Abstract
Objective:
MicroRNAs (miRNAs) are short non-coding RNAs that regulate genes and are both biomarkers and mediators of disease. We used small RNA (sRNA) sequencing and machine learning methodology to develop a miRNA panel to reliably differentiate between rheumatoid arthritis (RA) or systemic lupus erythematosus (SLE) and control subjects.
Methods:
Plasma samples from 167 RA and 91 control subjects frequency-matched for age, race and sex were used for sRNA sequencing. TIGER was used to analyze miRNAs. DESeq2 and random forest analyses were used to identify a prioritized list of miRNAs differentially expressed in patients with RA. Prioritized miRNAs were validated by quantitative PCR, and lasso and logistic regression were used to select the final panel of six miRNAs that best differentiated RA from controls. The panel was validated in a separate cohort of 12 SLE, 32 RA and 32 control subjects. Panel efficacy was assessed by area under the receiver operative characteristic curve (AUC) analyses.
Results:
The final panel included miR-22-3p, miR-24-3p, miR-96-5p, miR-134-5p, miR-140-3p, and miR-627-5p. The panel differentiated RA from control subjects in discovery (AUC=0.81) and validation cohorts (AUC=0.71), seronegative RA (AUC=0.84), RA remission (AUC=0.85), and SLE patients (AUC=0.80) versus controls. Pathway analysis showed upstream regulators and targets of panel miRNAs are associated with pathways implicated in RA pathogenesis.
Conclusion:
A miRNA panel identified by a bioinformatic approach differentiated between RA or SLE patients and control subjects. The panel may represent an autoimmunity signature, perhaps related to inflammatory arthritis, which is not dependent on active disease or seropositivity.
MicroRNAs are small non-coding RNAs that are important gene regulators and serve as biomarkers of disease. As gene regulators miRNAs can destabilize messenger RNAs (mRNAs) and block translation typically by binding to the 3’ untranslated region of the mRNA with a complementary seed region near the miRNA 5’ end (1, 2). miRNAs are found within cells, but also circulate in plasma protected from degradation by exosomes (3), microvesicles (4), lipoproproteins (5) and RNA-binding proteins (6). Moreover, miRNAs within these bodies can be transported to recipient cells (5) to regulate genes. Plasma miRNAs are stable in stored samples (7) and are more practical for use as biomarkers than miRNAs in specific cell types due to ease of isolation. Many studies, predominantly in cancer, show that miRNAs can be helpful diagnostic and prognostic biomarkers, particularly when used in a panel composed of multiple miRNAs (8-11).
We and others have found several plasma miRNAs which are differentially altered among patients with rheumatoid arthritis (RA) (12-14). However, most prior studies examined a few miRNAs with known relevant function using polymerase chain reaction (PCR) or used small arrays. Small RNA sequencing provides the ability to evaluate many more miRNAs in an unbiased fashion. Thus, small RNA sequencing could reveal novel miRNA signatures of RA and potentially provide mechanistic insights into disease pathogenesis. Our objective was to determine if a panel of miRNAs derived from small RNA sequencing could differentiate between patients with RA and control subjects, and if that panel validated in a separate RA cohort and was unique to RA or shared with another autoimmune disease: systemic lupus erythematosus (SLE). Additionally, we used pathway analysis to evaluate if these miRNAs have common disease-related upstream regulators which could affect their expression and if the miRNAs could affect RA-related pathways.
MATERIALS AND METHODS
Overview
We used a discovery cohort of RA and control subjects to perform small RNA sequencing for identification of candidate panel miRNAs. Potential candidate miRNAs differentiating RA from controls were prioritized using univariable differential expression analysis (R package ‘DESeq2’) and a multivariable random forest analysis. These prioritized candidates were validated by quantitative PCR (qPCR). Then, cross-validation lasso with logistic regression was used to further reduce validated miRNA candidates to a small panel of miRNAs which provided best discrimination between RA and control subjects based on area under the receiver operating characteristic curves (AUC). This reduced panel of miRNAs was then externally validated in an independent cohort of patients with RA and control subjects by qPCR. The panel was tested also in a small number of patients with SLE to see if it similarly differentiated between SLE and control subjects.
Study population
The discovery cohort included 167 patients with RA and 91 control subjects frequency-matched for age, race and sex from a prior cross-sectional study (15). The validation cohort included 32 patients with RA and 32 control subjects frequency-matched for age, race and sex from another prior crosssectional study (16), and twelve patients with SLE from another prior study (17).
