The particles most effective at inducing the freezing of water in the atmosphere are microbiological in origin; however, information on the species harboring this phenotype, their environmental distribution, and ecological sources are very limited. Analysis of precipitation collected over 2 years in Louisiana showed that INPs active at the warmest temperatures were sourced from terrestrial ecosystems and displayed behaviors that implicated specific bacterial taxa as the source of the ice nucleation activity. The abundance of biological INPs was highest in precipitation from winter storms and implied that their in-cloud concentrations were sufficient to affect the formation of ice and precipitation in nimbostratus clouds.
KEYWORDS: aeromicrobiology, bacteria, biological ice nuclei, environmental microbiology, meteorology, microbial ecology, precipitation
ABSTRACT
Airborne bacteria that nucleate ice at relatively warm temperatures (>−10°C) can interact with cloud water droplets, affecting the formation of ice in clouds and the residency time of the cells in the atmosphere. We sampled 65 precipitation events in southeastern Louisiana over 2 years to examine the effect of season, meteorological conditions, storm type, and ecoregion source on the concentration and type of ice-nucleating particles (INPs) deposited. INPs sensitive to heat treatment were inferred to be biological in origin, and the highest concentrations of biological INPs (∼16,000 INPs liter−1 active at ≥–10°C) were observed in snow and sleet samples from wintertime nimbostratus clouds with cloud top temperatures as warm as –7°C. Statistical analysis revealed three temperature classes of biological INPs (INPs active from −5 to –10°C, −11 to –12°C, and −13 to –14°C) and one temperature class of INPs that were sensitive to lysozyme (i.e., bacterial INPs, active from −5 to –10°C). Significant correlations between the INP data and abundances of taxa in the Bacteroidetes, Firmicutes, and unclassified bacterial divisions implied that certain members of these phyla may possess the ice nucleation phenotype. The interrelation between the INP classes and fluorescent dissolved organic matter, major ion concentrations (Na+, Cl−, SO42−, and NO3−), and backward air mass trajectories indicated that the highest concentrations of INPs were sourced from high-latitude North American and Asian continental environments, whereas the lowest values were observed when air was sourced from marine ecoregions. The intra- and extracontinental regions identified as sources of biological INPs in precipitation deposited in the southeastern United States suggests that these bioaerosols can disperse and affect meteorological conditions thousands of kilometers from their terrestrial points of origin.
IMPORTANCE The particles most effective at inducing the freezing of water in the atmosphere are microbiological in origin; however, information on the species harboring this phenotype, their environmental distribution, and ecological sources are very limited. Analysis of precipitation collected over 2 years in Louisiana showed that INPs active at the warmest temperatures were sourced from terrestrial ecosystems and displayed behaviors that implicated specific bacterial taxa as the source of the ice nucleation activity. The abundance of biological INPs was highest in precipitation from winter storms and implied that their in-cloud concentrations were sufficient to affect the formation of ice and precipitation in nimbostratus clouds.
INTRODUCTION
At temperatures warmer than –36°C, the phase change from water to ice requires the presence of impurities that serve as sites for ice nucleation (1–3). Dust aerosols have important roles in the troposphere by serving as ice-nucleating particles (INPs; see Table 1 for acronym usage) that contribute to ice formation in clouds, a prerequisite for snow and most rainfall (1, 4). However, atmospherically relevant mineral dust aerosols do not initiate freezing at temperatures warmer than –15°C (5), whereas certain microorganisms and biogenic molecules are very effective INPs and have ice-nucleating (IN) activities at temperatures of ≥–10°C (4, 6–10). For instance, some strains of Pseudomonas syringae express an outer membrane protein (InaZ) that structurally orders water molecules into an ice-like configuration, allowing the phase transition at temperatures as warm as –1.8°C (11–14). Since its initial discovery, the IN phenotype has been demonstrated in several other Gammaproteobacteria (6, 15–19) and a species of Firmicutes (20), as well as in certain fungi (21, 22), algae (23, 24), and pollens (10). High IN activities are associated with epiphytic bacterial communities on the leaves of deciduous plants (∼105 IN bacteria cm−2 [25]) and the microbiological decomposition of detritus in a variety of plant (9, 11, 26, 27), soil (28, 29), and aquatic (23, 30, 31) ecosystems. While various environments are recognized to harbor biological INPs and their presence in precipitation is well documented (7, 32–34), longitudinal data sets are lacking, and there is a need for studies that identify sources of atmospheric biological INPs and the conditions affecting their distribution in precipitation (5).
TABLE 1.
Reference list of acronym definitions in this study
| Acronym | Term | Definition |
|---|---|---|
| INP | Ice-nucleating particle | A particle which catalyzes the conversion of liquid or gaseous water to ice |
| IN | Ice nucleating | The act of nucleating ice |
| DOM | Dissolved organic matter | Soluble organic molecules, predominantly composed of carbon, nitrogen, oxygen, or phosphorus, that are dissolved within aqueous solutions |
| DOC | Dissolved organic carbon | Carbon fraction of DOM |
| EEMs | Excitation emission matrices | A three-dimensional scan produced by fluorescence spectroscopy which produces a contour plot with excitation and emission wavelengths on the x and y axis and intensity on the z axis; this method of chemical analysis provides qualitative information on the presence of fluorescent molecules within an aqueous solution |
| PARAFAC | Parallel factor | Statistical method used to analyze the output of EEMs and identify recurring areas of varying fluorescent intensities; PARAFAC analysis profiles aid in characterizing the dissolved organic matter present within an aqueous sample |
| MBL | Mixed boundary layer | Lower layer of the troposphere where turbulent mixing with the Earth’s surface occurs |
| HYSPLIT | Hybrid single-particle Lagrangian integrated trajectory | Atmospheric model created by the National Oceanic and Atmospheric Administration that can calculate the history of an air mass at any particular altitude within the atmosphere |
| LCL | Lifted condensation levels | Altitude within the troposphere at which condensation of water vapor occurs when air is forcibly lifted by mechanical means (frontal systems, convergence of air masses, or orographic lifting) |
| CCL | Convective condensation levels | Altitude within the troposphere at which condensation of water vapor occurs when air is lifted due to convection (heating and rising of a parcel of air in an unstable atmosphere) |
| EPA | Environmental Protection Agency | Agency of the USA which serves the purpose of protecting the environment |
| CEC | Commission for Environmental Cooperation | Agency established by Canada, Mexico, and the USA as part of the North American Agreement on Environmental Cooperation; this agency serves the purpose of protecting the environment by promoting cooperation between these nations and the public |
| EFA | Exploratory factor analysis | Multivariate statistical procedure which groups together similar variables based on the pattern of matrix data across all observations and all variables |
| MANOVA | Multivariate analysis of variance | Multivariate statistical procedure which accounts for variances within and between group values in order to determine whether there is any significant difference between the groups in question |
| OTU | Operational taxonomic unit | Group of related organisms based on a threshold of similarity among their DNA sequences; the taxonomic level of sampling, which is defined by the user, can correspond to bacterial populations, species, or genera |
The warm temperatures at which biological INPs initiate freezing coupled with their ubiquity in near-surface air (32, 35–37), cloud water (33, 38, 39), and precipitation (7, 32–34) implies that they could contribute to cloud ice formation under certain meteorological conditions (40). At cloud temperatures warmer than –10°C, biological INPs could affect ice formation directly or via secondary ice formation processes (1, 41–47). The interaction between biological INPs and cloud water may also indirectly affect climate by influencing cloud albedo, and consequently, the global radiative budget (48–51). Thus far, assessing the meteorological effects of biological INPs has been hampered by the shortage of data on their abundance and ice-nucleating activity in the atmosphere, which are important parameterizations in cloud modeling studies (40). There is also evidence that the ice nucleation phenotype could provide a selective advantage to aerially transported microbial populations, facilitating their removal from the atmosphere and delivery to the surface in precipitation (40). Well-studied IN-active bacteria, such as P. syringae, live epiphytically on plants and are phytopathogens, suggesting that IN activity could play an important role in dissemination to new hosts (52).
To improve understanding of the environmental factors affecting the type, abundance, and sources of biological INPs in precipitation, we collected microbiological, geochemical, and meteorological data from 65 precipitation events in Louisiana over a 2-year period. The purpose of this study was to determine if discernible patterns in the composition and concentration of biological INPs deposited in precipitation were associated with season, air mass history, cloud type, meteorological conditions, and precipitation chemistry. Multivariate statistical analysis revealed temperature-specific characteristics of the INP populations, identified explanatory variables that provide information on the atmospheric sources of the biological INPs, and detected specific meteorological conditions where they had high abundances in the precipitation. Our analysis implies that the bioaerosol sources could be quite distant from the deposition site (i.e., continental Asia and high-latitude North America) and indicated that terrestrial environments were the main source of warm-temperature biological INPs in the precipitation sampled. Moreover, our results show that the abundance of biological and bacterial INPs were strongly associated with the season, storm type, and the presence of specific bacterial taxa in the precipitation.
RESULTS
Total, biological, and bacterial INPs in Louisiana precipitation.
The data collected and analyzed during this study are provided in Data Set S1 in the supplemental material. Following Christner et al. (7), INPs in untreated samples are referred to as “total,” those sensitive to heating at 95°C for 10 min are inferred to be proteinaceous in origin and are classified as “biological,” and those inactivated by digestion with 3 mg ml−1 lysozyme at 22°C for 60 min are classified as “bacterial.”
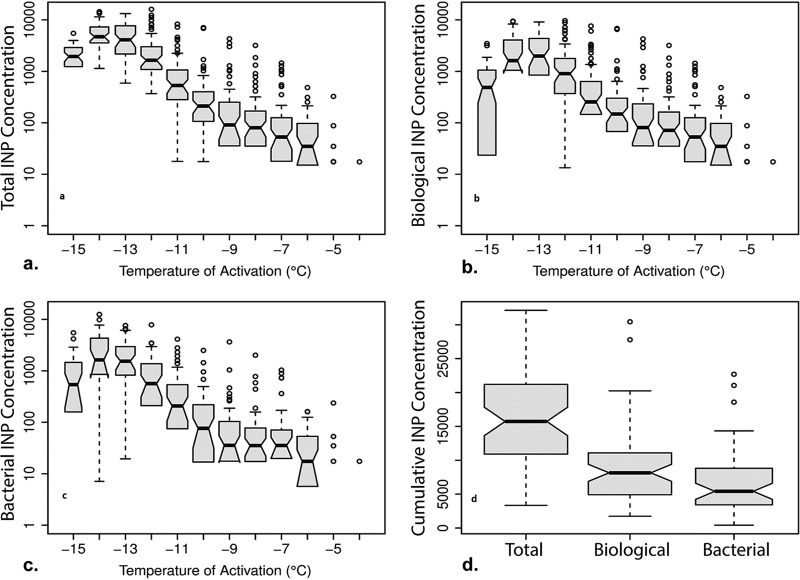
The warmest temperature at which ice nucleation occurred in all of the precipitation samples was –10°C, but freezing was detected at –4°C in six of the precipitation samples. The number and activity temperature of total, biological, and bacterial INPs varied significantly between precipitation events, with concentration values for a given temperature ranging 2 to 3 orders of magnitude (Fig. 1a to c). The cumulative concentrations of total, biological, and bacterial INPs active at –15°C averaged 16,200, 9,000, and 6,600 INPs liter−1 precipitation, respectively, with biological and bacterial INPs comprising 56% and 41%, respectively, of the total cumulative INPs (Fig. 1d and Table 2). Based on the total differential INP data, >95% of the INPs active at warmer than –10°C were inferred to be biological in origin (Table 2). At temperatures colder than –10°C, biological INPs were a maximum of 66% of the total differential INPs and 35% of the total differential INPs at –15°C (Table 2). On average, less than half of the INPs at temperatures colder than –8°C were bacterial, but they represented the vast majority of INPs that were active at >–8°C (Table 2).
FIG 1.
Concentrations of total, biological, and bacterial ice-nucleating particles (INPs) at activation temperatures of –4 to –15°C. The median and 95% confidence interval (CI) values are depicted by the black horizontal lines and dashed vertical lines, respectively; boxes represent the interquartile range; whiskers represent maximum and minimum values, excluding outliers; and open circles represent outliers. (a) Differential total INP concentrations, with sample sizes as follows: −4 to −12°C, n = 61; −13°C, n = 56; −14°C, n = 48; and −15°C, n = 44. (b) Differential biological INP concentrations, with sample sizes as follows: −4 to −12°C, n = 61; −13°C, n = 56; −14°C, n = 48; and −15°C, n = 44. (c) Differential bacterial INP concentrations, with sample sizes as follows: −4 to −12°C, n = 54; −13°C, n = 44; −14°C, n = 37; and −15°C, n = 34. (d) Cumulative concentrations of INPs active at temperatures of ≥−15°C for total, biological, and bacterial INPs.
TABLE 2.
