The centromere is the genetic locus responsible for kinetochore assembly and subsequent attachment to the mitotic spindle via kinetochore microtubules. The centromere was originally defined as a reference locus that allowed fungal geneticists to map the location and order of genes based on the frequency of first-division and second-division segregants in meiosis. It can be seen at the cytological level as the primary constriction in condensed chromosomes in mitotic cells (originally documented in 1882 (Flemming, 1882)). The first functional assay for centromere, mitotic stability of mini-chromosomes, was developed 100 years later by Clarke and Carbon in budding yeast (Clarke and Carbon, 1980). Sequence analysis revealed that a mere 125 bp was required for segregation of an entire chromosome (Fitzgerald-Hayes, Clarke, and Carbon, 1982). Isolation of centromeres from numerous organisms over the decades has revealed that budding yeast centromeres are deceptively simple. Centromeres range from point centromeres in S. cerevisiae and other budding yeasts to megabases of complex hierarchical arrays of simple sequence repeats, known as regional centromeres, in fission yeast, plants, and animals. As the field wades its way through the sequence complexity of these centromere DNA repeats, the function of centromeric chromatin remains somewhat enigmatic. Here, we examine the centromere’s biophysical attributes to identify conserved centromeric properties in the face of apparent sequence diversity.
Centromeres vs. Pericentromeres
The kinetochore is the protein complex that physically links dynamic microtubules to the chromosome at the centromere. The centromere is thus the chromosomal location of kinetochore assembly. However, the kinetochore-based definition of the centromere is inconsistent when comparing regional and point centromeres. Regional centromeres are based on the entire DNA sequence containing multiple kinetochore assembly sites as well as portions of DNA where no kinetochore assembly takes place. Point centromere size is strictly defined as the length of the DNA sequence to which a single kinetochore assembles. The field is left with the seeming disparity that centromere DNA size spans 4-5 orders of magnitude across phylogeny (see Table 1). The amount of DNA in regional centromeres on which kinetochores assemble is a small fraction of what is considered the centromere.
Table 1.
Scaling of sister kinetochore separation across phylogeny. Kinetochore separation is the distance between sister kinetochores observed in a number of organisms O. tauri (Gan, Ladinsky, and Jensen, 2011); budding yeast, S. cerevisiae (Lawrimore et al., 2016; Yeh et al., 2008); worm, C. albicans (Maddox et al., 2006); fission yeast, S. pombe (Ding, McDonald, and McIntosh, 1993); fly, D. melanogaster (Venkei et al., 2012); and human, H. sapiens (Salmon et al., 1976)]. Centromere DNA size is defined as the region of DNA required for segregation function or physically located at the primary constriction in condensed mitotic chromosomes. Centromere DNA spans 4-5 orders of magnitude in size from yeast to human, while sister kinetochore separation scales by a factor of 2, from the smallest single-cell eukaryote to multicellular organisms.
| Centromere DNA size | Kinetochore separation in mitosis | |
|---|---|---|
| O. Tauri | ~0.1 kb |  |
| S. cerevisiae | 0.125 kb |  |
| C. albicans | 3-4 kb | ~800 nm |
| S. pombe | 10 kb | ~1,000 nm |
| D. melanogaster | 200-500 kb | ~1,000 nm |
| H. sapiens | 500-1,500 kb |  |
In organisms with regional centromeres, pericentromeric chromatin flanking the centromere is rich in repeated sequences and characterized as constitutive heterochromatin. In organisms with point centromeres and little to no repetitive DNA, the pericentromere in more nuanced. The 30-50 kb of DNA flanking the point centromere is enriched in cohesin and condensin (Blat and Kleckner, 1999; D’Ambrosio et al., 2008; Megee et al., 1999). Furthermore, Histone H4 is hypoacetylated in the 1 kb region flanking the centromere in budding yeast (Choy et al., 2011; Choy et al., 2012), as in fly and human centromeres and pericentromeres (Sullivan and Karpen, 2004). Chromatin in the vicinity of the centromere is therefore distinguishable from bulk chromatin throughout phylogeny. The pericentromere in point centromere organisms is analogous to the centromere in regional centromere organisms.
Shared characteristics of pericentromere/centromeres arise when considering these regions from a spatial perspective. In budding yeast centromeres from all 16 chromosomes are clustered into a disc of ~50 nm x 250 nm at microtubules plus-ends (Dhatchinamoorthy et al., 2019; Lawrimore et al., 2016; Lawrimore et al., 2015; Stephens et al., 2013; Yeh et al., 2008). Furthermore using fluorescent reporter operator arrays (FROS), an 8-kb tetO array inserted 0.5 kb from CEN11 and a 10-kb lacO array 1.8 kb from CEN15 exhibited coordination motion and stretching (Stephens et al., 2013). The motion of the two pericentric arrays was significantly more correlated during metaphase than G1, and the correlated motion was lost upon disruption of proper condensin function (Stephens et al., 2013). Recently, reconstituted condensin has been shown to bind DNA in trans (Terakawa et al., 2017), suggesting that condensin can both extrude loops and crosslink DNA. Reduction of pericentric cohesin resulted in loss of coordinated stretching of the arrays in metaphase (Stephens et al., 2013). Thus, while point centromeres are on individual chromosomes, the pericentromeres of all 16 budding yeast chromosomes are knit together into a highly looped and coordinated network (Lawrimore et al., 2016; Lawrimore et al., 2015; Stephens et al., 2013). For the purpose of comparing the spatial organization of the budding yeast pericentromere to regional centromeres we consider the pericentromere as the chromatin network spanning the distance between separated sister kinetochores in metaphase (Figure 1).
Figure 1.
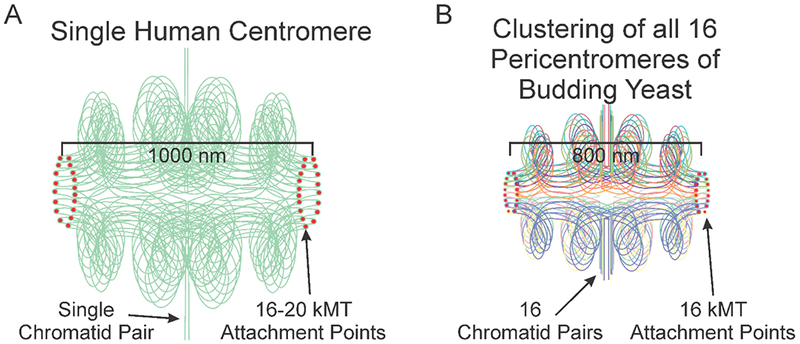
Depictions of a single human centromere and yeast pericentromere. A) A schematic of 2 sister chromatids at the centromere. One pair of sisters are aligned along the vertical axis. Within the centromere multiple loops are folded intramolecularly and biorient relative to one another. The apex of a loop is positioned at the kinetochore plus-ends (red dote). The sister chromatids are in green, highlighting the proposal that 16-20 loops emanate from one pair of sister chromatids in mammals (or organisms with multiple microtubule attachment sites) (Aze et al., 2016; Bloom and Costanzo, 2017), versus the clustering of loops from individual chromosomes (16 in budding yeast) in organisms with unit attachment sites. B) A schematic of the 16 sister pericentromeres in budding yeast. Pairs of sister chromatids are aligned along the vertical axis (vertical straight lines). In the pericentromere (horizontal, bounded by red kinetochore attachment sites), each sister centromere is folded into an intramolecular C-loop (Yeh et al., 2008) that biorient relative to one another that contains several subloops (Stephens et al., 2011). The apex of each C-loop (the 125 bp centromere) is positioned at the kinetochore microtubule plus-ends (red dot). The sister chromatids are color-coded (evident in the colors at the kinetochore microtubule attachment site). The schematics scaled by centromere separation, i.e. the cluster of 16 bi-oriented kinetochores in yeast occupy roughly the same volume as one mammalian kinetochore.
With this definition, the pericentromere can contain genes at a density indistinguishable from the entire genome (e.g. budding yeast) or can be comprised of highly repeated satellite DNA (mammals). The identification of the greater than 100 kinetochore proteins allows for direct visualization of sister kinetochores in a variety of organisms during metaphase. In the budding yeast S. cerevisiae, sister kinetochores of all 16 chromosomes are clustered in a cylindrical array around the central spindle (Figure 1)(Dhatchinamoorthy et al., 2019; Lawrimore et al., 2016; Yeh et al., 2008). The spindle is 1.5 microns in length, kinetochore microtubules are ~350 nm long, yielding sister kinetochore separation of ~800 nm (Pearson et al., 2001)(Table 1). In the regional centromeres of S. pombe, kinetochores of the 3 chromosomes are also clustered around the 3 micron spindle in early metaphase, with sister kinetochore separation of 800 to 1040 nm (Ding, McDonald, and McIntosh, 1993). The common roundworm, C. elegans, exhibits yet another strategy to organize the centromere. Organisms with holocentric centromeres, such as C. elegans have microtubule attachments along the entire length of the condensed chromosome (Melters et al., 2012). The distance between the separated sister kinetochores in these worms is ~1000 nm (Maddox et al., 2006). Holocentricity has evolved numerous times in both plants and animals (Melters et al., 2012). In several of these species where kinetochores were measured, the sister kinetochores are separated by about 1 micron (flower moths, (Wolf, 1994)). In flies, D. melanogaster (Venkei et al., 2012), Megaselia scalaris (Wolf et al., 1994), horses (Carbone et al., 2006), and human (Salmon et al., 1976), sister kinetochores are also separated by 1 micron. Even in the the smallest eukaryote, O. tauri, sister kinetochore separation is 400 nm (Gan, Ladinsky, and Jensen, 2011)(Table 1).
Sequence and Protein Composition of the Pericentromere
Many organisms have active genes within a few hundred base pairs from their point centromeres and therefore have not been considered to have classic “constitutive heterochromatin” within their pericentromeres. However, other common features of spatially defined pericentromeres, such as enrichment of cohesin and condensin, histone modifications, or density of DNA loops, yield a more conserved portrait of pericentromeres across phylogeny. The structural maintenance of chromosome proteins (SMCs) are enriched in the pericentromeres in a number of organisms (D’Ambrosio et al., 2008; Eckert, Gravdahl, and Megee, 2007; Kim and Sauro, 2012). These proteins play key roles in the organization of pericentromeric loops. Centromere-specific histones and histone modifications are also characteristic of pericentromeric heterochromatin. In yeast, this includes H3S10P (Castellano-Pozo et al., 2013), hypoacetylated H4 (Choy et al., 2012), and TSM (tension sensing motif) Lys42, Gly44, and Thr45 of Histone H3 (Deng and Kuo, 2018; Luo et al., 2016); in flies, fission yeast, and mammals, H3K9 (di-, di-, and tri-methylation, respectively (Noma, Allis, and Grewal, 2001)). Perhaps the most notable histone modification of pericentromeric chromatin is the incorporation of a centromere-specific histone H3 variant, CENP-A (Palmer et al., 1987).