Recruitment and study procedures were described previously (15-17), and will be detailed briefly. For the discovery and validation cohorts, subjects were 18 years of age or older. Patients with RA met classification criteria for RA (18), patients with SLE met classification criteria for SLE (19), and control subjects did not have a diagnosis of inflammatory autoimmune disease. Additionally, in the RA validation cohort, which was originally studied to examine the relationship between RA and structural and functional cardiac abnormalities, participants with current or prior heart failure, ischemic cardiovascular disease, structural cardiac disease, atrial fibrillation, estimated glomerular filtration rate <60ml/min, gadolinium hypersensitivity, pregnancy or breast feeding, or inability to have MRI were excluded from the study (16). Studies were approved by the Vanderbilt Institutional Review Board (IRB# 000567, 120314, and 990111) and all subjects gave written informed consent.
Clinical and laboratory Information
We collected clinical information and laboratory measurements as previously described (15, 16). RA disease activity was determined by the 28 joint count disease activity score (DAS28) using erythrocyte sedimentation rate (ESR) (20). High-sensitivity C-reactive protein (CRP) concentrations and ESR were measured by the Vanderbilt University Medical Center Clinical Laboratory.
Small RNA sequencing and microRNA alignments
Total RNA was extracted from stored plasma using Total RNA Purification Kits (Norgen). Libraries were prepared using TruSeq Small RNA Library Preparation Kits (Illumina). RNA extractions and library preparations were performed with both RA and control subject samples in each batch. Libraries were assessed for quality and size selected for approximately 128 to 157 nucleotides in length including adaptors by Pippin Prep (Sage Science) in the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core facility.
The cDNA libraries were sequenced using an Illumina NextSeq500 instrument by the VANTAGE core facility. TIGER (“Tools for Integrative Genome analysis of Extracellular sRNAs”), an in-house small RNA sequencing analysis pipeline (21), was used to analyze sequence data. In brief, high quality reads were demultiplexed using Illumina’s CASAVA 1.8 pipeline and 3’ adapters were trimmed using Cutadapt (22). Reads shorter than 16 nucleotides after adapter trimming were discarded. Three non-templated nucleotide addition (NTA) isoforms of each sRNA read were generated by removing 1, 2 or 3 bases from 3’ terminal. All four isoforms, including three NTA isoforms and original read, were aligned to the human genome (hg19) using Bowtie (23) allowing one mismatch. A sRNA read was identified as a miRNA if its mapped starting position matches any of the first 3 positions from the 5’ end of a miRNA based on the miRNA genome coordinates from miRBase (version 21,http://www.mirbase.org).
MicroRNA analyses
To obtain a prioritized group of miRNAs that were potential candidates for the panel we used both DESeq2 (24), which is Wald test based but can adjust for batch effect and other covariates; and random forest analysis, which is a machine learning method, to capture a prioritized list of candidate miRNAs for the panel. We assumed that many of the miRNAs would overlap, but that using both would help broaden potential candidates.
Sequencing read counts were normalized to total sRNA reads sequenced which passed quality control (reads per total read). Differentially expressed miRNAs were compared by DESeq2 (24), adjusting for age, race, sex, and batch effect. Benjamini-Hochberg adjusted p-values were used to select the 15 most significantly differentially expressed miRNAs (based on P value) for qPCR validation and further model development.
Random forest analyses, which allows for nonlinear relationship between disease status and miRNAs, were conducted to select miRNAs that are most important in separating RA and control subjects. The cross-validated prediction performance of models was evaluated with sequentially reduced number of predictors (ranked by variable importance) via a nested cross-validation procedure. Based on importance score, the top 15 miRNAs were selected for qPCR validation and further model development.
Using qPCR-based plasma concentrations of the miRNAs, we used lasso regularization with logistic regression to select a parsimonious final miRNA panel that maximized discrimination between RA and controls. This panel of miRNAs was validated using qPCR in a separate cohort of 12 patients with SLE, 32 patients with RA and 32 control subjects. Panel efficacy was assessed by area under the receiver operating characteristic curve (AUC) analyses.
qPCR validation
The same plasma samples used for small RNA sequencing were also used for qPCR validation. A cocktail of three C. elegans miRNA mimics (cel-miR-39, cel-miR-54, and cel-miR238; Qiagen) was added after the initial lysis step as a spike-in standard for normalizing RNA extraction efficiency. cDNA was prepared using qScript microRNA cDNA synthesis kits (Quantabio). Individual PCR assays (Quanta), and PerfeCTa SYBR green supermix for iQ (Quantabio) were used for qPCR. Plasma miRNA concentrations were determined from standard dilution curves of a DNA mimic and normalized to the spike-in standard. Samples were excluded from analysis if the Ct values of the spike-in standard exceeded one Ct from the median.
General statistical methods
Descriptive statistics were calculated as median [interquartile range] for continuous variables, and frequency and proportions for categorical variables. Wilcoxon’s rank sum tests were used to compare continuous variables and Pearson’s chi-square test to compare categorical variables. PCR-based miRNA concentrations were log-transformed in models due to skewness and are presented as geometric mean (95% confidence interval). Fold difference of the PCR-based miRNA concentrations was the fold difference of the geometric mean. Spearman correlation was used to assess the correlation of continuous variables.
