the two P450s are positioned in opposite directions, Arg190 and Arg84 are located in similar positions at the entrance and play similar roles. In the present study, the kcat/Km ratio calculated for the R70A/R190A double mutant of CYP105D7 was found to have increased only 1.35-fold; however, the conversion rate in vivo was increased almost 9-fold compared with the wild type. Our results, therefore, confirm the importance of the arginine residues in the substrate access channel and binding pocket, especially for the binding and hydroxylation of hydrophobic aromatic compounds such as steroids. Such mutational information supports the approach of future rational and semirational engineering of P450s using site-directed and saturation mutagenesis.
To investigate the efficiency of electron transfer between CYP105D7 and different redox partners, we tested three redox partners, Pdx/Pdr, RhFRED, and FdxH/FprD. RhFRED as a self-sufficient redox partner conferred enhanced catalytic activity and product selectivity when partnered with P450 PikC and MycG (28, 29). FdxH/FprD were considered optimal endogenous electron transfer proteins for CYP105D7 activity based on in vitro experiments (30). In the present study, the RhFRED fusion system and the native redox partner system FdxH/FprD did not increase the activity of CYP105D7 in vivo but slightly increased its activity in vitro compared with Pdx/Pdr. There were no correlations between the in vivo and in vitro levels of enzyme activity obtained with the different redox partners. These findings are similar to those observed for CYP154C3 from S. griseus, a P450 capable of catalyzing the 16α-hydroxylation of steroids (5).
In this study, we mainly performed biotransformation using E. coli BL21(DE3) cells because they are more suitable for use in industrial steroid synthesis than enzymatic reactions, which require high enzyme stability, NAD(P)H cost, substrate concentrations of steroids, and so on (21). However, the whole-cell system also has some bottlenecks that must be overcome. Although the R70A/R190A mutant exhibited a conversion rate for progesterone of almost 39%, the yield of products obtained only was about 10% (7.6 mg from 79 mg). As for low yield of steroids, we speculated that uptake of substrates and the pumping of products might be the major causes. For improving product recovery efficiency, we plan to try to add surfactant of CTAB (cetyltrimethylammonium bromide) (35) and toe use S. avermitilis as a host system (36) in future work.
Substrate‐promiscuous P450s are ideal candidates for protein engineering to achieve specific outcomes through a small number of mutations. For instance, by using rational mutagenesis to introduce larger residues instead of small or medium-sized residues into the active site of CYP3A4, one can successfully alter and/or improve the regio- and stereoselectivity of this enzyme (3). In our functional and structural study of CYP105D7, this P450 was shown to have broad substrate specificity, a large active site, and structural flexibility. These features are very similar to those of human CYP3A4 and are common to promiscuous enzymes (37). To the best of our knowledge, CYP105D7 is a typical microbial substrate‐promiscuous P450 and has potential as a biocatalyst, especially for synthesis of 2β- and 16β-hydroxylated steroids, which is difficult to achieve by chemical synthesis. For industrial application of CYP105D7, further improvements are needed in conversion efficiency, selectivity, production processes, and so on.
MATERIALS AND METHODS
Materials.
(±)-Naringenin, (±)-eriodictyol, diclofenac sodium, and 4′-hydroxydiclofenac were obtained from Sigma-Aldrich (St. Louis, MO, USA). All steroid substrates and 5-aminolevulinic acid hydrochloride (5-ALA) were purchased from Aladdin (Shanghai, China). DNA polymerase, restriction enzymes, and T4 DNA ligase were bought from TaKaRa (Dalian, China). Oligonucleotide and sequence analyses were performed by Sangon Biotech (Shanghai, China). All other chemicals were of the highest grade and from standard sources.
Heterologous cloning, expression, and purification of CYP105D7 wild-type, mutants, and redox partners.