Average differential concentrations of total, biological, and bacterial INPs
| Temp of activation (°C) | Total INP concn (mean ± SE) (liter−1 precipitation)a | Biological INP concn (mean ± SE) (liter−1 precipitation)b | % of total INP | Bacterial INP concn (mean ± SE) (liter−1 precipitation)c | % of total INP |
|---|---|---|---|---|---|
| −4 | 2 ± 1 | 2 ± 1 | 100 | 2 ± 1 | 100 |
| −5 | 11 ± 6 | 11 ± 6 | 100 | 10 ± 5 | 91 |
| −6 | 61 ± 11 | 60 ± 11 | 98 | 35 ± 6 | 57 |
| −7 | 174 ± 41 | 169 ± 41 | 97 | 108 ± 31 | 62 |
| −8 | 231 ± 64 | 223 ± 64 | 97 | 118 ± 42 | 51 |
| −9 | 349 ± 103 | 332 ± 101 | 95 | 160 ± 72 | 46 |
| −10 | 520 ± 161 | 460 ± 155 | 88 | 211 ± 54 | 41 |
| −11 | 1,030 ± 156 | 679 ± 121 | 66 | 523 ± 119 | 51 |
| −12 | 2,700 ± 341 | 1,530 ± 227 | 57 | 941 ± 153 | 35 |
| −13 | 5,200 ± 464 | 2,890 ± 326 | 56 | 2,060 ± 277 | 40 |
| −14 | 5,650 ± 496 | 2,800 ± 359 | 50 | 2,830 ± 480 | 50 |
| −15 | 2,090 ± 190 | 724 ± 119 | 35 | 1,020 ± 218 | 49 |
| Cumulative INP | 16,200 ± 954 | 9,020 ± 773 | 56 | 6,620 ± 660 | 41 |
Total INP was as follows: −4 to –12°C, n = 61; –13°C, n = 56; –14°C, n = 48; and –15°C, n = 44.
Biological INP was as follows: −4 to –12°C, n = 61; –13°C, n = 56; –14°C, n = 48; and –15°C, n = 44.
Bacterial INP was as follows: −4 to –12°C, n = 54; –13°C, n = 44; –14°C, n = 37; and –15°C, n = 34.
The concentrations for each category of INP increased exponentially with decreasing temperature to approximately –14°C (Fig. 1a to c). For 15 of the precipitation samples, all of the 96 wells being tested were frozen before cooling to –15°C, indicating high concentrations of INPs in the sample. Since at least one of the wells must remain unfrozen to calculate the INP concentration (53), abundances at the lowest temperatures could not be directly determined for these samples. Accordingly, the data reported at –13 to –15°C in Fig. 1 are missing observations for samples that had the highest INP concentrations at these temperatures. Specifically, the total differential INP data (n = 65) lacked values at –15°C (n = 3), ≤–14°C (n = 8), and ≤–13°C (n = 4), the biological differential INP data (n = 65) lacked values at –15°C (n = 4), ≤–14°C (n = 6), and ≤–13°C (n = 6), and the bacterial differential INP data (n = 56) lacked values at –15°C (n = 3), ≤–14°C (n = 7), and ≤–13°C (n = 7). To use observations with missing INP data in subsequent statistical analyses, multiple imputation was performed to estimate the values at these colder temperatures based on the pattern of available data (Table S1) (81, 82). Satisfactory relative efficiency values were produced for all INP types using 5 imputations and a monotone regression method (82). The descriptive statistics for the INP concentration data are listed in Table 3.
TABLE 3.
Descriptive statistics of numerical variables measured in this study
| Statistica | Total INP concn (liter−1 precipitation)b | Biological INP concn (liter−1 precipitation)b | Bacterial INP concn (liter−1 precipitation)b | Cell concn (liter−1 precipitation) | pH | Conductivity (μS cm−1) | DOC (ppm) | Concn (μM) of: |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cl− | NO3− | SO42− | Na+ | ||||||||
| n | 65 | 65 | 56 | 63 | 56 | 55 | 47 | 32 | 32 | 32 | 32 |
| Mean | 16,200 | 9,020 | 6,620 | 6.41E5 | 6.39 | 15.4 | 1.59 | 108 | 23.1 | 13 | 162 |
| SD | 6,349 | 5,108 | 3,628 | 9.54E5 | 0.83 | 8.95 | 1.12 | 104 | 14.1 | 7.8 | 101 |
| Upper 95% CI | 16,232 | 9,271 | 7,154 | 8.81E5 | 6.61 | 17.82 | 1.92 | 145 | 28.2 | 17.8 | 198 |
| Lower 95% CI | 16,133 | 9,192 | 7,094 | 4.00E5 | 6.17 | 12.98 | 1.26 | 70.1 | 18.0 | 12.2 | 125 |
| Median | 17,268 | 8,914 | 6,689 | 225,012 | 6.45 | 14.3 | 1.32 | 65.4 | 18.1 | 12.3 | 143 |
| Variance | 40,316,117 | 26,086,560 | 13,159,623 | 9.10E11 | 0.69 | 80.15 | 1.27 | 10,831 | 199 | 60 | 10,192 |
| IQR | 11,911 | 8,560 | 4,805 | 549,505 | 1.08 | 12.42 | 1.37 | 157 | 17.8 | 6.5 | 145 |
CI, confidence interval; IQR, interquartile range.
Cumulative concentration at –15°C.
Exploratory factor analysis of the INP data.
Exploratory factor analysis (EFA) was used to examine correlations between variables (Data Set S1) and the differential INP concentrations active from –4 to –15°C (Table 4). EFA is a statistical method that identifies patterns in multivariate data sets by searching for latent variables (referred to as factors), which are identified based on shared variance between the measured variables (83). In other words, the factors identified by EFA are measurements of intercorrelation between measured variables in the data set. Factor “loading” is the correlation between the factor and measured variable. A high-positive loading value (1.0 ≥ x ≥ 0.5) of a variable indicates that its variance in the data set is mostly explained by the factor onto which it is loading high. As such, a loading cutoff value of >0.50 was used to determine the variables retained for each factor (Table 4). The total variance of the factor is an estimation of how well the factor explains variance in the data set, which can be viewed as an indication of the influence the latent variable had on the measured variables. The analysis for total and bacterial INPs produced two significant factors, whereas biological INPs produced three significant factors (Table 4). Total INP concentrations between −5 and –11°C positively correlated with factor 1, and those between −11 and –14°C positively correlated with factor 2; however, the factor loadings for –4 and –15°C were not significant. The biological INP factor pattern produced results similar to those of the total INP factor pattern, wherein INP concentrations between –5 and –10°C loaded onto factor 1 for biological INPs, −13 and –14°C loaded onto factor 2, –11 and –12°C loaded onto factor 3, and –4 and –15°C did not load significantly onto any factor. For bacterial INPs, factor 1 positively correlated with INP concentrations between −5 and –10°C, whereas factor 2 positively correlated with INP concentrations at –4 and –12°C and negatively correlated with INP concentrations at –14°C. Due to the small number of observations for freezing at –4°C (n = 6), bacterial factor 2 (–4 and –12°C) was dropped from subsequent analyses. For simplicity, each factor is referred to here by INP type (total, biological [bio], or bacterial [bac]), with the temperature range as the subscript (e.g., factor 1 for the total INPs active between –5 to –11°C is “total−5 to −11”).
TABLE 4.
Results of exploratory factor analysis for total, biological, and bacterial INPsa
| Temp of activation (°C) | Results for each factor by INP type |
||||||
|---|---|---|---|---|---|---|---|
| Total |
Biological |
Bacterial |
|||||
| Factor 1, total−5 to −11 | Factor 2, total−11 to −14 | Factor 1, bio−5 to −10 | Factor 2, bio−13 to −14 | Factor 3, bio−11 to −12 | Factor 1, bac−5 to −10 | Factor 2, bac−4 and −12 | |
| −4 | 0.69 | ||||||
| −5 | 0.51 | 0.50 | 0.57 | ||||
| −6 | 0.77 | 0.80 | 0.69 | ||||
| −7 | 0.79 | 0.76 | 0.72 | ||||
| −8 | 0.91 | 0.91 | 0.70 | ||||
| −9 | 0.79 | 0.76 | 0.52 | ||||
| −10 | 0.75 | 0.67 | 0.65 | ||||
| −11 | 0.60 | 0.60 | 0.68 | ||||
| −12 | 0.72 | 0.78 | 0.54 | ||||
| −13 | 0.85 | 0.87 | |||||
| −14 | 0.75 | 0.92 | −0.56 | ||||
| −15 | |||||||
| % varianceb | 35.4 | 21.7 | 32.7 | 15.4 | 13.2 | 30.2 | 13.8 |
Factors at each temperature were interpreted by examining the factor loadings of each variable. Variable loadings of ≥|0.50| (i.e., accounts for ≥25% of overlap between variable and factor variance) were retained.
Indicates the proportion of variance each factor accounts for in the INP data.
INP factor concentrations correlate with the physical, chemical, and microbiological data.
The average (± standard error of the mean [SEM]) concentration of DNA-containing cells in the 65 precipitation events was (6.4 ± 9.5) × 105 cells liter−1, and the statistical average and data range for the chemical and physical measurements are provided in Table 3. Cell concentrations correlated positively and significantly with the warm INP concentrations (total−5 to −11, bio−5 to −10, and bac−5 to −10; Table 5). Precipitation pH correlated significantly with total−5 to −11 (Pearson’s r = 0.45) but not with any other factor. While NO3− and SO42− concentrations did not significantly correlate with any factor, bac−5 to −10 correlated negatively and significantly with Na+ concentration (Pearson’s r = −0.42), and Cl− concentration correlated negatively and significantly with total−5 to −11 (Table 5). Dissolved organic carbon (DOC) concentration and precipitation conductivity (Data Set S1) did not correlate with any of the INP factors.
TABLE 5.
Correlations of INP factors with physical and chemical measurements of the precipitation, shown as Pearson correlation coefficients calculated between factors of EFA and measured variables of precipitationa
| INP factor | Cell abundance (DNA-containing cells liter−1 precipitation) | pH | Conductivity (μS cm−1) | Concn (μM) of: |
DOC (ppm) | Fluorescent organic intensity (RU) by componentb: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cl− | Na+ | NO3− | SO42− | C1 | C2 | C3 | |||||
| total−5 to −11 | 0.45* | 0.39* | 0.26 | −0.41* | −0.34 | −0.03 | 0.29 | −0.01 | 0.48* | 0.23 | −0.01 |
| total−11 to −14 | 0.25 | 0.19 | 0.23 | 0.07 | 0.07 | 0.17 | 0.23 | −0.002 | 0.27 | −0.01 | −0.08 |
| bio−5 to −10 | 0.43* | 0.35 | 0.20 | −0.35 | −0.31 | −0.08 | 0.30 | −0.01 | 0.47* | 0.14 | −0.07 |
| bio−13 to −14 | 0.06 | −00.1 | 0.27 | 0.25 | 0.34 | 0.26 | 0.30 | 0.24 | 0.30 | 0.01 | 0.03 |
| bio−11 to −12 | 0.29 | 0.25 | 0.22 | −0.20 | −0.21 | 0.18 | 0.21 | 0.14 | 0.39* | 0.40* | 0.15 |
| bac−5 to −10 | 0.54** | 0.29 | 0.17 | −0.37 | −0.42* | −0.18 | 0.20 | −0.07 | 0.46* | 0.20 | −0.08 |
Significance levels of Pearson correlation coefficients are as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. The number of samples of each were the following: cell abundance, n = 61; pH and conductivity, n = 52; ionic concentrations, n = 31; DOC and C1 to C3, n = 43.
RU, Raman units.
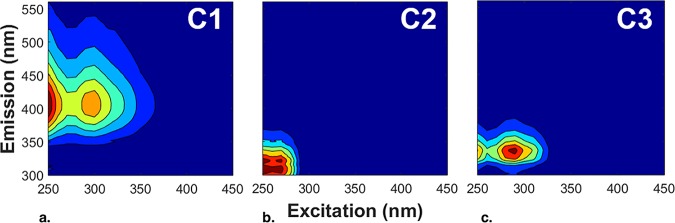
Parallel factor (PARAFAC) analysis revealed three components from the excitation emission matrices of all precipitation samples (Fig. 2a to c). PARAFAC component 1 (C1) correlated significantly (Pearson’s r = 0.39) with all the warmer INP factors (Table 5) and showed maximum fluorescence in two regions, one typically associated with the humic-like dissolved organic matter (DOM) of terrestrial soils and plants (excitation/emission wavelengths, 250 nm/400 to 440 nm) and the other associated with biologically influenced humic-like DOM of marine and freshwater environments. PARAFAC C2, representing tyrosine- and tannin-like fluorescence, correlated with bio−11 to −12, while PARAFAC C3 (associated with tryptophan-like fluorescence) did not correlate with any factors (Table 5). Tyrosine- and tryptophan-like components (of C2 and C3, respectively) represent amino acid-like constituents of the DOC pool that are indicative of DOC produced through microbial processes (84–86, 88). There were also significant correlations of PARAFAC C1 fluorescence with NO3− and SO42− concentrations but not with Cl− or Na+ concentrations.
FIG 2.