Sample size determination
For sRNA sequencing the discovery cohort sample size of 167 RA and 91 control subjects offered approximately 99% power to detect miRNAs which were >1.5 fold altered in RA versus control subjects assuming detection across all samples of approximately 500 miRNAs with a 5% false discovery rate (https://cqs.mc.vanderbilt.edu/shiny/RnaSeqSampleSize/). This sample size also gave approximately 99% power to detect an AUC ≥0.65 based on PCR-based plasma miRNA concentrations.
For the validation cohort, a sample size of 32 patients with RA and 32 controls provided approximately 90% power to detect and AUC ≥0.7 based on PCR-based plasma miRNA concentrations.
Pathway analysis
We separately evaluated upstream regulators of the panel miRNAs and downstream targets regulated by the panel miRNAs using Ingenuity Pathway Analysis (Qiagen, Version 01-07). For evaluation as upstream regulators, we selected direct and indirect regulators of each mature miRNA and its precursor using all available data. For evaluation of downstream targets of the panel miRNAs, a target was included in analysis if it was previously experimentally validated or is a highly predicted target of the miRNA. We assessed canonical pathways and functional analyses.
RESULTS
Clinical characteristics
The discovery cohort included 167 patients with RA and 91 control subjects. The groups were of similar age, race and sex (Table 1). The RA validation cohort included 32 patients with RA and 32 control subjects of similar age, race and sex (Supplemental Table 1). Compared to the discovery cohort disease activity was lower (DAS28 score 3.89 vs 2.80 units), and there were fewer seropositive individuals in the validation cohort (69% vs 54% positive for rheumatoid factor). The SLE validation cohort included 12 patients with SLE (Supplemental Table 2).
Table 1.
Subject characteristics- discovery cohort
| RA (N=167) | Control (N=91) | P | |
|---|---|---|---|
| Age, years | 54 [45, 63] | 53 [44, 59] | 0.35 |
| Race, #Caucasian | 148 (89) | 77 (85) | 0.49 |
| Sex, #female | 114 (68) | 57 (63) | 0.36 |
| Disease duration, years | 3 [2, 18] | - | - |
| DAS28 score, units | 3.89 [2.63, 4.9] | - | - |
| Tender joints, # | 2 [0, 7] | - | - |
| Swollen joints, # | 3 [0, 8] | - | - |
| ESR, mm/hr | 16 [7, 36] | - | - |
| hsCRP, mg/L | 4 [1.2, 11.0] | 0.5 [0.2, 1.7] | <0.001 |
| RF positive, # | 115 (69) | - | - |
| Methotrexate, # | 117 (71) | - | - |
| Leflunomide, # | 29 (17) | - | - |
| Hydroxychloroquine, # | 42 (25) | - | - |
| Corticosteroids, # | 89 (53) | 2 (2) | <0.001 |
| Anti-TNF, # | 33 (20) | - | - |
Data are presented as median [interquartile range] or number (%). DAS28=disease activity based on 28 joints using erythrocyte sedimentation rate, ESR=erythrocyte sedimentation rate, hsCRP= high sensitivity C-reactive protein, RF= rheumatoid factor (data available on 28 patients with RA). CCP= anti-cyclic citrullinated peptide antibody (data available on 17 patients with RA).
Significantly altered miRNAs based on sRNA sequencing comparing RA vs control subjects – discovery cohort
Among the 262 reliably detected plasma miRNAs, 175 were significantly altered in RA compared to control subjects after adjusting for age, race, sex and batch and FDR. Among these, 110 were ≥1.5 fold altered in RA compared to control subjects (Figure 1). Most of these miRNAs were increased in RA plasma, and one miRNA, miR-3168, was significantly decreased (−1.73-fold decreased, P=5.0E-03, Padj=1.2E-02).
Figure 1.
Volcano plot displaying differential plasma miRNA expression in the discovery cohort (n=167 RA vs n=91 control subjects). Small RNA sequencing was analyses by DESeq2 and adjusted for age, race, sex and batch and multiple comparisons. Among these 110 were ≥1.5 fold altered in RA compared to control subjects. Each dot represents an individual miRNA and the larger the dot the more abundant the miRNA is. Red indicates increased in RA and blue indicates increased in control subjects.