The physical nature and distribution of the CENP-A nucleosome is another enigma of the centromere/pericentromere. Reports run the gamut from tetrameric or octameric nucleosomes, short or tall, and right or left-handed wraps of DNA around the nucleosome, (reviewed in (Black and Cleveland, 2011; Verdaasdonk and Bloom, 2011)). It is important to distinguish CENP-A molecules at the kinetochore, versus CENP-A molecules in the pericentromere between the sister kinetochores (distal from microtubule plus-ends). In S. cerevisiae, even with their point centromeres, there are additional CENP-A (Cse4) molecules relative to the 125 bp of CEN DNA (Haase et al., 2013; Hoffmann et al., 2017; Lawrimore, Bloom, and Salmon, 2011). In mammals, the bulk of CENP-A is in the pericentromere region, not at the chromosome surface. Based on the dimensions of the mammalian centromere, we can estimate the surface area to volume ratio. A cylinder that approximates the shape of the two regional centromeres joined together by cohesin of 1 μm in height, defined by the distance between sister kinetochores, and a 0.25 μm radius (Cherry et al., 1989) has a surface-area-to-volume ratio of 1:10. Since kinetochores reside on the surface, i.e. the two ends of the cylinder, the kinetochore proximal surface of the mammalian centromere is ~1/5th of the surface area. If CENP-A is randomly positioned within the mammalian centromere, only 2 – 10% of pericentromeric CENP-A molecules will be proximal to microtubule plus-ends in mitosis. Interestingly, functional assays for CENP-A indicate that only about 10% of the endogenous levels are sufficient to promote chromosome segregation (Liu et al., 2006). This organization of centromeric chromatin is present in S. cerevisiae during metaphase as the pericentromeres of all 16 chromosomes cluster together to form a cylinder of similar dimensions to a single mammalian centromere (Figure 1)(Lawrimore et al., 2016; Yeh et al., 2008; Stephens et al., 2013).
What properties of CENP-A account for its residence at centromeres across phyla? Several physical studies indicate that the CENP-A nucleosomes are more compacted and rigid (Black et al., 2004; Miell et al., 2013). In contrast, functional studies using magnetic tweezers do not find that CENP-A confers any more or less resistance to force (Kim et al., 2016). There is a great deal of interest in the handedness of the DNA as it wraps around the CENP-A nucleosome. A physiological assay for nucleosome configuration is the state of supercoiling in vivo. Linking number reflects the twist (the number of times the strands cross) and the writhe (the number of times the duplexes cross) of the DNA helix. Since linking number is preserved following extraction of proteins, it can accurately be determined in isolated DNA. Furthermore, in the case of budding yeast, very small circular DNA molecules (~2.5 kb) can be introduced into cells, significantly enhancing the accuracy in the measurements (fewer number of total turns in small circles enhance the ability to distinguish changes of 1 or 2 in linking number). Using these small circles, it has been found that there is a positive (right-handed) wrap of centromere DNA around the Cse4/CENP-A-containing nucleosomes (Bloom, Amaya, and Yeh, 1984; Bloom, Fitzgerald-Hayes, and Carbon, 1983; Diaz-Ingelmo et al., 2015; Furuyama and Henikoff, 2009). In the context of regional centromeres, one must acknowledge that there may be heterogeneity in CENP-A structure within the pericentromere. The studies in yeast are restricted to a single nucleosome known to be imbedded in the kinetochore at the site of the microtubule plus-end (Joglekar, Bloom, and Salmon, 2009). In mammals, the bulk of CENP-A is not on the surface of the chromosome, and not proximal to microtubule plus-ends. Numerous studies documenting the canonical octameric CENP-A containing nucleosomes (Dechassa et al., 2011; Hasson et al., 2013; Nechemia-Arbely et al., 2017; Padeganeh et al., 2013) do not exclude the existence of a small subpopulation (2-10%) with differing properties. The inherent variance in bulk methodologies including biochemical, crystallographic, or microscopic measurements preclude ruling out rare, but functionally active subsets. Considering the thermal fluctuations of these complexes, both the DNA and the histone tetramer are more than capable of flipping from positive to negative forms (Hamiche et al., 1996; Vlijm et al., 2017), indicative of flexibility in chirality of nucleosomes. Several recent studies highlight the dynamic properties of CENP-A containing nucleosomes (Bui et al., 2012; Falk et al., 2016; Malik et al., 2018). It is likely that the bulk of CENP-A in pericentromeric chromatin within regional centromeres is organized into a canonical nucleosome. Based on the studies in budding yeast, we propose that the fraction of CENP-A molecules at the surface of the pericentromeres, imbedded in the kinetochore at microtubule plus-ends are dynamic, readily flipped to a right-handed wrap, perhaps stabilized by microtubule-based pulling forces or tension generated by the centromeric chromatin spring. As described below, the pericentromere is far from uniform in its organization.
In addition to cohesin and condensin enrichment and centromere-specific histone variants and modifications, proteins involved in DNA replication and repair also influence centromere synthesis and function. Centromere replication can be binned into early replicating point centromeres (budding yeast, (Feng et al., 2009; Pohl, Brewer, and Raghuraman, 2012)), early replicating regional centromeres (fission yeast, (Hayashi et al., 2009)) and late replicating regional centromeres (mammalian cells, (Shelby, Monier, and Sullivan, 2000)). Regional centromeres face the additional challenge of extensive repeats of simple sequence DNA. Repeat sequences are notoriously problematic for replication machinery due to their propensity to adopt alternative configurations (e.g. hairpins, plectonemes, etc.) (Branzei and Foiani, 2010). Interestingly, both the non-repetitive point centromeres and regional centromeres challenge the smooth progression of the replication fork. In budding yeast, replication forks stall through the 125 bp CEN DNA element (Greenfeder and Newlon, 1992; Hodgson, Calzada, and Labib, 2007), in a TOF1 (topoisomerase interacting factor 1), but not MRC1-dependent fashion (Hodgson, Calzada, and Labib, 2007). In fission yeast, transcriptional stalling contributes to centromere chromatin assembly (Catania, Pidoux, and Allshire, 2015). While it is not possible to measure replication through the microtubule plus-end proximal DNA in a regional centromere, replication forks stall as they traverse repetitive regions of the genome (Mizuno et al., 2009; Voineagu et al., 2008; Voineagu et al., 2009). Accordingly, repeat-rich pericentromeric chromatin is sensitive to replication stress-induced breaks and rearrangements (Simi et al., 1998). Thus, while centromeres are “strong” in their ability to resist microtubule-based pulling forces, they are “fragile” from the perspective of genome integrity.
It comes as no surprise that centromeres are enriched in various repair and recombination proteins. In C. albicans, the homologous repair proteins Rad51 and Rad52 physically bind CENP-A (Cse4) and have been proposed to regulate CENP-A deposition (Mitra et al., 2014). Reconstitution of centromeric chromatin with X. laevis extracts onto BAC chromosomes containing human centromeric α-satellite DNA allowed Aze et al., (Aze et al., 2016) to identify the proteome associated with centromere DNA. In addition to SMCs, DNA repair proteins such as mismatch repair (MSH2-6, MRX, XRCC1, PICH helicase and MUS81 endonuclease) were found. Most interestingly was the under-representation of several key proteins, such as the single-stranded DNA binding protein RPA and ATR activator TopBP1. These studies point to a potentially distinct biochemical repertoire of repair proteins within the pericentromeric region of the chromosome.
Pericentromeric-specific biochemistry (signaling and recombination)
Replication blocks are frequently encountered throughout the genome. These blocks are monitored by the ATR checkpoint kinase that prevents fork collapse and DNA breakage (Iyer and Rhind, 2017). A recent study estimated there are about 500 ATR dependent sites in the mouse and human genome (Shastri et al., 2018). Based on an average spacing of ~50 kb/origin, these blockage sites occur at a frequency of about 1 per every 200 origins. In the pericentromere and other regions rich in repeat sequences, the frequency of these encounters is likely significantly higher. This poses an interesting paradox. The checkpoint is activated at polymerase pause sites, but in regions of high pausing, this could lead to a futile cycle, inordinately delaying cell cycle progression. Two new findings indicate that the canonical ATR checkpoint is suppressed in the centromere (Aze et al., 2016), and instead, a mitotic specific and R-loop dependent ATR checkpoint operates within the centromere (Kabeche et al., 2018). In addition to the unique set of repair proteins in the centromere proteome (Aze et al., 2016), Aze et al. also showed that the positively supercoiled topology of pericentromeric chromatin contributed to the suppression of ATR activation. They noticed that replicated centromere DNA formed single strand bubbles, which, due to positive supercoiling, prevent cross-linking in their assay (Aze et al., 2016). To test the physiological relevance of supercoiling, they restored checkpoint signaling to the reconstituted centromeres through the inhibition of topoisomerase activity. Kabeche et al. (Kabeche et al., 2018) also found a non-canonical ATR pathway that contributes to genome integrity. Kabeche et al. (2018) propose that ATR is recruited through RPA to centromeric R-loops. In this way, canonical ATR can be suppressed, allowing for centromere DNA replication. Mitotic ATR signaling through R-loops reveals an alternative mechanism to monitor centromere DNA replication or kinetochore assembly that avoids direct detection of fork progression. In addition, the mitotic ATR pathway may shed light on potential functions of centromere transcription in these processes.
Meiotic recombination is suppressed at centromeres and pericentromeres throughout phylogeny (Blitzblau et al., 2007; Buhler, Borde, and Lichten, 2007; Mahtani and Willard, 1998). As the sequence and protein composition of the pericentromere becomes accessible, we can define the source of this suppression. In budding yeast, it is further possible to separate control of recombination between pericentromere repeats from potentially centromere- or kinetochore-specific sources, due the single-copy nature of the budding yeast pericentromere. Several studies have identified factors, including Zip1 (a synaptonemal protein) and Sgs1 (a DNA helicase important for repair), that regulate the level of recombination in the pericentromere and chromosome arms (Chen et al., 2008; Rockmill, Voelkel-Meiman, and Roeder, 2006). Centromere-specific factors were identified in a study of pericentromeric recombination in various cohesin loading mutants (Vincenten et al., 2015). Ctf19 is a member of the CCAN, one of several kinetochore complexes, and proximal to the chromatin. Ctf19 has been shown to recruit cohesin to the centromere and is responsible for the 3-fold enrichment of pericentromeric cohesin (Hinshaw et al., 2017). In the absence of ctf19, there is a 20-fold increase in recombination within the pericentromere (Vincenten et al., 2015). Vincenten et al., (2015) were able to distinguish Ctf19’s function in cohesin recruitment in suppression of DSB vs cross-over (CO) formation. Interestingly, suppression of DSB formation was independent of cohesin, while cohesin was largely responsible for suppression of pericentromeric crossovers. Cohesin has also been shown to suppress COs in vegetatively growing cells (Covo et al., 2014). These studies point to the ability of cells to exert spatial control of pericentromeric recombination.