Results of fluorescent dissolved organic matter (DOM) excitation-emission matrices data investigated using parallel factor (PARAFAC) analysis. (a to c) The three fluorescent DOM PARAFAC components (component 1 [more complex, humic-like materials] [a], component 2 [tyrosine- and tannin-like materials] [b], and component 3 [tryptophan-like materials] [c]) identified in 43 Louisiana precipitation events from 2013 to 2015 are consistent with DOM from terrestrial and aquatic environments.
INP factors correlate with season, cloud type, and air mass history.
Multivariate analysis of variance (MANOVA) was conducted to investigate if INP concentrations varied based on the history of air mass interactions with the mixed boundary layer (MBL). The results (Table S2 and Fig. 3) indicated that total, biological, and bacterial INP concentrations were significantly different based on air mass origin and interactions with the MBL (P < 0.05). Air mass trajectory interactions with the East Asia ecoregions (Fig. 4) produced the highest concentrations of INPs for all factors (Fig. 3). While most of the East Asia trajectories interacted with the temperate and taiga regions of Asia, one event (on 5 March 2015) did interact with the arid deserts and xeric shrublands of China and Mongolia. Tukey’s honest significant difference post hoc analysis indicated that for the total−5 to −11, bio−5 to −10, and bac−5 to −10 factors, the high northern latitudes ecoregion had INP concentrations statistically similar to those for the East Asia ecoregion (Table S2). The eastern woodlands and wetlands ecoregion and deserts and semiarid highlands ecoregion produced the lowest concentrations of total−11 to −14, bio−11 to −12, and bac−5 to −10 (Fig. 3). In general, the Pacific, North Atlantic, and South Atlantic maritime ecoregions were inferred to be minor sources of all INP classes. Trajectory analysis indicated that none of the air masses originating from the tropical forest ecoregion during this study interacted with the MBL or surface.
FIG 3.
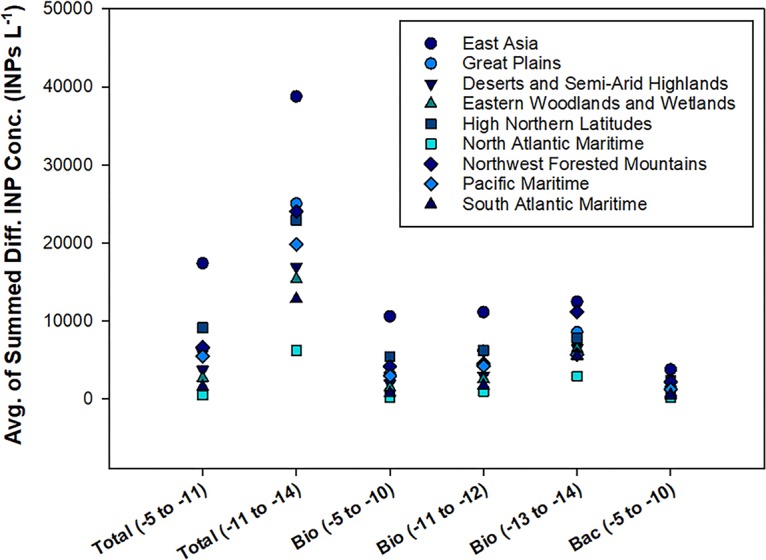
Average INP factor concentrations as a function of ecoregion classification. Multivariate analysis of variance indicated that average INP concentrations differed based on the interactions of air masses with the various ecoregions. This plot is showing the average of summed differential INP concentrations (y axis) for each INP factor for each ecoregion. For example, bac (–5 to −10) is the average value of the summed differential INP concentrations at −5, –6, –7, –8, –9, and –10°C for each ecoregion.
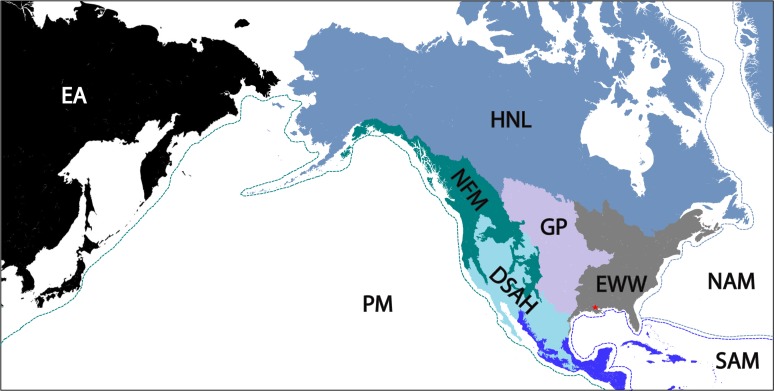
FIG 4.
Location and extent of the source ecoregions relevant in this study. These ecoregions of North America were defined by the EPA, USGS, and CEC, as described in the text. The source ecoregion was used to classify interaction patterns between backwards air mass trajectories and the mixed boundary layer for each precipitation event (Fig. S1). Location of the primary sampling site (Baton Rouge, LA) is indicated by the red star. EA, eastern Asia; PM, Pacific maritime; NFM, northwest forested mountains; HNL, high northern latitudes; DSAH, desert and semiarid highlands; GP, Great Plains; EWW, eastern woodlands and wetlands; NAM, North Atlantic maritime; SAM, South Atlantic maritime.
To validate trends unmasked from the INP and air mass data analysis, DOM PARAFAC component intensities were analyzed with backward air mass trajectories (Table S3). Overall, the fluorescent DOM components showed differences between air mass trajectories that interacted with central continental environments (i.e., the Great Plains, eastern woodlands and wetlands, and deserts and semiarid highlands), northern and more distant continental environments (East Asia, the high northern latitudes, and the northwest forested mountains), and marine environments (Pacific maritime and South Atlantic maritime) (Fig. S2). Since none of the storm air mass histories analyzed originated from North Atlantic maritime environments, no PARAFAC data are available from this ecoregion. Significant correlations were identified for DOM PARAFAC components and select major ion concentrations, as follows: C1 to NO3−, r = 0.46 and ρ = 0.61, and SO42−, r = 0.58 and ρ = 0.66; C2 to NO3−, r = 0.36 and ρ = 0.42, and SO42−, r = 0.30 and ρ = 0.54; and C3 to NO3−, r = 0.33. None of the components correlated with Cl− or Na+ concentrations.
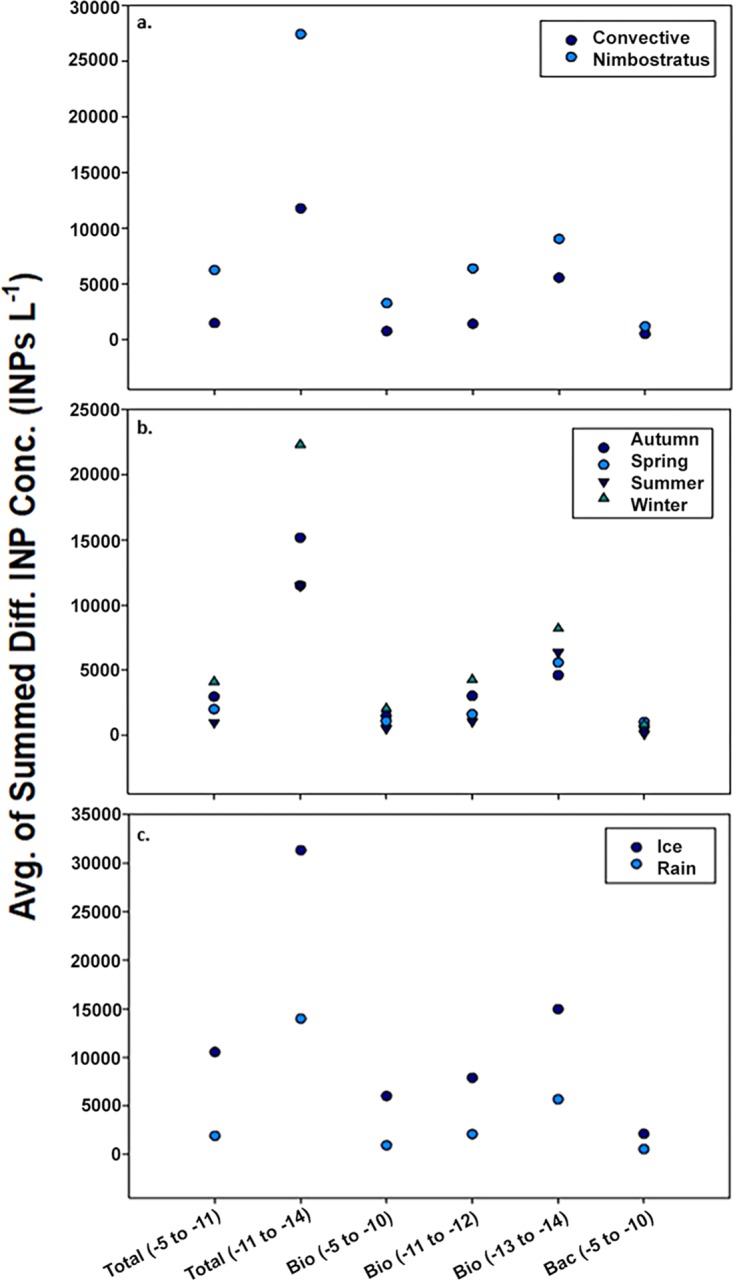
The concentration of all INP types correlated with season (MANOVA, P < 0.001; Table S4), and precipitation in winter contained the highest concentrations of all INPs (Fig. 5b). Highly significant differences in INP concentrations were associated with the type of mesoscale cloud system (i.e., stratiform versus convective) responsible for the precipitation (P < 0.05; Table S4), with stratiform systems containing the highest concentrations for all INP factors (Fig. 5a). A subset of the stratiform events with ice-containing precipitation (sleet or snow, n = 4) had the highest concentrations of all INP classes (Fig. 5c), and a MANOVA showed these differences were significant from those observed in rain samples (P < 0.05; Table S4). Several of the locally measured meteorological variables, including surface temperature and surface wind speed, correlated well (Pearson’s r > 0.30) with INP concentrations (Table S5).
FIG 5.
Average INP factor concentrations as a function of cloud type, season, and precipitation phase. (a to c) Multivariate analysis of variance indicated that average INP concentrations (Conc.) differed based on the cloud type (a), season (b), and precipitation type (c). The average of summed differential (Diff.) INP concentrations were determined as described in the Fig. 3 legend.
Correlations between abundances of bacterial operational taxonomic units and INPs.
Of the 65 samples collected for DNA extraction, 20 of the samples were excluded from further analysis because amplification of the 16S rRNA gene failed or they were associated with trials in which spurious amplicons were generated in the procedural controls. The composition of bacterial assemblages in 45 of the precipitation events was assessed by comparing the amplified 16S rRNA genes from the samples, and a total of 60,289 operational taxonomic units (OTUs) were identified. This analysis revealed that 1,425 OTUs had statistically significant positive correlations to the INP factors. Of these 1,425 OTUs, only those which had reads accounting for >0.1% of the total number of reads across all samples were retained for further analysis (Table S6). Spearman’s ρ correlations indicated that OTUs classified within the Bacteroidetes had the most significant correlations with all INP factors, especially those within the Cytophagales order. Several of the Cytophagales taxa (unidentified genus from Cytophagaceae, Hymenobacter, and Flexibacter) showed the highest and most significant correlations with total−5 to −11, bio−5 to −10, and bac−5 to −10 INPs (Table S7). Additionally, the family Rikenellaceae from the order Bacteroidales, the family env.OPS_17 from the order Sphingobacteriales, and the genus Segetibacter from the order Chitinophagales contained highly significant correlations with all INP classes except for bio−11 to −12.
Firmicutes and unclassified divisions also had a relatively large number of significant positive correlations with the INP factors (Table S6). Within the Firmicutes, OTUs significantly correlating with all warm classes of INPs (total−5 to −11, bio−5 to −10, and bac−5 to −10) were limited to the Carnobacteriaceae. A broader phylogenetic range of OTUs positively correlated with the colder INP factors (total−11 to −14, bio−11 to −12, and bio−13 to −14), and most of these taxa affiliated with the Bacteroidetes, Firmicutes, and Proteobacteria, but there were also representatives from the phyla Cyanobacteria, Planctomycetes, Spirochaetes, and Verrucomicrobia (Table S6). Overall, a smaller number of OTUs correlated with the bacterial INP factors, and those that did belonged to the Bacteroidetes, Proteobacteria, or unclassified divisions. Many of the OTUs (e.g., Hymenobacter, Sphingobacteriales, Cytophagaceae, and Flexibacter) correlating with the warmer INP factors (total−5 to −11, bio−5 to −10, and bac−5 to −10) had significantly higher abundances in precipitation from nimbostratus clouds (n = 16) and during the winter (n = 16; Fig. S3 and Table S7).
