Serotyping is the basis of public health surveillance of Salmonella. It remains a first-line subtyping method even as surveillance continues to be transformed by whole-genome sequencing. SeqSero allows the integration of Salmonella serotyping into a whole-genome-sequencing-based laboratory workflow while maintaining continuity with the classic serotyping scheme. SeqSero2, informed by extensive testing and application of SeqSero in the United States and other countries, incorporates important improvements and updates that further strengthen its application in routine and large-scale surveillance of Salmonella by whole-genome sequencing.
KEYWORDS: Salmonella, serotype, whole-genome sequencing, WGS
ABSTRACT
SeqSero, launched in 2015, is a software tool for Salmonella serotype determination from whole-genome sequencing (WGS) data. Despite its routine use in public health and food safety laboratories in the United States and other countries, the original SeqSero pipeline is relatively slow (minutes per genome using sequencing reads), is not optimized for draft genome assemblies, and may assign multiple serotypes for a strain. Here, we present SeqSero2 (github.com/denglab/SeqSero2; denglab.info/SeqSero2), an algorithmic transformation and functional update of the original SeqSero. Major improvements include (i) additional sequence markers for identification of Salmonella species and subspecies and certain serotypes, (ii) a k-mer based algorithm for rapid serotype prediction from raw reads (seconds per genome) and improved serotype prediction from assemblies, and (iii) a targeted assembly approach for specific retrieval of serotype determinants from WGS for serotype prediction, new allele discovery, and prediction troubleshooting. Evaluated using 5,794 genomes representing 364 common U.S. serotypes, including 2,280 human isolates of 117 serotypes from the National Antimicrobial Resistance Monitoring System, SeqSero2 is up to 50 times faster than the original SeqSero while maintaining equivalent accuracy for raw reads and substantially improving accuracy for assemblies. SeqSero2 further suggested that 3% of the tested genomes contained reads from multiple serotypes, indicating a use for contamination detection. In addition to short reads, SeqSero2 demonstrated potential for accurate and rapid serotype prediction directly from long nanopore reads despite base call errors. Testing of 40 nanopore-sequenced genomes of 17 serotypes yielded a single H antigen misidentification.
IMPORTANCE Serotyping is the basis of public health surveillance of Salmonella. It remains a first-line subtyping method even as surveillance continues to be transformed by whole-genome sequencing. SeqSero allows the integration of Salmonella serotyping into a whole-genome-sequencing-based laboratory workflow while maintaining continuity with the classic serotyping scheme. SeqSero2, informed by extensive testing and application of SeqSero in the United States and other countries, incorporates important improvements and updates that further strengthen its application in routine and large-scale surveillance of Salmonella by whole-genome sequencing.
INTRODUCTION
Routine and prospective application of whole-genome sequencing (WGS) continues to transform public health surveillance of Salmonella (1, 2), one of the most prevalent foodborne pathogens worldwide (36). Although the detection and investigation of Salmonella outbreaks are increasingly reliant on WGS-based subtyping methods, such as genome-wide multilocus sequence typing (MLST) and single nucleotide polymorphism (SNP) analysis (4), Salmonella serotype determination remains a routine practice in public health laboratories because it is still integral to surveillance and outbreak investigations. Salmonella serotypes are defined by two surface structures, O antigen and H antigen. More than 2,600 Salmonella serotypes have been described in the White-Kauffmann-Le Minor scheme (5, 6), though a much smaller number are commonly reported. Molecular methods for serotype determination that are based on genes responsible for serotype antigens (7, 8), including the rfb gene cluster, fliC, and fljB, provide continuity with the well-established scheme for phenotypic serotypes.
We developed SeqSero to allow identification of Salmonella serotypes from WGS data and launched its Web application (denglab.info/SeqSero) in 2015 (9). Compared with other WGS-based tools for Salmonella serotype determination (1, 10), SeqSero is unique because it (i) relies on characterizing genetic determinants of Salmonella serotype without consulting any surrogate markers, such as MLST types, and (ii) predicts serotypes directly from raw sequencing reads without time-consuming genome assembly. As sequencing platforms that produce short sequencing reads (<1,000 bp) are predominantly used for WGS-aided surveillance of microbial infectious agents, including that of Salmonella (4), rapid serotype prediction directly from raw reads supports the continued use of serotype as a first-line assay for Salmonella surveillance.
SeqSero is routinely used in public health, food safety, and research laboratories in the United States and other countries (11–14). The Web-based instance of SeqSero alone has received and analyzed more than 45,000 Salmonella genomes as of September 2019. It is also accessible through the Center for Genomic Epidemiology (cge.cbs.dtu.dk/services/SeqSero/) and on the BioNumerics software platform (applied-maths.com/bionumerics), which is widely available in public health laboratories. However, the original SeqSero (here termed SeqSero1) has several limitations. First, the raw read workflow of SeqSero1 is relatively slow (several minutes per genome), as it requires three consecutive rounds of read mapping followed by a final round of BLAST analysis. Second, the genome assembly workflow of SeqSero1 is not optimized to handle low-quality draft genomes; for example, fragmented or incorrect assemblies can hinder extraction and identification of serotype determinants because SeqSero1 uses in silico PCR to extract serotype determinant sequences from genome assemblies. Third, SeqSero1 was incapable of recognizing more than one wzx or wzy allele (O antigen determinant) or more than two flagellin alleles (H antigen determinant) in a query genome, which may indicate potential interserotype contamination in the sequencing data. Finally, SeqSero1 does not identify Salmonella species and subspecies and some serotype variants. Identification to the subspecies level informs Salmonella serotype determination because the same antigenic profile can be found in different species or subspecies. Variants that share the same antigens, such as the Salmonella enterica subsp. enterica serotype Paratyphi B pathotypes (15), require additional characterization to determine serotype (denglab.info/SeqSero/supple). Incorporation of sequence features responsible for such phenotypes can help differentiate such variants.
In this study, we created SeqSero2 (github.com/denglab/SeqSero2 and denglab.info/SeqSero2) by transforming the serotype prediction algorithms and developing new functions to better support routine and large-scale surveillance of Salmonella using WGS.
RESULTS
SeqSero2 pipeline.