A comparative study of the properties of centromeric and nucleolar repeat arrays
The repeated array of rDNA within the nucleolus is perhaps the best-studied case of differential recombinational control within nuclear subcompartments. In 1981 Zamb and Petes showed that rDNA recombination was independent of the major protein involved in homologous recombination, Rad52 (Zamb and Petes, 1981). Subsequently, this was attributed to the ability of a large array of repeats to compensate for the lack of rad52 (Ozenberger and Roeder, 1991). The repeat array has ample opportunity for interaction between sister strands even in the absence of repair proteins. A key discovery relating to spatial control was made when Torres-Rosell (Torres-Rosell et al., 2007) used cell biological approaches to show that repair foci containing Rad52 were excluded from the nucleolus. Torres et al., (2007) inserted a DNA double-strand cut site within a rDNA repeat. Upon induction of the I-SceI enzyme, the severed DNA moved out of the nucleolus, where it complexed with Rad52 foci in the nucleoplasm for DNA repair.
The parallel between a unique suite of repair proteins within the pericentromere (Aze et al., 2016; Kabeche et al., 2018; Janssen et al., 2019) and the nucleolus (Torres-Rosell et al., 2007) is one of just many functionalities in common between the two. One of the important functionalities relative to centromere biology is the tendency of repeated sequences to self-organize, a feature that is often marked by spatial segregation and frequent self-interaction (Hult et al., 2017). This kind of self-interaction lends itself to intra-molecular interactions, otherwise known as loops. Tandem repeat sequences are more prone to fold into hairpins and loops due to breathing of the helices and transient intra- or inter-molecular interactions between single strands of complementary sequences. The temporal feature of fork pausing through centromere repeat arrays or tandem arrays of loops will only increase the duration of transient interactions and serve to enhance the occurrence of a network of loops.
Pericentromeric loops in budding yeast, formed by the SMC protein complex condensin (Lawrimore et al., 2015; Stephens et al., 2013; Lawrimore et al., 2018; Stephens et al., 2011), are key to both the mechanical properties of the centromere and the mechanisms by which tension is generated between sister chromatids. Recombination between centromere DNA repeats through loop formation that stabilizes specific secondary structures has also been proposed (McFarlane and Humphrey, 2010). In mammalian centromeres, direct observation of loops of centromeric α-satellite DNA as demonstrated in (Aze et al., 2016) reflects the organization of centromeric chromatin and chromosome arms in mitosis (Gibcus et al., 2018).
The loops in the reconstituted Xenopus extract system were found to be positively supercoiled (Aze et al., 2016), a feature that may be related to their function in signaling (see above) as well as to mechanical aspects of mitosis. From the mechanical perspective, positive supercoils confer several properties that directly promote segregation. Positive supercoiling plays a role in promoting chromosome segregation through centromere decatenation in yeast (Baxter et al., 2011) and mammalian cells (Bizard et al., 2019). Using small circular centromere plasmids, Baxter et al. (2011) showed that positive supercoiling was able to bias topoisomerase II to decatenate versus relax replicated circles. This function required condensin, which itself drives positive supercoiling (Kimura et al., 1999). Bizard et al. (2019) showed that the PICH (Plk1-interacting checkpoint helicase) helicase extrudes DNA loops, redistributing the torsional stress in a way that leads to relaxation of the negatively supercoiled substrate via Top3A, which translates into positive supercoils after release of the loop (Bizard et al., 2019). DNA topoisomerase III is enriched within S. pombe centromeres, where it has also been hypothesized to control centromere topology (Norman-Axelsson et al., 2013).
Positively supercoiled molecules are more resistant to mechanical stress (Allemand et al., 1998; Lipfert, Klijnhout, and Dekker, 2010; Strick, Bensimon, and Croquette, 1999). Using magnetic tweezers, Strick et al., (Strick, Bensimon, and Croquette, 1999) showed that positively supercoiled DNA exhibits a non-linear response to force. At low force regimes (< 0.5 pN) the DNA molecules contract against pulling forces if they are positively or negatively supercoiled. As negative supercoils relax, the molecule can be extended, and as positive turns are introduced, the molecule contracts. In contrast, at higher force regime (1 pN), negatively supercoiled and relaxed molecules extend to the same length, while positive supercoils contract into plectonemic structures as a function of additional positive turns. In other words, positively supercoiled DNA is more resistive to extensional force between 0.5 and 3 pN than negatively coiled DNA (Fig. 2). 1-3 pN is well within the force regime these molecules experience in mitosis (Akiyoshi et al., 2010; Asbury et al., 2006). This attribute of positively supercoiled DNA, contracting upon increased force, reflects additional mechanisms that cells may exploit to tune the pericentromeric spring properties. At 3 pN or greater, a transition of positively supercoiled DNA to an extremely extended form (75% longer than b-form, 2.6 bp/turn) is observed (Allemand et al., 1998; Lipfert, Klijnhout, and Dekker, 2010; Strick, Bensimon, and Croquette, 1999) (Fig. 2). This highly extended form of DNA snaps into a conformation where the phosphate backbone is interwound, with the bases exposed (originally proposed by Pauling (Pauling and Corey, 1953)). Again, molecular mechanisms that provide greater flexibility in force-extension within the pericentric or centromeric regions of chromosomes are likely to be exploited in ways that lead to a robust and highly accurate process of chromosome segregation.
Figure 2.
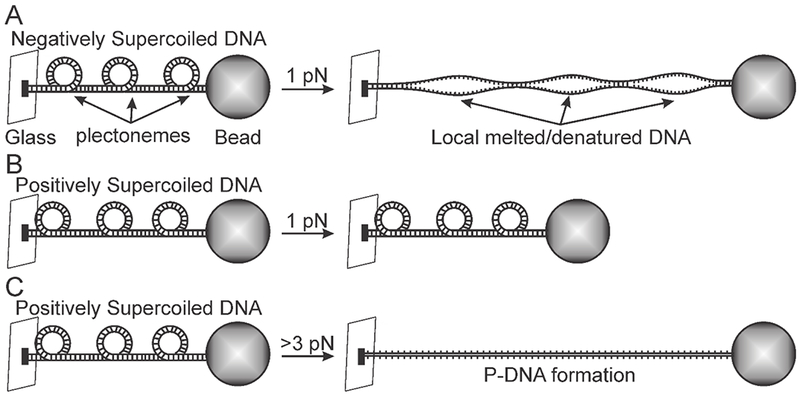
Positively supercoiled DNA is more resistant to extension then negatively supercoiled DNA.
A) A depiction of a single molecule experiment with magnetic tweezers. Negatively supercoiled DNA is pulled with 1 pN of force and is extended. Upon extension local regions of the DNA melt/denature. B) Positively supercoiled DNA is pulled with 1 pN of force but resists extension. C) Positively supercoiled DNA is pulled with greater than 3 pN of force and is extended. Upon extension the DNA is reorganized into Pauling DNA (P-DNA).
Finally, a consequence of intimate strand pairing in regions of high repeat density, such as α-satellite repeats in regional centromeres, is their tendency to phase separate (Hall, Ostrowski, and Mekhail, 2019). While a great deal of attention has been devoted to liquid-phase separation, another well-known phenomena is polymer-phase separation (Erdel and Rippe, 2018; Rubinstein and Colby, 2003). Polymer phase separation is driven through condensin-based dynamic protein cross-linking (Hult et al., 2017), or dynamic strand annealing between α-satellite repeats within regional centromeres. The recognition of subdomains within the nucleus has spawned a flurry of studies (Bergeron-Sandoval, Safaee, and Michnick, 2016; Weber, 2017). These findings beg the question as to whether nuclear subdomains confine a set of discrete biochemical reactions. It has been known for decades that centromeres and pericentromeres are regions of reduced meiotic recombination (Mather, 1936; Mather, 1939). With the full repertoire of kinetochore and centromere proteins we are in a position to dissect the protein contributions responsible for distinctive recombination rates at the centromere. The occurrence of double-strand DNA breaks (DSB’s) is reduced, but not to an extent that accounts for reduction of recombination at centromere (Blitzblau et al., 2007; Buhler, Borde, and Lichten, 2007). Thus it has been suggested that repair choice within the pericentromere might be distinct from choices made along chromosome arms (Chen et al., 2008). Toward this end, Vincenten et al., (Vincenten et al., 2015) found that the level of cohesin within the pericentromere dictates the choice between intra- versus inter- homology in meiosis (Kuhl and Vader, 2019). In addition, Covo et al., (Covo et al., 2014; Covo et al., 2010) have demonstrated that cohesin exerts a similar choice for biasing the repair outcomes to those between sister chromatids. Thus, the pericentromere is likely to reflect a unique nuclear compartment engaged in directing repair outcomes in addition to its role in segregation.
Mitotic function of pericentromeric chromatin in S. cerevisiae
Above, we have argued for a spatial-based, expanded definition of a pericentromere that defines any chromatin that lies between separated sister kinetochores as pericentromeric. Since the kinetochores are clustered together in S. cerevisiae and the pericentromeres of different chromosomes display coordinated motion and stretching (Stephens et al., 2013) , we propose that the ensemble of all budding yeast pericentromeric chromatin, containing 16 kinetochore microtubule attachments, can be considered a single regional centromere (Figure 1). For clarity, we will refer to this ensemble region as the pericentromere. Given the similarities between the yeast pericentromere and regional centromere, we believe the functional significance of the pericentromere in S. cerevisiae is applicable to regional centromeres. In order to address this significance, we will need to highlight the role pericentromeric chromatin plays in the process of chromosome segregation.
When observed from a cellular perspective, chromosome segregation appears simple and elegant. The process of chromosome segregation is often presented as follows: Chromosomes are replicated and held together by the ring-like SMC protein complex, cohesin. Upon mitotic entry, chromosomes are compacted by another ring-like SMC protein complex, condensin. Replicated chromosomes are attached to the mitotic spindle via the kinetochore. Sister chromatids, joined by cohesin, are pulled in opposing directions by kinetochore microtubules emanating from the opposite pole of a bi-polar spindle, resulting in sister chromatin bi-orientation. Proper bi-orientation of a sister chromatid pair results in tension between the sister kinetochores, resulting in a cascade of protein modifications effectively damping the spindle checkpoint that delays cell cycle progression until the chromosome is ready for segregation. Once all sister chromatids are bi-oriented, cohesin is cleaved, the mitotic spindle elongates and one copy of each chromosome is passed to the mother and daughter cells.
A key aspect of this narrative is tension, both within the kinetochore and across the pericentromeric chromatin upon sister chromatid bi-orientation. Tension is the key to error detection and to the proper timing of cohesin cleavage (Biggins, 2013; Bloom and Yeh, 2010; Joglekar, 2016; Joglekar and Kukreja, 2017; Musacchio and Salmon, 2007). Members of the COMA kinetochore complex (Ctf19) are phosphorylated by the Dbf4-dependent-kinase (DDK) and together with the cohesin loader Scc2/4 facilitate cohesin loading at budding yeast centromeres (Hinshaw et al., 2017). Sister chromatin cohesion, via the cohesin complex, resists the mitotic pulling forces, thereby generating the tension that is transmitted to the kinetochore. However, several discoveries suggest cohesion via a ring-like protein complex that directly links sister chromatids at the centromere cannot be the mechanism for tension between sisters. The observation that sister kinetochores are 1 micron apart (Table 1) is incompatible with a 40 nm protein ring holding one or two chromatids as the basis for cohesion. Thus a more complex organization of cohesin within the pericentromere is required to understand how tension is achieved across micron rather than nanometer scale. Below, we discuss the biophysical results that describe how pericentromeric chromatin is organized into a highly looped and cross-linked region, resembling a bottle-brush, capable of altering tension in the pericentromere to regulate chromosome segregation.