DISCUSSION
During this 2-year study, observations from a range of storm origins and types provided the opportunity to examine linkages between biological INPs in precipitation and geographic source, season, cloud lifetime and precipitation development, and the meteorologically influenced dispersal of microorganisms in the atmosphere. The key results from this analysis can be summarized as follows: (i) the sampled precipitation contained several “classes” of INPs with distinct behaviors; the highest concentrations of INPs likely originated from (ii) terrestrial regions geographically distant from Louisiana and (iii) nimbostratus clouds, ice-phase precipitation, and winter storms; (iv) certain bacterial taxa correlated significantly with the INP concentrations; and (v) the concentrations of INPs observed in the precipitation implied sufficient abundances in the atmosphere to affect precipitation production in certain cloud types. The significance and implications for each of these findings are discussed in greater detail below.
(i) INP “classes” identified in Louisiana precipitation. The EFA grouped differential INP concentrations by temperature of activation (Table 4), implying that the sampled precipitation contained distinct types or classes of INPs. Since the total−5 to −11, bio−5 to −10, and bac−5 to −10 factors were highly correlated to each other (r = 0.83 to 0.96, ρ = 0.83 to 0.94; Table S9), they are likely capturing similar aspects of the warm temperature IN activities. Based on the properties of known IN species and biological INPs (4, 11, 87), the total−5 to −11, bio−5 to −10, and bac−5 to −10 factors may represent activities conveyed by intact bacterial cells that harbor IN proteins or heat-sensitive compounds. Indeed, cell concentration correlated significantly with the warm INP factors (total−5 to −11, bio−5 to −10, and bac−5 to −10; Table 5). All of the warm INP factors also correlated significantly with PARAFAC component C1 (Table 5), which is known to be associated with terrestrially derived DOM (84–86, 89). Taken together, these results suggest that the warmest temperature INPs in the precipitation were largely bacterial in origin, conveyed by proteinaceous IN activity, and aerosolized from terrestrial environments. That no correlation existed for bio−11 to −12 or bio−13 to −14 with pH is interesting considering that proteinaceous IN activity in certain fungi is heat sensitive but unaffected by low pH (21). As such, the “colder” classes of biological INPs identified could correspond to fungi possessing IN proteins (22) or DOM with IN activity (28, 29).
It is important to note that our method for classifying INPs provides a conservative estimate of those from biological and bacterial sources (7). For example, the nonproteinaceous IN material of some pollens and bacteria is resistant to heating at 100°C (10, 20, 90) and therefore would not be identified as “biological” by our method. Likewise, the use of lysozyme sensitivity to diagnose bacterial INPs should underestimate their concentration because disparate cell wall compositions allow some bacteria to resist its hydrolytic activity. Nevertheless, the significant correlation (Pearson’s r = 0.54) between cell abundance and lysozyme-sensitive INPs (Table 5) implies that any bias introduced by this approach was relatively uniform across samples.
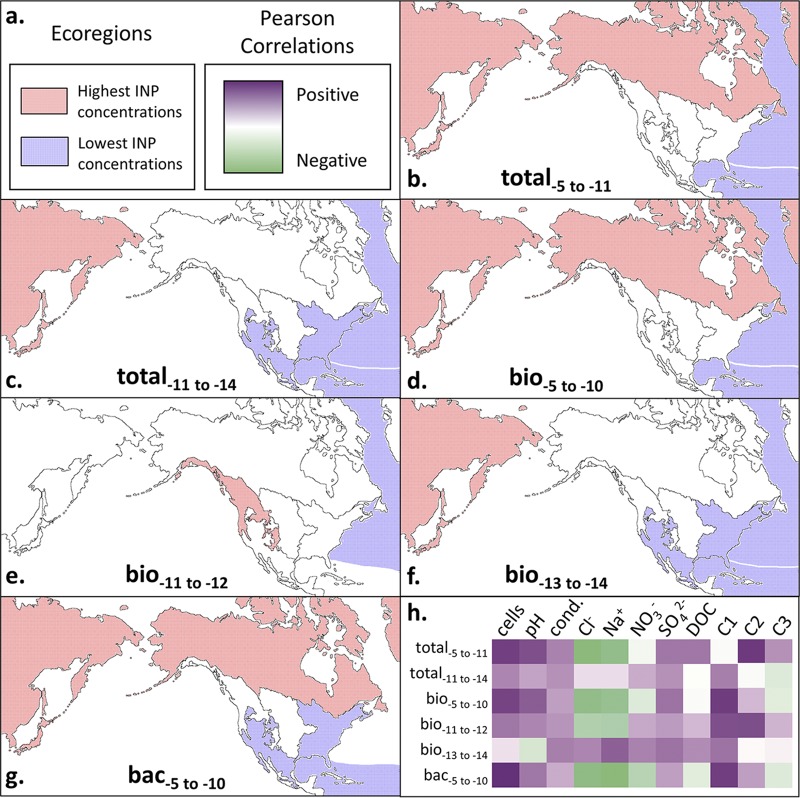
(ii) Potential geographic origins of INP classes in Louisiana precipitation. Previous studies have detected high abundances of biological INPs in plant and soil environments (26, 91) and the emission of IN strains of P. syringae and Erwinia herbicola from agricultural crops (92, 93). Our analysis revealed that air masses interacting with the MBL in midlatitude continental regions replete with agricultural, woodland, and herbaceous ecosystems (eastern woodland and wetlands and the Great Plains ecoregions; Fig. 4) did not subsequently produce precipitation with the highest concentrations of total−5 to −11, bio−5 to −10, and bac−5 to −10 INPs observed (Fig. 6b, d, and g). In fact, air masses from the eastern woodland and wetlands ecoregion produced the lowest INP concentrations in the precipitation from all nonmaritime locations (Fig. 6c, f, and g). East Asia, the northwest forested mountains, and/or the high northern latitudes ecoregions were the largest sources of all INP classes (Fig. 6). Although few data are available, a recent study implicates high northern latitude aerosols in North America as sources of biological INPs (94). The identification of soil-dwelling taxa (for example, Hymenobacter spp. are ubiquitous in soils, especially desert soils [95–102]) that correlate well with the warm INP factors also suggests that terrestrial soils were the most likely sources of the warm-temperature INPs.
FIG 6.
Summary of trends in ice-nucleating particle (INP) data with source ecoregion and physical, chemical, and microbiological properties of the precipitation. Ecoregions as depicted in Fig. 4 are outlined by black solid lines. (a) Key for INP concentrations in the ecoregion for panels b to g and Pearson correlation coefficents for panel h are shown. (b to g) Ecoregions that correlated significantly with total−5 to −11 (b), total−11 to −14 (c), bio−5 to −10 (d), bio−11 to −12 (e), bio−13 to −14 (f), and bac−5 to −10 (g) INP concentrations are shown. (h) Heat plot of Pearson correlation coefficients between INP concentrations and various precipitation measurements, as follows (units): cell abundance (cells liter−1 precipitation); conductivity (cond.; μS cm−1); chloride, sodium, nitrate, and sulfate (μM); DOC, dissolved organic carbon (parts per million); C1, PARAFAC component C1 (Raman units [R.U.]); C2, PARAFAC component C2 (A.U.); and C3, PARAFAC component C3 (A.U.).
Nearly all the INP factors correlated significantly (P < 0.05) with DOM PARAFAC C1 (Table 5 and Fig. 6h). The region of maximum fluorescence in C1 (excitation [Ex]/emission [Em] maxima, 250 nm/410 nm) is known to represent constituents of terrestrially derived DOM from plants, soils, estuaries, wastewater, and agricultural catchments (84–86, 89). Fluorescence at higher Ex/Em wavelength maxima of C1 (300 nm/410 nm) exhibited intensities similar to the “M peak” described by Coble et al. (84) in shallow eutrophic marine waters from the Gulfs of Mexico and Maine, transitional waters of the Puget Sound (103), and freshwater environments (89, 104, 105). DOM associated with C1 typically consists of higher-molecular-weight aromatic chemical species, characteristic of material that has been highly processed by biological and geochemical activities (89, 104, 105). However, that the C1 component did not correlate with the Cl− concentration in precipitation suggests that its source was not marine. This contention is further supported by the significant positive correlation of C1 with NO3− (r = 0.46 and ρ = 0.61), which is an indicator of a terrestrial aerosol source (64). Together, these results support the contention that continental environments were the main source of warm INPs in the precipitation analyzed.
Aircraft-based measurements in air masses above the Sierra Nevadas have shown high concentrations of biological INPs in clouds and precipitation containing desert dust from Asia (106). Additionally, studies have also shown that the intercontinental transport of microorganisms as aerosols is possible (106–109). This together with the observation of terrestrial DOM (i.e., PARAFAC component C1) in air masses originating from Asia implies an extracontinental source for the INPs in these precipitation samples (Fig. 6). The same can be said for the high northern latitudes ecoregion; however, we are not aware of prior studies that have examined geographic regions in the tundra or northern forests of North America as atmospheric sources of biological INPs.
The bio−11 to −12 factor is the only class of INPs that has a significant correlation (r = 0.40) with the tannin- and tyrosine-like DOM chemical species associated with PARAFAC C2. Previous studies have identified C2 in microbially produced DOM from terrestrial (89, 110) and freshwater (89) ecosystems. Additionally, the bio−11 to −12 factor is the only one that did not have the highest concentrations when air masses interacted with the surface in eastern Asia or the high northern latitudes. Instead, its highest concentrations were observed in storms that originated from the forest mountains ecoregion (Fig. 6e).
(iii) INP concentration as a function of season and meteorology. Winter storms contained the highest concentrations of nearly all the INP factors (Fig. 5b and Data Set S1), supporting the hypothesis that seasonal changes in meteorological patterns and/or the source ecosystems of bioaerosols influenced the abundance of INPs in the precipitation. A considerable fraction of OTUs that correlate with the INP data had higher abundances in winter precipitation (Fig. S3 and Table S7) and are related to psychrotolerant or psychrophilic species, suggesting a linkage between cold adaptation, lower winter temperatures, and IN activity. This deduction is also supported by surface temperature being inversely correlated with the warmest INP classes (total−5 to −11, bio−5 to −10, and bac−5 to −10; Table S5). An alternative explanation for this finding is the possible inactivation of biological INPs while descending through the atmosphere or in the collection cans at the surface during summer. If this was the case, the actual number and activity of INPs at cloud heights may be underestimated based on inferences from summer rain samples. However, negative correlations between INP concentration and the sample collection time (amount of time precipitation samples sat in collection cans before being processes) during the summer months were not observed, suggesting that the data were not affected by this potential sampling artifact. In fact, the only significant correlation to collection time observed was positive and associated with the bio−13 to−14 data (r = 0.648, P = 0.005). Some IN active pollens release their ice-nucleating macromolecules into solution when immersed in water (10), providing one possible explanation for this curious result.
A subset of the wintertime nimbostratus events contained all the extreme outlying INP data (Fig. 1), had the highest concentrations for all factors, had significantly higher concentrations of INPs than winter convective storms (Fig. 5b), and were significantly enriched for OTUs that correlated with the INP data (Fig. S3). There were only four observations of ice-containing precipitation (snow or sleet) during this study, and these samples had the highest INP concentrations and activity observed (Fig. 5c). While it is tempting to suggest this result as evidence for the involvement of biological INPs in ice-phase precipitation, it may also be explained by differences in below-cloud scavenging efficiencies between snowflakes and raindrops. However, previous work documented distinct microbial assemblages in snow versus the air mass from which it precipitated (32), suggesting that snowflakes may be relatively inefficient at scrubbing bacterium-sized aerosols from the atmosphere. Although there are a number of uncertainties regarding aerosol scavenging in snow versus rain (111–113), cell concentrations in the snow and sleet were not significantly different from those observed in rain samples, implying similar scavenging processes regardless of precipitation phase.
(iv) Potential phylogenetic and geographic sources of ice-nucleating bacteria. The 16S rRNA gene sequences from 45 of the 65 precipitation events were successfully amplified, sequenced, and analyzed (69). Of the OTUs that correlate significantly with INP concentrations (Table S6), many are related to bacterial taxa documented in soils, plant ecosystems, air, and precipitation (69, 114, 115). Although there were no significant trends between INPs and the known ice-nucleating genera Pseudomonas or Pantoea, there are significant correlations between OTUs from Xanthomonadaceae (i.e., containing the IN species Xanthomonas campestris) and the total−5 to −11, total−11 to −14, and bio−5 to −10 factors. Taxa in the phylum Bacteroidetes had the highest correlations with INP concentrations and were abundant in the samples (Table S6), including OTUs from the Sphingobacteriales and Cytophagales. Several genera from these orders have been reported in desert soils (Hymenobacter [100, 116, 117]), snow and hail (Hymenobacter and genera from Sphingobacteriales [118, 119]), and air samples collected over the northwest forested mountain/desert and semiarid highlands ecoregions and Asia (Flexibacter, Segetibacter, Hymenobacter, and genera from Sphingobacteriales [37, 118, 120–122]). The strong correlations between Bacteroidetes OTUs and the warm INP factors (total−5 to −11, bio−5 to −10, and bac−5 to −10) are noteworthy because ice-nucleating activity has not been documented within this phylum. In particular, the Hymenobacter and Segetibacter taxa were strongly correlated with the total−5 to −11, total−11 to −14, bio−5 to −10, and bac−5 to −10 INP data, and members of these genera have also been reported in air (36, 37) and precipitation containing Saharan and Asian dust (118). In fact, Meola et al. (118) showed that similar Cytophagaceae phylotypes in European snow sequences coincided exclusively with the presence of Saharan dust. Several of the OTUs strongly correlated with the INP data (Table S6) classified within groups of human- and animal-associated bacteria (e.g., Rikenellaceae, Blautia spp., and Roseburia spp. [55, 123, 124]), indicating an anthropogenic and/or agricultural influence on the microbial composition of the precipitation analyzed. There were also strong correlations between the INP data and numerous unclassified taxa (Table S6), supporting the possibility for the presence of ice-nucleating activity in these poorly characterized bacterial lineages. Considering that correlations may be illusory, exploration of species in these genera requires further investigation to determine if they possess species with the IN phenotype.