The top 15 differentially expressed plasma miRNAs as determined each by DESeq2 and by random forest analysis are listed in Table 2. Twelve of the 15 miRNAs were common to both analytic approaches (miR-3615, miR-22-3p, miR-502-3p, miR-345-5p, miR-29c-3p, miR-221-3p, miR-140-3p, miR-30e-5p, miR-501-3p, miR-22-5p, miR-127-3p, miR-134-5p). Additionally, 3 miRNAs were identified each by DESeq2 only (miR-99b-5p, miR-130a-3p, miR-21-3p), and 3 by random forest only (miR-627-5p, miR-24-3p, miR-96-5p).
Table 2.
PCR validation of top miRNA candidates from discovery cohort sRNA sequencing
| Plasma fM concentration presented as geometric mean (95% CI) | ||||
|---|---|---|---|---|
| RA | Control | Fold diff | P | |
| miR-3615 | 0.34 (0.28, 0.41) | 0.25 (0.17, 0.35) | 1.38 | 4.69E-01 |
| miR-22-3p | 58.1 (45.3, 74.5) | 15.1 (10.8, 21) | 3.85 | 8.45E-12 |
| miR-502-3p | N/A | N/A | N/A | N/A |
| miR-345-5p | 3.08E-2 (1.63E-02, 5.81E-02) | 7.75E-3 (2.95E-03, 2.04E-02) | 3.98 | 1.72E-04 |
| miR-29c-3p | 1.46 (1.18, 1.8) | 0.56 (0.41, 0.77) | 2.59 | 5.32E-09 |
| miR-99b-5p | 1.50 (0.86, 2.61) | 0.99 (0.48, 2.05) | 1.51 | 7.27E-04 |
| miR-221-3p | 9.96 (7.48, 13.2) | 3.04 (2.15, 4.31) | 3.27 | 6.78E-09 |
| miR-140-3p | 1.22 (1.0, 1.49) | 0.25 (0.17, 0.37) | 4.93 | 2.12E-13 |
| miR-130a-3p | 5.00 (3.51, 7.12) | 1.27 (0.81, 1.97) | 3.94 | 8.56E-08 |
| miR-30e-5p | 4.23 (3.51, 5.11) | 3.24 (2.49, 4.21) | 1.31 | 2.39E-02 |
| miR-501-3p | N/A | N/A | N/A | N/A |
| miR-22-5p | 1.18 (0.891, 1.56) | 0.39 (0.25, 0.61) | 2.99 | 1.25E-07 |
| miR-127-3p | 5.17 (4.7, 5.68) | 5.53 (4.9, 6.24) | −1.08 | 4.88E-01 |
| miR-21-3p | 1.21E-02 (4.56E-03, 3.23E-02) | 6.97E-04 (1.33E-04, 6.64E-03) | 17.4 | 4.08E-03 |
| miR-134-5p | 1.45E-01 (6.38E-02, 3.35E-01) | 6.76E-03 (1.64E-03, 2.80E-02) | 21.6 | 5.11E-07 |
| miR-627-5p | 7.18E-02 (2.99E-02, 1.72E-01) | 4.39E-03 (9.58E-04, 2.01E-02) | 16.3 | 2.73E-04 |
| miR-24-3p | 3.49 (2.35, 5.18) | 1.32 (0.85, 2.06) | 2.64 | 1.19E-06 |
| miR-96-5p | 13.9 (12.9, 14.9) | 15.2 (13.6, 17) | −1.10 | 1.85E-01 |
N/A indicates that the miRNA was too low for reliable detection by PCR. fM=femtomolar concentration. Fold diff= fold difference comparing the geometric mean of qPCR-based miRNA concentration in RA vs control subjects.
PCR validation of altered miRNAs
The top candidate miRNAs (18 total) were measured by qPCR in the discovery cohort. Two of the miRNAs (miR-502-3p and miR-501-3p) were too low in abundance to be assayed reliably by qPCR. All but two (miR-127-3p and miR-96-5p) of the remaining miRNAs were significantly increased among patients with RA compared to control subjects using PCR-based concentrations of miRNAs (Table 2).
miRNA panel development
Using lasso variable selection with logistic regression of the qPCR-based concentrations of miRNAs in the discovery cohort, the following miRNAs were chosen for the panel so as to include the fewest miRNAs that discriminated RA from control subjects: miR-22-3p, miR-24-3p, miR-96-5p, miR-134-5p, miR-140-3p, miR-627-5p. The panel had an AUC=0.81 (95% CI: 0.75, 0.87; P<0.001) for differentiating RA and control subjects. The panel was similarly robust among those with seropositive RA AUC=0.79 (95% CI: 0.73, 0.86; P<0.001), seronegative RA AUC=0.84 (95% CI: 0.77, 0.91; P<0.001), RA in remission (DAS28 score<2.6(25)) (AUC=0.85 (95% CI: 0.78, 0.92; P<0.001) and high RA disease activity (DAS28>5.1) (AUC=0.79 (95% CI: 0.70, 0.88; P<0.001).