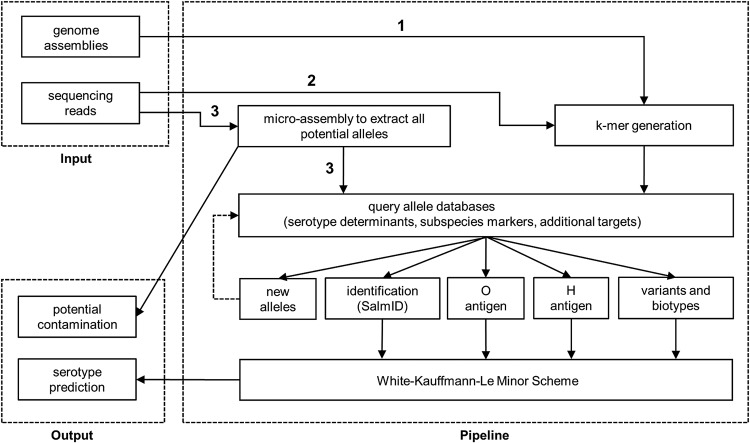
The major components and workflows of SeqSero2 are outlined in Fig. 1. Detailed information and algorithmic explanation of the pipeline are in Materials and Methods and Fig. 2.
FIG 1.
The major components and workflows of SeqSero2. Genome assemblies (1) or raw sequencing reads (2) are inputs for the k-mer-based algorithms for serotype determinants. The microassembly workflow (3) is used for serotype prediction, new allele identification, and contamination detection.
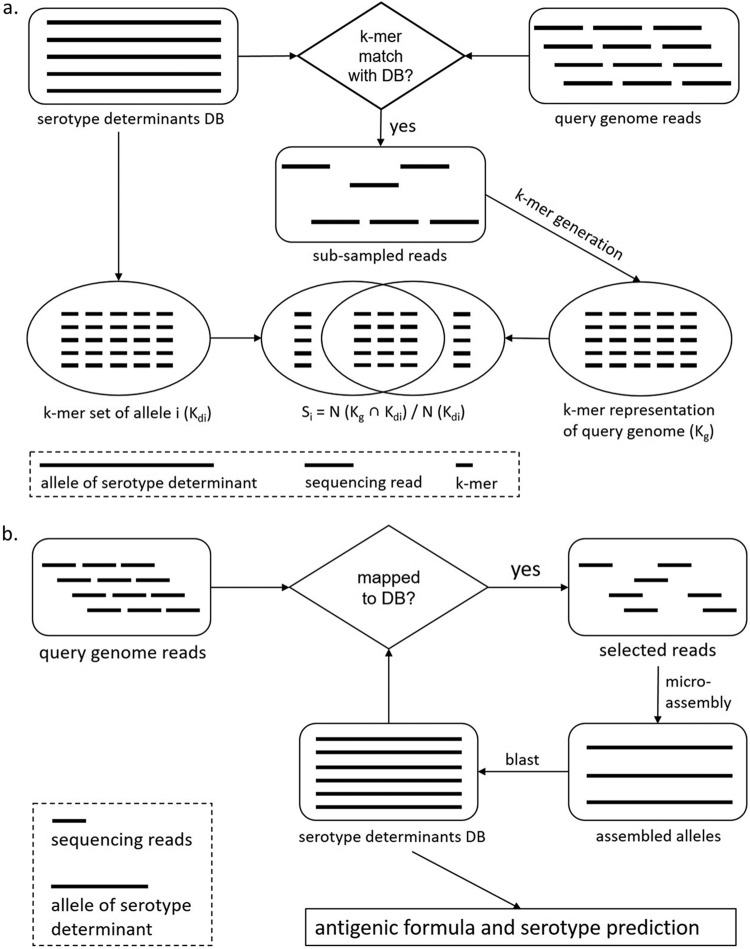
FIG 2.
Schematic overview of SeqSero2 algorithms. (a) The k-mer-based workflow for raw sequencing reads. (b) The microassembly workflow.
Serotype prediction accuracy using Illumina short reads.
The overall performance of SeqSero2, including that of its three workflows (raw reads k-mer, allele microassembly, and genome assembly), was evaluated by analyzing 2,280 human clinical isolates submitted to the National Antimicrobial Resistance Monitoring System (NARMS) at the Centers for Disease Control and Prevention (CDC) in 2015. These isolates represented 117 distinct serotypes (Table S1). Serotype prediction accuracy of each workflow is summarized in Table 1. The serotypes of 2,190 isolates (96.1%) were correctly predicted by all three workflows. A total of 90 isolates (3.9%) yielded at least one serotype prediction by any workflow that was discordant with the confirmed NARMS serotype. The majority of these discordant predictions (n = 60) were generated by the raw read k-mer or the assembly k-mer workflows. Among the three workflows, the allele microassembly workflow produced the most accurate serotype predictions, with a 98.7% concordance with phenotypic serotyping results.
TABLE 1.
Summary of SeqSero2 prediction results using 3 workflows
| Prediction result | Raw read k-mer (no. [%]) | Allele microassembly (no. [%]) | Genome assembly (no. [%]) |
|---|---|---|---|
| Expected serotypea | 2,198 (96.4%) | 2,250 (98.7%) | 2,246 (98.5%) |
| Unexpected serotypeb | 73 (3.2%) | 19 (0.8%) | 23 (1.0%) |
| Partial or no serotypec | 9 (0.4%) | 11 (0.5%) | 11 (0.5%) |
| Results of all tests | 2,280 | 2,280 | 2,280 |
The predicted serotype was consistent with the serotype identified by phenotypic methods.
The predicted serotype was inconsistent with the serotype identified by phenotypic methods.
Some or all of the expected serotype determinants were not detected.
Additional markers were used to differentiate specific serotypes or serotype variants (Table 2) for 989 isolates (Table S1). Specifically, using the allele microassembly workflow, 48 isolates were distinguished as Paratyphi B or Paratyphi B var. L(+) tartrate(+); 433 of 439 S. enterica subsp. enterica serotype Enteritidis isolates were identified to have the sdf marker gene (3); 66 of 389 S. enterica subsp. enterica Typhimurium or 1,4,[5],12:i:− isolates were found to carry a previously described mutation that can result in an O5-negative (O5−) variant (previously known as variant Copenhagen) (16); and 112 of 113 isolates belonging to the O13 group were differentiated into either O22 and O23 serotypes, which are indistinguishable by SeqSero1.