The taxonomic data and chemical composition of the precipitation also provide information to aid in assessing the potential ecological sources of the INPs sampled. For instance, the warmer classes of INPs (total−5 to −11, bio−5 to −10, and bac−5 to −10 factors) were positively correlated with taxa associated with desert and arctic soils (15, 96, 100, 116–118) and DOM typically associated with terrestrial plant, soil, and freshwater ecosystems (89, 105) but were negatively correlated with the Cl− concentration. This implies that the most efficient INPs may have been sourced from desert and high northern latitude continental environments as opposed to marine habitats (Fig. 6), which is supported by hybrid single-particle Lagrangian integrated trajectory (HYSPLIT) back-trajectory analysis that showed air masses interacting with the East Asia and high northern latitude continental surfaces contained the highest INP concentrations (Fig. 6b to g and Table S2). In contrast, OTUs related to known marine taxa (e.g., Oceanospirillaceae) correlated with colder classes of INPs (total−11 to −14, bio−11 to −12, and bio−13 to −14), suggesting marine environments as their source.
(v) Potential implications for biological INPs on meteorological processes. While ice-containing clouds produce the majority of precipitation on a global scale (56), there are large uncertainties regarding ice formation in mixed-phase clouds, the abundances of biological INPs at cloud altitudes, and their involvement in precipitation generation (5). To investigate the possibility that biological INPs have sufficient abundances to affect ice formation in cloud water droplets, we used the precipitation data to estimate their in-cloud abundances. Below-cloud scavenging complicates efforts to directly relate precipitation to cloud water composition, and therefore, we estimated boundary conditions following the approach of Petters and Wright (57).
Modeled simulations of summer and springtime convective clouds over North America imply that as few as 1 INP m−3 (of air) active at –10°C could produce the observed rates of precipitation (48, 58). At temperatures of ≥–10°C, the Louisiana precipitation samples averaged cumulative concentrations of 1,300 total INPs liter−1, 1,100 biological INPs liter−1, and 600 bacterial INPs liter−1 of precipitation (Fig. 1). Based on these data, we estimated the relative abundance of INPs within a cubic meter of developing cumulonimbus cloud (i.e., convective cloud) and stratocumulus cloud (i.e., stratiform). Over continental regions, cumulonimbus clouds typically contain cloud droplet sizes in the range of 12 to 16 μm at concentrations of approximately 500 cm−3 (59, 60). Assuming a median cumulonimbus cloud droplet size of 14 μm (44), the average cumulative concentration of INPs active at –10°C in precipitation would represent approximately 0.5 to 1 INP m−3 of air, which is similar to modeled values affecting precipitation rates (1 INP m−3 [48]). When considering the highest observed INP abundances (16,100 total, 15,900 biological, and 5,900 bacterial INPs liter−1 of precipitation, active at ≥–10°C; Fig. 1), concentrations as high as 5 to 10 INPs m−3 of cumulonimbus cloud are inferred.
The mechanisms responsible for primary and secondary ice formation in nimbostratus clouds differ substantially from those of convective clouds (43, 46, 50, 51). The Wegener-Bergeron-Findeisen (WBF) process is thought to be the dominant process of ice formation in mixed-phase clouds with updraft speeds of <2 m s−1 (convective clouds typically contain updraft speeds of >2 m s−1), which is consistent with nimbostratus cloud conditions (41, 61, 62). Hence, it is possible that the WBF process allows efficient removal of warm-temperature INPs from a nimbostratus cloud via ice formation and subsequent precipitation initiation. Although INPs active at temperatures warmer than –9°C are also important for the Hallett-Mossop mechanism, this process requires updraft speeds of >3 m s−1 for riming to work effectively and may only be relevant to convective clouds (46). Stratus clouds that form over continents typically contain fewer (∼250 cm−3) and smaller cloud droplets than do convective clouds, with average diameters of ∼10 μm (60). Based on these meteorological parameters, we infer concentrations in stratus clouds of 0.1 to 0.2 INP m−3 based on the average biological INP concentrations and 0.8 to 2 INP m−3 using the highest INP concentrations observed. To our knowledge, INP concentration thresholds for precipitation formation in stratus or stratus-like clouds have not been constrained within an aerosol-cloud modeling framework. As our data show that biological INPs were at their highest concentrations in precipitation from stratus-like clouds, and ice-phase stratiform precipitation in particular (Fig. 5), such modeling efforts represent fertile territory for further study.
It is important to note that there are a number of uncertainties inherent to our estimates of cloud INP concentrations based on data from precipitation, including the actual cloud droplet sizes, the cloud thermodynamic properties, and the contribution of below-cloud scavenging. With respect to below-cloud scavenging, aerosols the size of most bacteria (0.5 to 1.5 μm) are typically referred to as the “scavenging gap” in aerosol size distributions because particles of this size are not efficiently scrubbed by raindrops regardless of the rainfall rate (63). Previous studies have shown similar concentrations of INPs in cloud water and in precipitation at ground level (54, 57), suggesting that below-cloud scrubbing may have a negligible role in altering the composition of bacterium-sized INPs in raindrops.
Conclusion.
Despite the fact that biological INPs have been studied for nearly 50 years, little is known about the microbiological and ecological sources of these particles to the atmosphere. In this study, we analyzed precipitation from 65 storms originating from air masses that had traversed regions of Asia, North America, the Pacific Ocean, the Atlantic Ocean, and the Gulf of Mexico (Fig. 4), providing new observations to assess the geographic sources of INPs deposited with precipitation in the southeastern United States. Various studies have detected biological INPs in marine waters and concluded that the ocean is a major atmospheric source of these bioaerosols (23, 30, 31, 91). However, we observed that prestorm air masses that interacted with marine environments generated precipitation with the lowest concentrations of INPs observed (Fig. 6). In contrast, the concentrations of all INP classes were highest when air masses had interacted with continental ecoregions. The continental origin of these bioaerosols was supported by positive correlation with the abundance of DOM and bacterial taxa typically associated with terrestrial ecosystems and negative correlation to marine aerosol proxies (Fig. 6h). These results lead to the conclusion that forest and soil ecosystems are important year-round atmospheric sources of biological INPs to precipitation in this subtropical region. Unexpectedly, our analysis implicated the high northern latitudes and eastern Asia ecoregions as sources that produced the highest concentrations of INPs in Louisiana precipitation (Fig. 6), corroborating similar observations in the western United States (106).
Multiple studies have confirmed the ubiquity of biological INPs in the atmosphere and precipitation (5, 7, 39), but the meteorological conditions under which they may have roles in affecting cloud ice formation and precipitation have remained speculative. We observed the highest concentrations of total, biological, and bacterial INPs in precipitation associated with winter storms composed of nimbostratus-like cloud formations that originated from the western United States (Fig. 6). Since nimbostratus clouds are low-level, relatively warm clouds (67), biological INPs would be in a favorable position to affect precipitation processes if sufficiently abundant. Indeed, our estimates from precipitation samples imply that INPs originating from warm clouds (tops warmer than –15°C) were at concentrations predicted to influence ice formation and precipitation. The role of biological INPs in meteorological processes may be especially relevant to stratus-like clouds where updrafts are minimal and the WBF process is significant for precipitation production. Considering that low temperature and nutrient limitation affect the IN activity of P. syringae (73), metabolically active microorganisms are present in cloud droplets (74), and liquid water can persist for days in nimbostratus clouds at temperatures conducive to metabolism (10°C to –25°C [67, 75]), conditions within a nimbostratus cloud might even promote in situ expression of the ice nucleation phenotype.
Bacterial ice nucleation has been thought to be limited to certain species of Gammaproteobacteria (i.e., P. syringae, Pseudomonas viridiflava, Pseudomonas fluorescens, Pantoea agglomerans, and Xanthomonas campestris) but recently was expanded to include a member of the phylum Firmicutes (20). This raises the possibility that the ice nucleation phenotype may be distributed in a broader phylogenetic range of bacteria than previously appreciated. We identified a number of phyla (Acidobacteria, Bacteriodetes, Chlorobi, Cyanobacteria, Firmicutes, Planctomycetes, Proteobacteria, Spirochaetes, and Verrucomicrobia) that may contain unrecognized lineages of ice-nucleating bacteria, providing a motivation for future studies to explore the presence of this phenotype in these taxa. Improved understanding of the diversity of ice-nucleating bacteria and their atmospheric sources, transport, and role in hydrologic cycling would lend valuable insight into their capacity to disperse aerially and participate in landscape-atmospheric feedbacks that have meteorological consequences (76).
MATERIALS AND METHODS
Precipitation sampling.
With the exception of one sleet storm sampled in Alexandria, LA (31.3113°N, 92.4451°W) during March 2015, 64 precipitation events (61 rain, 1 sleet, and 2 snow events) were sampled between May 2013 and July 2015 from the roof of a six-floor building (∼20 m above ground level) on the Louisiana State University campus in Baton Rouge, LA (30.4145°N, 91.1783°W). The precipitation was sampled at ambient temperatures by direct collection in 10 120-liter galvanized cans that were lined with clean, sterile 94 by 122-cm polypropylene bags (Fisher Scientific, Pittsburgh, PA). A total of 11 cans (10 for samples and one as a procedural control) were used to collect samples for each precipitation event. The procedural control remained sealed for the duration of the precipitation event, and then 3 liters of sterile deionized water that was poured into the can was collected in a manner identical to the samples. A minimum of 3 liters of rain or snow water equivalent was amalgamated from three collection cans for each sample. Precipitation from Alexandria was collected using seven cans (six for samples and one as a procedural control). For 14 of the low-accumulation events, the precipitation collected in all 10 cans was pooled for DNA extraction. Immediately following each precipitation event, the material collected was transferred to sterile 9-liter carboys and stored at 4°C in the dark until processed. Processing typically occurred within ∼1 h, but in certain cases, the samples were processed up to 48 h after collection.
Precipitation samples for the measurement of dissolved organic carbon (DOC), fluorescent dissolved organic matter (DOM), and major ions (NO3−, SO42−, Na+, and Cl−) were collected separately from those used for microbiological analysis in a borosilicate glass funnel and bottle that was thoroughly cleaned by washing with detergent, soaked in 10% HCl for 30 min, rinsed with ultrapure deionized water (18.2 MΩ), and subjected to combustion at 400°C for 4 h. Following collection, the samples were filtered through precombusted GF/F filters (Whatman, Inc.), stored frozen in the dark at –20°C, and analyzed within 6 months.
Quantification of INPs and cells.
Immersion freezing assays were performed as described previously (7) but with the following modifications: no filter concentration was performed, and 200-μl aliquots (rain or snow water equivalent) of the precipitation sample were placed into each well of a 96-well plate and sealed with adhesive film. Triplicates of each sample and experimental treatment were tested over a temperature range of −4 to –15°C in 0.5°C increments using a Neslab RTE 7 series refrigerated ethylene glycol bath (Thermo Scientific, Waltham, MA). The number of wells frozen at each temperature was recorded, and the differential concentrations (the number of INPs activated at a specific temperature) and cumulative concentrations (the number of INPs activated at all temperatures warmer than a given temperature) of INPs were calculated by the method of Vali (53).
Each precipitation sample was tested by analyzing triplicate preparations that were either untreated, heated for 10 min at 95°C, or incubated with 3 mg ml−1 lysozyme for 1 h prior to the immersion freezing assay. The data and related calculations were used to assess the total (i.e., untreated), biological (i.e., heat-sensitive), and bacterial (i.e., lysozyme-sensitive) INP contents of each sample. Given that only proteinaceous INPs may be sensitive to heat denaturation and that not all bacteria are sensitive to lysozyme, this method should be viewed as a conservative estimate for INPs of biological or bacterial origin.
DNA-containing cells were stained with SYBR gold (Invitrogen, Carlsbad, CA) and counted using epifluorescence microscopy (BX51-TRF; Olympus, Center Valley, PA), according to the method of Christner et al. (68). Triplicate measurements from each precipitation event were analyzed.
Amplification and sequencing of 16S rRNA genes.