The panel had similar performance across other subgroups of patients compared to control subjects. This included: RA patients with disease duration <1 year (N=29) AUC=0.80 (95% CI: 0.72, 0.89; P<0.001), those not taking biologic DMARDs (bDMARDs) (N=133) AUC=0.81 (95% CI: 0.75, 0.87; P<0.001), those taking bDMARDs (N=33) AUC=0.80 (95%CI: 0.71, 0.88; P<0.001), those not taking conventional synthetic DMARDs (csDMARDs) or bDMARD (N=19) AUC=0.90 (95%CI: 0.83, 0.96; P<0.001), those taking any csDMARD or bDMARD (N=150) AUC=0.80 (0.73, 0.86; P<0.001), those not receiving any csDMARD, bDMARD or corticosteroid (N= 13) AUC= 0.89 (95%CI: 0.83, 0.96; P<0.001), and those taking either csDMARD or bDMARD or corticosteroid only (N=153) compared to control subjects AUC=0.80 (95% CI: 0.74, 0.86; P<0.001).
Validation
The panel of six miRNAs was robust in the separate RA validation cohort (AUC=0.71) (95% CI: 0.58, 0.84; P=0.004). Similarly, in the validation cohort, the panel differentiated between seropositive (AUC=0.73; 95% CI: 0.58, 0.87; P=0.01) or seronegative RA (AUC=0.73; 95% CI: 0.57, 0.89; P=0.02) vs control subjects.
We additionally measured the panel in 12 patients with SLE and compared model performance to control subjects. The panel also differentiated SLE patients from controls subjects (AUC=0.80 (95% CI: 0.65, 0.96; P=0.001)), but was not significantly different comparing patients with SLE to RA (AUC=0.63 (95% CI: 0.44, 0.82; P=0.13)).
Relationship between miRNA components of the panel and disease-related variables
Three of the miRNAs were weakly associated with RA disease activity by DAS28 score in the discovery cohort (miR-24-3p: Rho= −0.16, P=0.04; miR-96-5p: Rho= 0.16, P=0.04; miR-140-3p: Rho=−0.16, P=0.05); however, these significant associations were not observed in the RA validation cohort.
What are the upstream regulators of these miRNAs in the panel?
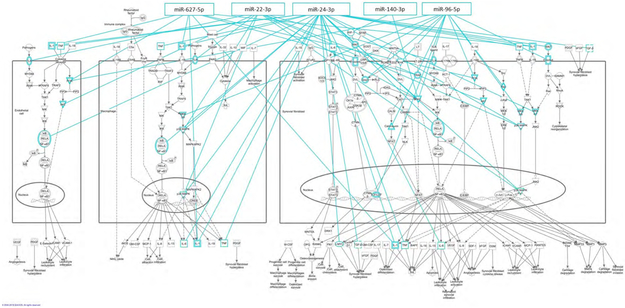
We examined upstream regulators of the miRNAs included in the panel to determine if there are commonalities in regulation between the miRNAs which would promote their ability to be used as a RA miRNA signature. There was little information regarding upstream regulators of miR-627-5p or its precursor at the time of analysis, thus this miRNA was excluded from pathway analysis. The top identified function that the upstream regulators possess related to invasion of cells (P=7.89E-27), for which 35 of the upstream regulators were included (Figure 2). Additionally, these upstream regulators are involved in cell death (P=7.11E-22). The top overlapping canonical pathway was role of macrophages, fibroblasts and endothelial cells in RA with 11 overlapping molecules (Figure 2).
Figure 2.
Upstream regulators of RA panel miRNAs are involved in invasion of cells and RA-specific pathways. Using Ingenuity Pathway Analysis of upstream regulators of the mature and precursor miRNAs, the top related functional pathway was related to invasion of cells and the top related canonical pathway was role of macrophages, fibroblasts, and endothelial cells in RA.
miRNA panel pathway targets
Because circulating miRNAs can be delivered to cells with functional consequences (5) or could reflect cellular processes, we evaluated if experimentally validated and highly predicted targets of panel miRNAs are involved in RA pathways. There was little information regarding miR-134-5p function at the time of analysis, thus this miRNA was excluded from pathway analysis. Among the top canonical pathways, role of osteoblasts, osteoclasts and chondrocytes in RA (Supplemental Figure) and role of macrophages, fibroblasts and endothelial cells in RA (Figure 3) were third (29 molecules) and sixth (28 molecules) respectively in the number of molecules in the pathway which the miRNA panel may target. Other top canonical pathways include molecular mechanisms of cancer (52 genes), G-protein coupled receptor signaling (30 molecules), protein kinase A signaling (29 molecules) and axonal guidance signaling (28 molecules).
Figure 3.