TABLE 2.
Additional markers for differentiating specific serotypes and variants
| Target serotype(s) | Marker | Description | Reference or source |
|---|---|---|---|
| S. Paratyphi B pathotypes | SNP in STM3356 | STM3356 is required for tartrate fermentation; an SNP that inactivates this gene is found in commonly circulating typhoidal pathotype strains | Malorny et al. (29) |
| S. Enteritidis | sdf gene | sdf is found in commonly circulating strains of S. Enteritidis but not in S. Gallinarum | Agron et al. (3) |
| O5− strains of Salmonella serotype Typhimurium | Deletion in oafA | 7-bp deletion in gene responsible for O5− phenotype | Hauser et al. (16) |
| Serogroup O13 serotypes | galE | Different alleles appear to be markers for ancillary O22 (NCBI accession no. NZ_LS483489) and O23 (accession no. NZ_CP029041) | This study |
Performance comparison between SeqSero2 and SISTR using Illumina short reads.
The NARMS data set was also analyzed using the Salmonella In Silico Typing Resource (SISTR) (10), a Salmonella serotype prediction tool that requires assembled genomes. SISTR uses both antigen identification and core genome MLST (cgMLST) for serotype prediction. Out of 2,280 genomes analyzed, 55 isolates yielded a prediction that was discordant with the confirmed NARMS serotype by antigen identification and 52 isolates by cgMLST, resulting concordances of 97.6% and 97.7%, respectively (Table S1). The final SISTR serotype prediction, a composite of both results, was incorrect for 47 isolates, giving a concordance of 97.9% (Table S1). In comparison, SeqSero2 made discordant predictions for 34 isolates by genome assembly, 30 isolates by allele microassembly, and 82 isolates by raw read k-mer, giving concordances of 98.5%, 98.7%, and 96.4%, respectively. Among the aforementioned comparisons, serotype prediction based on antigen identification from genome assemblies is the most direct comparison between the two tools because that is the only analysis they have in common. SeqSero2 made 38% fewer incorrect predictions in this comparison (34 versus 55).
Performance comparison between SeqSero2 and SeqSero1 using Illumina short reads.
To compare serotype prediction by SeqSero2 and SeqSero1, we analyzed the same set of Salmonella genomes that was used to evaluate SeqSero1 (9) (see Materials and Methods). Results from both platforms are summarized in Table 3. The specific antigens determined for each genome are in Table S2.
TABLE 3.
Summary of SeqSero1 and SeqSero2 prediction results
| Result | No. of genomes (% of total) for: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Raw reads (CDC strains) |
Raw reads (GenomeTrakr strains) |
Genome assemblies |
||||||
| SeqSero2 (k-mer) | SeqSero2 (microassembly) | SeqSero1 | SeqSero2 (k-mer) | SeqSero2 (microassembly) | SeqSero1 | SeqSero2 | SeqSero1 | |
| Expected serotypea | 292 (95.1%) | 304 (99.0%) | 303 (98.7%) | 3,014 (94.0%) | 3,020 (94.2%) | 3,018 (94.1%) | 3,305 (94.1%) | 3,043 (86.6%) |
| Unexpected serotypeb | 13 (4.2%) | 1 (0.3%) | 2 (0.7%) | 174 (5.4%) | 165 (5.1%) | 167 (5.2%) | 186 (5.3%) | 184 (5.2%) |
| Partial or no serotypec | 2 (0.7%) | 2 (0.7%) | 2 (0.7%) | 19 (0.6%) | 22 (0.7%) | 22 (0.7%) | 23 (0.7%) | 287 (8.2%) |
| All results | 307 | 307 | 307 | 3,207 | 3,207 | 3,207 | 3,514d | 3,514 |
The predicted serotype was considered correct when the serotype antigens detected corresponded to the antigens detected by phenotypic methods.
Numbers represent serotype predictions inconsistent with the annotated serotype; the accuracy of the annotated serotype is unknown for GenomeTrakr strains and genome assemblies.
Some or all of the expected serotype determinants were not detected.
CDC strains (n = 307) and GenomeTrakr strains (n = 3,207) combined.
For genome assemblies, the overall serotype accuracy (i.e., percentage of expected serotype prediction, defined as correct identification of all serotype antigens) of SeqSero2 was substantially increased from that of SeqSero1 (94.1% versus 86.5%). Of the 287 genome assemblies with partial or no prediction by SeqSero1, 269 were caused by the failure of in silico PCR to extract fliC or fljB genes (Table S2). Of this subset of 269 genomes, 256 were correctly serotyped by SeqSero2 using assemblies. For raw sequencing reads, the accuracy of SeqSero2 was comparable to that of SeqSero1 (Table 3).
In the initial SeqSero1 analysis (9), some genomes were partially identified, yielding multiple possible serotype predictions. With SeqSero2, 607 of these 746 genomes were definitively assigned into 41 distinct serotypes (Table S2). Among the 607 isolates, 236 were distinguished through subspecies identification by SalmID (17), and 37 were differentiated through O22 and O23 characterization. Another 337 genomes represented seven serotype pairs from serogroup O8. The two serotypes in each of these pairs differ only by an O6 antigen. O8 serotypes that differ by only O6 have been shown to variably express O6 and are genetically indistinguishable (18); they have been combined in SeqSero2 (Table S3).
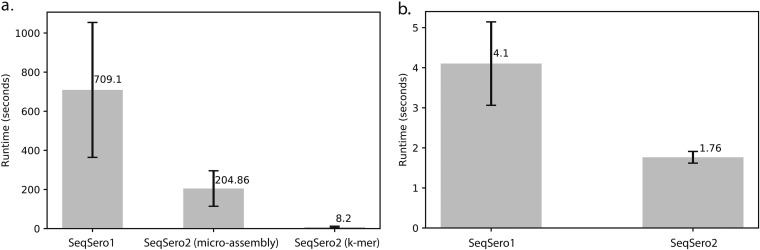
The average execution time per genome (see Materials and Methods for details) of SeqSero2 was <10 s and <2 s for raw reads (k-mer workflow) and draft assemblies, respectively, compared with about 540 s and about 10 s, respectively, for SeqSero1 (Fig. 3). The microassembly workflow of SeqSero2 for raw read analysis was substantially faster than the raw read workflow of SeqSero1 as well, averaging about 160 s per genome.