The case for complicating sister chromatin cohesion
In budding yeast, cohesin is three-fold enriched in pericentromeric chromatin during mitosis as measured by chromatin immunoprecipitation (Blat and Kleckner, 1999; D’Ambrosio et al., 2008; Glynn et al., 2004; Weber et al., 2004) and live-cell, quantitative fluorescence microscopy (Yeh et al., 2008). The default assumption is sister centromeres are physically proximal due to direct cohesion. However live-cell imaging using fluorescent reporters for centromere-proximal DNA (arrays of lac operator visualized with lac repressor-GFP fusion protein (Robinett et al., 1996; Straight et al., 1996)) reveals the opposite. Fluorescently labeled sister chromatids marked within their pericentromeres are separated in mitotic cells (Pearson et al., 2001; Goshima and Yanagida, 2000; He, Asthana, and Sorger, 2000; Tanaka et al., 1999). Moreover, fluorescently labeled CSE4/CENP-A appears as two-foci during mitosis (Pearson et al., 2003). As discussed above, the distance between separated centromeres is conserved across phylogeny (Table 1 (Bloom, 2014)). The extreme conservation of the distance of kinetochore separation indicates a fundamental mechanism of chromosome segregation.
Tension and the pericentromeric chromatin landscape
The pericentromere physically responds to changes in tension. The microtubule depolymerizing drug nocodazole is frequently used to synchronize budding yeast cells in metaphase of mitosis (Jacobs et al., 1988). High-doses of nocodazole, which is similar to the microtubule poison benomyl, collapse the mitotic spindle and reduce tension on the pericentromeric chromatin (Eckert, Gravdahl, and Megee, 2007). Simply reducing tension via low-dose treatment of benomyl results in the expansion of the distribution of pericentromeric cohesin and pericentromeric chromatin (Haase et al., 2012). Even with the spindle still intact, low doses of benomyl significantly decreases tension within the outer-kinetochore (Suzuki et al., 2016). Upon low-benomyl treatment, the distribution of the inner kinetochore proteins Cse4 and Ame1 are narrowed perpendicular to the spindle axis (Haase et al., 2012) and the dynamics of pericentromeric chromatin are reduced (Haase et al., 2012). Thus, the pericentromere is not a passive structure; it is responsive to changes in tension.
Alterations to the pericentromeric region are dependent on the spindle assembly checkpoint (Haase et al., 2012). One of the major spindle assembly checkpoint kinases, Bub1, is responsible for recruiting the protector of cohesin (shugoshin, Sgo1) to centromeres through phosphorylation of histone H2A at serine 121 (Kawashima et al., 2010). In budding yeast, Sgo1 and the phosphatase, Rts1PP2A ensure proper pericentromeric condensin and Ipl1AuroraB localization (Peplowska, Wallek, and Storchova, 2014). Sgo1 (Eshleman and Morgan, 2014; Nerusheva et al., 2014) and Bub1 (Nerusheva et al., 2014) themselves are removed from the pericentromeric region upon sister-chromatid bi-orientation. The kinase activity of Bub1, the phosphorylation of serine 121 of H2A, Sgo1, and RTS1 are all required for preventing premature silencing of the spindle assembly checkpoint in the absence of tension (Jin, Bokros, and Wang, 2017). The alterations to the pericentromeric chromatin were suppressed in the absence of Bub1 or Sgo1 or if S121 of histone H2A was rendered non-phosphorylatable (Haase et al., 2012). Thus, the pericentromere is actively responding to spatial and mechanical cues throughout metaphase using the Bub1/Sgo1/Rts1/H2A tension-sensing system.
Condensin in the pericentromere
The discovery that condensin can extrude loops on DNA (Ganji et al., 2018; Terakawa et al., 2017) suggests that the pericentromeric region may be more intricately involved in tension maintenance than simply acting as a tension sensor. Previous work had observed that the spindles of metaphase yeast cells are elongated when histone H3 is depleted (Bouck and Bloom, 2007), when pericentromeric cohesin was depleted, or when condensin was disrupted with the brn1-9 temperature sensitive allele of condensin’s kleisin subunit (Stephens et al., 2011; Bouck and Bloom, 2007). Cohesin depletion and condensin disruption, but not H3 depletion, resulted in more variable metaphase spindle lengths, suggesting cohesin and condensin stabilize the chromatin in some way (Stephens et al., 2011).
When imaging live budding yeast cells, the approximate position of the spindle can be determined by fluorescently labeling a component of the spindle pole body, e.g. Spc29-RFP or Spc42-RFP. During metaphase of mitosis, the duplicated spindle pole bodies (SPBs) will appear as two foci inside the mother cells. The two SPB foci can be used to establish a biologically relevant “spindle-axis.” By convention, the axis parallel to the spindle corresponds to the X-axis and the axis perpendicular to the spindle is the Y-axis. In wild-type cells, fluorescently labeled pericentromeric chromatin appears as a focus 90% of the time, and as an elongated or stretched signal, whose long axis is parallel to and aligned along the spindle axis 10% of the time (Stephens et al., 2011). Both depletion of pericentromeric cohesin or disruption of condensin resulted in over 50% of all pericentromeric chromatin signals to appear elongated or stretched. When fluorescently labeled, pericentromeric chromatin appears as a focus, those foci are radially displaced, either above or below, the spindle axis (Stephens et al., 2011). However, when the pericentromeric chromatin signal is stretched, the signals are less radially displaced from the spindle axis (Stephens et al., 2011). Thus, when pericentromeric chromatin is under sufficient tension to extend the signal, the chromatin localizes closer to the spindle axis. Therefore, 90% of pericentromeric chromatin is not under as much tension as a subset of chromatin near the spindle axis, indicative of heterogeneity in tension within the pericentromere (Figure 3).
Figure 3.
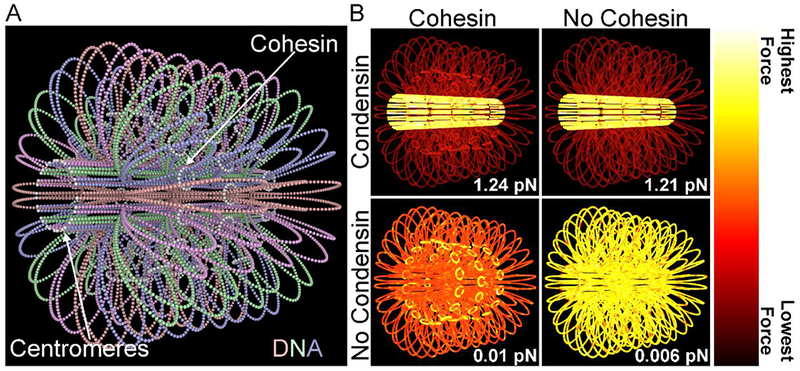
Polymer model of budding yeast pericentromere. A) A three-dimensional, polymer model of the budding yeast pericentromere. Cohesin are the white rings composed of 16 masses. The DNA is looped by the condensin complex (condensin not shown). Point centromeres are in white. The DNA is extended to its rest length to emphasize the highly looped nature of the DNA in the model. This visualization uses the simulation date from Lawrimore et al. (Lawrimore et al., 2016). B) Three-dimensional, polymer models of the budding yeast pericentromere with cohesin and condensin (Top Left), without cohesin and with condensin (Top Right), with cohesin and without condensin (Bottom Left), and without cohesin and without condensin (Bottom Right). Coloring of models is based on the average amount of force on each mass over the course of a simulation. The forces listed in the bottom right corner of the panels are the average entropic spring force on each point centromere. These are visualization of the simulations from Lawrimore et al. (Lawrimore et al., 2016) and tension data from Lawrimore et al. (Lawrimore et al., 2018).
Despite having similar physical localizations, budding yeast cohesin and condensin have a distinct geometric localization in the pericentromeric region (Stephens et al., 2011). Condensin is proximal to the spindle axis, co-localizing with the pericentromeric chromatin that appears extended when labeled, while cohesin is more radially displaced, co-localizing with the pericentromeric chromatin that appears as foci (Stephens et al., 2013; Lawrimore et al., 2018) . Given the disparate geometric positions of cohesin and condensin, it was hypothesized that chromatin in the pericentromeric region is highly looped, with condensin forming the loops proximal to the spindle microtubule axis (Lawrimore et al., 2016; Lawrimore et al., 2015; Yeh et al., 2008; Stephens et al., 2013; Lawrimore et al., 2018).
Pericentromeres are comprised of DNA loops
Fluorescently labeled pericentromeric chromatin is not static during metaphase in budding yeast. The two sister foci exhibit confined diffusion (Pearson et al., 2001; Goshima and Yanagida, 2000). Since pericentromeric foci that appear as foci are geometrically displaced relative to fluorescently labeled kinetochore proteins on the spindle axis (Anderson et al., 2009), the impact that kinetochore microtubule dynamics has on pericentromeric chromatin motion was called into question. Treatment of budding yeast cells with low-doses of the microtubule depolymerization drug, benomyl, reduces the dynamics of the microtubules five-fold (Pearson et al., 2003). However, the motion of pericentromeric chromatin foci was only reduced by 21%, not the expected 80% (Lawrimore et al., 2015). Close examination of the foci showed that the type of motion that both benomyl and depletion of microtubule plus-end binding protein Stu2 eliminated from the pericentromeric chromatin was not the random jostling motion, but the sporadic, directed motion parallel to the spindle (Lawrimore et al., 2015). Thus, the motion of the pericentromeric chromatin, which appear as foci, appears largely divorced from the fluctuations of kinetochore microtubules. Stretched signals of pericentromeric chromatin are localized proximal to the spindle axis (Stephens et al., 2011), suggesting that a subset of pericentromeric chromatin is under increased tension relative to the radially-displaced pericentromeric chromatin. To characterize the force exerted on pericentromeric chromatin, a fluorescently labeled, conditionally dicentric plasmid (Dewar et al., 2004) was monitored in live yeast cells during mitosis (Lawrimore et al., 2015). For metaphase cells, whose spindles were between 500 and 2000 nm, the 5.5-kb tetO/TetR-GFP array appeared elongated 35% of the time with a mean signal length of 630 nm. Surprisingly, cells treated with low doses of benomyl still exhibited elongated signals 28% of the time and had a mean signal length of 635 nm (Lawrimore et al., 2015). Thus, for metaphase cells, reduction in tension on the kinetochore (Suzuki et al., 2016), presumably from a drop in spindle force, did not strongly affect pericentromeric chromatin extension. Moreover, upon depletion of pericentromeric condensin, the pericentromeric regions of sister chromatid separate perpendicular to the spindle in 30% of the cells (Snider et al., 2014). These results suggest that an additional force besides microtubule-based pulling drives the separation of sister centromeres.