RA panel miRNAs can target RA-specific pathways. Using Ingenuity Pathway Analysis of experimentally validated or highly predicted targets of the panel miRNAs, one of the top related canonical pathways was role of macrophages, fibroblasts, and endothelial cells in RA.
DISCUSSION
We used plasma sRNA sequencing and bioinformatics approaches to develop a panel of miRNAs that reliably differentiates between patients with RA and control subjects. The panel included miR-22-3p, miR-24-3p, miR-96-5p, miR-134-5p, miR-140-3p, and miR-627-5p and was robust across seronegative and seropositive RA and RA of varying disease activity. The panel also differentiated between patients with SLE and control subjects but was not significantly different between patients with RA and SLE, suggesting that this panel represents an autoimmunity signature.
Strong evidence indicates that miRNAs as a class of sRNAs are important in RA. Dicer and Drosha, which are endoribonucleases involved in cleaving miRNAs to their mature form, are upregulated in peripheral blood mononuclear cells (PBMCs) of patients with RA (26). Moreover, activation of Dicer decreases tumor necrosis factor alpha (TNFα) production, suggesting a homeostatic role of miRNAs in RA. The upregulation of Dicer and Drosha in PBMCs from patients with RA likely explains why we observed an overall increase in miRNAs in plasma from patients with RA, since many plasma miRNAs derive from PBMCs and other blood cells (27). However, in RA not all cell types have increased expression of Dicer and the associated increased miRNA expression. For example, Dicer and miRNA expression were lower in RA synovial fibroblasts, leading to an exaggerated response to inflammatory stimuli and resistance to apoptosis (28). In addition to broad changes in miRNA production in RA, there are several individual miRNAs with biologic significance implicated in RA.
Several of the panel miRNAs have known associations with RA, either as biomarkers or known biology of RA. For example, miR-22-3p was increased significantly among ACPA positive individuals who subsequently progressed to RA (29). Also, lower concentrations of miR-22-3p before treatment were associated with response to adalimumab at 12 months in a randomized double-blinded placebo-controlled trial of 180 patients with early RA (30). miR-22-3p is decreased in fibroblast-like synoviocytes (FLS) from RA compared to osteoarthritis patients; lower concentrations of miR-22-3p promoted FLS proliferation (31). Thus miR-22-3p may be a driving force for the synovial hyperplasia characteristic of RA. miR-22-3p also regulates Th17 responses in emphysema (32) and drives hyperresponsive B cells in SLE (33). Moreover, we found that inhibition of miR-22-3p improves nephritis in a mouse model of SLE (manuscript In preparation).
Plasma concentrations of miR-24-3p were elevated in patients with RA compared to controls in several prior studies, and is a component of a prior RA plasma miRNA panel which we and others have proposed (12, 14). In response to IL-6, miR-24-3p increases and promotes plasma cell survival, an effect that could support autoreactive plasma cells in RA (34). miR-24-3p also plays a homeostatic role by dampening inflammation through nuclear factor-KB signaling pathway (35) and chitinase 3-like-1 (36) in the vasculature.
miR-140-3p has been most widely studied in osteoarthritis and participates in cartilage homeostasis (37); its concentrations are also decreased in synovial tissue from patients with RA compared to osteoarthritis (38). Intra-articular administration of pre-miR-140 (the precursor for miR-140-3p) reduced arthritis scores in mice with collagen induced arthritis by way of increasing synovial fibroblast apoptosis and reducing proliferation (38).
Three of the panel miRNAs (miR-96-5p, miR-134-5p, and miR-627-5p) have not been studied widely in RA. Thus, if we had limited selection of miRNAs for the panel to those previously studied in RA, these would have been overlooked. We believe we identified new miRNAs because of our methods which used sequencing rather than a preselected panel and random forest analysis to identify candidates, which has not been previously done in RA to our knowledge. miR-96-5p remained helpful to the model despite not being overall significantly altered in RA based on PCR data. Both miR-96-5p and miR-134-5p can target the KRAS signaling pathway (39, 40), which may affect T-cell activation thresholds, enabling responses to autoantigens (41). Calcitriol induced expression of miR-627 and miR-627 reduced proliferation of colorectal cancer cell line (42). Exposure of calcitrol to synoviocytes cultured from RA patients reduced cell proliferation and cytokine production (43). It is possible that miR-627 may be part of this mechanism.
Proposed pathways which promote or are altered by the miRNAs In the panel
Pathway analysis indicates that upstream regulators of panel miRNAs and that panel miRNA targets are involved in RA-related disease processes. This observation also provides proof-of-principle for the methodology we used to develop the panel. Use of non-biased techniques, such as random forest analysis, serves to find novel biomarkers and offer some insight into mechanisms of disease.