Initial testing showed that reliable DNA amplification typically required a minimum of 3 liters of precipitation (data not shown). Therefore, at least 3 liters of precipitation or sterile deionized water (for the procedural controls) was collected, filtered onto sterile 0.2-μm-pore-size 47-mm Supor polyethersulfone (PES) membrane filters (Pall Corp., Port Washington, NY), and stored at –80°C until processed. DNA was extracted from the filters and the procedural controls using the FastDNA Spin kit for soil (MP Biomedicals, Santa Ana, CA) and PowerSoil kit (Mo Bio Laboratories, Carlsbad, CA), with several modifications to the manufacturers’ protocols (Methods section of the supplemental material). The V4 region of the 16S rRNA gene was successfully PCR amplified using DNA extracted from 45 of the 65 precipitation events, and the amplicons were sequenced on a MiSeq platform (Illumina, Inc., San Diego, CA, USA). Contamination was assessed by attempting PCR amplification with the V4 primers on extracts from the procedural controls, as described by Aho et al. (69). The sequences obtained were analyzed using the mothur software (70) and aligned to a SILVA bacterial 16S rRNA gene reference alignment (71). Sequence library size was normalized by randomly sampling 49,263 sequences from each library, which was the number of sequences in the smallest library. Sequences were clustered into operational taxonomic units (OTUs) based on a sequence dissimilarity of ≤0.03. OTUs were classified using the Ribosomal Database Project Classifier implemented in the mothur software and assigned to a particular taxon if they classified with ≥80% confidence.
Inorganic and organic chemistry.
The conductivity and pH of the precipitation samples were measured using a multiparameter PCSTest probe (Oakton Instruments, Vernon Hills, IL). The concentrations of major ions and DOC were determined on aqueous samples that were filtered through precombusted 25-mm 0.7-μm-pore-size GF/F Whatman filters (GE Healthcare, Chicago, IL). Major ions were analyzed using a Dionex ICS-3000 ion chromatography system (Methods section of the supplemental material). DOC concentrations were measured with a GE Sievers 900 total organic carbon analyzer (Methods section of the supplemental material). Excitation emission matrices of the fluorescent dissolved organic matter (DOM) were generated on a Horiba Jobin Yvon Fluoromax-4 spectrofluorometer (Methods section of the supplemental material). The excitation emission matrices (EEMs) of the fluorescent DOM were modeled with the drEEM multivariate parallel factor analysis (PARAFAC) toolbox in Matlab, according to Murphy et al. (72).
Analysis of meteorological data and ecoregions.
Each storm was classified as a “stratus” or “convective” event, based on cloud structure. Cloud structure, height, and depth were estimated based on radar reflectivity, infrared satellite imagery, and radiosonde data (77, 78) (Methods section of the supplemental material). Backward trajectories (120 to 168 h) of air masses over the site at the time of each precipitation event were determined using the National Oceanic and Atmospheric Administration (NOAA) Air Resources hybrid single-particle Lagrangian integrated trajectory (HYSPLIT) model, accessed via the NOAA Air Resources Laboratory (ARL) Real-time Environmental Applications and Display sYstem (READY) website (https://www.ready.noaa.gov/HYSPLIT.php). For each precipitation event, six altitudes were chosen for backward-trajectory analysis based on the lifting mechanism responsible for cloud formation, as well as the level of cloud base and height (Fig. S1). Cloud base and height were determined using lifted condensation levels (LCL) and/or convective condensation levels (CCL), depending on whether the cloud was formed by mechanical or convective lifting mechanisms, respectively (Methods section of the supplemental material and Fig. S1a to d). Geographic locations where backward trajectories interacted with the Earth’s surface or the mixed boundary layer (MBL; i.e., the lowest layer of the troposphere where turbulent mixing with the Earth’s surface occurs) were graphed in R by using data from the “tdump.csv” hourly data files produced by HYSPLIT for each backward trajectory (Fig. S1e and f).
Trajectory interactions with the surface or MBL were categorized based on North American level 1 ecoregions (79, 80). Briefly, an ecoregion is defined by the Environmental Protection Agency and Commission for Environmental Cooperation as an ecological region that contains similar plant and animal communities, climate, geology, topography, hydrology, soil, land use, and natural resources (80). The most recent classification system defined 15 broad categories for level 1 ecoregions. To increase homoscedasticity and the number of degrees of freedom for downstream statistical analyses, several regions of similar ecology, geography, and climate were combined. For this analysis, the following level I ecoregions were combined: the Arctic Cordillera, Tundra, Taiga, Hudson Plain, and northern forests were combined and designated “high northern latitudes”; the northwestern forested mountains and Marine West Coast mountains were combined and designated “northwest forested mountains”; the North American deserts, Mediterranean California, southern semiarid highlands, and temperate Sierras were combined and designated “deserts and semiarid highlands”; and the “tropical dry forests” and “tropical wet forests” were combined and designated “tropical forests” (Fig. 4). Backward trajectories that interacted with the MBL in Asia occurred at latitudes above approximately 35°N and longitudes higher than 90°E and were therefore classified into a single ecoregion called “East Asia” (Fig. 4). The Great Plains and eastern U.S. ecoregions were not combined with other ecoregions and are referred to as “the Great Plains” and “eastern woodlands and wetlands,” respectively (Fig. 4). Marine regions were divided into Pacific Maritime, North Atlantic maritime, and South Atlantic maritime (Fig. 4).
Statistical analyses.
All statistical procedures (Methods section of the supplemental material) were performed using version 9.4 of the SAS System for Windows. Graphs and plots were produced using R software version 3.2.1 (the R Core Team, 2015).
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by National Science Foundation grants from the Division of Environmental Biology to B.C.C. and C.F.W. (grants 1241161 and 1241069, respectively). Partial support was also provided by the Institute of Food and Agricultural Sciences at the University of Florida.
We thank Huiming Bao and Christine Foreman for the use of laboratory instrumentation, James Geaghan for advice on statistical analyses, and Grace Hunt and Aubrey Gilliland for assistance with sampling. We acknowledge the NOAA Air Resources Laboratory (ARL) for the provision of the HYSPLIT transport and dispersion model and/or READY website (https://www.ready.noaa.gov/index.php).
The data presented in the manuscript have not been submitted for publication elsewhere, and we declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01567-19.
REFERENCES
- 1.Pruppacher HR, Klett JD. 2010. Microphysics of clouds and precipitation, 2nd ed. Springer Netherlands, Dordrecht, the Netherlands. [Google Scholar]
- 2.Möhler O, Demott PJ, Vali G, Levin Z. 2007. Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences 4:1059–1071. doi: 10.5194/bg-4-1059-2007. [DOI] [Google Scholar]
- 3.Vali G, DeMott PJ, Möhler O, Whale TF. 2015. Technical note: a proposal for ice nucleation terminology. Atmos Chem Phys 15:10263–10270. doi: 10.5194/acp-15-10263-2015. [DOI] [Google Scholar]
- 4.Murray BJ, O'Sullivan D, Atkinson JD, Webb ME. 2012. Ice nucleation by particles immersed in supercooled cloud droplets. Chem Soc Rev 41:6519–6554. doi: 10.1039/c2cs35200a. [DOI] [PubMed] [Google Scholar]
- 5.DeMott P, Prenni A. 2010. New directions: need for defining the numbers and sources of biological aerosols acting as ice nuclei. Atmos Environ 44:1944–1945. doi: 10.1016/j.atmosenv.2010.02.032. [DOI] [Google Scholar]
- 6.Lee RE, Warren GJ, Gusta LV. 1995. Biological ice nucleation and its applications. APS Press, St. Paul, MN. [Google Scholar]
- 7.Christner BC, Cai R, Morris CE, McCarter KS, Foreman CM, Skidmore ML, Montross SN, Sands DC. 2008. Geographic, seasonal, and precipitation chemistry influence on the abundance and activity of biological ice nucleators in rain and snow. Proc Natl Acad Sci U S A 105:18854–18859. doi: 10.1073/pnas.0809816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris CE, Sands DC, Glaux C, Samsatly J, Asaad S, Moukahel AR, Gonçalves FLT, Bigg EK. 2013. Urediospores of rust fungi are ice nucleation active at >−10°C and harbor ice nucleation active bacteria. Atmos Chem Phys 13:4223–4233. doi: 10.5194/acp-13-4223-2013. [DOI] [Google Scholar]
- 9.Weber CF. 2016. Polytrichum commune spores nucleate ice and associated microorganisms increase the temperature of ice nucleation activity onset. Aerobiologia 32:353–361. doi: 10.1007/s10453-015-9395-1. [DOI] [Google Scholar]
- 10.Pummer BG, Bauer H, Bernardi J, Bleicher S, Grothe H. 2012. Suspendable macromolecules are responsible for ice nucleation activity of birch and conifer pollen. Atmos Chem Phys 12:2541–2550. doi: 10.5194/acp-12-2541-2012. [DOI] [Google Scholar]
- 11.Maki LR, Galyan EL, Chang-Chien M-M, Caldwell DR. 1974. Ice nucleation induced by Pseudomonas syringae. Appl Microbiol 28:456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kajava AV, Lindow SE. 1993. A model of the three-dimensional structure of ice nucleation proteins. J Mol Biol 232:709–717. doi: 10.1006/jmbi.1993.1424. [DOI] [PubMed] [Google Scholar]
- 13.Lindow SE, Lahue E, Govindarajan AG, Panopoulos NJ, Gies D. 1989. Localization of ice nucleation activity and the iceC gene product in Pseudomonas syringae and Escherichia coli. Mol Plant Microbe Interact 2:262–272. doi: 10.1094/MPMI-2-262. [DOI] [PubMed] [Google Scholar]
- 14.Pandey R, Usui K, Livingstone RA, Fischer SA, Pfaendtner J, Backus EHG, Nagata Y, Fröhlich-Nowoisky J, Schmüser L, Mauri S, Scheel JF, Knopf DA, Pöschl U, Bonn M, Weidner T. 2016. Ice-nucleating bacteria control the order and dynamics of interfacial water. Sci Adv 2:e1501630. doi: 10.1126/sciadv.1501630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortazavi R, Attiya S, Ariya PA. 2015. Arctic microbial and next-generation sequencing approach for bacteria in snow and frost flowers: selected identification, abundance and freezing nucleation. Atmos Chem Phys 15:6183–6204. doi: 10.5194/acp-15-6183-2015. [DOI] [Google Scholar]
- 16.Hill TCJ, Moffett BF, DeMott PJ, Georgakopoulos DG, Stump WL, Franc GD. 2014. Measurement of ice nucleation-active bacteria on plants and in precipitation by quantitative PCR. Appl Environ Microbiol 80:1256–1267. doi: 10.1128/AEM.02967-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nejad P, Ramstedt M, Granhall U, Roos S, McIvor I. 2006. Biochemical characterization and identification of ice-nucleation-active (INA) willow pathogens by means of Biolog MicroPlate, INA gene primers and PCR-based 16S rRNA-gene analyses. J Plant Dis Prot 113:97–106. doi: 10.1007/BF03356165. [DOI] [Google Scholar]