The bacterial strains, plasmids, and primers used in this work are shown in Table S1 to S3 in the supplemental material. Expression systems for the P450s investigated were prepared as described before with a few modifications. A gene (sav7469; UniProt accession no. Q825I8) encoding CYP105D7 was digested with NdeI and XhoI and inserted in-frame with an N-terminal His6 tag into the pET28b vector digested with the same enzymes. Cloning of the CYP105D7 mutants was performed according to the manufacturer’s protocol accompanying a Sangon Biotech site-directed mutagenesis kit. E. coli BL21 CodonPlus (DE3) RIL was used as the heterologous host for plasmid pET28b encoding CYP105D7 or its mutants. Briefly, a single colony was grown in Luria-Bertani (LB) medium containing kanamycin (100 μg/ml) and chloramphenicol (34 μg/ml) overnight at 37°C. Then, the overnight culture was inoculated at 1:100 into Terrific Broth (TB) medium containing kanamycin (100 μg/ml), chloramphenicol (34 μg/ml), 0.475 mM FeCl3·6H2O, 0.5 mM 5-ALA, 1 mM vitamin B1, and 0.025% (vol/vol) trace element solution [2.0 g/liter (NH4)2MoO4·2H2O, 2.0 g/liter CoCl3·6H2O, 1.0 g/liter CaCl2·H2O, 1.0 g/liter CuCl2, 1.0 g/liter ZnCl2, 0.5 g/liter MnCl2·4H2O, 0.5 g/liter H3BO3, 27.0 g/liter FeCl3·6H2O, and 20 ml of concentrated hydrochloric acid]. The cultures were grown at 37°C in a rotary shaker until the optical density at 600 nm (OD600) reached 0.8. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the medium (0.2 mM), and the cultures were further incubated at 22°C for another 20 h at 180 rpm. The cells were collected by centrifugation at 6,000 rpm for 15 min at 4°C, and the cell pellets were stored at −80°C.
Expression of Pdx (UniProt accession no. P00259), Pdr (UniProt accession no. P16640), RhFRED (UniProt accession no. Q8KU27), and glucose-6-phosphate dehydrogenase (G6P-DH) was conducted as described previously (9). The fdxH and fprD genes were inserted into pET28b between the NcoI and XhoI restriction sites to express FdxH (UniProt accession no. Q825I7) and FprD (UniProt accession no. Q82BM9). The cells were incubated in TB medium containing kanamycin (100 μg/ml) or ampicillin (100 μg/ml) and 4% (vol/vol) glycerol at 37°C until the OD600 reached 0.8. Next, 0.2 mM IPTG was added to the medium, and the cells were incubated at 22°C for another 20 h at 180 rpm. The cells were collected by centrifugation at 6,000 rpm for 15 min at 4°C, and the cell pellets were stored at −80°C.
For protein extraction and purification, the cells were first resuspended in lysis buffer (50 mM NaH2PO4 [pH 7.4], 300 mM NaCl, 10% glycerol, 10 mM imidazole) and then subjected to sonication with a sonicator (JY92-IIN; Scientz, Ningbo, China) for 20 min at an output setting of 35% in an ice bath. The soluble protein fraction was harvested by centrifugation at 8,000 rpm for 60 min at 4°C, and then the cell extract was applied to a column of nickel-nitrilotriacetic acid (Ni-NTA) Superflow Agarose (Thermo Fisher Scientific, Rockford, IL, USA). The bound fraction was washed with wash buffer (50 mM NaH2PO4 [pH 7.4], 300 mM NaCl, 10% glycerol, 20 mM imidazole) to remove impurities. Next, the His6-tagged proteins were eluted with elution buffer (50 mM NaH2PO4 [pH 7.4], 300 mM NaCl, 10% glycerol, 300 mM imidazole). Finally, the eluted fractions were concentrated and the buffer was exchanged with desalting buffer (50 mM NaH2PO4 [pH 7.4], 0.1 mM ethylenediaminetetraacetic acid disodium salt [EDTA], 10% glycerol, 0.1 mM dithiothreitol [DTT]) using a 10 K Amicon ultracentrifugation filter (Millipore, Carrigtwohill, Ireland). Purified proteins were flash-frozen and stored at −80°C. The concentration of the P450 enzymes was determined by CO-bound reduced-difference spectroscopy (38). The concentrations of the redox partners were calculated by Bradford assay using bovine serum albumin (BSA) as the standard (39).
Construction of recombinant plasmids for heterologous expression.
To construct plasmid pET28b-sav7469-RhFRED, the RhFRED gene fragment was amplified by PCR from plasmid pET28b-mycG-RhFRED (28). Then, pET28b-sav7469 was digested with XhoI and ligated to the RhFRED gene with a One Step cloning kit (Vazyme, Nanjing, China). To construct pCDFDuet-fdxH-fprD, the fdxH and fprD genes were amplified from plasmids pET28b-fdxH and pET28b-fprD and inserted into the NdeI/XhoI and BamHI/HindIII restriction sites of pCDFDuet-1. Plasmids pET28b-sav7469 and pCDFDuet-fdxH-fprD were cotransformed into E. coli BL21(DE3) to acquire the final strain. The recombinant plasmid pET11-sav7469-pdx-pdr variants were constructed from a previously published plasmid, pET11-sav7469-pdx-pdr (16). The plasmid was subjected to double digestion at the NdeI and SpeI sites and then ligated with the variant gene fragments. All three kinds of recombinant plasmids described here were transformed into E. coli BL21(DE3) (see Fig. S1 in the supplemental material).