One possible explanation of the force that separates sister centromeres, the 120-bp sequence wrapped around the CSE4/CENP-A nucleosome in budding yeast, is the molecular crowding of the pericentric chromatin due to the high concentration of chromatin in a relatively small cellular volume. In budding yeast, the centromeres are clustered together due to their attachment to the microtubule spindle during metaphase of mitosis (Anderson et al., 2009). The geometric distribution of pericentromeric cohesin has been measured to adopt a “barrel-like” distribution of 550 nm in length (parallel to spindle) and 500 nm in diameter (perpendicular to the spindle) (Stephens et al., 2013). The estimated size of the pericentromeric enrichment of cohesin has been measured to be 20 – 50 kb surrounding the centromere (Blat and Kleckner, 1999; D’Ambrosio et al., 2008; Glynn et al., 2004; Weber et al., 2004). If we assume an average pericentromeric cohesin region size of 35 kb, there is approximately 10.2 Mb/μm3 of DNA in this geometric region. In contrast, the average DNA density inside a budding yeast nucleus, whose nuclear volume is approximately 2.91 μm3 (Jorgensen et al., 2007), is 4.8 Mb/μm3. Since chromatin in this region is more compact than in other regions of the genome, the chromatin may be balancing a compaction force against an expansion force driven by molecular crowding. The pericentromeric region expands radially, i.e. perpendicular to the spindle, upon histone depletion, pericentromeric cohesin depletion, and condensin disruption (Stephens et al., 2011). Depletion of pericentromeric condensin on a single chromosome, via tDNA deletion, also resulted in radial expansion of that chromosome’s pericentromeric region (Snider et al., 2014). As previously stated above, pericentromeric radial expansion occurs upon treatment of cells with low-doses of benomyl in a Bub1- and Sgo1-dependent manner (Haase et al., 2012). Whether the radial expansion of the pericentromeric region occurs through loss of tension or directly through loss of cohesin or condensin remains an open question. However, in silico experiments of the pericentromere demonstrate the vastly different effects of cohesin and condensin on the tension landscape of pericentromeric chromatin (Lawrimore et al., 2016).
Building a simulated pericentromere
Detailed measurements of the cohesin and condensin distributions from live cell fluorescent images (Stephens et al., 2013; Stephens et al., 2013; Stephens et al., 2013) and the position of fluorescently marked pericentromeric DNA allowed for the creation of three-dimensional model of the budding yeast pericentromere (Figure 3). A highly looped model of the pericentromere was simulated using the polymer dynamics simulator ChromoShake (Lawrimore et al., 2016). The cohered arm region of the sister chromatid was not simulated. Instead, the ends of the pericentromeric regions were directly joined to mimic the sister chromatin cohesion of the arm regions. Pinning two nonconsecutive beads formed chromatin loops, as a surrogate for the loop extrusion activity of condensin. The loops were cross-linked by passing two chromatin strands inside one ring to mimic the 40 nm cohesin protein ring. The 40 nm cohesin ring was large enough to where it could topologically encircle both strands and still passively diffuse. The simulated pericentromere lacks any ATP-dependent forces, including microtubule dynamics. The model recapitulated the motion and radial displacement of a 10-kb lacO/LacI-GFP array placed 1.8 kb from CEN15 of cells treated with sodium azide and deoxyglucose to deplete ATP, mimicking the lack of ATP-based forces in the model (Lawrimore et al., 2015). Previously, the motion of two fluorescent reporter operator systems (FROS) were placed in the pericentromeric region on different chromosomes. The motion of the two FROS arrays were correlated in untreated, metaphase budding yeast cells, but that correlated motion was lost in the condensin temperature sensitive mutant, brn1-9 (Stephens et al., 2013). In the simulated pericentromere, coordinated motion was lost upon elimination of condensin-mediated loops and upon elimination of cohesin-mediated crosslinks between chromatin loops (Lawrimore et al., 2015). Elimination of cohesin in the simulation did not alter the radial displacement of the simulated FROS arrays unless condensin was absent from the simulations (Lawrimore et al., 2016). Thus, both loop formation and chromatin crosslinking with cohesin can radially compress pericentromeric chromatin; although, cohesin’s ability to compact chromatin was masked by chromatin loops in the model.
Given that stretched pericentromeric FROS array signals are less radially displaced than signals that are isotropic foci, we queried the tension landscape using the simulated pericentromere model. When condensin-mediated chromatin loops are in present in the model, the tension is greater in the chromatin along the spindle axis versus in the chromatin that is radially displaced in loops (Figure 3B)(Lawrimore et al., 2018). When condensin-mediated loops are eliminated, tension is greatest in the cohesin rings (Figure 3B)(Lawrimore et al., 2018). When both cohesin and condensin-mediated loops are eliminated, tension is evenly distributed amongst the pericentromeric chromatin (Figure 3B)(Lawrimore et al., 2018). In the simulated pericentromere, the chromatin is not under any strain, meaning the polymer is initialized at rest length. Therefore, in the absence of any simulated thermal motion, there would be no inward force. However, when the pericentromere is exposed to thermal forces, there is a net inward force of 1.24 pN (Lawrimore et al., 2018). This inward force is reduced to 1.21 pN upon loss of cohesin (sister cohesion still intact), to 0.0097 pN upon loss of condensin, and to 0.0055 pN upon loss of both cohesin and condensin (Lawrimore et al., 2018). Polymers in a thermal bath coil into a shape of a given volume depending on a variety of factors such as polymer length, stiffness, and branched-configuration (Rubinstein and Colby, 2003). Just as a metal spring compressed or extended past its rest length will exert a net force, a compacted or extended polymer will exert a force to reach its entropically favored volume. Therefore, the 1.24 pN exerted by the simulated pericentromere is due to the entropic crowding forces, not due to direct extension of the polymer itself, i.e. the DNA strand.
Reconstitution experiments found that kinetochores bind to microtubules in a catch-bond fashion, with the longest kinetochore-microtubule bond lifetimes at 5 pN. Increasing the force to more than 8 pN or decreasing the force less than 2 pN halves the lifetime of the attachment (Akiyoshi et al., 2010). Therefore, the organization of pericentromeric chromatin into this polymer bottle brush configuration, without extending the chromatin beyond its rest length, may provide ~1/4th of the inward force needed to stabilize kinetochore-microtubule attachments.
Limitations in absolute tension measurements from chromatin motion
It will be informative to directly measure the stiffness of the centromere-spring in vivo. The strategies to date include measuring the rate and period of sister kinetochore oscillations (Jaqaman et al., 2010), thermal fluctuations of centromere-linked lac operator arrays (FROS arrays) (Chacon et al., 2014; Mukherjee et al., 2019) and thermal fluctuations of a centromere-protein tag (Harasymiw et al., 2019). Tandem arrays of binding sites for DNA-binding proteins have revolutionized our ability to visualize locus-specific chromatin dynamics (Robinett et al., 1996; Straight et al., 1996). However, an insight from the modeling is that the number of operator repeats influences both the slope and magnitude of motion (see Fig. 5 in (Lawrimore et al., 2018)). This is due to the inherent averaging as the arrays increase in size. Consider that the average motion of a crowd (many operators) may appear static, while the motion of an individual (single operator) will vary greatly. Similarly, Parry et al. (Parry et al., 2014) found both the diffusion constant (μm2/sec) and the scaling exponent (α, MSD) to be size dependent (Fig. S6C in (Parry et al., 2014)). The diffusion constant decreases with increasing size of the particle, while the scaling exponent increases with increasing size of the particle. These findings highlight the power of modeling to guide our intuition in a polymer world dominated by thermal motion and viscous forces.
Condensin loop extrusion and force balance
If the kinetochore-microtubule bond is a catch-bond with a dynamic range of 10-15 pN (Akiyoshi et al., 2010), how can the spindle, with dynamic microtubule plus ends, maintain such a narrow force range? One possibility is that the pericentromeric chromatin can dynamically react to changes in tension. The discovery that reconstituted condensin has the ability to form loops at a staggering rate of 1500 bp/s in vitro (~ 500 nm/s of B-form DNA), coupled with the observation that condensin’s extrusion rate is negatively correlated with substrate tension (Ganji et al., 2018), highlights the interplay between the motor and the substrate in forming the pericentromeric spring. However, pericentromeric condensin’s behavior in live cell experiments remains poorly understood.
A recent study recorded the signal lengths of a fluorescently labeled, conditionally dicentric plasmid over time to characterize how condensin affects the dynamics of pericentromeric chromatin (Figure 4)(Lawrimore et al., 2018). The signal lengths of unreplicated yet bi-oriented dicentric plasmids were pooled to generate a distribution of signal lengths and changes in signal length per 30 second timestep. In WT and pericentromeric cohesin depleted cells (via an mcm21∆ mutation), the signal length distributions exhibited a long right tail, indicating many compact signal lengths with only a few longer signal lengths in the population. In cells with a brn1-9 mutation, the signal length distribution was symmetric and exhibited greater signal lengths on average than WT or mcm21∆ cells (Lawrimore et al., 2018). Thus condensin appeared to directly compact pericentromeric chromatin in vivo.
Figure 4.