FIG 3.
Speed comparison between SeqSero1 and SeqSero2. (a) Comparison of run times for predicting serotypes from raw sequencing reads. Average number of seconds for analyzing a genome is shown for each workflow. BWA-MEM was used for read mapping for both SeqSero1 and the microassembly workflow of SeqSero2. (b) Comparison of run times for predicting serotypes from genome assemblies. Average number of seconds for analyzing a genome is shown for each workflow. Run time was defined as the elapsed real time (wall time) for predicting the serotype of a genome using a single processor.
Accessory tools in microassembly workflow.
The microassembly workflow provides additional information regarding sequence matches to the SeqSero serotype determinant databases, which can assist in detecting atypical results due to interserotype contamination or to divergent flagellin alleles. Besides serotype prediction, the microassembly workflow reports all assembled O and H antigen alleles, along with their sequence similarity scores to alleles in the serotype determinant databases (number of base matches/length of allele) in order to judge how good the matches are.
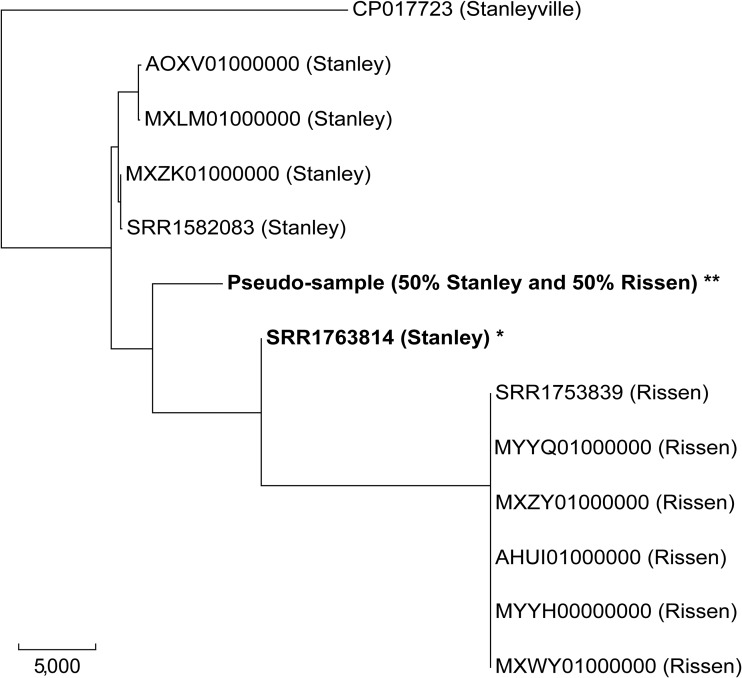
A genome contaminated with a second Salmonella serotype might be detected by the presence of 2 or more O antigen or 3 or more H antigen calls. For a genome annotated as S. enterica subsp. enterica serotype Stanley (SRA accession no. SRR1763814, antigenic formula 4:d:1,2), the microassembly analysis reported two wzx alleles representing O antigens O:7 and O:4, two fliC alleles representing antigens H:d and H:f,g, and one fljB allele representing antigen H:1,2. The extra O:7 and H:f,g alleles may be due to contamination by a strain with the antigenic formula 7:f,g:−, which corresponds to S. enterica subsp. enterica serotype Rissen. Contamination in SRR1763814 was further supported by phylogenetic analysis (Fig. 4). SRR1763814 clustered more closely to three serotype Rissen genomes; however, average SNP distance to the Rissen genomes (15,217 SNPs) was longer than that to three Stanley genomes (12,900 SNPs). An example of how the SeqSero2 output flags interserotype contamination can be found in File S1.
FIG 4.
Phylogenetic analysis based on SNPs of WGS samples with potential and artificial contaminations. The tree is rooted by an S. enterica subsp. enterica serotype Stanleyville strain as outgroup. Reference serotype S. Stanley, S. Rissen, and S. Stanleyville genomes are shown by their NCBI accession numbers. * indicates that a WGS sample was annotated as serotype Stanley but potential contamination from a serotype Rissen genome was detected. ** indicates that a pseudosample (6.0 Mb) was created by mixing sequencing reads from a reference serotype Stanley genome (SRA accession no. SRR1582083) and a reference serotype Rissen genome (SRA accession no. SRR1753839) at a 1:1 ratio. Bar, 5,000 SNPs.
To further evaluate the possibility of detecting interserotype contamination of different levels in sequencing data, we simulated contamination by creating pseudosamples that contained sequencing reads from different serotypes representing a broad range of serotype antigens. A total of 500 pseudosamples representing 443 distinct serotype combinations were created (see Materials and Methods for details). Simulated contamination was detected in 461 samples (92.2%). In the 39 samples where contamination was not detected, contaminant reads varied from 5% to 25% of the pseudosamples, covering the entire range of contamination ratios and showing no apparent overrepresentation of low ratios (Table S4).
Among the 2,280 NARMS isolates, 861 were determined by the allele microassembly workflow to have potential interserotype contamination (Table S1), although correct serotype prediction was still made for 845 isolates. Among the 90 isolates that resulted in at least one incorrect prediction by any of the three workflows of SeqSero2, 61 were found to carry potential interserotype contamination.
Potential interserotype contamination was also detected among some genomes, resulting in discordant serotype predictions between the raw read k-mer and the raw read microassembly workflows of SeqSero2. Among the 307 CDC strains used previously for evaluating SeqSero1 (9) and analyzed by both SeqSero1 and SeqSero2 in the current study (Table 2), 14 were incorrectly predicted by the raw read k-mer workflow but correctly predicted by the raw read microassembly workflow by SeqSero2. Ten of these 14 genomes had potential interserotype contamination detected by the raw read microassembly workflow (Table S2).