- 18.Ponder MA, Gilmour SJ, Bergholz PW, Mindock CA, Hollingsworth R, Thomashow MF, Tiedje JM. 2005. Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol Ecol 53:103–115. doi: 10.1016/j.femsec.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Vanderveer TL, Choi J, Miao D, Walker VK. 2014. Expression and localization of an ice nucleating protein from a soil bacterium, Pseudomonas borealis. Cryobiology 69:110–118. doi: 10.1016/j.cryobiol.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Failor KC, Schmale DG, Vinatzer BA, Monteil CL. 2017. Ice nucleation active bacteria in precipitation are genetically diverse and nucleate ice by employing different mechanisms. ISME J 11:2740–2753. doi: 10.1038/ismej.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fröhlich-Nowoisky J, Hill TCJ, Pummer BG, Yordanova P, Franc GD, Pöschl U. 2015. Ice nucleation activity in the widespread soil fungus Mortierella alpina. Biogeosciences 12:1057–1071. doi: 10.5194/bg-12-1057-2015. [DOI] [Google Scholar]
- 22.Pouleur S, Richard C, Martin J-G, Antoun H. 1992. Ice nucleation activity in Fusarium acuminatum and Fusarium avenaceum. Appl Environ Microbiol 58:2960–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knopf DA, Alpert PA, Wang B, Aller JY. 2011. Stimulation of ice nucleation by marine diatoms. Nature Geosci 4:88–90. doi: 10.1038/ngeo1037. [DOI] [Google Scholar]
- 24.D’souza NA, Kawarasaki Y, Gantz JD, Lee RE, Beall BFN, Shtarkman YM, Koçer ZA, Rogers SO, Wildschutte H, Bullerjahn GS, McKay R. 2013. Diatom assemblages promote ice formation in large lakes. ISME J 7:1632–1640. doi: 10.1038/ismej.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindow SE, Arny DC, Upper CD. 1978. Distribution of ice nucleation-active bacteria on plants in nature. Appl Environ Microbiol 36:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnell RC, Vali G. 1973. World-wide source of leaf-derived freezing nuclei. Nature 246:212–213. doi: 10.1038/246212a0. [DOI] [Google Scholar]
- 27.Lindow SE. 1983. The role of bacterial ice nucleation in frost injury to plants. Annu Rev Phytopathol 21:363–384. doi: 10.1146/annurev.py.21.090183.002051. [DOI] [Google Scholar]
- 28.O’Sullivan D, Murray BJ, Malkin TL, Whale TF, Umo NS, Atkinson JD, Price HC, Baustian KJ, Browse J, Webb ME. 2014. Ice nucleation by fertile soil dusts: relative importance of mineral and biogenic components. Atmos Chem Phys 14:1853–1867. doi: 10.5194/acp-14-1853-2014. [DOI] [Google Scholar]
- 29.Conen F, Morris CE, Leifeld J, Yakutin MV, Alewell C. 2011. Biological residues define the ice nucleation properties of soil dust. Atmos Chem Phys 11:9643–9648. doi: 10.5194/acp-11-9643-2011. [DOI] [Google Scholar]
- 30.Schnell RC, Vali G. 1975. Freezing nuclei in marine waters. Tellus 27:321–323. doi: 10.3402/tellusa.v27i3.9911. [DOI] [Google Scholar]
- 31.Wilson TW, Ladino LA, Alpert PA, Breckels MN, Brooks IM, Browse J, Burrows SM, Carslaw KS, Huffman JA, Judd C, Kilthau WP, Mason RH, McFiggans G, Miller LA, Nájera JJ, Polishchuk E, Rae S, Schiller CL, Si M, Temprado JV, Whale TF, Wong JPS, Wurl O, Yakobi-Hancock JD, Abbatt JPD, Aller JY, Bertram AK, Knopf DA, Murray BJ. 2015. A marine biogenic source of atmospheric ice-nucleating particles. Nature 525:234–238. doi: 10.1038/nature14986. [DOI] [PubMed] [Google Scholar]
- 32.Bowers RM, Lauber CL, Wiedinmyer C, Hamady M, Hallar AG, Fall R, Knight R, Fierer N. 2009. Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl Environ Microbiol 75:5121–5130. doi: 10.1128/AEM.00447-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joly M, Attard E, Sancelme M, Deguillaume L, Guilbaud C, Morris CE, Amato P, Delort A-M. 2013. Ice nucleation activity of bacteria isolated from cloud water. Atmos Environ 70:392–400. doi: 10.1016/j.atmosenv.2013.01.027. [DOI] [Google Scholar]
- 34.Monteil CL, Bardin M, Morris CE. 2014. Features of air masses associated with the deposition of Pseudomonas syringae and Botrytis cinerea by rain and snowfall. ISME J 8:2290–2304. doi: 10.1038/ismej.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowers RM, McLetchie S, Knight R, Fierer N. 2011. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J 5:601–612. doi: 10.1038/ismej.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia E, Hill TCJ, Prenni AJ, DeMott PJ, Franc GD, Kreidenweis SM. 2012. Biogenic ice nuclei in boundary layer air over two U.S. High Plains agricultural regions. J Geophys Res Atmos 117:D18209. doi: 10.1029/2012JD018343. [DOI] [Google Scholar]
- 37.Fierer N, Liu Z, Rodríguez-Hernández M, Knight R, Henn M, Hernandez MT. 2008. Short-term temporal variability in airborne bacterial and fungal populations. Appl Environ Microbiol 74:200–207. doi: 10.1128/AEM.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deguillaume L, Leriche M, Amato P, Ariya PA, Delort AM, Poschl U, Chaumerliac N, Bauer H, Flossmann A, Morris CE. 2008. Microbiology and atmospheric processes: chemical interactions of primary biological aerosols. Biogeosciences 5:1073–1084. doi: 10.5194/bg-5-1073-2008. [DOI] [Google Scholar]
- 39.Pratt KA, DeMott PJ, French JR, Wang Z, Westphal DL, Heymsfield AJ, Twohy CH, Prenni AJ, Prather KA. 2009. In situ detection of biological particles in cloud ice-crystals. Nature Geosci 2:398–401. doi: 10.1038/ngeo521. [DOI] [Google Scholar]
- 40.Morris CE, Conen F, Alex Huffman J, Phillips V, Pöschl U, Sands DC. 2014. Bioprecipitation: a feedback cycle linking Earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Glob Change Biol 20:341–351. doi: 10.1111/gcb.12447. [DOI] [PubMed] [Google Scholar]
- 41.Korolev AV, Isaac GA, Cober SG, Strapp JW, Hallett J. 2003. Microphysical characterization of mixed-phase clouds. Q J R Meteorol Soc 129:39–65. doi: 10.1256/qj.01.204. [DOI] [Google Scholar]
- 42.Khain A, Pokrovsky A, Pinsky M, Seifert A, Phillips V. 2004. Simulation of effects of atmospheric aerosols on deep turbulent convective clouds using a spectral microphysics mixed-phase cumulus cloud model. Part I: model description and possible applications. J Atmos Sci 61:2963–2982. doi: 10.1175/JAS-3350.1. [DOI] [Google Scholar]
- 43.Cantrell W, Heymsfield A. 2005. Production of ice in tropospheric clouds: a review. Bull Am Meteor Soc 86:795–808. doi: 10.1175/BAMS-86-6-795. [DOI] [Google Scholar]
- 44.Diehl K, Wurzler S. 2010. Air parcel model simulations of a convective cloud: bacteria acting as immersion ice nuclei. Atmos Environ 44:4622–4628. doi: 10.1016/j.atmosenv.2010.08.003. [DOI] [Google Scholar]
- 45.Mossop SC. 1976. Production of secondary ice particles during the growth of graupel by riming. Q J R Meteorol Soc 102:45–57. doi: 10.1002/qj.49710243104. [DOI] [Google Scholar]
- 46.Mossop SC. 1985. The origin and concentration of ice crystals in clouds. Bull Am Meteor Soc 66:264–273. doi:. [DOI] [Google Scholar]
- 47.Mignani C, Creamean JM, Zimmermann L, Alewell C, Conen F. 2019. New type of evidence for secondary ice formation at around −15°C in mixed-phase clouds. Atmos Chem Phys 19:877–886. doi: 10.5194/acp-19-877-2019. [DOI] [Google Scholar]
- 48.Zeng X, Tao W-K, Zhang M, Hou AY, Xie S, Lang S, Li X, Starr DO, Li X, Simpson J. 2009. An indirect effect of ice nuclei on atmospheric radiation. J Atmos Sci 66:41–61. doi: 10.1175/2008JAS2778.1. [DOI] [Google Scholar]
- 49.Phillips VTJ, Blyth AM, Brown PRA, Choularton TW, Latham J. 2001. The glaciation of a cumulus cloud over New Mexico. Q J R Meteorol Soc 127:1513–1534. doi: 10.1256/smsqj.57502. [DOI] [Google Scholar]
- 50.Phillips VTJ, Choularton TW, Illingworth AJ, Hogan RJ, Field PR. 2003. Simulations of the glaciation of a frontal mixed-phase cloud with the explicit microphysics model. Q J R Meteorol Soc 129:1351–1371. doi: 10.1256/qj.02.100. [DOI] [Google Scholar]
- 51.Khain AP, BenMoshe N, Pokrovsky A. 2008. Factors determining the impact of aerosols on surface precipitation from clouds: an attempt at classification. J Atmos Sci 65:1721–1748. doi: 10.1175/2007JAS2515.1. [DOI] [Google Scholar]
- 52.Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, Yan S, Dominguez H, Thompson BM. 2008. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2:321–334. doi: 10.1038/ismej.2007.113. [DOI] [PubMed] [Google Scholar]
- 53.Vali G. 1971. Quantitative evaluation of experimental results an [sic] the heterogeneous freezing nucleation of supercooled liquids. J Atmos Sci 28:402–409. doi:. [DOI] [Google Scholar]
- 54.Joly M, Amato P, Deguillaume L, Monier M, Hoose C, Delort A-M. 2014. Quantification of ice nuclei active at near 0°C temperatures in low-altitude clouds at the Puy de Dôme atmospheric station. Atmos Chem Phys 14:8185–8195. doi: 10.5194/acp-14-8185-2014. [DOI] [Google Scholar]
- 55.Kaakoush NO. 2015. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Microbiol 5:84. doi: 10.3389/fmicb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mülmenstädt J, Sourdeval O, Delanoë J, Quaas J. 2015. Frequency of occurrence of rain from liquid-, mixed-, and ice-phase clouds derived from A-Train satellite retrievals. Geophys Res Lett 42:6502–6509. doi: 10.1002/2015GL064604. [DOI] [Google Scholar]
- 57.Petters MD, Wright TP. 2015. Revisiting ice nucleation from precipitation samples. Geophys Res Lett 42:8758–8766. doi: 10.1002/2015GL065733. [DOI] [Google Scholar]
- 58.DeMott PJ, Prenni AJ, Liu X, Kreidenweis SM, Petters MD, Twohy CH, Richardson MS, Eidhammer T, Rogers DC. 2010. Predicting global atmospheric ice nuclei distributions and their impacts on climate. Proc Natl Acad Sci U S A 107:11217–11222. doi: 10.1073/pnas.0910818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hess M, Koepke P, Schult I. 1998. Optical properties of aerosols and clouds: the software package OPAC. Bull Am Meteor Soc 79:831–844. doi:. [DOI] [Google Scholar]
- 60.Rosenfeld D, Lensky IM. 1998. Satellite-based insights into precipitation formation processes in continental and maritime convective clouds. Bull Am Meteor Soc 79:2457–2476. doi:. [DOI] [Google Scholar]
- 61.Korolev AV, Mazin IP. 2003. Supersaturation of water vapor in clouds. J Atmos Sci 60:2957–2974. doi:. [DOI] [Google Scholar]
- 62.Korolev A. 2007. Limitations of the Wegener-Bergeron-Findeisen mechanism in the evolution of mixed-phase clouds. J Atmos Sci 64:3372–3375. doi: 10.1175/JAS4035.1. [DOI] [Google Scholar]
- 63.Jung CH, Bae SY, Kim YP. 2011. Approximated solution on the properties of the scavenging gap during precipitation using harmonic mean method. Atmos Res 99:496–504. doi: 10.1016/j.atmosres.2010.11.023. [DOI] [Google Scholar]
- 64.Wolff GT. 1984. On the nature of nitrate in coarse continental aerosols. Atmos Environ 18:977–981. doi: 10.1016/0004-6981(84)90073-8. [DOI] [Google Scholar]
- 65.Reference deleted.
- 66.Reference deleted.
- 67.Houze RA., Jr. 2014. Cloud dynamics. Academic Press, Cambridge, MA. [Google Scholar]
- 68.Christner BC, Royston-Bishop G, Foreman CM, Arnold BR, Tranter M, Welch KA, Lyons WB, Tsapin AI, Studinger M, Priscu JC. 2006. Limnological conditions in Subglacial Lake Vostok, Antarctica. Limnol Oceanogr 51:2485–2501. doi: 10.4319/lo.2006.51.6.2485. [DOI] [Google Scholar]
- 69.Aho K, Weber C, Christner B, Vinatzer B, Morris C, Joyce R, Werth J, Bayless-Edwards A, Lavender H, Schmale D. The spatiotemporal drivers of microbial composition and diversity in precipitation. Ecol Monogr, in press.