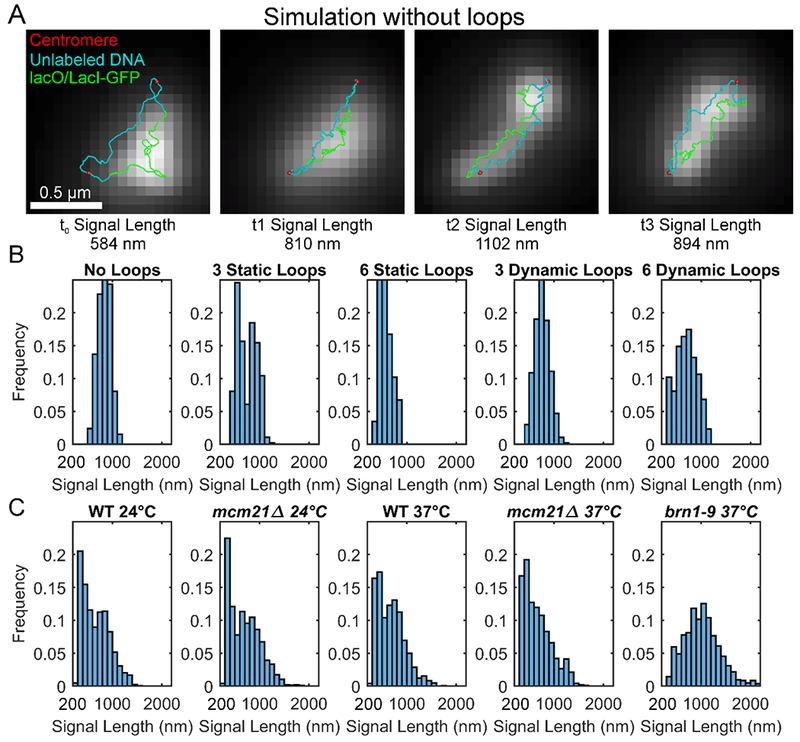
Direct comparison of simulated and experimental dicentric plasmid lengths during mitosis. A) Cartoons of the dicentric plasmid simulations without loops overlaying simulated images of the lacO/LacI-GFP array. The centromeres are in red. The unlabeled DNA is in blue. The lacO/LacI-GFP arrays are in green. The simulated images were constructed by converting the ChromoShake simulation output files (Lawrimore et al., 2016) into XML files that were in turn converted to image stacks using Microscrope Simulator 2 (Quammen et al., 2008). The simulated fluorescence images are maximum intensity projections. B) Histograms of the signal lengths of the simulated fluorescent images of the dicentric ChromoShake simulations. Each simulation condition, i.e. number and type of loops, is composed of 10 separate simulations. Each simulation either contained 0, 3, or 6 loops. These loops could be static, not increasing or decreasing in size, or dynamic, increasing or decreasing in size. The static loops were simulated by randomly joining non-consecutive bead together with a 10 nm spring. The dynamic loops were simulated using the RotoStep program (Lawrimore et al., 2017). In the RotoStep program, loops are unidirectionally extruded by simulated condensin complexes until tension on the substrate increases past a threshold causing loop dissociation, condensin translocation to a new region, and then loop extrusion resumes. The simulation data was previously published in Lawrimore et al. (Lawrimore et al., 2018). C) Histograms of experimental lacO/LacI-GFP signals on a dicentric plasmid during mitosis. The experimental data was previously published in Lawrimore et al. (Lawrimore et al., 2018).
The plasmid utilized in these experiments was approximately 11 kb, after excision of the autonomously replicating sequence (ARS) (Lawrimore et al., 2018). The authors estimated only 3-6 condensin complexes would be able to bind to a plasmid of this size. Whether 3-6 loop extrusion sites on a single 11 kb plasmid would be sufficient to dampen the plasmid’s thermal fluctuations was unclear. An additional complication was the differing behavior condensin was reported to exhibit when DNA was placed under tension. Terakawa et al. (Terakawa et al., 2017) reported that condensin on a taut substrate would translocate. Ganji et al. (Ganji et al., 2018) reported that condensin slowed its loop extrusion at low salt concentrations and would only translocate at higher salt concentrations. Lawrimore et al. (Lawrimore et al., 2018) modeled both possibilities by allowing condensin complexes to translocate when the tension became high on the substrate (dynamic condensin). The other condensin behavior was modeled by preventing randomly placed condensin complexes from extruding loops (static condensin), assuming the tension from bi-orientation of the two centromeres would cause sufficient tension to prevent loop extrusion. When 3 condensin complexes were in the simulation, both the dynamic and static models resulted in a decrease in average signal lengths compared to a simulation lacking condensin entirely. However, when 6 condensin complexes were in the simulation, the dynamic model resulted in an increase in the average signal lengths, while the static model resulted in a decrease in the average signal lengths (Figure 4). Furthermore, the simulated signal length distributions of dynamic condensin were symmetric, while the distributions of static condensin simulations (3 loops) were right skewed, mimicking experimental observations (Lawrimore et al., 2018). Thus, static condensin-mediated loops recapitulated the experimental observations.
In dynamic simulations of a stepping motor on a chromatin substrate (RotoStep), condensin loop extrusion is converted to condensin translocation on a taut substrate by simply destabilizing a single, strong DNA-condensin binding site, while leaving the motor activity intact (Lawrimore et al., 2017). Indeed, there are several potential sites on condensin that may interact with DNA, but in order for successful loop extrusion there must be a strong DNA binding site that anchors condensin to the DNA (see perspective on tethered inchworm model (Nichols and Corces, 2018)). Recent work showed the kleisin, Brn1, and the heat-repeat containing protein, Ycg1, work together to form a sequence independent DNA-binding groove that resembles a safety belt (Kschonsak et al., 2017). Lawrimore et al. (Lawrimore et al., 2018) reported the average signal length of the dicentric plasmid increased in brn1-9 and ycg1-2 temperature sensitive, condensin mutants. Live cell, fluorescent imaging of the condensin subunit SMC4 in brn1-9 yeast showed a much more homogenous SMC4-GFP signal with little pericentromeric or nucleolar enrichment. The SMC4-GFP signal also recovered faster after photobleaching (Lawrimore et al., 2018). The increased homogeneity and recovery rate could be due to decreased condensin binding affinity, dysregulation of condensin activity, increased chromosome mobility, loss of ATPase activity, or some combination of those factors. However, since the dicentric plasmid simulations with 6 dynamic condensin complexes, which translocate when the substrate is under tension (Lawrimore et al., 2018), result in longer and more variable dicentric plasmid simulations, mimicking the brn1-9 experimental results (Figure 4), it is possible that the mutations brn1-9 or ycg1-2 destabilize condensin’s main tether to DNA.
Consequences of Condensin and Cohesin Dysregulation
In this review, we have argued that the pericentromere of the budding yeast S. cerevisiae is structurally similar to regional centromeres. Both pericentromeres and regional centromeres contain multiple kinetochore microtubule attachments, exhibit similar sister kinetochore separation (800 nm vs 1000 nm), are in enriched in SMC protein complexes, exhibit alternative histones and histone modifications, are enriched with replication repair proteins, and exhibit suppression of meiotic recombination. However, the highly aberrant dynamics of the mitotic spindle and pericentric chromatin upon disruption of condensin suggest the key similarity may be the highly looped nature of chromatin in pericentromeres and regional centromeres. Mounting evidence from numerous studies demonstrates the pericentromere is remodeled upon loss of tension. Given the recent discovery that condensin can extrude loops at a rapid pace (Terakawa et al., 2017) (Ganji et al., 2018), altering the number, size, and physical properties of chromatin loops allows for tight regulation over the tension of the kinetochore.
The in vivo and in vitro studies with budding yeast condensin highlight the role chromatin loops may play in proper chromosome segregation. If the highly looped topology of chromatin in regional centromeres is important in humans, one can reasonably assume that disruption of the potential loop extruders in humans, condensin and/or cohesin, would be seen in human cancers. A recent review exploring the role of human condensin subunit dysregulation in cancer came to the startling conclusion that all subunits of human condensin I and II, except hCAP-D2, the non-SMC, heat-repeat subunit of condensin I, have been shown to be dysregulated or mutated in human cancers (Wang et al., 2018). Similarly, cohesin subunits are frequently mutated in human cancers (Hill, Kim, and Waldman, 2016). Cohesin’s HEAT-repeat subunit, STAG2/SA2, was found to be one of the 12 genes significantly mutated in at least 4 cancer types, putting STAG2/SA2 in the same category as the infamous cancer genes TP53, RB1, KRAS, and NRAS (Lawrence et al., 2014). The reconstitution experiments of budding yeast condensin (Terakawa et al., 2017; Ganji et al., 2018) have clearly shown condensin to be a powerful motor. Given loop extrusion’s proposed ability to shape the genome in both interphase and mitosis (Fudenberg and Mirny, 2012; Goloborodko et al., 2016; Nuebler et al., 2018), it is unsurprising that the dysregulation of cohesin and condensin would be detrimental to proper genome organization and segregation. In budding yeast, the chromosome mis-segregation and decompactions that occur in the temperature sensitive alleles brn1-9 (Lavoie et al., 2000) and ycg1-2 (Lavoie, Hogan, and Koshland, 2002) may be due to loss of condensin’s DNA anchor site formed by Brn1 and Ycg1 (Kschonsak et al., 2017). Therefore, discovering how loop extrusion is utilized by SMC complexes and regulated in human centromeres will lead to advances in understanding genome organization and chromosome segregation vital insights into cancer progression and treatment. The similarities between the pericentromere in S. cerevisiae and human regional centromeres make the humble budding yeast a valuable tool for exploring how chromatin can be harnessed to regulate chromosome segregation.