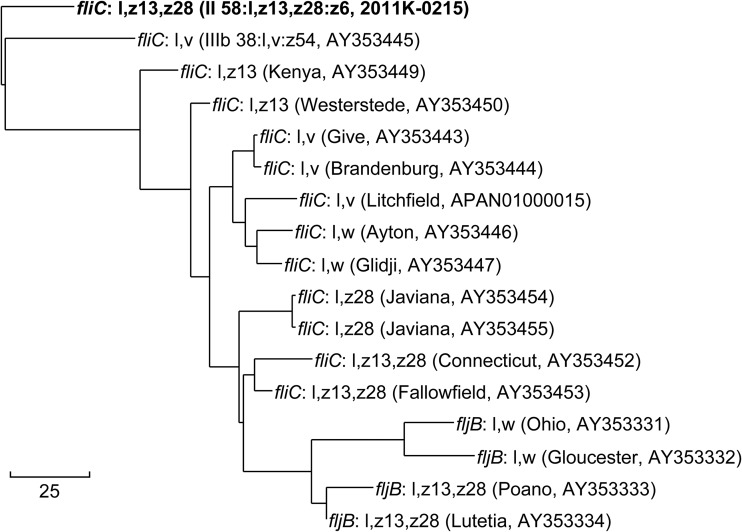
Misidentification of a flagellin allele by SeqSero2 can occur if a close relative to the query allele is not present in the database. For example, SeqSero1 and SeqSero2 misidentified the fliC allele as H:l,v in CDC strain 2011K-0215, which had been phenotypically identified as serotype II 58:l,z13,z28:z6. The fliC gene from 2011K-0215 generated by the microassembly workflow was distinct from all other L complex alleles in the serotype determinant database, including H:l,z13,z28 alleles from subspecies I serotypes (Fig. 5). The serotype of the strain was correctly predicted after adding the new allele to the serotype determinant database.
FIG 5.
Phylogenetic relationship between a new fliC allele (in bold) and alleles of related antigenic types in the serotype determinant database. Original serotype and NCBI accession number for each allele are shown in parentheses. Bar, 25 SNPs.
Serotype prediction from nanopore sequencing data.
In addition to the aforementioned analyses using Illumina sequencing data, we further tested SeqSero2 with nanopore sequencing data using 40 genomes of 17 serotypes that were publicly available (Table S7).
First, we directly analyzed raw nanopore reads without assembly through (i) the SeqSero2 raw read k-mer workflow and (ii) the SeqSero2 assembly workflow originally designed for genome assemblies from short sequencing reads, because long nanopore reads were equivalent to some assembled short-read contigs in length. The allele microassembly workflow was designed specifically for short reads and was not evaluated. The serotypes for 39 genomes determined by SeqSero2 using both raw reads and raw reads as an assembly were concordant with annotated serotypes (Table S7). The only discordant prediction was made for an annotated S. enterica subsp. enterica serotype Bareilly genome, for which both methods misidentified an H2:1,5 allele as H2:1,2. These two alleles are known to be similar to each other and are sometimes difficult to differentiate (9). For comparison, we also tested long nanopore reads as input for SISTR, even though SISTR requires assembled genomes for analysis. While nanopore reads were long enough to be analyzed by the genome assembly workflow of SeqSero2, they were inappropriate for analysis by SISTR, likely due to excessive base call errors. SISTR’s serotype predictions based on cgMLST were S. enterica subsp. enterica Choleraesuis for all but one annotated serotype S. Enteritidis genome, for which no prediction was made. SISTR’s serotype predictions based on antigen identification were concordant with the annotation for 27 of the 40 genomes (Table S7).
Second, we assembled nanopore reads to correct some base call errors and used the assembled genomes as input for the SeqSero2 genome assembly workflow and SISTR. With assembled genomes, all of SeqSero2’s predictions were concordant with annotation. Genome assembly substantially improved SISTR predictions, resulting in 23 results concordant with the annotation by cgMLST clustering and 36 by antigen identification (Table S7).
The direct use of nanopore raw reads for the SeqSero2 genome assembly workflow was substantially faster than any other methods tested. The per-genome turnaround time by this method varied by the size of sequencing data, ranging from 6 s (126-Mb raw reads) to 180 s (5,608-Mb raw reads) and averaging 4 s/100 Mb (Table S7). In comparison, the raw read k-mer workflow of SeqSero2 was about 18 times slower, averaging 73 s/100 Mb (1.8 to 70 min per genome). SeqSero2 and SISTR predictions by genome assembly took an average of 117 s/100 Mb (2 to 450 min per genome) just for the assembly. All of the run time measurements (clock time) were based on processing with a single core.
DISCUSSION
SeqSero2 represents a major algorithmic transformation for serotype determination. It uses a k-mer-based approach that unifies serotype determinant identification for both raw sequencing reads and genome assemblies. This approach replaced SeqSero1’s cumbersome handling of sequencing reads, which included multiple rounds of read mapping, and the older serotyping tool’s unreliable in silico PCR amplification of serotype determinants from draft genome assemblies. This change not only accelerates serotype prediction from all types of WGS data but also improves the prediction for genome assemblies. SeqSero1 was primarily designed for short sequencing reads and was not optimized for draft genome assemblies (9). A recent study compared only the genome assembly workflow of SeqSero1 with other genome assembly-based Salmonella serotype prediction tools (19); it showed that incomplete and inaccurate predictions by SeqSero1 were often caused by suboptimal or failed assembly of serotype determinant alleles. Similarly, we estimated that the majority of unexpected serotype predictions from genome assemblies by SeqSero1 were due to failures in extracting fliC or fljB alleles by in silico PCR (9). Using the k-mer-based approach, both genome assemblies and sequencing reads are treated in a similar manner regardless of their length difference. Because of this optimization, the genome assembly workflow of SeqSero2 is more accurate for serotype prediction and more tolerant of low-quality genome assemblies compared to that of SeqSero1.
SeqSero2 predicts serotypes in a matter of seconds directly from short raw sequencing reads, whereas de novo genome assembly, a prerequisite for other serotype prediction tools and methods (1, 10), takes 60 to 240 min per genome under the same computational conditions (a single core). Accurate serotype predictions can be obtained by SeqSero2 almost instantaneously after sequencing runs. This efficiency not only allows substantial time saving for processing large amounts of genomes but also helps smaller projects with limited computational resources by bypassing genome assembly if not otherwise needed.