- 70.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Horn DJV, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy KR, Stedmon CA, Graeber D, Bro R. 2013. Fluorescence spectroscopy and multi-way techniques. PARAFAC. Anal Methods 5:6557–6566. doi: 10.1039/c3ay41160e. [DOI] [Google Scholar]
- 73.Nemecek-Marshall M, LaDuca R, Fall R. 1993. High-level expression of ice nuclei in a Pseudomonas syringae strain is induced by nutrient limitation and low temperature. J Bacteriol 175:4062–4070. doi: 10.1128/jb.175.13.4062-4070.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sattler B, Puxbaum H, Psenner R. 2001. Bacterial growth in supercooled cloud droplets. Geophys Res Lett 28:239–242. doi: 10.1029/2000GL011684. [DOI] [Google Scholar]
- 75.Doyle S, Dieser M, Broemsen E, Christner B. 2012. General characteristics of cold-adapted microorganisms, p 103–125. In Miller RV, Whyte LG (ed), Polar microbiology: life in a deep freeze. ASM Press, Washington, DC. [Google Scholar]
- 76.Morris CE, Soubeyrand S, Bigg EK, Creamean JM, Sands DC. 2017. Mapping rainfall feedback to reveal the potential sensitivity of precipitation to biological aerosols. Bull Am Meteor Soc 98:1109–1118. doi: 10.1175/BAMS-D-15-00293.1. [DOI] [Google Scholar]
- 77.Anagnostou EN. 2004. A convective/stratiform precipitation classification algorithm for volume scanning weather radar observations. Met Apps 11:291–300. doi: 10.1017/S1350482704001409. [DOI] [Google Scholar]
- 78.Biggerstaff MI, Listemaa SA. 2000. An improved scheme for convective/stratiform echo classification using radar reflectivity. J Appl Meteor 39:2129–2150. doi:. [DOI] [Google Scholar]
- 79.Omernik JM, Griffith GE. 2014. Ecoregions of the conterminous United States: evolution of a hierarchical spatial framework. Environ Manage 54:1249–1266. doi: 10.1007/s00267-014-0364-1. [DOI] [PubMed] [Google Scholar]
- 80.Commission for Environmental Cooperation. 1997. Ecological regions of North America: toward a common perspective. Commission for Environmental Cooperation, Montreal, Canada: http://www3.cec.org/islandora/en/item/1701-ecological-regions-north-america-toward-common-perspective-en.pdf. [Google Scholar]
- 81.Li P, Stuart EA, Allison DB. 2015. Multiple imputation: a flexible tool for handling missing data. JAMA 314:1966–1967. doi: 10.1001/jama.2015.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan Y. 2005. Multiple imputation for missing data: concepts and new development. SAS, Inc, Rockville, MD. [Google Scholar]
- 83.Yong AG, Pearce S. 2013. A beginner’s guide to factor analysis: focusing on exploratory factor analysis. Tutor Quant Methods Psychol 9:79–94. doi: 10.20982/tqmp.09.2.p079. [DOI] [Google Scholar]
- 84.Coble PG. 1996. Characterization of marine and terrestrial DOM in seawater using excitation-emission matrix spectroscopy. Mar Chem 51:325–346. doi: 10.1016/0304-4203(95)00062-3. [DOI] [Google Scholar]
- 85.Coble PG, Green SA, Blough NV, Gagosian RB. 1990. Characterization of dissolved organic matter in the Black Sea by fluorescence spectroscopy. Nature 348:432–435. doi: 10.1038/348432a0. [DOI] [Google Scholar]
- 86.Coble PG, Del Castillo CE, Avril B. 1998. Distribution and optical properties of CDOM in the Arabian Sea during the 1995 Southwest Monsoon. Deep sea research part II. Top Stud Oceanogr 45:2195–2223. doi: 10.1016/S0967-0645(98)00068-X. [DOI] [Google Scholar]
- 87.Gurian-Sherman D, Lindow SE. 1993. Bacterial ice nucleation: significance and molecular basis. FASEB J 7:1338–1343. doi: 10.1096/fasebj.7.14.8224607. [DOI] [PubMed] [Google Scholar]
- 88.Parlanti E, Wörz K, Geoffroy L, Lamotte M. 2000. Dissolved organic matter fluorescence spectroscopy as a tool to estimate biological activity in a coastal zone submitted to anthropogenic inputs. Org Geochem 31:1765–1781. doi: 10.1016/S0146-6380(00)00124-8. [DOI] [Google Scholar]
- 89.Stedmon CA, Markager S. 2005. Resolving the variability in dissolved organic matter fluorescence in a temperate estuary and its catchment using PARAFAC analysis. Limnol Oceanogr 50:686–697. doi: 10.4319/lo.2005.50.2.0686. [DOI] [Google Scholar]
- 90.Pummer BG, Budke C, Augustin-Bauditz S, Niedermeier D, Felgitsch L, Kampf CJ, Huber RG, Liedl KR, Loerting T, Moschen T, Schauperl M, Tollinger M, Morris CE, Wex H, Grothe H, Pöschl U, Koop T, Fröhlich-Nowoisky J. 2015. Ice nucleation by water-soluble macromolecules. Atmos Chem Phys 15:4077–4091. doi: 10.5194/acp-15-4077-2015. [DOI] [Google Scholar]
- 91.Schnell RC, Vali G. 1976. Biogenic ice nuclei: part i. terrestrial and marine sources. J Atmos Sci 33:1554–1564. doi:. [DOI] [Google Scholar]
- 92.Lindemann J, Constantinidou HA, Barchet WR, Upper CD. 1982. Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Appl Environ Microbiol 44:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Constantinidou HA, Hirano SS, Baker LS, Upper CD. 1990. Atmospheric dispersal of ice nucleation-active bacteria: the role of rain. Phytopathology 80:934–937. doi: 10.1094/Phyto-80-934. [DOI] [Google Scholar]
- 94.Creamean JM, Kirpes RM, Pratt KA, Spada NJ, Maahn M, de Boer G, Schnell RC, China S. 2018. Marine and terrestrial influences on ice nucleating particles during continuous springtime measurements in an Arctic oilfield location. Atmos Chem Phys 18:18023–18042. doi: 10.5194/acp-18-18023-2018. [DOI] [Google Scholar]
- 95.Buczolits S, Denner EB, Kämpfer P, Busse H-J. 2006. Proposal of Hymenobacter norwichensis sp. nov., classification of “Taxeobacter ocellatus,” “Taxeobacter gelupurpurascens” and “Taxeobacter chitinovorans” as Hymenobacter ocellatus sp. nov., Hymenobacter gelipurpurascens sp. nov. and Hymenobacter chitinivorans sp. nov., respectively, and emended description of the genus Hymenobacter Hirsch et al. 1999. Int J Syst Evol Microbiol 56:2071–2078. doi: 10.1099/ijs.0.64371-0. [DOI] [PubMed] [Google Scholar]
- 96.Hirsch P, Ludwig W, Hethke C, Sittig M, Hoffmann B, Gallikowski CA. 1998. Hymenobacter roseosalivarius gen. nov., sp. nov. from continental Antarctic soils and sandstone: bacteria of the Cytophaga/Flavobacterium/Bacteroides line of phylogenetic descent. Syst Appl Microbiol 21:374–383. doi: 10.1016/S0723-2020(98)80047-7. [DOI] [PubMed] [Google Scholar]
- 97.Hoang V-A, Kim Y-J, Nguyen NL, Yang D-C. 2013. Hymenobacter ginsengisoli sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol 63:661–666. doi: 10.1099/ijs.0.039719-0. [DOI] [PubMed] [Google Scholar]
- 98.Kim K-H, Im W-T, Lee S-T. 2008. Hymenobacter soli sp. nov., isolated from grass soil. Int J Syst Evol Microbiol 58:941–945. doi: 10.1099/ijs.0.64447-0. [DOI] [PubMed] [Google Scholar]
- 99.Lee J-J, Srinivasan S, Lim S, Joe M, Lee SH, Kwon SA, Kwon YJ, Lee J, Choi JJ, Lee HM, Auh YK, Kim MK. 2014. Hymenobacter swuensis sp. nov., a gamma-radiation-resistant bacteria isolated from mountain soil. Curr Microbiol 68:305–310. doi: 10.1007/s00284-013-0478-3. [DOI] [PubMed] [Google Scholar]
- 100.Reddy GS, Garcia-Pichel F. 2013. Description of Hymenobacter arizonensis sp. nov. from the southwestern arid lands of the United States of America. Antonie Van Leeuwenhoek 103:321–330. doi: 10.1007/s10482-012-9812-1. [DOI] [PubMed] [Google Scholar]
- 101.Srinivasan S, Joo ES, Lee J-J, Kim MK. 2015. Hymenobacter humi sp. nov., a bacterium isolated from soil. Antonie Van Leeuwenhoek 107:1411–1419. doi: 10.1007/s10482-015-0436-0. [DOI] [PubMed] [Google Scholar]
- 102.Zhang D-C, Busse H-J, Liu H-C, Zhou Y-G, Schinner F, Margesin R. 2011. Hymenobacter psychrophilus sp. nov., a psychrophilic bacterium isolated from soil. Int J Syst Evol Microbiol 61:859–863. doi: 10.1099/ijs.0.023465-0. [DOI] [PubMed] [Google Scholar]
- 103.Fellman JB, Hood E, Spencer R. 2010. Fluorescence spectroscopy opens new windows into dissolved organic matter dynamics in freshwater ecosystems: a review. Limnol Oceanogr 55:2452–2462. doi: 10.4319/lo.2010.55.6.2452. [DOI] [Google Scholar]
- 104.Murphy KR, Stedmon CA, Waite TD, Ruiz GM. 2008. Distinguishing between terrestrial and autochthonous organic matter sources in marine environments using fluorescence spectroscopy. Mar Chem 108:40–58. doi: 10.1016/j.marchem.2007.10.003. [DOI] [Google Scholar]
- 105.Balcarczyk KL, Jones JB, Jaffé R, Maie N. 2009. Stream dissolved organic matter bioavailability and composition in watersheds underlain with discontinuous permafrost. Biogeochemistry 94:255–270. doi: 10.1007/s10533-009-9324-x. [DOI] [Google Scholar]
- 106.Creamean JM, Suski KJ, Rosenfeld D, Cazorla A, DeMott PJ, Sullivan RC, White AB, Ralph FM, Minnis P, Comstock JM, Tomlinson JM, Prather KA. 2013. Dust and biological aerosols from the Sahara and Asia influence precipitation in the western U.S. Science 339:1572–1578. doi: 10.1126/science.1227279. [DOI] [PubMed] [Google Scholar]
- 107.Smith DJ, Timonen HJ, Jaffe DA, Griffin DW, Birmele MN, Perry KD, Ward PD, Roberts MS. 2013. Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl Environ Microbiol 79:1134–1139. doi: 10.1128/AEM.03029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kellogg CA, Griffin DW. 2006. Aerobiology and the global transport of desert dust. Trends Ecol Evol 21:638–644. doi: 10.1016/j.tree.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 109.Griffin DW, Kellogg CA, Garrison VH, Shinn EA. 2002. The global transport of dust: an intercontinental river of dust, microorganisms and toxic chemicals flows through the Earth’s atmosphere. Am Sci 90:228–235. doi: 10.1511/2002.3.228. [DOI] [Google Scholar]
- 110.Milne PJ, Zika RG. 1993. Amino acid nitrogen in atmospheric aerosols: occurrence, sources and photochemical modification. J Atmos Chem 16:361–398. doi: 10.1007/BF01032631. [DOI] [Google Scholar]
- 111.Kyrö E-M, Grönholm T, Vuollekoski H, Virkkula A, Kulmala M, Laakso L. 2009. Snow scavenging of ultrafine particles: field measurements and parameterization. Boreal Environ Res 14:527–538. [Google Scholar]
- 112.Paramonov M, Grönholm T, Virkkula A. 2011. Below-cloud scavenging of aerosol particles by snow at an urban site in Finland. Boreal Environ Res 16:304–320. [Google Scholar]
- 113.Zhang L, Wang X, Moran MD, Feng J. 2013. Review and uncertainty assessment of size-resolved scavenging coefficient formulations for below-cloud snow scavenging of atmospheric aerosols. Atmos Chem Phys 13:10005–10025. doi: 10.5194/acp-13-10005-2013. [DOI] [Google Scholar]
- 114.Bowers RM, Sullivan AP, Costello EK, Collett JL, Knight R, Fierer N. 2011. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl Environ Microbiol 77:6350–6356. doi: 10.1128/AEM.05498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bowers RM, McCubbin IB, Hallar AG, Fierer N. 2012. Seasonal variability in airborne bacterial communities at a high-elevation site. Atmos Environ 50:41–49. doi: 10.1016/j.atmosenv.2012.01.005. [DOI] [Google Scholar]
- 116.Zhang L, Dai J, Tang Y, Luo X, Wang Y, An H, Fang C, Zhang C. 2009. Hymenobacter deserti sp. nov., isolated from the desert of Xinjiang, China. Int J Syst Evol Microbiol 59:77–82. doi: 10.1099/ijs.0.000265-0. [DOI] [PubMed] [Google Scholar]
- 117.Zhang Q, Liu C, Tang Y, Zhou G, Shen P, Fang C, Yokota A. 2007. Hymenobacter xinjiangensis sp. nov., a radiation-resistant bacterium isolated from the desert of Xinjiang, China. Int J Syst Evol Microbiol 57:1752–1756. doi: 10.1099/ijs.0.65033-0. [DOI] [PubMed] [Google Scholar]
- 118.Meola M, Lazzaro A, Zeyer J. 2015. Bacterial composition and survival on Sahara dust particles transported to the European Alps. Front Microbiol 6:1454. doi: 10.3389/fmicb.2015.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Šantl-Temkiv T, Finster K, Dittmar T, Hansen BM, Thyrhaug R, Nielsen NW, Karlson UG. 2013. Hailstones: a window into the microbial and chemical inventory of a storm cloud. PLoS One 8:e53550. doi: 10.1371/journal.pone.0053550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weon H-Y, Kwon S-W, Son J-A, Kim S-J, Kim Y-S, Kim B-Y, Ka J-O. 2010. Adhaeribacter aerophilus sp. nov., Adhaeribacter aerolatus sp. nov. and Segetibacter aerophilus sp. nov., isolated from air samples. Int J Syst Evol Microbiol 60:2424–2429. doi: 10.1099/ijs.0.018374-0. [DOI] [PubMed] [Google Scholar]
- 121.Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci U S A 104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Buczolits S, Denner EBM, Vybiral D, Wieser M, Kämpfer P, Busse H-J. 2002. Classification of three airborne bacteria and proposal of Hymenobacter aerophilus sp. nov. Int J Syst Evol Microbiol 52:445–456. doi: 10.1099/00207713-52-2-445. [DOI] [PubMed] [Google Scholar]
- 123.Graf J. 2014. The family Rikenellaceae, p 857–859. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer Berlin Heidelberg, Berlin, Germany. [Google Scholar]
- 124.Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, McLellan SL. 2015. A single genus in the gut microbiome reflects host preference and specificity. ISME J 9:90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.