In vitro biotransformation assays of CYP105D7 activity.
The enzyme reconstitution assay system contained 10 μM P450, 50 μM Pdx, 10 μM Pdr (or 50 μM FdxH and 10 μM FprD or 50 μM RhFRED), 5 U G6P-DH, 10 mM glucose-6-phosphate, 0.1 mM substrates (stock solution dissolved in methanol or ethanol), and 1 μl of 100 mM NADPH mixed in 100 μl of desalting buffer. The reaction was initiated by the addition of NADPH at 30°C for 12 h and stopped by adding equivalent volumes of ethyl acetate three times to obtain the extract. The products were dried and analyzed by high-performance liquid chromatography (HPLC) using an Agilent 1260 spectrometer (Agilent Technologies, Santa Clara, CA, USA). The compounds were separated using an Agilent Zorbax SB-C18 column (4.6 by 250 mm, 5-μm pore size) with a methanol-water or acetonitrile-water mixture (containing 0.1% trifluoroacetic acid) at a flow rate of 1 ml/min. The HPLC conditions were as follows: a linear gradient of solvent B (methanol) from 35% to 100% for 20 min and 100% solvent B for 8 min followed by 35% solvent B for 8 min. The eluates were monitored in the range of 190 to 390 nm, and the column temperature was kept constant at 30°C. The injection volume of the compounds was 20 μl, and a negative control was used (no P450 added) to prove the P450 dependency of the reactions. To estimate the proportional amounts of product formation, the peak areas of each product and substrate were calculated by using their own λmax values (absorption maxima).
In vivo CYP105D7 reconstitution assays.
The E. coli BL21(DE3) cells harboring pET11-sav7469-pdx-pdr, pET28b-sav7469-RhFRED, and pET28b-sav7469/pCDFDuet-fdxH-fprD were cultured overnight at 37°C in LB medium containing antibiotics (ampicillin, kanamycin and streptomycin, respectively). The cells were diluted 1:100 in 10 ml M9 medium (5× M9 salts, 1% Casamino Acids, 0.4% glucose, 0.1 mM CaCl2, 1 mM MgCl2, 0.1 mM FeCl3, 100 μg/ml ampicillin) and were incubated at 37°C. When the OD600 reached 0.8, protein expression was initiated by the addition of 0.2 mM IPTG and 0.5 mM 5-ALA and further incubation at 22°C for 20 h at 200 rpm. After incubation, the cells were harvested by centrifugation at 6,000 rpm for 15 min. The cell pellets were washed twice with buffer (50 mM Na2HPO4 [pH 7.2], 1 mM EDTA, 2 mM DTT, 10% glycerol) and suspended in the same buffer (1 ml). Substrates were dissolved in methanol or ethanol and added to reach a final concentration of 0.1 mM. Reactions were performed at 30°C for 12 h at 200 rpm and were stopped by adding equal volumes of ethyl acetate. This was performed three times for extraction. Product analysis was performed using the same method as that described for the in vitro assays.
Kinetic characterization of CYP105D7 and its variants.
To determine the Km and kcat parameters for testosterone hydroxylation, the reactions were assayed in a reaction solution containing 10 μM P450s, 10 μM Pdr, and 50 μM Pdx in buffer (50 mM NaH2PO4 [pH 7.4], 0.1 mM EDTA, 10% glycerol, 0.1 mM DTT) at 30°C for 5 min, and testosterone was added to reach a concentration of 0 to 150 μM. The reactions were initiated by the addition of 1 μl 100 mM NADPH to the final volume of 100 μl and were later stopped by the addition of an equivalent volume of methanol after 10 min of incubation at 30°C. The products were analyzed by HPLC using the same method as that described for the bioconversion assay. The values of Vmax, Km, and kcat were measured by plotting the substrate consumption rate versus the substrate concentration using a hyperbolic fit (Michaelis-Menten kinetics) and analysis with GraphPad Prism 7.0 software (La Jolla, CA, USA).