References
- 1.Flemming W (1882). Zellsubstanz, Kern und Zelltheilung (Cell substance, nucleus and cell division). Leipzig, Vogel, 1882 [Google Scholar]
- 2.Clarke L, Carbon J. (1980). Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature, 287, 504–9 [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald-Hayes M, Clarke L, Carbon J. (1982). Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell, 29, 235–44 [DOI] [PubMed] [Google Scholar]
- 4.Blat Y, Kleckner N. (1999). Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell, 98, 249–59 [DOI] [PubMed] [Google Scholar]
- 5.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. (2008). Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes & development, 22, 2215–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Megee PC, Mistrot C, Guacci V, Koshland D. (1999). The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell, 4, 445–50 [DOI] [PubMed] [Google Scholar]
- 7.Choy JS, Acuña R, Au W-C, Basrai MA. (2011). A Role for Histone H4K16 Hypoacetylation in Saccharomyces cerevisiae Kinetochore Function. Genetics, 189, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choy JS, Mishra PK, Au WC, Basrai MA. (2012). Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim Biophys Acta, 1819, 776–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan BA, Karpen GH. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol, 11, 1076–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhatchinamoorthy K, Unruh JR, Lange JJ, Levy M, Slaughter BD, Gerton JL. (2019). The stoichiometry of the outer kinetochore is modulated by microtubule-proximal regulatory factors. J Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrimore J, Aicher JK, Hahn P, Fulp A, Kompa B, Vicci L, Falvo M, Taylor RM 2nd, Bloom K. (2016). ChromoShake: a chromosome dynamics simulator reveals that chromatin loops stiffen centromeric chromatin. Molecular biology of the cell, 27, 153–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrimore J, Vasquez PA, Falvo MR, Taylor RM 2nd, Vicci L, Yeh E, Forest MG, Bloom K. (2015). DNA loops generate intracentromere tension in mitosis. The Journal of cell biology, 210, 553–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens AD, Quammen CW, Chang B, Haase J, Taylor RM 2nd, Bloom K. (2013). The spatial segregation of pericentric cohesin and condensin in the mitotic spindle. Mol Biol Cell, 24, 3909–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. (2008). Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol, 18, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens AD, Haggerty RA, Vasquez PA, Vicci L, Snider CE, Shi F, Quammen C, Mullins C, Haase J, Taylor RM 2nd, Verdaasdonk JS, Falvo MR, Jin Y, Forest MG, Bloom K. (2013). Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J Cell Biol, 200, 757–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC. (2017). The condensin complex is a mechanochemical motor that translocates along DNA. Science [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson CG, Maddox PS, Salmon ED, Bloom K. (2001). Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol, 152, 1255–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding R, McDonald KL, McIntosh JR. (1993). Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol, 120, 141–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melters DP, Paliulis LV, Korf IF, Chan SW. (2012). Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res, 20, 579–93 [DOI] [PubMed] [Google Scholar]
- 20.Maddox PS, Portier N, Desai A, Oegema K. (2006). Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc Natl Acad Sci U S A, 103, 15097–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf KW. (1994). The unique structure of Lepidopteran Spindls. Int Rev Cytol, 152, 1–48 [Google Scholar]
- 22.Venkei Z, Przewloka MR, Ladak Y, Albadri S, Sossick A, Juhasz G, Novak B, Glover DM. (2012). Spatiotemporal dynamics of Spc105 regulates the assembly of the Drosophila kinetochore. Open Biol, 2, 110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf KW, Mitchell A, Nicol L, Jeppesen P. (1994). Analysis of centromere structure in the fly Megaselia scalaris (Phoridae, Diptera) using CREST sera, anti-histone antibodies, and a repetitive DNA probe. Biol Cell, 80, 11–23 [DOI] [PubMed] [Google Scholar]
- 24.Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P, Raudsepp T, Chowdhary BP, Guerin G, Archidiacono N, Rocchi M, Giulotto E. (2006). Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics, 87, 777–82 [DOI] [PubMed] [Google Scholar]
- 25.Salmon ED, Goode D, Maugel TK, Bonar DB. (1976). Pressure-induced depolymerization of spindle microtubules. III. Differential stability in HeLa cells. J Cell Biol, 69, 443–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan L, Ladinsky Mark S, Jensen Grant J. (2011). Organization of the Smallest Eukaryotic Spindle. Current Biology, 21, 1578–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckert CA, Gravdahl DJ, Megee PC. (2007). The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes & development, 21, 278–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KH, Sauro HM. (2012). In search of noise-induced bimodality. BMC Biol, 10, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, Garcia-Muse T, Aguilera A. (2013). R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Molecular cell, 52, 583–90 [DOI] [PubMed] [Google Scholar]
- 30.Deng X, Kuo MH. (2018). Tripartite Chromatin Localization of Budding Yeast Shugoshin Involves Higher-Ordered Architecture of Mitotic Chromosomes. G3 (Bethesda), 8, 2901–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J, Deng X, Buehl C, Xu X, Kuo MH. (2016). Identification of Tension Sensing Motif of Histone H3 in Saccharomyces cerevisiae and Its Regulation by Histone Modifying Enzymes. Genetics, 204, 1029–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noma K, Allis CD, Grewal SI. (2001). Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science, 293, 1150–5 [DOI] [PubMed] [Google Scholar]
- 33.Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol, 104, 805–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Black BE, Cleveland DW. (2011). Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell, 144, 471–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdaasdonk JS, Bloom K. (2011). Centromeres: unique chromatin structures that drive chromosome segregation. Nature reviews Molecular cell biology, 12, 320–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haase J, Mishra PK, Stephens A, Haggerty R, Quammen C, Taylor RM 2nd, Yeh E, Basrai MA, Bloom K. (2013). A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr Biol, 23, 1939–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann G, Samel-Pommerencke A, Weber J, Cuomo A, Bonaldi T, Ehrenhofer-Murray AE. (2017). A role for CENP-A/Cse4 phosphorylation on serine 33 in deposition at the centromere. FEMS Yeast Research, 18 [DOI] [PubMed] [Google Scholar]
- 38.Lawrimore J, Bloom KS, Salmon ED. (2011). Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol, 195, 573–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cherry LM, Faulkner AJ, Grossberg LA, Balczon R. (1989). Kinetochore size variation in mammalian chromosomes: an image analysis study with evolutionary implications. Journal of cell science, 92 ( Pt 2), 281–9 [DOI] [PubMed] [Google Scholar]
- 40.Liu ST, Rattner JB, Jablonski SA, Yen TJ. (2006). Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol, 175, 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr., Cleveland DW. (2004). Structural determinants for generating centromeric chromatin. Nature, 430, 578–82 [DOI] [PubMed] [Google Scholar]
- 42.Miell MD, Fuller CJ, Guse A, Barysz HM, Downes A, Owen-Hughes T, Rappsilber J, Straight AF, Allshire RC. (2013). CENP-A confers a reduction in height on octameric nucleosomes. Nat Struct Mol Biol, 20, 763–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SH, Vlijm R, van der Torre J, Dalal Y, Dekker C. (2016). CENP-A and H3 Nucleosomes Display a Similar Stability to Force-Mediated Disassembly. PLoS One, 11, e0165078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bloom K, Amaya E, Yeh E, Eds., Centromeric DNA structure in yeast chromatin, in Molecular Biology of the Cytoskeleton, Cold Spring Harbor Laboratory Press, 1984 [Google Scholar]
- 45.Bloom KS, Fitzgerald-Hayes M, Carbon J. (1983). Structural analysis and sequence organization of yeast centromeres. Cold Spring Harbor symposia on quantitative biology, 47 Pt 2, 1175–85 [DOI] [PubMed] [Google Scholar]
- 46.Diaz-Ingelmo O, Martinez-Garcia B, Segura J, Valdes A, Roca J. (2015). DNA Topology and Global Architecture of Point Centromeres. Cell reports, 13, 667–77 [DOI] [PubMed] [Google Scholar]
- 47.Furuyama T, Henikoff S. (2009). Centromeric nucleosomes induce positive DNA supercoils. Cell, 138, 104–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joglekar AP, Bloom K, Salmon ED. (2009). In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol, 19, 694–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. (2011). Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nature communications, 2, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. (2013). The octamer is the major form of CENP-A nucleosomes at human centromeres. Nature structural & molecular biology, 20, 687–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nechemia-Arbely Y, Fachinetti D, Miga KH, Sekulic N, Soni GV, Kim DH, Wong AK, Lee AY, Nguyen K, Dekker C, Ren B, Black BE, Cleveland DW. (2017). Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J Cell Biol, 216, 607–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Padeganeh A, Ryan J, Boisvert J, Ladouceur AM, Dorn JF, Maddox PS. (2013). Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr Biol, 23, 764–9 [DOI] [PubMed] [Google Scholar]
- 53.Hamiche A, Carot V, Alilat M, De Lucia F, O’Donohue MF, Revet B, Prunell A. (1996). Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: Potential flipping of the protein from a left- to a right-handed superhelical form. Proc Natl Acad Sci U S A, 93, 7588–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vlijm R, Kim SH, De Zwart PL, Dalal Y, Dekker C. (2017). The supercoiling state of DNA determines the handedness of both H3 and CENP-A nucleosomes. Nanoscale, 9, 1862–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. (2012). Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell, 150, 317–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falk SJ, Lee J, Sekulic N, Sennett MA, Lee TH, Black BE. (2016). CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat Struct Mol Biol, 23, 204–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malik N, Dantu SC, Shukla S, Kombrabail M, Ghosh SK, Krishnamoorthy G, Kumar A. (2018). Conformational flexibility of histone variant CENP-A(Cse4) is regulated by histone H4: A mechanism to stabilize soluble Cse4. The Journal of biological chemistry, 293, 20273–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng W, Bachant J, Collingwood D, Raghuraman MK, Brewer BJ. (2009). Centromere replication timing determines different forms of genomic instability in Saccharomyces cerevisiae checkpoint mutants during replication stress. Genetics, 183, 1249–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pohl TJ, Brewer BJ, Raghuraman MK. (2012). Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLoS Genet, 8, e1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H. (2009). The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nature cell biology, 11, 357–62 [DOI] [PubMed] [Google Scholar]
- 61.Shelby RD, Monier K, Sullivan KF. (2000). Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol, 151, 1113–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Branzei D, Foiani M. (2010). Maintaining genome stability at the replication fork. Nature reviews Molecular cell biology, 11, 208–19 [DOI] [PubMed] [Google Scholar]
- 63.Greenfeder SA, Newlon CS. (1992). Replication forks pause at yeast centromeres. Molecular and cellular biology, 12, 4056–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hodgson B, Calzada A, Labib K. (2007). Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell, 18, 3894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Catania S, Pidoux AL, Allshire RC. (2015). Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet, 11, e1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. (2009). Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes & development, 23, 2876–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. (2008). Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci U S A, 105, 9936–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. (2009). Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol, 16, 226–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simi S, Simili M, Bonatti S, Campagna M, Abbondandolo A. (1998). Fragile sites at the centromere of Chinese hamster chromosomes: a possible mechanism of chromosome loss. Mutat Res, 397, 239–46 [DOI] [PubMed] [Google Scholar]
- 70.Mitra S, Gomez-Raja J, Larriba G, Dubey DD, Sanyal K. (2014). Rad51-Rad52 mediated maintenance of centromeric chromatin in Candida albicans. PLoS genetics, 10, e1004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. (2016). Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nature cell biology, 18, 684–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iyer DR, Rhind N. (2017). The Intra-S Checkpoint Responses to DNA Damage. Genes, 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shastri N, Tsai Y-C, Hile S, Jordan D, Powell B, Chen J, Maloney D, Dose M, Lo Y, Anastassiadis T, Rivera O, Kim T, Shah S, Borole P, Asija K, Wang X, Smith KD, Finn D, Schug J, Casellas R, Yatsunyk LA, Eckert KA, Brown EJ. (2018). Genome-wide Identification of Structure-Forming Repeats as Principal Sites of Fork Collapse upon ATR Inhibition. Molecular Cell, 72, 222–38.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kabeche L, Nguyen HD, Buisson R, Zou L. (2018). A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science, 359, 108–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. (2007). Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol, 17, 2003–12 [DOI] [PubMed] [Google Scholar]
- 76.Buhler C, Borde V, Lichten M. (2007). Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS biology, 5, e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mahtani MM, Willard HF. (1998). Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res, 8, 100–10 [DOI] [PubMed] [Google Scholar]