Limitations
This study had some limitations. Both discovery and validation cohorts were predominantly patients with established disease (defined as ≥6 months duration (44)); thus, the panel may not differentiate individuals with RA at the time of diagnosis or pre-RA states from control subjects. The panel was robust among those with disease duration <1 year and among those not on DMARDs, however. We did not design the study to develop a diagnostic panel for RA when other inflammatory autoimmune diseases are under consideration, but initial testing of the panel in a small set of patients with SLE suggests that the panel may represent an autoimmunity signature. Future studies examining the panel in a variety of inflammatory autoimmune diseases at varied levels of activity would be helpful to determine if these miRNAs compose an inflammatory autoimmune disease signature rather than an RA signature. The strong relationship between the miRNAs and annotated RA pathways was reassuring, but there are limitations to pathway analysis. In general RA has more extensive annotated pathways than many other inflammatory autoimmune diseases, so it is possible that we do not see as strong a relationship with other inflammatory autoimmune diseases because there are insufficient annotated pathways.
Conclusion
A miRNA panel identified by bioinformatics approaches was able to differentiate between patients with RA and control subjects with reproducibility. Many of the upstream regulators of the miRNAs and many of the miRNA targets regulate RA-related pathways. The panel may represent an autoimmunity signature which is not dependent on active disease.
Supplementary Material
Acknowledgments
Funding: Veterans Health Administration CDA IK2CX001269, Arthritis Foundation Delivering on Discovery grant, Alpha Omicron Pi, NIH Grants: P60 AR056116, P01 HL116263 and CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Disclosures: None
References
- 1.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of micrornas on protein output. Nature 2008;455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. Micrornas: Target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007, 9:654–9. [DOI] [PubMed] [Google Scholar]
- 4.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microrna expression in human peripheral blood microvesicles. PloS one 2008;3:e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Micrornas are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating micrornas independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating micrornas as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Zhou X, Zhang Y, Zhu X, Liu H. Systematic review and meta-analysis of diagnostic accuracy of mirnas in patients with pancreatic cancer. Dis Markers 2018;2018:6292396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating micrornas as potential cancer biomarkers: The advantage and disadvantage. Clin Epigenetics 2018;10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free micrornas in cancer. Nat Rev Clin Oncol 2014;11:145–56. [DOI] [PubMed] [Google Scholar]
- 11.Alevizos I, Illei GG. Micrornas as biomarkers in rheumatic diseases. Nat Rev Rheumatol 2010;6:391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ormseth MJ, Solus JF, Vickers KC, Oeser AM, Raggi P, Stein CM. Utility of select plasma microrna for disease and cardiovascular risk assessment in patients with rheumatoid arthritis. J Rheumatol 2015;42:1746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long H, Wang X, Chen Y, Wang L, Zhao M, Lu Q. Dysregulation of micrornas in autoimmune diseases: Pathogenesis, biomarkers and potential therapeutic targets. Cancer Lett 2018;428:90–103. [DOI] [PubMed] [Google Scholar]
- 14.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, et al. Plasma and synovial fluid micrornas as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther 2010;12:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung CP, Oeser A, Raggi P, Gebretsadik T, Shintani AK, Sokka T, et al. Increased coronary-artery atherosclerosis in rheumatoid arthritis: Relationship to disease duration and cardiovascular risk factors. Arthritis Rheum 2005;52:3045–53. [DOI] [PubMed] [Google Scholar]
- 16.Bradham W, Ormseth MJ, Elumogo C, Palanisamy S, Liu CY, Lawson MA, et al. Absence of fibrosis and inflammation by cardiac magnetic resonance imaging in rheumatoid arthritis patients with low to moderate disease activity. J Rheumatol 2018;45:1078–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asanuma Y, Oeser A, Shintani AK, Turner E, Olsen N, Fazio S, et al. Premature coronary-artery atherosclerosis in systemic lupus erythematosus. N Engl J Med 2003;349:2407–15. [DOI] [PubMed] [Google Scholar]
- 18.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The american rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. [DOI] [PubMed] [Google Scholar]
- 19.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1982;25:1271–7. [DOI] [PubMed] [Google Scholar]
- 20.Prevoo ML, van 't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum 1995;38:44–8. [DOI] [PubMed] [Google Scholar]