Large-scale production, purification, and characterization of products.
Large-scale cultivation was performed as described for the in vivo biotransformation method with slight modifications regarding the structural determination of the products. For structural determinations, the preculture (10 ml) was inoculated into M9 medium (1 liter) containing 100 μg/ml ampicillin, 5× M9 salts, 1% Casamino Acids, 0.4% glucose, 0.1 mM CaCl2, 1 mM MgCl2, and 0.1 mM FeCl3 in an Erlenmeyer flask followed by incubation at 37°C and 180 rpm. When the OD600 reached 0.8, 0.2 mM IPTG and 0.5 mM 5-ALA were added followed by further incubation at 22°C for 20 h at 180 rpm. After incubation, the cells were harvested by centrifugation at 6,000 rpm for 15 min. The cell pellets were washed twice and resuspended with buffer (50 mM Na2HPO4 [pH 7.2], 1 mM EDTA, 2 mM DTT, 10% glycerol). Each substrate was dissolved in methanol or ethanol and was added to the cell suspensions to reach a final concentration of 0.5 mM. The reactions were performed at 30°C and 180 rpm for 36 h and stopped by adding equal volumes (100 ml) of ethyl acetate and were conducted three times for extraction. After extraction, the organic phases were evaporated to dryness and purified by SiO2 column chromatography and HPLC with a Welch Ultimate XB-C18 column (Welch, Shanghai, China) (10 by 250 mm, 5-μm pore size). Nuclear magnetic resonance (NMR) spectra were obtained with a JNM-ECZ600R/S1 device (JEOL, Tokyo, Japan), and high-resolution electrospray ionization mass spectrometry (HRESIMS) data were acquired on an Agilent 1260-6230 time of flight liquid chromatography mass spectrometry (TOF LC/MS) spectrometer (Agilent Technologies).
Molecular docking analysis.
A model structure of R70A/R190A was built using the SWISS-MODEL server (40). The crystal structure of CYP105D7 (PDB ID 4UBS) was used as the template. Molecular docking analysis was performed with the CDOCKER tool of DS 3.5 (Discovery Studio 3.5; Accelrys Co., Ltd.) (41).
Supplementary Material
ACKNOWLEDGMENTS
We thank Shengying Li of Shandong University, who generously contributed the plasmid containing the RhFRED gene.
This work was supported by the fundamental research fund of the National Natural Science Foundation (grant number 81402810) and the research startup fund of Zhejiang Sci-Tech University (grant number 18042236-Y).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01530-19.
REFERENCES
- 1.Rudolf JD, Chang CY, Ma M, Shen B. 2017. Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function. Nat Prod Rep 34:1141–1172. doi: 10.1039/c7np00034k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guengerich FP, Munro AW. 2013. Unusual cytochrome P450 enzymes and reactions. J Biol Chem 288:17065–17073. doi: 10.1074/jbc.R113.462275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiavini P, Cheong KJ, Moitessier N, Auclair K. 2017. Active site crowding of cytochrome P450 3A4 as a strategy to alter its selectivity. Chembiochem 18:248–252. doi: 10.1002/cbic.201600546. [DOI] [PubMed] [Google Scholar]
- 4.Gupta RD. 2016. Recent advances in enzyme promiscuity. Sustain Chem Process 4:2. doi: 10.1186/s40508-016-0046-9. [DOI] [Google Scholar]
- 5.Makino T, Katsuyama Y, Otomatsu T, Misawa N, Ohnishi Y. 2014. Regio- and stereospecific hydroxylation of various steroids at the 16α position of the D ring by the Streptomyces griseus cytochrome P450 CYP154C3. Appl Environ Microbiol 80:1371–1379. doi: 10.1128/AEM.03504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flück CE, Mullis PE, Pandey AV. 2010. Reduction in hepatic drug metabolizing CYP3A4 activities caused by P450 oxidoreductase mutations identified in patients with disordered steroid metabolism. Biochem Biophys Res Commun 401:149–153. doi: 10.1016/j.bbrc.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Du L, Li F, Zhang X, Qu Z, Han L, Li Z, Sun J, Qi F, Yao Q, Sun Y, Geng C, Li S. 2018. Mechanistic insights into interactions between bacterial class I P450 enzymes and redox partners. ACS Catal 8:9992–10003. doi: 10.1021/acscatal.8b02913. [DOI] [Google Scholar]