The k-mer workflow for raw sequencing reads employs subsampling of sequencing reads to achieve superior speeds of analysis. Allele matching is based on k-mer similarity of the subsampled reads and alleles in the serotype determinant database (see Materials and Methods). Compared with the microassembly workflow, in which antigen sequences assembled from a query genome are directly aligned to alleles in the serotype determinant database, k-mer matching appears to be less robust when it comes to atypical alleles or serotype contamination in query genomes due to its higher sensitivity to allelic divergence. This algorithmic difference likely accounted for the higher serotype prediction accuracy by the microassembly workflow observed in this study.
Sample contamination detection using the microassembly workflow is a good quality control tool for both in-house sequencing and public data sourcing. Sample contamination can occur during culturing, DNA extraction and preparation, or sequencing. WGS is typically performed in a multiplex format in which multiple isolates are sequenced together. Mixed DNA from different samples has been identified as the predominant source of errors for postsequencing detection of sequence variants (20). Our analysis of 3,514 publicly available Salmonella genomes suggests that 3% of them may have potential interserotype contamination. We recommend contamination screening for sequenced and downloaded Salmonella genomes prior to using the genomes. It should be noted that SeqSero2 will not detect contamination from the same serotype or from a non-Salmonella organism. Also, contamination detection requires the presence of serotype determinant sequences, e.g., rfb, fliC, or fljB, from the contaminant genome. At low levels of contamination, such reads may be absent.
Unlike other Salmonella serotype prediction tools that consult surrogate markers, such as phylogenetic clustering with a reference genome in SISTR, to achieve or improve serotype identification (1, 10), SeqSero1’s design of only interrogating antigenic determinants of serotype contributes to the ambiguity in serotype identification (19). Specifically, in some cases, multiple-serotype predictions are generated by SeqSero1 for one query genome because the particular O and H antigen combination may indicate serotypes that belong to different subspecies or serotypes that require additional characterization for full identification. Lack of definitive serotype assignment in such cases has been substantially alleviated by SeqSero2 through the addition of an identification tool (SalmID) and additional targets that differentiate certain variants.
Horizontal gene transfer is common in Salmonella (21); this transfer includes serotype determinants and can result in distinct linages having the same complement of serotype antigens or, conversely, in the same lineage having two different “serotypes” (22, 23). In such cases, relying on or incorporating phylogenetic markers for serotype identification may cause results inconsistent with the classic, phenotypic definition of Salmonella serotype. SeqSero2 adheres to the genetic determinants for the phenotypic markers of Salmonella serotypes. By targeting these determinants, SeqSero2 provides continuity with historical data sets based on serotype and facilitates communication using our serotype-based understanding of Salmonella epidemiology.
Further evidence that targeting serotype determinants alone without consulting surrogate markers is robust enough for serotype prediction came from the comparison between SeqSero2 and SISTR, as the latter tool resorts to phylogenetic markers (cgMLST) in addition to serotype determinants. The overall performances of SeqSero2 and SISTR were similar to each other.
Nanopore sequencing has been preliminarily investigated for Salmonella serotype prediction and SNP subtyping (24). It is advantageous for generating long sequencing reads and supporting real-time data analysis but is limited by its higher base call error rates than those of Illumina sequencing platforms (24). We demonstrated that SeqSero2 has potential for accurate and rapid serotype prediction directly from nanopore sequencing reads without genome assembly. This capability was enabled by SeqSero2’s k-mer-based algorithm, which takes advantage of both long nanopore reads as if they were assemblies from short reads and the algorithm’s tolerance of base call errors. Without base call correction by genome assembly, such errors hindered cgMLST analysis in SISTR, causing no correct serotype prediction by cgMLST clustering. Similarly, the base call errors were excessive for antigen identification by SISTR, leading to at least one incorrect antigen call in 32.5% of the tested genomes. In comparison, SeqSero2’s performance was mostly unaffected by these error-prone reads even without genome assembly (correct serotype prediction for 39 of the 40 genomes), which is particularly promising for supporting real-time Salmonella characterization through nanopore sequencing.
MATERIALS AND METHODS
Genomes.
Two data sets were used to analyze SeqSero2 (v1.0.2) performance on short sequencing reads. (i) Genomes from 2,280 strains submitted to CDC NARMS in 2015 were used to assess overall performance (Table S1). NARMS performs surveillance for antimicrobial resistance in Salmonella (https://www.cdc.gov/narms/index.html); every 20th isolate, along with serotype information, is submitted by state and local health departments. Since all strains are in our collection, we also investigated and arbitrated discordant results between SeqSero2 and the state-submitted serotype. (ii) Genomes that had been used to test SeqSero1 (9) were used to evaluate the accuracy and speed of SeqSero2 in comparison to SeqSero1 (Table S2). To test SeqSero2 on long nanopore reads, a total of 40 nanopore-sequenced genomes of 17 serotypes were downloaded from NCBI and analyzed (Table S7). This set included all of the nanopore-sequenced Salmonella genomes with a sequencing coverage over 25× that were available from NCBI at the time of this study.
Genome sequencing and assembly.
Strains were sequenced on the MiSeq and HiSeq platforms using procedures previously described (25). De novo assembly from raw sequencing reads was performed for all genomes using SPAdes (26). Nanopore-sequenced genomes were assembled by using minimap2 and miniasm (27), followed by an error correction step using Racon (28).
Microbial identification.
Microbial identification from a query genome was determined using the open access software tool SalmID (github.com/hcdenbakker/SalmID). SalmID uses a k-mer approach to differentiate Salmonella species and subspecies based on invA and rpoB. It was validated using 132 strains representing all Salmonella species and subspecies (Table S5).
Additional markers.