- 21.Allen RM, Zhao S, Ramirez Solano MA, Zhu W, Michell DL, Wang Y, et al. Bioinformatic analysis of endogenous and exogenous small rnas on lipoproteins. J Extracell Vesicles 2018;7:1506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, et al. Deep sequencing reveals differential expression of micrornas in favorable versus unfavorable neuroblastoma. Nucleic Acids Res 2010;38:5919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fransen J, Creemers MC, Van Riel PL. Remission in rheumatoid arthritis: Agreement of the disease activity score (das28) with the ara preliminary remission criteria. Rheumatology (Oxford) 2004;43:1252–5. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Yuan M, Song L, Zhang X, Geng Q, Zhang H, et al. Expression of dicer in rheumatoid arthritis is associated with disease activity and balances the production of tnfalpha. Mol Med Rep 2017;16:1590–5. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating micrornas: A cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alsaleh G, Nehmar R, Bluml S, Schleiss C, Ostermann E, Dillenseger JP, et al. Reduced dicer1 expression bestows rheumatoid arthritis synoviocytes proinflammatory properties and resistance to apoptotic stimuli. Arthritis Rheumatol 2016;68:1839–48. [DOI] [PubMed] [Google Scholar]
- 29.Ouboussad L, Hunt L, Hensor EMA, Nam JL, Barnes NA, Emery P, et al. Profiling micrornas in individuals at risk of progression to rheumatoid arthritis. Arthritis research & therapy 2017;19:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krintel SB, Dehlendorff C, Hetland ML, Horslev-Petersen K, Andersen KK, Junker P, et al. Prediction of treatment response to adalimumab: A double-blind placebo-controlled study of circulating microrna in patients with early rheumatoid arthritis. Pharmacogenomics J 2016;16:141–6. [DOI] [PubMed] [Google Scholar]
- 31.Lin J, Huo R, Xiao L, Zhu X, Xie J, Sun S, et al. A novel p53/microrna-22/cyr61 axis in synovial cells regulates inflammation in rheumatoid arthritis. Arthritis Rheumatol 2014;66:49–59. [DOI] [PubMed] [Google Scholar]
- 32.Lu W, You R, Yuan X, Yang T, Samuel EL, Marcano DC, et al. The microrna mir-22 inhibits the histone deacetylase hdac4 to promote t(h)17 cell-dependent emphysema. Nat Immunol 2015; 16:1185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu XN, Ye YX, Niu JW, Li Y, Li X, You X, et al. Defective pten regulation contributes to b cell hyperresponsiveness in systemic lupus erythematosus. Sci Transl Med 2014;6:246ra99. [DOI] [PubMed] [Google Scholar]
- 34.Gabler J, Wittmann J, Porstner M, Renz H, Jack HM, Abram M, et al. Contribution of microrna 24 3p and erk1/2 to interleukin-6-mediated plasma cell survival. Eur J Immunol 2013;43:3028–37. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, Li Y, Liu G, Qi X, Cao X. Microrna-24 inhibits the proliferation and migration of endothelial cells in patients with atherosclerosis by targeting importin-alpha3 and regulating inflammatory responses. Exp Ther Med 2018;15:338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maegdefessel L, Spin JM, Raaz U, Eken SM, Toh R, Azuma J, et al. Mir-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat Commun 2014; 5:5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R, Ma J, Yao J. Molecular mechanisms of the cartilage-specific microrna-140 in osteoarthritis. Inflamm Res 2013;62:871–7. [DOI] [PubMed] [Google Scholar]
- 38.Peng JS, Chen SY, Wu CL, Chong HE, Ding YC, Shiau AL, et al. Amelioration of experimental autoimmune arthritis through targeting of synovial fibroblasts by intraarticular delivery of micrornas 140-3p and 140-5p. Arthritis Rheumatol 2016;68:370–81. [DOI] [PubMed] [Google Scholar]
- 39.Ress AL, Stiegelbauer V, Winter E, Schwarzenbacher D, Kiesslich T, Lax S, et al. Mir-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patients. Mol Carcinog 2015;54:1442–50. [DOI] [PubMed] [Google Scholar]
- 40.Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong FY, et al. Mir-134: A human cancer suppressor? Mol Ther Nucleic Acids 2017;6:140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh K, Deshpande P, Li G, Yu M, Pryshchep S, Cavanagh M, et al. K-ras gtpase- and b-raf kinase-mediated t-cell tolerance defects in rheumatoid arthritis. Proc Natl Acad Sci U S A 2012;109:E1629–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padi SK, Zhang Q, Rustum YM, Morrison C, Guo B. Microrna-627 mediates the epigenetic mechanisms of vitamin d to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice. Gastroenterology 2013;145:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huhtakangas JA, Veijola J, Turunen S, Karjalainen A, Valkealahti M, Nousiainen T, et al. 1,25(oh)2d3 and calcipotriol, its hypocalcemic analog, exert a long-lasting anti-inflammatory and anti-proliferative effect in synoviocytes cultured from patients with rheumatoid arthritis and osteoarthritis. J Steroid Biochem Mol Biol 2017;173:13–22. [DOI] [PubMed] [Google Scholar]
- 44.Singh JA, Saag KG, Bridges SL Jr., Akl EA, Bannuru RR, Sullivan MC, et al. 2015 american college of rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Care Res 2016;68:1–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.