- 8.Moody SC, Loveridge EJ. 2014. CYP105—diverse structures, functions and roles in an intriguing family of enzymes in Streptomyces. J Appl Microbiol 117:1549–1563. doi: 10.1111/jam.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao Q, Ma L, Liu L, Ikeda H, Fushinobu S, Li S, Xu LH. 2017. Hydroxylation of compactin (ML-236B) by CYP105D7 (SAV_7469) from Streptomyces avermitilis. J Microbiol Biotechnol 27:956–964. doi: 10.4014/jmb.1610.10079. [DOI] [PubMed] [Google Scholar]
- 10.Ba L, Li P, Zhang H, Duan Y, Lin Z. 2013. Semi-rational engineering of cytochrome P450sca-2 in a hybrid system for enhanced catalytic activity: insights into the important role of electron transfer. Biotechnol Bioeng 110:2815–2825. doi: 10.1002/bit.24960. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe I, Nara F, Serizawa N. 1995. Cloning, characterization and expression of the gene encoding cytochrome P-450sca-2 from Streptomyces carbophilus involved in production of pravastatin, a specific HMG-CoA reductase inhibitor. Gene 163:81–85. doi: 10.1016/0378-1119(95)00394-l. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi K, Sugimoto H, Shinkyo R, Yamada M, Ikeda S, Ikushiro S, Kamakura M, Shiro Y, Sakaki T. 2008. Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1. Biochemistry 47:11964–11972. doi: 10.1021/bi801222d. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto H, Shinkyo R, Hayashi K, Yoneda S, Yamada M, Kamakura M, Ikushiro S, Shiro Y, Sakaki T. 2008. Crystal structure of CYP105A1 (P450SU-1) in complex with 1α,25-dihydroxyvitamin D3. Biochemistry 47:4017–4027. doi: 10.1021/bi7023767. [DOI] [PubMed] [Google Scholar]
- 14.Takamatsu S, Xu LH, Fushinobu S, Shoun H, Komatsu M, Cane DE, Ikeda H. 2011. Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis. J Antibiot (Tokyo) 64:65–71. doi: 10.1038/ja.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Yao Q, Ma Z, Ikeda H, Fushinobu S, Xu L-H. 2016. Hydroxylation of flavanones by cytochrome P450 105D7 from Streptomyces avermitilis. J Mol Catal B Enzym 132:91–97. doi: 10.1016/j.molcatb.2016.07.001. [DOI] [Google Scholar]
- 16.Xu LH, Ikeda H, Liu L, Arakawa T, Wakagi T, Shoun H, Fushinobu S. 2015. Structural basis for the 4’-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase. Appl Microbiol Biotechnol 99:3081–3091. doi: 10.1007/s00253-014-6148-y. [DOI] [PubMed] [Google Scholar]
- 17.Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, Lacaze P, Watterson S, Griffiths SJ, Spann NJ, Meljon A, Talbot S, Krishnan K, Covey DF, Wenk MR, Craigon M, Ruzsics Z, Haas J, Angulo A, Griffiths WJ, Glass CK, Wang Y, Ghazal P. 2013. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 38:106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dangi B, Kim KH, Kang SH, Oh TJ. 2018. Tracking down a new steroid-hydroxylating promiscuous cytochrome P450: CYP154C8 from Streptomyces sp. W2233-SM Chembiochem 19:1066–1077. doi: 10.1002/cbic.201800018. [DOI] [PubMed] [Google Scholar]
- 19.Lee GY, Kim DH, Kim D, Ahn T, Yun CH. 2015. Functional characterization of steroid hydroxylase CYP106A1 derived from Bacillus megaterium. Arch Pharm Res 38:98–107. doi: 10.1007/s12272-014-0366-9. [DOI] [PubMed] [Google Scholar]
- 20.Dangi B, Lee CW, Kim K-H, Park S-H, Yu E-J, Jeong C-S, Park H, Lee JH, Oh T-J. 2019. Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures. FEBS J 286:1683–1699. doi: 10.1111/febs.14729. [DOI] [PubMed] [Google Scholar]
- 21.Bracco P, Janssen DB, Schallmey A. 2013. Selective steroid oxyfunctionalisation by CYP154C5, a bacterial cytochrome P450. Microb Cell Fact 12:95. doi: 10.1186/1475-2859-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatri Y, Ringle M, Lisurek M, von Kries JP, Zapp J, Bernhardt R. 2016. Substrate hunting for the myxobacterial CYP260A1 revealed new 1α-hydroxylated products from C-19 steroids. Chembiochem 17:90–101. doi: 10.1002/cbic.201500420. [DOI] [PubMed] [Google Scholar]