- 78.Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. (2008). Global analysis of the meiotic crossover landscape. Developmental cell, 15, 401–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rockmill B, Voelkel-Meiman K, Roeder GS. (2006). Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics, 174, 1745–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vincenten N, Kuhl LM, Lam I, Oke A, Kerr AR, Hochwagen A, Fung J, Keeney S, Vader G, Marston AL. (2015). The kinetochore prevents centromere-proximal crossover recombination during meiosis. Elife, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hinshaw SM, Makrantoni V, Harrison SC, Marston AL. (2017). The Kinetochore Receptor for the Cohesin Loading Complex. Cell, 171, 72–84 e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Covo S, Puccia CM, Argueso JL, Gordenin DA, Resnick MA. (2014). The sister chromatid cohesion pathway suppresses multiple chromosome gain and chromosome amplification. Genetics, 196, 373–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zamb TJ, Petes TD. (1981). Unequal sister-strand recombination within yeast ribosomal DNA does not require the RAD 52 gene product. Curr Genet, 3, 125–32 [DOI] [PubMed] [Google Scholar]
- 84.Ozenberger BA, Roeder GS. (1991). A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol, 11, 1222–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. (2007). The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nature cell biology, 9, 923–31 [DOI] [PubMed] [Google Scholar]
- 86.Janssen A, Colmenares SU, Lee T, Karpen GH. (2019). Timely double-strand break repair and pathway choice in pericentromeric heterochromatin depend on the histone demethylase dKDM4 A. Genes & development, 33, 103–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hult C, Adalsteinsson D, Vasquez PA, Lawrimore J, Bennett M, York A, Cook D, Yeh E, Forest MG, Bloom K. (2017). Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res, 45, 11159–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lawrimore J, Doshi A, Friedman B, Yeh E, Bloom K. (2018). Geometric partitioning of cohesin and condensin is a consequence of chromatin loops. Molecular biology of the cell, 29, 2737–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stephens AD, Haase J, Vicci L, Taylor RM 2nd, Bloom K. (2011). Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol, 193, 1167–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McFarlane RJ, Humphrey TC. (2010). A role for recombination in centromere function. Trends in genetics : TIG, 26, 209–13 [DOI] [PubMed] [Google Scholar]
- 91.Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, Mirny LA, Dekker J. (2018). A pathway for mitotic chromosome formation. Science, 359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragon L. (2011). Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science, 331, 1328–32 [DOI] [PubMed] [Google Scholar]
- 93.Bizard AH, Allemand JF, Hassenkam T, Paramasivam M, Sarlos K, Singh MI, Hickson ID. (2019). PICH and TOP3A cooperate to induce positive DNA supercoiling. Nat Struct Mol Biol, 26, 267–74 [DOI] [PubMed] [Google Scholar]
- 94.Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. (1999). 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell, 98, 239–48 [DOI] [PubMed] [Google Scholar]
- 95.Norman-Axelsson U, Durand-Dubief M, Prasad P, Ekwall K. (2013). DNA topoisomerase III localizes to centromeres and affects centromeric CENP-A levels in fission yeast. PLoS Genet, 9, e1003371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Allemand JF, Bensimon D, Lavery R, Croquette V. (1998). Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc Natl Acad Sci U S A, 95, 14152–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lipfert J, Klijnhout S, Dekker NH. (2010). Torsional sensing of small-molecule binding using magnetic tweezers. Nucleic Acids Res, 38, 7122–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Strick TR, Bensimon D, Croquette V. (1999). Micro-mechanical measurement of the torsional modulus of DNA. Genetica, 106, 57–62 [DOI] [PubMed] [Google Scholar]
- 99.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. (2010). Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature, 468, 576–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. (2006). The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A, 103, 9873–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pauling L, Corey RB. (1953). A Proposed Structure For The Nucleic Acids. Proc Natl Acad Sci U S A, 39, 84–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hall AC, Ostrowski LA, Mekhail K. (2019). Phase Separation as a Melting Pot for DNA Repeats. Trends in genetics : TIG [DOI] [PubMed] [Google Scholar]
- 103.Erdel F, Rippe K. (2018). Formation of Chromatin Subcompartments by Phase Separation. Biophys J, 114, 2262–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rubinstein M, Colby RH. (2003). Polymer Physics. Oxford, Oxford University Press [Google Scholar]
- 105.Bergeron-Sandoval LP, Safaee N, Michnick SW. (2016). Mechanisms and Consequences of Macromolecular Phase Separation. Cell, 165, 1067–79 [DOI] [PubMed] [Google Scholar]
- 106.Weber SC. (2017). Sequence-encoded material properties dictate the structure and function of nuclear bodies. Curr Opin Cell Biol, 46, 62–71 [DOI] [PubMed] [Google Scholar]
- 107.Mather K (1936). The determination of position in crossing-over. Journal of Genetics, 33, 207–35 [Google Scholar]
- 108.Mather K (1939). Crossing over and Heterochromatin in the X Chromosome of Drosophila Melanogaster. Genetics, 24, 413–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kuhl LM, Vader G. (2019). Kinetochores, cohesin, and DNA breaks: Controlling meiotic recombination within pericentromeres. Yeast, 36, 121–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Covo S, Westmoreland JW, Gordenin DA, Resnick MA. (2010). Cohesin Is limiting for the suppression of DNA damage-induced recombination between homologous chromosomes. PLoS Genet, 6, e1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stephens AD, Snider CE, Haase J, Haggerty RA, Vasquez PA, Forest MG, Bloom K. (2013). Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J Cell Biol, 203, 407–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Biggins S (2013). The composition, functions, and regulation of the budding yeast kinetochore. Genetics, 194, 817–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bloom K, Yeh E. (2010). Tension management in the kinetochore. Curr Biol, 20, R1040–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joglekar AP. (2016). A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Joglekar AP, Kukreja AA. (2017). How Kinetochore Architecture Shapes the Mechanisms of Its Function. Curr Biol, 27, R816–R24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Musacchio A, Salmon ED. (2007). The spindle-assembly checkpoint in space and time. Nature reviews Molecular cell biology, 8, 379–93 [DOI] [PubMed] [Google Scholar]
- 117.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. (2004). Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS biology, 2, E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. (2004). The kinetochore is an enhancer of pericentric cohesin binding. PLoS biology, 2, E260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. (1996). In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. The Journal of cell biology, 135, 1685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Straight AF, Belmont AS, Robinett CC, Murray AW. (1996). GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol, 6, 1599–608 [DOI] [PubMed] [Google Scholar]
- 121.Goshima G, Yanagida M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell, 100, 619–33 [DOI] [PubMed] [Google Scholar]
- 122.He X, Asthana S, Sorger PK. (2000). Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell, 101, 763–75 [DOI] [PubMed] [Google Scholar]
- 123.Tanaka T, Cosma MP, Wirth K, Nasmyth K. (1999). Identification of cohesin association sites at centromeres and along chromosome arms. Cell, 98, 847–58 [DOI] [PubMed] [Google Scholar]
- 124.Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. (2003). Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell, 14, 4181–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bloom KS. (2014). Centromeric Heterochromatin: The Primordial Segregation Machine. Annual review of genetics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jacobs CW, Adams AE, Szaniszlo PJ, Pringle JR. (1988). Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol, 107, 1409–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. (2012). Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr Biol, 22, 471–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Suzuki A, Badger BL, Haase J, Ohashi T, Erickson HP, Salmon ED, Bloom K. (2016). How the kinetochore couples microtubule force and centromere stretch to move chromosomes. Nature cell biology, 18, 382–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. (2010). Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science, 327, 172–7 [DOI] [PubMed] [Google Scholar]
- 130.Peplowska K, Wallek AU, Storchova Z. (2014). Sgo1 regulates both condensin and Ipl1/Aurora B to promote chromosome biorientation. PLoS Genet, 10, e1004411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Eshleman HD, Morgan DO. (2014). Sgo1 recruits PP2A to chromosomes to ensure sister chromatid bi-orientation during mitosis. Journal of cell science, 127, 4974–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nerusheva OO, Galander S, Fernius J, Kelly D, Marston AL. (2014). Tension-dependent removal of pericentromeric shugoshin is an indicator of sister chromosome biorientation. Genes & development, 28, 1291–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jin F, Bokros M, Wang Y. (2017). Premature Silencing of the Spindle Assembly Checkpoint Is Prevented by the Bub1-H2A-Sgo1-PP2A Axis in Saccharomyces cerevisiae. Genetics, 205, 1169–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. (2018). Real-time imaging of DNA loop extrusion by condensin. Science, 360, 102–05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC. (2017). The condensin complex is a mechanochemical motor that translocates along DNA. Science, 358, 672–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bouck DC, Bloom K. (2007). Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol, 17, 741–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Anderson M, Haase J, Yeh E, Bloom K. (2009). Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell, 20, 4131–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dewar H, Tanaka K, Nasmyth K, Tanaka TU. (2004). Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature, 428, 93–7 [DOI] [PubMed] [Google Scholar]
- 139.Snider CE, Stephens AD, Kirkland JG, Hamdani O, Kamakaka RT, Bloom K. (2014). Dyskerin, tRNA genes, and condensin tether pericentric chromatin to the spindle axis in mitosis. Journal of Cell Biology, 207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jorgensen P, Edgington NP, Schneider BL, Rupeš I, Tyers M, Futcher B. (2007). The Size of the Nucleus Increases as Yeast Cells Grow. Molecular Biology of the Cell, 18, 3523–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jaqaman K, King EM, Amaro AC, Winter JR, Dorn JF, Elliott HL, McHedlishvili N, McClelland SE, Porter IM, Posch M, Toso A, Danuser G, McAinsh AD, Meraldi P, Swedlow JR. (2010). Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. The Journal of Cell Biology, 188, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chacon JM, Mukherjee S, Schuster BM, Clarke DJ, Gardner MK. (2014). Pericentromere tension is self-regulated by spindle structure in metaphase. The Journal of cell biology, 205, 313–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mukherjee S, Sandri BJ, Tank D, McClellan M, Harasymiw LA, Yang Q, Parker LL, Gardner MK. (2019). A Gradient in Metaphase Tension Leads to a Scaled Cellular Response in Mitosis. Developmental cell, 49, 63–76 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harasymiw LA, Tank D, McClellan M, Panigrahy N, Gardner MK. (2019). Centromere mechanical maturation during mammalian cell mitosis. Nat Commun, 10, 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Parry Bradley R, Surovtsev Ivan V, Cabeen Matthew T, O’Hern Corey S, Dufresne Eric R, Jacobs-Wagner C. (2014). The Bacterial Cytoplasm Has Glass-like Properties and Is Fluidized by Metabolic Activity. Cell, 156, 183–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lawrimore J, Friedman B, Doshi A, Bloom K. (2017). RotoStep: a chromosome dynamics simulator reveals mechanisms of loop extrusion. Cold Spring Harbor symposia on quantitative biology, LXXXII. [DOI] [PubMed] [Google Scholar]
- 147.Nichols MH, Corces VG. (2018). A tethered-inchworm model of SMC DNA translocation. Nat Struct Mol Biol, 25, 906–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kschonsak M, Merkel F, Bisht S, Metz J, Rybin V, Hassler M, Haering CH. (2017). Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell, 171, 588–600.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang H-Z, Yang S-H, Li G-Y, Cao X. (2018). Subunits of human condensins are potential therapeutic targets for cancers. Cell Division, 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hill VK, Kim J-S, Waldman T. (2016). Cohesin mutations in human cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer, 1866, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature, 505, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fudenberg G, Mirny LA. (2012). Higher-order chromatin structure: bridging physics and biology. Current opinion in genetics & development, 22, 115–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goloborodko A, Imakaev MV, Marko JF, Mirny L. (2016). Compaction and segregation of sister chromatids via active loop extrusion. eLife, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. (2018). Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proceedings of the National Academy of Sciences, 115, E6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lavoie BD, Tuffo KM, Oh S, Koshland D, Holm C. (2000). Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Molecular biology of the cell, 11, 1293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lavoie BD, Hogan E, Koshland D. (2002). In vivo dissection of the chromosome condensation machinery. The Journal of Cell Biology, 156, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bloom K, Costanzo V. (2017). Centromere Structure and Function. Progress in molecular and subcellular biology, 56, 515–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Quammen CW, Richardson AC, Haase J, Harrison BD, Taylor RM, Bloom KS. (2008). FluoroSim: A Visual Problem-Solving Environment for Fluorescence Microscopy. Eurographics Workshop Vis Comput Biomed, 2008, 151–58 [DOI] [PMC free article] [PubMed] [Google Scholar]