Additional markers were used to differentiate certain serotypes, pathotypes, and variants of particular serotypes (Table 2). (i) Two pathotypes of serotype Paratyphi B have been described, a gastrointestinal pathotype and a typhoidal pathotype (29); we targeted an SNP associated with the inability to ferment tartrate in the typhoidal pathotype in order to differentiate the pathotypes (29). (ii) S. enterica subsp. enterica serotype Gallinarm is a nonmotile, bird-adapted serotype that is rare in the United States. It possesses a nonexpressed fliC allele (g,m) that makes it indistinguishable from serotype Enteritidis using genetic methods for serotype determination. We used sdf (3) to identify commonly circulating strains of serotype Enteritidis. (iii) oafA encodes an acetyltransferase that is responsible for ancillary O antigen 5 in some group O4 serotypes; a 7-bp deletion inactivates this gene in many serotype Typhimurium strains, resulting in an O5− phenotype (16). We targeted this marker to detect strains carrying the 7-bp deletion since it is a useful epidemiologic marker (16). It is important to note that this marker identifies only some phenotypically O5− strains; no conclusions can be drawn about O5 status if it is not detected. (iv) Serotypes within group O13 are differentiated by ancillary antigens O22 and O23. Differences in galE (UDP-galactose 4-epimerase) between O22-positive (O22+) and O23+ rfb regions (GenBank accession no. NZ_LS483489 and NZ_CP029041) were targeted to differentiate O22+ and O23+ serotypes.
k-mer-based serotype prediction from raw sequencing reads and genome assemblies.
A k-mer-based workflow was developed for serotype prediction directly from raw sequencing reads (Figure 2a). Specifically, unique short sequences (i.e., k-mers; k = 27 bases by default) were derived from each allele in the serotype determinant (wzx and wzy genes for O antigen; fliC and fljB genes for H antigen) database (Table S6). These k-mers formed a new serotype determinant database (Kd) in which a particular serotype determinant allele (i) was represented by a set of k-mers (Kdi). Sequencing reads were subsampled from a query genome by collecting reads whose middle k-mer matched any sequence in the k-mer-based serotype determinant database. The subsampling, similar to that of StringMLST (30), continued until the total length of sampled reads reached a default threshold of 4,000,000 bases. Subsampled reads were converted into a set of k-mers to represent the input genome (Kg). A similarity score (Si) for a particular serotype determinant allele (i) was calculated using the following formula:
where the function N enumerates k-mers in a k-mer set. The type of O or H antigen of the query genome was determined by the type of the corresponding allele that yielded the highest similarity score. The final serotype of the query genome was called by consulting the White-Kauffmann-Le Minor scheme (5, 6).
A similar k-mer-based workflow was developed for rapid serotype prediction from genome assemblies. Each contig of a genome assembly was converted into pseudoreads by breaking the contig into tandemly contiguous substrings with a default length of 60 bases. The rest of the workflow overlapped with that of the sequencing reads, as previously described, by treating the substrings as pseudoreads.
Microassembly of serotype determinants for serotype determination.
A schematic of the microassembly approach is presented in Fig. 2b. The microassembly workflow assembles serotype determinants, i.e., wzx and wzy for O antigen identification and fliC and fljB for H antigen identification. All sequencing reads from a query genome were mapped to the serotype determinant database using BWA-MEM (31). Reads that had been mapped to alleles in the database were extracted and assembled by SPAdes (26). The resulting contigs were then aligned back to the serotype determinant database by BLAST (32). The type of O or H antigen of the query genome was determined by the type of the corresponding allele that yielded the highest BLAST similarity. The final serotype of the query genome was called by consulting the White-Kauffmann-Le Minor scheme.
Speed comparison between SeqSero1 and SeqSero2.
The speeds of SeqSero2 and SeqSero1 were compared by recording their respective run times in analyzing the 307 Salmonella genomes sequenced by the CDC (9) under the same computational conditions, including a single processing core. Run time was defined as the elapsed real time (wall time) for predicting the serotype of a genome using a single processor.
Contamination detection.
The microassembly workflow for serotype prediction was also used to detect potential interserotype contamination in WGS data. Potential contamination is indicated when more than one O antigen allele or more than two H antigen alleles were assembled from a WGS sample.
Simulation of interserotype contamination.
WGS samples with simulated interserotype contamination were created by combining two genomes (G1 and G2) of different serotypes from the set of 307 genomes sequenced by the CDC (9). For each pair of genomes, G2 was designated the contaminant. Sequencing reads from G2 replaced 5% to 25% of G1 reads to create the pseudosample. Sequencing reads involved in the replacement were randomly sampled from the two genomes. Less than 2% contamination may occur in Illumina sequencing data due to index misassignment or index hopping (33).
Serotype prediction by SISTR.
SISTR command line tool v1.0.2 (github.com/phac-nml/sistr_cmd) was used to analyze genome assemblies of 2,280 Illumina-sequenced genomes in the NARMS data set and raw reads and genome assemblies of 40 nanopore-sequenced genomes.
Phylogenetic analysis.
Phylogenetic analysis of genomes was performed by Parsnp (34). Comparison and phylogenetic clustering of fliC and fljB alleles was performed by Molecular Evolutionary Genetics Analysis version 7.0 (MEGA7) (35).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Broad Agency Announcement Contract by the Food and Drug Administration (HHSF223201610061C) and by a Hatch project (1006141) from the USDA National Institute of Food and Agriculture.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01746-19.