- 23.Jóźwik IK, Kiss FM, Gricman Ł, Abdulmughni A, Brill E, Zapp J, Pleiss J, Bernhardt R, Thunnissen A. 2016. Structural basis of steroid binding and oxidation by the cytochrome P450 CYP109E1 from Bacillus megaterium. FEBS J 283:4128–4148. doi: 10.1111/febs.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Du L, Chen H, Sun Y, Huang S, Zheng X, Kim ES, Li S. 2015. Reconstitution of the in vitro activity of the cyclosporine-specific P450 hydroxylase from Sebekia benihana and development of a heterologous whole-cell biotransformation system. Appl Environ Microbiol 81:6268–6275. doi: 10.1128/AEM.01353-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munro AW, Girvan HM, McLean KJ. 2007. Variations on a (t)heme–novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat Prod Rep 24:585–609. doi: 10.1039/B604190F. [DOI] [PubMed] [Google Scholar]
- 26.Munro AW, Girvan HM, McLean KJ. 2007. Cytochrome P450–redox partner fusion enzymes. Biochim Biophys Acta 1770:345–359. doi: 10.1016/j.bbagen.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Roberts GA, Grogan G, Greter A, Flitsch SL, Turner NJ. 2002. Identification of a new class of cytochrome P450 from a Rhodococcus sp. J Bacteriol 184:3898–3908. doi: 10.1128/jb.184.14.3898-3908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Liu Y, Yan J, Cao S, Bai F, Yang Y, Huang S, Yao L, Anzai Y, Kato F, Podust LM, Sherman DH, Li S. 2014. New reactions and products resulting from alternative interactions between the P450 enzyme and redox partners. J Am Chem Soc 136:3640–3646. doi: 10.1021/ja4130302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Podust LM, Sherman DH. 2007. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J Am Chem Soc 129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey BP, Lee N, Choi KY, Kim JN, Kim EJ, Kim BG. 2014. Identification of the specific electron transfer proteins, ferredoxin, and ferredoxin reductase, for CYP105D7 in Streptomyces avermitilis MA4680. Appl Microbiol Biotechnol 98:5009–5017. doi: 10.1007/s00253-014-5525-x. [DOI] [PubMed] [Google Scholar]
- 31.Pandey BP, Roh C, Choi KY, Lee N, Kim EJ, Ko S, Kim T, Yun H, Kim BG. 2010. Regioselective hydroxylation of daidzein using P450 (CYP105D7) from Streptomyces avermitilis MA4680. Biotechnol Bioeng 105:697–704. doi: 10.1002/bit.22582. [DOI] [PubMed] [Google Scholar]
- 32.Kille S, Zilly FE, Acevedo JP, Reetz MT. 2011. Regio- and stereoselectivity of P450-catalysed hydroxylation of steroids controlled by laboratory evolution. Nat Chem 3:738–743. doi: 10.1038/nchem.1113. [DOI] [PubMed] [Google Scholar]
- 33.Tian W, Chen C, Lei X, Zhao J, Liang J. 2018. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agematu H, Matsumoto N, Fujii Y, Kabumoto H, Doi S, Machida K, Ishikawa J, Arisawa A. 2006. Hydroxylation of testosterone by bacterial cytochromes P450 using the Escherichia coli expression system. Biosci Biotechnol Biochem 70:307–311. doi: 10.1271/bbb.70.307. [DOI] [PubMed] [Google Scholar]
- 35.Bagherinejad MR, Korbekandi H, Tavakoli N, Abedi D. 2012. Research in pharmaceutical sciences. Res Pharm Sci 7:79–85. [PMC free article] [PubMed] [Google Scholar]
- 36.Choi K-Y, Jung E, Yun H, Yang Y-H, Kim B-G. 2014. Engineering class I cytochrome P450 by gene fusion with NADPH-dependent reductase and S. avermitilis host development for daidzein biotransformation. Appl Microbiol Biotechnol 98:8191–8200. doi: 10.1007/s00253-014-5706-7. [DOI] [PubMed] [Google Scholar]
- 37.Ekroos M, Sjögren T. 2006. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci U S A 103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378. [PubMed] [Google Scholar]
- 39.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 40.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. 2003. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model 21:289–307. doi: 10.1016/S1093-3263(02)00164-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.