REFERENCES
- 1.Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, Tewolde R, Schaefer U, Jenkins C, Dallman TJ, de Pinna EM, Grant KA, Salmonella Whole Genome Sequencing Implementation Group . 2016. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4:e1752. doi: 10.7717/peerj.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inns T, Ashton PM, Herrera-Leon S, Lighthill J, Foulkes S, Jombart T, Rehman Y, Fox A, Dallman T, DE Pinna E, Browning L, Coia JE, Edeghere O, Vivancos R. 2017. Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol Infect 145:289–298. doi: 10.1017/S0950268816001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agron PG, Walker RL, Kinde H, Sawyer SJ, Hayes DC, Wollard J, Andersen GL. 2001. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl Environ Microbiol 67:4984–4991. doi: 10.1128/AEM.67.11.4984-4991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng X, den Bakker HC, Hendriksen RS. 2016. Genomic epidemiology: whole-genome-sequencing-powered surveillance and outbreak investigation of foodborne bacterial pathogens. Annu Rev Food Sci Technol 7:353–374. doi: 10.1146/annurev-food-041715-033259. [DOI] [PubMed] [Google Scholar]
- 5.Grimont PAD, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Centre for Reference and Research on Salmonella, Paris, France. [Google Scholar]
- 6.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, Weill FX. 2010. Supplement 2003–2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J Clin Microbiol 45:3323–3334. doi: 10.1128/JCM.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McQuiston JR, Waters RJ, Dinsmore BA, Mikoleit ML, Fields PI. 2011. Molecular determination of H antigens of Salmonella by use of a microsphere-based liquid array. J Clin Microbiol 49:565–573. doi: 10.1128/JCM.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VP, Nash JH, Taboada EN. 2016. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timme RE, Sanchez Leon M, Allard MW. 2019. Utilizing the public GenomeTrakr database for foodborne pathogen traceback. Methods Mol Biol 1918:201–212. doi: 10.1007/978-1-4939-9000-9_17. [DOI] [PubMed] [Google Scholar]
- 12.Moreno LZ, Gomes VTM, Moreira J, de Oliveira CH, Peres BP, Silva APS, Thakur S, La Ragione RM, Moreno AM. 2018. First report of mcr-1-harboring Salmonella enterica serovar Schwarzengrund isolated from poultry meat in Brazil. Diagn Microbiol Infect Dis 93:376–379. doi: 10.1016/j.diagmicrobio.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Carrera PM, Kantarjian HM, Blinder VS. 2018. The financial burden and distress of patients with cancer: understanding and stepping-up action on the financial toxicity of cancer treatment. CA Cancer J Clin 68:153–165. doi: 10.3322/caac.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bale J, Meunier D, Weill FX, dePinna E, Peters T, Nair S. 2016. Characterization of new Salmonella serovars by whole-genome sequencing and traditional typing techniques. J Med Microbiol 65:1074–1078. doi: 10.1099/jmm.0.000325. [DOI] [PubMed] [Google Scholar]
- 15.Connor TR, Owen SV, Langridge G, Connell S, Nair S, Reuter S, Dallman TJ, Corander J, Tabing KC, Le Hello S, Fookes M, Doublet B, Zhou Z, Feltwell T, Ellington MJ, Herrera S, Gilmour M, Cloeckaert A, Achtman M, Parkhill J, Wain J, De Pinna E, Weill FX, Peters T, Thomson N. 2016. What’s in a name? Species-wide whole-genome sequencing resolves invasive and noninvasive lineages of Salmonella enterica serotype Paratyphi B. mBio 7:e00527-16. doi: 10.1128/mBio.00527-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauser E, Junker E, Helmuth R, Malorny B. 2011. Different mutations in the oafA gene lead to loss of O5-antigen expression in Salmonella enterica serovar Typhimurium. J Appl Microbiol 110:248–253. doi: 10.1111/j.1365-2672.2010.04877.x. [DOI] [PubMed] [Google Scholar]
- 17.den Bakker HC, Zhang S. 2018. SalmID: first Zenodo release (version 0.122). doi: 10.5281/zenodo.1409766. [DOI]
- 18.Mikoleit M, Van Duyne MS, Halpin J, McGlinchey B, Fields PI. 2012. Variable expression of O:61 in Salmonella group C2. J Clin Microbiol 50:4098–4099. doi: 10.1128/JCM.01676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yachison CA, Yoshida C, Robertson J, Nash JHE, Kruczkiewicz P, Taboada EN, Walker M, Reimer A, Christianson S, Nichani A, PulseNet Canada Steering Committee, Nadon C. 2017. The validation and implications of using whole genome sequencing as a replacement for traditional serotyping for a national Salmonella reference laboratory. Front Microbiol 8:1044. doi: 10.3389/fmicb.2017.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porwollik S, McClelland M. 2003. Lateral gene transfer in Salmonella. Microbes Infect 5:977–989. doi: 10.1016/S1286-4579(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 22.Deng X, Desai PT, den Bakker HC, Mikoleit M, Tolar B, Trees E, Hendriksen RS, Frye JG, Porwollik S, Weimer BC, Wiedmann M, Weinstock GM, Fields PI, McClelland M. 2014. Genomic epidemiology of Salmonella enterica serotype Enteritidis based on population structure of prevalent lineages. Emerg Infect Dis 20:1481–1489. doi: 10.3201/eid2009.131095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugarel M, Cook PW, den Bakker HC, Harhay D, Nightingale KK, Loneragan GH. 2019. Complete genome sequences of four Salmonella enterica strains (including those of serotypes Montevideo, Mbandaka, and Lubbock) isolated from peripheral lymph nodes of healthy cattle. Microbiol Resour Announc 8. doi: 10.1128/MRA.01450-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyeon JY, Li S, Mann DA, Zhang S, Li Z, Chen Y, Deng X. 2018. Quasimetagenomics-based and real-time-sequencing-aided detection and subtyping of Salmonella enterica from food samples. Appl Environ Microbiol 84:e02340-17. doi: 10.1128/AEM.02340-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anonymous. 2016. Laboratory standard operating procedure for PulseNet Nextera XT library prep and run setup for Illumina MiSeq. https://www.cdc.gov/pulsenet/pdf/PNL32-MiSeq-Nextera-XT.pdf.
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaser R, Sovic I, Nagarajan N, Sikic M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malorny B, Bunge C, Helmuth R. 2003. Discrimination of d-tartrate-fermenting and -nonfermenting Salmonella enterica subsp. enterica isolates by genotypic and phenotypic methods. J Clin Microbiol 41:4292–4297. doi: 10.1128/jcm.41.9.4292-4297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta A, Jordan IK, Rishishwar L. 2017. stringMLST: a fast k-mer based tool for multilocus sequence typing. Bioinformatics 33:119–121. doi: 10.1093/bioinformatics/btw586. [DOI] [PubMed] [Google Scholar]
- 31.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 13033997v2 [q-bioGN] https://arxiv.org/abs/1303.3997.
- 32.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Illumina. 2017. Effects of index misassignment on multiplexing and downstream analysis. https://www.illumina.com/content/dam/illumina-marketing/documents/products/whitepapers/index-hopping-white-paper-770-2017-004.pdf?linkId=36607862.
- 34.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease 'Burden of Illness' Studies . 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.